
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Genet., 26 August 2022
Sec. Genetics of Common and Rare Diseases
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.964098
This article is part of the Research TopicGenetic and Environmental Factors in the Occurrence of Paediatric Disorders – Volume IIView all 10 articles
 Jianlong Zhuang1†
Jianlong Zhuang1† Junyu Wang1†
Junyu Wang1† Qi Luo2
Qi Luo2 Shuhong Zeng1
Shuhong Zeng1 Yu’e Chen3
Yu’e Chen3 Yuying Jiang1
Yuying Jiang1 Xinying Chen1
Xinying Chen1 Yuanbai Wang1
Yuanbai Wang1 Yingjun Xie4,5*
Yingjun Xie4,5* Gaoxiong Wang6*
Gaoxiong Wang6* Chunnuan Chen7*
Chunnuan Chen7*Background: Lethal multiple pterygium syndrome (LMPS) is a rare autosomal recessive inherited disorder typically characterized by intrauterine growth retardation, multiple pterygia, and flexion contractures.
Case presentation: We herein report a Chinese case with a history of three adverse pregnancies demonstrating the same ultrasonic phenotypes, including increased nuchal translucency, edema, fetal neck cystoma, reduced movement, joint contractures, and other congenital features. Whole-exome sequencing (WES) revealed novel compound heterozygous variants in the CHRNA1 gene NM_000079.4: c.[1128delG (p.Pro377LeufsTer10)]; [505T>C (p.Trp169Arg)] in the recruited individual, and subsequent familial segregation showed that both parents transmitted their respective mutation.
Conclusion: For the first time, we identified an association between the CHRNA1 gene and the recurrent lethal multiple pterygium syndrome (LMPS) in a Chinese family. This finding may also enrich the mutation spectrum of the CHRNA1 gene and promote the applications of WES technology in etiologic diagnosis of ultrasound anomalies in prenatal examination.
The acetylcholine receptor (AChR) is a member of the superfamily of transmitter-gated ion channels and plays a critical role in controlling electrical signals between nerves and skeletal muscle cells. In the embryonic development, AChR consists of one β (CHRNB), one δ (CHRND), one γ (CHRNG), and two α (CHRNA1) subunits, but after a gestational age of 33 weeks, the γ subunit is replaced by an ε (CHRNE) subunit (Hesselmans et al., 1993). The α subunit of the muscle acetylcholine receptor encoded by CHRNA1 gene is known as the main target of pathogenic autoantibodies in autoimmune myasthenia gravis.
CHRNA1 (MIM 100690), CHRND (MIM 100720), CHRNG (MIM 100730), RAPSN (MIM 601592), DOK7 (MIM 610285), CNTN1 (MIM 600016), and SYNE1 (MIM 608441) gene mutations would lead to fetal akinesia deformation sequence and/or multiple pterygium syndrome (MPS) (Chen, 2012), a rare autosomal recessive inherited disorder mainly manifested as arthrogryposis multiplex congenita, pterygia of the neck, fingers, and antecubital, popliteal, and intercrural areas, developmental delay, and facial, vertebral, and genital anomalies (Penchaszadeh and Salszberg, 1981; Ramer et al., 1988). The prevalence of MPS remains uncertain and is supposed to be less than 1/100,000, as reported by a previous study (Mohtisham et al., 2019). MPS is typically divided into prenatally lethal and nonlethal types (Barros et al., 2012; Chen, 2012). The nonlethal form of MPS is also known as Escobar syndrome. The lethal multiple pterygium syndrome (LMPS) is a rare autosomal recessive inherited disorder characterized by intrauterine growth retardation, multiple pterygia, and flexion contractures, causing severe arthrogryposis and fetal akinesia (Vogt et al., 2008; Joshi et al., 2016; Mohtisham et al., 2019). In addition, although the most common inheritance model is autosomal recessive, the autosomal dominant and X-linked inheritance has also been reported (Tolmie et al., 1987; Meyer-Cohen et al., 1999; Chong et al., 2015). Usually, fetuses with LMPS would result in spontaneous miscarriage or stillbirth (Nazari et al., 2019).
In this study, we used whole-exome sequencing (WES) to make a diagnosis of genetic etiology diagnosis in a Chinese family with increased nuchal translucency, fetal edema, fetal neck cystoma, reduced movement, and joint contractures and identified two novel compound heterozygous variants in the CHRNA1 gene in the fetus, which would lead to LMPS. This study may broaden the spectrum of CHRNA1 gene variants that lead to LMPS and provide valuable data for application of prenatal WES technology and genetic consultation.
In this study, a Chinese family with a history of three adverse pregnancies was recruited. The couple denied consanguineous marriage and any related inherited history. This study was approved by the ethics committee of Quanzhou Women’s and Children’s Hospital (2020 No. 31). The three fetuses in this family all had similar ultrasonic abnormalities. Among them, ultrasound of the first pregnancy in the first trimester showed that the fetus had fetal systemic edema, and the pregnant woman and her family chose to terminate the pregnancy without further genetic etiology testing. Subsequently, the woman had the second pregnancy, and ultrasound examination results elicited that the fetus had edema, increased nuchal translucency, neck water sac tumor, reduced movement, and abnormal posture (joint contractures) (Figure 1), and stillbirth occurred at the gestational age of 18+5 weeks in the second pregnancy. Upon informed consent from the family, we collected the fetal specimen of the second pregnancy for further cytogenetic and molecular genetic analyses. The third pregnancy of the woman also showed a similar ultrasonic phenotype. Although no stillborn occurred in the second trimester, the family still chose to terminate the pregnancy.
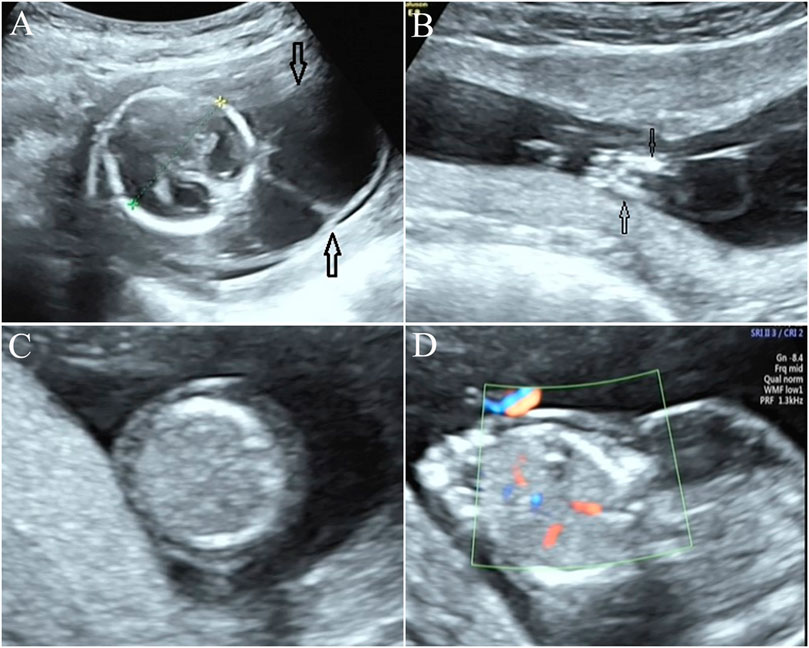
FIGURE 1. Prenatal ultrasonic examination results of the fetus. (A) Ultrasound examination elicited neck water sac tumor in the fetus. (B) Approach, flexion, and fixation of both lower limbs were observed in the fetus, indicating joint contractures. (C,D): the trunk skin layer is obviously thickened, and the echo is reduced, and diagnosed as fetal edema.
No obvious chromosomal abnormalities and copy number variants were detected by karyotype analysis and chromosomal microarray analysis in the fetus of the second pregnancy, as well as their parents who exhibit normal clinical phenotypes.
WES technology was performed for further genetic etiology in the recruited fetus. A novel frameshift variant in exon 8 compound with a novel missense variant in exon 5 in the CHRNA1 gene NM_000079.4: c.[1128delG (p.Pro377LeufsTer10)]; [505T>C (p.Trp169Arg)] was detected in the recruited fetus by WES technology detection, which was inherited from their parents, respectively (Figure 2 and Supplementary Table 1). In the third pregnancy, the compound heterozygous variants were also detected by other hospitals using WES technology. At present, the c.1128delG (p.Pro377LeufsTer10) variant was absent in the gnomAD (http://gnomad-sg.org/, accession date: 28 June 2022), dbSNP (https://www.ncbi.nlm.nih.gov/snp/?term=, accession date: 28 June 2022), 1000 genomes project (http://browser.1000genomes.org/, accession date: 28 June 2022), PubMed (https://pubmed.ncbi.nlm.nih.gov, accession date: 28 June 2022), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accession date: 28 June 2022) (Table 1), and HGMD (http://www.hgmd.cf.ac.uk/ac/index.php, accession date: 28 June 2022) databases and was not found in the local database as well, but it was interpreted as a likely pathogenic variant (PVS1, PM2_Supporting) according to the ACMG (The American College of Medical Genetics and Genomics, ACMG) guidelines (Richards et al., 2015). In addition, the c.505T>C (p.Trp169Arg) variant was also absent in the databases mentioned earlier, and online computer-aided analysis predictions (http://159.226.67.237/sun/varcards/welcome/index) suggest that this variant is more likely to affect the protein structure/function (damaging score: 0.87). According to the ACMG guidelines (Richards et al., 2015), the c.505T>C (p.Trp169Arg) variant was interpreted as variant of uncertain significance (PM3, PM2_Supporting, and PP3).
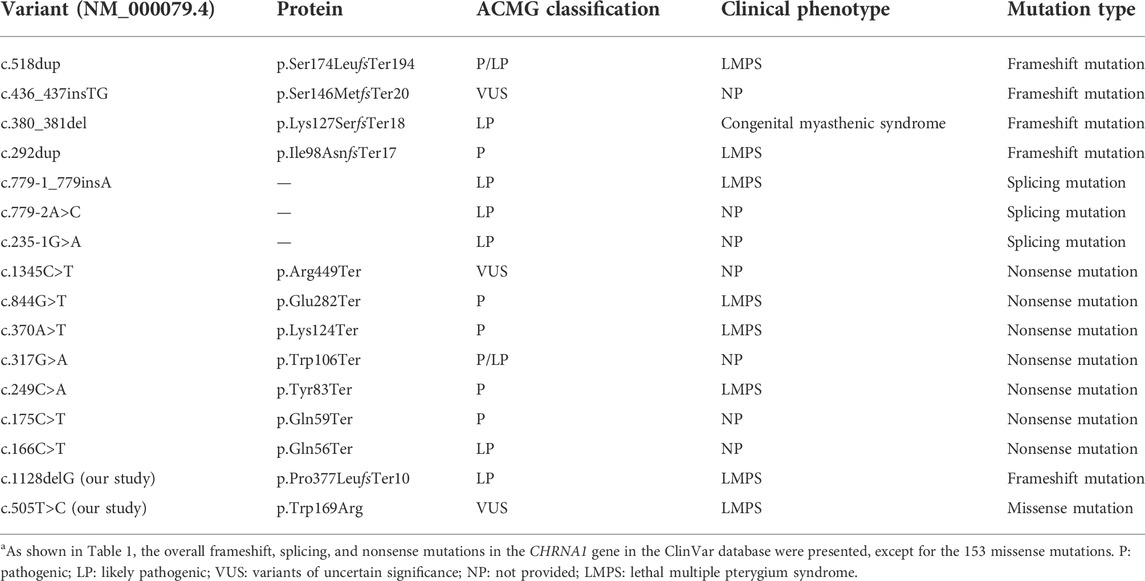
TABLE 1. Variants of the CHRNA1 gene and related clinical findings in the ClinVar database.a
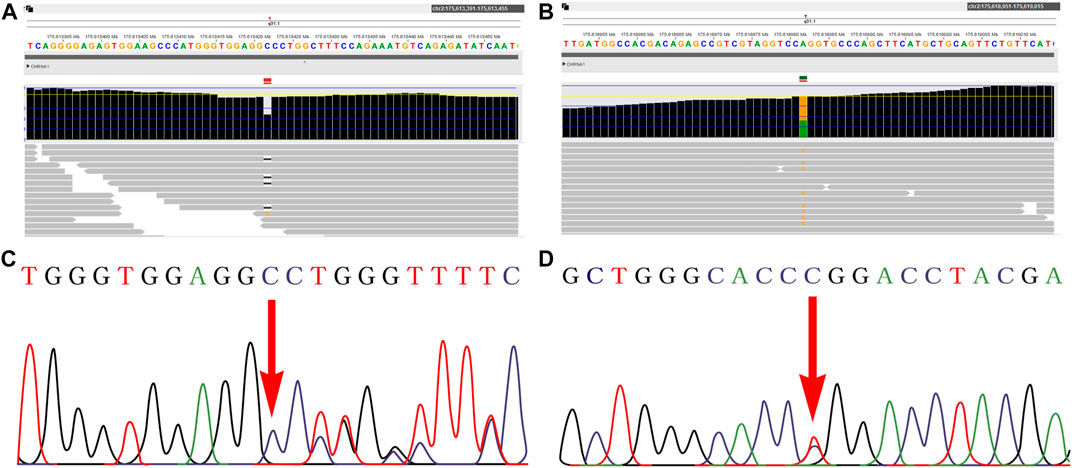
FIGURE 2. Novel variants of the CHRNA1 gene identified in the fetus. A–B: WES detection results demonstrated novel compound heterozyous variants in the CHRNA1 gene NM_000079.4: c.[1128delG (p.Pro377LeufsTer10)]; [505T>C (p.Trp169Arg)] in the fetus. C–D: both variants were further confirmed by Sanger sequencing.
In the clinical practice, chromosomal microarray analysis (CMA) or copy number variation sequencing has been increasingly used to assess the genetic cause in miscarriage and stillbirth (Sahlin et al., 2014; Martinez-Portilla et al., 2019; Marquès et al., 2020; Zhang et al., 2021). A recent meta-analysis of seven studies involving 903 stillborn fetuses demonstrated a 4% incremental yield of pathogenic copy number variants of CMA over karyotyping, among which 22q11.21 deletion was the most common variant responsible for stillbirth (Martinez-Portilla et al., 2019). However, more than half of them could not get a clear genetic diagnosis. WES has obvious advantages in the detection of monogenic diseases and is, therefore, suggested to be used in prenatal genetic etiology diagnosis in fetuses with ultrasonic structural abnormalities (Lord et al., 2019; Petrovski et al., 2019). In the present study, we detected two novel CHRNA1 gene mutations in the stillbirth fetus that may result in LMPS by WES.
Previous studies have indicated the application value of WES in identifying the genetic etiology for pregnancy loss or stillbirth (Fu et al., 2018; Demetriou et al., 2019; Zhao et al., 2021). More studies have identified CHRNA1 mutations in fetuses with recurrent pregnancy loss or stillbirth using WES. A systematic review of 50 studies (Colley et al., 2019) reported a range of candidate genes (CHRNA1, DYNC2H1, and RYR1) that may induce pregnancy loss. In addition, a recent study reviewed 15 articles of 74 families including 279 reported recurrent pregnancy loss, identified 34 candidate pathogenic variants in 19 genes including CHRNA1 gene by exome sequencing, and recommended that trio-based exome sequencing can be performed in cases with recurrent pregnancy loss and with normal parental karyotypes (Robbins et al., 2019). In addition, a novel mutation in CHRNA1 was identified by exome sequencing, which was suggested as the cause for recurrent fetal loss, and they hypothesized that exome sequencing could disclose the underlying autosomal recessive mutations in families with recurrent fetal loss (Shamseldin et al., 2013). In the present case study, we identified two compound heterozyous variants in the CHRNA1 gene, which further confirms the application value of WES in genetic analysis of recurrent pregnancy loss or stillbirth.
The nicotinic acetylcholine receptor (AChR) has five subunits of four different types: two alpha subunits and one each of beta, gamma (or epsilon), and delta subunits, which control electrical signaling between nerve and muscle cells by opening and closing a gate (Miyazawa et al., 2003). Among them, CHRNA1 encodes two alpha subunits, playing an important role in maintaining the AChR structure. As exhibited in the OMIM database, heterozygous mutations of CHRNA1 can cause autosomal dominant congenital slow-channel myasthenic syndrome and congenital fast-channel myasthenic syndrome, while homozygous mutations or compound heterozygous mutations of CHRNA1 would lead to autosomal recessive LMPS. A previous study identified homozygous mutations in the CHRNA1 gene in patients from two families with LMPS (Michalk et al., 2008). Among them, the homozygous c.761G>T (p.Arg234Leu) mutation was identified in both fetuses in family 1 with similar prenatal ultrasonic features including growth delay, edema, cystic hygroma, decreased movements, and joint contractures. They observed that the first fetus of family 1 was a stillbirth at a gestational age of 24 weeks, and the second fetus was terminated at a gestational age of 20 weeks. Like family 1, similar intrauterine problems were displayed in both fetuses of family 2, who carried the homozygous mutation of c.117-133dup17 (p.His25ArgfsX19) in the CHRNA1 gene. In addition, a study conducted by Dickinson et al. (2016) found that homozygous lethal mutation was observed in the CHRNA1-knockout mice. In the present study, compound heterozygous variants of NM_000079.4: c.[1128delG (p.Pro377LeufsTer10)]; [505T>C (p.Trp169Arg)] in the fetuses were detected, who manifested intrauterine edema, increased nuchal translucency, neck water sac tumor, and joint contractures, which are consistent with the intrauterine clinical features of LMPS.
At present, neither of two novel CHRNA1 variants identified in this study has been reported in the databases or the literature. Among them, the missense variant was classified as the variant of uncertain significance according to the ACMG guidelines. Given the consistence of the clinical phenotypes in the fetuses with LMPS and the genotype–phenotype segregation present in the family, we believe that both variants detected in CHRNA1 could be pathogenic variants and may lead to LMPS. However, future functional studies are required to clarify the molecular mechanism.
In conclusion, an etiologic diagnosis was conducted successfully in a Chinese family with recurrent fetal edema, fetal neck cystoma, and joint contractures by WES. This is the first study reporting the identification of two novel compound heterozyous variants in the CHRNA1 gene that may lead to LMPS. Our findings may broaden the spectrum of CHRNA1 gene mutations that result in LMPS and provide valuable data for the application of the prenatal WES technology and genetic consultation.
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Institutional Ethics Committee of Quanzhou women’s and children’s hospital to the commencement of the study (2020No. 31). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
JZ and JW wrote the article; XC, YC, and YJ recruited the participants and performed clinical consultation; YW, QL, and SZ performed the karyotype analysis and analyzed the data; GW, CC, and YX revised and polished the article. All authors approved the final article.
This research was sponsored by Fujian Provincial Health Technology Project (2020QNB045) and Quanzhou City Science and Technology Program of China (2020C026R).
The authors wish to express our appreciation to the Fujian Provincial Health Commission and Quanzhou Science and Technology Bureau for funding this work. They also express their appreciation to the patient who participated in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.964098/full#supplementary-material
Barros, F. S., Araujo Júnior, E., Rolo, L. C., and Nardozza, L. M. (2012). Prenatal diagnosis of lethal multiple pterygium syndrome using two-and three-dimensional ultrasonography. J. Clin. Imaging Sci. 2, 65. doi:10.4103/2156-7514.103055
Chen, C. P. (2012). Prenatal diagnosis and genetic analysis of fetal akinesia deformation sequence and multiple pterygium syndrome associated with neuromuscular junction disorders: a review. Taiwan. J. Obstet. Gynecol. 51 (1), 12–17. doi:10.1016/j.tjog.2012.01.004
Chong, J. X., Burrage, L. C., Beck, A. E., Marvin, C. T., McMillin, M. J., Shively, K. M., et al. (2015). Autosomal-dominant multiple pterygium syndrome is caused by mutations in MYH3. Am. J. Hum. Genet. 96 (5), 841–849. doi:10.1016/j.ajhg.2015.04.004
Colley, E., Hamilton, S., Smith, P., Morgan, N. V., Coomarasamy, A., and Allen, S. (2019). Potential genetic causes of miscarriage in euploid pregnancies: a systematic review. Hum. Reprod. Update 25 (4), 452–472. doi:10.1093/humupd/dmz015
Demetriou, C., Chanudet, E., GOSgene, , Topf, M., Thomas, A. C., Bitner-Glindzicz, M., et al. (2019). Exome sequencing identifies variants in FKBP4 that are associated with recurrent fetal loss in humans. Hum. Mol. Genet. 28 (20), 3466–3474. doi:10.1093/hmg/ddz203
Dickinson, M. E., Flenniken, A. M., Ji, X., Teboul, L., Wong, M. D., White, J. K., et al. (2016). High-throughput discovery of novel developmental phenotypes. Nature 537 (7621), 508–514. doi:10.1038/nature19356
Fu, M., Mu, S., Wen, C., Jiang, S., Li, L., Meng, Y., et al. (2018). Whole-exome sequencing analysis of products of conception identifies novel mutations associated with missed abortion. Mol. Med. Rep. 18 (2), 2027–2032. doi:10.3892/mmr.2018.9201
Hesselmans, L. F., Jennekens, F. G., Van den Oord, C. J., Veldman, H., and Vincent, A. (1993). Development of innervation of skeletal muscle fibers in man: relation to acetylcholine receptors. Anat. Rec. 236 (3), 553–562. doi:10.1002/ar.1092360315
Joshi, T., Noor, N. N., Kural, M., and Tripathi, A. (2016). Lethal multiple pterygium syndrome. J. Fam. Med. Prim. Care 5 (2), 477–478. doi:10.4103/2249-4863.192361
Lord, J., McMullan, D. J., Eberhardt, R. Y., Rinck, G., Hamilton, S. J., Quinlan-Jones, E., et al. (2019). Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet 393 (10173), 747–757. doi:10.1016/S0140-6736(18)31940-8
Marquès, B., Benitez, L., Peguero, A., Madrigal, I., Gomez, O., Figueras, F., et al. (2020). Cytogenetic investigation in 136 consecutive stillbirths: Does the tissue type Affect the success rate of chromosomal microarray analysis and karyotype? Fetal diagn. Ther. 47 (4), 315–320. doi:10.1159/000505399
Martinez-Portilla, R. J., Pauta, M., Hawkins-Villarreal, A., Rial-Crestelo, M., Paz Y Mino, F., Madrigal, I., et al. (2019). Added value of chromosomal microarray analysis over conventional karyotyping in stillbirth work-up: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 53 (5), 590–597. doi:10.1002/uog.20198
Meyer-Cohen, J., Dillon, A., Pai, G. S., and Conradi, S. (1999). Lethal multiple pterygium syndrome in four male fetuses in a family: evidence for an X-linked recessive subtype? Am. J. Med. Genet. 82 (1), 97–99. doi:10.1002/(sici)1096-8628(19990101)82:1<97::aid-ajmg22>3.0.co;2-g
Michalk, A., Stricker, S., Becker, J., Rupps, R., Pantzar, T., Miertus, J., et al. (2008). Acetylcholine receptor pathway mutations explain various fetal akinesia deformation sequence disorders. Am. J. Hum. Genet. 82 (2), 464–476. doi:10.1016/j.ajhg.2007.11.006
Miyazawa, A., Fujiyoshi, Y., and Unwin, N. (2003). Structure and gating mechanism of the acetylcholine receptor pore. Nature 423 (6943), 949–955. doi:10.1038/nature01748
Mohtisham, F. S., Sallam, A., and Shawli, A. (2019). Lethal multiple pterygium syndrome. BMJ Case Rep. 12 (5), e229045. doi:10.1136/bcr-2018-229045
Nazari, T., Rashidi-Nezhad, A., Ganji, M., Rezaei, Z., Talebi, S., Ghasemi, N., et al. (2019). Utilization of Whole exome sequencing in lethal form of multiple pterygium syndrome: identification of mutations in embryonal subunit of acetylcholine receptor. Int. J. Mol. Cell. Med. 8 (4), 258–269. doi:10.22088/IJMCM.BUMS.8.4.258
Penchaszadeh, V. B., and Salszberg, B. (1981). Multiple pterygium syndrome. J. Med. Genet. 18 (6), 451–455. doi:10.1136/jmg.18.6.451
Petrovski, S., Aggarwal, V., Giordano, J. L., Stosic, M., Wou, K., Bier, L., et al. (2019). Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet 393 (10173), 758–767. doi:10.1016/S0140-6736(18)32042-7
Ramer, J. C., Ladda, R. L., and Demuth, W. W. (1988). Multiple pterygium syndrome. an overview. Am. J. Dis. Child. 142 (7), 794–798. doi:10.1001/archpedi.1988.02150070108039
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17 (5), 405–424. doi:10.1038/gim.2015.30
Robbins, S. M., Thimm, M. A., Valle, D., and Jelin, A. C. (2019). Genetic diagnosis in first or second trimester pregnancy loss using exome sequencing: a systematic review of human essential genes. J. Assist. Reprod. Genet. 36 (8), 1539–1548. doi:10.1007/s10815-019-01499-6
Sahlin, E., Gustavsson, P., Liedén, A., Papadogiannakis, N., Bjareborn, L., Pettersson, K., et al. (2014). Molecular and cytogenetic analysis in stillbirth: results from 481 consecutive cases. Fetal diagn. Ther. 36 (4), 326–332. doi:10.1159/000361017
Shamseldin, H. E., Swaid, A., and Alkuraya, F. S. (2013). Lifting the lid on unborn lethal Mendelian phenotypes through exome sequencing. Genet. Med. 15 (4), 307–309. doi:10.1038/gim.2012.130
Tolmie, J. L., Patrick, A., and Yates, J. R. (1987). A lethal multiple pterygium syndrome with apparent X-linked recessive inheritance. Am. J. Med. Genet. 27 (4), 913–919. doi:10.1002/ajmg.1320270418
Vogt, J., Harrison, B. J., Spearman, H., Cossins, J., Vermeer, S., ten Cate, L. N., et al. (2008). Mutation analysis of CHRNA1, CHRNB1, CHRND, and RAPSN genes in multiple pterygium syndrome/fetal akinesia patients. Am. J. Hum. Genet. 82 (1), 222–227. doi:10.1016/j.ajhg.2007.09.016
Zhang, X., Huang, Q., Yu, Z., and Wu, H. (2021). Copy number variation characterization and possible candidate genes in miscarriage and stillbirth by next-generation sequencing analysis. J. Gene Med. 23 (12), e3383. doi:10.1002/jgm.3383
Keywords: whole-exome sequencing, chromosomal microarray analysis, CHRNA1, lethal multiple pterygium syndrome, stillbirth
Citation: Zhuang J, Wang J, Luo Q, Zeng S, Chen Y, Jiang Y, Chen X, Wang Y, Xie Y, Wang G and Chen C (2022) Case Report: Novel compound heterozygous variants in CHRNA1 gene leading to lethal multiple pterygium syndrome: A case report. Front. Genet. 13:964098. doi: 10.3389/fgene.2022.964098
Received: 08 June 2022; Accepted: 02 August 2022;
Published: 26 August 2022.
Edited by:
Muhammad Jawad Hassan, National University of Medical Sciences (NUMS), PakistanReviewed by:
Amjad Khan, Université de Strasbourg, FranceCopyright © 2022 Zhuang, Wang, Luo, Zeng, Chen, Jiang, Chen, Wang, Xie, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunnuan Chen, Y2hlbmNodW5udWFuMTk4M0BhbGl5dW4uY29t; Gaoxiong Wang, d2FuZ2dhb3hpb25nMjAxM0AxNjMuY29t; Yingjun Xie, eGlleWp1bkBtYWlsMi5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.