
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 21 November 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.963964
This article is part of the Research Topic Molecular basis of epigenetic regulation in cancer therapies View all 21 articles
 Miao Jiang1†
Miao Jiang1† Yongliang Jia2†
Yongliang Jia2† Jinming Han1
Jinming Han1 Jianxiang Shi3
Jianxiang Shi3 Chang Su1
Chang Su1 Rui Zhang1
Rui Zhang1 Menglu Xing1
Menglu Xing1 Shuiling Jin1*
Shuiling Jin1* Hong Zong1*
Hong Zong1*Objective: Studies have demonstrated an association between somatic POLE exonuclease domain mutations (EDMs) and the prognosis of colorectal cancer (CRC). However, the prognostic value of POLE non-EDMs remains unclear. This retrospective study aimed to explore the possible relationships between POLE mutation subtypes and CRC prognosis.
Methods: The 272 CRC patients from the First Affiliated Hospital of Zhengzhou University (ZZ cohort) and 499 CRC patients from The Cancer Genome Atlas database (TCGA cohort) were retrospectively collected. The cases were divided into subgroups based on POLE mutation sites and microsatellite instability (MSI) status. The continuous variables were compared among three subgroups with Kruskal-Wallis tests. Pairwise comparisons between three groups were performed by Bonferroni correction method, and adjusted p < 0.05 was considered statistically significant. The categorical variables were compared with Chi-square test and Fisher’s exact test. The Kaplan—Meier curves and Cox regression models were conducted to evaluate prognostic values of POLE mutations.
Results: In the ZZ cohort, POLE EDMs (2.6%) were significantly associated with younger age (p = 0.018) and localized in the left colon (p = 0.001). POLE non-EDMs were significantly associated with MSI-high status (p < 0.001) and localization in the right colon (p = 0.001). In the TCGA cohort, the tumor mutation burden (TMB) of both POLE EDM tumors (p < 0.001) and POLE non-EDM tumors (p < 0.001) was significantly higher than that of POLE wild-type (WT) tumors. A similar trend was observed in the ZZ cohort, although there were no significant differences. In the ZZ cohort, the POLE EDM group had higher progression-free survival (PFS) (p = 0.002) and overall survival (OS) (p = 0.042) than the POLE non-EDM group and POLE WT group. We also report one CRC patient harboring a germline POLE mutation who received camrelizumab and exhibited long-term stable disease.
Conclusion: Both POLE-EDMs and POLE non-EDMs were associated with significantly increased TMB in CRC and may be biomarkers for CRC treatment and prognosis. Current evidence does not support an effect of POLE non-EDMs on PFS and OS. A significant association between POLE EDMs and improved PFS and OS may exist, but future studies with larger sample sizes are needed. Entire coding region of the POLE gene should be screened.
The global incidence and mortality of colorectal cancer (CRC) rank first among gastrointestinal cancers (Sung et al., 2021). The poor prognosis of CRC is mainly due to its insidious onset, as approximately 25% of patients have metastasized CRC at the time of diagnosis, resulting in limited treatment options (Andre et al., 2015; Bryan et al., 2018). CRC is a highly heterogeneous cancer that develops mainly by affecting the expression and behavior of genes related to cell growth and differentiation (Fearon, 2011). In recent years, increasing studies have indicated that mutations in the DNA polymerase gene POLE mutation may be important for guiding CRC management, and are a potential biomarker for treatment and prognosis (Huhns et al., 2020).
The nuclear DNA replication-repair-associated polymerases Pol α, Polδ and Polε all belong to the polymerase B family (Doublie and Zahn, 2014). During replication, the main function of POLε is to lengthen the leading strand. The catalytic subunit of POLε has 5′ to 3′ DNA polymerase activity and 3′ to 5′ exonuclease activity and is capable of the timely removal of erroneous bases generated during replication to ensure the fidelity of DNA replication. This catalytic subunit is encoded by POLE (Henninger and Pursell, 2014). In 2012, The Cancer Genome Atlas (TCGA) exome sequencing project conducted a complete genome analysis of 224 CRC cases and showed that POLE mutation is closely related to an ultra-hypermutated phenotype (TMB >100 mut/Mb) (Cancer Genome Atlas, 2012). Subsequently, several studies have shown that CRC patients carrying POLE mutations often have TMB and infiltration of immune cells in tumors (Forgó et al., 2020; Picard et al., 2020). The aggregation of epitopes in tumors makes them more susceptible to immune checkpoint inhibitors (ICIs). To date, microsatellite instability-high (MSI-H)/deficient mismatch repair (dMMR) is the only widely recognized specific biomarker related to the positive effect of ICIs in CRC treatment (Andre et al., 2020). However, only 5% of CRC patients have MSI-H/dMMR (Battaglin et al., 2018). POLE mutations have the potential to serve as a specific biomarker to screen for candidates who may benefit from ICIs. In addition, similar to MSI in nonmetastatic CRC, POLE mutations also imply lower recurrence and metastasis rates. For stage II CRC patients whose need for adjuvant therapy is still controversial, POLE mutations indicate a better prognosis and may be important evidence for guiding treatment decisions.
In the predictions of treatment and prognosis of CRC, somatic POLE mutations have been reported to be a promising candidate biomarker. However, most studies have focused on POLE exonuclease domain mutations (EDMs) or individual mutation points. The significance of POLE non-EDMs in CRC remains unclear. Thus, this retrospective study investigated the clinical characteristics and prognostic value of POLE mutation subtypes in a real-world dataset. A similar analysis was carried out in a TCGA dataset, and the results of the two cohorts are compared and discussed.
The Chinese cohort included 272 CRC patients treated at The First Affiliated Hospital of Zhengzhou University (ZZ cohort) between January 2016 and December 2020. The latest follow-up date was 1 March 2021. All patients were pathologically diagnosed with primary CRC by tissue biopsy and underwent NGS. Ethics committee approval was obtained from the institutional research ethics board (NO. 2021-KY-1040-002). Data from 499 CRC patients in the TCGA database (PanCancer Atlas) (TCGA cohort) were downloaded (15 January 2022) and included in the statistical analysis (http://www.cbioportal.org/). Patients with insufficient information, including POLE status and follow-up information, were excluded. The following factors were extracted for statistical analysis: age, sex, MSI status, pathology, tumor location, tumor differentiation, clinical stage at the time of diagnosis, depth of tumor invasion, lymph node metastases, and hazard factors.
The genomic profiling was conducted by a hybridization capture-based NGS assay using a commercial panel consisting of 520 cancer-associated genes (OncoScreen Plus, Burning Rock Biotech), spanning 1.64 Mb of the human genome (Wang et al., 2022). Tissue DNA was fragmented using Covaris M220 (Covaris, MA, United States) followed by end repair, adapter ligation and purification of fragments with sizes between 200 and 400 base pairs. Fragment size and quality were assessed with high-sensitivity DNA kit using Bioanalyzer 2100 (Agilent Technologies, CA, United States). Subsequently, the Indexed samples were sequenced on the NovaSeq 6000 platform (Illumina, Inc., CA, United States) with 150-base pair read lengths.
Sequence data were analyzed using the Burning Rock analysis system. Concisely, raw reads were aligned to the reference human genome (hg19) using Burrows-Wheeler Aligner (version 0.7.10). Variant calling was implemented using VarScan (version 2.4.3) with the following filtering steps to retain high-confidence variants: loci with depths ≥100, at least eight supporting reads for single nucleotide variations (SNVs), at least two and five supporting reads for Indel variants. Single nucleotide polymorphisms (SNPs) were all removed.
TMB was defined as the number of non-synonymous variants per megabase of genome examined, and was estimated using the OncoScreen Plus panel (OncoScreen plus, Burning Rock, Guangzhou, China) with a total size of 1.003 Mb of coding regions. Hotspot variants, copy number variations, structural variants, and germline SNPs are not counted.
MSI status of tumor and plasma samples was determined using a read-count-distribution-based approach that utilizes a given set of repeat lengths of coverage as the prime characteristic of each microsatellite locus. A locus is classified as unstable if more than 30% of the total number of microsatellite markers in the sample is below this threshold.
Overall survival (OS) was defined as the time from histological diagnosis of CRC to death. Progression-free survival (PFS) was defined as the time from first-line therapy to the first tumor progression or recurrence. The end date was defined as the date of the last follow-up visit if there was no cancer recurrence or death. Continuous variables are described as the mean and standard deviation or the median and the interquartile range. Categorical variables are described with frequencies and percentages. The continuous variables (age at diagnosis, TMB) were compared among POLE EDM, POLE non-EDM and POLE WT groups with Kruskal-Wallis tests. Pairwise comparisons between three groups were performed using the Bonferroni correction method, and an adjusted p value < 0.05 was considered statistically significant. The same method was used to compare TMB levels among POLE non-EDM (MSI-L/MSS), POLE WT (MSI-H) and POLE WT (MSI-L/MSS) groups. The categorical variables were compared among POLE EDM, POLE non-EDM and POLE WT groups with Chi-square test and Fisher’s exact test. Survival function curves were generated using the Kaplan-Meier method (Ying et al., 2021). Survival differences among groups were evaluated by the log-rank test. Univariate and multivariate Cox regression models were employed to evaluate the prognostic value of POLE mutations for OS and PFS(Burke et al., 2017). All statistical analyses were performed with SPSS version 23.0 software (IBM, Chicago, IL). A two-tailed p value < 0.05 was considered statistically significant.
The protein distribution of POLE mutations is shown in Figure 1. In the ZZ cohort, the somatic POLE mutation rate was 7.7% (21 out of 272), including 2.6% (7 out of 272) of POLE EDMs and 5.1% (14 out of 272) of POLE non-EDMs. Five of the seven POLE EDMs were known pathogenic mutations (V411 L in 1 case, P286R in 4 cases). A mutation of unknown significance (E396 fs) was detected in 2 cases (Figure 1A). The location and genetic characteristics of each POLE mutation are shown in Table 1. Compared with POLE WT tumors, POLE non-EDM tumors were mainly MSI-H (p < 0.001). Most POLE EDM tumors were MSI-L/MSS; however, the difference was not significant.
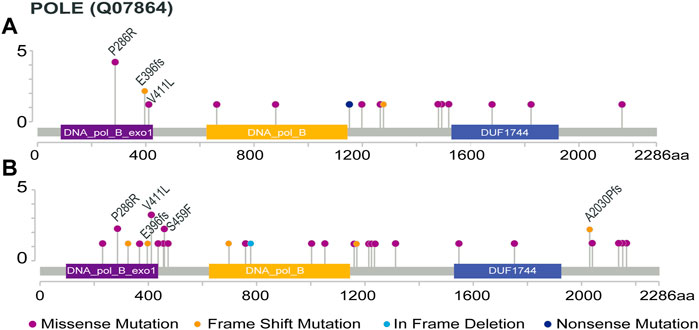
FIGURE 1. Protein distribution of POLE mutations. POLE ED including 86 to 427 amino acids (http://pfam.xfam.org/protein/DPOE1_HUMAN). (A) POLE mutations in the ZZ cohort, except 2 cases with intronic deletion in exon 25 and 1 case with long fragment insertion in exon 29 (the specific amino acid sites are unknown); (B) POLE mutations in the TCGA cohort. Recurring protein changes are labeled.
In the TCGA cohort, the somatic POLE mutation rate was 6.6% (33 out of 499), including 1.8% (9 out of 499) of POLE EDMs and 4.8% (24 out of 499) of POLE non-EDMs. The 9 POLE EDMs comprised 5 known pathogenic POLE mutations (P286R, 2 cases; V411 L, 3 case). E396fs was also detected in one case (Figure 1B). Compared with POLE WT tumors, POLE non-EDM tumors were mainly MSI-H, and POLE EDM tumors were mainly MSI-L/MSS (p < 0.001).
In the TCGA cohort, compared with POLE WT tumors, both POLE EDM tumors (median TMB = 115.3 mut/Mb, p < 0.001) and POLE non-EDM tumors (median TMB = 64.2 mut/Mb, p < 0.001) exhibited a significantly increased TMB. A similar trend was observed in the ZZ cohort; however, in the pairwise comparisons, the Bonferroni corrected p values indicated no significant difference between each pair of groups (p > 0.05).
Given that POLE non-EDM tumors are mostly MSI-H (ZZ cohort p < 0.001; TCGA cohort p < 0.001), this study further compared the TMB level among the POLE non-EDM (MSI-L/MSS), POLE WT (MSI-H) and POLE WT (MSI-L/MSS) subgroups to explore whether the high TMB in the POLE non-EDM group should be attributed to POLE non-EDM or MSI-H status. Since only 1 case with POLE non-EDM (MSI-L/MSS), in the ZZ cohort, the difference of TMB levels among groups including POLE non-EDM (MSI-L/MSS), POLE WT (MSI-H) and POLE WT (MSI-L/MSS) were only explored in the TCGA cohort. The results showed that both the POLE non-EDM (MSI-L/MSS) group (median TMB = 93.2 mut/Mb, p = 0.015) and the POLE WT (MSI-H) group (median TMB = 36.4 mut/Mb) (p < 0.001) had significant higher TMB levels than the POLE WT (MSI-L/MSS) group (median TMB = 3.3 mut/Mb). The first two groups had similar TMB levels (p = 0.613), and both tended to be hypermutated phenotypes (Table 2).
In the ZZ cohort, the POLE EDM group had younger age at diagnosis (p = 0.018) and more frequent left-sided tumor localization (p = 0.002). Right-sided tumor localization was more frequent in the POLE non-EDM group (p = 0.001). Most POLE EDM tumors were diagnosed at an early stage and had a low risk of recurrence. Among POLE EDM tumors, 3 at stage II (42.9%, p = 0.536), 5 at pT3 (71.4%, p = 0.744), 5 at N0 (71.4%, p = 0.728), 6 at M0 (85.7%, p = 0.247) and 6 had no hazard factors (85.7%, p = 0.181).
In the TCGA cohort, POLE EDM mostly occurred in male patients (p = 0.013). Patients with POLE non-EDM tumors more frequent had right-sided tumor localizations (p = 0.010) and adenocarcinoma histology (p = 0.004). Among POLE EDM tumors, there were 6 at stage II (66.6%, p = 0.207), 7 at pT3 (77.7%, p = 0.334),7 at N0 (77.7%, p = 0.305) and 8 at M0 (88.9%, p = 0.805). The detailed clinicopathological features of patients in the ZZ cohort and TCGA cohort are summarized in Table 2.
All patients were divided into 3 subgroups: the POLE EDMs, POLE non-EDMs and POLE WT groups. In the ZZ cohort, the 272 CRC patients were followed for a median of 16.8 months. Since no patients in the POLE EDM group had progressed by the last follow-up, the median PFS was not reached. Based on the stratified log-rank test, the PFS rate of the POLE EDM group was significantly higher than that of the POLE non-EDM (median = 22.0 months, χ2 = 5.407, p = 0.020) and POLE WT groups (median = 14.6 months, χ2 = 8.830, p = 0.003) (Figure 2A). The OS of the POLE EDM group and POLE non-EDM group were not reached. There was no significant difference in OS among these three groups (p = 0.056) (Figure 2B).
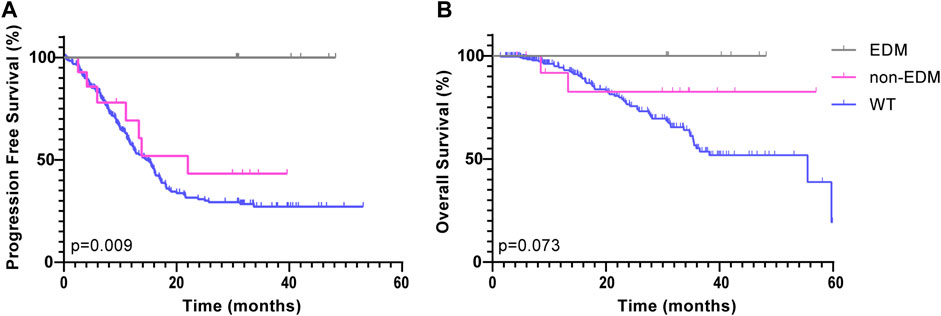
FIGURE 2. Kaplan-Meier survival curves of patients in the ZZ cohort according to POLE mutation status. PFS (A) and OS (B) of POLE EDMs, non-EDMs and WT patients in the ZZ cohort.
POLE WT group was further subdivided into the POLE WT (MSI-L/MSS) subgroup and the POLE WT (MSI-H) subgroup. Based on the Kaplan-Meier analysis, both the POLE EDM group (χ2 = 9.845, p = 0.002) and the POLE WT (MSI-H) group (χ2 = 7.036, p = 0.008) showed improved PFS compared to the POLE WT (MSI-L/MSS) group (median = 13.3 months) (Figure 3A). In analyses that used OS as the end point, the POLE EDM group (χ2 = 4.125, p = 0.042) and the POLE WT (MSI-H) group (χ2 = 6.032, p = 0.014) also showed better outcomes than the POLE WT (MSI-L/MSS) group (median = 38.2 months) (Figure 3B). The prognosis of the POLE EDM group and POLE WT (MSI-H) group was similar. The PFS and OS of the POLE EDM group and POLE WT (MSI-H) group were not reached.
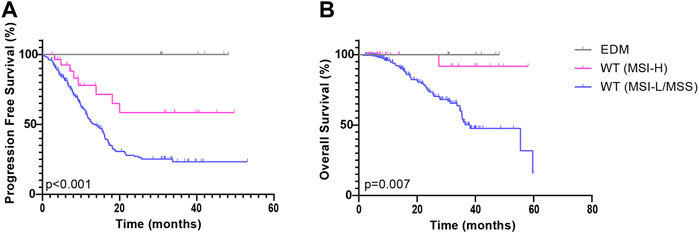
FIGURE 3. Kaplan-Meier survival curves of patients in the ZZ cohort according to POLE mutation and MMR status. PFS (A) and OS (B) of patients in the POLE EDM, POLE WT (MSI-H) and POLE WT (MSI-L/MSS) groups.
In this study, POLE non-EDM tumors were mostly MSI-H (ZZ cohort p < 0.001; TCGA cohort p < 0.001). To exclude the effect of the interaction of MSI and POLE non-EDMs on PFS, the Kaplan—Meier survival curves of the POLE non-EDM (MSI-L/MSS) group, POLE WT (MSI-H) group, and POLE WT (MSI-L/MSS) group were compared with pairwise comparisons to explore the influence of POLE non-EDMs alone. In the ZZ cohort, PFS exhibited similar trends in the POLE non-EDM (MSI-L/MSS) group (median = 13.3 months, χ2 = 0.131, p = 0.718) and POLE WT (MSI-L/MSS) group (median = 13.3 months) (Figure 4A). Similarly, here was no significant difference between the POLE non-EDM (MSI-L/MSS) group (median = 13.3 months, χ2 = 1.361, p = 0.243) and the POLE WT (MSI-L/MSS) group (median = 38.2 months) on OS (Figure 4B). With the univariate and multivariate Cox regression models, POLE EDM and POLE non-EDM were both prognostic protective factors (HR<1) without statistical significance levels (Table 3, Supplementary Table S1). Distant metastasis and advanced clinical stage (stage III-IV) were independent risk factors for shortened PFS while age ≥60 and poor differentiation (G3) were independent risk factors for shortened OS.
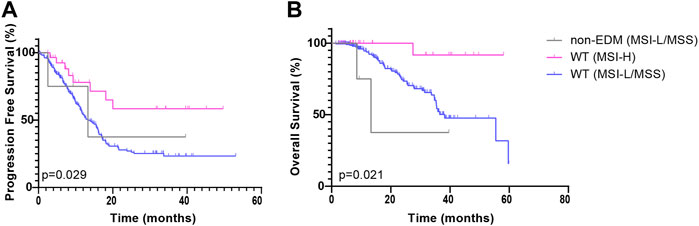
FIGURE 4. Kaplan-Meier survival curves of patients in the ZZ cohort according to POLE mutation and MMR status. PFS (A) and OS (B) of patients in the POLE non-EDM, POLE WT (MSI-H) and POLE WT (MSI-L/MSS) groups.
In the TCGA cohort, the median follow-up periods of 499 CRC patients was 22.0 months. The similar analyses were also performed among these three subgroups in the TCGA cohort; however, there were no significant differences on PFS and OS among the groups. With the univariate and multivariate Cox regression models of the TCGA cohort, POLE EDM and POLE non-EDM were both prognostic risk factors (HR>1) without statistical significance levels (Table 3, Supplementary Table S2). Lymph node metastasis and pT4 were independent risk factors for shortened PFS. Distant metastasis, pT4 and age >60 years are independent risk factors for shortened OS.
Polymerase proof-reading associated polyposis and Lynch-like syndrome are inherited cancer susceptibility syndromes associated with germline POLE mutations. (Elsayed et al., 2015; Vande Perre et al., 2019). Such patients often progressively develop CRC or extraintestinal tumors (Bellido et al., 2016). Identifying germline POLE mutations may help to understand the pathogenesis of CRC, reduce the morbidity and mortality, and guide treatment. The germline POLE mutations identified in CRC patients published from 2017 to 2020 are summarized in Table 4. There were two relatively rare cases in which p. V411L was previously described as a somatic hotspot alteration and p. V474I was located outside the ED. This study also reports a rare case.
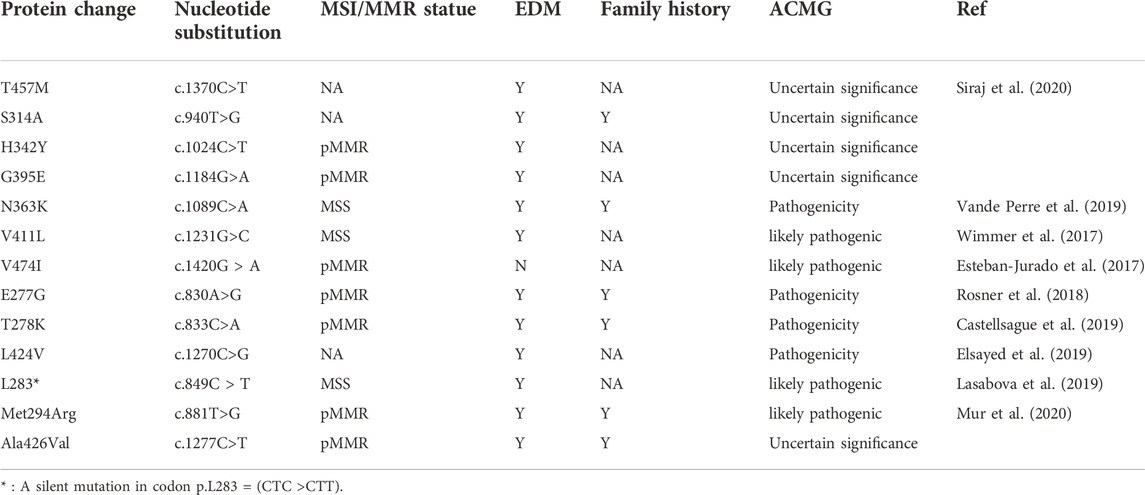
TABLE 4. Summary of germline POLE mutations in colorectal cancer reported in published articles (2017–2020).
A male patient with abdominal pain, abdominal distention, and difficulty defecating was referred to our center in December 2019. Medical imaging examination and tissue biopsy suggested bowel obstruction and rectal adenocarcinoma with multiple lymph node metastases. He received first-line treatment with an oxaliplatin plus capecitabine regimen. He developed adrenal metastasis 3 months later and was treated with bevacizumab. However, rectal occupation progressed soon after this addition, so the above regimen was stopped. Treatment with “FOLFIRI + bevacizumab” began on 16 April 2020; however, the effect was poor. The tumor continued to progress, and the patient presented with liver metastasis 2 months later. NGS results of a 41-gene panel suggested the presence of a POLE mutation (exon 45, S2084 fs), KRAS mutation (G12S), TP53 (R2084 fs) and MSS. The administration of anlotinib and camrelizumab began on 8 June 2020 and was continued until the last follow-up (6 December 2021), with no progression observed (PFS >18 months) (Figures 5, 6). The POLE mutation was an inherited germline mutation located outside the ED; this variant has not been previously identified in a large population database. According to the ACMG 2015 guidelines, this variant was evaluated as a hereditary variant with possible pathogenicity. The patient had MSS but received sustained long-term benefit from immunotherapy. It is believed that POLE mutation may be used to predict the response to ICIs.
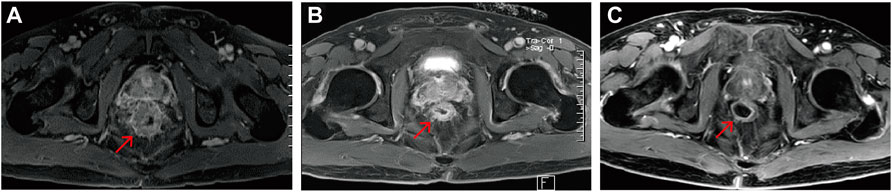
FIGURE 5. Rectal magnetic resonance images of the POLE-mutant rectal cancer patient receiving camrelizumab and anlotinib. (A) Pre-immunotherapy; (B) 7 months post—immunotherapy; (C) 18 months post—immunotherapy. Red arrows indicate the same rectal tumor lesion.
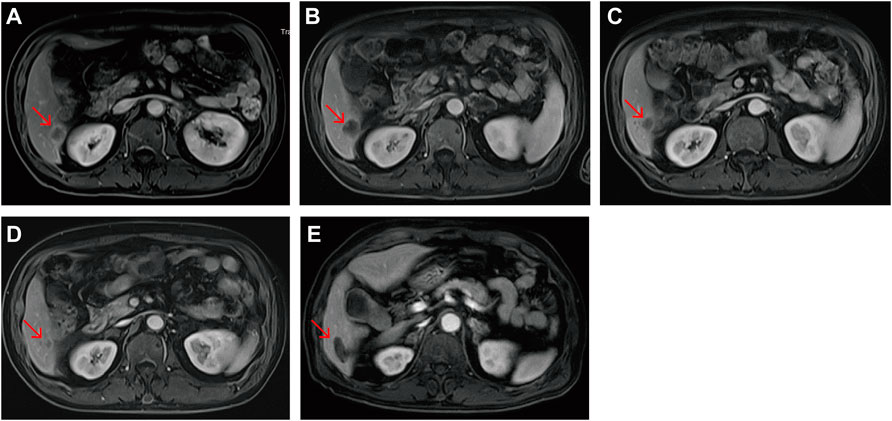
FIGURE 6. Liver metastasis magnetic resonance images of the POLE-mutant rectal cancer patient receiving camrelizumab and anlotinib. (A) Pre-immunotherapy; (B) 1 month post—immunotherapy; (C) 3 months post—immunotherapy; (D) 7 months post—immunotherapy; (E) 9 months post—immunotherapy. The image of the metastasis 2 months after argon-helium cryoablation. Red arrows indicate the same liver metastases.
This study included mutations inside and outside of the POLE exonuclease domain. We aimed to explore the molecular pathological features and prognostic value of different POLE mutation subtypes. As previously reported, in the Asian population, POLE EDMs are mainly found in the left colon and relatively young CRC patients (Hino et al., 2019; Hu et al., 2021), whereas POLE non-EDMs are more common in the right colon.
Somatic POLE mutations were evenly located throughout the POLE gene with no apparent tendency to cluster as shown in Figure 1. In the ZZ cohort, the frequency of POLE EDMs was 2.6%, while the frequency of POLE non-EDMs was 5.1%; in the TCGA cohort, the frequencies were 1.8% and 4.8%, respectively. This finding is consistent with the previously reported frequency of POLE mutations in CRC (Campbell et al., 2017). Although the frequency of POLE mutations is low, its unique high immunogenicity has attracted widespread attention.
Tumors harboring POLE EDMs often manifest with a high TMB, which is associated with an enhanced intertumoral immune response and better outcome (Llosa et al., 2015). This discovery was first reported in the TCGA whole-exome sequencing project in 2012 and is a critical first step for moving treatment of toward precision therapy. In addition, some tumors harboring only POLE non-EDMs also exhibited elevated mutation burdens, such as C810 and E978. This study showed that both POLE EDMs and POLE non-EDMs were associated with significantly increased TMB (Table 2). Since the POLE EDM tumors in this study were mostly MSI-L/MSS, and the POLE non-EDM tumors were mostly MSI-H, we analyzed the data again after excluding the interference of MSI status and still reached the same conclusion. POLE EDM tumors tended to have ultra-hypermutated phenotypes (TMB>100 mut/Mb), and POLE non-EDM tumors tended to have hypermutated phenotypes (TMB>10 mut/Mb). Although consistent with previous reports that POLE EDMs are predominantly MSS, this study identified 3 cases of CRC harboring both POLE E396fs and MSI-H (ZZ cohort, 2 cases; TCGA cohort, 1 case) (Stenzinger et al., 2014; Kawai et al., 2021). All 3 cases were stage II CRC with prolonged PFS. The significance of this mutation merits further study. This study indicated that mutation location is not a determining factor for the predictive value of POLE mutations. Thus, it is necessary to thoroughly assess POLE mutations throughout the coding region.
In tumors with MSI/dMMR or POLE mutations, the production of new antigens is caused by a large accumulation of nonsynonymous substitution and/or frameshift mutations. Major histocompatibility complexs can present these new antigens to the immune system, thereby enhancing the immune system’s attack on tumor cells. In recent years, several patients with both POLE EDMs and MSS have been reported to obtain clinical benefit from ICI treatment (Guerra et al., 2017; Keenan et al., 2021). A study of a cohort of 295 patients with stage II CRC indicated that POLE mutant tumors have significantly elevated mutation levels (Domingo et al., 2016). These patients have a better prognosis and may not require adjuvant treatment. Studies have indicated that the predicted amount of new antigens in MSI/dMMR tumors is 10–50 times those in MSS tumors, and in POLE mutant tumors produce 15 times the amount of new antigens compared to that of MSI/dMMR tumors (Shinbrot et al., 2014; Howitt et al., 2015). Therefore, the prognosis and treatment response of CRC patients with POLE mutations may be improved and enhanced.
MSI and POLE mutations have similar effects on tumors. To exclude the influence of MSI status and thus determine the prognostic value of POLE mutation itself, this study conducted 3 subgroup analyses according to POLE mutation and MSI status. Additionally, the prognostic value of MSI status and POLE mutation was compared.
First, this study divided all patients into three groups: the POLE EDM, POLE non-EDM and POLE WT groups. In the ZZ cohort, we found that POLE EDM tumors were less prone to recurrence or progression than POLE WT tumors (Figure 2A). POLE non-EDM tumors did not show a PFS advantage. Moreover, no difference in OS was observed among the groups (Figure 2B). Subsequently, we divided the patients into POLE EDM, POLE WT (MSI-H) and POLE WT (MSI-L/MSS) subgroups. In the ZZ cohort, POLE EDM and POLE WT (MSI-H) tumors had better OS and PFS outcomes than POLE WT (MSI-L/MSS) tumors (Figure 3). POLE EDMs and MSI-H status had similar roles in improving the prognosis of CRC. Finally, we divided the patients into POLE non-EDM (MSI-L/MSS), POLE WT (MSI-H) and POLE WT (MSI-L/MSS) subgroups. POLE non-EDM tumors did not show improvement or deterioration of PFS or OS in the ZZ cohort (Figure 4). Based on the above subgroup analyses, POLE EDMs and MSI-H statue improve clinical outcomes to a similar degree. Currently, POLE non-EDMs do not demonstrate this beneficial effect.
In the univariate and multivariate Cox regression models, POLE EDMs and POLE non-EDMs were both protective factors for PFS and OS prolongation (HR<1) in the ZZ cohort but did not reach statistical significance levels (Table 3). We considered that the accuracy and validity of the Cox regression model was reduced due to the high proportion of censored data for most patients who did not reach the clinical outcome of PFS or OS.
In this study, the above 3 subgroup analyses were also performed in the TCGA cohort; however, POLE mutations did not show an effect on the PFS or OS outcomes. Paradoxically, POLE mutation may be a risk factor for reduce PFS and OS in the TCGA cohort (HR > 1). It should be noted that the cases in the TCGA cohort were diagnosed from 1998 to 2013. However, the clinical application of ICIs has only gradually been realized in the past 5 years. POLE mutations and MSI-H statue are both factors closely related to the effect of immunotherapy. Therefore, the above contradictory results are likely related to the application of ICIs. In addition, it is worth noting that only 12 (2.4%) cases in the TCGA dataset were Asian, and the differences between ethnic groups cannot be ignored.
Somatic POLE mutations have the potential to guide personalized treatment, thereby improving clinical outcomes. The discovery of germline POLE mutations is highly important for reducing the incidence of CRC. Esteban et al. reported a germline POLE mutation (V474I) located outside the ED (Esteban-Jurado et al., 2017). This study also identified a potentially pathogenic germline POLE non-EDM (S2084 fs). This metastatic rectal cancer patient progressed rapidly after early treatment but obtained continued benefits after receiving camrelizumab and anlotinib (PFS >18 months). Interestingly, after two cycles of application of this regimen, MRI scans showed that the metastasis in the right lower lobe of the liver first increased and then gradually decreased and remained stable after continuous administration (Figure 6). We suggested the efficiency of ICI treatment should not be evaluated too soon after application due to the temporary increase in reactivity.
This study excluded the effect of MSI status on tumors and extended the scope of the study to the entire region of the POLE gene. We fully analyzed the clinico-molecular pathological features of POLE EDM tumors and POLE non-EDM tumors and the prognostic impact of POLE mutation subtypes from different aspects. In addition, this study compared the difference between the effects of POLE mutation and MSI status on CRC. Our study also had a few limitations. First, patients with POLE mutations had a high survival rate and PFS rate, and the insufficient follow-up time resulted in insufficient statistical power for some subgroups. We need to continue to closely follow-up with these patients. Second, the total number of POLE mutation was small, and additional studies are required to verify the applicability of the findings in this study. Third, racial differences in the clinical characteristics and prognosis of CRC patients with POLE mutations should be explored further in future studies.
In conclusion, both POLE EDMs and POLE non-EDMs were associated with significantly increased TMB in CRC, which is an important biomarker for CRC treatment and prognosis. It is also necessary to study the entire region of the POLE gene. POLE EDMs may be significantly associated with prolonged PFS and OS; however, the evidence is currently insufficient. Future studies need larger sample sizes to provide more data. The current data do not support the impact of POLE non-EDMs on CRC prognosis. Future studies need to eliminate the interference caused by ethnicity and treatment to analyze the specific role of POLE genes more accurately.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
MJ conceived and designed this paper. Study implementation and feasibility analysis for JH and JS. YJ analyzed and explained the results. SJ modified the later versions. HZ and SJ revised the pictures and the manuscript.
This study was conducted with support from the Health Commission of Henan Province (Nos. SBGJ202102136 and SBGJ202102137). The work of YJ was supported by Zhengzhou University (Grant No. 32212456), Henan Institute of Medical and Pharmacological Sciences (Grant No. 2021BP0113), and Henan Science and Technology Department (Grant No. 222102310721).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.963964/full#supplementary-material
ACMG, American College of Medical Genetics and Genomics; CRC, colorectal cancer; dMMR, deficient mismatch repair; ED, exonuclease domain; EDMs, exonuclease domain mutations; ICIs, immune checkpoint inhibitors; MSI, microsatellite instability; MSS, microsatellite stability; mCRC, metastasis colorectal cancer; NA, not assessable; OS, overall survival; PFS, progression-free survival; pMMR, proficient mismatch repair; TCGA, The Cancer Genome Atlas; TMB, tumor mutation burden; 5-FU, 5-Fluorouracil; WT, wild-type; Y, yes; N, no.
Andre, T., de Gramont, A., Vernerey, D., Chibaudel, B., Bonnetain, F., Tijeras-Raballand, A., et al. (2015). Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: Updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J. Clin. Oncol. 33 (35), 4176–4187. doi:10.1200/JCO.2015.63.4238
Andre, T., Shiu, K. K., Kim, T. W., Jensen, B. V., Jensen, L. H., Punt, C., et al. (2020). Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 383 (23), 2207–2218. doi:10.1056/NEJMoa2017699
Battaglin, F., Naseem, M., Lenz, H. J., and Salem, M. E. (2018). Microsatellite instability in colorectal cancer: Overview of its clinical significance and novel perspectives. Clin. Adv. Hematol. Oncol. 16 (11), 735–745.
Bellido, F., Pineda, M., Aiza, G., Valdes-Mas, R., Navarro, M., Puente, D. A., et al. (2016). POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: Review of reported cases and recommendations for genetic testing and surveillance. Genet. Med. 18 (4), 325–332. doi:10.1038/gim.2015.75
Bryan, S., Masoud, H., Weir, H. K., Woods, R., Lockwood, G., Smith, L., et al. (2018). Cancer in Canada: Stage at diagnosis. Health Rep. 29 (12), 21–25.
Burke, D. L., Ensor, J., and Riley, R. D. (2017). Meta-analysis using individual participant data: One-stage and two-stage approaches, and why they may differ. Stat. Med. 36 (5), 855–875. doi:10.1002/sim.7141
Campbell, B. B., Light, N., Fabrizio, D., Zatzman, M., Fuligni, F., de Borja, R., et al. (2017). Comprehensive analysis of hypermutation in human cancer. Cell 171 (5), 1042–1056. doi:10.1016/j.cell.2017.09.048
Cancer Genome Atlas, N. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 (7407), 330–337. doi:10.1038/nature11252
Castellsague, E., Li, R., Aligue, R., Gonzalez, S., Sanz, J., Martin, E., et al. (2019). Novel POLE pathogenic germline variant in a family with multiple primary tumors results in distinct mutational signatures. Hum. Mutat. 40 (1), 36–41. doi:10.1002/humu.23676
Domingo, E., Freeman-Mills, L., Rayner, E., Glaire, M., Briggs, S., Vermeulen, L., et al. (2016). Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: A retrospective, pooled biomarker study. Lancet. Gastroenterol. Hepatol. 1 (3), 207–216. doi:10.1016/s2468-1253(16)30014-0
Doublie, S., and Zahn, K. E. (2014). Structural insights into eukaryotic DNA replication. Front. Microbiol. 5, 444. doi:10.3389/fmicb.2014.00444
Elsayed, F. A., Kets, C. M., Ruano, D., van den Akker, B., Mensenkamp, A. R., Schrumpf, M., et al. (2015). Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur. J. Hum. Genet. 23 (8), 1080–1084. doi:10.1038/ejhg.2014.242
Elsayed, F. A., Tops, C. M. J., Nielsen, M., Ruano, D., Vasen, H. F. A., Morreau, H., et al. (2019). Low frequency of POLD1 and POLE exonuclease domain variants in patients with multiple colorectal polyps. Mol. Genet. Genomic Med. 7 (4), e00603. doi:10.1002/mgg3.603
Esteban-Jurado, C., Giménez-Zaragoza, D., Muñoz, J., Franch-Expósito, S., Álvarez-Barona, M., Ocaña, T., et al. (2017). POLE and POLD1 screening in 155 patients with multiple polyps and early-onset colorectal cancer. Oncotarget 8 (16), 26732–26743. doi:10.18632/oncotarget.15810
Fearon, E. R. (2011). Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 6, 479–507. doi:10.1146/annurev-pathol-011110-130235
Forgó, E., Gomez, A., Steiner, D., Zehnder, J., and Longacre, T. (2020). Morphological, immunophenotypical and molecular features of hypermutation in colorectal carcinomas with mutations in DNA polymerase ε (POLE). Histopathology 76 (3), 366–374. doi:10.1111/his.13984
Guerra, J., Pinto, C., Pinto, D., Pinheiro, M., Silva, R., Peixoto, A., et al. (2017). POLE somatic mutations in advanced colorectal cancer. Cancer Med. 6 (12), 2966–2971. doi:10.1002/cam4.1245
Henninger, E. E., and Pursell, Z. F. (2014). DNA polymerase epsilon and its roles in genome stability. IUBMB Life 66 (5), 339–351. doi:10.1002/iub.1276
Hino, H., Shiomi, A., Kusuhara, M., Kagawa, H., Yamakawa, Y., Hatakeyama, K., et al. (2019). Clinicopathological and mutational analyses of colorectal cancer with mutations in the POLE gene. Cancer Med. 8 (10), 4587–4597. doi:10.1002/cam4.2344
Howitt, B. E., Shukla, S. A., Sholl, L. M., Ritterhouse, L. L., Watkins, J. C., Rodig, S., et al. (2015). Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 1 (9), 1319–1323. doi:10.1001/jamaoncol.2015.2151
Hu, H., Cai, W., Wu, D., Hu, W., Dong Wang, L., Mao, J., et al. (2021). Ultra-mutated colorectal cancer patients with POLE driver mutations exhibit distinct clinical patterns. Cancer Med. 10 (1), 135–142. doi:10.1002/cam4.3579
Huhns, M., Nurnberg, S., Kandashwamy, K. K., Maletzki, C., Bauer, P., and Prall, F. (2020). High mutational burden in colorectal carcinomas with monoallelic POLE mutations: Absence of allelic loss and gene promoter methylation. Mod. Pathol. 33 (6), 1220–1231. doi:10.1038/s41379-019-0430-6
Kawai, T., Nyuya, A., Mori, Y., Tanaka, T., Tanioka, H., Yasui, K., et al. (2021). Clinical and epigenetic features of colorectal cancer patients with somatic POLE proofreading mutations. Clin. Epigenetics 13 (1), 117. doi:10.1186/s13148-021-01104-7
Keenan, B. P., K, V. A. N. L., Khilnani, A. D., Fidelman, N., Behr, S. C., Atreya, C. E., et al. (2021). Molecular and radiological features of microsatellite stable colorectal cancer cases with dramatic responses to immunotherapy. Anticancer Res. 41 (6), 2985–2992. doi:10.21873/anticanres.15080
Lasabova, Z., Kalman, M., Holubekova, V., Grendar, M., Kasubova, I., Jasek, K., et al. (2019). Mutation analysis of POLE gene in patients with early-onset colorectal cancer revealed a rare silent variant within the endonuclease domain with potential effect on splicing. Clin. Exp. Med. 19 (3), 393–400. doi:10.1007/s10238-019-00558-7
Llosa, N. J., Cruise, M., Tam, A., Wicks, E. C., Hechenbleikner, E. M., Taube, J. M., et al. (2015). The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 5 (1), 43–51. doi:10.1158/2159-8290.CD-14-0863
Mur, P., Garcia-Mulero, S., Del Valle, J., Magraner-Pardo, L., Vidal, A., Pineda, M., et al. (2020). Role of POLE and POLD1 in familial cancer. Genet. Med. 22 (12), 2089–2100. doi:10.1038/s41436-020-0922-2
Picard, E., Verschoor, C. P., Ma, G. W., and Pawelec, G. (2020). Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front. Immunol. 11, 369. doi:10.3389/fimmu.2020.00369
Rosner, G., Gluck, N., Carmi, S., Bercovich, D., Fliss-Issakov, N., Ben-Yehoyada, M., et al. (2018). POLD1 and POLE gene mutations in jewish cohorts of early-onset colorectal cancer and of multiple colorectal adenomas. Dis. Colon Rectum 61 (9), 1073–1079. doi:10.1097/DCR.0000000000001150
Shinbrot, E., Henninger, E. E., Weinhold, N., Covington, K. R., Göksenin, A. Y., Schultz, N., et al. (2014). Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 24 (11), 1740–1750. doi:10.1101/gr.174789.114
Siraj, A. K., Bu, R., Iqbal, K., Parvathareddy, S. K., Masoodi, T., Siraj, N., et al. (2020). POLE and POLD1 germline exonuclease domain pathogenic variants, a rare event in colorectal cancer from the Middle East. Mol. Genet. Genomic Med. 8 (8), e1368. doi:10.1002/mgg3.1368
Stenzinger, A., Pfarr, N., Endris, V., Penzel, R., Jansen, L., Wolf, T., et al. (2014). Mutations in POLE and survival of colorectal cancer patients--link to disease stage and treatment. Cancer Med. 3 (6), 1527–1538. doi:10.1002/cam4.305
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Vande Perre, P., Siegfried, A., Corsini, C., Bonnet, D., Toulas, C., Hamzaoui, N., et al. (2019). Germline mutation p.N363K in POLE is associated with an increased risk of colorectal cancer and giant cell glioblastoma. Fam. Cancer 18 (2), 173–178. doi:10.1007/s10689-018-0102-6
Wang, M., Chen, X., Dai, Y., Wu, D., Liu, F., Yang, Z., et al. (2022). Concordance study of a 520-gene next-generation sequencing-based genomic profiling assay of tissue and plasma samples. Mol. Diagn. Ther. 26 (3), 309–322. doi:10.1007/s40291-022-00579-1
Wimmer, K., Beilken, A., Nustede, R., Ripperger, T., Lamottke, B., Ure, B., et al. (2017). A novel germline POLE mutation causes an early onset cancer prone syndrome mimicking constitutional mismatch repair deficiency. Fam. Cancer 16 (1), 67–71. doi:10.1007/s10689-016-9925-1
Ying, J., Yang, L., Yin, J. C., Xia, G., Xing, M., Chen, X., et al. (2021). Additive effects of variants of unknown significance in replication repair-associated DNA polymerase genes on mutational burden and prognosis across diverse cancers. J. Immunother. Cancer 9 (9), e002336. doi:10.1136/jitc-2021-002336
Keywords: colorectal cancer, POLE mutation, polymerase epsilon, immunotherapy, prognosis
Citation: Jiang M, Jia Y, Han J, Shi J, Su C, Zhang R, Xing M, Jin S and Zong H (2022) Distinct clinical pattern of colorectal cancer patients with POLE mutations: A retrospective study on real-world data. Front. Genet. 13:963964. doi: 10.3389/fgene.2022.963964
Received: 08 June 2022; Accepted: 25 October 2022;
Published: 21 November 2022.
Edited by:
Susana Romero-Garcia, Universidad Nacional Autónoma de México, MexicoReviewed by:
Zhonglin Zhu, Fudan University, ChinaCopyright © 2022 Jiang, Jia, Han, Shi, Su, Zhang, Xing, Jin and Zong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuiling Jin, ZmNjamluc2xAenp1LmVkdS5jbg==; Hong Zong, ZmNjem9uZ2hAenp1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.