- 1Department of Population Genetics, Institute of Food Biotechnology and Genomics, National Academy of Sciences of Ukraine, Kyiv, Ukraine
- 2Department of Cell Biology and Biotechnology, Institute of Food Biotechnology and Genomics, National Academy of Sciences of Ukraine, Kyiv, Ukraine
- 3Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany
- 4M. M. Gryshko National Botanic Garden of National Academy of Sciences of Ukraine, Kyiv, Ukraine
- 5Department of Genomics and Molecular Biotechnology, Institute of Food Biotechnology and Genomics, National Academy of Sciences of Ukraine, Kyiv, Ukraine
Cytokinin dehydrogenase/oxidase (CKX) enzymes play a key role in regulating cytokinin (CK) levels in plants by degrading the excess of this phytohormone. CKX genes have proven an attractive target for genetic engineering, as their silencing boosts cytokinin accumulation in various tissues, thereby contributing to a rapid increase in biomass and overall plant productivity. We previously reported a similar effect in finger millet (Eleusine coracana) somaclonal lines, caused by downregulation of EcCKX1 and EcCKX2. However, the CKX gene family has numerous representatives, especially in allopolyploid crop species, such as E. coracana. To date, the entire CKX gene family of E. coracana and its related species has not been characterized. We offer here, for the first time, a comprehensive genome-wide identification and analysis of a panel of CKX genes in finger millet. The functional genes identified in the E. coracana genome are compared with the previously-identified genes, EcCKX1 and EcCKX2. Exon-intron structural analysis and motif analysis of FAD- and CK-binding domains are performed. The phylogeny of the EcCKX genes suggests that CKX genes are divided into several distinct groups, corresponding to certain isotypes. Finally, the phenotypic effect of EcCKX1 and EcCKX2 in partially silencing the SE7 somaclonal line is investigated, showing that lines deficient in CKX-expression demonstrate increased grain yield and greater bushiness, enhanced biomass accumulation, and a shorter vegetation cycle.
Introduction
Plant growth regulation depends on the balance of phytohormones, among which cytokinins play an important role. Cytokinins are plant hormones that not only take part in cell proliferation and differentiation, but also in the establishment of plant architecture. They regulate many developmental processes, strongly influence grain yield (Zhang et al., 2012; Jameson and Song, 2016; Chen et al., 2020b; Dash and Rai, 2022), and impact the plant signaling pathways of biotic and abiotic stresses (Werner et al., 2006; Blume et al., 2017). Cytokinin homeostasis is regulated by members of several multigene families through a balance between biosynthesis [isopentenyl transferase (IPT)], activation [Lonely Guy (LOG)], inactivation (O-glucosyl transferase), re-activation (β-glucosidase), and degradation [cytokinin oxidase/dehydrogenase (CKX)] (Werner et al., 2006; Jameson and Song, 2016). In particular, CKX genes encode the cytokinin oxidase/dehydrogenase enzymes, which catalyze the irreversible degradation of cytokinin. Such enzymes are responsible for the balance of cytokinins in the plants, and for the control of cytokinin-dependent processes (Werner et al., 2006; Dash and Rai, 2022).
It should be noted that CKXs are encoded by a relatively small gene family in different plant species, such as rice (Ashikari et al., 2005; Mameaux et al., 2012), Arabidopsis (Galuszka et al., 2007), maize (Gu et al., 2010; Mameaux et al., 2012; Zalabák et al., 2014), foxtail millet (Mameaux et al., 2012; Wang et al., 2014), barley, wheat, sorghum, and brachypodium (Mameaux et al., 2012), Chinese cabbage (Liu et al., 2013), potato (Suttle et al., 2014), oilseed rape (Liu et al., 2018), forage legume barrel medic (Medicago truncatula) (Wang et al., 2021), soybean (Liu et al., 2021; Nguyen et al., 2021), cowpea (Liu et al., 2021) and apple (Liu et al., 2022). Following phylogenetic, molecular, and comparative analyses of CKX families in sequenced grass species, such as rice, Brachypodium distachyon, sorghum, maize and foxtail millet, as well as members identified from wheat and barley transcriptomes⁄genomes, the phylogenetic analyses identified four Poaceae CKX clades (Mameaux et al., 2012). Comparative analysis showed that such phylogenetic groupings of CKX can largely be explained by a combination of local gene duplication and a whole-genome duplication event that preceded their speciation. Recently, the evolutionary origin of CKXs has been analyzed and attempts have been made to understand their function in relation to their structure in different organisms (Dabravolski and Isayenkov, 2021).
Even monocots and dicots show significant differences in cytokinin metabolite composition (Jiskrová et al., 2016). Inhibition of CKXs can lead to increased plant productivity, and thereby increased crop yields (Zhang et al., 2012; Jameson and Song, 2016; Szala et al., 2020). These changed features were detected by us earlier in finger millet (Eleusine coracana) (Radchuk et al., 2012). We found that downregulation of two CKX genes (i.e., EcCKX1, EcCKX2) in the obtained somaclonal variants of E. coracana leads to the formation of a dwarf and highly productive phenotype of finger millet (Radchuk et al., 2012). As a consequence of this gene downregulation, attenuated degradation of cytokinins in such E. coracana lines leads to higher cytokinin levels, increased accumulation, and stimulation of meristem activity, resulting in the production of more inflorescences and seed setting (Radchuk et al., 2012; Yemets et al., 2020). Similar data for rice were previously reported by Ashikari et al. (2005), where the authors found that a decrease in OsCKX2 expression leads to the accumulation of cytokinin in the meristems of inflorescences, an increase in the number of reproductive organs, and, as a result, an increase in grain yield. Other studies confirm such an effect: the silencing ckx2 in barley, for example (Zalewski et al., 2014), with overexpression of this gene leading to a non-flowering phenotype (Mrízová et al., 2013). This gene is described as having the same function in A. thaliana (Li et al., 2013) and also in the development of seed pods in B. napus (Liu et al., 2018).
Given the role of cytokinins in plant development and productivity, the identification and regulation of CKX gene expression in main crops is extremely important from the perspective of increasing yield and food security. Due to the ability of E. coracana to grow in arid and semi-arid regions of Central Africa and India, as well as in tropical regions (more than 25 countries in Africa and Asia), its ability to withstand abiotic and biotic (resistance to pathogens) stresses (Gupta et al., 2017); its nutritional value (the grains are rich in amino acids, such as methionine and lysine; minerals such as calcium, iron, zinc, phosphorus, and potassium; vitamins; and fibers), and its good grain storage properties, finger millet is one of the most important crops for sustainable agriculture in developing countries, and a very valuable food resource. Finger millet is gluten free and therefore can be included in the diet of patients suffering from celiac disease (Pagano, 2006). In addition, finger millet can still potentially be used as animal fodder (Gupta et al., 2017), and for bioethanol production (Yemets et al., 2020). To date, there is no accurate information on the global cultivation of this crop, although India is known to be the largest producer, with finger millet grown on 1.19 million hectares and yielding over 1,660 kg/ha (Sood et al., 2019).
The availability of finger millet genomic resources (Hittalmani et al., 2017; Hatakeyama et al., 2018; Zhang et al., 2019b; Bančič et al., 2021; Pendergast et al., 2022) makes it possible to analyze different groups of genes and estimate practical methods for molecular breeding and biotechnological improvement of this crop (Antony Ceasar et al., 2018; Pandian and Ramesh, 2019; Sood et al., 2019). Certainly, the identification and characterization of all CKX family members in E. coracana is a very important tool for achieving these practical goals. In this paper, we present for the first time the comprehensive genome-wide identification and analysis of a panel of CKX genes in finger millet. These functional genes, identified in the E. coracana genome, are compared with the two previously-identified genes, EcCKX1 and EcCKX2. Exon-intron structural analysis, and a motif analysis of the FAD- and CK-binding domains are carried out. Study of the phylogeny of EcCKX genes establishes that CKX genes are divided into several distinct groups, according to their specific isotypes. Finally, the phenotypic effect of partial silencing of EcCKX1 and EcCKX2 in the somaclonal line of SE7 is investigated, suggesting that lines deficient in CKX-expression manifest increased grain yield, higher bushiness, enhanced biomass accumulation, and a shorter vegetation cycle. The bioinformatic data obtained will prove useful for further manipulation of finger millet CKX genes so as to improve crop quality and yield.
Materials and methods
Initial identification and analysis of cytokinin dehydrogenase/oxidase genes in the E. coracana genome
The initial search for CKX genes in the E. coracana genome was conducted using a series of BLAST searches of the Eleusine coracana annotation release v1.1 (genome ID: 560), which is deposited in the Phytozome v13 database, and which contains the most recent and complete E. coracana genome assembly and annotation. Personal permission to use the genome data for the current study was obtained from Prof. Katrien M. Devos, as the current release of the E. coracana annotation has a “Reserved Analysis” status, limiting its use for publication.
The BLAST algorithm was used to search the translated nucleotide databases, and the coding regions of the OsCKX and AtCKX genes (Swarbreck et al., 2008) were used as queries. In addition, sequences of two previously identified genes, EcCKX1 (HE800184.1) and EcCKX2 (HE800185.1) of E. coracana, were used for the genomic search (Radchuk et al., 2012). Search parameters were: E-value threshold—1e−5, comparison matrix—BLOSUM62, and word length—3 (Henikoff and Henikoff, 1992; Altschul, 1993). We analyzed the results and discarded short and insignificant hits.
Information on E. coracana CKX genes, including location, genomic coordinates, sequence ID, genomic sequence, protein sequence, and coding sequence (CDS), was acquired from the Phytozome v13 database. Orthologous information for the identified genes was retrieved from the KEGG database (https://www.genome.jp).
The genomic organization and synteny of cytokinin dehydrogenase/oxidase genes
A multiple alignment CDS sequence of the CKX genes was performed using the MUSCLE algorithm (Edgar, 2004). Representations of the exon-intron structure of the genes were obtained using the Gene Structure Display Server (http://gsds.gao-lab.org/) (Hu, 2015).
The domain organization of CKX peptides was analyzed using the Pfam tool (https://pfam.xfam.org/) (Mistry et al., 2021), which allowed us to confirm the presence or absence of key FAD- and CK- binding domains. Calculation of the rate of non-synonymous substitution (KA), the rate of synonymous substitutions (KS), and their ratio (KA/KS), were performed in the TBtools v1.0971 software (Chen et al., 2020a), using CDS sequences of the identified EcCKX.
The organization of CK-responsive cis-elements was analyzed in the 2 kbp upstream regions of the identified EcCKX genes (taking into account the initiation codon), using the PLACE v30.0 tool (https://www.dna.affrc.go.jp/PLACE) (Higo et al., 1999), and the results were compared with the search results in a similar database—PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Further data on the presence of particular cis-elements motifs were filtered to identify only the CK-responsive elements. The structure of the protein domain organization, and the allocation of CK-responsive cis-elements were visualized using TBtools v1.0971 software (Chen et al., 2020a).
Syntenic relationships between homeologous CKX genes from different subgenomes of E. coracana were analyzed in TBtools v1.0971 software (Chen et al., 2020a), using the MCScanX algorithm (Wang et al., 2012). The results were further visualized as a circos plot.
To explore the syntenic relationships of E. coracana orthologous CKX genes with O. sativa species, the genome data and the gene annotation files of rice (GCA_001433935.1 assembly) were also downloaded from the NCBI database. The synteny analyzing dual plot graphs were constructed using the dual synteny plotter function in TBtools, while inter-genome synteny was inferred using the MCScanX algorithm.
Phylogenetic analysis
Phylogenetic analysis was performed using MEGAX (Kumar et al., 2018). Respective amino acid sequences of CKX proteins from different species (Supplementary Table S1), including E. coracana, were aligned using the MUSCLE algorithm (Edgar, 2004). The CKX protein sequences of Arabidopsis thaliana and Oryza sativa were retrieved from the KEGG genome database, while those of Setaria italica, Hordeum vulgare, Populus trichocarpa, and Prunus persica were obtained from the Phytozome v13 database.
Initial isotype determination of EcCKX genes was performed using cds sequences as the initial data set, which was then compared to the previously described EcCKX1 (HE800184.1) and EcCKX2 (HE800185.1) (Radchuk et al., 2012). This “guide” tree was constructed using the neighbor joining (NJ) method with the default software settings. The NJ tree was generated with a bootstrap support of 1,000 replicates (Supplementary Figure S1).
Before the maximum likelihood (ML) phylogenetic tree was constructed, the best substitution model analysis was performed. The Jones-Taylor-Thornton model with gamma distribution rate and invariant sites (JTT + G + I) was chosen as optimal (Jones et al., 1992). The selected model was used to infer a phylogenetic tree using the ML estimation method. The initial tree was derived using neighbor-joining analysis, following the nearest-neighbor-interchange heuristic method. The number of discrete gamma categories was four; the treatment for gaps and missing sites was “use all sites.” The statistical confidence of tree topology was assessed using a bootstrap test with 1,000 replicates.
Plant material, growth conditions, and agro-morphological evaluation
Field evaluation of the characteristics of finger millet somaclonal mutants was carried out using approx. 100 plants per wild-type (variety Tropikanka) and SE7 (registered now in Ukraine as Yaroslav-8 variety) finger millet lines. Approximately 15–25 seeds were planted in 20-cm diam. pots in a greenhouse at the Leibniz Institute of Plant Genetics and Crop Plant Research (Gatersleben, Germany) in the late April. Plants were grown for 2 weeks in a greenhouse and then some of them were transplanted to a field in Gatersleben in May. Spacing between individual plants was 20 cm × 20 cm. Plant height and yields from a random 25 plants were measured in late September 2011. Additional field trials were conducted in Kyiv, Ukraine, at the National Botanical Garden experimental field, under the conditions described earlier (Rakhmetova et al., 2020). Plants were harvested manually and the following parameters were recorded at the plant maturity stage: plant height, 1000-grain weight, number of primary and secondary stems per plant, dry matter content (w/w), dry biomass yield, and seed yield. The obtained data were also extrapolated to rate per hectare.
Tissue sampling for expression analyses
For quantitative reverse transcription-PCR (qRT-PCR) and northern blot analyses of different tissues, total RNA was isolated 4 and 5 days after imbibition (4 and 5 DAI), from the young leaves of seedlings at the tillering stage, and from the old leaves of maturating plants. In addition, tissue samples of the developing inflorescence were taken at three stages: Stage A at a length of 2–3 cm; Stage B at 3–4 cm; and Stage C at > 4 cm. All samples were collected at least in triplicate from biologically-independent plant material.
RNA extraction
Total RNA was extracted from different tissues of the wild type and SE7 mutant of finger millet using Trizol reagent (Invitrogen). To do this, 100 mg of tissue was ground in liquid nitrogen, mixed with 1 ml of preheated Trizol (60°C) for 5 min, and centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant was transferred into a new tube, mixed with 0.2 ml of chloroform for 2 min, and centrifuged at 13,000 rpm for 10 min at 4°C. The aqueous phase was transferred into a new tube and mixed with 0.6 vol. of isopropanol, incubated at room temperature for 10 min, and then centrifuged at 13,000 rpm for 10 min at 4°C. The pellet was rinsed once with cold 70% ethanol and dissolved in 100 µl of distilled water. The isolated RNA was treated with RNase-free DNase (Qiagen), purified using a RNeasy plant mini kit (Qiagen), and used for the synthesis of cDNA, quantitative RT-PCR, and cDNA array.
Expression analyses
The procedure for cDNA synthesis was identical to that described previously (Radchuk et al., 2012). cDNA fragments of the EcCKX1.A/B and EcCKX2.A/B genes were further amplified from different tissues of wild-type and mutant lines of finger millet by RT-PCR, using gene-specific primers selected from conserved regions of the corresponding sequences. For distinguishing of EcCKX1.A/B genes, the following primers were used: EcCKX1.A/B-For—5′-TGCGCCTCGACGGCCATTTCAG-3′ and EcCKX1. A/B-Rev—5′-GGATCGTACGATTCGCCCTTCC-3′, while for EcCKX2.A/B genes — EcCKX2.A/B-For—5′-CACCACCATCGCTGCGTCCAGT-3′ and EcCKX2.A/B-polyT—5′-GTTGGGTNTTTTTTTTTTTTTTTTTTT-3´. The actin gene was an internal control, for which primers targeting barley actin were used: qactin_u—5′-ATGGTGGGGATGGGGCAGAAG-3′ and qvactin_r—5′-CTCCTCCGGGGCAACACGAA-3´. Northern blot analysis of the EcCKX genes transcript was performed according to the previously-described procedure (Conrad et al., 2007). For qRT-PCR, as described above, 5 µg of the total RNA isolated were used for reverse transcription by SuperScript III reverse transcriptase (Invitrogen), with an oligo (dT) primer. The resulting cDNAs were used as templates for qRT-PCR assays, which were performed as previously described (Radchuk et al., 2011). The PCR efficiency was assessed using LinRegPCR software (Ramakers et al., 2003). Analysis of RT-PCR data was performed according to the procedure described in our previous publication (Radchuk et al., 2012).
Statistical processing of the data
The obtained data were statistically processed using OriginPro 2019b software. Deviations of all means were calculated as a standard deviation (SD). To reveal the significance of differences in various parameters between the studied genotypes, one-way ANOVA was used, which included the calculation of Fisher’s least significant differences (LSDs). The LSDs were used to determine homogeneous groups for values of specific morphological or productive parameters at different level of significance, p < 0.05, p < 0.01, and p < 0.001.
Results
The cytokinin dehydrogenase/oxidase gene family in finger millet
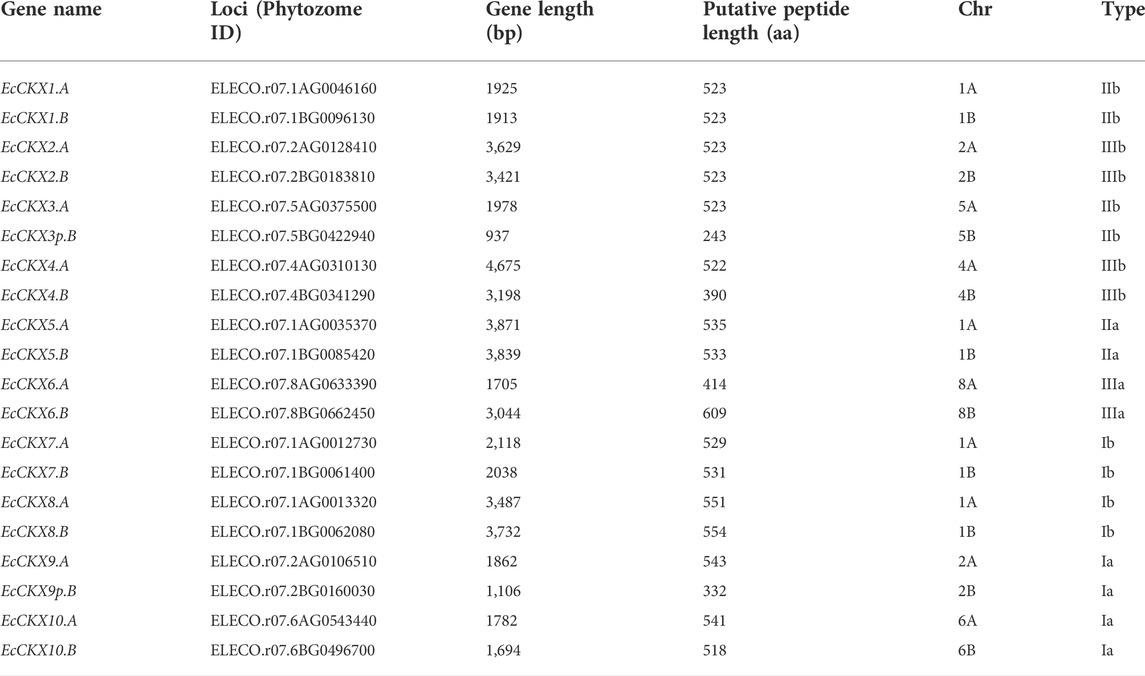
As the result of a thorough search of the finger millet genome, deposited in the Phytozome v13.0 database, 20 representatives of the CKX gene family were identified (Table 1). The length of the genes varied from 937 to 3,871 bp, and the length of the encoding peptides ranged from 243 to 609 aa. MUSCLE alignment of the previously-described EcCKX1 and EcCKX2 (Radchuk et al., 2012) against cds of the identified EcCKX panel revealed the existing orthologs of EcCKX1 and EcCKX2 in the finger millet genome, which were then named EcCKX1.A/B and EcCKX2.A/B, respectively (Supplementary Figure S1). Types of the encoded EcCKX proteins were determined on the basis of the phylogenetic analysis that will be described separately.
All identified EcCKX were present in doublets and were allocated on homologous chromosomes from subgenomes A and B of E. coracana. As this species has an allotetraploid nature, all gene pairs were defined as homologous. In the vast majority of cases, the homologous genes possessed similar gene length and encoded highly identical peptides (92.55%–98.85%). Two genes, EcCKX3p.B and EcCKX9p.B, were of significantly reduced length, 937 bp and 1,106 bp, respectively, compared to other EcCKX, and thus encoded shortened peptides, 243 aa and 332 aa, respectively. It is also worth noting that four EcCKX, encoding different isotypes of this enzyme, were located on the same 1A chromosome (and their four homeologs on 1B). EcCKX1.A and EcCKX5.A were located relatively close at 7.11 Mbp, while EcCKX7.A and EcCKX8.A—were only 710 kbp apart. Also, EcCKX1.A/B and EcCKX5.A/B both belong to type II of CKXs, while both EcCKX7.A/B and EcCKX8.A/B are type I members. Co-location of pairs of these genes on the same chromosome could possibly correspond to their potentially paralogous nature.
Exon-intron structure of EcCKX, and domain distribution in their peptides
Next, the exon-intron structure of the identified EcCKX (Figure 1), as well as the domain organization of the encoded EcCKX peptides were investigated (Figure 2). The number of exons was not conserved among members of the CKX gene family. Type I members, such as EcCKX9.A and EcCKX8.A/B, had three exons, while others, EcCKX7.A/B and EcCKX10.A/B, had only two exon structures, most likely due to the reduction of the intron2. On the other hand, most representatives of type II and III had five exons, with a few exceptions, such as EcCKX6.A and EcCKX6.B, which had 3- and 4-exon structures respectively. EcCKX3p.B and EcCKX9p.B genes, which showed a significant reduction in their length, lacked several exons in their structure. For example, EcCKX3p.B had only exon1 and exon2, while exons3-5 were eliminated. At the same time, exon1 was lost in EcCKX9p.B if compared to its homeolog, EcCKX9.A.
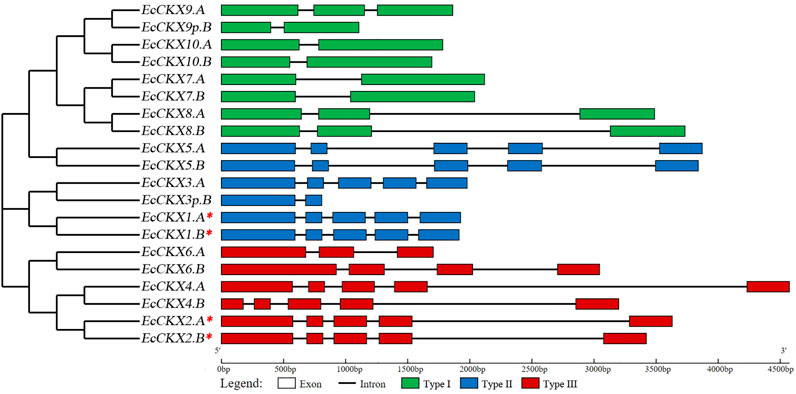
FIGURE 1. Exon-intron structure of the identified CKX genes within the E. coracana genome. The NJ phylogenetic tree was constructed with bootstrap support of 1,000 iterations (all branches had support of 65% and higher), based on cds sequences of the identified genes. A red asterix denotes genes identical to previously sequenced and characterized EcCKX1 and EcCKX2.
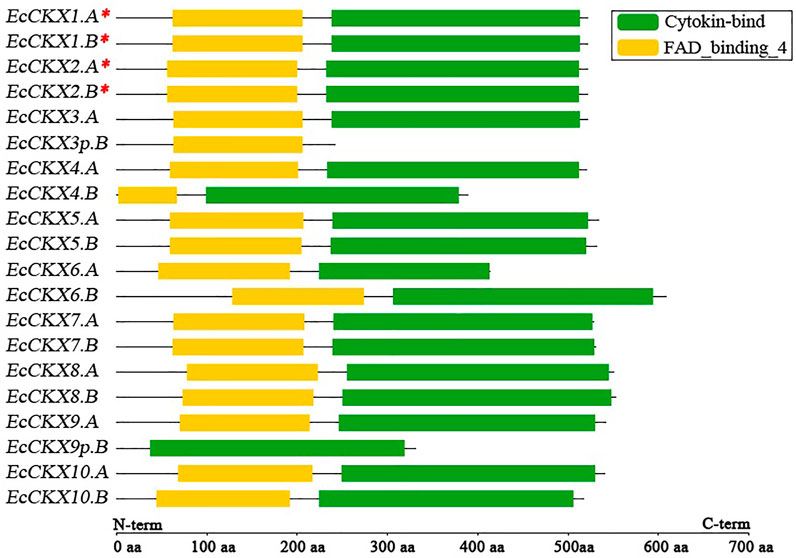
FIGURE 2. Putative motif/domain distribution in 20 translated peptide sequences, derived from identified EcCKX genes. A red asterix denotes genes identical to previously sequenced and characterized EcCKX1 and EcCKX2.
Typically, CKX proteins should contain two major domains (motifs) in their structure, which are responsible for the FAD- and CK-binding activity of the enzymes. Analysis of the translated peptides of the identified EcCKX revealed the presence and location of these crucial structural elements (Figure 2). Most of the identified genes contained both FAD- and CK-binding domains, despite the significant difference in the exon-intron structure of the genes. Usually, the FAD-binding domain is located above the CK-binding ones. Notably, EcCKX6.B, which had an enlarged exon1, also had a longer variable sequence, upstream to the FAD-binding domain, while the EcCKX6.B peptide had the longest sequence of 609 amino acids—almost 55 amino acids longer than the typical EcCKX protein, and 195 amino acids longer than the peptide of its homeolog EcCKX6.A.
It is also worth noting that EcCKX1.A/B and EcCKX2.A/B, orthologs of the previously-described EcCKX1 and EcCKX2, had a conserved protein length of 523 amino acids and domain organization, despite the significant sequence differences that led to their classification into distinct CKX types. Most of the homeologous pairs of genes have preserved the identical domain organization of their peptides. However, such representatives as EcCKX3p.B lacked the CK-binding domain, which correlates well with the loss of exons3-5 of this gene. Similarly, the peptide of EcCKX9p.B lacked the FAD-binding domain, while the gene did not contain exon1, which should be present in a normal gene, e.g., in its homeolog. Based on these findings, it is hypothesized that EcCKX3p.B and EcCKX9p.B may be pseudogenes that potentially faced selective pressure after the allopolyploidization of the Eleusine species, leading to the origin of E. coracana.
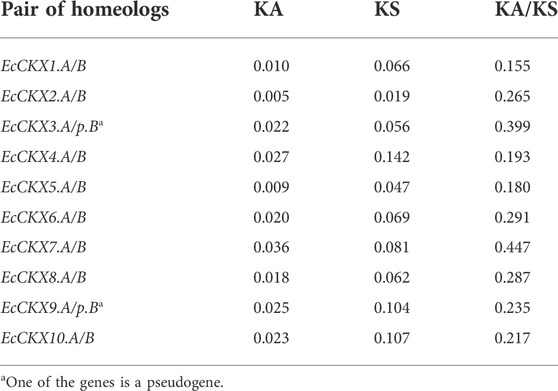
Further, analyses were performed of the rate of non-synonymous substitution (KA), the rate of synonymous substitutions (KS), and their ratio (KA/KS) (Table 2). All of the EcCKX homeologous pairs showed extremely low KA values—in a range of 0.005–0.036—while the rate of synonymous substitutions was higher: KS = 0.019–0.142. Interestingly, EcCKX2.A/B possessed the lowest rates of substitutions (KA = 0.005; KS = 0.019) among all identified EcCKX. EcCKX7.A/B showed the highest rate of non-synonymous substitutions (KA = 0.036), which led to the highest KA/KS ratio—0.447. All homeologous pairs of EcCKX showed KA/KS values lower than 1 (in a range of 0.155–0.447), meaning that all of these duplicated genes were under purifying or stabilizing selection. EcCKX1. A/B possessed the lowest value of KA/KS at 0.155. Such low values of KA/KS ratios among EcCKX homeologs may suggest that the duplicated genes will be further preserved in their current state, and not face loss of function or neofunctionalization.
Organization of cis-elements in upstream regions of EcCKX
To identify the putative cis-acting regulatory elements, the 2 kbp upstream sequences (from the start codon) were analyzed for all sets of the identified EcCKX (Figure 3). The upstream regions of various EcCKXs appear to be rich in various cis-regulatory elements, responsible for inducing expression in response to a wide range of factors. However, from the perspective of this study, the most interesting elements are those involved in cytokinin-mediated induction of expression. The PLACE database provides for analysis of only four known and confirmed cis-elements associated with CK-response: ARR1 (PLACE ID: S000454), AS1LIKECSHPRA (S000260), CPBCSPOR (S000491), and CYTOSITECSHPRA (S000260). Although the sequences of these cis-elements may vary depending on their type, all of them have a common tetranucleotide motif -NGAT- (more often -AGAT-). Only two types of cis-acting regulatory elements were identified in EcCKX: CPBCSPOR (-TATTAGN-, containing an inverted NGAT motif at 3′) and ARR1 (NGATT), with the latter being the most common. No AS1LIKECSHPRA or CYTOSITECSHPRA were detected.
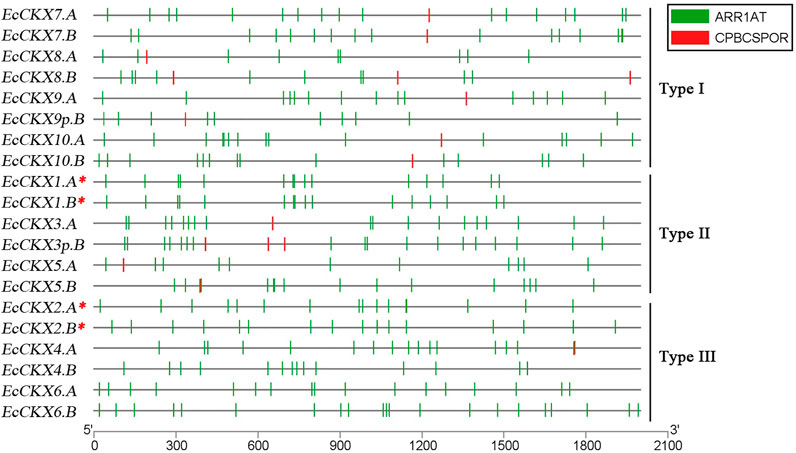
FIGURE 3. Presence of cis-elements, responsible for CK-mediated expression induction, in the 2 kb upstream region of the identified EcCKX genes of finger millet. A red asterix denotes genes identical to previously sequenced and characterized EcCKX1 and EcCKX2.
On average, all identified EcCKXs contained 9–21 ARR1 and 1–3 CPBCSPOR elements in the 2 kbp upstream regions. It is interesting to note that type III members contained almost no CPBCSPOR elements in their upstream regions, except for EcCKX4. A, which had CPBCSPOR at position 1754–1759 bp, and overlapped another ARR1 element located at 1758–1762 bp. Often ARR1 elements were closely located, formed clusters (containing 2–3 ARR1 elements), or overlapped each other. However, the presence of two overlapping ARR1 (one straight and one inverted element), was identified in the upstream region of EcCKX10.A [463–473 (+) and 473–477 (-)], thus forming a -NGATTTAGN- palindromic sequence motif. For most of the CKX genes, the presence of a double placement of ARR1 elements in the upstream region (2–4 bp away from each other) was found, but their sequence had same direction, thus not forming palindromic motifs.
Different types of CKX genes had different combinations of regulatory cis-elements. For example, type I EcCKX had the lowest number of ARR1 elements, ranging from 9 to 17 ARR1 motifs per promoter region of each gene, whereas all type I genes, without exception, contained one to three CPBCSPOR motifs. For example, type I EcCKX had the lowest number of ARR1 elements, ranging from 9 to 17 ARR1 motifs per promoter region of each gene, whereas all type I genes contained from one to three CPBCSPOR motifs, without exception. In contrast, type III genes had the highest number of ARR1 regulatory elements in the upstream region (14–21 per gene), whereas CPBCSPOR motifs were absent. Only gene EcCKX4. had one CPBCSPOR element, which, however, overlapped the ARR1 motif sequence. Some of the genes had clusters of ARR1 motifs, where on a 100 bp sequence three or more ARR1 elements were present (e.g., EcCKX6.B).
Not all homeologous pairs shared the same patterns of ARR1 motif distribution. The structure of the upstream regions of EcCKX1.A/B was almost identical, while in the case of EcCKX2.A/B, it was similar only in a certain range—approx. 900–1,200 bp, etc. EcCKX3p.B displayed a very similar pattern of CK-responsive cis-elements to that of EcCKX3.A, whereas another possible pseudogene, EcCKX9p.B, had a distinct pattern of ARR1 allocation from its functional homeolog. Despite its pseudogenic nature, EcCKX9p.B had 10 ARR1, which is comparable to the upstream regions of other functional genes. It is unknown whether they were influenced by the process of gene disruption (possible loss of exon1 and its associated regions), or arose due to stochastic causes.
Phylogenetic analysis of the cytokinin dehydrogenase/oxidase gene family
The next step was to analyze the phylogenic relationships between the identified EcCKX and members of this gene family in other species (Figure 4). In order to perform this analysis, 83 CKX amino acid sequences were aligned, and further phylogenic analysis was performed using the maximum likelihood (ML) method, which allowed us to obtain a tree with high confidence at all crucial branching nodes (bootstrap values for main nodes were 85 and higher). The results allowed us to conclude that CKX proteins can be divided into three main types, which include sequences of both monocotyledonous and dicotyledonous species. Seven functional EcCKX proteins and one reduced (EcCKX9p.B) were associated with type I CKX. Among the type I proteins, the vast majority came from monocot species, in which type I panel proteins expanded rapidly compared to dicot species. Dicot proteins (e.g., AtCKX2, AtCKX3, and AtCKX4) formed a separate branch of CKX, specific only to this taxonomic group (named Dicot type I). At the same time, monocot CKX proteins formed four minor clades, which could be grouped into subtype Ia of monocots belonging to EcCKX9.A, EcCKX10.A/B, and subtype Ib of monocots containing EcCKX7.A/B and EcCKX8.A/B.
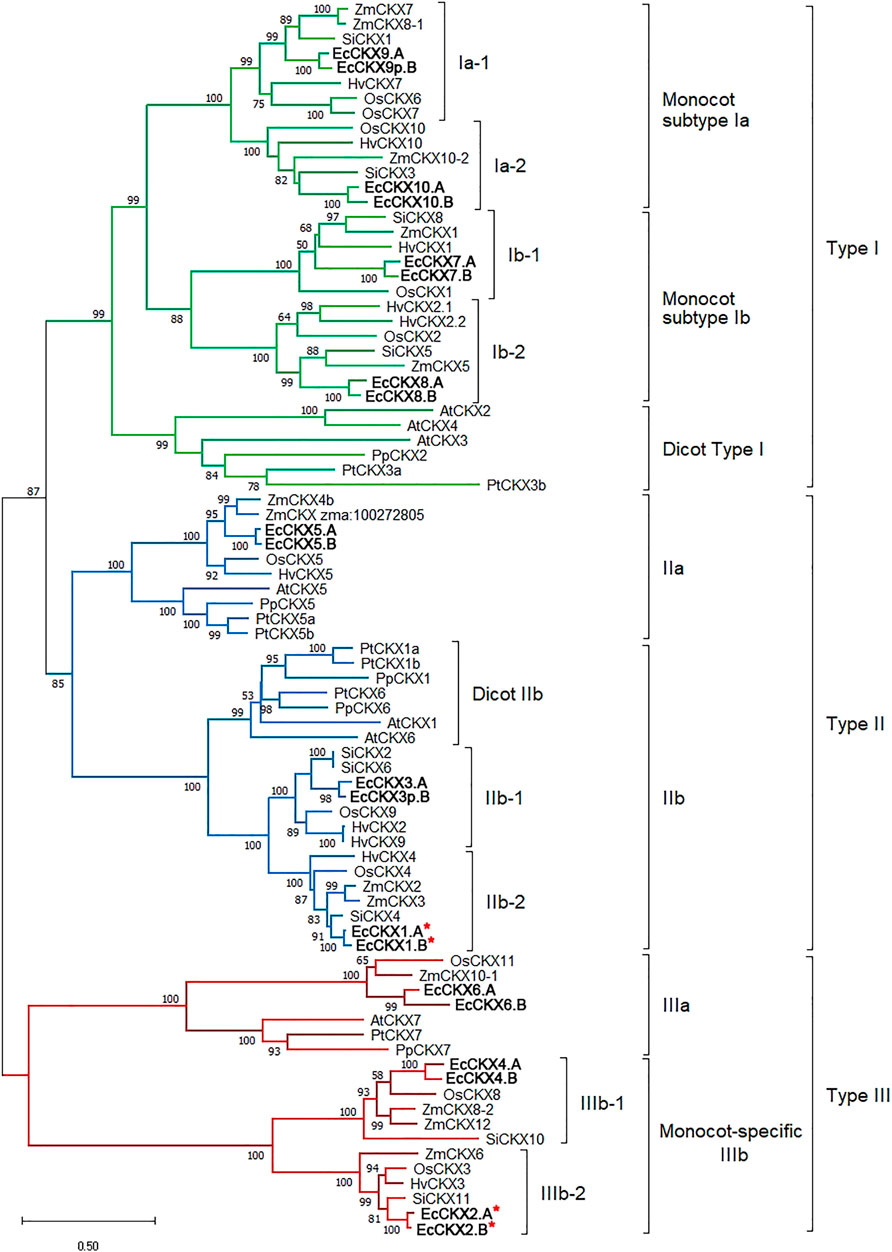
FIGURE 4. Unrooted ML phylogenetic tree for dicot (A. thaliana, P. persica, and P. trichocarpa) and monocot (O. sativa, Z. mays, H. vulgare, and S. italica) CKX proteins, as well as translated peptides of identified genes in E. coracana genome. The tree was constructed with bootstrap support of 1,000 iterations (50% and higher values are displayed). A red asterix denotes genes identical to previously sequenced and characterized EcCKX1 and EcCKX2.
Proteins of each minor clade Ia-1, Ia-2, Ib-1, and Ib-2, represent groups of sub-orthologs, as each clade contains the respective proteins from the analyzed monocot species: O. sativa, Z. mays, S. italica, H. vulgare, and E. coracana. This allows us to assume that diversification of these minor subtypes of CKX in monocots occurred at least before the emergence of the Poaceae species. Such diversity could potentially result from local (in the case of EcCKX7.A/B and EcCKX8.A/B) or genome-wide duplication.
Type II of CKX is comprised of two major subtypes: IIa and IIb, which can be further divided into three minor clades of Dicot-specific IIb, and Monocot IIb-1 and IIb-2. Similar to Type I, the diversity of the monocot CKX was higher than in the dicot. However, in the case of Type 2 CKX, dicot proteins did not group distinctly, but were present in subtype IIa (AtCKX5), IIb (AtCKX1, and AtCKX6) subtype clades. This indicates that EcCKX5.A/B appears to be an ancient paralog in relation to representatives of the IIb subtype (EcCKX1.A/B and EcCKX3.A), if the close allocation of EcCKX5.A/B and EcCKX1.A/B on the same chromosome is taken into account.
Proteins of Type III were separated into two branches. The primary branch (IIIa) included several monocot proteins and their orthologs from dicots (e.g., AtCKX7). EcCKX6.A/B were also attributed to this subtype. The second branch, subtype IIIb, contained only monocot CKX, and formed two minor clades representing distinct lineages of CKX peptide paralogs: IIIb-1 and IIIb-2, which included EcCKX4.A/B and EcCKX2.A/B, respectively. The previously-identified EcCKX1 and EcCKX2 were attributed to different types of CKX, despite their similarity in protein length, genomic structure, and potentially similar function. It is also worth noting that the branches within the IIb and IIIb clades were the shortest observed, compared to other subtypes of CKX, suggesting that EcCKX1 and EcCKX2 orthologs are conserved in monocots (at least among the Poaceae species). However, the fact that these genes were not eliminated, despite their paralogous nature, may indicate that multiple copies of particular EcCKX have undergone sub-functionalization, or that an increased number of their copies confer a selective advantage.
Synteny of EcCKX
Furthermore, interchromosomal synteny of finger millet CKX was inferred (Figures 5, 6), which is of additional interest, since E. coracana is considered an allotetraploid species and has an expanded panel of EcCKX. Figure 6 shows the syntenic relations between the identified EcCKX genes. Thus, all the genes formed syntenic pairs with their homeologs from A and B subgenomes, which further confirms the hypothesis that the rapid expansion of the CKX gene family in E. coracana is the result of an allopolyploidization event. Only genes from Type I formed pairs, not only between homeologs, but also with paralogs of the same type. For example. EcCKX10.A/B formed syntenic pairs with EcCKX9.A. This suggests that the sequences of these genes remained sufficiently conserved, and that they were recognized as possible duplicates during the synteny analysis. No syntenic pairs between genes of different CKX isotypes of Type II or III were detected.
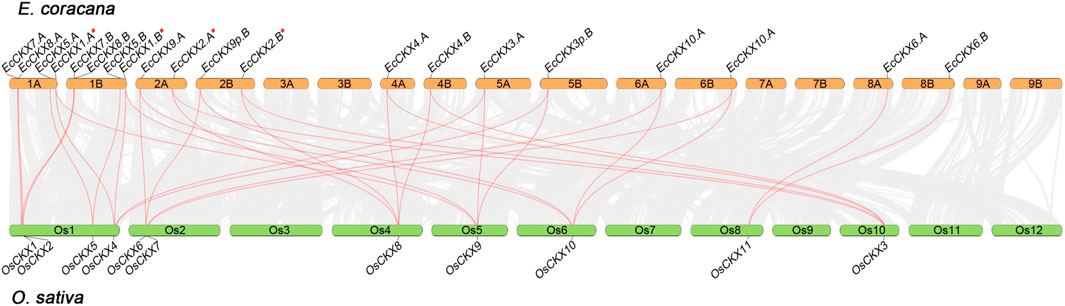
FIGURE 5. Synteny analysis of CKX genes between E. coracana and O. sativa species. The collinear blocks between O. sativa and E. coracana species are shown with gray lines, while syntenic pairs of CKX genes are highlighted in red. The chromosome number is indicated above each respective chromosome. A red asterix denotes genes identical to previously sequenced and characterized EcCKX1 and EcCKX2.
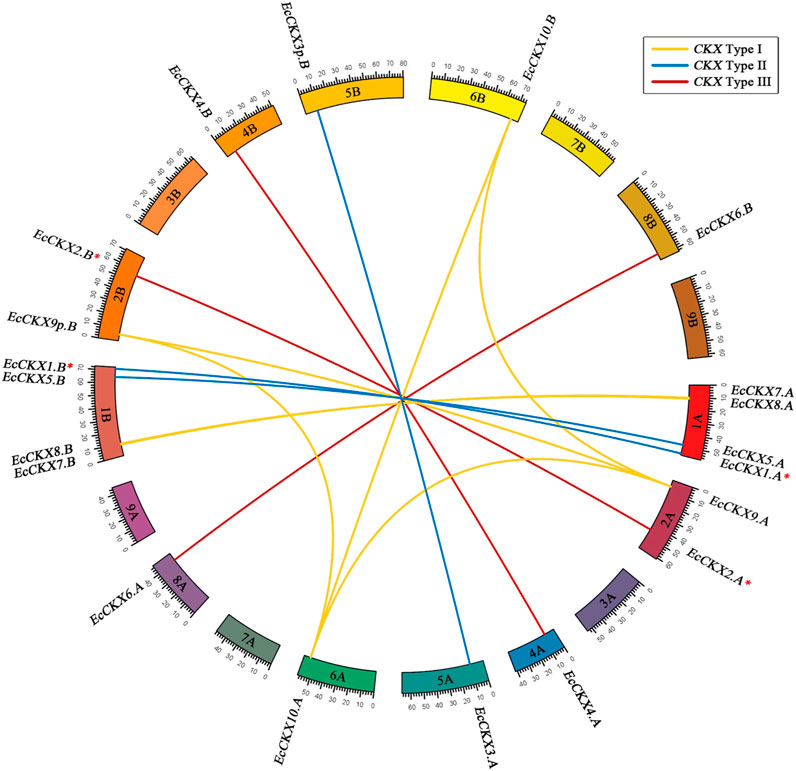
FIGURE 6. Synteny analysis of interchromosomal relationships of EcCKX of various types from different subgenomes of E. coracana. Syntenic gene pairs are colored according to the type of CKX, to which they are attributed. A red asterix denotes genes identical to previously sequenced and characterized EcCKX1 and EcCKX2.
Intergenomic synteny between E. coracana and the model crop species O. sativa made it possible to identify orthologs of EcCKX (Figure 5). The potential duplicates EcCKX7.A/B and EcCKX8A/B share the same loci on chromosomes 1A and 1B of finger millet, similar to their orthologs OsCKX1 and OsCKX2 in the rice genome. At the same time, the orthologs of EcCKX1.A/B and EcCKX5.A/B—OsCKX5 and OsCKX4—were located distantly on Os1 in the rice genome and formed syntenic pairs with only one of their direct orthologs in E. coracana. Taking into account the significant phylogenetic distance between the EcCKX1.A/B-OsCKX5 and EcCKX5.A/B-OsCKX4 clades, it may be concluded that the lineages of these genes did not arise as a result of a recent duplication event.
At the same time, members of Type III, EcCKX2.A/B and EcCKX4.A/B, each formed syntenic pairs with both of their orthologs in rice: OsCKX3 and OsCKX8. Unlike the previous case, this finding confirms the paralogous nature of the relatively recent duplicates EcCKX2 and EcCKX4. Similarly, members of Type III, EcCKX9.A/B and EcCKX10.A/B, formed double pairs with OsCKX6 and OsCKX7. It is worth noting that OsCKX6 and OsCKX7 are located relatively close on the Os2 chromosome, which may indicate the origin of these paralogs from a tandem duplication event.
Apart from the other genes, EcCKX6.A/B formed a syntenic pair only with OsCKX11. Both of these genes belong to the sub-Type IIIa (Figure 5), the clade which also contains dicot orthologs. This confirms that the present gene of this CKX isotype was evolutionarily preserved and diversified from other Type III genes, at least in the last common ancestor of dicots and monocots.
Expression profiling of EcCKX
Using the previously confirmed CKX-deficient lines of E. coracana somaclonal mutants (Radchuk et al., 2012; Bayer et al., 2014), we performed an expression profiling of EcCKX and EcCKX1.A/B genes, in particular of EcCKX2.A/B (Figure 7). As mentioned above, homeologous pairs of EcCKX1.A/B and EcCKX2.A/B encode highly conserved peptide products with sequence similarities up to 97.7% and 98.85%, respectively. It is the highest degree of conservation among all identified EcCKX homeolog pairs. Since the transcripts of these genes are hard to distinguish and are highly conserved, the expression of EcCKX1.A/B and EcCKX2.A/B was analyzed jointly for both homologous copies (Figure 7B).
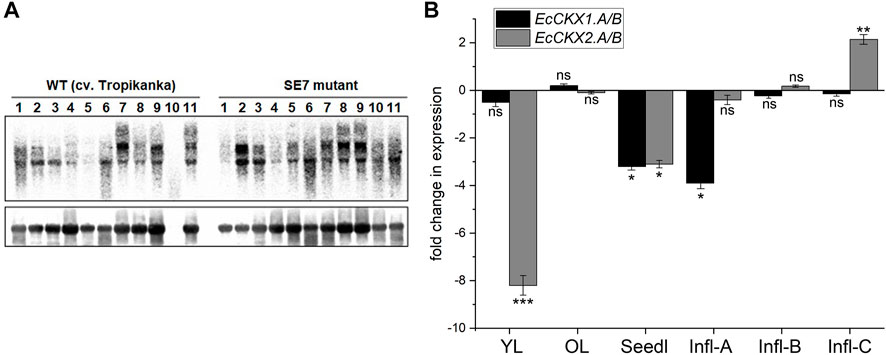
FIGURE 7. Expression profiling of EcCKX genes in the E. coracana mutant SE7 line. (A)—Northern blot of different tissues and developing stages of wild type and mutant SE7 finger millet plants hybridized with the EcCKX sequence as a probe (above): 1—node; 2—upper leaf; 3—lower leaf; 4—internode; 5—young root; 6—seedling 4 DAI; 7—developing panicle, < 1 cm in length; 8—developing panicle, 2–3 cm; 9—developing panicle, 3–4 cm; 10—developing panicle, > 4 cm; 11—seedling, 5 DAI. The signals were quantified by sequential hybridization with a ribosomal fragment as probe (below). (B)—Transcript profiling of the EcCKX1. A/B and EcCKX2. A/B genes in the developing inflorescences and young leaves of finger millet determined by quantitative RT-PCR analysis: YL—young leaf; OL—old leaf; Seedl—seedling 5 DAI; Infl-A/B/C—inflorescence tissue at stage A, B, and C. Data shown as fold change of upregulation of downregulation of respective gene in SE7 mutant line, compared to WT: ns—difference not statistically significant, *—significant at p < 0.05; **—p < 0.01; ***—p < 0.001.
The northern blot array allowed us to determine the total level of EcCKX transcripts in different E. coracana tissues (Figure 7A). For example, the upper leaves of the SE7 line were found to have higher levels of EcCKX transcripts than the WT line. Similarly, young roots of SE7 plants had higher levels of EcCKX transcripts compared to the WT, while no significant differences in EcCKX levels were found at 4 and 5 DAI in either line. However, at early stages of panicle development, SE7 plants had a lower level of EcCKX expression than the WT, whereas at later stages, the total levels of expression were comparable, or the EcCKX expression in SE7 could even be higher. The total EcCKX expression in the nodes and internodes of SE7 was lower than in the WT.
Previously, total transcriptomic analysis revealed the genes responsible for the semi-dwarf and bushy phenotype of SE7 (Radchuk et al., 2012). In this study, a detailed analysis of the expression of EcCKX1.A/B and EcCKX2.A/B reveals patterns of changes in the expression of these genes in the SE7 mutant, especially given that the SE7 has normal rates of CK synthesis (Radchuk et al., 2012). The most dramatic decrease in EcCKX2.A/B expression is observed in young leaves of SE7 (more than 8-fold), compared to the wild type, whereas the levels of EcCKX1.A/B remain unchanged (Figure 7B). At the same time, these genes exhibit no significant changes in expression in old leaves. However, in seedlings (5 DAI), the expression of both EcCKX1.A/B and EcCKX2.A/B decreases 3.1 to 3.2-fold, whereas analysis of total EcCKX expression shows no significant difference. It is possible that during germination the total level of these CK-degrading enzymes remains high, thus partially compensating for the deficiency of the EcCKX1.A/B and EcCKX2.A/B isotypes, possibly through the increased expression of their paralogs.
Most interestingly, EcCKX1.A/B expression levels change during early panicle development, while no differences in EcCKX2.A/B expression are found in the SE7 and WT lines. The transcript levels of EcCKX1.A/B are found to be nearly 3.9-fold lower in SE7 inflorescences during the early inflorescence development (stage A). It is noteworthy that at later stages the expression of EcCKX1.A/B, as well as that of EcCKX2.A/B, remains unchanged. However, in the later stage of inflorescence development (stage C), EcCKX2.A/B shows a 2.13-fold increase in expression. This correlates well with the observed increase in the total EcCKX level of transcripts at late stages of inflorescence development. Perhaps this observation can serve as a partial confirmation of the stage-dependent specificity of the expression of these two genes. Most likely, the early stages of panicle growth, such as stage A, are the key phases, in which high levels of CK determine subsequent panicle/inflorescence size, thus shaping crop grain productivity.
Phenotypic changes in cytokinin dehydrogenase/oxidase-deficient somaclonal mutants of E. coracana
For field evaluation of the phenotype, around 100 wild type and SE7 finger millet plants were grown for 2 weeks in a greenhouse and then transplanted in May to a field in Gatersleben, Germany, using 20 cm plant spacing. Plant height and the yields from 25 plants were measured in late September. For greenhouse phenotype evaluation, 25 SE7 mutant and wild-type plants were grown in a greenhouse during the same period (Figure 8).
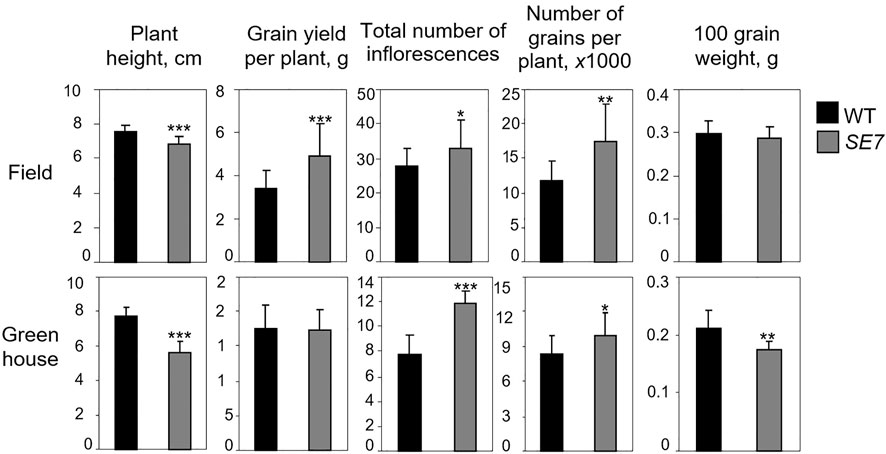
FIGURE 8. Productivity trait difference of field-grown (upper panel) and greenhouse-grown (lower panel) SE7 somaclonal mutant plants compared to the wild type of finger millet. Statistically significant differences are marked with *—significant at p < 0.05; **—p < 0.01; ***—p < 0.001.
SE7 somatic mutant plants grown in both the field and the greenhouse were characterized by significantly lower plant height (Figure 8) than the wild type, whereas greenhouse-grown plants differed more. The total number of inflorescences per plant also increased, indicating that more vegetative meristems were formed in SE7 plants (Figure 8). Surprisingly, greenhouse-grown plants did not show any significant difference in this parameter, probably due to altered growing conditions. Also, the CKX-deficient mutant line had a shortened vegetation cycle.
The mutant plants also produced more seeds per plant compared to the wild type, indicating extensive production of generative meristems in the SE7 line in response to higher levels of CK, which were not degraded (Figure 8). Increased numbers of inflorescences and flowers led to a significantly higher grain yield per plant in field conditions. However, this effect was less significant among the greenhouse-grown plants, since under such conditions, both lines produced more inflorescences, and thus showed higher grain productivity (Figure 8). The less significant difference in total plant productivity in the greenhouse could be explained by the fact that greenhouse-grown SE7 plants had a significantly lower grain mass. In contrast, 100-grain weight was not significantly different between field-grown SE7 and WT plants. It can therefore be concluded that the increase in the number of inflorescences is a key factor in determining the increased grain productivity (per plant) of the SE7 line (Figure 8).
Following the successful comparison of SE7 plant performance, and confirmation that the increased plant productivity is also found in field-grown plants, a full-scale field trial was conducted using the SE7 line and its WT (Figure 9). The most prominent features of the obtained somaclonal line SE7 were significantly reduced height, compared to the parental genotype cv. Tropikanka, and a much greater bush density, due to an increased number of primary and lateral stems. However, the somaclones did not show the reduced grain size. It was established that the SE7 line had a reduced height of almost 40%, compared to the WT (60.4±8.8 v. 100.1±5.5 cm, Figure 9A). Similar to the comparison shown in Figure 8, the weight of 1,000 grains manifest no significant change during field trials (Figure 9B).
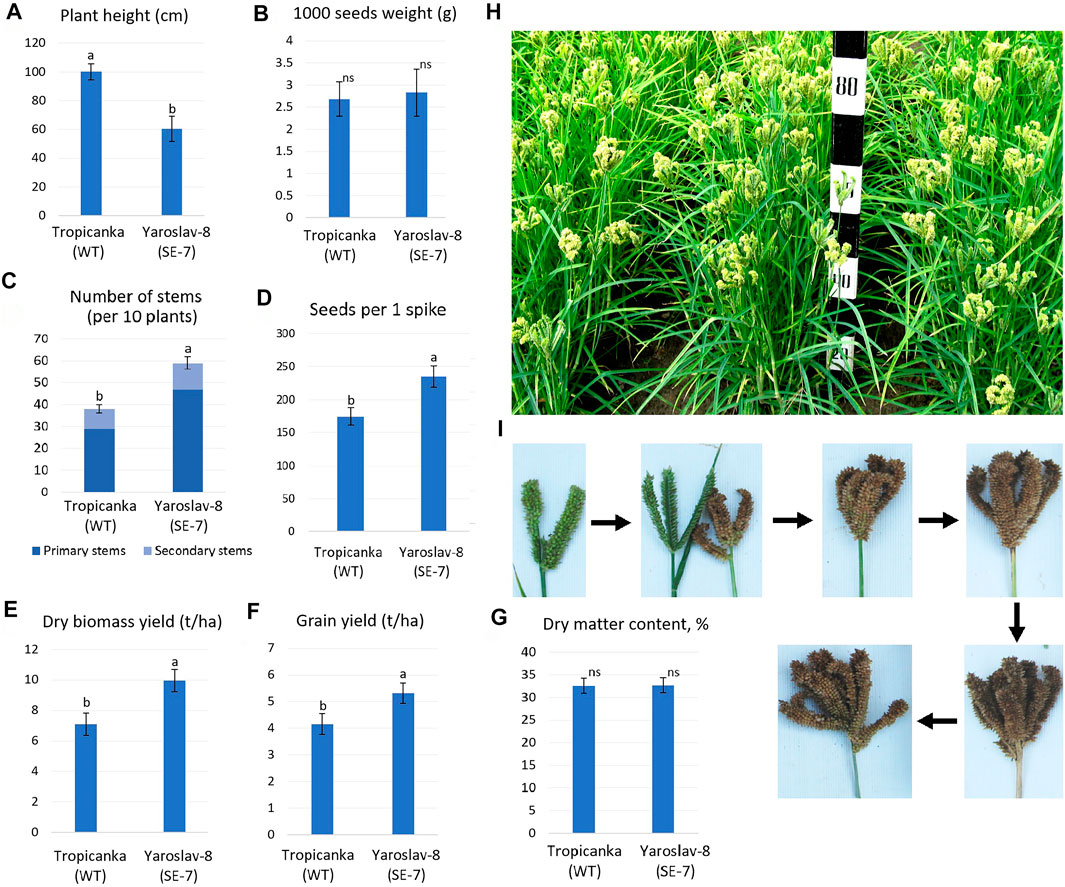
FIGURE 9. Performance of the SE7 mutant line in comparison with the WT during field trials. (A–D), (G)—measurements of different productivity traits; (E,F)—grain and biomass yield; (H)— plants of the SE7 line during maturation; (I)—different stages of finger millet panicle maturation. Different letters within each parameter (A–G): a, b—significantly different values; ns—difference not significant (at p < 0.05, Fischer LSD).
At the same time, the SE7 had a significantly greater number of primary stems (48±2.9 v. 29±1.9 in WT), while the number of secondary stems was similar for both the SE7 and WT—nine on average. The number of grains per spike increased by 34.3% in the SE7 line (236±16 vs. 175±13), compared to the WT (Figures 9C,D). From the concluding observations of two independent studies, it can be summarized that the key factors shaping the increased productivity of the CKX-deficient line are an increase in the number of inflorescences (due to the larger number of primary stems), and an increase in the number of seeds per panicle, with grain weight not significantly affecting the resulting plant productivity.
The resulting grain productivity of the SE7 line reached 5,320 kg/ha on average, 27.9% higher than the WT, which produced 4,159 kg/ha of grain (Figure 9F). In addition, the SE7 line showed increased levels of biomass accumulation, which is associated with the previously-identified downregulation of CKX. The SE7 line showed almost 10 t/ha of straw (absolutely dry mass), which is higher than that of the WT at 7.1 t/ha. Such a substantial increase in biomass productivity is most likely associated with the appearance of additional stems.
Discussion
A total of 20 EcCKXs were identified in the latest E. coracana assembly. Two of all the identified genes are considered pseudogenes, as they lack important structural domains: FAD- and CK-binding domains. The rapid expansion of the CKX gene family in finger millet can be associated with the evolutionary history of this species, which includes at least one allopolyploidization event giving rise to modern E. coracana (Zhang et al., 2019a; 2019b). EcCKXs are represented by complete homeologous pairs, indicating that the majority of these duplicated genes were not eliminated, but most likely passed through subfunctionalization. Using the example of Brassica rapa, it is shown that CKX, resulting from segmental duplications, tend to change their expression patterns and thus potentially gain new functions (Liu et al., 2013). It is obvious that in such a case, the duplicated genes are not eliminated or pseudogenized over time.
Other species closely related to E. coracana contain a lower number of CKX in their genomes. For example, S. italica has only 11 CKX genes (Wang et al., 2014); H. vulgare has 13–14 genes (Zalewski et al., 2014); O. sativa has 11; Brachypodium distachyon has 13; Sorghum bicolor has 12; and Z. mays has 16 (Mameaux et al., 2012). Genomes of dicot species can contain 7 CKX genes, as is observed in A. thaliana, while some species can contain a lower number of CKX genes, such as P. persica which has only 6; or more, e.g., 8 genes in P. trichocarpa (Immanen et al., 2013). Polyploid dicots possess much larger panels of CKX genes, for example Glycine max has 16 genes (Liu et al., 2021), while polyploid Brassica species have a much larger gene family compared to diploids. While B. rapa has only 12 CKX genes (Liu et al., 2013), amphidiploid B. napus contains 23 (Liu et al., 2018), while B. oleracea accounts for up to 36 CKX genes (Zhu et al., 2022).
Analysis of cis-regulatory elements make it possible to reveal the patterns of distribution of CK-responsive elements within the identified genes. Understanding of the diversity of CK-responsive cis-elements is currently limited to only a few validated motifs, such as ARR1 (AGAT [T,C] motif) (Brenner and Schmülling, 2015). However, it is evident that the expression of CKX is specifically induced by CK (Curaba et al., 2014), hence, new types of CK-responsive cis-elements have yet to be described. In this study we find unconventional motif organization, such as the presence of palindromic ARR1s or overlapping inverted ARR1-CPBCSPOR motifs. However, it is important to note that in silico identification of cis-regulatory elements, especially short-sequence motifs (such as ARR1, discussed above), is not enough for prediction of the binding site of functional proteins involved in transcriptional regulation (Hernandez-Garcia and Finer, 2014). Despite the presence of numerous copies of short-sequence motifs in the genome, only a small part of them are accessible for protein binding and further regulation of gene expression (Hardison and Taylor, 2012). Similarly, as analyzed in our study, ARR1 elements consist of 3–4 nucleotides. It may be expected that only a few of the identified cis-elements might be involved in CK-mediated expression of EcCKX. This could be a question for further separate research.
It is not clear what the role of such a broad diversity of these genes is in different taxa. However, the so-called “ancient” CKXs (which have direct orthologs in non-angiosperm species) have recently been found to preferentially degrade cis-zeatin (cZ)-type cytokinins, while non-ancient (relatively recent duplicates) mostly target N6-(Δ2-isopentenyl) adenines (iPs) and trans-zeatins (tZs) (Wang et al., 2021). In addition, it has been hypothesized that non-ancient CKX and their substrates (iP) may regulate the development of plant organs, especially flowers, and may regulate stress responses (Wang et al., 2021). Rapidly expanded type I CKXs belong to such non-ancient type genes, since their entire group is sub-orthologous to AtCKX4, which is considered one of the remarkable representatives of recent CKXs. Similarly, groups IIa, IIb, and IIIb are also orthologous to such non-ancient CKX, including EcCKX1.A/B and EcCKX2.A/B (widely discussed in this study), the deficiency of which results in increased inflorescence/panicle. By contrast, the phylogenetic group IIIa is orthologous with AtCKX7—the gene of ancient-type CKX. Genes EcCKX6.A/B belong to this group. Notably, this CKX homeolog pair has very strict orthology with OsCKX11 of rice, unlike other EcCKX. In a detailed study on Poaceae CKX, a clade containing OsCKX11 orthologs, clustered as basal to all monocot CKXs (Mameaux et al., 2012), further supports the assignment to “ancient” type genes.
Measurement of CK species in finger millet identifies iP as the major form of CKs in developing inflorescences of this species (Radchuk et al., 2012). When we analyzed the somaclonal variant SE7, produced previously (Baer et al., 2007), the levels of iP and the iP precursor riboside phosphate were strongly increased early in inflorescences of this genotype, but did not change compared to the wild type later on (Radchuk et al., 2012). We hypothesized that increased iP levels are probably due to impaired degradation, as evidenced by lower EcCKX expression in young inflorescences. Such disturbed CK homeostasis, brought about by either reduced CK degradation or biosynthesis, can stimulate plant productivity. As observed in the present study, effects of EcCKX deficiency in the SE7 mutant may contribute to an understanding of the phenotypic effects caused by CKX downregulation. Analyzed EcCKX1.A/B and EcCKX2.A/B were preferentially down-regulated in actively growing and generative tissues, leading to rapid increase of grain and biomass productivity of the mutant. Increased plant productivity of the SE7 line may be beneficial not only for food production, but may also open ways to use finger millet for biofuel production, in particular, the production of second-generation bioethanol (Yemets et al., 2020; Blume et al., 2021).
Initially, down-regulated CKX expression was shown to decrease CK-degradation activity and increase CK levels in rice, leading to an increase of seed numbers per plant and 1000-grain weight (Ashikari et al., 2005). The same evidence was then obtained in experiments with RNAi-based silencing of the HvCKX1 in barley, resulting in reduced cytokinin oxidase/dehydrogenase levels and higher plant productivity (Zalewski et al., 2010). This is confirmed in other publications by Zalewski et al. (2012; 2014).
Later, HvCKX1 and HvCKX9, predominantly expressed in the aleurone layer of maturing grains and leaf vasculature, were used for stable Agrobacterium tumefaciens-mediated transformation of the barley cultivar Golden Promise (Mrízová et al., 2013). As a result, constitutive overexpression of these two genes independently induced morphological changes in barley plants and prevented their transition to flowering. In all obtained transgenic lines, roots proliferated more rapidly and root-to-shoot ratios were higher than in wild-type plants. Depleted CK levels during early phases of development are restored by down-regulation of endogenous CKX and reinforced de novo biosynthesis of CKs. When barley plants were transformed with Arabidopsis AtCKX1 under a mild root-preferred promoter, the obtained transgenic lines were found to have higher drought tolerance then the control (Pospíšilová et al., 2016). These transgenic plants can maintain higher yield parameters following revitalization compared to wild type plants under stress conditions. Knockdown of the inflorescence meristem-specific cytokinin oxidase in rice (OsCKX2) regulates primary flowering activity, and modulates rice grain yield under normal conditions as well as under abiotic stress conditions by controlling cytokinin levels (Joshi et al., 2018).
Several recent studies have demonstrated that RNAi-based silencing of CKX leads to increased grain yields in other cereals. In particular, a significantly increased grain number per spike was found as the effect of TaCKX2.4 silencing by RNAi in wheat (Li et al., 2018). Coordinated effects of TaCKX1 silencing on the expression of other TaCKX, wheat spike phytohormone levels, and yield-related traits were demonstrated in silenced T2 lines (Jabłoński et al., 2020). In the moss Physcomitrella patens, an evolutionarily early divergent plant, it was shown that under normal growth conditions overexpression of PpCKX1 caused many phenotypic changes at different developmental stages, and that increased growth of the rhizoid could affect these changes. In addition, evidence is provided that the PpCKX1-overexpressing plants show enhanced dehydration and salt stress tolerance. Taken together, the authors suggested that PpCKX1 plays a regulatory role in the development and adaptation to abiotic stresses of this evolutionarily early land plant species (Hyoung et al., 2020).
Finally, knockout mutation of HvCKX1 by CRISPR/Cas9 editing in barley had a limited effect on yield productivity. However, significant reductions in CKX enzyme activity in young spikelets and 10-day-old roots corresponded to greater root length, number of root hairs, and increased surface area (Gasparis et al., 2019). In contrast, the roots of ckx3 knockout mutants were smaller. The observed changes suggest that the knockout of a single CKX in barley may not be sufficient to disrupt cytokinin homeostasis or increase grain yields. These authors then demonstrated that underlying mechanisms of co-regulation of the expression of TaCKX family members were similar in different spring wheat cultivars, but, depending on the content and composition of phytohormones, regulation of yield-related traits was differentially affected (Jablonski et al., 2021). On the other hand, identification and characterization of all members of the CKX family, not only in E. coracana, but in closely-related species with already characterized genomes and transcriptomes (Zhang et al., 2019a; Zhang et al., 2019b; Hall et al., 2021), may provide more information about functional features of different CKX types.
Successful application of genome editing technics, targeting CKX genes in different taxa, will require a comprehensive understanding of genomic organization, phylogenetic classification, and evolution of CKX genes, in order to efficiently affect all paralogous copies with similar function and achieve desirable effects of genetic engineering. The greater diversity of CKX in monocots compared to dicots, and the relationship between different metabolic pathways of different forms of cytokinins, remains unclear. Understanding of these matters may become an important tool for targeted crop design to ensure global food security in the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
RB, AY, and YB wrote the original draft of the manuscript and contributed to the conception and design of the study. VK, VR, and DR contributed to particular sections of the manuscript. RB, AY, VK, VR, and DR performed the collection of the experimental data, while RB and YB analyzed and interpreted the obtained results. RB, VK, and VR performed the statistical analysis. All authors listed made a substantial and direct intellectual contribution to the work, and approved it for publication.
Acknowledgments
We are grateful to Prof. Katrien M. Devos, who generously provided permission to use the E. coracana sequence data produced under NSF-BREAD award IOS-1543901. We acknowledge the suggestions by Dr. V. Kyrylenko (Institute of Food Biotechnology and Genomics, National Academy of Sciences of Ukraine, Kyiv), aimed at improving the language of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.963789/full#supplementary-material
References
Altschul, S. F. (1993). A protein alignment scoring system sensitive at all evolutionary distances. J. Mol. Evol. 36 (3), 290–300. doi:10.1007/BF00160485
Antony Ceasar, S., Maharajan, T., Ajeesh Krishna, T. P., Ramakrishnan, M., Victor Roch, G., Lakkakula, S., et al. (2018). Finger millet [Eleusine coracana (L.) Gaertn.] improvement: Current status and future interventions of whole genome sequence. Front. Plant Sci. 9. doi:10.3389/fpls.2018.01054
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takasi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309 (5735), 741–745. doi:10.1126/science.1113373
Baer, G. Y., Yemets, A. I., Stadnichuk, N. A., Rakhmetov, D. B., and Blume, Ya.B. (2007). Somaclonal variability as a source for creation of new varieties of finger millet (Eleusine coracana (L.) Gaertn.). Cytol. Genet. 41, 204–208. doi:10.3103/S0095452707040020
Bančič, J., Odeny, D. A., Ojulong, H. F., Josiah, S. M., Buntjer, J., Gaynor, R. C., et al. (2021). Genomic and phenotypic characterization of finger millet indicates a complex diversification history. bioRxiv [Preprint]. doi:10.1101/2021.04.06.438618
Bayer, G. Y., Yemets, A. I., and Blume, Y. B. (2014). Obtaining the transgenic lines of finger millet Eleusine coracana (L.). with dinitroaniline resistance. Cytol. Genet. 48, 139–144. doi:10.3103/S0095452714030025
Blume, R. Y., Yemets, A. I., Rakhmetov, D. B., and Blume, Y. B. (2021). “Somaclonal ckx-downregulated line of finger millet as a feedstock for sustainable bioethanol production,” in European biomass conference and exhibition proceedings. Editors P. Mauguin, N. Scarlat, and A. Grassi (Florence: ETA-Florence Renewable Energies), 81–84.
Blume, Ya.B., Krasylenko, Yu.A., and Yemets, A. I. (2017). “The role of plant cytoskeleton on phytohormone signaling under abiotic and biotic stresses,” in Mechanism of plant hormone signaling under stress: A functional genomic frontier. Editor G. Pandey (Hoboken: Wiley-Blackwell), 2, 127–185. doi:10.1002/9781118889022.ch23
Brenner, W. G., and Schmülling, T. (2015). Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front. Plant Sci. 6, 29. doi:10.3389/fpls.2015.00029
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2020b). Cytokinin dehydrogenase: A genetic target for yield improvement in wheat. Plant Biotechnol. J. 18 (3), 614–630. doi:10.1111/pbi.13305
Conrad, K., Motyka, V., and Schlüter, T. (2007). Increase in activity, glycosylation and expression of cytokinin oxidase/dehydrogenase during the senescence of barley leaf segments in the dark. Physiol. Plant. 130, 572–579. doi:10.1111/j.1399-3054.2007.00914.x
Curaba, J., Singh, M. B., and Bhalla, P. L. (2014). miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 65 (6), 1425–1438. doi:10.1093/jxb/eru002
Dabravolski, S. A., and Isayenkov, S. V. (2021). Evolution of the cytokinin dehydrogenase (CKX) domain. J. Mol. Evol. 89 (9-10), 665–677. doi:10.1007/s00239-021-10035-z
Dash, P. K., and Rai, R. (2022). Green revolution to grain revolution: Florigen in the frontiers. J. Biotechnol. 343, 38–46. doi:10.1016/j.jbiotec.2021.10.002
Edgar, R. C. (2004). Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi:10.1093/nar/gkh340
Galuszka, P., Popelková, H., Werner, T., Frébortová, J., Pospíšilová, H., Mik, V., et al. (2007). Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J. Plant Growth Regul. 26, 255–267. doi:10.1007/s00344-007-9008-5
Gasparis, S., Przyborowski, M., Kała, M., and Nadolska-Orczyk, A. (2019). Knockout of the HvCKX1 or HvCKX3 gene in barley (Hordeum vulgare L.) by RNA-guided Cas9 nuclease affects the regulation of cytokinin metabolism and root morphology. Cells 8 (8), 782. doi:10.3390/cells8080782
Gu, R., Fu, J., Guo, S., Duan, F., Wang, Z., Mi, G., et al. (2010). Comparative expression and phylogenetic analysis of maize cytokinin dehydrogenase/oxidase (CKX) gene family. J. Plant Growth Regul. 29, 428–440. doi:10.1007/s00344-010-9155-y
Gupta, S. M., Arora, S., Mirza, N., Pande, A., Lata, C., Puranik, S., et al. (2017). Finger millet: A “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Front. Plant Sci. 8, 643. doi:10.3389/fpls.2017.00643
Hall, N. D., Patel, J. D., McElroy, J. S., and Goertzen, L. R. (2021). Detection of subgenome bias using an anchored syntenic approach in Eleusine coracana (finger millet). BMC Genomics 22, 175. doi:10.1186/s12864-021-07447-y
Hardison, R., and Taylor, J. (2012). Genomic approaches towards finding cis-regulatory modules in animals. Nat. Rev. Genet. 13, 469–483. doi:10.1038/nrg3242
Hatakeyama, M., Aluri, S., Balachadran, M. T., Sivarajan, S. R., Patrignani, A., Grüter, S., et al. (2018). Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. 25 (1), 39–47. doi:10.1093/dnares/dsx036
Henikoff, S., and Henikoff, J. G. (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U. S. A. 89 (22), 10915–10919. doi:10.1073/pnas.89.22.10915
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217-218, 109–119. doi:10.1016/j.plantsci.2013.12.007
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 (1), 297–300. doi:10.1093/nar/27.1.297
Hittalmani, S., Mahesh, H. B., Shirke, M. D., Biradar, H., Uday, G., Aruna, Y. R., et al. (2017). Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 18, 465. doi:10.1186/s12864-017-3850-z
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). Gsds 2.0: An upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi:10.1093/bioinformatics/btu817
Hyoung, S., Cho, S. H., Chung, J. H., So, W. M., Cui, M. H., and Shin, J. S. (2020). Cytokinin oxidase PpCKX1 plays regulatory roles in development and enhances dehydration and salt tolerance in Physcomitrella patens. Plant Cell. Rep. 39, 419–430. doi:10.1007/s00299-019-02500-3
Immanen, J., Nieminen, K., Duchens Silva, H., Rodríguez Rojas, F., Meisel, L. A., Silva, H., et al. (2013). Characterization of cytokinin signaling and homeostasis gene families in two hardwood tree species: Populus trichocarpa and Prunus persica. BMC Genomics 14, 885. doi:10.1186/1471-2164-14-885
Jablonski, B., Bajguz, A., Bocian, J., Orczyk, W., and Nadolska-Orczyk, A. (2021). Genotype-dependent effect of silencing of TaCKX1 and TaCKX2 on phytohormone crosstalk and yield-related traits in wheat. Int. J. Mol. Sci. 22 (21), 11494. doi:10.3390/ijms222111494
Jabłoński, B., Ogonowska, H., Szala, K., Bajguz, A., Orczyk, W., and Nadolska-Orczyk, A. (2020). Silencing of TaCKX1 mediates expression of other TaCKX genes to increase yield parameters in wheat. Int. J. Mol. Sci. 21 (13), 4809. doi:10.3390/ijms21134809
Jameson, P. E., and Song, J. (2016). Cytokinin: A key driver of seed yield. J. Exp. Bot. 67 (3), 593–606. doi:10.1093/jxb/erv461
Jiskrová, E., Novák, O., Pospíšilová, H., Holubová, K., Karády, M., Galuszka, P., et al. (2016). Extra- and intracellular distribution of cytokinins in the leaves of monocots and dicots. Nat. Biotechnol. 3, 735–742. doi:10.1016/j.nbt.2015.12.010
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi:10.1093/bioinformatics/8.3.275
Joshi, R., Sahoo, K. K., Tripathi, A. K., Kumar, R., Gupta, B. K., Pareek, A., et al. (2018). Knockdown of an inflorescence meristem-specific cytokinin oxidase - OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell. Environ. 41 (5), 936–946. doi:10.1111/pce.12947
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Li, J., Nie, X., Tan, J. L. H., and Berger, F. (2013). Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 110 (38), 15479–15484. doi:10.1073/pnas.1305175110
Li, Y. L., Song, G. Q., Gao, J., Zhang, S. J., Zhang, R. Z., Li, W., et al. (2018). Enhancement of grain number per spike by RNA interference of cytokinin oxidase 2 gene in bread wheat. Hereditas 155, 33. doi:10.1186/s41065-018-0071-7
Liu, L.-M., Zhang, H.-Q., Cheng, K., and Zhang, Y.-M. (2021). Integrated bioinformatics analyses of PIN1, CKX, and yield-related genes reveals the molecular mechanisms for the difference of seed number per pod between soybean and cowpea. Front. Plant Sci. 12, 749902. doi:10.3389/fpls.2021.749902
Liu, P., Zhang, C., Ma, J. Q., Zhang, L. Y., Yang, B., Tang, X. Y., et al. (2018). Genome-wide identification and expression profiling of cytokinin oxidase/dehydrogenase (CKX) genes reveal likely roles in pod development andstress responses in oilseed rape (Brassica napus L.). Genes. 9 (3), 168. doi:10.3390/genes9030168
Liu, Y., Wang, X., Wang, X., Gao, W., and You, C. (2022). Identification and functional characterization of apple MdCKX5.2 in root development and abiotic stress tolerance. Horticulturae 8, 62. doi:10.3390/horticulturae8010062
Liu, Z., Lv, Y., Zhang, M., Liu, Y., Kong, L., Zou, M., et al. (2013). Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14, 594. doi:10.1186/1471-2164-14-594
Mameaux, S., Cockram, J., Thiel, T., Steuernagel, B., Stein, N., Taudien, S., et al. (2012). Molecular, phylogenetic and comparative genomic analysis of the cytokinin oxidase/dehydrogenase gene family in the Poaceae. Plant Biotechnol. J. 10 (1), 67–82. doi:10.1111/j.1467-7652.2011.00645.x
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49 (D1), D412–D419. doi:10.1093/nar/gkaa913
Mrízová, K., Jiskrová, E., Vyroubalová, Š., Novák, O., Ohnoutková, L., Pospíšilová, H., et al. (2013). Overexpression of cytokinin dehydrogenase genes in barley (Hordeum vulgare cv. Golden Promise) fundamentally affects morphology and fertility. PLoS ONE 8 (11), e79029. doi:10.1371/journal.pone.0079029
Nguyen, H. N., Kambhampati, S., Kisiala, A., Seegobin, M., and Emery, R. J. N. (2021). The soybean (Glycine max L.) cytokinin oxidase/dehydrogenase multigene family; Identification of natural variations for altered cytokinin content and seed yield. Plant Direct 5, e00308. doi:10.1002/pld3.308
Pandian, S., and Ramesh, M. (2019). Decoding of finger millet genome: A milestone of millet genomics. Signal Transduct. Insights 8, 117864341882054. doi:10.1177/1178643418820541
Pendergast, T. H., Qi, P., Odeny, D. A., Dida, M. M., and Devos, K. M. (2022). A high-density linkage map of finger millet provides QTL for blast resistance and other agronomic traits. Plant Genome 15, e20175. doi:10.1002/tpg2.20175
Pospíšilová, H., Jiskrová, E., Vojta, P., Mrízová, K., Kokáš, F., Majeská Čudejková, M., et al. (2016). Transgenic barley overexpressing a cytokinin dehydrogenase gene shows greater tolerance to drought stress. Nat. Biotechnol. 33, 692–705. doi:10.1016/j.nbt.2015.12.005
Radchuk, V., Radchuk, R., Pirko, Y., Vankova, R., Gaudinova, A., Korkhovoy, V., et al. (2012). A somaclonal line SE7 of finger millet (Eleusine coracana) exhibits modified cytokinin homeostasis and increased grain yield. J. Exp. Bot. 63 (15), 5497–5506. doi:10.1093/jxb/ers200
Radchuk, V., Weier, D., Radchuk, R., Weschke, W., and Weber, H. (2011). Development of maternal seed tissue in barley is mediated by regulated cell expansion and cell disintegration and coordinated with endosperm growth. J. Exp. Bot. 62, 1217–1227. doi:10.1093/jxb/erq348
Rakhmetova, S. O., Vergun, O. M., Blume, R. Y., Bondarchuk, O. P., Shymanska, O. V., Tsygankov, S. P., et al. (2020). Ethanol production potential of sweet sorghum in North and Central Ukraine. Open Agric. J. 14, 321–338. doi:10.2174/1874331502014010321
Ramakers, C., Ruijter, J. M., Deprez, R. H. L., and Moorman, A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi:10.1016/s0304-3940(02)01423-4
Sood, S., Joshi, D. C., Chandra, A. K., and Kumar, A. (2019). Phenomics and genomics of finger millet: Current status and future prospects. Planta 250, 731–751. doi:10.1007/s00425-019-03159-6
Suttle, J. C., Huckle, L. L., Lu, S., and Knauber, D. C. (2014). Potato tuber cytokinin oxidase/dehydrogenase genes: Biochemical properties, activity, and expression during tuber dormancy progression. J. Plant Physiol. 171 (6), 448–457. doi:10.1016/j.jplph.2013.11.007
Swarbreck, D., Wilks, C., Lamesch, P., Berardini, T. Z., Garcia-Hernandez, M., Foerster, H., et al. (2008). The Arabidopsis information resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36, D1009–D1014. doi:10.1093/nar/gkm965
Szala, K., Ogonowska, H., Lugowska, B., Zmijewska, B., Wyszynska, R., Dmochowska-Boguta, M., et al. (2020). Different sets of TaCKX genes affect yield-related traits in wheat plants grown in a controlled environment and in field conditions. BMC Plant Biol. 20, 496. doi:10.1186/s12870-020-02713-9
Wang, C., Wang, H., Zhu, H., Ji, W., Hou, Y., Meng, Y., et al. (2021). Genome-wide identification and characterization of cytokinin oxidase/dehydrogenase family genes in Medicago truncatula. J. Plant Physiol. 256, 153308. doi:10.1016/j.jplph.2020.153308
Wang, Y., Liu, H., and Xin, Q. (2014). Genome-wide analysis and identification of cytokinin oxidase/dehydrogenase (CKX) gene family in foxtail millet (Setaria italica). Crop J. 2 (4), 244–254. doi:10.1016/j.cj.2014.05.001
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Werner, T., Köllmer, I., Bartrina, I., Holst, K., and Schmülling, T. (2006). New insights into the biology of cytokinin degradation. Plant Biol. 8 (3), 371–381. doi:10.1055/s-2006-923928
Yemets, A., Blume, R., Rakhmetov, D., and Blume, Y. (2020). Finger millet as a sustainable feedstock for bioethanol production. Open Agric. J. 14 (1), 257–272. doi:10.2174/1874331502014010257
Zalabák, D., Galuszka, P., Mrízová, K., Podlešáková, K., Gu, R., and Frébortová, J. (2014). Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol. Biochem. 74, 283–293. doi:10.1016/j.plaphy.2013.11.020
Zalewski, W., Galuszka, P., Gasparis, S., Orczyk, W., and Nadolska-Orczyk, A. (2010). Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 61, 1839–1851. doi:10.1093/jxb/erq052
Zalewski, W., Gasparis, S., Boczkowska, M., Rajchel, I. K., Kala, M., Orczyk, W., et al. (2014). Expression patterns of HvCKX genes indicate their role in growth and reproductive development of barley. PLoS ONE 9, e115729. doi:10.1371/journal.pone.0115729
Zalewski, W., Orczyk, W., Gasparis, S., and Nadolska-Orczyk, A. (2012). HvCKX2 gene silencing by biolistic or Agrobacterium-mediated transformation in barley leads to different phenotypes. BMC Plant Biol. 12, 206. doi:10.1186/1471-2229-12-206
Zhang, H., Hall, N., Goertzen, L. R., Bi, B., Chen, C. Y., Peatman, E., et al. (2019a). Development of a goosegrass (Eleusine indica) draft genome and application to weed science research. Pest Manag. Sci. 75 (10), 2776–2784. doi:10.1002/ps.5389
Zhang, H., Hall, N., Goertzen, L. R., Chen, C. Y., Peatman, E., Patel, J., et al. (2019b). Transcriptome analysis reveals unique relationships among Eleusine species and heritage of Eleusine coracana. G3-Genes Genom. Genet. 9 (6), 2029–2036. doi:10.1534/g3.119.400214
Zhang, L., Zhao, Y. L., Gao, L. F., Zhao, G. Y., Zhou, R. H., Zhang, B. S., et al. (2012). TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 195 (3), 574–584. doi:10.1111/j.1469-8137.2012.04194.x
Zhu, M., Wang, Y., Lu, S., Yang, L., Zhuang, M., Zhang, Y., et al. (2022). Genome-wide identification and analysis of cytokinin dehydrogenase/oxidase (CKX) family genes in Brassica oleracea L. reveals their involvement in response to Plasmodiophora brassicae infections. Hortic. Plant J. 8 (1), 68–80. doi:10.1016/j.hpj.2021.05.003
Keywords: cytokinin dehydrogenase/oxidase enzymes, EcCKX genes, phylogeny, gene expression, Eleusine coracana, finger millet, food security
Citation: Blume R, Yemets A, Korkhovyi V, Radchuk V, Rakhmetov D and Blume Y (2022) Genome-wide identification and analysis of the cytokinin oxidase/dehydrogenase (ckx) gene family in finger millet (Eleusine coracana). Front. Genet. 13:963789. doi: 10.3389/fgene.2022.963789
Received: 07 June 2022; Accepted: 05 August 2022;
Published: 27 September 2022.
Edited by:
Jitender Singh, Sardar Vallabhbhai Patel University of Agriculture and Technology, IndiaReviewed by:
Jose Sebastian, Indian Institute of Science Education and Research Berhampur (IISER), IndiaPranav Pankaj Sahu, Czech Academy of Sciences, Czechia
Copyright © 2022 Blume, Yemets, Korkhovyi, Radchuk, Rakhmetov and Blume. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rostyslav Blume, Ymx1bWUucm9zdGlzbGF2QGdtYWlsLmNvbQ==
‡ORCID: Rostyslav Blume, orcid.org/0000-0003-4936-1803; Alla Yemets, orcid.org/0000-0001-6887-0705; Vitaliy Korkhovyi, orcid.org/0000-0002-4803-9970; Volodymyr Radchuk, orcid.org/0000-0002-0472-6605; Dzhamal Rakhmetov, orcid.org/0000-0001-7260-3263; Yaroslav Blume, orcid.org/0000-0001-7078-7548
†These authors have contributed equally to this work and share first authorship
 Rostyslav Blume
Rostyslav Blume Alla Yemets
Alla Yemets Vitaliy Korkhovyi2‡
Vitaliy Korkhovyi2‡ Volodymyr Radchuk
Volodymyr Radchuk Yaroslav Blume
Yaroslav Blume