- 1Department of Bacteriology, Immunology, and Mycology, Faculty of Veterinary Medicine, Suez Canal University, Ismailia, Egypt
- 2Department of Virology, Animal Health Research Institute (AHRI), Agriculture Research Center (ARC), Giza, Egypt
- 3Department of Biology, College of Science, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
- 4Department of Hygiene and Zoonoses, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
- 5Department of Biology, College of Science, King Khalid University, Abha, Saudi Arabia
- 6Animal Health Research Institute(AHRI), Mansoura laboratory Branch, Agriculture Research Center (ARC), Giza, Egypt
- 7Department of Theriogenology, Faculty of Veterinary Medicine, South Valley University, Qena, Egypt
- 8Department of Animal Medicine, Faculty of Veterinary Medicine, Suez Canal University, Ismailia, Egypt
This study investigated the prevalence, antibiogram, virulence, extended-spectrum β-lactamases (ESBLs), and non-β-lactam encoding genes of Proteus species isolated from infected dogs in Ismailia province, Egypt. The study was conducted on 70 fecal swabs collected from dogs with diarrhea for bacteriological identification of Proteus spp. The positive isolates were evaluated for antibiotic susceptibility, molecular tests of virulence, ESBLs, and non-β-lactam encoding genes. Prevalence of Proteus spp. was 35.7% (25/70), including Proteus mirabilis (n = 23) and Proteus vulgaris (n = 2). The Proteus spp. prevalence revealed diversity, higher in males than females, in ages < 12 weeks. Investigation of antimicrobial resistance was found against penicillin and amoxicillin (100%), amoxicillin–clavulanic acid (32%), cephalosporins: cefotaxime and ceftazidime (36%), and monobactam: aztreonam (28%) as ESBLs, in addition to tetracycline (32%) and trimethoprim sulfamethoxazole (100%). The strains retrieved by PCR revealed ureC, zapA, and rsbA virulence genes with variant prevalence as 92%, 60%, and 52%, respectively. In addition, the recovered strains contained ESBL genes with a dramatic variable prevalence of 100%, 92%, 36%, and 32%, to blaTEM, blaSHV, blaCTX-M, and blaOXA-1, respectively, and non β-lactam encoding genes with a prevalence of 100%, 48%, 44%, 20%, and 12%, to sul1, tetA, intI1, qnrA, and aadA1. Moreover, 28% (7/25) of recovering strains were MDR (multidrug-resistant) up to four classes of antimicrobials, and 48% (12/25) of the examined strains were MDR up to three antimicrobial classes. In conclusion, to the best of our knowledge, our study could be the first report recording MDR Proteus spp. in dogs in Egypt.
1 Introduction
Proteus species are Gram-negative bacilli belonging to the Enterobacteriaceae family, which is widespread in both the human and animal gastrointestinal tracts, especially the members of the genus Proteus (Hegazy, 2016). It is the ultimate example of an opportunistic nosocomial pathogen (Jacobsen and Shirtliff, 2011). P. mirabilis, P. vulgaris, P. penneri, P. hauseri, P. terrae, and P. cibarius, with the unidentified genomic species, are included currently within the Proteus genus (Manos and Belas, 2006; Drzewiecka, 2016). P. mirabilis pathogens were isolated from humans, dogs, monkeys, pigs, sheep, goats, raccoons, cats, rodents, and other mammals. It is considered a part of the normal microbiota of the mammalian intestinal tract (Guentzel, 1996). In addition, P. mirabilis and P. vulgaris are widely disseminated in the environment, occurring in contaminated water, sewage, and soil. It is critical to decompose the animals’ organic substances (Rózalski et al., 1997).
The antimicrobial-resistant strains of Proteus from companion animals were recorded. Their infectious nature results from their resistance to antipathogenic chemicals, enabling them to still be intact in their environment (Hola et al., 2012). The developed antimicrobial resistance leads to failure in treatment associated with an adverse impact on the animal’s welfare. Bacterial pathogens with resistance properties against antimicrobials can have substantial public health consequences for human beings if isolates are transmitted from pets to their owners (Guardabassi et al., 2004; Lloyd, 2007; Algammal et al., 2020). The resistance to extended-spectrum β-lactam is mostly related to the production of extended-spectrum β-lactamases; ESBLs would lyse many β-lactam antimicrobial agents like penicillins, different generations of cephalosporins, and carbapenems (Magiorakos et al., 2012; Rubin and Pitout, 2014; Algammal et al., 2020; Algammal et al., 2021b). The chemical backbone of β-lactam antibiotics is the -lactam ring, which can be broken by these enzymes (Fadare and Okoh, 2021). The resistance of ESBL producers to many antibiotic classes is a major concern in clinical settings as it renders the treatment of individuals infected with these bacteria difficult and frequently impossible (Rawat and Nair, 2010). Resistance to sulfonamides usually develops primarily due to dihydropteroate synthase (DHPS) enzyme or their mutations carrying folP gene involved in the biosynthesis of the nucleotide or via the development of alternative genes of DHPS, such as sul1, sul2, and sul3, showing low affinity to sulfonamides (Perreten and Boerlin, 2003; Yun et al., 2012). Sulfonamides are chemotherapeutic drugs that act as competitive inhibitors of the folP gene-encoded DHPS. Widespread sulfonamide resistance is mostly caused by plasmid- and integron-borne sul1-3 genes that code encoding mutant DHPS enzymes that do not bind to sulfonamides (Sánchez-Osuna et al., 2019). Tetracycline-resistant genes are most commonly found on conjugated plasmids and transposons. On the other hand, other isolates have the necessary genes encoded on their chromosomes (Guillaume et al., 2000; Oppegaard et al., 2001). Efflux pumps, ribosome protection, and enzymatic deactivation are the critical mechanisms of tetracycline resistance acquired by acquiring tet genes. The aad A family of genes encodes aminoglycoside-3″-adenylyltransferase (AAD), which promote aminoglycosides (White and Rawlinson, 2001). The quinolone-resistant Enterobacteriaceae are growing worldwide. The qnr genes enhance resistance to nalidixic acid and reduce ciprofloxacin susceptibility (Crump et al., 2003). Class 1 integrons are frequently linked to multidrug resistance due to their ability to acquire or eliminate several antimicrobial resistance gene cassettes (Chen et al., 2017). Awareness of bacterial resistance against the antimicrobials, among the isolates of canine Proteus spp., is essential from a veterinary standpoint and an international public health perspective (Harada et al., 2014).
Currently, the pathogenic mechanism of Proteus-associated diarrhea remains unknown. It is necessary and useful to investigate the intestinal pathogenicity of Proteus, which can cause diarrhea. Few published studies have elucidated the pathogenic mechanism of Proteus diarrhea until now (Gong et al., 2019). Proteus spp. invasiveness is related to several virulent factors, and virulence genes encoded in plasmids regulate these virulent factors (Manos and Belas, 2006). Swarming on solid surfaces is a prominent characteristic property of Proteus spp. Although several genes were linked with swarming, the rsbA gene is a necessary swarming phenomenon for swarming control (Rather, 2005). Urease is an essential biomarker in Proteus infection for developing bladder and kidney stones. Many urease genes, like ureC, are essential to the urease enzyme development process (Li and Mobley, 2002). The zap gene product is encoded by many genes essential for protease production, especially zapA for regulating the expression of IgA protease during differentiation between the swimmer and swarmer cells (Walker et al., 1999). Urease and protease are considered diagnostic and differential aspects that describe this genus member from other Enterobacteriaceae members (Ali and Yousif, 2015).
Since the risky infection of Proteus spp. in dogs in Egypt has not been comprehensively clarified yet, this study aimed to clarify the prevalence, antibiogram, virulence genes, and ESBLs and non-β-lactam encoding genes of different Proteus spp. isolates to explore the potential hazard of Proteus spp. in dogs.
2 Materials and methods
2.1 Ethical approval
The study was conducted in accordance with the applicable rules and regulations of the Animal Ethics Review Committee of Suez Canal University (AERC-SCU) in Egypt. The experimental methods and laboratory work followed bacterial isolation, biosafety, and quality standards. In this study, no animals were experimentally used; however, dog handling and samples were applied with the owners’ consent.
2.2 Study period and location
This study has been performed in a private small animal clinic in the Ismailia governorate, Egypt, during the period from April to September 2020.
2.3 Animals and clinical examination
Moreover, 70 diseased puppies of different breeds and sex aged between 3–6 months were examined in our study. All the admitted dogs were suffering from diarrhea. Clinical examinations of such cases were performed as described by Côté et al. (2015). Fecal swabs placed in peptone water (Oxoid, United Kingdom) were randomly and aseptically obtained from the diseased animals and transmitted directly to the laboratory for bacteriological examination.
2.4 Isolation of Proteus spp. for identification
The swabs were supplemented in peptone water (Oxoid, United Kingdom) for 24 h at 37°C, then enriched broth was passed on XLD (xylose lysine deoxycholate) agar, 5% sheep blood agar, MacConkey agar, and TSI agar (triple sugar iron) (Oxoid, United Kingdom) by using a sterile bacterial loop, and then incubated at 37°C for 24–48 h. Bacterial identification was performed based on the culture characteristics, swarming activity, hemolytic activity, morphological characteristics with Gram’s staining, and suspicious colonies of bacteria were biochemically identified by catalase, H2S generation, urease, methyl red, citrate utilization, oxidase, and lactose fermentation. As previously stated, the indole test was applied to differentiate between P. mirabilis and P. vulgaris (Quinn et al., 2002).
2.4.1 Susceptibility of the antimicrobial agents
Antimicrobial susceptibility for Proteus spp. isolates was performed using the Mueller–Hinton agar (Oxoid, United Kingdom) involved in the disc diffusion method, in which 11 (n = 11) antimicrobial agents were used: amoxicillin (AMX; 10 μg), penicillin (P; 10 U), amoxicillin–clavulanic acid (AMC; 30 μg), ceftazidime (CAZ; 30 μg), cefotaxime (CTX; 30 μg), aztreonam (ATM; 30 μg), norfloxacin (NOR; 10 μg), nalidixic acid (NA; 30 μg), gentamicin (gen; 10 μg), tetracycline (TE; 10 μg), and sulfamethoxazole–trimethoprim (SXT; 25 μg) (Oxoid, United Kingdom). E. coli-ATCC 35218 was used as a quality control strain. The isolates were tested for ESBLs using cephalosporin indicators CAZ (30 μg) and CTX (30 μg) (Oxoid, United Kingdom). The diameter of inhibition zones was calculated as < 22 mm and < 27 mm, respectively (CLSI, 2017). The phenotype resistance patterns were classified as pan drug-resistant (PDR) and are resistant to all used antimicrobial agents; extensive drug-resistant (XDR) species is resistant to one agent in all but two antimicrobial classes, multidrug-resistant (MDR) species is resistant to one agent in three antimicrobial classes or drug-resistant (DR) species is resistant to less than one agent in three antimicrobial classes as stated by Magiorakos et al. (2012). The multiple antibiotic resistance index (MARI) was calculated following this mathematical equation: MARI = c/d. The “c’’ indicates the sum of the antimicrobial agents against which the bacteria exhibited resistance, while “d’’ indicates the total of those agents used.
2.4.2 DNA extraction
Bacterial genomic DNA was extracted from the retrieved Proteus spp. isolates using the QIAamp DNA Mini Kit (QIAGEN Sciences Inc., Germantown, MD, United States/Cat. No. ID 51326) according to the manufacturer’s instruction manual.
2.4.3 Virulence-determinant, extended-spectrum β-lactamases, and non-β-lactam encoding gene detection
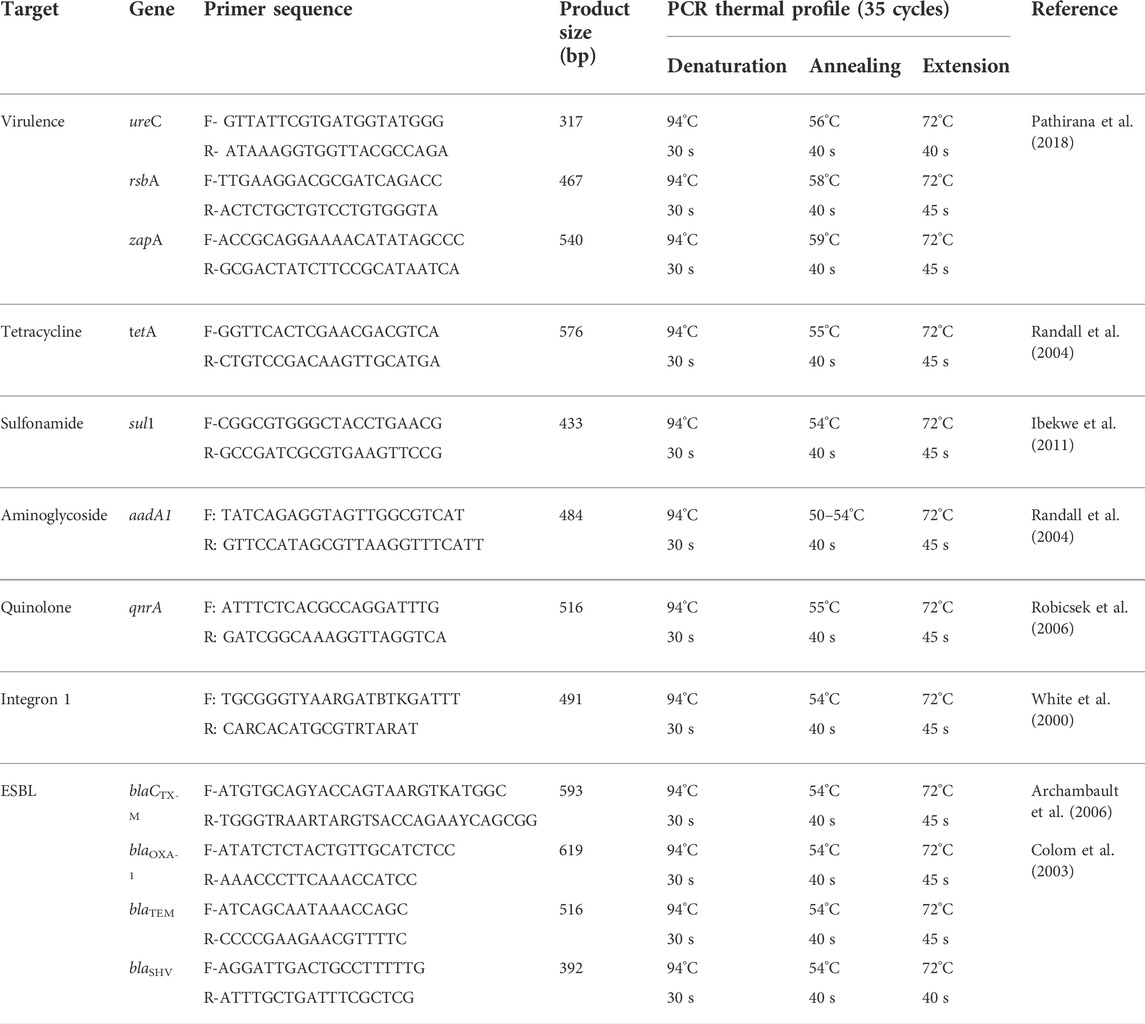
Moreover, 25 positive isolates of DNA extracts were used for virulence-determinant genes by using the PCR targeting (ureC, zapA, and rsbA) genes, as previously conducted (Pathirana et al., 2018); ESBL genes (blaTEM, blaCTX-M, blaSHV, and blaOXA-1), as previously mentioned (Colom et al., 2003; Sharma et al., 2009); and non-β-lactam encoding genes for sulfonamides (sul1), tetracyclines (tetA), aminoglycosides (aadA1), quinolones (qnrA), and integrons (intI1), as previously illustrated (White et al., 2000; Randall et al., 2004; Robicsek et al., 2006; Ibekwe et al., 2011). All genes’ detection of PCR experiments in this study were carried out with a 50 μl total reaction volume as 5 μl of PCR buffer (10×), 1 μl 200 μM (from each dNTP of 10 mM dNTP mix), 4 μl of the bacterial template, 0.4 μl Taq DNA polymerase (5 U/L), 30 pmol per primer, and H2O was added up to 0.05 ml. The PCR cycling conditions and oligonucleotide primers (Thermo Fisher Scientific, United States) are presented in Table 1. Positive control strains, provided by the Animal Health Research Institute (AHRI), Dokki, Egypt, and the negative template control (NTC) were used in all assays. The amplified PCR products were visualized against a 100-pb DNA marker by agar gel electrophoresis on 1.5% agarose containing ethidium bromide 0.5 g/ml.
2.4.4 Statistical data analyses
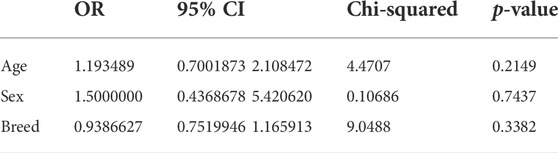
Data were analyzed by the chi-squared test according to Feldman et al. (2008) by R software (version 4.0.2, www.r-project.org), and the differences between groups were considered significant at p < 0.05. Visualization of Proteus isolates according to the virulence and resistance genes with a heatmap supported by hierarchical clustering (dendrogram) was performed according to Kolde (2019), and cor and ggcorrplot packages were used to identify the correlation among the various antimicrobial agents. Fisher’s exact test and odds ratio [confidence interval (CI) = 0.95%] were determined among sex, age, and breed of examined dogs.
3 Results
3.1 Clinical manifestation presentation in dogs
All examined dogs showed an increased body temperature higher than or equal to 39.5°C, lethargy, anorexia, depression, dehydration, and foul-smelling watery bloody diarrhea. The record increase in body temperature progressed to subnormal later in the late stage of disease; also, the intensity of dehydration increased in parallel to diarrhea.
3.2 Phenotype and prevalence of the Proteus spp. in examined samples
Moreover, 25 Proteus spp. isolates were isolated from 70 fecal swabs obtained from sick dogs exhibiting diarrhea, depending on the phenotypic characteristics of colonies, microscopic morphology, and biochemical markers. On XLD, the colonies were red with a black center but appeared pale (non-lactose fermenter) on MacConkey agar, black on TSI (H2S producer), and hemolytic on the blood agar with clearly detected swarming activity. The biochemical tests of those isolates were positive for catalase, H2S generation, urease, methyl red, and citrate utilization. In contrast, a negative response to oxidase, lactose fermentation, and indole was detected, but the Voges–Proskauer tests were positive for Proteus spp. The indole test was used to differentiate between P. vulgaris and P. mirabilis as it was positive for P. vulgaris and negative for P. mirabilis. The overall prevalence of Proteus spp. was 35.7% (25/70) as 92% (23/25) vs. 8% (2/25) for P. mirabilis and P. vulgaris, respectively. In addition, 25.7% of the examined diseased animals (n = 18) are infected with other bacterial pathogens, including, E. coli (13/70, 18.6%), Klebsiella pneumoniae (4/70, 5.7%), and Pseudomonas aeruginosa (1/70, 1.4%).
3.3 Risk factors for Proteus spp. prevalence in dogs
Prevalence of Proteus spp. varied with sex, age, and breed of examined dogs. It was higher in males, 64% (16/25) than in females 36% (9/25). It showed 60% (15/25) in dogs less than 12 weeks old rather than those of 4-, 5-, and 6 months old; 16% (4/25), 4% (1/25), and 20% (5/25), respectively. Concerning breeds, the prevalence rates showed 32% (8/25) in German Shepherd, 20% (5/25) in Rottweiler, 12% (3/25) in both Golden Retriever and 12% (3/25) Cane Corso, 8% (2/25) in each of Husky, 8% (2/25) Griffon, and 8% (2/25) Pitbull. Statistically, no significant difference was detected among the retrieved isolates according to age, gender, and breed, as exhibited in Table 2 and Figure 1.
3.4 Antibiotic-resistant phenotypes of retrieved strains
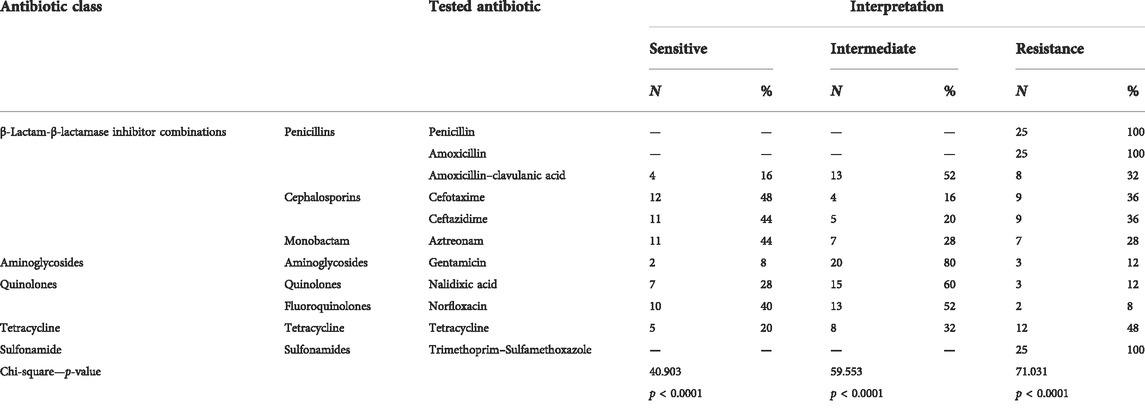
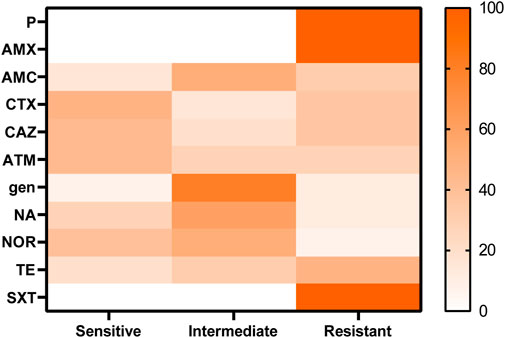
Susceptibility of the antimicrobial agents of the recovered Proteus spp. isolates revealed resistance to penicillins: penicillin and amoxicillin (100%), amoxicillin and clavulanic acid (32%); cephalosporins: cefotaxime and ceftazidime (36%); and monobactam: aztreonam (28%) as extended-spectrum β-lactamases, tetracycline (32%), and sulfonamides: trimethoprim sulfamethoxazole (100%), while both gentamicin and nalidixic acid showed low resistance (12%), as shown in Table 3; Figure 2. A significant difference was detected in the resistance of recovered isolates to the antibiotics tested (p < 0.05).
3.5 Antimicrobial agents’ correlation
Positive correlations were detected between amoxicillin, penicillin, and trimethoprim–sulfamethoxazole, between cefotaxime and ceftazidime, and also between cefotaxime, ceftazidime, and amoxicillin and clavulanic acid that confirm the relation between ESBL agents and finally between nalidixic acid, norfloxacin, and gentamicin, as illustrated in Figure 3.
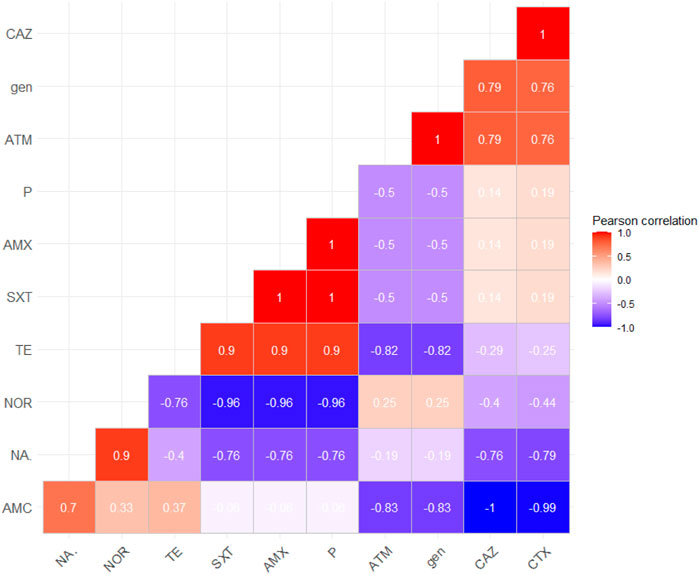
FIGURE 3. Correlation between antimicrobial agents; the red color detects the positive correlation, the darker red denotes a stronger positive correlation coefficient (0.5:1), the blue color detects negative correlation, and the darker blue detects a stronger negative correlation coefficient (−0.5:−1). AMX, amoxicillin; P, penicillin; AMC, amoxicillin–clavulanic acid; CAZ, ceftazidime; CTX, cefotaxime; ATM, aztreonam; NOR, norfloxacin; NA, nalidixic acid; gen, gentamicin; TE, tetracycline; SXT, sulfamethoxazole–trimethoprim.
3.6 Distribution of the virulence-determinant factors among the examined isolates
PCR confirmed that those examined strains harbored three virulence genes ureC, zapA, and rsbA with variable prevalence as 92% (23/25), 60% (15/25), and 52% (13/25), respectively (Table 4; Figure 4). No significant difference was detected in prevalence of the virulence-determinant genes among the different Proteus spp. (p < 0.05).
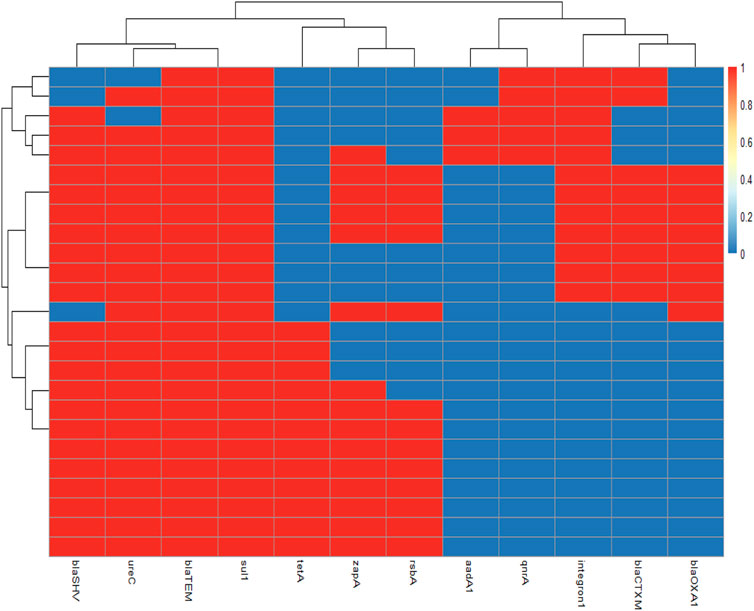
FIGURE 4. Overall distribution of virulence genes and antibiotic resistance profiles in Proteus isolates is represented by a heatmap supported by hierarchical clustering (dendrogram). The red color shows the presence of a gene, whereas the blue color shows the absence of a gene.
3.7 Detection of potential extended-spectrum β-lactamase genes
Phenotypically, ESBLs showed 32% (9/25) of the recovered isolates. However, the genes of antimicrobial resistance, blaTEM, blaSHV, blaCTX-M, and blaOXA-1, as detected in the isolates by PCR with specific predicted bands, were successfully amplified, proving that the examined strains contained ESBL genes with a dramatic variable prevalence as 100% (25/25), 92% (22/25), 36% (9/25), and 32% (8/25), respectively (Table 5; Figure 4). Significant differences among the ESBL gene prevalence of the recovered strains were detected (p < 0.05).
3.8 Detection of integron class 1 and non-extended-spectrum β-lactamase genes
The integron (intI1) was detected at a prevalence of 44% (11/25), and the sulfonamide antimicrobial resistance gene (sul1) detected in Proteus spp. isolates showed a prevalence of 100% (25/25), while the tetracycline resistance gene (tetA) was detected at a prevalence of 48% (12/25), the quinolone resistance gene (qnrA) was detected at a prevalence of 20% (5/25), and the aminoglycoside resistance gene (aadA1) was detected in a prevalence of 12% (3/25) (Table 5; Figure 4). A significant difference was recorded in the sulfonamide or tetracycline gene prevalence in the recovered strains.
3.9 The relation between the phenotypic- and genotypic-resistant patterns among Proteus spp. isolates
The present findings found that 28% (7/25) of recovered isolates were multidrug-resistant to five antimicrobial families, penicillins: amoxicillin and ampicillin, cephalosporins: cefotaxime and ceftazidime, β-Lactam-β-lactamase inhibitor combination: amoxicillin–clavulanic acid, sulfonamides: trimethoprim–sulfamethoxazole, and tetracyclines: tetracycline, in response to the following resistance genes blaTEM, blaSHV, blaOXA-1, blaCTX-M, and sul1. In addition, 48% (12/25) of those isolates were resistant to three antimicrobial families; penicillins: amoxicillin and ampicillin, sulfonamides: trimethoprim–sulfamethoxazole, and also tetracyclines: doxycycline, in response to blaTEM, blaSHV, tetA, and sul1 genes. Furthermore, 12% (3/25) of the tested strains revealed MDR to four antimicrobial families aminoglycoside: gentamicin, penicillins: amoxicillin and ampicillin, quinolones: nalidixic acid, and sulfonamides: trimethoprim–sulfamethoxazole, with the resistant genes such as blaTEM, blaSHV, and sul1 (Table 6; Figure 4). Multiple antibiotic resistance index (MARI) values in this study (≥0.2) exhibited resistance patterns of recovered isolates obtained from a high-risk contamination level (Table 6).
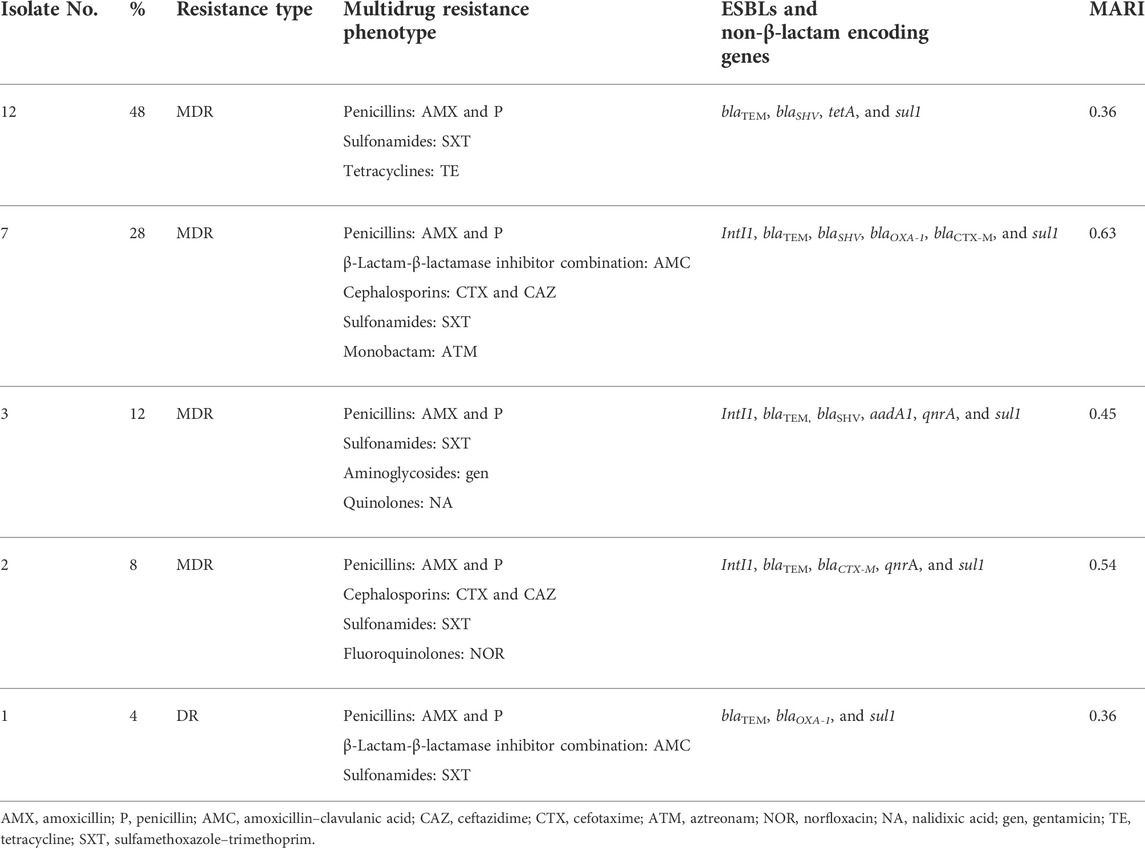
TABLE 6. Distribution of phenotypic resistance antibiotics and antimicrobial resistance genes among the examined isolates.
4 Discussion
Studies in human medicine investigated the microbial activities of Proteus (Chen et al., 2012; Decôme et al., 2020) but still need to be unveiled in veterinary medicine (Wong et al., 2015). The present study is a novel investigation of prevalence, PCR-based virulence, antimicrobial drug resistance, and ESBL genes contributing to pathogenicity of the Proteus spp. isolated from canine fecal samples in Egypt. Few studies are concerning the emergency of the Proteus spp. in canines. In this study, the overall prevalence of Proteus spp. in feces of clinically diseased dogs was 37.5%. Two species were identified, with the most prevalent species being P. mirabilis, followed by P. vulgaris. This is less than the 13.6% infection rate of Proteus spp. in dogs with urinary tract diseases in Thailand (Amphaiphan et al., 2021). In another study, P. mirabilis was detected in 22.7% of dogs’ urinary samples in the United Kingdom (Fonseca et al., 2021). In addition, domestic dogs in Nigeria had a 28% prevalence of Proteus spp. in their feces (Obioma et al., 2020). The phenotypic attributes of recovered Proteus spp. strains were unambiguous and revealed a high degree of agreement between them: red-colored and centrally black colonies on the XLD agar, pale colonies (non-lactose fermenter) on the MacConkey agar, black colonies on TSI agar, and characteristic swarming activity of the colonies (characteristics for Proteus spp.). Catalase, H2S, urease, methyl red, and citrate utilization are all positive for the recovered isolates. However, oxidase, lactose fermentation, Voges–Proskauer, and indole tests are all negative, except for P. vulgaris, which is positive for indole. Our results agreed with findings reported by Reich et al. (2013) and Lei et al. (2016). The conventional biochemical tests are necessary for the differentiation of Proteus and Morganella, and some tests are used for differentiation between them as swarming is characteristic for Proteus spp. and also H2S (O’Hara et al., 2000).
According to our findings, the recovered isolates showed resistance to penicillins, cephalosporins, monobactams (extended-spectrum β-lactamases), sulfonamides, and tetracycline, while both gentamicin and nalidixic acid showed low resistance. MARI values were ≥ 0.2, so it exhibited multiple resistance patterns, denoting that the recovered isolates were taken from high-risk contamination and that P. mirabilis from dogs’ isolates has high MARI (Zhang et al., 2018). Small animal bacterial infections were frequently treated with broad-spectrum antimicrobials such as penicillins and tetracyclines. The antimicrobials’ misuse in small animals and the ability of P. mirabilis to acquire antimicrobial-resistant genes from other pathogens are the main causes of MDR strains (Algammal et al., 2021b).
Unfortunately, antimicrobial resistance was more common in P. mirabilis from dogs compared to those in human isolates (Siebor and Neuwirth, 2013). Prevalence of ampicillin resistance increased than predicted (71%) (Harada et al., 2014), which was most likely caused by TEM penicillinases (Hordijk et al., 2013). Resistance to trimethoprim/sulfamethoxazole, quinolones, and aminoglycosides to P. mirabilis restricts the therapeutic options of such antimicrobials used in the treatment of dogs infected with P. mirabilis (Wong et al., 2015).
The PCR proved that the recovered Proteus spp. strains from dogs are highly virulent as they harbored ureC, zapA, and rsbA virulence genes exhibiting prevalence as 92, 60, and 52%, respectively. The ureC gene causes urine pH to rise, resulting in the formation of stones (Armbruster et al., 2018). The Proteus spp. demonstrated in our study had 92% ureC amplification, indicating a higher frequency of ureC than other genes, thus playing a pivotal role in the virulence of Proteus depending on this gene. The ureC gene was identified in approximately 96.6% of human-infected urinary tract isolates (Ali and Yousif, 2015). In previous studies from ducks, the ureC gene was found in 100% of Proteus isolates (Algammal et al., 2021b). In addition, Proteus spp. lacked rsbA gene which encodes the characteristic swarming activity of Proteus and expresses a membrane sensor for promoting the extracellular polysaccharides (Walker et al., 1999; Algammal et al., 2021a). The current report showed that 15 isolates (60%) had the zapA gene, which codes for protease enzyme production, and ZapA-protease could degrade IgG, IgA1, and IgA2, controlled by the zapA gene (Pathirana et al., 2018; Algammal et al., 2021b).
When ESBLs are present, infectious bacteria develop resistance to lactamase, demonstrating the negative effect of the variant therapeutics (Schultz et al., 2017). ESBL-positive and -negative isolates showed different AMR rates. The mechanism of increased non-β-lactam resistance in ESBL-producing strains is unknown. ESBLs hydrolyze broad-spectrum lactam antibiotics, such as penicillins and cephalosporins in addition to piperacillin. Enterobacteriales frequently produce ESBLs. According to our results, all ESBL-positive isolates had blaTEM gene. P. mirabilis, identified in the hospitalized patients’ samples, frequently contained ESBLs of the TEM type (Ahn et al., 2017; Rajivgandhi et al., 2018). The blaCTX-M gene also exhibits resistance to cephalosporins, ceftazidime, and cefotaxime. The blaSHV gene is commonly detected in members of the Enterobacteriaceae family (Alonso et al., 2017), which confirms our findings. Furthermore, according to our findings, the OXA-1-positive isolates showed a significantly higher prevalence than that stated in a French study (Bonnet et al., 2002). The blaOXA-1 gene promotes piperacillin and cephalosporin resistance. The blaOXA-1 and blaCTX-M genes work together protecting P. mirabilis from lactam-lactamase inhibitor combinations (Schultz et al., 2017). Concurrently, Proteus is also resistant to sulfonamides and tetracyclines due to sul1 and tetA genes (Cadena et al., 2018; Algammal et al., 2021a). Class 1 integrons were connected with ESBL-producing isolates more frequently than non-ESBL-producing isolates (Chen et al., 2017). Some researchers suggest that the ESBL genes and integron are frequently connected with the qnr gene (Hopkins et al., 2007; Sharma et al., 2009). When bacteria are subjected to selective pressure by sulfonamides, the presence of sul1 genes with class 1 integrons provides a useful tool for the maintenance and subsequent development of resistance to other antimicrobial agents (Antunes et al., 2005).
Animal companions could be as reservoirs of Proteus, especially P. mirabilis, for human infection according to previous research that found human and animals infections with closely-related bacterium strains (Marques et al., 2019). P. mirabilis showed high resistance levels against the antimicrobial agents, making it a potential health threatening factor to human health due to its high elimination frequency through diarrhea in dogs.
5 Limitations and future recommendations
Future work is recommended to perform appropriate identification by molecular techniques like MALDI-TOF/16S rRNA gene sequencing to realize the clonal relatedness of the obtained strains.
6 Conclusion
Eventually, this report study could be the first study of MDR in Proteus spp. isolated from Egyptian dogs. P. mirabilis was a highly recorded prevalence in dogs, which was associated with diarrhea. The ureC, zapA, and rsbA virulence genes are commonly detected in the Proteus isolates obtained from infected dogs. The isolated Proteus strains showed multidrug resistance (MDR) property against penicillins, β-lactamases, sulfonamides, cephalosporins, quinolones, aminoglycosides, and tetracyclines. They often carried the blaOXA-1, blaTEM, blaCTX-M, blaSHV, intI1, tetA, aadA1, qnrA, and sul1 resistance genes. The potential hazards and health risks of the Proteus infection in dogs need more investigation. Therefore, we encourage continuous epidemiological data collection with the antimicrobial testing of susceptibility in both humans and animals and testing variant antibiotics used in human or veterinary medicinal fields, either.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the Animal Ethics Review Committee of Suez Canal University (AERC-SCU) in Egypt.
Author contributions
RE-T and AM: conceptualization and study design. RE-T, AM, and EA: conducted the experiments. TE, AM, and RE-T: drafted the manuscript. RE-T, AHA, and AEA: carried out the statistical analysis and investigation. NA, DN, SKA, AM, AHA, and EME: data validation, data accuracy, and supervision. AM and TE: performed the clinical examination of diseased cases and sampling. RE-T, AHA, AM, SKA, and TE: wrote the original draft. EA, DN, SKA, and NA: critically revised the manuscript. All authors have revised and approved the final manuscript.
Acknowledgments
The authors express their appreciation to Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R153), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. Also, the authors extend their appreciation to the deanship of Scientific Research at King Khalid University, Abha KSA for supporting this work under grant number (R.G.P 1-100/43). We also acknowledge Animal Health Research Institute (AHRI), Dokki, Egypt, provides positive control strains used in the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RE declared a shared affiliation with the authors DN and TE to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahn, J. Y., Ann, H. W., Jeon, Y., Ahn, M. Y., Oh, D. H., Kim, Y. C., et al. (2017). The impact of production of extended-spectrum β-lactamases on the 28-day mortality rate of patients with Proteus mirabilis bacteremia in Korea. BMC Infect. Dis. 17 (1), 327–410. doi:10.1186/s12879-017-2431-8
Algammal, A. M., Enany, M. E., El-Tarabili, R. M., Ghobashy, M. O. I., and Helmy, Y. A. (2020). Prevalence, antimicrobial resistance profiles, virulence and enterotoxins-determinant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens 9 (5), E362. doi:10.3390/pathogens9050362
Algammal, A. M., Hashem, H. R., Al-otaibi, A. S., Alfifi, K. J., El-dawody, E. M., Mahrous, E., et al. (2021a). Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol. 21 (1), 237. doi:10.1186/s12866-021-02287-y
Algammal, A. M., Hashem, H. R., Alfifi, K. J., Hetta, H. F., Sheraba, N. S., Ramadan, H., et al. (2021b). atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 11 (1), 9476. doi:10.1038/s41598-021-88861-w
Ali, H. H., and Yousif, M. G. (2015). Detection of some virulence factors genes of Proteus mirablis that. Int. J. Adv. Res. 3 (1), 156
Alonso, C. A., Michael, G. B., Li, J., Somalo, S., Simón, C., Wang, Y., et al. (2017). Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J. Antimicrob. Chemother. 72 (6), 1589–1596. doi:10.1093/JAC/DKX024
Amphaiphan, C., Yano, T., Som-in, M., Kungwong, P., Wongsawan, K., Pusoonthornthum, R., et al. (2021). Antimicrobial drug resistance profile of isolated bacteria in dogs and cats with urologic problems at Chiang Mai University Veterinary Teaching Hospital, Thailand (2012–2016). Zoonoses Public Health 68 (5), 452–463. doi:10.1111/zph.12832
Antunes, P., Machado, J., Sousa, J. C., and Peixe, L. (2005). Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49 (2), 836–839. doi:10.1128/aac.49.2.836-839.2005
Archambault, M., Petrov, P., Hendriksen, R. S., Asseva, G., Bangtrakulnonth, A., Hasman, H., et al. (2006). Molecular characterization and occurrence of extended-spectrum β-lactamase resistance genes among Salmonella enterica serovar corvallis from Thailand, Bulgaria, and Denmark. Drug Resist. 12, 192–198. doi:10.1089/mdr.2006.12.192
Armbruster, C. E., Mobley, H. L. T., and Pearson, M. M. (2018). Pathogenesis of Proteus mirabilis infection. EcoSal Plus 8 (1). doi:10.1128/ecosalplus.ESP-0009-2017
Bonnet, R., Marchandin, H., Chanal, C., Sirot, D., Labia, R., De Champs, C., et al. (2002). Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46 (6), 2004–2006. doi:10.1128/AAC.46.6.2004-2006.2002
Cadena, M., Durso, L. M., Miller, D. N., Waldrip, H. M., Castleberry, B. L., Drijber, R. A., et al. (2018). Tetracycline and sulfonamide antibiotic resistance genes in soils from Nebraska organic farming operations, doi:10.3389/fmicb.2018.01283
Chen, C. M., Lai, C. H., Wu, H. J., and Wu, L. T. (2017). Genetic characteristic of class 1 integrons in proteus mirabilis isolates from urine samples. Biomed. (Taipei) 7 (2), 9. doi:10.1051/bmdcn/2017070202
Chen, C. Y., Chen, Y. H., Lu, P. L., Lin, W. R., Chen, T. C., and Lin, C. Y. (2012). Proteus mirabilis urinary tract infection and bacteremia: Risk factors, clinical presentation, and outcomes. J. Microbiol. Immunol. Infect. = Wei mian yu gan ran za zhi 45 (3), 228–236. doi:10.1016/J.JMII.2011.11.007
CLSI (2017). Clinical and laboratory standards Institute : Performance standards for antimicrobial susceptibility testing.” in 27th ed 424 informational supplement. CLSI
Colom, K., Perez, J., Alonso, R., Fernandez-Aranguiz, A., Larino, E., and Cisterna, R. (2003). Simple and reliable multiplex PCR assay for detection of blaTEM, bla(SHV) and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 223 (2), 147–151. doi:10.1016/S0378-1097(03)00306-9
Côté, E., Edwards, N. J., Ettinger, S. J., Fuentes, V. L., MacDonald, K. A., Scansen, B. A., et al. (2015). Management of incidentally detected heart murmurs in dogs and cats. J. Am. Vet. Med. Assoc. 246 (10), 1076–1088. doi:10.2460/javma.246.10.1076
Crump, J. A., Barrett, T. J., Nelson, J. T., and Angulo, F. J. (2003). Reevaluating fluoroquinolone breakpoints for Salmonella enterica serotype Typhi and for non-Typhi salmonellae. Clin. Infect. Dis. 37 (1), 75–81. doi:10.1086/375602
Decôme, M., Cuq, B., Fairbrother, J. H., Gatel, L., and Conversy, B. (2020). Clinical significance of proteus mirabilis bacteriuria in dogs, risk factors and antimicrobial susceptibility. Can. J. Veterinary Res. 84 (4), 252
Drzewiecka, D. (2016). Significance and roles of Proteus spp. bacteria in natural environments. Microb. Ecol. 72 (4), 741–758. doi:10.1007/S00248-015-0720-6
Fadare, F. T., and Okoh, A. I. (2021). Distribution and molecular characterization of ESBL, pAmpC beta-lactamases, and non-beta-lactam encoding genes in Enterobacteriaceae isolated from hospital wastewater in Eastern Cape Province, South Africa. PLoS One 16 (7), e0254753. doi:10.1371/journal.pone.0254753
Feldman, I., Rzhetsky, A., and Vitkup, D. (2008). Network properties of genes harboring inherited disease mutations. Proc. Natl. Acad. Sci. U. S. A. 105 (11), 4323–4328. doi:10.1073/pnas.0701722105
Fonseca, J. D., Mavrides, D. E., Graham, P. A., and McHugh, T. D. (2021). Results of urinary bacterial cultures and antibiotic susceptibility testing of dogs and cats in the UK. J. Small Anim. Pract. 62, 1085–1091. doi:10.1111/JSAP.13406
Gong, Z., Shi, X., Bai, F., He, X., Zhang, H., Li, Y., et al. (2019). Characterization of a novel diarrheagenic strain of Proteus mirabilis associated with food poisoning in China. Front. Microbiol. 10, 2810. doi:10.3389/fmicb.2019.02810
Guardabassi, L., Schwarz, S., and Lloyd, D. H. (2004). Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54 (2), 321–332. doi:10.1093/JAC/DKH332
Guentzel, M. N. (1996). Escherichia, klebsiella, enterobacter, serratia, sitrobacter, and proteus. Editors S. Baron. Med. Microbiol.. 4th Edn. Galveston (TX): University of Texas Medical Branch at Galveston Chapter 26. Available at: https://www.ncbi.nlm.nih.gov/books/NBK8035/
Guillaume, G., Verbrugge, D., Chasseur-Libotte, M.-L., Moens, W., and Collard, J.-M. (2000). PCR typing of tetracycline resistance determinants (Tet A-E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 32 (1), 77–85. doi:10.1111/J.1574-6941.2000.TB00701.X
Harada, K., Niina, A., Shimizu, T., Mukai, Y., Kuwajima, K., Miyamoto, T., et al. (2014). Phenotypic and molecular characterization of antimicrobial resistance in Proteus mirabilis isolates from dogs. J. Med. Microbiol. 63, 1561–1567. doi:10.1099/JMM.0.081539-0
Hegazy, W. A. H. (2016). Diclofenac inhibits virulence of Proteus mirabilis isolated from diabetic foot ulcer. Afr. J. Microbiol. Res. 10 (21), 733–743. doi:10.5897/AJMR2016.8043
Hola, V., Peroutkova, T., and Ruzicka, F. (2012). Virulence factors in Proteus bacteria from biofilm communities of catheter-associated urinary tract infections. FEMS Immunol. Med. Microbiol. 65 (2), 343–349. doi:10.1111/J.1574-695X.2012.00976.X
Hopkins, K. L., Wootton, L., Day, M. R., and Threlfall, E. J. (2007). Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J. Antimicrob. Chemother. 59 (6), 1071–1075. doi:10.1093/jac/dkm081
Hordijk, J., Schoormans, A., Kwakernaak, M., Duim, B., Broens, E., Dierikx, C., et al. (2013). High prevalence of fecal carriage of extended-spectrum β-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 4 (AUG), 242. doi:10.3389/fmicb.2013.00242
Ibekwe, A. M., Murinda, S. E., and Graves, A. K. (2011). Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS One 6 (6), e20819. doi:10.1371/journal.pone.0020819
Jacobsen, S. M., and Shirtliff, M. E. (2011). Proteus mirabilis biofilms and catheter-associated urinary tract infections. Virulence 2 (5), 460–465. doi:10.4161/VIRU.2.5.17783
Lei, C.-W., Zhang, A.-Y., Wang, H.-N., Liu, B.-H., Yang, L.-Q., and Yang, Y.-Q. (2016). Characterization of SXT/R391 integrative and conjugative elements in Proteus mirabilis isolates from food-producing animals in China. Antimicrob. Agents Chemother., 60(3), 1935–1938. doi:10.1128/AAC.02852-15
Li, X., and Mobley, H. L. T. (2002). Vaccines for Proteus mirabilis in urinary tract infection. Int. J. Antimicrob. Agents 19 (6), 461–465. doi:10.1016/S0924-8579(02)00102-4
Lloyd, D. H. (2007). Reservoirs of antimicrobial resistance in pet animals. Clin. Infect. Dis. 45, S148–S152. doi:10.1086/519254
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 (3), 268–281. doi:10.1111/j.1469-0691.2011.03570.x
Manos, J., and Belas, R. (2006). The genera Proteus, providencia, and Morganella. Undefined, 245–269. doi:10.1007/0-387-30746-X_12
Marques, C., Belas, A., Aboim, C., Trigueiro, G., Cavaco-Silva, P., Gama, L. T., et al. (2019). Clonal relatedness of Proteus mirabilis strains causing urinary tract infections in companion animals and humans. Vet. Microbiol. 228, 77–82. doi:10.1016/J.VETMIC.2018.10.015
O'Hara, C. M., Brenner, F. W., and Miller, J. M. (2000). Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin. Microbiol. Rev. 13 (4), 534–546. doi:10.1128/CMR.13.4.534
Obioma, A., Nnenna, I., and Christian, A. (2020). Antibiogram pattern of potential pathogenic bacteria associated with domestic dog faecal matter in port harcourt metropolis. J. Biomed. Res. Environ. Sci. 1 (2), 039–047. doi:10.37871/jels1118
Oppegaard, H., Steinum, T. M., and Wasteson, Y. (2001). Horizontal transfer of a multi-drug resistance plasmid between coliform bacteria of human and bovine origin in a farm environment. Appl. Environ. Microbiol. 67 (8), 3732–3734. doi:10.1128/AEM.67.8.3732-3734.2001
Pathirana, H. N. K. S., De Silva, B. C. J., Wimalasena, S. H. M. P., Hossain, S., and Heo, G. J. (2018). Comparison of virulence genes in proteus species isolated from human and pet turtle. Iran. J. Vet. Res. 19 (1), 48–52. doi:10.22099/ijvr.2018.4768
Perreten, V., and Boerlin, P. (2003). A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47 (3), 1169–1172. doi:10.1128/AAC.47.3.1169-1172.2003
Quinn, P. J., Markey, B. K., Carter, M. E., Donnelly, W. J. C., and Leonard, F. C. (2002). Veterinary microbiology and microbial disease. Blackwell science.
Rajivgandhi, G., Maruthupandy, M., and Manoharan, N. (2018). Detection of TEM and CTX-M genes from ciprofloxacin resistant Proteus mirabilis and Escherichia coli isolated on urinary tract infections (UTIs). Microb. Pathog. 121, 123–130. doi:10.1016/J.MICPATH.2018.05.024
Randall, L. P., Cooles, S. W., Osborn, M. K., Piddock, L. J., and Woodward, M. J. (2004). Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 53 (2), 208–216. doi:10.1093/jac/dkh070
Rather, P. N. (2005). Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7 (8), 1065–1073. doi:10.1111/J.1462-2920.2005.00806.X
Rawat, D., and Nair, D. (2010). Extended-spectrum β-lactamases in Gram negative bacteria. J. Glob. Infect. Dis. 2 (3), 263–274. doi:10.4103/0974-777x.68531
Reich, F., Atanassova, V., and Klein, G. (2013). Extended-spectrum β-lactamase– and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg. Infect. Dis. 19 (8), 1253–1259. doi:10.3201/EID1908.120879
Robicsek, A., Jacoby, G. A., and Hooper, D. C. (2006). The worldwide emergence of plasmid-mediated quinolone resistance. Lancet. Infect. Dis. 6 (10), 629–640. doi:10.1016/S1473-3099(06)70599-0
Rózalski, A., Sidrocyzx, Z., Kotelko, K., and RozAlski, A. (1997). Potential virulence factors of Proteus bacilli. Microbiol. Mol. Biol. Rev. 61 (1), 65–89. doi:10.1128/MMBR.61.1.65-89.1997
Rubin, J. E., and Pitout, J. D. D. (2014). Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet. Microbiol. 170 (1-2), 10–18. doi:10.1016/J.VETMIC.2014.01.017
Sánchez-Osuna, M., Cortés, P., Barbé, J., and Erill, I. (2019). Origin of the mobile di-hydro-pteroate synthase gene determining sulfonamide resistance in clinical isolates. Front. Microbiol. 9, 3332. doi:10.3389/fmicb.2018.03332
Schultz, E., Cloeckaert, A., Doublet, B., Madec, J. Y., and Haenni, M. (2017). Detection of SGI1/PGI1 elements and resistance to extended-spectrum cephalosporins in proteae of animal origin in France. Front. Microbiol. 8 (JAN), 32. doi:10.3389/fmicb.2017.00032
Sharma, A., Bora, C. R., Chaurasia, R. K., and Sahu, V. (2009). Antibiotic susceptibility and genetic analysis of Vibrio species isolated from reverine environment. Curr. Res. Bacteriol. 2 (2), 36–49. doi:10.3923/crb.2009.36.49
Siebor, E., and Neuwirth, C. (2013). Emergence of Salmonella genomic island 1 (SGI1) among Proteus mirabilis clinical isolates in Dijon, France. J. Antimicrob. Chemother. 68 (8), 1750–1756. doi:10.1093/JAC/DKT100
Walker, K. E., Moghaddame-Jafari, S., Lockatell, C. V., Johnson, D., and Belas, R. (1999). ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 32 (4), 825–836. doi:10.1046/J.1365-2958.1999.01401.X
White, P. A., McIver, C. J., Deng, Y., and Rawlinson, W. D. (2000). Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182 (2), 265–269. doi:10.1111/j.1574-6968.2000.tb08906.x
White, P. A., and Rawlinson, W. D. (2001). Current status of the aadA and dfr gene cassette families. J. Antimicrob. Chemother. 47 (4), 495–496. doi:10.1093/jac/47.4.495
Wong, C., Epstein, S. E., and Westropp, J. L. (2015). Antimicrobial susceptibility patterns in urinary tract infections in dogs (2010-2013). J. Vet. Intern. Med. 29 (4), 1045–1052. doi:10.1111/JVIM.13571
Yun, M. K., Wu, Y., Li, Z., Zhao, Y., Waddell, M. B., Ferreira, A. M., et al. (2012). Catalysis and sulfa drug resistance in dihydropteroate synthase. Sci. (New York, N.Y.) 335 (6072), 1110–1114. doi:10.1126/SCIENCE.1214641
Zhang, P. L. C., Shen, X., Chalmers, G., Reid-Smith, R. J., Slavic, D., Dick, H., et al. (2018). Prevalence and mechanisms of extended-spectrum cephalosporin resistance in clinical and fecal enterobacteriaceae isolates from dogs in Ontario, Canada. Vet. Microbiol. 213, 82–88. doi:10.1016/J.VETMIC.2017.11.020
Keywords: antimicrobial resistance, virulence genes, MDR, Proteus, dog, diarrhea, prevalence, Egypt
Citation: El-Tarabili RM, Ahmed EM, Alharbi NK, Alharbi MA, AlRokban AH, Naguib D, Alhag SK, El Feky TM, Ahmed AE and Mahmoud AE (2022) Prevalence, antibiotic profile, virulence determinants, ESBLs, and non-β-lactam encoding genes of MDR Proteus spp. isolated from infected dogs. Front. Genet. 13:952689. doi: 10.3389/fgene.2022.952689
Received: 25 May 2022; Accepted: 02 September 2022;
Published: 06 October 2022.
Edited by:
Danila De Vito, University of Bari Medical School, ItalyReviewed by:
Ping Wei, Guangxi University, ChinaRasha Elkenany, Mansoura University, Egypt
Mangesh Vasant Suryavanshi, Lerner Research Institute, United States
Copyright © 2022 El-Tarabili, Ahmed, Alharbi, Alharbi, AlRokban, Naguib, Alhag, El Feky, Ahmed and Mahmoud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reham M. El-Tarabili, cmloYW1fdGFyYWJpbHlAdmV0LnN1ZXouZWR1LmVn; Elsayyad M. Ahmed, ZWxzYXl5YWRAYWhyaS5jb20=
 Reham M. El-Tarabili
Reham M. El-Tarabili Elsayyad M. Ahmed
Elsayyad M. Ahmed Nada K. Alharbi
Nada K. Alharbi Maha A. Alharbi3
Maha A. Alharbi3 Doaa Naguib
Doaa Naguib Ahmed Ezzat Ahmed
Ahmed Ezzat Ahmed