- 1Medical Genetics and Ophthalmic Genomics Unit, National Eye Institute, National Institutes of Health, Bethesda, MD, United States
- 2Bioinformatics Group, National Eye Institute, National Institutes of Health, Bethesda, MD, United States
- 3Section on Epithelial and Retinal Physiology and Disease, National Eye Institute, National Institutes of Health, Bethesda, MD, United States
- 4Pediatric, Developmental and Genetic Ophthalmology, National Eye Institute, National Institutes of Health, Bethesda, MD, United States
The macula and fovea comprise a highly sensitive visual detection tissue that is susceptible to common disease processes like age-related macular degeneration (AMD). Our understanding of the molecular determinants of high acuity vision remains unclear, as few model organisms possess a human-like fovea. We explore transcription factor networks and receptor-ligand interactions to elucidate tissue interactions in the macula and peripheral retina and concomitant changes in the underlying retinal pigment epithelium (RPE)/choroid. Poly-A selected, 100 bp paired-end RNA-sequencing (RNA-seq) was performed across the macular/foveal, perimacular, and temporal peripheral regions of the neural retina and RPE/choroid tissues of four adult Rhesus macaque eyes to characterize region- and tissue-specific gene expression. RNA-seq reads were mapped to both the macaque and human genomes for maximum alignment and analyzed for differential expression and Gene Ontology (GO) enrichment. Comparison of the neural retina and RPE/choroid tissues indicated distinct, contiguously changing gene expression profiles from fovea through perimacula to periphery. Top GO enrichment of differentially expressed genes in the RPE/choroid included cell junction organization and epithelial cell development. Expression of transcriptional regulators and various disease-associated genes show distinct location-specific preference and retina-RPE/choroid tissue-tissue interactions. Regional gene expression changes in the macaque retina and RPE/choroid is greater than that found in previously published transcriptome analysis of the human retina and RPE/choroid. Further, conservation of human macula-specific transcription factor profiles and gene expression in macaque tissues suggest a conservation of programs required for retina and RPE/choroid function and disease susceptibility.
Introduction
The retina is a thin tissue of the posterior eye (range: 166.9 ± 20.9 μm–271.4 ± 19.6 μm (Grover et al., 2010) essential for processing light and transmitting information to the brain through the optic nerve. It is composed of multiple layers, which contain seven cell classes including rod and cone photoreceptors, in apposition with the retinal pigment epithelium (RPE), which is underlaid by the choroid. The choroid is a vascular structure that supports the outer retina, the RPE is necessary for maintaining healthy photoreceptors, and rods mediate dark-adapted vision while cones are responsible for color vision (Purves, 2001; Nickla and Wallman, 2010). The relative composition of cell types varies across the retina, and provides visual function specializations, such as high acuity color vision at the fovea, which is cone photoreceptor rich and rod depleted. Furthermore, the neural retinal layers change in thickness and morphology, including the inner and outer nuclear layers of the photoreceptors, whereas the choroidal layer has increased thickness at the macula but the morphology of the entire layer remains relatively unchanged (Bagci et al., 2008; Mori et al., 2016). Many genes have varied expression in different cell types and tissue layers, with 819 genes implicated in heritable retinal degenerations as of 2022 (RetNet, https://sph.uth.edu/RetNet/) (Daiger, 1998; Holt et al., 2015). While retinal degeneration may initiate in a single tissue layer, eventually the disease progresses across all tissue layers (Di Pierdomenico et al., 2017). Importantly, retinal manifestations in many diseases are region specific. For example, Stargardt disease and Best disease primarily affect the macula and cause loss of central vision first, whereas retinitis pigmentosa initiates in the rod-rich periphery (Walia and Fishman, 2009; MacDonald and Lee, 2013; Jones et al., 2018; Lamin et al., 2019). We hypothesize that this location-specific disease susceptibility of the retina is due to cellular specialization and region-dependent molecular interactions, which we can assess by regional layer-specific gene expression profiling of healthy retinae.
Previous investigations of the transcriptomic landscape of the human retina support a significant differential gene expression between the retina and the RPE/choroid/sclera tissue layers as well as across macular and peripheral regions, as described by Li et al. (2014). Comparison of gene expression between the nasal and temporal peripheries of retina and RPE/choroid/sclera tissues indicated little to no variability, yet macular and peripheral profiles remained distinct in each tissue (Li et al., 2014). Gene expression across the retina has shown to be consistent with the spatial distribution of photoreceptors and ganglion cells, and RPE-specific genes appear to be enriched in the periphery over the macula as outlined by Whitmore et al. (2014). Further characterization of gene expression between foveal and peripheral retina has been conducted using single-cell RNA sequencing in both primate (Macaca fascicularis) by Peng et al. (2019) and human retina by Voigt et al. (2019a). These studies showed variability in both cell type distribution and gene expression between foveal and peripheral retina (Voigt et al., 2019a; Peng et al., 2019). While it has been well established that regional differences exist across tissue layers in the primate retina, the molecular drivers of these differences remain unclear.
Here we generate an RNA-seq dataset from adult Rhesus macaque (Macaca mulatta) eyes that reflect the changing photoreceptor composition of the contiguous macular/foveal, perimacular, and peripheral regions to determine tissue- and location-dependent differential gene expression of the neural retina and RPE/choroid. The use of matched retina and RPE/choroid tissue biopsies in this analysis with an additional perimacular sample provides a contiguous assessment of gene expression across location as opposed to a binary macular vs. periphery comparison and allows for a more comprehensive look at patterns of gene expression and their related pathways and ontologies. We additionally explore conservation of these shared and distinct pathways through a meta-analysis comparing our macaque datasets to previously published human data. Finally, we examine gene regulation via transcription factors and tissue-tissue interactions using ligand/receptor analysis to understand differences in the macular and peripheral retina and the affiliated changes in the RPE/choroid.
Materials and methods
Animal care
All experimental protocols were approved by the National Eye Institute Animal Care and Use Committee. Procedures were performed in accordance with the United States Public Health Service policy on the humane care and use of laboratory animals.
Tissue acquisition
Postmortem eyes were enucleated from two adult Rhesus monkeys (M. mulatta), aged 13 and 18 years. Two-millimeter punch biopsies were removed from macular/foveal, perimacular, temporal, and nasal regions of the retina. Following the biopsy, the tissues were separated into layers of neural retina and RPE/choroid using microdissection techniques. Biopsies were optimized for smallest diameter in other macaque samples in order to obtain the minimal number of cells required for RNA-sequencing. Optimization was performed by measuring total RNA yield in different sized biopsy samples as below and requiring a minimum of 200 ng as input for RNA-seq.
RNA extraction
Total RNA was isolated from fresh tissues using the PicoPure™ RNA Isolation Kit (Applied Biosystems, Waltham, Massachusetts, United States) following mortar and pestle tissue homogenization. Extracted RNA samples were checked for quality using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) and only samples with RIN value > 6.5 were used (Range 1–10).
RNA sequencing
Extracted RNA was prepared into 100 bp libraries for paired-end, poly-A-selected RNA sequencing on the Illumina HiSeq4000 at the NIH Intramural Sequencing Center. The raw sequence data is available at the Gene Expression Omnibus under accession GSE194285.
Bioinformatic analysis
Briefly, reads were quantified against the macaque genome Mmul_8.0.1 using Salmon pseudo-alignment transcript quantification tool (version 0.13.0; Patro et al., 2017) (Patro et al., 2017), imported into R with tximport (Soneson et al., 2015) and analyzed for differential expression using DESeq2, using the Rhesus macaque sample as a covariate (Love et al., 2014). Transcript reads were additionally aligned to the human genome (GRCH38, gencode version 30 (Harrow et al., 2012), to supplement any unquantified genes. To select the gene name for each macaque transcript, we used the HGNC Comparison of Orthology Predictions (Yates, 2016) data, aggregated by the hcop R package (https://github.com/stephenturner/hcop). We counted the number of sources supporting each gene ID to transcript and selected the gene ID with the most support, as detailed in HYPERLINK "https://github.com/davemcg/macaque_macula_RNA-seq/blob/master/analysis/01_QC_prelim_analysis.Rmd" \o "https://github.com/davemcg/macaque_macula_RNA-seq/blob/master/analysis/01_QC_prelim_analysis.Rmd"https://github.com/davemcg/macaque_macula_RNA-seq/blob/master/analysis/01_QC_prelim_analysis.Rmdhttps://github.com/davemcg/macaque_macula_RNA-seq/blob/master/analysis/01_QC_prelim_analysis.Rmd.
Genes were considered to be differentially expressed based on IHW (Ignatiadis et al., 2016) corrected p-value < 0.05. Gene lists were then assigned gene ontologies using the clusterProfiler “enrichGO” function (Yu et al., 2012).
For the heatmap visualizations, we used length-scaled transcripts per million (TPM) quantification from tximport scaled by library size with the edgeR (Robinson et al., 2010) “calcNormFactors” function. The heatmaps were made with the R package ComplexHeatmap (Gu et al., 2016).
Human single cell transcriptome data from the scEiaD resource at plae.nei.nih.gov was searched for machine predicted RPE cells from fovea or peripheral punches. We found 217 cells, 159 from the fovea and 58 from the periphery, across four studies (Voigt et al., 2019b; Cowan et al., 2020; Voigt et al., 2020; Yan et al., 2020; Swamy et al., 2021). The Seurat object was downloaded on 14-01-2022 from plae.nei.nih.gov and subsetted to these 217 cells. The Seurat object was converted to a SingleCellExperiment object to run wilcox and t tests with the scran “findMarkers” test and used the study as a covariate. Genes were kept which had both a wilcox AUC (area under curve) greater than 0.7 (1 is best), an abs (log2) fold change greater than 0.7, and a FDR corrected p-value < 0.01. For the overlap testing we kept genes that had a padj < 0.01 in each comparison (single cell human RPE, bulk human RPE-choroid, or bulk macaque RPE-choroid) and the direction of the fold change between fovea and peripheral was the same. The venn diagram was created with the R package eulerr (https://cran.r-project.org/web/packages/eulerr/citation.html).
For the exact commands used in the analysis of this data, we make our Snakemake (Köster and Rahmann, 2012) reproducible workflow available at https://github.com/davemcg/macaque_macula_RNA-seq.
Results
We performed RNA-seq on 2 mm retina and RPE/choroid tissues of 4 adult Rhesus macaque eyes in each of the central macular (foveal), perimacular, and peripheral regions (Supplementary Figure S1). Tissue dissections and RNA extractions were optimized for minimal input samples (<1 μg) to maximize spatial resolution. Principle component analysis (PCA) visualization indicates that samples group primarily by tissue. Foveal/macular and peripheral samples generally cluster furthest from each other, and perimacular samples cluster in between. Tissue from the nasal retina was also obtained and these samples cluster closest with perimacular samples. However, these were excluded from the analysis, as we were interested contiguous changes from the fovea/macula through the perimacula to the periphery (Supplementary Figure S2). We then assessed overall gene expression by tissue layer and location as well as differential gene expression between foveal/macular samples and other peripheral samples (Table 1).

TABLE 1. A summary of the differential gene expression (padj < 0.05) in retina and RPE/choroid by location.
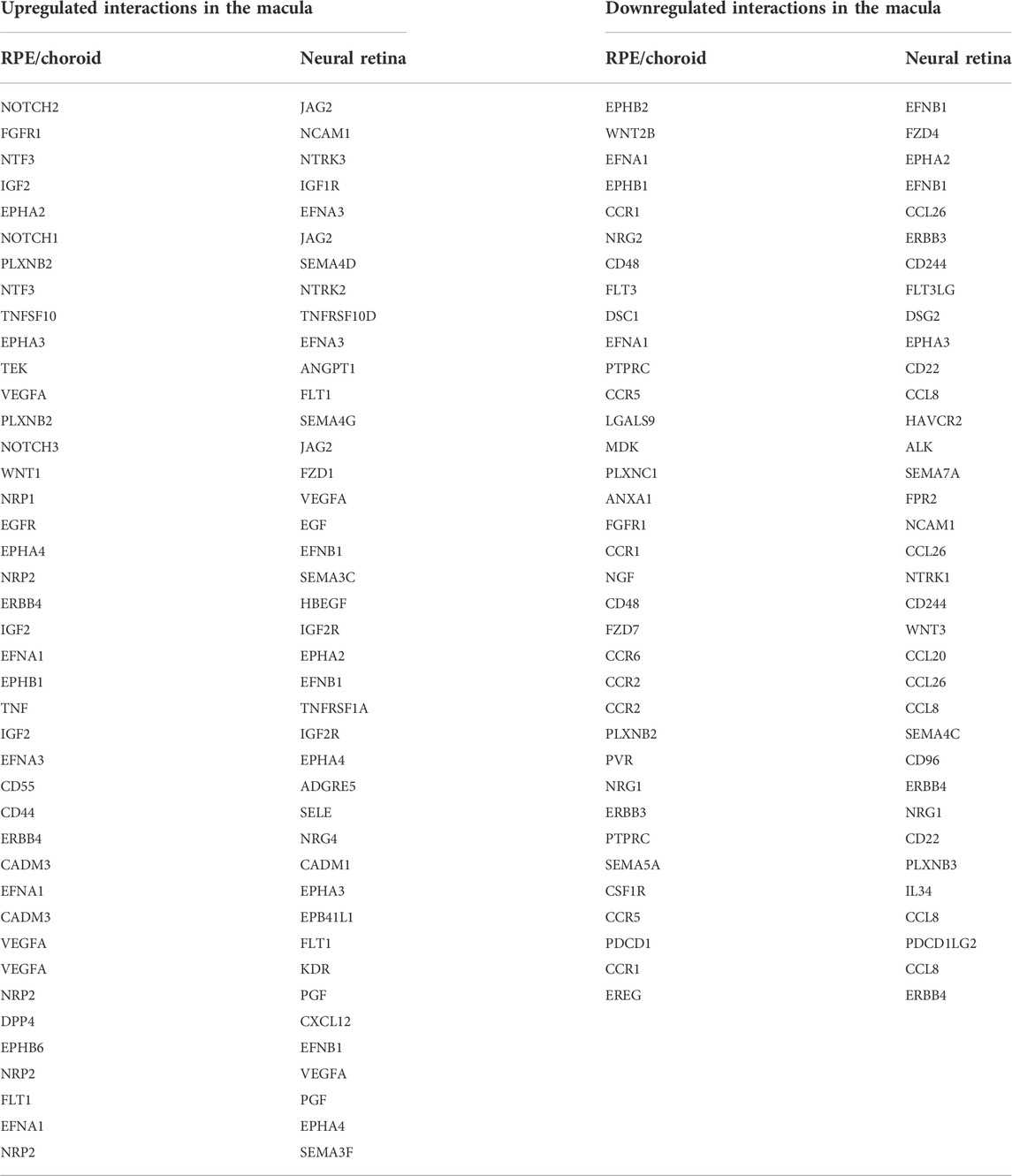
Analysis of the foveal/macular versus peripheral regions in the retina and RPE/choroid depicted distinct sets of differentially expressed genes in each tissue layer, with 9644 and 7925 differentially expressed genes in the retina and RPE/choroid, respectively (Padj < 0.05) (Table 1). Many of the significantly differentially expressed genes are known to have tissue-specific roles in development and disease (Figures 1A,B). Retinal diseases also manifest uniquely at particular regions of the RPE or retina. For example, Age-Related Macular Degeneration (AMD) is thought to begin in the macular RPE (Boulton and Dayhaw-Barker, 2001), Late Onset Retinal Degeneration manifests in the peripheral RPE (Milam et al., 2000), Retinitis Pigmentosa causes photoreceptor loss in the peripheral retina (Jones et al., 2018), and finally, Cone-Rod Dystrophies primarily affect photoreceptors in the central retina (Hamel, 2007).
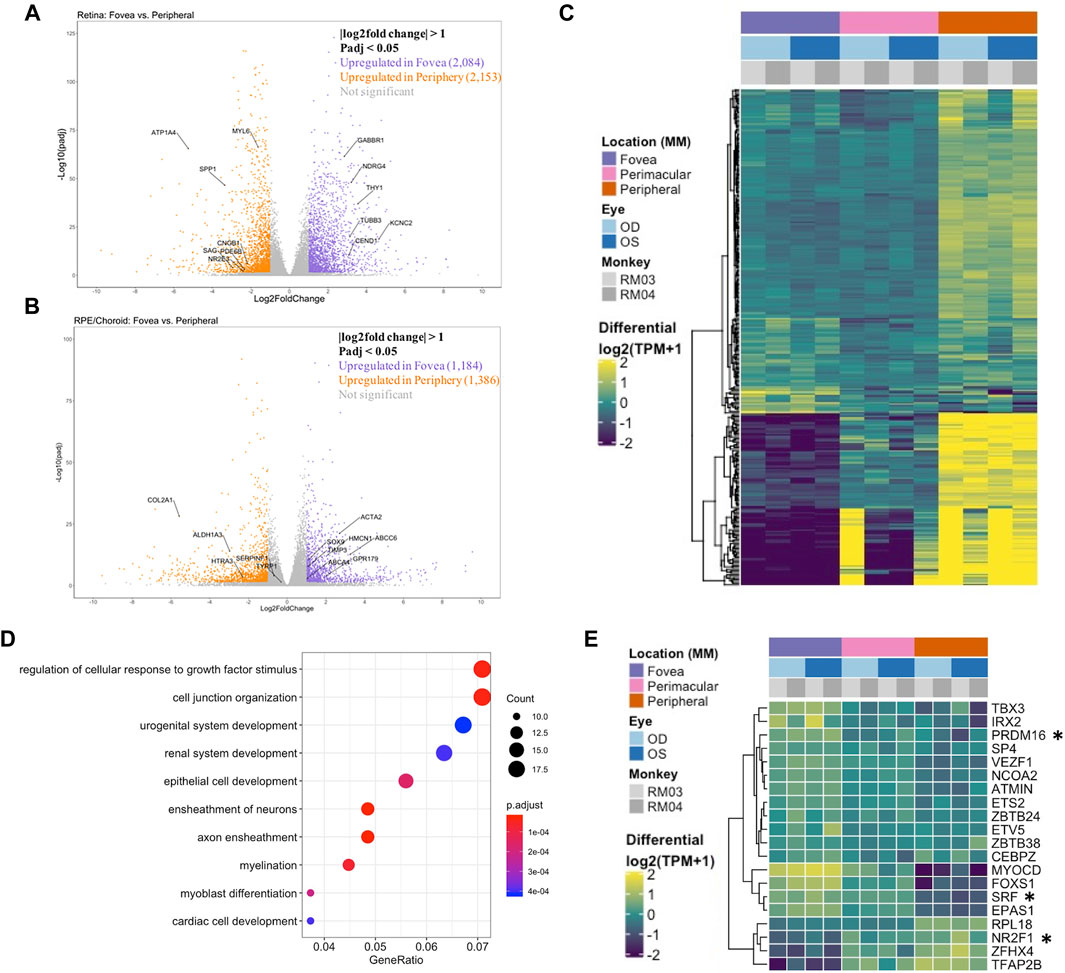
FIGURE 1. (A) A volcano plot depicting differentially expressed genes between foveal and peripheral tissues in the neural retina. Colored genes show significant up- or downregulation with several labeled genes having known tissue-specific roles in development and disease including PDE6B. (B) A volcano plot showing RPE fovea vs. periphery differential expression, with key genes labeled, including ABCA4 and TIMP3. (C) Unfiltered differential expression analysis of contiguously changing genes from fovea to perimacula to periphery in the RPE/choroid exhibits distinct blocks of lowly expressed genes in the fovea with a progressive increase in expression in peripheral tissues as well as blocks of genes with progressive downregulation moving outwards through the tissue despite less obvious morphologic changes in the tissue as compared to neural retina. (D) Top 10 enriched gene ontology (GO) terms from the RPE/choroid differentially expressed gene set include cell junction organization and epithelial cell development. VEGF is enriched in multiple of these ontologies and has increased expression in the macula over the periphery. (E) A subset of genes identified as differentially expressed transcription factors by location in the RPE/choroid. Changes in transcriptional regulators can be a factor in driving gene expression changes. *These transcription factors have published associations with the respective tissue in the literature.
In order to confirm that the differentially expressed genes across locations further reflect cellular composition of different tissues, we used expression patterns of published rod-enriched genes as identified in Holt et al. (2015) and Mustafi et al. (2016). We observed correlated gene expression between eyes and animals, as well as a progressive increase in rod-enriched gene expression from the fovea/macula to the perimacula and then to the periphery, reflecting changes in neural retina composition (Supplementary Figure S3A). In this list of rod enriched genes, we find that our peripheral retina samples are 2.8 fold enriched in rod signal relative to the fovea/macula (t-test p < 3.5 e-12). The perimacular region was enriched in rod signal by 1.7 fold over the fovea/macula (p < 1.1 e-09). The enrichment of cone gene expression between the fovea/macula and peripheral retina samples was 1.6 fold (p < 0.02) (Supplementary Figure S3B; Supplementary Table S1). Among the RPE samples the enrichment of rod and cone markers was less substantial and not significant (Supplementary Figure S3C).
Next, we performed an unfiltered analysis of the differentially expressed, contiguously changing genes from fovea/macula to perimacula to periphery in the RPE/choroid. This analysis highlights stepwise changes due to changing photoreceptor composition in the cone-predominant macula, rod-predominant periphery, and admixed perimacula, and how these morphological changes in the neural retina may affect changing gene expression in the RPE/choroid. Differential expression analysis revealed distinct blocks of genes with low expression in the fovea/macula that increase moving outward through the perimacula to the periphery, and other genes that are highly expressed in the fovea/macula but progressively decrease moving outwards (Figure 1C). We observe enrichment for gene ontologies including cell junction organization and epithelial cell development, as well as urogenital and renal system development (Figure 1D).
Next, we sought to understand the transcriptional regulators driving the spatial and inter-tissue differences in gene expression. Using a list of all known transcription factors in humans, we identified a subset which is differentially expressed by location in each of the retina (Supplementary Figure S4B) and RPE/choroid datasets (Figure 1E). After conducting a literature review, we further identified transcription factors with known associations in the respective tissues. In the retina and RPE, distinct sets of fovea/macula -enriched and periphery-enriched transcription were elucidated. Our data shows three transcription factors, NFRF1, IRX2, and ZFHX4, to be differentially expressed in both neural retina and RPE/choroid and show a similar pattern in independent human data curated from eyeIntegration (Swamy and McGaughey, 2019) (Supplementary Figure S5). Interestingly, ZFHX4 appears to be enriched in the macular neural retina and the peripheral RPE/choroid, whereas NFRF1 and IRX2 expression is present in both tissues.
We then analyzed tissue-tissue interactions using ligand and receptor characterizations from the CellPhoneDB database (Efremova et al., 2020). After converting protein names from the database to gene names via the Uniprot conversion tool and merging the data with our location-based differential expression, we generated lists of distinct up- and down-regulated interactions in which both interacting partners in each tissue were either up- or downregulated in the fovea/macula over the periphery (Table 2). Pathway analysis on these gene lists shows enrichment in the upregulated interactors for cell adhesion pathways (Figures 2A,C) and that downregulated interactors were highly enriched for Wnt signaling in kidney disease and the ErbB signaling pathway, both of which are heavily involved in tissue development, including ocular development (Figures 2B,D; Supplementary Figures S7C,D). Additionally, receptors involved in chemokine signaling were downregulated in the fovea/macula (Figure 2B).
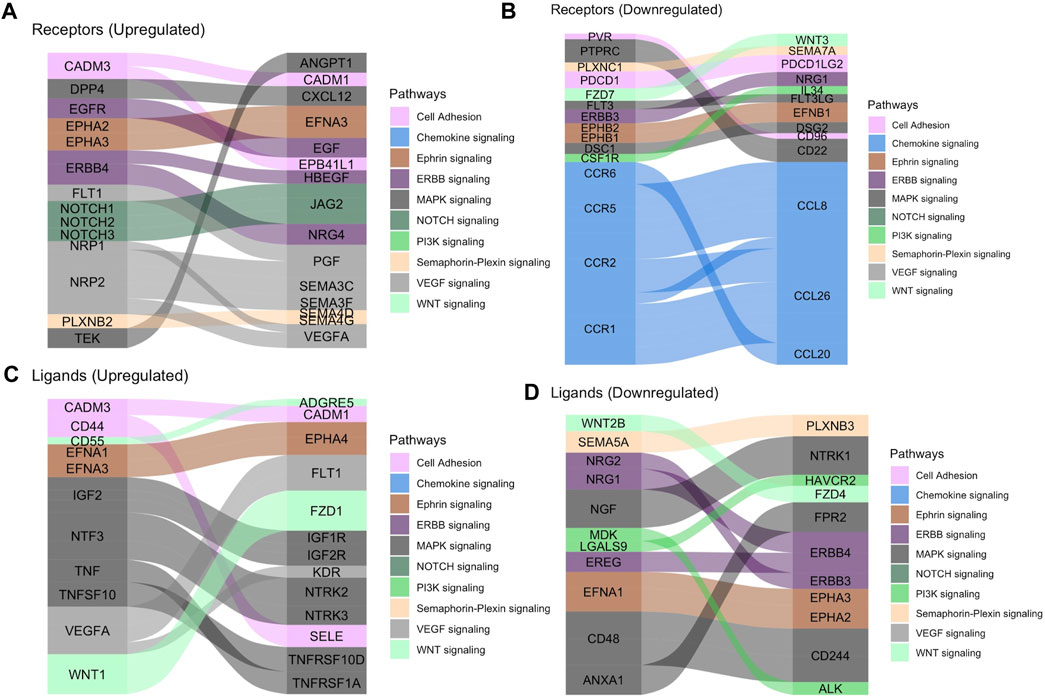
FIGURE 2. Tissue-tissue interactions showing receptor/ligand pairs in the central retina. (A) Upregulated macular receptors in the RPE/choroid are linked to their upregulated ligand in the macular neural retina. Interactors are grouped by signaling pathway and the width is scaled by average log2 fold change. (B) Downregulated macular receptors in the RPE/choroid and their respective downregulated ligand in the macular neural retina. (C) Upregulated macular ligands in the RPE/choroid are linked to their upregulated receptor in the macular neural retina. (D) Downregulated macular ligands in the RPE/choroid and their corresponding downregulated receptors in the macular neural retina.
To determine the conservation of location-specific differential expression of transcriptional regulators across fovea/macula to periphery, we performed a meta-analysis comparing our macaque data to previously published human datasets (Li et al., 2014; Whitmore et al., 2014). We see generally conserved patterns of expression from fovea/macula to periphery in neural retina and to a lesser extent, RPE/choroid (Figures 3A,B). We performed the same analysis on unfiltered gene lists from the human and macaque and found approximately 700 genes that follow similar differential expression patterns from fovea/macula to periphery in both species. (Figure 3A; Supplementary Table S1). GO terms for this list of conserved genes involve neuronal connectivity and cellular morphogenesis (Figure 3C), similar to the GO terms for the macaque retina data seen previously in this study. In one case, we see a distinct block of macula-specific transcription factors in the Rhesus monkey neural retina including HR, MYC, EBF3, AR, EBF2, EBF1, TWIST1, IRX2, POU4F2, IRX1, and SHOX2 that seem to also correlate to macula-specific transcription factors in the human data (Supplementary Figure S6B). Overall, this supports a conservation of gene expression and function across monkey and human species.
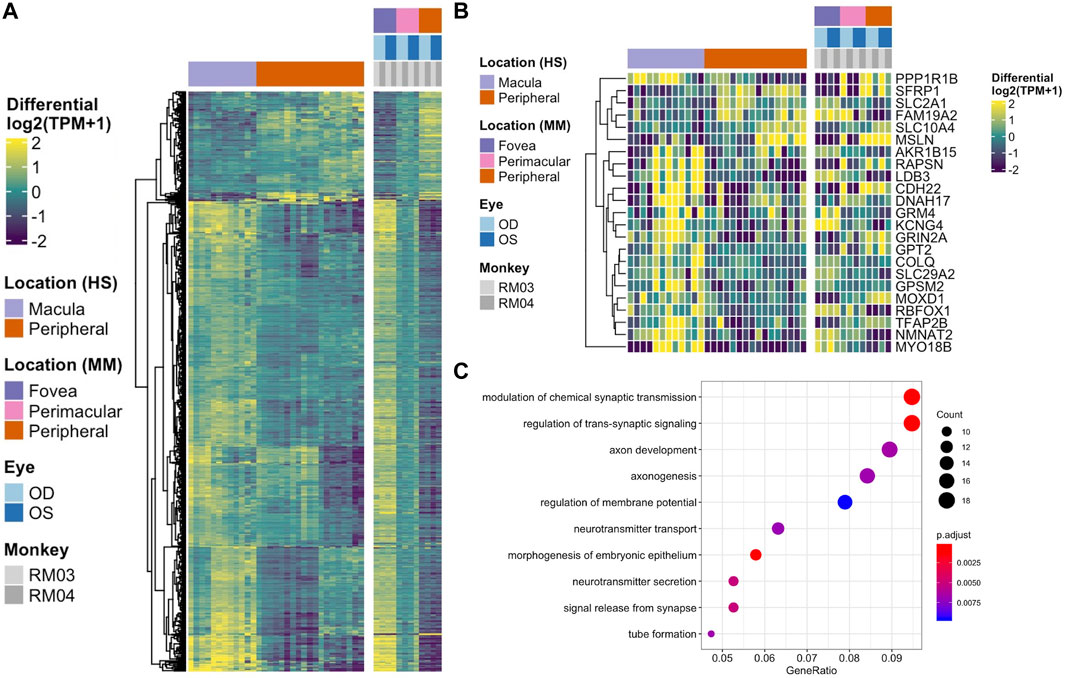
FIGURE 3. (A) Heatmap displaying the comparison of differential gene expression in the neural retina between published data from a human macula vs. periphery set (Li et al., 2014; Whitmore et al., 2014) and the macaque data presented here, with 664 genes following similar expression patterns. (B) A comparison of conserved differential expression to the human studies in the RPE/choroid tissue. Fewer genes follow similar patterns of differential expression. (C) GO enrichment for conserved differentially expressed genes between human and macaque datasets in the neural retina. Terms including neuronal connectivity and cellular morphogenesis are highly enriched.
To test whether there was any correspondence between our macaque tests and an independent single cell RPE cells taken from either the fovea/macula or periphery, we extracted 219 cells from the scEiaD resource at plae.nei.nih.gov. We found ten genes that overlap between our macaque resource and the scEiaD resource: CXCL14, GNG11, IVNS1ABP, PMEL, S100B, SCG5, SRSF3, SULF1, VIM, and WFDC1. Of these ten overlapping genes, CXCL4, SULF4, and WFDC1 are shown to be enriched in the macula, and S100B, PMEL, and SCG5 are enriched in the periphery (Figure 4).
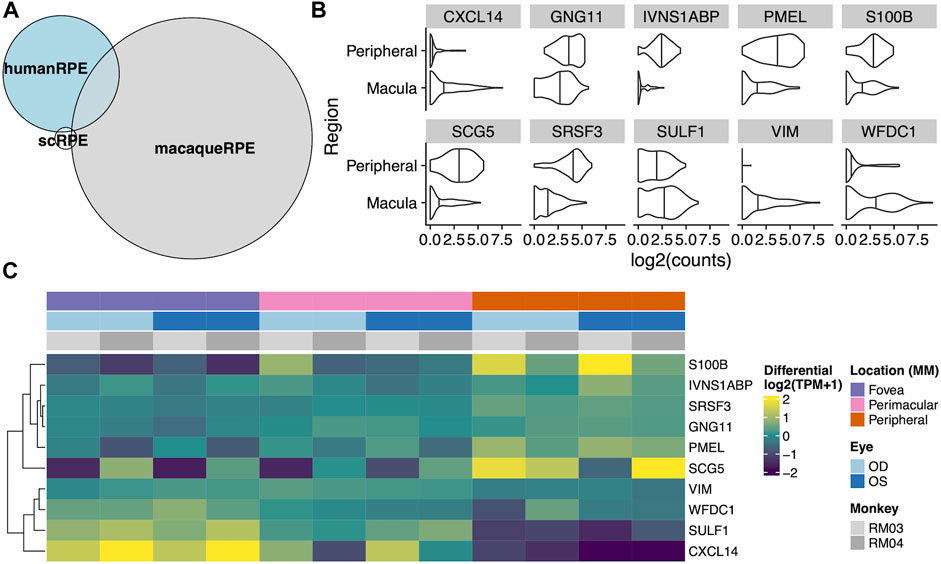
FIGURE 4. (A) Venn diagram of overlaps between differentially expressed genes between the fovea and periphery in three systems: Human single cell RPE, bulk human RPE-choroid, and bulk macaque RPE-choroid. (B) Violin plot of the single cell expression with the ten genes which overlap between the human single cell RPE testing and bulk macaque RPE-choroid testing. (C) Heatmap of the same ten genes in our macaque RPE-choroid punches.
Discussion
Using the Rhesus macaque as a model to study the transcriptome landscape of higher primate retina, we conducted RNA-sequencing across neural retina and RPE/choroid tissue layers. The small RNA punch size and bulk processing of the tissues allowed for high resolution and sensitivity of transcript quantification due to the decreased likelihood of variability from extended amplification, gene dropouts, and biological noise. Furthermore, it allows for the ability to profile rare and low-expressed transcripts as compared to single-cell RNAseq such as the 10x Genomics Chromium platform, in which only the top 20%–30% of expressed genes are captured, or Drop-seq, which requires a more manual approach. Unlike previous studies, our approach included the perimacular retina to define contiguously changing gene expression across the retinal landscape, thereby reflecting the stepwise differences in the ratio of cone and rod composition. By doing this, we isolated the critical pathways regulating changes in retinal connectivity and RPE/choroid function necessary to support different photoreceptor mosaics.
The macaque gene expression changes observed in our data are orthogonally confirmed in human datasets. Furthermore, our findings also indicate conservation of contiguously changing gene expression patterns from the central to peripheral retina between Rhesus monkeys and humans, establishing a more available model system for future human transcriptomic studies. Rhesus monkeys offer an accessible model in which conditions of tissue acquisition can be further controlled, including age, optimization of biopsy and RNA extraction, and reduced time from enucleation to tissue dissection and RNA extraction.
PCA of gene expression data by tissue type predicts the distinct nature of the neural retina and RPE/choroid tissues, and further clustering by location confirms there are transcriptomic differences between regions of the retina. The grouping of transcriptomes by animal highlights the genetic variability observed between individuals. The lower number of differentially expressed genes in fovea/macula vs. perimacular comparisons versus fovea/macula vs. periphery indicates a continuum of gene expression. The highly variable patterns of gene expression across the neural retina corresponds to the changing composition of cell types based on location.
In contrast, the presence of distinct changing RPE/choroid gene expression suggests variability in the cell processes occurring across the retina in the RPE and choroid. Due to the relatively unchanged morphology of these tissues across the retina save for increased RPE diameter and choroidal thickness in the macula, the contiguous changes in gene expression observed may be a factor in driving changes in cell processes across the entire retina. GO terms enriched for the contiguously changing genes in the retina involved neuron projection and axon development as well as regulation of cell morphogenesis, supporting known findings that there are differences in circuitry and cone packing across the neural retina (Jones et al., 2018). Furthermore, the changing gene expression across the RPE/choroid is enriched for GO terms such as cell junction organization and epithelial cell development, supporting reported findings that RPE diameter is smaller in the fovea/macula as compared to the periphery (R Sparrrow et al., 2010). It is known that RPE cells form tight junctions, creating the outer blood-retinal barrier. These tight junctions serve as regulators of cell proliferation, polarity, and transport, as well as transducers of signals responsible for regulating cell size and shape (Zou et al., 2020). Therefore, genes involved in cell junction organization and epithelial cell development may play a role in varied cell sizes across the RPE. GO terms involving urogenital and renal system development are also enriched in the RPE/choroid gene list, and interestingly, there are several heritable genetic conditions that affect both the eye and kidney, and there is also a link between chorioretinal thinning and chronic kidney disease, thought to be due to inflammation and endothelial dysfunction (Balmforth et al., 2016; Paterson et al., 2020). In addition to observing greater levels of gene expression variability across location in the RPE-choroid than expected, several disease-relevant genes known to play roles in various ocular disorders including TIMP3 [various retinopathies (Dewing et al., 2020)], ABCA4 [Stargardt disease (Walia and Fishman, 2009)], and TYRP1 [oculocutaneous albinism (Simeonov et al., 2013)], were found to be significantly differentially expressed by location.
Differential expression of particular transcription factors, including neural retina- and RPE/choroid show conserved patterns of expression between macaque and human datasets, with IRX2 being upregulated in the macula. Other differentially expressed transcription factors that show conservation with past human studies include FOXI3, which is upregulated in the periphery, and POU4F2, POU4F1, as well as its target, RIT2, all of which are enriched in the macula in both monkey and human datasets7. Additional transcription factors following similar gene expression patterns across species in the RPE/choroid include VEZF1 and NR2F1, which are thought to be involved in retinal and vascular development (Tang et al., 2010; Zou et al., 2010). Identifying conserved location-specific transcription factors can provide insight into the regulatory landscape of the two tissues and how they relate to patterns of differential gene expression. It may be inferred then, that these changes in global gene expression across location and tissue, driven by distinct sets of transcription factors, likely reflect location-specific cellular processes and interactions.
Beyond patterns of transcriptional regulators, location-based tissue-tissue interactions are also indicative of specific pathways being up- or downregulated by location, suggesting specific roles in different locations of the tissues. The enrichment for Wnt signaling in kidney disease in the downregulated interactions in the macula may be connected to the GO enrichment for renal development in the RPE/choroid. Furthermore, ErbB signaling and peptide G-protein coupled receptor signaling are required for retinal development, with ErbB involved in neural crest development and adult pigment formation (Budi et al., 2008) and GCPR signaling involved in light processing in photoreceptors (Martemyanov, 2014). The tissue-tissue interaction analysis suggests these pathways are upregulated in the peripheral tissues of the eye. Certain pathways were enriched for tissue-tissue interactions both up- and downregulated in the macula, including Hippo signaling. Hippo-YAP signaling is known to play roles in both ocular development as well as disease, as it regulates retinogenesis, photoreceptor cell differentiation, and retinal vascular development, but misregulation also has associations with coloboma and optic fissure closure, uveal melanoma, and retinal degeneration (Lee et al., 2018). The downregulation of chemokine receptors observed in the fovea/macula may suggest a higher sensitivity to chemokine signaling in this region during disease, as AMD is known to be associated with upregulated levels of chemokine signaling (Newman et al., 2012). The widespread enrichment of this pathway informs the many roles it plays in regulating the retinal landscape across location.
The three-way comparison of single cell, bulk human, and Rhesus macaque data showed many more differentially expressed genes detected in our macaque dataset than the other two sequencing methods. It remains unclear whether this subset of differentially expressed genes without parent conservation in our dataset is identifying a Rhesus macaque-specific set of genes or if some of these genes are novel identifications due to the higher resolution of sequencing achieved in this dataset. Further investigation including functional studies are necessary to determine this. Furthermore, identifying differentially expressed genes in the Rhesus monkey data that intersect with the single cell data allows us to identify known RPE-specific genes that are conserved between the datasets including VIM (Zou et al., 2020), and PMEL (Reyes et al., 2020), which we have shown to change contiguously by location. Of the three macula-enriched genes in the overlapping set, CXCL14 and WFDC1 have been previously identified as enriched in the macula compared to peripheral retina via RNA microarray (Radeke et al., 2007), and WFDC1 enrichment in the macula was independently confirmed by both expression and immunostaining (van Soest et al., 2007). As such, our data shows the known macula-enrichment of WFDC1 to be conserved. As a serine protease inhibitor, mediator of endothelial cell migration and promoter of angiogenesis, its enrichment in the RPE and specifically, the macula, may suggest a role for WFDC1 in age-related macular degeneration (Zhu et al., 2021), in which neovascularization beneath the macula is characteristic (Radeke et al., 2007). Additionally, S100B, a gene encoding a calcium-binding protein which we found enriched in the peripheral RPE has been linked with glaucoma, which can result in loss of peripheral vision (Kuehn et al., 2018).
Considering the differential gene expression patterns across contiguous regions of the neural retina and especially the RPE/choroid allows for a combined approach in which we assessed the drivers of change as well as gross changes in gene expression by location. We identified highly enriched gene ontologies associated with each tissue location and layer, highlighted sets of contiguously changing transcription factors, determined important tissue-tissue interactions that highlight various up- and downregulated location-specific pathways, and examined the conservation of gene expression patterns across multiple independent studies. In addition to the previous RNA-sequencing studies performed on human and non-human primate retinas, the data and findings presented here provide valuable resources for future studies aimed at identifying regional specialization of the retina and understanding disease mechanisms.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was reviewed and approved by NEI IACUC.
Author contributions
AM, DM, TF, and RH performed data analysis and wrote the manuscript; CZ, SY, JL, and AM performed experiments and collected the data; all authors read and approved the final manuscript.
Funding
This work was supported by National Eye Institute intramural funds.
Acknowledgments
We would like to thank Bob Wurtz, Bruce Cumming, James Cavanaugh, Mitch Smith of the Laboratory of Sensorimotor Research, National Eye Institute for providing non-human primate samples. We would also like to thank the NEI/NIMH Animal Program, including Jim Raber and Ginger Tansey, the National Institutes of Health Intramural Sequencing Center, where RNA sequencing was performed, and Sheldon Miller for helpful comments and advice about the experiments and manuscript. The contents of this manuscript have been submitted for preprint to biorxiv.org, (Mungale, 2022).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.949449/full#supplementary-material
References
Bagci, A. M., Shahidi, M., Ansari, R., Blair, M., Blair, N. P., and Zelkha, R. (2008). Thickness profiles of retinal layers by optical coherence tomography image segmentation. Am. J. Ophthalmol. 146 (5), 679–687. doi:10.1016/j.ajo.2008.06.010
Balmforth, C., van Bragt, J. J., Ruijs, T., Cameron, J. R., Kimmitt, R., Moorhouse, R., et al. (2016). Chorioretinal thinning in chronic kidney disease links to inflammation and endothelial dysfunction. JCI insight 1 (20), e89173. doi:10.1172/jci.insight.89173
Boulton, M., and Dayhaw-Barker, P. (2001). The role of the retinal pigment epithelium: Topographical variation and ageing changes. Eye 15 (3), 384–389. doi:10.1038/eye.2001.141
Budi, E. H., Patterson, L. B., and Parichy, D. M. (2008). Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation.
Cowan, C. S., Renner, M., De Gennaro, M., Gross-Scherf, B., Goldblum, D., Hou, Y., et al. (2020). Cell types of the human retina and its organoids at single-cell resolution. Cell. 182 (6), 1623–1640.e34. e34. doi:10.1016/j.cell.2020.08.013
Daiger, S. (1998). Data services and software for identifying genes and mutations causing retinal degeneration. Investig. Ophthalmol. Vis. Sci. 39 (S295).
Dewing, J. M., Carare, R. O., Lotery, A. J., and Ratnayaka, J. A. (2020). The Diverse Roles of TIMP-3: Insights into degenerative diseases of the senescent retina and brain. Cells 9 (1), 39. doi:10.3390/cells9010039
Di Pierdomenico, J., Garcia-Ayuso, D., Pinilla, I., Cuenca, N., Vidal-Sanz, M., Agudo-Barriuso, M., et al. (2017). Early events in retinal degeneration caused by rhodopsin mutation or pigment epithelium malfunction: Differences and similarities. Front. Neuroanat. 11, 14. doi:10.3389/fnana.2017.00014
Efremova, M., Vento-Tormo, M., Teichmann, S. A., and Vento-Tormo, R. (2020). CellPhoneDB: Inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15 (4), 1484–1506. doi:10.1038/s41596-020-0292-x
Grover, S., Murthy, R. K., Brar, V. S., and Chalam, K. V. (2010). Comparison of retinal thickness in normal eyes using Stratus and Spectralis optical coherence tomography. Investig. Ophthalmol. Vis. Sci. 51 (5), 2644–2647. doi:10.1167/iovs.09-4774
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32 (18), 2847–2849. doi:10.1093/bioinformatics/btw313
Hamel, J.-F. (2007). Les ruines du progrès chez walter benjamin: Anticipation futuriste, fausse reconnaissance et politique du présent. pr. 35 (2), 7–14. doi:10.7202/017462ar
Harrow, J., Frankish, A., Gonzalez, J. M., Tapanari, E., Diekhans, M., Kokocinski, F., et al. (2012). Gencode: The reference human genome annotation for the ENCODE project. Genome Res. 22 (9), 1760–1774. doi:10.1101/gr.135350.111
Holt, R., Brown, L., Broadgate, S., Butler, R., Jagannath, A., Downes, S., et al. (2015). Identification of rod- and cone-specific expression signatures to identify candidate genes for retinal disease. Exp. Eye Res. 132, 161–173. doi:10.1016/j.exer.2015.01.002
Ignatiadis, N., Klaus, B., Zaugg, J. B., and Huber, W. (2016). Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13 (7), 577–580. doi:10.1038/nmeth.3885
Jones, B. W., Marc, R. E., and Pfeiffer, R. L. (2018). Retinal degeneration, remodeling and plasticity.
Köster, J., and Rahmann, S. (2012). Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 28 (19), 2520–2522. doi:10.1093/bioinformatics/bts480
Kuehn, S., MeiBner, W., Grotegut, P., Theiss, C., Dick, H. B., and Joachim, S. C. (2018). Intravitreal S100B injection leads to progressive glaucoma like damage in retina and optic nerve. Front. Cell. Neurosci. 12, 312. doi:10.3389/fncel.2018.00312
Lamin, A., Oakley, J. D., Dubis, A. M., Russakoff, D. B., and Sivaprasad, S. (2019). Changes in volume of various retinal layers over time in early and intermediate age-related macular degeneration. Eye 33 (3), 428–434. doi:10.1038/s41433-018-0234-9
Lee, M., Goraya, N., Kim, S., and Cho, S. H. (2018). Hippo-yap signaling in ocular development and disease. Dev. Dyn. 247 (6), 794–806. doi:10.1002/dvdy.24628
Li, M., Jia, C., Kazmierkiewicz, K. L., Bowman, A. S., Tian, L., Liu, Y., et al. (2014). Comprehensive analysis of gene expression in human retina and supporting tissues. Hum. Mol. Genet. 23 (15), 4001–4014. doi:10.1093/hmg/ddu114
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi:10.1186/s13059-014-0550-8
Martemyanov, K. A. (2014). G protein signaling in the retina and beyond: The cogan lecture. Investig. Ophthalmol. Vis. Sci. 55 (12), 8201–8207. doi:10.1167/iovs.14-15928
Milam, A. H., Curcio, C. A., Cideciyan, A. V., Saxena, S., John, S. K., Kruth, H. S., et al. (2000). Dominant late-onset retinal degeneration with regional variation of sub-retinal pigment epithelium deposits, retinal function, and photoreceptor degeneration. Ophthalmology 107 (12), 2256–2266. doi:10.1016/s0161-6420(00)00419-x
Mori, K., Kanno, J., and Gehlbach, P. L. (2016). Retinochoroidal morphology described by wide-field montage imaging of spectral domain optical coherence tomography. Retina 36 (2), 375–384. doi:10.1097/IAE.0000000000000703
Mungale, A. (2022). Transcriptional mapping of the macaque retina and RPE-choroid reveals conserved inter-tissue transcription drivers and signaling pathways. Cold Spring Harbor: bioRxiv, 2022.
Mustafi, D., Kevany, B. M., Bai, X., Golczak, M., Adams, M. D., Wynshaw-Boris, A., et al. (2016). Transcriptome analysis reveals rod/cone photoreceptor specific signatures across mammalian retinas. Hum. Mol. Genet. 25 (20), 4376–4388. doi:10.1093/hmg/ddw268
Newman, A. M., Gallo, N. B., Hancox, L. S., Miller, N. J., Radeke, C. M., Maloney, M. A., et al. (2012). Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 4 (2), 16–18. doi:10.1186/gm315
Nickla, D. L., and Wallman, J. (2010). The multifunctional choroid. Prog. Retin. Eye Res. 29 (2), 144–168. doi:10.1016/j.preteyeres.2009.12.002
Paterson, E. N., Ravindran, M. L., Griffiths, K., Le Velly, C. A., Cardwell, C. C., McCarter, R. V., et al. (2020). Association of reduced inner retinal thicknesses with chronic kidney disease. BMC Nephrol. 21 (1), 37–12. doi:10.1186/s12882-019-1679-1
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14 (4), 417–419. doi:10.1038/nmeth.4197
Peng, Y.-R., Shekhar, K., Yan, W., Herrmann, D., Sappington, A., Bryman, G. S., et al. (2019). Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell. 176 (5), 1222–1237.e22. e22. doi:10.1016/j.cell.2019.01.004
Purves, D. (2001). “Neuroscience,” in Sunderland (ma) sinauer associates. 2nd edition (Sunderland: Types of Eye Movements and Their Functions).
R Sparrrow, J., Hicks, D., and P Hamel, C. (2010). The retinal pigment epithelium in health and disease. Curr. Mol. Med. 10 (9), 802–823. doi:10.2174/156652410793937813
Radeke, M. J., Peterson, K. E., Johnson, L. V., and Anderson, D. H. (2007). Disease susceptibility of the human macula: Differential gene transcription in the retinal pigmented epithelium/choroid. Exp. Eye Res. 85 (3), 366–380. doi:10.1016/j.exer.2007.05.006
Reyes, A. P., Petrus-Reurer, S., Padrell Sanchez, S., Kumar, P., Douagi, I., Bartuma, H., et al. (2020). Identification of cell surface markers and establishment of monolayer differentiation to retinal pigment epithelial cells. Nat. Commun. 11 (1), 1609–1615. doi:10.1038/s41467-020-15326-5
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. doi:10.1093/bioinformatics/btp616
Simeonov, D. R., Wang, X., Wang, C., Sergeev, Y., Dolinska, M., Bower, M., et al. (2013). DNA variations in oculocutaneous albinism: An updated mutation list and current outstanding issues in molecular diagnostics. Hum. Mutat. 34 (6), 827–835. doi:10.1002/humu.22315
Soneson, C., Love, M. I., and Robinson, M. D. (2015). Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. F1000Research4, 1521. doi:10.12688/f1000research.7563.2
Swamy, V., and McGaughey, D. (2019). Eye in a disk: eyeIntegration human pan-eye and body transcriptome database version 1.0. Investig. Ophthalmol. Vis. Sci. 60 (8), 3236–3246. doi:10.1167/iovs.19-27106
Swamy, V. S., Fufa, T. D., Hufnagel, R. B., and McGaughey, D. M. (2021). Building the mega single cell transcriptome ocular meta-atlas. Oxford: bioRxiv.
Tang, K., Xie, X., Park, J. I., Jamrich, M., Tsai, S., and Tsai, M. J. (2010). COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development 137 (5), 725–734. doi:10.1242/dev.040568
van Soest, S., de Wit, G. M. J., Essing, A. H. W., ten Brink, J. B., Kamphuis, W., de Jong, P. T. V. M., et al. (2007). Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Mol. Vis. 13, 1608–1617.
Voigt, A. P., Mulfaul, K., Mullin, N. K., Flamme-Wiese, M. J., Giacalone, J. C., Stone, E. M., et al. (2019). Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 116 (48), 24100–24107. doi:10.1073/pnas.1914143116
Voigt, A. P., Whitmore, S. S., Mulfaul, K., Chirco, K. R., Giacalone, J. C., Flamme-Wiese, M. J., et al. (2020). Bulk and single-cell gene expression analyses reveal aging human choriocapillaris has pro-inflammatory phenotype. Microvasc. Res. 131, 104031. doi:10.1016/j.mvr.2020.104031
Voigt, A., Whitmore, S. S., Flamme-Wiese, M. J., Riker, M. J., Wiley, L. A., Tucker, B. A., et al. (2019). Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res. 184, 234–242. doi:10.1016/j.exer.2019.05.001
Walia, S., and Fishman, G. A. (2009). Natural history of phenotypic changes in Stargardt macular dystrophy. Ophthalmic Genet. 30 (2), 63–68. doi:10.1080/13816810802695550
Whitmore, S. S., Wagner, A. H., DeLuca, A. P., Drack, A. V., Stone, E. M., Tucker, B. A., et al. (2014). Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp. Eye Res. 129, 93–106. doi:10.1016/j.exer.2014.11.001
Yan, W., Peng, Y. R., van Zyl, T., Regev, A., Shekhar, K., Juric, D., et al. (2020). Cell atlas of the human fovea and peripheral retina. Sci. Rep. 10 (1), 9802–9817. doi:10.1038/s41598-020-66092-9
Yates, B. (2016). Genenames. Org: The HGNC and VGNC resources in 2017. Oxford: Nucleic acids research, gkw1033.
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics a J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Zhu, S., Ye, L., Bennett, S., Xu, H., He, D., and Xu, J. (2021). Molecular structure, gene expression and functional role of WFDC1 in angiogenesis and cancer. Cell. biochem. Funct. 39, 588–595. doi:10.1002/cbf.3624
Zou, H., Shan, C., Liu, J., Yang, N., and Zhao, J. (2020). Polarity and epithelial-mesenchymal transition of retinal pigment epithelial cells in proliferative vitreoretinopathy. PeerJ 8, e10136. doi:10.7717/peerj.10136
Keywords: transcriptome, macula, fovea, retina, RPE
Citation: Mungale A, McGaughey DM, Zhang C, Yousaf S, Liu J, Brooks BP, Maminishkis A, Fufa TD and Hufnagel RB (2022) Transcriptional mapping of the macaque retina and RPE-choroid reveals conserved inter-tissue transcription drivers and signaling pathways. Front. Genet. 13:949449. doi: 10.3389/fgene.2022.949449
Received: 21 May 2022; Accepted: 01 November 2022;
Published: 25 November 2022.
Edited by:
Mariya Moosajee, Faculty of Brain Sciences, University College London, United KingdomReviewed by:
Hala El-Bassyouni, National Research Centre, EgyptHelen Louise May-Simera, Johannes Gutenberg University Mainz, Germany
Copyright © 2022 Mungale, McGaughey, Zhang, Yousaf, Liu, Brooks, Maminishkis, Fufa and Hufnagel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert B. Hufnagel, cm9iZXJ0Lmh1Zm5hZ2VsQG5paC5nb3Y=
†These authors have contributed equally to this work and share first authorship
 Ameera Mungale
Ameera Mungale David M. McGaughey2†
David M. McGaughey2† Sairah Yousaf
Sairah Yousaf Robert B. Hufnagel
Robert B. Hufnagel