- 1ICAR-Indian Institute of Soybean Research, Indore, India
- 2ICAR-Indian Institute of Maize Research, Ludhiana, India
- 3Department of Plant Pathology, Punjab Agricultural University, Ludhiana, India
- 4ICAR-Indian Agricultural Research Institute, New Delhi, India
- 5National Agri-Food Biotechnology Institute, Mohali, India
- 6Mountain Research Centre for Field Crops, Sher-e-Kashmir University of Agricultural Sciences and Technology, Srinagar, Jammu and Kashmir, India
Soybean is one of the largest sources of protein and oil in the world and is also considered a “super crop” due to several industrial advantages. However, enhanced acreage and adoption of monoculture practices rendered the crop vulnerable to several diseases. Phytophthora root and stem rot (PRSR) caused by Phytophthora sojae is one of the most prevalent diseases adversely affecting soybean production globally. Deployment of genetic resistance is the most sustainable approach for avoiding yield losses due to this disease. PRSR resistance is complex in nature and difficult to address by conventional breeding alone. Genetic mapping through a cost-effective sequencing platform facilitates identification of candidate genes and associated molecular markers for genetic improvement against PRSR. Furthermore, with the help of novel genomic approaches, identification and functional characterization of Rps (resistance to Phytophthora sojae) have also progressed in the recent past, and more than 30 Rps genes imparting complete resistance to different PRSR pathotypes have been reported. In addition, many genomic regions imparting partial resistance have also been identified. Furthermore, the adoption of emerging approaches like genome editing, genomic-assisted breeding, and genomic selection can assist in the functional characterization of novel genes and their rapid introgression for PRSR resistance. Hence, in the near future, soybean growers will likely witness an increase in production by adopting PRSR-resistant cultivars. This review highlights the progress made in deciphering the genetic architecture of PRSR resistance, genomic advances, and future perspectives for the deployment of PRSR resistance in soybean for the sustainable management of PRSR disease.
Introduction
Soybean (Glycine max (L.) Merrill) is an important legume crop that fulfills the substantial demand for food and feed globally. It is high in protein and oil content and also serves as a source of nutraceuticals such as bioflavonoids, lecithins, phytosterols, saponins, and tocopherols. Its oil is mainly used for domestic purposes; however, recent trends move toward the use of soybean oil as biodiesel to decrease reliance on fossil fuels (Mofijur et al., 2013). Approximately 70% of soybean’s economic value is for its meal, of which 97% is consumed as livestock and poultry feed (Raghuvanshi and Bisht 2010). The demand for soybean in the international market is increasing due to interest in functional food and the use of various soybean seed constituents and by-products in a wide array of specific industrial products (Kumawat et al., 2016). Globally, soybean is grown in an area of 122.6 million hectares (mha) with an annual average production of 336.6 million tons (mt) (USDA, 2020). The leading producers of soybean are Brazil, the United States, China, Argentina, and India (USDA, 2020). Like other food crops, soybean production is being challenged by various forms of abiotic and biotic stresses. The remarkable growth in the number of major diseases and their area has been observed in the past 50 years; subsequently, it negatively affects soybean production worldwide. The impact of diseases on soybean may be cited to the fact that the average annual economic loss due to soybean diseases in the US reached nearly $4.55 billion based on an investigation from 1996 to 2016 (Bandara et al., 2020). This increase in the number and spread of diseases can be attributed to enhanced acreage in new un-adapted regions and monoculture practices resulting in high pathogen density. Various factors governing the disease severity and economic losses include the pathogen type, plant tissue under attack, affected number of plants, the severity of an attack, pathogen-favoring environment, host plant vulnerability, plant stress level, and crop development stages (Hartman and Hill, 2010).
Among the various soybean diseases, Phytophthora root and stem rot (PRSR), caused by the soil-borne pathogen Phytophthora sojae (Kaufmann and Gerdemann) (oomycete pathogen), is the second most important economic disease after soybean cyst nematodes in the world. Earlier, P. sojae was part of the Phytophthora megasperma species complex which causes rot diseases in plants (Lin et al., 2021). Phytophthora sansomeana is identified as another causal agent for root rot in soybean, and partial resistance to P. sansomeana in soybean has been observed (Lin et al., 2021). Like P. sojae, P. sansomeana is also part of the P. megasperma complex. However, stem and root rot caused by P. sansomeana is not included in this review to keep the article length in check. PRSR drastically limits the yields of soybean globally as losses caused by it range between 10 and 40% or complete yield loss in some scenarios (Xiao et al., 2011; Zhang et al., 2013a). In the last few decades, Phytophthora root and stem rot resistance has been characterized by many researchers (Ryley et al., 1998; Dorrance et al., 2003; Grau et al., 2004; Sugimoto et al., 2006; Dorrance et al., 2008). In the United States, a loss of nearly 20.5 million tons was reported from 1996 to 2014, with an average annual loss of over 1.1 million tons due to this pathogen (Allen et al., 2017). P. sojae was first reported in Indiana state of the United States in 1948 (Kaufmann, 1957). Later, it spread to the major soybean growing areas of the United States, particularly in the pathogen-favoring environment of the Northern United States (Dorrance and Schmitthenner 2000). In addition to the United States, PRSR has been reported in other soybean-producing continents, namely, Asia, Africa, Australia, and Europe. The occurrence and development of PRSR are facilitated by poorly drained clay soils, low temperatures, and high rains (Kaufmann, 1957; Han et al., 2008). PRSR is generally characterized by the damping-off of seedlings and rotting of roots in adult plants (Tyler, 2007), and affected plants exhibit red–brown water-soaked lesions, wilting, and chlorosis, which in the case of extreme severity leads to mortality (Schmitthenner 1985; Dorrance et al., 2003).
P. sojae has abundant pathogenic diversity, and complete and partial resistance reactions have been reported for this pathogen. PRSR is being managed by cultivars with one or two dominant resistance genes for Phytophthora sojae named “Rps” (Jang and Lee 2020). However, the Rps genes are race-specific and useful as introgression of such genes is easy; but partial resistance has its own advantage for long-term protection. The first resistance gene against soybean P. sojae (named Rps1a) was identified in the 1950s (Bernard et al., 1957). Later, with the advent of sequencing technology and development of abundant simple sequence repeat (SSR) markers (Song et al., 2010), molecular linkage mapping gained pace, and nearly 30 Rps genes have been identified to date (Lin et al., 2013; Sun et al., 2014a; Ping et al., 2016; Niu et al., 2017; Zhong et al., 2019; Zhong et al., 2019; Jiang et al., 2020). In addition to SSR markers, a large number of single nucleotide polymorphisms (SNPs) and insertion/deletion markers for fine genetic mapping and molecular breeding have also been studied in different mapping populations (Li et al., 2016a; Li et al., 2017a). Functional characterization of identified genes has also gained progress in the recent past (Fan et al., 2015; Fan et al., 2017; Jang and Lee, 2020; Zhou et al., 2022). Few transcriptomic studies also uncovered molecular pathways in response to P. sojae infection (Guo et al., 2011; Lin et al., 2014). Newly identified genes for PRSR resistance serve as a good source for modern breeding programs to improve the resistance of cultivars to PRSR disease. Furthermore, the identified quantitative disease resistance loci (QDRL) can be employed in gene stock mining for the identification of novel alleles. This review aims to provide the current progress and future perspectives on genetics and genomics-assisted studies of P. sojae R-genes/QDRL and their utilization in soybean improvement.
Disease management, pathogenic diversity, and potential genetic resources
PRSR is a serious concern today as it causes a significant yield loss in soybean production. Current PRSR control strategies include applications of various fungicides (Anderson and Buzzell, 1992), improving soil drainage systems (Schmitthenner, 1985), tillage systems (Workneh et al., 1998), application of calcium-containing fertilizers (Sugimoto et al., 2010), and the use of resistant varieties (Schmitthenner, 1999; Dorrance et al., 2003). Germplasm screening-based identification of resistant genotypes and development of PRSR-resistant soybean cultivars is the most effective and sustainable approach for minimizing yield losses (Burnham et al., 2003).
Currently, the management of PRSR is largely dependent on resistant cultivars, having one or more resistance genes. For understanding, pathogen race refers to a pathogen’s ability to cause disease in its host (Anderson et al., 2010); in other words, the pathogen race attacks certain resistance genes (Dorrance et al., 2016), and this kind of resistance is accompanied by several mechanisms including effector-triggered immunity (ETI), where R gene products in the host is recognized (directly/indirectly) by specific pathogen effectors termed avirulence (Avr) proteins (Li et al., 2021). Till now, nine Avr genes of P. sojae have been cloned (Yang et al., 2019). Soybean R genes whose products recognize P. sojae Avr effectors and trigger Phytophthora resistance are known as Rps (resistance to P. sojae) genes (Tyler and Gijzen, 2014). For understanding, avirulence 1c (Avr1c) gene in P. sojae confers the resistance by Rps 1c gene in soybean populations; the K105 amino-acid residue in Avr gene is the main determinant of the avirulence of Avr1c that interacts with Rps gene (Yang et al., 2019). So Rps genes have the potential to combat PRSR, but they are race-specific; therefore, they would be operational against limited P. sojae isolates, and each Rps gene often remains effective for about 8–15 years, which leads to the emergence of new isolates after a certain period (Schmitthenner 1985; Dorrance et al., 2003).
The diversity of the P. sojae population has been investigated in the United States and Canada since the 1960s, and most of the P. sojae isolates were determined based on studies with 15 host-differentials (Dorrance et al., 2004). During the 1980s, Rps genes 1a, 1d, and 1k have been widely exploited to combat PRSR losses; however, the emergence of new isolates and enhancement in virulence lead to the evolution of more than 55 races reported against eight soybean differentials (Rps1a, 1b, 1c, 1d, 1k, 2, 3a, 6, and 7 genes) (Abney et al., 1997; Grau et al., 2004). A total of 213 virulent pathotypes were identified from 873 isolates of the North Central United States (Dorrance et al., 2016).
In China, after the identification of P. sojae in the Heilongjiang region in 1991 (Su and Shen 1993), the incidence of the pathogen was reported mainly in the Inner Mongolia Autonomous Region, Xinjiang Uygur Autonomous Region, and Fujian Province till 2015 (Wen and Chen 2002; Chen et al., 2004; Liu et al., 2006; Xiao et al., 2011). During 2005–2007 in Heilongjiang Province, a total of 96 isolates were collected and investigated, which revealed that four out of the eight races had new pathotypes (Zhang et al., 2010).
PRSR was first reported in Hokkaido, Japan, in 1977 (Tsuchiya, 1990). Tsuchiya (1990) found the genetic differences between American and Japanese isolates during the investigation of 49 Japanese isolates and 55 known American P. sojae races. Sugimoto et al. (2006) collected 51 isolates from Hyogo in Japan and identified four new races. More than 100 P. sojae isolates were reported from 14 different regions for 14 Rps genes including Rps1a, 1b, 1c, 1d, 1k, 3b, 7, and 8; among them, Rps1d and 1k were determined as the most promising resistance genes (Moriwaki 2010). Similarly, in Brazil, P. sojae was found to be different from other regions; 17 pathotypes were determined based on 37 Brazilian isolates, which were genetically different from the previously reported ones (Costamilan et al., 2013). Subsequently, Rps1a, 1c, and 1k were highly utilized in Brazilian soybean breeding programs, whereas Rps1a and 1c were not effective in the United States (Costamilan et al., 2013). In South Korea, PRSR was first reported 2 decades ago (Jee et al., 1998). Kang et al. (2019) reported genetic differences among the pathotypes of Korean P. sojae isolates. Thus, based on these facts, it is a prerequisite to collect P. sojae isolates from several regions/fields and to assess them with differential varieties.
A number of genetic sources for P. sojae resistance have been identified and utilized to map Rps genes and to develop resistant cultivars through different breeding strategies (Table 1). Similarly, numerous genetic resources for incomplete or partial resistance for P. sojae have been utilized for genetic studies in the form of breeding lines and introgression (Table 2). Dorrance and Schmitthenner (2000) evaluated over 1,000 accessions from USDA germplasm accessions and found 162 accessions to be resistant to three races (7, 17, and 25). In addition to this, they also reported partial resistance in 55.5% of the 887 accessions for P. sojae. Kang et al. (2019) evaluated the Rps resistance against four isolates in 20 popular varieties of South Korea, while Daewon was identified as a resistant cultivar.
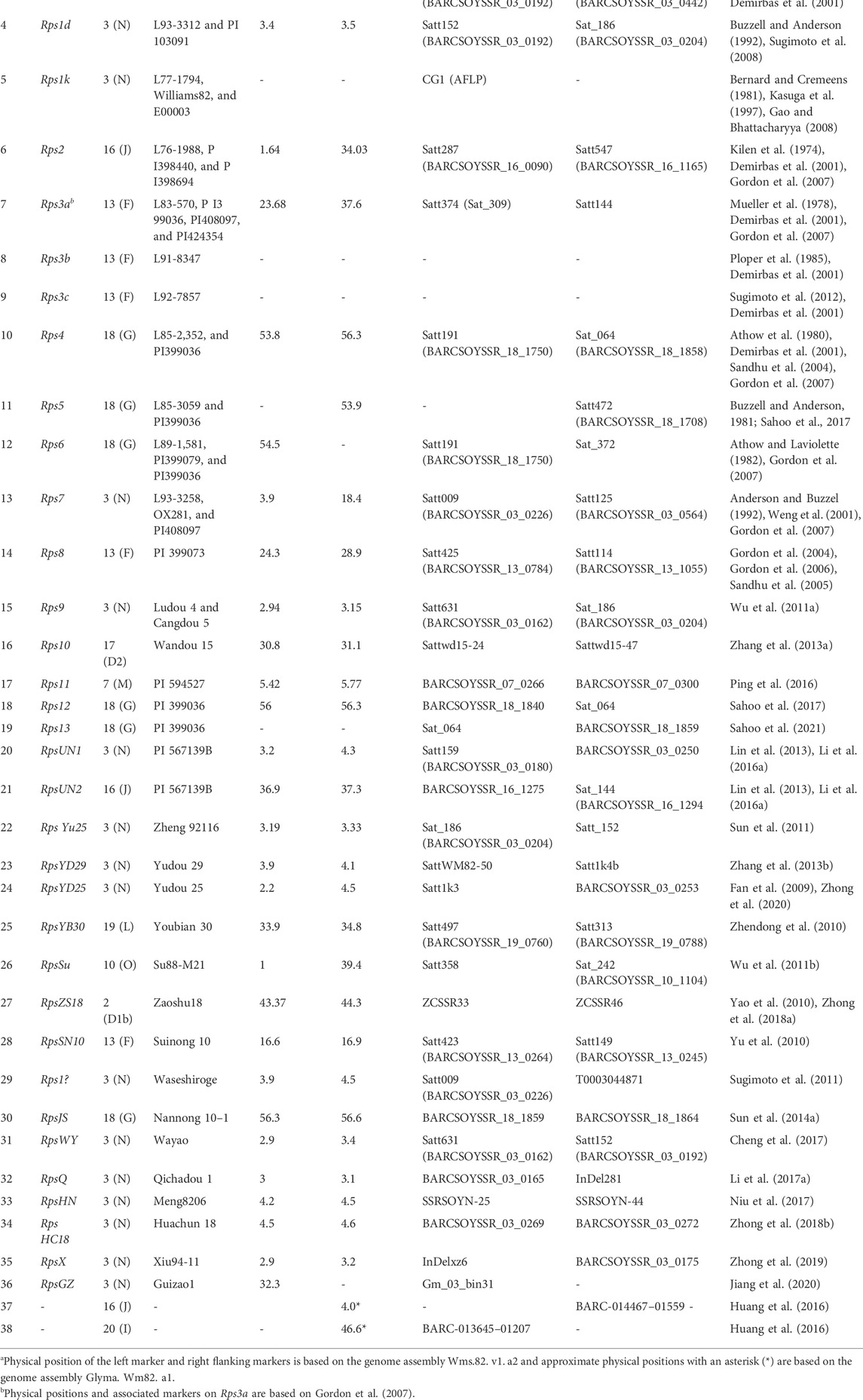
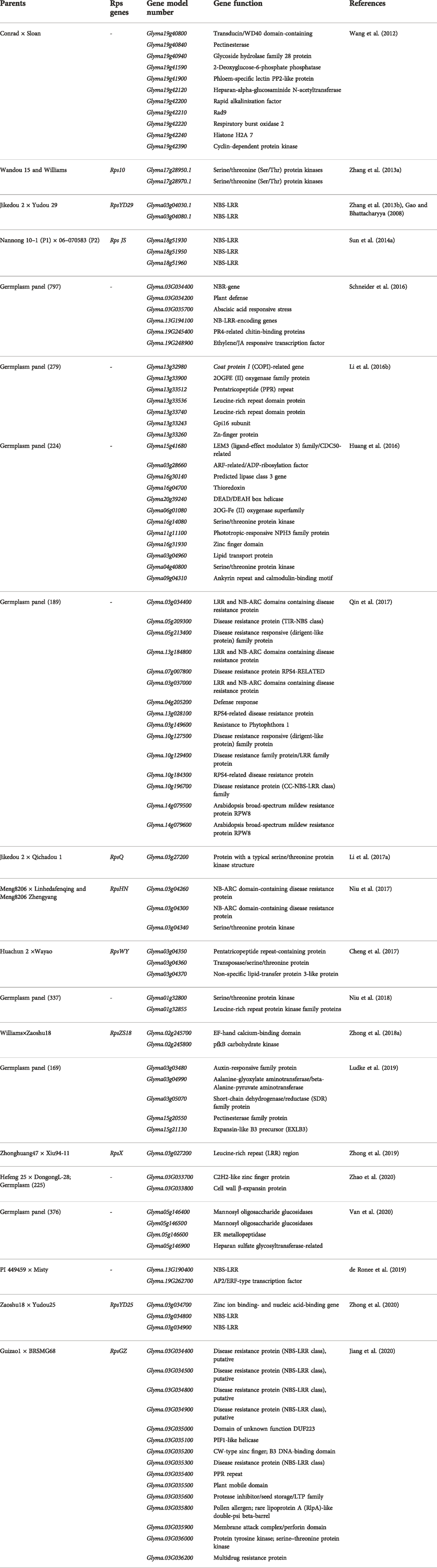
TABLE 1. Details of P. sojae-resistant genes (Rps), their source, chromosomal positions, and associated markers.
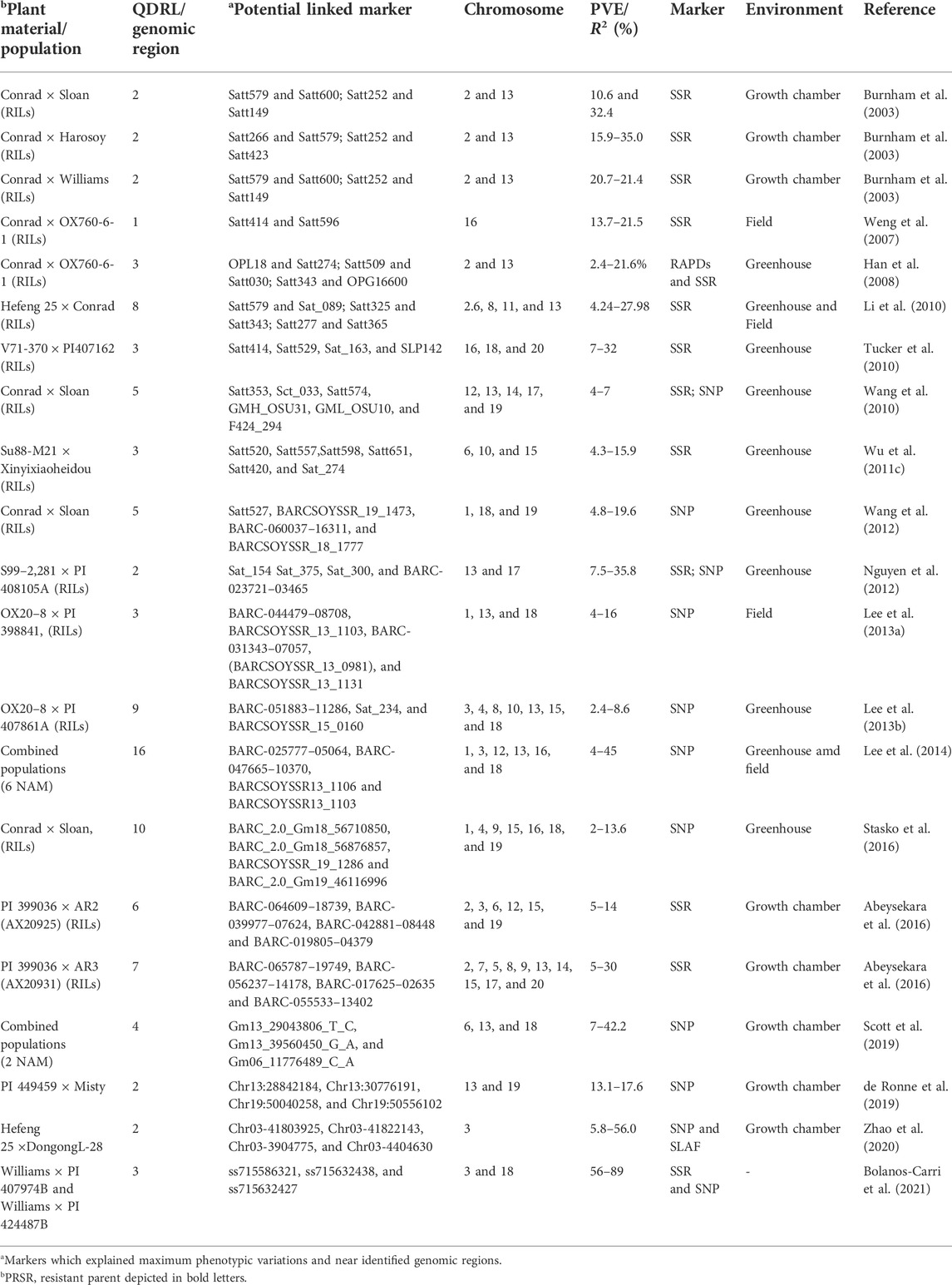
TABLE 2. Details of QDRL for P. sojae resistance identified through the bi-parental mapping approach in soybean.
Genetics of complete resistance versus partial resistance
There are two types of resistance to P. sojae reported in soybean, namely, complete resistance and partial resistance (Sugimoto et al., 2012). Complete resistance is race-specific and exhibits a single dominant resistance gene (Rps) that provides immunity or near immunity, whereas partial resistance is controlled by major and minor genes, and it restricts pathogen colonization and spread (Dorrance et al., 2003, 2004; Sugimoto et al., 2012). Previous studies over the last 2–3 decades identified both complete and partial resistance to P. sojae (Burnham et al., 2003; Dorrance et al., 2004; Sugimoto et al., 2012; Jang and Lee 2020).
During the single dominant gene resistance mechanism against P. sojae, expressed products of Rps genes interact with those of P. sojae through a gene-for-gene interaction and prevent disease development in plants (Hartwig et al., 1968; Schmitthenner, 1999; Burnham et al., 2003; Fan et al., 2009). There are very few reports explaining the detailed expressed products of Rps genes; Gao et al. (2005) mentioned the role of coiled-coil–nucleotide-binding–leucine-rich repeat (CC-NB-LRR)-type proteins in the case of Rps1-k locus ,and Li et al. (2021) demonstrated that E3 ligase GmPUB1 protein is required for the interaction of P. sojae effector protein Avr1b with the resistance of Rps1b and Rps1k in soybean. As an example, during an investigation of the inheritance pattern of Rps genes, Li et al. (2017b) used detached-petiole and hypocotyl inoculation methods in F2 and F2:3 populations derived from a cross “Zhonghuang47” × “Xiu94-11.” A segregation ratio of 3:1 for the resistance and the susceptible reaction indicated a single dominant gene for P. sojae resistance in their study. All the Rps genes provide race-specific and complete resistance with the exception of Rps2, which provides incomplete resistance (Mideros et al., 2007).
Genes for complete resistance
To the best of our knowledge, more than 30 Rps genes/alleles have been reported and are present on 10 different chromosomes in soybean (Table 1). Most of the Rps loci are located on chromosome 3 (14 genes), followed by chromosome 18 (6 genes) and chromosome 13 (5 genes). The Rps genes on these three chromosomes constitute approximately 70% of the total Rps genes reported (Figure 1). Rps1 (with five different alleles, Rps1a, Rps1b, Rps1c, Rps1d, and Rps1k), Rps7, Rps9, RpsYu25, RpsYD29, RpsYD25, RpsUN1, RpsWY, RpsQ, RpsHC18, RpsX, RpsHN, and RpsGZ and an unnamed Rps gene (Rps1?) were mapped on the short arm of chromosome 3 (Figure 1; Table 1). Similarly, Rps4, Rps5, Rps6, Rps12, Rps13, and RpsJS are located on chromosome 18; Rps2, RpsUN2, and one unknown Rps are located on chromosome 16; Rps3 (three alleles, Rps3a, Rps3b, and Rps3c) and RpsSN10 which was linked with Rps8 were mapped on chromosome 13. Furthermore, the remaining genes, namely, RpsZS18, Rps11, RpsSu, Rps10, and RpsYB30, and an unnamed Rps were identified on chromosomes 2, 7, 10, 17, 19, and 20, respectively (Sandhu et al., 2004; Sandhu et al., 2005; Gordon et al., 2007; Yao et al., 2010; Yu et al., 2010; Wu et al., 2011b; Zhang et al., 2013a; Lin et al., 2013; Sun et al., 2014a; Li et al., 2016a; Huang et al., 2016; Ping et al., 2016; Sahoo et al., 2017; Sahoo et al., 2021). A few genomic regions were repeatedly found in many mapping studies using bi-parent populations. For example, on chromosome 3, a genomic region of ∼2 Mb was found to be a hot spot, where major resistance was identified in over 10 investigations using different resistance sources (Figure 1). Zhong et al. (2019) identified RpsX in soybean cultivar Xiu94-11; subsequently, it was revealed that RpsX was located in the 242-kb genomic region spanning the RpsQ locus on chromosome 3. Zhong et al. (2020) fine-mapped RpsYD25 in 1127 F3:4 families derived from “Zaoshu18” and “Yudou25;” subsequently, 7 out of 178 soybean genotypes containing RpsYD25 were identified using five co-segregated SSR markers. Recently, Jiang et al. (2020) have fine-mapped RpsGZ to a 367.371-kb genomic region on chromosome 3 in recombinant inbred lines (RILs) derived from a cross of the resistant cultivar “Guizao1” and the susceptible cultivar “BRSMG68.” Sahoo et al. (2017) identified Rps12 on chromosome 18 in an RIL population developed by crossing the P. sojae resistant cultivar “PI399036” with the susceptible “AR2” line, and this gene was mapped at 2.2 cM proximal to the NBSRps4/6-like sequence that co-segregated with the Phytophthora resistance genes Rps4 and Rps6.
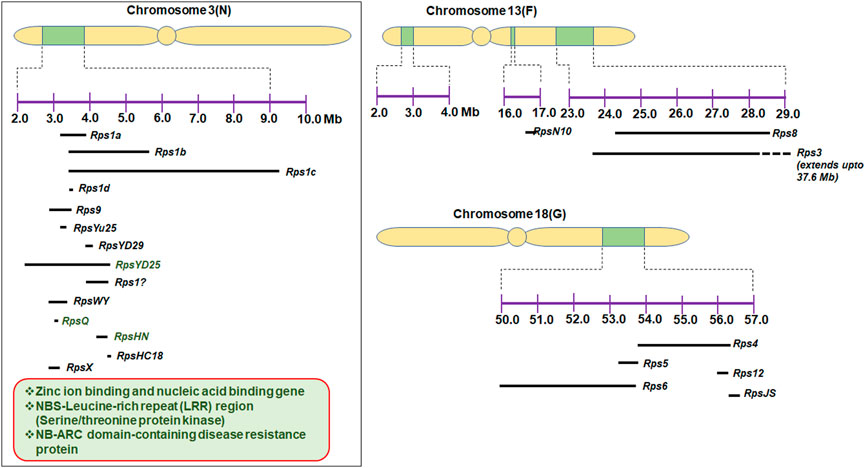
FIGURE 1. Genomic regions of chromosomes 3, 13, and 18, where more than 20 Rps genes were mapped; some potential characterized candidate genes are also depicted on chromosome 3.
In general, Rps gene efficacy is limited to 8–15 years (Grau et al., 2004; Sugimoto et al., 2012). Therefore, continuous efforts are required for the identification of new Rps genes and for the development of PRSR-tolerant cultivars. Under the conditions of high disease pressure, cultivars with complete resistance are far more effective than those having partial resistance to P. sojae (Schmitthenner, 1999; Dorrance et al., 2003). Contradictorily, partial resistance conferred by many QDRL has been found to be durable compared to complete resistance (single Rps gene) in the United States where P. sojae races evolve at a much faster rate to knock down even the most effective Rps genes (Dorrance et al., 2003). This indicates the significance of both complete resistance and partial resistance to P. sojae in different situations.
Quantitative disease resistance loci for partial resistance
Partial resistance to P. sojae is a quantitative trait which is usually race non-specific and provides long-term resistance stability against the pathogen (Schmitthenner 1985; Dorrance et al., 2003; Dorrance et al., 2008; Wang et al., 2012). Partial resistance has moderate to high heritability and thus can be improved through selection pressure. For stable and durable management of PRSR, partial resistance along with complete resistance (Rps genes) may be used, as both types of resistance have different mechanisms to respond to PRSR.
Usually, the levels of partial resistance are evaluated using lesion length measurement, root rot score, tray test, inoculum layer test, or field evaluation (Tooley and Grau 1984; Dorrance et al., 2008). The development of cultivars with increased levels of partial or incomplete resistance needs the identification and characterization of novel resources of partial resistance.
Partial resistance or field resistance to P. sojae is governed by several genomic regions called quantitative trait loci (QTL or alternatively termed QDRL), each contributing a certain magnitude of resistance (Scott et al., 2019). There are a number of resources that have been utilized for mapping QDRL for partial resistance to P. sojae (Table 2). Extensive mapping studies using two contrasting parents in soybean reported about more than 90 QDRL for partial resistance to P. sojae (Table 2; Figure 2). Later on, the large confidence interval spanning genomic regions against P. sojae was further narrowed down through fine-mapping to pinpoint the exact position of QDRL (Huang et al., 2016; Karhoff et al., 2019). The cultivar “Conrad” that does not exhibit Rps genes but shows high partial resistance has been extensively used in QDRL mapping, identifying over 35 QDRL using different bi-parental populations (Burnham et al., 2003; Weng et al., 2007; Han et al., 2008; Li et al., 2010; Wang et al., 2010; Wang et al., 2012; Stasko et al., 2016). Some common QDRL were identified in a “Conrad” × “Sloan” population against three isolates of P. sojae, showing that a common resistance mechanism may occur in response to the individual inoculated isolates (Stasko et al., 2016). The detailed list of recent mapping studies using bi-parental populations leading to the identification of major QDRL along with significant markers imparting partial resistance to P. sojae is given in Table 2. Although over 15 QDRL explained more than 10% phenotypic variance (PV), the majority of QDRL explained <10% of the PV for partial resistance toward PRSR (Table 2) (Burnham et al., 2003; Weng et al., 2007; Han et al., 2008; Li et al., 2010; Nguyen et al., 2012; Lee et al., 2013a; Lee et al., 2014; Abeysekara et al., 2016; de Ronne et al., 2019; Scott et al., 2019). Apart from RILs, nested association mapping (NAM) populations have also been used to map QDRL associated with PRSR (Lee et al., 2014). Recently, Scott et al. (2019) have carried out inoculation of two Soy-NAM populations with P. sojae isolate Win371 for the identification of major QDRL (Figure 2). Four major QDRL were identified by Abeysekara et al. (2016) using RILs derived from “AX20925” (PI 399036 × AR2) and “AX20931” (PI 399036 × AR3). In the latest study, Zhao et al. (2020) identified quantitative trait nucleotides (QTNs) explaining up to 56% PV on chromosome 3 using RILs derived from crossing “DongnongL-28” and “Hefeng 25.”
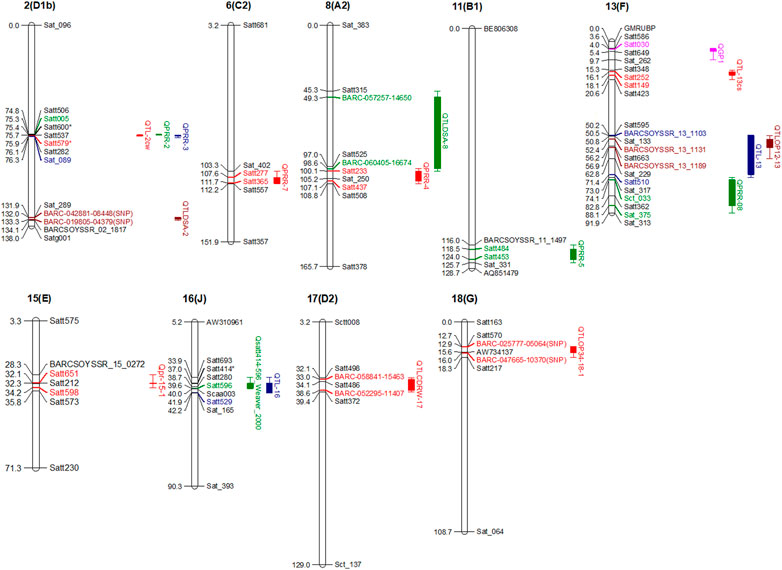
FIGURE 2. Major QDRL (phenotypic variations explaining (PVE) more than 10%) identified for P. sojae resistance along with their flanking makers and chromosomal locations.
Alternatively, another approach, genome-wide association studies (GWAS), provide high-resolution mapping than the traditional bi-parental mapping strategy. In a SSR-based association mapping study among 214 soybean accessions, four SSR alleles, viz., Satt634-133, Satt634-149, Sat_222-168, and Satt301-190, were found to be significantly associated with P. sojae partial resistance (Sun et al., 2014b). Similarly, in another GWAS on resistance to 11 P. sojae isolates involving 224 germplasm accessions, Huang et al. (2016) identified 14 marker–trait associations for PRSR resistance including five novel loci. In USDA soybean germplasm, significant associations were detected for 28 SNPs located on chromosomes 3, 13, and 18 (Chang et al., 2016). The updated information on all GWAS conducted on soybean against PRSR is given in Table 3. The majority of association studies identified SNPs explaining small variations (minor QDRL); however, some of the studies (Ludke et al., 2019; Rolling et al., 2020; Zhao et al., 2020) identified major QDRL explaining variations for PRSR. Ludke et al. (2019) conducted a SNP-based GWAS on 169 soybean cultivars for P. sojae resistance and identified four QDRL on two chromosomes (two each on chromosomes 3 and 15). Interestingly, the identified genomic regions were found to be co-localized with already known and annotated resistance genes. Recently, Rolling et al. (2020) analyzed QDRL in 478 and 495 plant introductions (PIs) against P. sojae isolates OH.121 and C2.S1, respectively, and 24 significant associated SNPs were identified. Five QDRL identified in this study were found to be co-localized with P. sojae meta-QDRL identified from previous bi-parental mapping studies (Rolling et al., 2020). Using available disease phenotypic information, Van et al. (2020) identified 75 novel QTNs using 16 panels consisting of 2,233 soybean accessions. The identified SNPs linked to QDRL can be used in marker-assisted selection for introgression and stacking of partial PRSR resistance loci for imparting durable resistance.
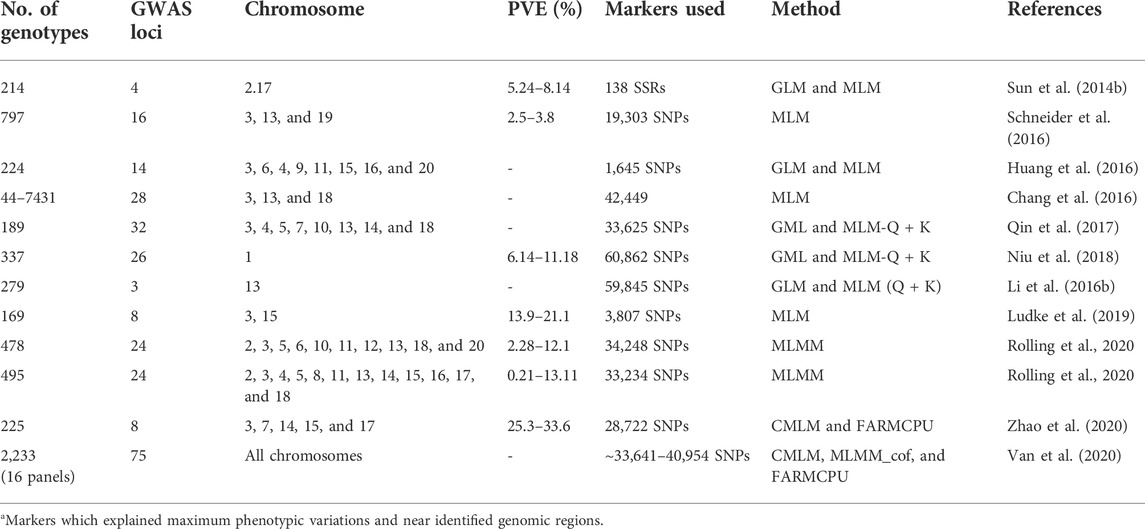
TABLE 3. Details of genomic regions associated with P. sojae resistance identified through the association mapping/GWAS approach in soybean.
Candidate genes for Phytophthora root and stem rot resistance
Characterization of putative genes imparting resistance to P. sojae has also been progressed. Graham et al. (2002) characterized the sequence of the Rps2 genomic region. Rps2 locus sequences included 16 resistance gene homologs with similarities to the TIR/NBD/LRR family of disease resistance genes, a leucine zipper protein, four gene sequences with similarities to Ca2+-binding domains of a calmodulin gene, and three genes with homology to an NtPRp27-like protein (Graham et al., 2002). Sequencing of the Rps1k locus identified a coiled-coil–nucleotide-binding site–leucine-rich repeat (CC-NBS-LRR)-type gene (Bhattacharyya et al., 2005). Further characterization of Rps1k by bacterial artificial chromosome (BAC) sequencing revealed the presence of two nucleotide-binding site–leucine-rich repeat (NBS-LRR)-encoding genes (Rps1k-1 and Rps1k-2) (Gao et al., 2005; Gao and Bhattacharyya, 2008; Sandhu et al., 2009). Sandhu et al. (2004) demonstrated that the deletion of NBSRps4/6 in mutant M1 is correlated with the loss of Rps4 function. With the availability of a complete reference genome sequence, genomic regions of different Rps regions were analyzed for the identification of candidate resistance genes. A list of putative candidate genes for P. sojae resistance is given in Table 4 along with their gene annotations. The maximum number of candidate genes reported are from chromosome 3 (Zhang et al., 2013a; Zhang et al., 2013b; Sun et al., 2014a; Li et al., 2017a; Cheng et al., 2017; Niu et al., 2017; Zhong et al., 2018a; Zhong et al., 2019; Jiang et al., 2020; Zhong et al., 2020). Some of these genes, viz., zinc ion binding- and nucleic acid-binding genes, NB-ARC domain-containing disease resistance proteins, and NBS-LRR genes, were functionally analyzed (Li et al., 2017a; Niu et al., 2017; Zhong et al., 2020) (Figure 1). Li et al. (2016a) reported multiple copies of R-gene-type annotation in RpsUN1 and UN2. Li et al. (2016b) conducted GWAS in an association panel of 279 accessions and identified seven candidate genes on chromosome 13 that are reported to govern natural variations for partial resistance to P. sojae. Unlike Li et al. (2016a), non-NBS-LRR types of genes have also been proposed as candidates for another Rps allele on chromosome 3 (Cheng et al., 2017). Cheng et al. (2017) identified candidate genes against P. sojae using the high-throughput genome-wide sequencing approach by mapping 3,469 recombination bins in RILs. This study revealed the localization of RpsWY gene in bin 401 (on chromosome 3). Bin 401 was found to contain three genes, namely, pentatricopeptide repeat-containing protein, transposase/serine/threonine protein, and non-specific lipid-transfer protein 3-like protein. Sahoo et al. (2017) also reported several NBS-LRR-like genes in genetic investigations of Rps12. Jiang et al. (2020) and Zhong et al. (2020) also reported NBS-LRR and zinc ion-binding genes as candidate genes by fine mapping of RpsYD25 and RpsGZ. Though reference genome sequencing can provide information on the majority of genes present in the identified genomic region, de novo sequencing of the haplotype carrying the target Rps gene is important to identify candidate genes.
Rps 11 showed resistance to 12 races of P. sojae; therefore, it is a broad-spectrum resistance gene (Wang et al., 2021). Wang et al. (2021) sequenced the genome of “PI 594527” by long-read sequencing, and the assembled genome sequence identified that the Rps11 locus was present in a genomic region harboring a cluster of 12 NLR genes of a single origin in soybean. Fine mapping and gene expression analysis pinpointed a 27.7-kb NLR gene (Wang et al., 2021). Genetic transformation of an Rps11-coding DNA sequence in a susceptible soybean genotype conferred a resistant phenotype. Pan-genome analysis revealed that Rps11 is a unique gene in “PI 594527” and does not have any other allelic copy in the other genotypes. The isolation of Rps11 will help soybean breeders accelerate the improvement of broad-spectrum resistance to P. sojae in soybean. The unique structural features of Rps11 make it a suitable model to investigate the resistance mechanism to further improve high-yielding cultivars.
Transcriptomic studies on PRSR
Recent developments in the genomics of P. sojae and soybean have made this pathosystem a model to understand molecular bases underpinning plant–oomycete interactions (Guo et al., 2011). Furthermore, transcriptomics of PRSR resistance in soybean has been extensively carried out to study the candidate genes and the role of biochemical pathways involved in conferring resistance. Through microarray analysis, genes governing pathogenesis-related proteins and enzymes involved in phytoalexin biosynthesis were found to be upregulated and reached a peak at 24 dpi. On the other hand, genes encoding lipoxygenases and peroxidases were found to be downregulated during the infection process (Moy et al., 2004).
To gain deep insights into the molecular basis of resistance to P. sojae, differential gene expression in response to P. sojae infection in the cultivar “Suinong 10” was studied by Xu et al. (2012). A total of eight transcripts were found to be upregulated in the treated plants as compared to those of the control. These transcripts are responsible for enzymes involved in the phytoalexin biosynthesis pathway and pathogenesis-related proteins and some defense response-related proteins such as phenylalanine ammonia-lyase, WRKY transcription factor 31, isoflavone reductase, pleiotropic drug resistance protein 12, and major allergen Pru ar 1 (Xu et al., 2012). Molecular responses induced by different Rps genes and the association of phytohormone signaling pathways with disease reactions to P. sojae infection were studied by Lin et al. (2014). Transcriptome analysis on 10 near-isogenic lines (NILs) (Rps1-a, 1-b, 1-c, 1-k, Rps3-a, 3-b, 3-c, Rps4, 5, and 6, each in the genetic background of “Williams”) and the susceptible genotype, “Williams” during pre- and post-inoculation was carried out to identify differentially expressed genes (DEGs) across different treatments (Lin et al., 2014). A total of 5,806 incompatible interaction genes (IIGs) were identified by comparing DEGs between “Williams” and NILs, and 1,139 compatible interaction genes (CIGs) were identified in “Williams.” Of these 5,806 IIGs, 23 were found to be common across 10 NILs and are mostly associated with biotic and abiotic stress responses, suggesting the overlap of molecular responses induced by different Rps genes. Two NPR-1-like IIGs, Glyma02g45260 and Glyma14g03510, were involved in mediating the SA signaling pathway during an incompatible reaction, suggesting the role of the SA pathway in genetic resistance. Several JAZ-like proteins that repress the jasmonic acid (JA) pathway were found, such as IIGs and/or CIGs. These proteins were downregulated in NILs and were upregulated in “Williams.” Also, a JAR1 homolog, Glyma07g06370 that activates the JA signaling pathway, was upregulated during the susceptible reaction in Williams. Genes that repress the ethylene (ET) pathway were found to be downregulated in NILs and upregulated in “Williams,” suggesting that the ET pathway was repressed during the susceptible host reaction in “Williams” and activated in NILs during the incompatible reaction. In addition, three BAK1 homolog IIGs that activate brassinosteroid (BR) signaling were found to be upregulated in NILs, suggesting the role of the BR signaling pathway during defense against P. sojae.
Role of transcription factors
Transcription factors (TFs) are master switches for regulating the expression of genes and controlling several signaling pathways (Chattopadhyay et al., 2019) and also play a vital role in different defense mechanisms in different plant species against different phytopathogens. In soybean, several TFs have been identified for their role in regulating genes and pathways involved in resistance to P. sojae. A bHLH (basic helix–loop-helix) transcription factor associated with resistance to P. sojae was functionally characterized through its hypo- and hyper-expression in a resistant soybean genotype, “L77-1863,” and was designated as GmPIB1. GmPIB1 represses the expression of the GmSPOD1 gene by directly binding to its promoter. Through RNAi assay, it was found that GmSPOD1 is involved in the production of reactive oxygen species (ROS) during P. sojae infection. Hence, the role of GmPIB1 TF in P. sojae resistance through reduced ROS production has been established (Cheng et al., 2018). Several ethylene-responsive element binding factor (ERF) transcription factors are linked with disease resistance in different plants (Gu et al., 2000; Song et al., 2005). An ERF-associated amphiphilic repression (EAR) motif-containing ERF TF, GmERF5, conferring resistance to P. sojae through the positive regulation of pathogenesis-related (PR) genes, PR10, PR1-1, and PR10-1, has been identified (Dong et al., 2015). A TF gene, GmWRKY40, was found to impart resistance in soybean to P. sojae and acts as a positive regulator of ROS accumulation and the JA signaling pathway (Cui et al., 2019). A transcription factor, GmMYB29A2, was found to impart resistance to P. sojae infection in soybean through the regulation of glyceollin I accumulation (Jahan et al., 2020). WRKY transcription factor 31 identified in response to P. sojae infection (Xu et al., 2012) was functionally characterized through overexpression and RNAi silencing (Fan et al., 2017). Gene GmWRKY31 interacts with GmHDL56 and jointly engages in the activation of GmNPR1, which in turn manifests resistance during the Suinong 10–P. sojae interaction. Another TF, GMERF113, was isolated from “Suinong 10” and characterized for its response to P. sojae infection in a susceptible genotype “Dongnog 5.” The overexpression of GMERF113 in this genotype resulted in an enhanced resistance level and expression of pathogenesis-related genes, PR1 and PR10-1. Thus, the role of GMERF113 in the defense mechanism through positive regulation of these two pathogenesis-related genes has been well-demonstrated (Zhao et al., 2017).
Role of enzymes and proteins
Fan et al. (2015) studied the expression of class 10 protein Gly m 4l and found its role in the resistance to P. sojae. Zhang et al. (2017) identified a phenylalanine ammonia-lyase (PAL) gene family member, GmPAL2.1, to be linked with resistance to P. sojae through reverse genetics. The role of enzyme class 4-coumarate: CoA ligase (4CL) in plant defense against pathogens has been investigated extensively (Ehlting et al., 1999). A member of the 4CL (enzyme 4-coumarate: CoA ligase) gene family, GmPI4L, identified in soybean is associated with resistance to P. sojae infection through the enhanced production of glyceollins, genistein, and daidzein in soybean, laying the foundation for the enzymatic basis for resistance to this pathogen (Chen et al., 2019). The mediator complex is a part of RNA polymerase II, which acts as a regulatory element of the transcription process. A mediator subunit, GmMED16-1 in soybean, through its silencing, was found to govern P. sojae by modulating the transcription of NPR1, PR1a, and PR5 genes (Xue et al., 2019).
Role of miRNAs
MicroRNAs (miRNAs) are also known to be regulated under defense mechanisms in several plant species. Guo et al. (2011) revealed the role of miRNA in P. sojae resistance. Wong et al. (2014) identified miR393 and miR166, as triggered by heat-inactivated P. sojae hyphae, suggesting their roles in soybean basal defense. Knockdown of miR393 led to the increased susceptibility of soybean to P. sojae. The expression of iso-flavonoid synthesis genes was drastically reduced in miR393 knockdown roots, suggesting that miR393 promotes soybean defense against P. sojae.
Molecular breeding for resistance to P. sojae
Soybean witnessed a significant improvement in yields over the past 60 years through conventional breeding approaches. Soybean yields were estimated to improve at the rate of 23 kg/ha/annum (Specht et al., 1999), and Wilcox (2001) reported an increase of 60% in seed yields over the past 60 years in the United States of America. The significant increase in yields has been witnessed mainly due to the toiling efforts of conventional breeding-based public sector soybean breeding programs. But considering the limitations of conventional breeding methods for P. sojae resistance improvement, further progress for yield enhancement is stagnated at the global level. The stagnated progress due to P. sojae infection can be further brought back to an accelerated track by the adoption of MAS and genomics-aided approaches in the PRSR resistance soybean breeding programs.
Molecular markers ranging from hybridization (RFLP and AFLP) and polymerase chain reaction-based markers (SSRs) to sequencing-based markers (SNPs) have been used to a greater extent for high-resolution mapping as well as for fine mapping of genomic regions governing the resistance to P. sojae (Table 2).
The identified major genomic regions for P. sojae resistance can be introgressed into elite soybean cultivars through the use of genomics-assisted breeding techniques, viz., marker-assisted backcross breeding (MABB), marker-assisted recurrent selection (MARS), marker-assisted gene pyramiding (MAGP), and genomic selection (GS). The identified major QDRL can be targeted for introgression into elite cultivars using the MABB approach (Ribaut and Ragot, 2007; Choudhary et al., 2019). Selection of Rps gene for introgression is also very important as it depends on particular regions of cultivation. Dorrance et al. (2016) estimated pathotype variability in 11 different states of the US with 873 isolates and concluded that Rps 6 and Rps 8 are more effective against the majority of isolates collected from northern regions. Several efforts have been made for the introgression of single-gene (Rps)-mediated resistance into soybean cultivars for controlling PRSR (Roth et al., 2020). Six of these genes (Rps1a, Rps1b, Rps1c, Rps1k, Rps3a, Rps 6, and Rps3a) already exist in commercial varieties and provide disease management against Phytophthora root and stem rot (Roth et al., 2020), which were transferred with the help of conventional approaches. In Japan, “Hyogo Prefecture,” the black-seeded PRSR-resistant line, was used as the donor for introgression and for the development of resistant cultivars (Sugimoto et al., 2010). Although plant breeders use MAS-based approaches mainly for transferring Rps genes in soybean (Li et al., 2010; Ramalingam et al., 2020), due to high disease pressure, rapid evolution in the pathotypes of P. sojae has been witnessed over the past 3 decades, hence making vertical resistance ineffective. This forced the plant breeders to target partial resistance for the effective and sustainable management of PRSR (Schmitthenner, 1985). Studies on mapping QDRL dissected the genetic basis of partial resistance to P. sojae and revealed small-to-moderate effect QDRL, many of which individually explained less than 10% of phenotypic variance for PRSR in a population (Table 2). The difficulty of identifying small-effect QDRL in small mapping populations can be resolved by deploying joint linkage QDRL analysis of multiple populations (Lander and Kruglyak, 1995; Beavis 1998). Although relatively less, a good number of major QDRL have been mapped for PRSR partial resistance in soybean (Figure 2).
The utilization of MABB is restricted to the introgression of major QDRL only, which have more PVE (phenotypic variance explanation) percentages and limited localization in the genome, as it is very difficult to follow a large number of QDRL during introgression programs. Hence, other molecular breeding approaches such as MAGP, MARS, and genomic selection can serve as a good alternative for accumulating favorable QDRL (minor and major effects) for PRSR resistance. Pyramiding of PRSR-resistant QDRL was demonstrated by Li et al. (2010) by targeting seven consistent QDRL (detected across multiple environments) from two different donors (“Conrad” and “Hefeng 25”). Limited efforts of QDRL stacking for PRSR resistance revealed a significant increase in the tolerance level of introgressed lines, and the tolerance level against PRSR was found to be positively correlated with the number of QDRL stacked (Li et al., 2010). Recently, Karhoff et al. (2019) demonstrated the genetic gains from selections of a major QTL for partial resistance to P. sojae. The introgression of a resistance allele from the respective “PI 427105B” and “PI 427106” improved the genetic levels of resistance to P. sojae by ∼20% and ∼40%, respectively, and the yield by 13%–29% under diseased conditions (Karhoff et al., 2019). These are a few examples of PRSR resistance introgression through molecular breeding, demonstrating the fruitful results of genetic and genomic mapping for PRSR resistance. Dorrance et al. (2016) emphasized on stacking of Rps genes with strong partial resistance for limiting the loss caused by PRSR. With the new genomics-assisted breeding approaches, it will be practically more feasible and applicable in stacking of major genes for complete resistance and multiple QDRL of partial resistance for imparting sustainable PRSR resistance in soybean cultivars.
Genome editing for understanding PRSR resistance
Not only naturally available and induced mutations are the source for introducing new resistance genes in crop improvement programs, but also genetic engineering and gene editing (genome editing) are techniques that enable precise and targeted modifications. Now, gene-editing technologies are gaining momentum for crop improvement as they are more similar to the widely accepted “mutation breeding” technology.
CRISPR/Cas9 gene editing is particularly useful in deciphering the plant–pathogen interaction and understanding effector-triggered immunity. Pathogen avirulence (Avr) effectors interplay with corresponding plant resistance (R) proteins and activate robust immune responses in the host plant. Avr4/6, an RxLR effector gene of P. sojae, which is recognized by soybean R-genes (Rps6 and Rps4), was edited using CRISPR/Cas9 technology to study its possible role in pathogenicity (Fang and Tyler 2016). This study validated the contribution of Avr4/6 in pathogen recognition by soybean R-gene loci, Rps4 and Rps6. Ochola et al. (2020) engineered the promoter region of PsAvr3b gene which is recognized by Rps3b, and mutants with low PsAvr3b expression successfully colonized soybean plants carrying the cognate R-gene Rps3b. Wang L et al. (2020) edited PsSu(z)12 gene associated with effector locus Avr1b. PsSu(z)12 is epigenetically governed and encodes a core subunit of the H3K27me3 methyltransferase complex. CRISPR/Cas9-mediated H3K27me3 depletion within the Avr1b genomic region was correlated with impaired Avr1b gene silencing, and the mutants lost their ability to evade immune recognition by soybeans carrying Rps1b (Wang P et al., 2020). Tan et al. (2020) studied knockout mutants of P. sojae generated via the CRISPR/Cas9 system for the PsGH7a (GH7 family cellobiohydrolase) gene, and the mutants were found to have reduced virulence on susceptible soybean as compared to wild-type strain “P6497.” It is expected that in the future, the CRISPR/Cas9 system coupled with other genomic techniques will be an important approach to create disease-resistant cultivars that can withstand biotic stresses (Kumar et al., 2020).
Challenges and future perspectives
The urgency and significance of P. sojae-resistant cultivar development can be realized from its vast spread and rapid occurrence of the disease across soybean-growing areas. This demands a strong emphasis on strengthening P. sojae resistance soybean breeding programs globally. Although significant progress has been made through the utilization of race-specific resistance genes (Rps genes), the rapid evolution of pathotypes in P. sojae resulted in resistance breakdown. This problem was quickly assessed by soybean breeders and, hence, shifted the focus to partial resistance (horizontal resistance) which provides relatively broad and highly durable resistance. Extensive genetic and genomics studies identified several major genes and QDRL for P. sojae resistance. The Rps-linked markers can be utilized in the selection of genotypes having PRSR resistance genes in early stages, and subsequent backcrossing will enable the rapid development of PRSR-resistant soybean cultivars. Marker-assisted breeding approaches such as MAGP can help in pyramiding vertical and horizontal resistance by the utilization of major resistance genes and QDRL identified in different genetic backgrounds. This strategy of combining complete and partial resistance in the same cultivars will prove to be the most effective approach in the near future. Soybean breeders need to continuously identify novel and unique resistance genes to cope with the emerging new pathotypes (Sugimoto et al., 2011). Though it is challenging to incorporate a large number of genes and QDRL from multiple genetic backgrounds into a single background using MABB, MARS and genomic selection can be used in resistance breeding programs to incorporate all PRSR resistance loci for durable resistance. It will be useful to mine the germplasm and geographical regions with enormous diversity for the presence of resistance to prevailing P. sojae pathotypes. For example, soybean germplasm collections in the Republic of Korea have greater variability for resistance to P. sojae for specific Rps loci, as well as partial resistance (Dorrance and Schmitthenner, 2000), and can be used for incorporating durable resistance through large-scale breeding programs. Emerging approaches such as gene discovery through re-sequencing, proteomics, metabolomics, RNA-seq, and exome sequencing of soybean and its wild relatives need to be exploited at a broader level. Furthermore, the QTL-seq approach will likely augment the rapid identification of novel QDRL and advancement of selected progenies for cultivar improvement (Zhang et al., 2018). The CRISPR/Cas9-mediated identification of effector-triggered immunity and R-gene editing is a highly targeted approach for the understanding and rapid development of PRSR resistance. Thus, different “Omics” approaches may be employed to explore the plant defense mechanisms in plant–pathogen interactions along with a gene-editing approach. In addition to the genetic improvement of cultivars for PRSR resistance, other alternative approaches need to be adopted and integrated to achieve prolonged resistance. Such approaches include the identification of effective compounds such as calcium that could help control the PRSR to certain levels (Sugimoto et al., 2010). Since the roots are primary targets for PRSR infection, the extensive comparative study of root traits in wild relatives or resistant cultivars to those of susceptible cultivars will help in the identification of certain target traits for phenotyping and resistance management. For such studies, phenotyping platforms that help in better visualization of root system architecture should be given high priority. The combined approach of genetic resistance, integrated disease management, and climate-smart agronomic practices can pave the path for the sustainable management of PRSR in soybean.
Author contributions
SC and MC initiated the idea and drafted the manuscript. PB, VN, GK, and JC performed the literature search and conducted data analysis. SW, HS, MR, and SG critically revised the manuscript.
Acknowledgments
The authors thank the Indian Council of Agricultural Research and the Department of Biotechnology, Govt. of India, for encouragement and support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abeysekara, N. S., Matthiesen, R. L., Cianzio, S. R., Bhattacharyya, M. K., and Robertson, A. E. (2016). Novel sources of partial resistance against Phytophthora sojae in soybean PI 399036. Crop Sci. 56, 2322–2335. doi:10.2135/cropsci2015.09.0578
Abney, T., Melgar, J., Richards, T., Scott, D., Grogan, J., and Young, J. (1997). New races of Phytophthora sojae with Rps 1-d virulence. Plant Dis. 81, 653–655. doi:10.1094/PDIS.1997.81.6.653
Allen, T. W., Bradley, C. A., Sisson, A. J., Byamukama, E., Chilvers, M. I., Coker, C. M., et al. (2017). Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 18, 19–27. doi:10.1094/PHP-RS-16-0066
Anderson, J. P., Gleason, C. A., Foley, R. C., Thrall, P. H., Burdon, J. B., and Singh, K. B. (2010). Plants versus pathogens: An evolutionary arms race. Funct. Plant Biol. 37 (6), 499–512. doi:10.1071/FP09304
Anderson, T. R., and Buzzell, R. I. (1992). Diversity and frequency of races of Phytophthora megasperma f. sp. glycinea in soybean fields in Essex County, Ontario, 1980-1989. Plant Dis. 76, 587–589. doi:10.1094/PD-76-0587
Athow, K. L., Laviolette, F. A., and Mueller, E. H. (1980). A new major gene for resistance to Phytophthora megasperma var sojae in soybean. Phytopathology 70, 977–980. doi:10.1094/phyto-70-977
Athow, K. L., and Laviolette, F. A. (1982). Rps 6, a major gene for resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology 72, 1564–1567. doi:10.1094/phyto-72-1564
Bandara, A. Y., Weerasooriya, D. K., Bradley, C. A., Allen, T. W., and Esker, P. D. (2020). Dissecting the economic impact of soybean diseases in the United States over two decades. PLOS one 15 (4), e0231141. doi:10.1371/journal.pone.0231141
Beavis, W. D. (1998). “QTL analyses: Power, precision, and accuracy,” in Molecular dissection of complex traits. Editor A. H. Paterson (Boca Raton, US App: CRC Press), 145–162.
Bernard, R. L., and Cremeens, C. R. (1981). An allele at the rps1 locus from the variety 'Kingwa. Soybean Genet. Newsl. 8, 14. Available at: https://lib.dr.iastate.edu/soybean.genetics/vol8/iss1/14.
Bernard, R. L., Smith, P. E., Kaufmann, M. J., and Schmitthenner, A. F. (1957). Inheritance of resistance to Phytophthora root and stem rot in the soybean 1. Agron. J. 49, 391. doi:10.2134/agronj1957.00021962004900070016x
Bhattacharyya, M. K., Narayanan, N. N., Gao, H. O., Santra, D. K., Salimath, S. S., Kasuga, T., et al. (2005). Identification of a large cluster of coiled coil-nucleotide binding site–leucine rich repeat-type genes from the Rps1 region containing Phytophthora resistance genes in soybean. Theor. Appl. Genet. 111, 75–86. doi:10.1007/s00122-005-1993-9
Bolanos-Carriel, C., Batnini, A., McHale, L. K., and Dorrance, A. (2021). Identification of resistance loci towards Phytophthora sojae (Rps) in South Korean soybean plant introductions 407974B and 424487B. Crop Sci 62, 275–285. doi:10.1002/csc2.20596
Burnham, K. D., Dorrance, A. E., VanToai, T. T., and St Martin, S. K. (2003). Quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Crop Sci. 43, 1610–1617. doi:10.2135/cropsci2003.1610
Buzzell, R. I., and Anderson, T. R. (1981). Another major gene for resistance to Phytophthora megasperma var. sojae in soybean. Soybean Genet. Newsl. 18, 30–33. Available at: http://lib.dr.iastate.edu/soybeangenetics/vol8/iss1/10.
Buzzell, R. I., and Anderson, T. R. (1992). Inheritance and race reaction of a new soybean Rps1 allele. Plant Dis. 76, 600–601. doi:10.1094/PD-76-0600
Chang, H. X., Lipka, A. E., Domier, L. L., and Hartman, G. L. (2016). Characterization of disease resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Phytopathology 106 (10), 1139–1151. doi:10.1094/PHYTO-01-16-0042-FI
Chattopadhyay, A., Purohit, J., Tiwari, K. K., and Deshmukh, R. (2019). Targeting transcription factors for plant disease resistance: Shifting paradigm. Curr. Sci. 117 (10), 1598–1607. doi:10.18520/cs/v117/i10/1598-1607
Chen, Q., Weng, Q., Wang, Y., and Zheng, X. (2004). Identification and sequencing of ribosomal DNA-ITS of Phytophthora sojae in Fujian. Acta Phytopathol. Sin. 34, 112–116.
Chen, X., Fang, X., Zhang, Y., Wang, X., Zhang, C., Yan, X., et al. (2019). Overexpression of a soybean 4-coumaric acid: coenzyme A ligase (GmPI4L) enhances resistance to Phytophthora sojae in soybean. Funct. Plant Biol. 46, 304–313. doi:10.1071/FP18111
Cheng, Q., Dong, L., Gao, T., Liu, T., Li, N., Wang, L., et al. (2018). The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 69 (10), 2527–2541. doi:10.1093/jxb/ery103
Cheng, Y., Ma, Q., Ren, H., Xia, Q., Song, E., Tan, Z., et al. (2017). Fine mapping of a Phytophthora-resistance gene RpsWY in soybean (Glycine max L.) by high-throughput genome-wide sequencing. Theor. Appl. Genet. 130, 1041–1051. doi:10.1007/s00122-017-2869-5
Choudhary, M., Wani, S. H., Kumar, P., Bagaria, P. K., Rakshit, S., Roorkiwal, M., et al. (2019). QTLian breeding for climate resilience in cereals: Progress and prospects. Funct. Integr. Genomics 19, 685–701. doi:10.1007/s10142-019-00684-1
Costamilan, L. M., Clebsch, C. C., Soares, R. M., Seixas, C. D. S., Godoy, C. V., and Dorrance, A. E. (2013). Pathogenic diversity of Phytophthora sojae pathotypes from Brazil. Eur. J. Plant Pathol. 135, 845–853. doi:10.1007/s10658-012-0128-9
Cui, X., Yan, Q., Gan, S., Xue, D., Wang, H., Xing, H., et al. (2019). GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L, enhanced the resistance to. BMC Plant Biol. 19, 598. doi:10.1186/s12870-019-2132-0
de Ronne, M., Labbé, C., Lebreton, A., Sonah, H., Deshmukh, R., Jean, M., et al. (2019). Integrated QTL mapping, gene expression and nucleotide variation analyses to investigate complex quantitative traits: A case study with the soybean– Phytophthora sojae interaction. Plant Biotechnol. J. 18, 1492–1494. doi:10.1111/pbi.13301
Demirbas, A., Rector, B. G., Lohnes, D. G., Fioritto, R. J., Graef, G. L., Cregan, P. B., et al. (2001). Simple sequence repeat markers linked to the soybean Rps genes for Phytophthora resistance. Crop Sci. 41, 1220–1227. doi:10.2135/cropsci20014141220x
Dong, L., Cheng, Y., Wu, J., Cheng, Q., Li, W., Fan, S., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66 (9), 2635–2647. doi:10.1093/jxb/erv078
Dorrance, A. E., Berry, S. A., Anderson, T. R., and Meharg, C. (2008). Isolation, storage, pathotype characterization, and evaluation of resistance for Phytophthora sojae in soybean. Plant Health Prog. 10, 1094–1101. doi:10.1094/PHP-2008-0118-01-DG
Dorrance, A. E., Jia, H., and Abney, T. S. (2004). Evaluation of soybean differentials for their interaction with Phytophthora sojae. Plant Health Prog. 5, 9. doi:10.1094/PHP-2004-0309-01-RS
Dorrance, A. E., McClure, S. A., and St Martin, S. K. (2003). Effect of partial resistance on Phytophthora stem rot incidence and yield of soybean in Ohio. Plant Dis. 87, 308–312. doi:10.1094/PDIS2003873308
Dorrance, A. E., and Schmitthenner, A. F. (2000). New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis. 84, 1303–1308. doi:10.1094/PDIS200084121303
Dorrance, A., Kurle, J., Robertson, A., Bradley, C., Giesler, L., Wise, K., et al. (2016). Pathotype diversity of Phytophthora sojae in eleven states in the United States. Plant Dis. 100, 1429–1437. doi:10.1094/PDIS-08-15-0879-RE
Ehlting, J., Büttner, D., Wang, Q., Douglas, C. J., Somssich, I. E., and Kombrink, E. (1999). Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionary classes in angiosperms. Plant J. 19, 9–20. doi:10.1046/j1365-313X199900491x
Fan, A., Wang, X., Fang, X., Wu, X., and Zhu, Z. (2009). Molecular identification of Phytophthora resistance gene in soybean cultivar yudou 25. A. A. S. 35, 1844–1850. doi:10.3724/SPJ1006200901844
Fan, S., Dong, L., Han, D., Zhang, F., Wu, J., Jiang, L., et al. (2017). GmWRKY31 and GmHDL56 enhances resistance to Phytophthora sojae by regulating defense-related gene expression in soybean. Front. Plant Sci. 8, 781. doi:10.3389/fpls.2017.00781
Fan, S., Jiang, L., Wu, J., Dong, L., Cheng, Q., Xu, P., et al. (2015). A novel pathogenesis related class 10 protein glym 4l, increases resistance upon Phytophthora sojae infection inSoybean (Glycine max [L] merr). PLoS ONE 10 (10), e0140364. doi:10.1371/journal.pone.0140364
Fang, Y., and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in theoomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17, 127–139. doi:10.1111/mpp12318
FAOSTAT (2019). Available at: https://www.fao.org/faostat/en/#data/QCL.
Gao, H., Narayanan, N. N., Ellison, L., and Hattacharyya, M. K. (2005). Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant. Microbe. Interact. 18, 1035–1045. doi:10.1094/MPMI-18-1035
Gao, H. Y., and Bhattacharyya, M. K. (2008). The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 8, 29. doi:10.1186/1471-2229-8-29-2229-8-29
Gordon, S. G., St Martin, S. K., and Dorrance, A. E. (2004). Mapping Rps8, a gene for resistance to Phytophthora root and stem rot in soybean. Madison, WI: Crop Sci Soc America Annu Mtg, ASA-CSA-SS, 320.
Gordon, S. G., St Martin, S. K., and Dorrance, A. E. (2006). Rps 8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci. 46, 168–173. doi:10.2135/cropsci200404-0024
Gordon, S., Kowitwanich, K., Pipatpongpinyo, W., St Martin, S., and Dorrance, A. (2007). Molecular marker analysis of soybean plant introductions with resistance to Phytophthora sojae. Phytopathology 97, 113–118. doi:10.1094/PHYTO-97-0113
Graham, M. A., Marek, L. F., and Shoemaker, R. C. (2002). Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 162, 1961–1977. doi:10.1093/genetics/162.4.1961
Grau, C. R., Dorrance, A. E., Bond, J., and Russin, J. S. (2004). “Fungal diseases,” in Soybeans: Improvement, production, and uses. Editors H. R. Boerma, and J. E. Specht. 3rd edn. (American Soc Agron MA), 679–763.
Gu, Y. Q., Yang, C., Thara, V. K., Zhou, J., and Martin, G. B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12, 771–786. doi:10.1105/tpc.12.5.771
Guo, N., Ye, W. W., Wu, X. L., Shen, D. Y., Wang, Y. C., Xing, H., et al. (2011). Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome 54 (11), 954–958. doi:10.1139/g11-050
Han, Y., Teng, W., Yu, K., Poysa, V., Anderson, T., Qiu, L., et al. (2008). Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica 162, 231–239. doi:10.1007/s10681-007-9558-4
Hartman, G. L., and Hill, C. B. (2010). “Diseases of soybean and their management,” in The soybean: Botany, production, and uses. Editor G. Singh (Cambridge, MA: CAB International), 276–299.
Hartwig, E. E., Keeling, B. L., and Edwards, C. J. (1968). Inheritance of reaction to Phytophthora rot in the soybean. Crop Sci. 8, 634–636. doi:10.2135/cropsci1968.0011183x000800050042x
Huang, J., Guo, N., Li, Y., Sun, J., Hu, G., Zhang, H., et al. (2016). Phenotypic evaluation and genetic dissection of resistance to Phytophthora sojae in the Chinese soybean mini core collection. BMC Genet. 17, 85. doi:10.1186/s12863-016-0383-4
Jahan, M. A., Harris, B., Lowery, M., Infante, A. M., Percifield, R. J., and Kovinicha, N. (2020). Glyceollin transcription factor GmMYB29A2 regulates soybean resistance to Phytophthora sojae. Plant Physiol. 183, 530–546. doi:10.1104/pp1901293Available at: wwwplantphysiolorg/cgi/doi/101104/pp1901293.
Jang, I. H., and Lee, S. (2020). A review and perspective on soybean (Glycine max L) breeding for the resistance to Phytophthora sojae in Korea. Plant Breed. Biotechnol. 8 (2), 114–130. doi:10.9787/pbb.2020.8.2.114
Jee, H., Kim, W., and Cho, W. (1998). Occurrence of Phytophthora root rot on soybean (Glycine max) and identification of the causal fungus RDA. J. Crop Prot. 40, 16–22.
Jiang, B., Cheng, Y., Cai, Z., Li, M., Jiang, Z., Ma, R., et al. (2020). Fine mapping of a Phytophthora-resistance locus RpsGZ in soybean using genotyping-by-sequencing. BMC Genomics 21 (1), 280. doi:10.1186/s12864-020-6668-z
Kang, I. J., Kang, S., Jang, I. H., Jang, Y. W., Shim, H. K., Heu, S., et al. (2019). Identification of new isolates of Phytophthora sojae and the reactions of Korean soybean cultivars following hypocotyl inoculation. Plant Pathol. J. 35, 698–704. doi:10.5423/PPJ.NT.09.2019.0249
Karhoff, S., Lee, S., Mian, R., Ralston, T. I., Niblack, T. L., Dorrance, A. E., et al. (2019). Phenotypic characterization of a major quantitative disease resistance locus for partial resistance to Phytophthora sojae. Crop Sci. 59, 968–980. doi:10.2135/cropsci2018.08.0514
Kasuga, T., Salimath, S. S., Shi, J., Gijzen, M., Buzzell, R. I., and Bhattacharyya, M. K. (1997). High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol. Plant. Microbe. Interact. 10 (9), 1035–1044. doi:10.1094/mpmi.1997.10.9.1035
Kaufmann, M. J. (1957). Root and stem rot of soybean caused by Phytophthora sojae n. sp. Champaign: University of Illinois at Urbana-Champaign.
Kilen, T. C., Hartwig, E. E., and Keeling, B. L. (1974). Inheritance of a second major gene for resistance to Phytophthora rot in soybeans 1. Crop Sci. 14 (2), 260–262. doi:10.2135/cropsci1974.0011183x001400020027x
Kumar, K., Gambhir, G., Dass, A., Tripathi, A. K., Singh, A., Jha, A. K., et al. (2020). Genetically modified crops: Current status and future prospects. Planta 251 (4), 91–27. doi:10.1007/s00425-020-03372-8
Kumawat, G., Gupta, S., Ratnaparkhe, M. B., Maranna, S., and Satpute, G. K. (2016). QTLomics in Soybean: a way forward for translational genomics and breeding. Frontiers in Plant Science, 7–1852. doi:10.3389/fpls.2016.01852
Lander, E., and Kruglyak, L. (1995). Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 11 (3), 241–247. doi:10.1038/ng1195-241
Lee, S., Mian, M. A., McHale, L. K., Wang, H., Wijeratne, A. J., Sneller, C. H., et al. (2013a). Novel quantitative trait loci for partial resistance to Phytophthora sojae in soybean PI 398841. Theor. Appl. Genet. 126 (4), 1121–1132. doi:10.1007/s00122-013-2040-x
Lee, S., Mian, M. A., Sneller, C. H., Wang, H., Dorrance, A. E., and McHale, L. K. (2014). Joint linkage QTL analyses for partial resistance to Phytophthora sojae in soybean using six nested inbred populations with heterogeneous conditions. Theor. Appl. Genet. 127, 429–444. doi:10.1007/s00122-013-2229-z
Lee, S., Rouf Mian, M. A., McHale, L. K., Sneller, C. H., and Dorrance, A. E. (2013b). Identification of quantitative trait loci conditioning partial resistance to Phytophthora sojae in soybean PI 407861A. Crop Sci. 53 (3), 1022–1031. doi:10.2135/cropsci2012.10.0578
Li, L., Guo, N., Niu, J., Wang, Z., Cui, X., Sun, J., et al. (2016b). Loci and candidate gene identification for resistance to Phytophthora sojae via association analysis in soybean [Glycine max (L.) Merr.]. Mol. Genet. Genomics 291 (3), 1095–1103. doi:10.1007/s00438-015-1164-x
Li, L., Lin, F., Wang, W., Ping, J., Fitzgerald, J. C., Zhao, M., et al. (2016a). Fine mapping and candidate gene analysis of two loci conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 129 (12), 2379–2386. doi:10.1007/s00122-016-2777-0
Li, S., Hanlon, R., Wise, H., Pal, N., Brar, H., Liao, C., et al. (2021). Interaction of Phytophthora sojae effector Avr1b with E3 ubiquitin ligase GmPUB1 is required for recognition by soybeans carrying Phytophthora resistance rps1-b and rps1-k genes. Front. Plant Sci. 12, 725571. doi:10.3389/fpls.2021.725571
Li, X., Han, Y., Teng, W., Zhang, S., Yu, K., Poysa, V., et al. (2010). Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ‘Conrad’and ‘Hefeng 25. Theor. Appl. Genet. 121 (4), 651–658. doi:10.1007/s00122-010-1337-2
Li, Y., Sun, S., Zhong, C., Wang, X., Wu, X., and Zhu, Z. (2017a). Genetic mapping and development of co-segregating markers of RpsQ, which provides resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 130 (6), 1223–1233. doi:10.1007/s00122-017-2883-7
Li, Y., Sun, S., Zhong, C., and Zhu, Z. (2017b). Detached-petiole inoculation method to evaluate Phytophthora root rot resistance in soybean plants. Crop Pasture Sci. 68 (6), 555–560. doi:10.1071/cp17158
Lin, F., Li, W., McCoy, A. G., Gao, X., Collins, P. J., Zhang, N., et al. (2021). Molecular mapping of quantitative disease resistance loci for soybean partial resistance to Phytophthora sansomeana. Theor. Appl. Genet. 134 (7), 1977–1987. doi:10.1007/s00122-021-03799-x
Lin, F., Zhao, M., Baumann, D. D., Ping, J., Sun, L., Liu, Y., et al. (2014). Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC genomics 15 (1), 18–13. doi:10.1186/1471-2164-15-18
Lin, F., Zhao, M., Ping, J., Johnson, A., Zhang, B., Abney, T. S., et al. (2013). Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor. Appl. Genet. 126 (8), 2177–2185. doi:10.1007/s00122-013-2127-4
Liu, F. P., Gao, Z. M., Wang, X. F., Yu, C., and Pan, Y. M. (2006). On identification and biological characteristics of the causal organism of Phytophthora root rot of soybean in Anhui. J. Guangxi Agric. Biol. Sci. 25, 48–51.
Ludke, W. H., Schuster, I., Silva, F. L. D., Montecelli, T. D. N., Soares, B. D. A., Oliveira, A. B. D., et al. (2019). SNP markers associated with soybean partial resistance to Phytophthora sojae. Crop Breed. Appl. Technol. 19, 31–39. doi:10.1590/1984-70332019v19n1a05
Mideros, S., Nita, M., and Dorrance, A. E. (2007). Characterization of components of partial resistance, Rps2, and root resistance to Phytophthora sojae in soybean. Phytopathology 97, 655–662. doi:10.1094/PHYTO-97-5-0655
Mofijur, M., Atabani, A. E., Masjuki, H. A., Kalam, M. A., and Masum, B. M. (2013). A study on the effects of promising edible and non-edible biodiesel feedstocks on engine performance and emissions production: A comparative evaluation. Renew. Sustain. energy Rev. 23, 391–404. doi:10.1016/j.rser.2013.03.009
Moriwaki, J. (2010). Aiming at the construction of the race distinction system of Japanese Phytophthora sojae. Plant Prot. Jpn. 64, 508–510.
Moy, P., Qutob, D., Chapman, B. P., Atkinson, I., and Gijzen, M. (2004). Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant. Microbe. Interact. 17 (10), 1051–1062. doi:10.1094/MPMI.2004.17.10.1051
Mueller, E. H., Athow, K. L., and Laviolette, F. A. (1978). Inheritance of resistence to four physiologic races of Phytophthora, Phytophthora megasperma Var. sojae Phytopathol 68, 1318–1322.
Nguyen, V. T., Vuong, T. D., VanToai, T., Lee, J. D., Wu, X., Mian, M. R., et al. (2012). Mapping of quantitative trait loci associated with resistance to Phytophthora sojae and flooding tolerance in soybean. Crop Sci. 52 (6), 2481–2493. doi:10.2135/cropsci2011.09.0466
Niu, J., Guo, N., Sun, J., Li, L., Cao, Y., Li, S., et al. (2017). Fine mapping of a resistance gene RpsHN that controls Phytophthora sojae using recombinant inbred lines and secondary populations. Front. Plant Sci. 8, 538. doi:10.3389/fpls.2017.00538
Niu, J., Guo, N., Zhang, Z., Wang, Z., Huang, J., Zhao, J., et al. (2018). Genome-wide SNP-based association mapping of resistance to Phytophthora sojae in soybean (Glycine max (L.) Merr.). Euphytica 214 (10), 187–189. doi:10.1007/s10681-018-2262-8
Ochola, S., Huang, J., Ali, H., Shu, H., Shen, D., Qiu, M., et al. (2020). Editing of an effector gene promoter sequence impacts plant‐Phytophthora interaction. J. Integr. Plant Biol. 62 (3), 378–392. doi:10.1111/jipb.12883
Ping, J., Fitzgerald, J. C., Zhang, C., Lin, F., Bai, Y., Wang, D., et al. (2016). Identification and molecular mapping of Rps11, a novel gene conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 129 (2), 445–451. doi:10.1007/s00122-015-2638-2
Ploper, L. D., Athow, K. L., and Laviolette, F. A. (1985). A new allele at theRps3Locus for resistance toPhytophthora megaspermaf. sp.glycineain soybean. Phytopathology 75 (6), 690–694. doi:10.1094/phyto-75-690
Qin, J., Song, Q., Shi, A., Li, S., Zhang, M., and Zhang, B. (2017). Genome-wide association mapping of resistance to Phytophthora sojae in a soybean [Glycine max (L.) Merr.] germplasm panel from maturity groups IV and V. PLoS One 12 (9), e0184613. doi:10.1371/journal.pone.0184613
Raghuvanshi, R. S., and Bisht, K. (2010). “Uses of soybean: Products and preparation,” in The soybean: Botany, production, and uses CABI international. Editor G. Singh. (Wallingford, UK: CABI International, 404–426.
Ramalingam, J., Alagarasan, G., Savitha, P., Lydia, K., Pothiraj, G., Vijayakumar, E., et al. (2020). Improved host-plant resistance to Phytophthora rot and powdery mildew in soybean (Glycine max (L.) Merr.). Sci. Rep. 10 (1), 13928–14011. doi:10.1038/s41598-020-70702-x
Ribaut, J. M., and Ragot, M. (2007). Marker-assisted selection to improve drought adaptation in maize: The backcross approach, perspectives, limitations, and alternatives. J. Exp. Bot. 58 (2), 351–360. doi:10.1093/jxb/erl214
Rolling, W., Lake, R., Dorrance, A. E., and McHale, L. K. (2020). Genome-wide association analyses of quantitative disease resistance in diverse sets of soybean [Glycine max (L.) Merr.] plant introductions. PLoS ONE. 15 (3). doi:10.1371/journal.pone.0227710
Roth, M. G., Webster, R. W., Mueller, D. S., Chilvers, M. I., Faske, T. R., Mathew, F. M., et al. (2020). Integrated management of important soybean pathogens of the United States in changing climate. J. Integr. Pest Manag. 11 (1), 17. doi:10.1093/jipm/pmaa013
Ryley, M., Obst, N., Irwin, J., and Drenth, A. (1998). Changes in the racial composition of Phytophthora sojae in Australia between1979 and 1996. Plant Dis. 82, 1048–1054. doi:10.1094/PDIS.1998.82.9.1048
Sahoo, D. K., Abeysekara, N. S., Cianzio, S. R., Robertson, A. E., and Bhattacharyya, M. K. (2017). A novel Phytophthora sojae resistance Rps12 gene mapped to a genomic region that contains several Rps genes. PloS one 12 (1), e0169950. doi:10.1371/journal.pone.0169950
Sahoo, D. K., Das, A., Huang, X., Cianzio, S., and Bhattacharyya, M. K. (2021). Tightly linked Rps12 and Rps13 genes provide broad-spectrum Phytophthora resistance in soybean. Sci. Rep. 11 (1), 16907–16913. doi:10.1038/s41598-021-96425-1
Sandhu, D., Gao, H., Cianzio, S., and Bhattacharyya, M. K. (2004). Deletion of a disease resistance nucleotide-binding-site leucine-rich-repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics 168 (4), 2157–2167. doi:10.1534/genetics.104.032037
Sandhu, D., Schallock, K. G., Rivera-Velez, N., Lundeen, P., Cianzio, S., and Bhattacharyya, M. K. (2005). Soybean Phytophthora resistance gene Rps8 maps closely to the Rps3 region. J. Hered. 96 (5), 536–541. doi:10.1093/jhered/esi081
Sandhu, D., Tasma, I., Frasch, R., and Bhattacharyya, M. K. (2009). Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol. 9 (1), 105–114. doi:10.1186/1471-2229-9-105
Schmitthenner, A. F. (1999). “Phytophthora rot of soybean,” in Compendium of soybean diseases. Editors G. L. Hartman, J. B. Sinclair, and J. C. Rupe. 4th edn (USA: The American Phytopathological Society (APS) Press), 39–42.
Schmitthenner, A. F. (1985). Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 69, 362–368doi. doi:10.1094/pd-69-362
Schneider, R., Rolling, W., Song, Q., Cregan, P., Dorrance, A. E., and McHale, L. K. (2016). Genome-wide association mapping of partial resistance to Phytophthora sojae in soybean plant introductions from the Republic of Korea. BMC genomics 17 (1), 607–614. doi:10.1186/s12864-016-2918-5
Scott, K., Balk, C., Veney, D., McHale, L. K., and Dorrance, A. E. (2019). Quantitative disease resistance loci towards Phytophthora sojae and three species of Pythium in six soybean nested association mapping populations. Crop Sci. 59 (2), 605–623. doi:10.2135/cropsci2018.09.0573
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 (8), 2384–2396. doi:10.1105/tpc.105.033043
Song, Q., Jia, G., Zhu, Y., Grant, D., Nelson, R. T., Hwang, E. Y., et al. (2010). Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1. 0) in soybean. Crop Sci. 50 (5), 1950–1960. doi:10.2135/cropsci2009.10.0607
Specht, J. E., Hume, D. J., and Kumudini, S. V. (1999). Soybean yield potential—A genetic and physiological perspective. Crop Sci. 39 (6), 1560–1570. doi:10.2135/cropsci1999.3961560x
Stasko, A. K., Wickramasinghe, D., Nauth, B. J., Acharya, B., Ellis, M. L., Taylor, C. G., et al. (2016). High-density mapping of resistance QTL toward Phytophthora sojae, Pythium irregulare, and Fusarium graminearum in the same soybean population. Crop Sci. 56 (5), 2476–2492. doi:10.2135/cropsci2015.12.0749
Su, Y. C., and Shen, C. Y. (1993). The discovery and biological characteristic studies of Phytophthora megasperma f. sp. glycinea on soyabean in China. Acta Phytopathol. Sin. 23 (4), 341–347.
Sugimoto, T., Kato, M., Yoshida, S., Matsumoto, I., Kobayashi, T., Kaga, A., et al. (2012). Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed. Sci. 61, 511–522. doi:10.1270/jsbbs61511
Sugimoto, T., Watanabe, K., Yoshida, S., Aino, M., Furiki, M., Shiono, M., et al. (2010). Field application of calcium to reduce Phytophthora stem rot of soybean, and calcium distribution in plants. Plant Dis. 94, 812–819. doi:10.1094/PDIS-94-7-0812
Sugimoto, T., Yoshida, S., Aino, M., Watanabe, K., Shiwaku, K., and Sugimoto, M. (2006). Race distribution of Phytophthora sojae on soybean in Hyogo. J. Gen. Plant Pathol. 72, 92–97. doi:10.1007/s10327-005-0255-7
Sugimoto, T., Yoshida, S., Kaga, A., Hajika, M., Watanabe, K., Aino, M., et al. (2011). Genetic analysis and identification of DNA markers linked to a novel Phytophthora sojae resistance gene in the Japanese soybean cultivar. Waseshiroge. Euphytica 182 (1), 133–145. doi:10.1007/s10681-011-0525-8
Sugimoto, T., Yoshida, S., Watanabe, K., Aino, M., Kanto, T., Maekawa, K., et al. (2008). Identification of SSR markers linked to the Phytophthora resistance gene Rps1‐d in soybean. Plant Breed. 127 (2), 154–159. doi:10.1111/j.1439-0523.2007.01440.x
Sun, J., Guo, N., Lei, J., Li, L., Hu, G., and Xing, H. (2014b). Association mapping for partial resistance to Phytophthora sojae in soybean (Glycine max (L) Merr). J. Genet. 93, 355–363. doi:10.1007/s12041-014-0383-y
Sun, J., Li, L., Zhao, J., Huang, J., Yan, Q., Xing, H., et al. (2014a). Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean [Glycine max (L.) Merr.]. Theor. Appl. Genet. 127 (4), 913–919. doi:10.1007/s00122-014-2266-2
Sun, S., Wu, X. L., Zhao, J. M., Wang, Y. C., Tang, Q. H., Yu, D. Y., et al. (2011). Characterization and mapping of RpsYu25, a novel resistance gene to Phytophthora sojae. Plant Breed. 130 (2), 139–143. doi:10.1111/j1439-0523201001794x
Tan, X., Hu, Y., Jia, Y., Hou, X., Xu, Q., Han, C., et al. (2020). A conserved glycoside hydrolase family 7 cellobiohydrolase PsGH7a of Phytophthora sojae is required for full virulence on soybean. Front. Microbiol. 11, 1285. doi:10.3389/fmicb.2020.01285
Tooley, P. W., and Grau, C. R. (1984). Field characterization of rate-reducing resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology 74 (10), 1201–1208. doi:10.1094/phyto-74-1201
Tsuchiya, S. (1990). Classification of race of Phytophthora sojae and screening of resistant soybean cultivars to Phytophthora stem rot. Ann. Phytopathol. Soc. Jpn. 56, 144.
Tucker, D. M., Saghai Maroof, M. A., Mideros, S., Skoneczka, J. A., Nabati, D. A., Buss, G. R., et al. (2010). Mapping quantitative trait loci for partial resistance to Phytophthora sojae in a soybean interspecific cross. Crop Sci. 50 (2), 628–635. doi:10.2135/cropsci2009.03.0161
Tyler, B. M., and Gijzen, M. J. (2014). “The Phytophthora sojae genome sequence foundation for a revolution,” in Genomics of plant-associated fungi and oomycetes: Dicot pathogens. Editors R. A. Dean, A. Lichens-Park, and C. Kole (Heidelberg: Springer), 133–157. doi:10.1007/978-3-662-44056-8_7
Tyler, B. M. (2007). Phytophthora sojae: Root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8 (1), 1–8. doi:10.1111/j.1364-3703.2006.00373.x
USDA (2020). Oilseeds - world markets and trade, a USDA publication. SW Washington DC: USDA. Available at: https://wwwfasusdagov/data/oilseeds-world-markets-and-trade.
Van, K., Rolling, W., Biyashev, R. M., Matthiesen, R. L., Abeysekara, N. S., Robertson, A. E., et al. (2020). Mining germplasm panels and phenotypic datasets to identify loci for resistance to Phytophthora sojae in soybean. doi:10.1002/tpg2.20063
Wang, H., Waller, L., Tripathy, S., St Martin, S. K., Zhou, L., Krampis, K., et al. (2010). Analysis of genes underlying soybean quantitative trait loci conferring partial resistance to Phytophthora sojae. Plant Genome 3 (1), 23–40. doi:10.3835/plantgenome2009.12.0029
Wang, H., Wijeratne, A., Wijeratne, S., Lee, S., Taylor, C. G., St Martin, S. K., et al. (2012). Dissection of two soybean QTL conferring partial resistance to Phytophthora sojae through sequence and gene expression analysis. BMC genomics 13 (1), 428–523. doi:10.1186/1471-2164-13-428
Wang L, L., Chen, H., Li, J., Shu, H., Zhang, X., Wang, Y., et al. (2020). Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Res. 48 (4), 1790–1799. doi:10.1093/nar/gkz1160
Wang P, P., Wu, H., Zhao, G., He, Y., Kong, W., Zhang, J., et al. (2020). Transcriptome analysis clarified genes involved in resistance to Phytophthora capsici in melon. PloS one 15 (2), e0227284. doi:10.1371/journalpone0227284
Wang, W., Chen, L., Fengler, K., Bolar, J., Llaca, V., Wang, X., et al. (2021). A giant NLR gene confers broad-spectrum resistance to Phytophthora sojae in soybean. Nat. Commun. 12, 6263. doi:10.1038/s41467-021-26554-8
Wen, J., and Chen, H. (2002). Races and virulence phenotypes of Phytophthora sojae in Heilongjiang province and Mongolia Region of China. Chinese Journal of Oil Crop Sciences 24, 63–66.
Weng, C., Yu, K., Anderson, T. R., and Poysa, V. (2007). A quantitative trait locus influencing tolerance to Phytophthora root rot in the soybean cultivar ‘Conrad. Euphytica 158 (1), 81–86. doi:10.1007/s10681-007-9428-0
Weng, C., Yu, K., Anderson, T. R., and Poysa, V. (2001). Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J. Hered. 92 (5), 442–446. doi:10.1093/jhered/925442
Wilcox, J. R. (2001). Sixty years of improvement in publicly developed elite soybean lines. Crop Sci. 41 (6), 1711–1716. doi:10.2135/cropsci20011711
Wong, J., Gao, L., Yang, Y., Zhai, J., Arikit, S., Yu, Y., et al. (2014). Roles of small RNA s in soybean defense against Phytophthora sojae infection. Plant J. 79 (6), 928–940. doi:10.1111/tpj.12590
Workneh, F., Yang, X. B., and Tylka, G. L. (1998). Effect of tillage practices on vertical distribution of Phytophthora sojae. Plant Dis. 82 (11), 1258–1263. doi:10.1094/PDIS.1998.82.11.1258
Wu, X., Zhang, B., Shi, S., Zhao, J., Feng, Y., Na, G., et al. (2011a). Identification, genetic analysis and mapping of resistance to Phytophthora sojae of Pm28 in soybean. Agr. Sci. China 10, 1506–1511. doi:10.1016/S1671-2927(11)60145-4
Wu, X., Zhou, B., Sun, S., Zhao, J., Chen, S., Gai, J., et al. (2011b). Genetic analysis and mapping of resistance to Phytophthora sojae of Pm14 in soybean. Scientia Agricultura Sinica 44, 456–460.
Wu, X., Zhou, B., Zhao, J., Guo, N., Zhang, B., Yang, F., et al. (2011c). Identification of quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Plant Breed. 130, 144–149. doi:10.1111/j.1439-0523.2010.01799.x
Xiao, J. T., Miao, M. I. A. O., Gao, K., Xiang, G. L., Yang, S., and Dong, S. M. (2011). Genetic diversity of Phytophthora sojae in China based on RFLP. Sci. Agric. Sin. 44 (20), 4190–4198. doi:10.3864/j.issn.0578-1752.2011.20.007
Xu, P. F., Wu, J. J., Xue, A., Li, W. B., Chen, W. Y., Lai, W. E. I., et al. (2012). Differentially expressed genes of soybean during infection by Phytophthora sojae. J. Integr. Agric. 11 (3), 368–377. doi:10.1016/s2095-3119(12)60021-5
Xue, D., Guo, N., Zhang, X. L., Zhao, J. M., Bu, Y. P., Jiang, D. L., et al. (2019). Genome-wide analysis reveals the role of mediator complex in the soybean—phytophthora sojae interaction. Int. J. Mol. Sci. 20 (18), 4570. doi:10.3390/ijms20184570
Yang, J., Wang, X., Guo, B., Huang, J., Ye, W., Dong, S., et al. (2019). Polymorphism in natural alleles of the avirulence gene Avr1c is associated with the host adaptation of Phytophthora sojae. Phytopathol. Res. 1 (1), 28–13. doi:10.1186/s42483-019-0035-5
Yao, H. Y., Wang, X. M., Wu, X. F., Xiao, Y. N., and Zhu, Z. D. (2010). Molecular mapping of Phytophthora resistance gene in soybean cultivar Zaoshu18. J. Plant Genet. Resour. 11 (2), 213–217.
Yu, A., Xu, P., Wang, J., Zhang, S., Wu, J., Li, W., and Jiang, L. (2010). Genetic analysis and SSR mapping of gene resistance to Phytophthora sojae race 1 in soybean cv Suinong 10. Chin. J. Oil Crop Sci. 32 (4), 462–466.
Zhang, C., Wang, X., Zhang, F., Dong, L., Wu, J., Cheng, Q., et al. (2017). Phenylalanine ammonia-lyase2. 1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 7 (1), 7242–7313. doi:10.1038/s41598-017-07832-2
Zhang, J., Xia, C., Duan, C., Sun, S., Wang, X., Wu, X., et al. (2013a). Identification and candidate gene analysis of a novel Phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PloS one 8 (7), e69799. doi:10.1371/journal.pone.0069799
Zhang, J., Xia, C., Wang, X., Duan, C., Sun, S., Wu, X., et al. (2013b). Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor. Appl. Genet. 126 (6), 1555–1561. doi:10.1007/s00122-013-2073-1
Zhang, S., Xu, P., Wu, J., Xue, A. G., Zhang, J., Li, W., et al. (2010). Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis. 94 (1), 87–91. doi:10.1094/PDIS-94-1-0087
Zhang, X., Wang, W., Guo, N., Zhang, Y., Bu, Y., Zhao, J., et al. (2018). Combining QTL-seq and linkage mapping to fine map a wild soybean allele characteristic of greater plant height. BMC genomics 19 (1), 226–312. doi:10.1186/s12864-018-4582-4
Zhao, X., Bao, D., Wang, W., Zhang, C., Jing, Y., Jiang, H., et al. (2020). Loci and candidate gene identification for soybean resistance to Phytophthora root rot race 1 in combination with association and linkage mapping. Mol. Breed. 40 (10), 100–112. doi:10.1007/s11032-020-01179-9
Zhao, Y., Chang, X., Qi, D., Dong, L., Wang, G., Fan, S., et al. (2017). A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front. Plant Sci. 8, 299. doi:10.3389/fpls.2017.00299
Zhendong, Z., Yunlong, H., Xiaoming, W., Junbin, H., and Xiaofei, W. (2010). Molecular identification of a novel Phytophthora resistance gene in soybean. Eur. PMC 33, 154–157.
Zhong, C., Li, Y., Sun, S., Duan, C., and Zhu, Z. (2019). Genetic mapping and molecular characterization of a broad-spectrum Phytophthora sojae resistance gene in Chinese soybean. Int. J. Mol. Sci. 20 (8), 1809. doi:10.3390/ijms20081809
Zhong, C., Sun, S., Li, Y., Duan, C., and Zhu, Z. (2018b). Next-generation sequencing to identify candidate genes and develop diagnostic markers for a novel Phytophthora resistance gene, RpsHC18, in soybean. Theor. Appl. Genet. 131 (3), 525–538. doi:10.1007/s00122-017-3016-z
Zhong, C., Sun, S., Yao, L., Ding, J., Duan, C., and Zhu, Z. (2018a). Fine mapping and identification of a novel Phytophthora root rot resistance locus RpsZS18 on chromosome 2 in soybean. Front. Plant Sci. 9, 44. doi:10.3389/fpls.2018.00044
Zhong, C., Sun, S., Zhang, X., Duan, C., and Zhu, Z. (2020). Fine mapping, candidate gene identification and co-segregating marker development for the Phytophthora root rot resistance gene RpsYD25. Front. Genet. 11, 799. doi:10.3389/fgene.2020.00799
Keywords: soybean, Phytophthora, disease resistance, gene stacking, sustainable management, genomic approaches
Citation: Chandra S, Choudhary M, Bagaria PK, Nataraj V, Kumawat G, Choudhary JR, Sonah H, Gupta S, Wani SH and Ratnaparkhe MB (2022) Progress and prospectus in genetics and genomics of Phytophthora root and stem rot resistance in soybean (Glycine max L.). Front. Genet. 13:939182. doi: 10.3389/fgene.2022.939182
Received: 08 May 2022; Accepted: 21 October 2022;
Published: 14 November 2022.
Edited by:
Aamir Raina, Aligarh Muslim University, IndiaReviewed by:
Yingpeng Han, Northeast Agricultural University, ChinaAsela Wijeratne, University of Memphis, United States
Copyright © 2022 Chandra, Choudhary, Bagaria, Nataraj, Kumawat, Choudhary, Sonah, Gupta, Wani and Ratnaparkhe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Milind B. Ratnaparkhe, bWlsaW5kLnJhdG5hcGFya2hlQGljYXIuZ292Lmlu
†These authors share first authorship
 Subhash Chandra1†
Subhash Chandra1† Vennampally Nataraj
Vennampally Nataraj Giriraj Kumawat
Giriraj Kumawat Jeet Ram Choudhary
Jeet Ram Choudhary Humira Sonah
Humira Sonah Sanjay Gupta
Sanjay Gupta Shabir Hussain Wani
Shabir Hussain Wani Milind B. Ratnaparkhe
Milind B. Ratnaparkhe