
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet. , 12 October 2022
Sec. RNA
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.938672
 Yafei Xiao1†
Yafei Xiao1† Mengyuan Qiu2†
Mengyuan Qiu2† Cong Tan3†
Cong Tan3† Wanting Huang4
Wanting Huang4 Shaowen Hu1
Shaowen Hu1 Xiaowei Jiang5
Xiaowei Jiang5 Mingjie Guo6
Mingjie Guo6 Congcong Wang1
Congcong Wang1 Jingyu Liang3
Jingyu Liang3 Yimei Wu3
Yimei Wu3 Mengmeng Li1
Mengmeng Li1 Quanying Li1
Quanying Li1 Changjiang Qin1*
Changjiang Qin1*As the third most common cancer and the second leading cause of cancer death worldwide, colorectal cancer (CRC) poses a serious threat to people’s health. In recent years, circRNA has been widely reported as a new biomarker in CRC, but a comprehensive summary and analysis is lacking. This study aims to evaluate the diagnostic, therapeutic and prognostic significance of circRNAs in CRC by systematically analysing their expression patterns, biological functions and clinical significance in CRC. The literature on circRNA in CRC was searched in the PubMed database and included for analysis after screening according to strict inclusion and exclusion criteria. The UALCAN online tool was used to obtain host gene expression data. The miRTargetLink 2.0 was used to predict target genes for miRNAs action in CRC patients. Cytoscape was used to construct circRNA-miRNA-mRNA interaction networks. From the 236 included papers, we identified 217 circRNAs and their associated 108 host genes and 145 miRNAs. Among the 145 miRNAs, 27 miRNAs had no corresponding target genes. After prediction of target genes and differential analysis, a total of 25 target genes were obtained and a circRNA-miRNA-mRNA interaction network was constructed. Among the 217 circRNAs, 74 were associated with diagnosis, 160 with treatment and 51 with prognosis. And 154 of them function as oncogenes while 58 as tumour suppressor genes. In addition, these circRNAs include 32 exosomal circRNAs, which have unique advantages as biomarkers. In total, we summarize and analyze the expression patterns, biological functions and clinical significance of circRNAs in CRC. In addition, we constructed some new circRNA-miRNA-mRNA regulatory axes based on the miRNAs sponged by circRNAs.
Colorectal cancer, as the third most common cancer and the second leading cause of cancer death worldwide, is a serious threat to people’s life and health with a total of 1,931,590 cases of CRC diagnosed and 935,173 deaths worldwide in 2021 (Siegel et al., 2021). Although the overall 5-year survival rate has been improving over the past few decades, the survival rate for patients with advanced CRC is only about 20%, compared with up to 90% for patients with early-stage CRC (Zeng et al., 2018; Shen et al., 2020). Unfortunately, about 56% of CRC patients have advanced cancer at the time of diagnosis (Millas et al., 2015; Siegel et al., 2017). At present, the commonly used clinical methods of tumour detection, such as ultrasound, X-ray, CT, MRI, endoscopy and nuclear imaging, can only detect lesions visible to the naked eye. When the asymptomatic lumps gradually grow to a size, that is, perceived by oneself, some of the tumours are already in the middle or late stages, and some have already metastasized, thus numerous patients have lost the best treatment period. Tumour marker is considered to be one of the best methods for the early detection of asymptomatic microfocal tumours.
Non-coding RNAs (ncRNAs) are a class of endogenous RNAs that are involved in the regulation of gene expression. In recent years, the regulatory role of ncRNAs in a variety of pathophysiological processes has received extensive attention (Anastasiadou et al., 2018). Circular RNA (circRNA) is a novel endogenous non-coding RNA (ncRNA) molecule that can be reversely spliced to produce a circular structure (Liu et al., 2017a). It can regulate gene expression at the transcriptional or post-transcriptional level by sponging at acting as microRNA (miRNA) and are involved in regulating many important biological processes (Cortes-Lopez and Miura, 2016). circRNA is a closed RNA molecule without 5′ cap structure and 3′ poly tail. Circular structure makes it highly resistant to RNA exonucleases and highly conserved evolutionarily (Louis et al., 2019). Similar to the case of exosomes, circRNA was initially considered to be a non-functional product (Sanger et al., 1976). However, with the development of technology, numerous studies have shown that circRNA plays an irreplaceable role in various cancer biological processes. circRNA acts via exosome packaging on distant tissues and cells to regulate various signaling mechanisms in tumors (Meng et al., 2017; Liu et al., 2019). Exosomal circRNA has recently received more attention and is considered to be one of the most promising biomarkers for the future.
circRNA is of stability, specificity, universality and conservation and may be used as a tumour marker and potential therapeutic target in clinical applications (Jeck et al., 2013; Zhou et al., 2018). Although there are many related studies on circRNAs in CRC, these studies are scattered and lack systematic organization and summary. Therefore, this study summarizes and analyzes the expression patterns, biological functions and clinical significance of circRNAs (including exosomal circular RNAs) and their corresponding miRNAs, host genes and target genes in CRC. In addition, given that some miRNAs in the circRNA regulatory axis do not have corresponding target genes, we made some predictions based on the corresponding software and organized them into circRNA-miRNA-mRNA regulatory axis. To our knowledge, this article is the first in CRC field to summarize and systematically analyze circRNA as diagnostic, therapeutic and prognostic markers in such a comprehensive manner, which we hope will shed a light on future research.
The PubMed database, the most commonly used information resource in the biomedical field, provides comprehensive searches on specific topics and is free and easy to use (Falagas et al., 2008). To screen circRNAs as diagnostic, therapeutic and prognostic markers for CRC, we searched the PubMed database (https://pubmed.ncbi.nlm.nih.gov/) on 4 December 2021. Search strategy: (“circRNA” OR “circular RNA” OR “has_circ”) AND (“colorectal cancer” OR “colon cancer” OR “colorectal carcinoma” OR “carcinoma of large intestine " OR “CRC” OR “colonic neoplasms” OR “colorectal neoplasms” OR “rectal neoplasms”). Eligible studies should meet the following criteria: 1) patients with a gold standard definitive diagnosis of CRC; 2) independent original studies assessing differential circRNA expression in CRC tissue; 3) as a diagnostic, therapeutic or prognostic marker for CRC. Exclusion criteria were as follows: 1) reviews, meta-analyses and pure bioinformatics; 2) case reports; 3) no clinical samples; 4) no circRNA studies; 5) non-human studies: cell lines or mice. The screened circRNAs were then categorized and summarized according to their roles in CRC. Besides, we categorized circRNAs as therapeutic markers that were not clearly indicated as CRC biomarkers but were associated with cancer proliferation, invasive metastasis, and progression. Two authors (Y.X. and S.H.) independently extracted data from the included studies using a standardized table that included the following items: name of circRNA, expression (up- or down-regulated), host gene, target gene, sponged miRNA, sample size, study method, mechanism, regulate pathways, in CRC, function, prognosis, biomarker type. All assessments were conducted independently by two investigators to ensure accurate study inclusion. The checklist of circRNA-related literatures and results of assessment are shown in Supplementary Table S1. The flow chart of this study is shown in Figure 1.

FIGURE 1. Flow-chart diagram of this study. (k: number of records. ONC: oncogenes. TS: tumour suppressor gene).
All expression data of host genes were obtained from the TCGA database. The UALCAN online tool is one of the tools that enables in-depth analysis of TCGA data (Chandrashekar et al., 2017). UALCAN is open and free to use at http://ualcan.path.uab.edu.
The miRTargetLink 2.0 was used to predict target genes for the action of miRNAs in human CRC patients and is available for free at https://www.ccb.uni-saarland.de/mirtargetlink2 (Kern et al., 2021). Cytoscape (https://cytoscape.org/) is a free JAVA-based tool capable of mapping biomolecular interaction networks (Shannon et al., 2003). circRNA-miRNA-mRNA interaction network maps are constructed by Cytoscape software.
From the initial search of 382 literatures, 236 literatures were finally included after removing 146 ineligible literatures according to the exclusion criteria. We identified 217 circRNAs biomarkers associated with CRC from these and analyzed their potential role in diagnosis, treatment and prognosis. By removing duplicates, 108 host genes and 145 miRNAs were screened (Supplementary Table S2). Moreover, we analyzed the expression patterns of these host genes and predicted target genes of miRNAs sponged by circRNAs. Finally, a circRNA-miRNA-mRNA interaction network was constructed based on the predicted target genes. Notably, because of the special existence of the exosomal circRNA, we have analysed it separately.
circRNAs are steady-state byproducts of partial fragment splicing of host genes (Kristensen et al., 2019). A total of 108 host genes were collected, and 92 genes were available in the TCGA database on the UALCAN platform for expression profiles (Supplementary Table S3). Significant differences with p < 0.05 in expression between normal and cancerous tissues were found for 78 genes. Among them, two host genes were down-regulated (FMN2 and NOX4) and the remaining 76 genes were up-regulated. What’s more, the expression levels of 19 host genes differed more than 2-fold change between tumour and normal tissues, 17 host genes of which were up-regulated and two host genes of which were down-regulated (Table 1, Supplementary Table S3). Although circRNAs are derived from the host gene (Lv et al., 2018), we did not observe a clear relationship between circRNAs and their host genes in terms of expression patterns.
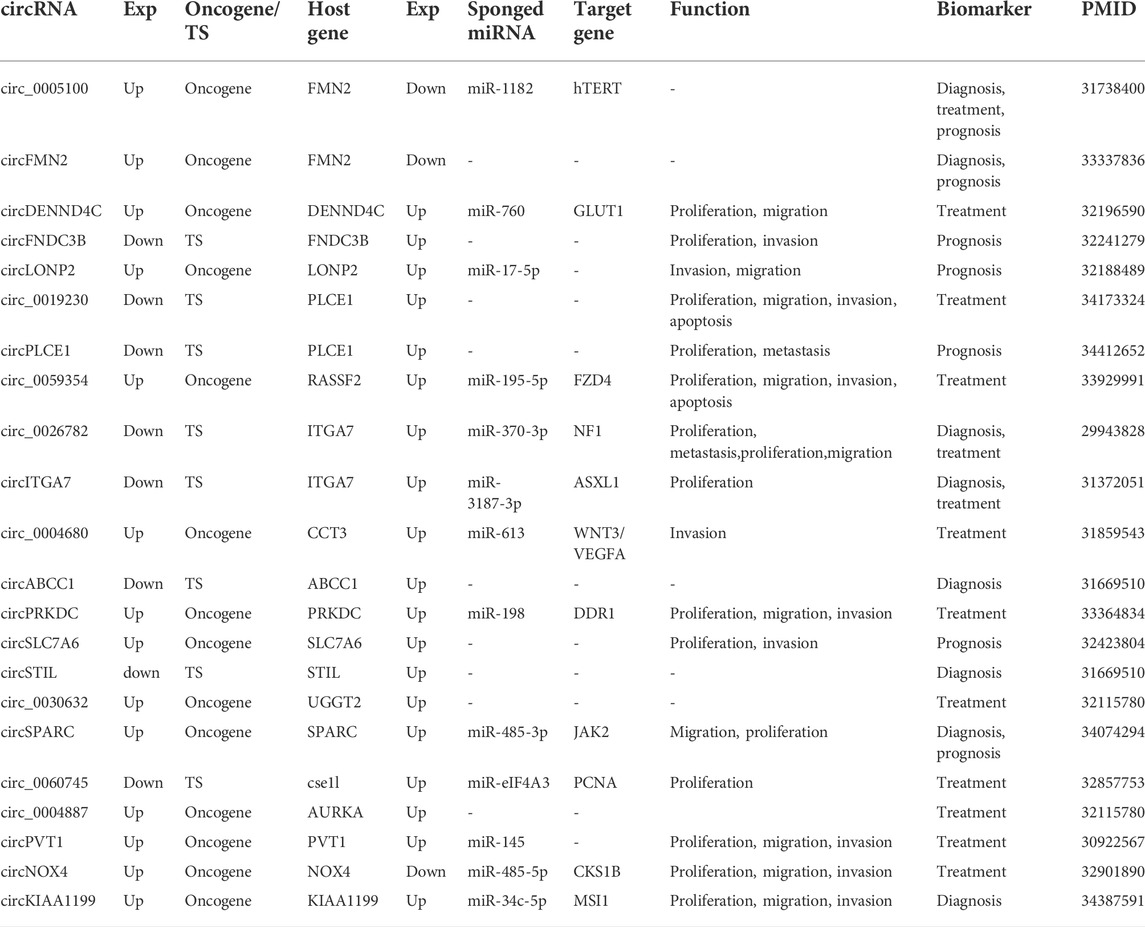
TABLE 1. Description of circRNAs in colorectal cancer (Host genes expression with p < 0.05 and >2-fold change (fold-change >2 or <0.5) in cancer tissues compared with normal tissues.).
MicroRNAs (miRNAs) are a class of non-coding RNAs consisting of approximately 22 nucleotides that bind to the 3ʹ-untranslated region (3ʹUTR) of their target genes to induce or repress the translational expression process (Zhang et al., 2017a). circRNA is a novel non-coding RNA with a covalently closed continuous loop, which makes it more stable than linear microRNAs with 3′ and 5′ ends (Nigro et al., 1991; Jeck et al., 2013; Memczak et al., 2013; Chen and Yang, 2015). circRNA can act as miRNA sponge and regulate gene expression (Zhang et al., 2017b; Panda, 2018; Chen et al., 2020a). As miRNA sponges, circRNAs are most notable for their competitive endogenous RNA (ceRNA) role (Zhang et al., 2019a). In the ceRNA network, circRNAs can bind to miRNAs through their miRNA binding sites (also known as miRNA response elements [MREs]), thereby regulating the mRNAs of the corresponding miRNAs’ target genes and attenuating the repressive effects of miRNAs on target genes (Hansen et al., 2013; Memczak et al., 2013; Xi et al., 2021). circRNA-miRNA-mRNA network plays a key role in cancer and non-cancer pathways (Jin et al., 2016; Yang et al., 2019), such as bladder cancer (Huang et al., 2016), colorectal cancers (Weng et al., 2017), osteosarcoma (Liu et al., 2017b), and human cartilage degeneration (Liu et al., 2016). Among the circRNAs we collected, more than a quarter of circRNAs exerted tumor suppressor or tumor-promoting effects in colorectal cancer through the ceRNA mechanism. For example, circVAPA can act as sponge of miR-125a to suppress colorectal cancer cell growth process by regulating miR-125a/CREB5 axisplay (Zhang et al., 2020a). circ_103809 can exert tumor suppressor effect through miR-532-3p/FOXO4 axis (Bian et al., 2018) (Supplementary Table S2, Figure 2).
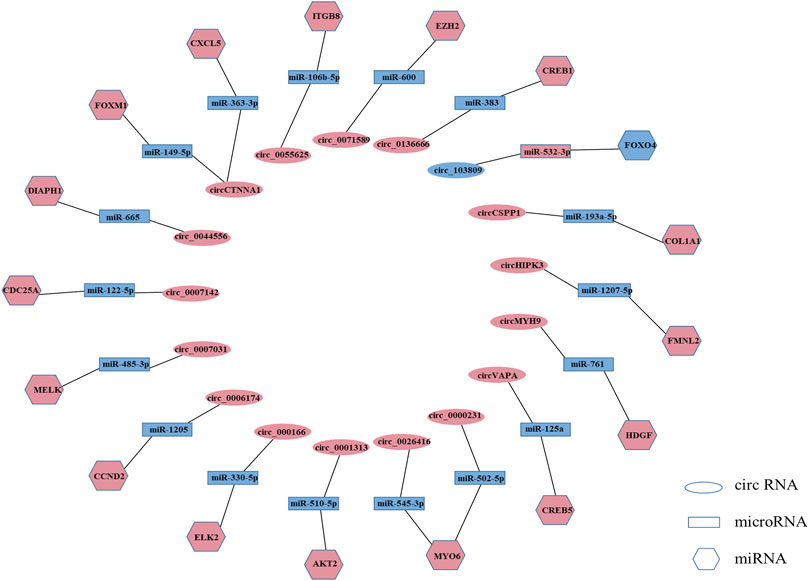
FIGURE 2. Regulatory mechanisms of circRNAs in colorectal cancer. (Light coral represents high expression, and blue represents low expression. Only circRNAs that were reported repeatedly are listed in this figure).
Some circRNAs have corresponding miRNAs, but lack the corresponding target genes. Among the 145 miRNAs, 27 miRNAs have no responsive target genes. The predicted target genes of these miRNAs were conducted on the miRTargetLink 2.0 online platform, and finally 3567 target genes were obtained. After excluding the weakly correlated target genes, only 496 target genes were left. Among these target genes, 70 target genes regulated by at least two miRNAs were identified (Figure 3A, Supplementary Table S4).
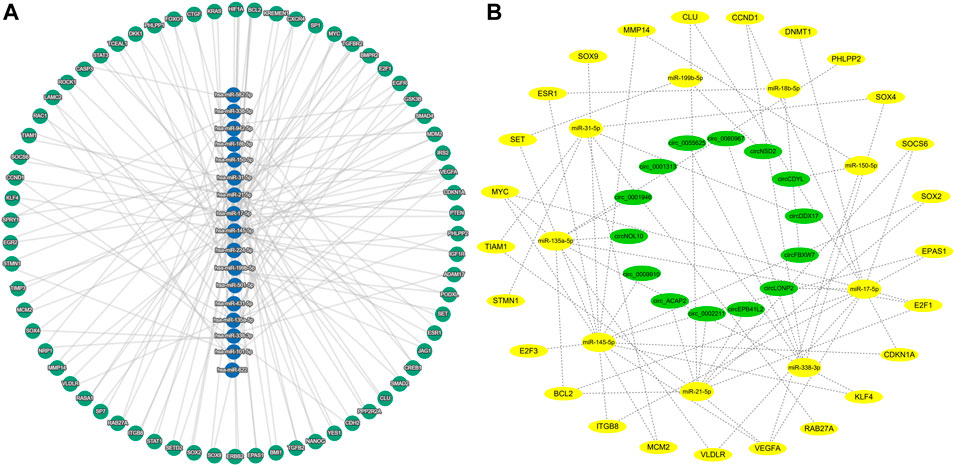
FIGURE 3. Predicted target genes and construction of circRNA-miRNA-mRNA network (A) miRNA and predicted target genes regulated by at least two miRNAs. (B) circRNA-miRNA-mRNA network in colorectal cancer.
70 target genes were analyzed in the UALCAN platform, and 25 target genes were finally obtained by eliminating the ineligible genes (p > 0.05 and 0.5 < Fold-change <2) (Supplementary Table S5). The miRNAs corresponding to these 25 target genes were identified in Supplementary Table S4, and the corresponding circRNAs were then found in the Supplementary Table S2 according to these miRNAs. Based on the circRNA-miRNA-mRNA correspondence, the network map was generated in the Cytoscape software (Figure 3B).
There is growing evidence that circRNAs play an important role in the development of CRC as oncogenes or tumour suppressor genes. circRNAs are involved in different processes of tumour pathogenesis, including cell proliferation, migration, invasion, apoptosis, metastasis, epithelial mesenchymal transition (EMT) and cell cycle (Yin et al., 2020; Zhang et al., 2021a; Su et al., 2022). A summary analysis of the literature revealed that most circRNAs were associated with cancer cell proliferation (n = 64), invasion (n = 48), migration (n = 49) and apoptosis (n = 18), while a few cyclic RNAs were involved in cancer cell metastasis (n = 9), cell cycle (n = 5) and EMT (n = 3) (Figure 4). Of the 217 circRNAs we studied, 154 functioned as oncogenes, 58 tumour suppressor genes and five controversial genes (some publications reported them as oncogenes, while others reported them as tumour suppressors), which are circ_0000338, circ_0060745, circ_103,809, circCCDC66, circZNF609.
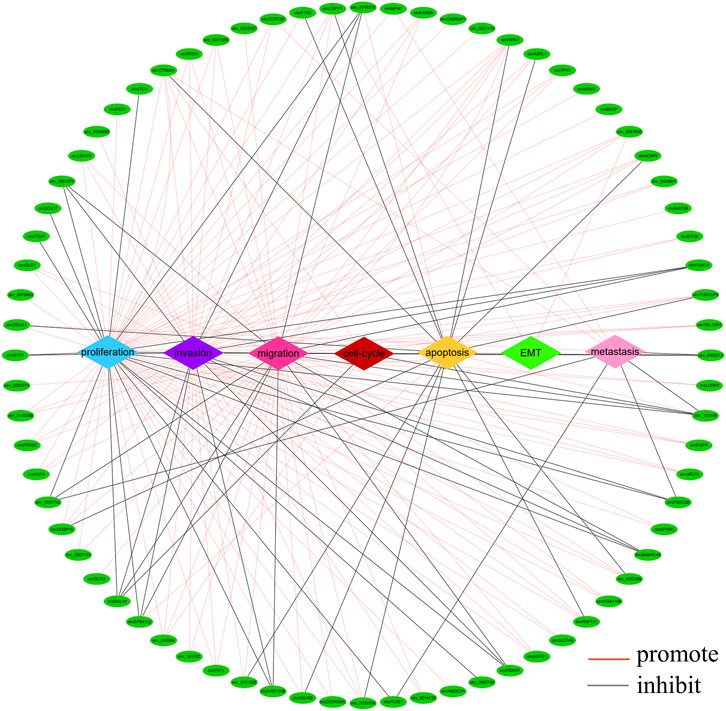
FIGURE 4. Network of aberrantly expressed circRNAs in diverse cellular functions. (The figure shows only circRNAs with a p < 0.05 in host gene expression).
Some circRNAs are up-expressed in CRC tissues and cells, and play an oncogenic role by regulating downstream target genes or activating certain signaling pathways. In 154 oncogenes, 23 circRNAs were repeatedly studied, among which the most frequently studied were circ_0007142 (Zhu et al., 2019; Yin et al., 2020; Wen et al., 2021) and circCSPP1 (Wang et al., 2020a; Li et al., 2021; Xi et al., 2021).
circ_0007142 was found to be significantly upregulated in CRC and is associated with poor differentiation and lymphatic metastasis. Overexpressed circ_0007142 promotes proliferation, migration, and invasion of CRC cells by targeting miR-103a-2-5p/PARP-1, and subsequently activating Wnt/β-Catenin pathway (Zhu et al., 2019). Another article indicated that circ_0007142 also regulated miR-455-5p/SGK1 to inhibit cell apoptosis of CRC (Yin et al., 2020). In addition, Yin et al. revealed the third way for circ_0007142 to play an oncogenic role through miR-122-5p/CDC25A (Wen et al., 2021).
A study has demonstrated that the upregulation of circCSPP1 in CRC tissues and cells promoted cell proliferation, migration, invasion, and inhibited apoptosis by regulating miR-431/LASP1 axis (Li et al., 2021). Moreover, activation of the circCSPP1/miR-193a-5p/COL1A1 facilitated EMT and liver metastasis (Wang et al., 2020a). Xi et al. (2021) confirmed the conclusion in Doxorubicin-resistant CRC cells, and found upregulated circCSPP1 reduced doxorubicin sensitivity through miR-944/FZD7 axis.
In addition to oncogenes, 58 circRNAs are down-regulated in CRC and have a suppressive effect on tumour proliferation, invasion and migration. Two of them have been repeatedly reported, which are circFBXW758 (Lu et al., 2019; Xu et al., 2021) and circFNDC3B (Pan et al., 2020; Zeng et al., 2020).
circFBXW7 is reduced in CRC patients and exerts cancer-inhibiting effects, suppressing cell proliferation, migration and invasion in CRC. Mechanically, low expression of circFBXW7 prohibited the progression of CRC through NEK2, mTOR, and PTEN pathways (Lu et al., 2019).To understand the effect of circFBXW7 on chemoresistance, Xu et al. (2021) transferred circFBXW7 into oxaliplatin-resistant CRC cells. They found that circFBXW7 increased oxaliplatin-induced apoptosis and inhibited oxaliplatin-induced EMT, increasing the sensitivity of drug-resistant cells to oxaliplatin.
It has been shown that circFNDC3B is downregulated in CRC patients who have a poor prognosis with shorter OS than patients with overexpressed circFNDC3B. circFNDC3B could encode a novel protein circFNDC3B-218aa to inhibit the proliferation, invasion and migration of CRC cells by reducing the expression of Snail (Pan et al., 2020). Besides, circFNDC3B could also modulate CRC growth, angiogenesis and liver metastasis by circFNDC3B/miR-97-5p/TIMP3 axis (Zeng et al., 2020). These studies suggested that circFNDC3B may be a potential prognosis and therapeutic biomarker for CRC.
In addition, we found that the current research on circRNAs mainly focused on oncogenes, and less research is performed on tumour suppressor genes. Oncogenes mainly function through AKT, JAK2/STAT3, AKT/mTOR, HIF-1α, Hippo-YAP, NF-κB, p53/EMT, PI3K/AKT, ROS and Wnt/β-Catenin pathways, and tumor suppressor genes mainly function through MAPK, NF-κB, PI3K/AKT, PTEN/AKT, Ras and Wnt/β-Catenin pathways (Figure 5). Notably, the most studied pathway for both oncogenes and tumour suppressor genes is the Wnt/β-Catenin pathway. The Wnt/β-Catenin pathway plays an integral role in embryogenesis and adult homeostasis in vivo (Nusse and Clevers, 2017). Aberrant activation of this pathway is associated with growth-related diseases and cancers, particularly as a key promoter of CRC development and progression (Vallee and Lecarpentier, 2018; Bian et al., 2020).
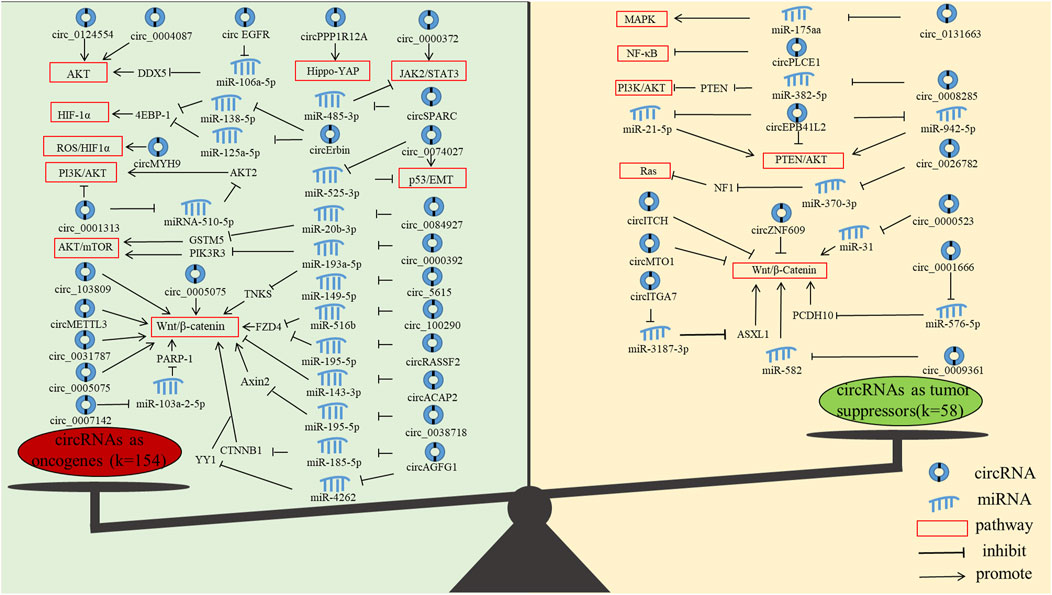
FIGURE 5. The molecular functions of circRNAs in CRC progression. (The figure shows only circRNAs with signaling pathways.)
Of the 217 circRNAs, 82 were associated with diagnosis, 154 with treatment and 43 with prognosis. Among these circRNAs, 30 circRNAs were repeatedly reported (≥2). Among them, six circRNAs have been reported several times as diagnostic markers (circ_0000338, circ_0001178, circ_0006174, circ_0007142, circ_103,809, circZNF609); 20 circRNAs have been reported repeatedly as treatment markers (circCSPP1, circCCDC66, circZNF609 etc.) and two circRNAs have been reported repeatedly as prognosis markers (circ_0005075, circ_100,876) (Supplementary Table S6). We next focused on those circRNAs reported repeatedly.
There are six circRNAs (circ_0000338, circ_0001178, circ_0006174, circ_0007142, circ_103,809, circZNF609) that have been repeatedly reported as diagnostic markers. Among them, circ_0001178, circ_0006174, and circ_0007142 were all found overexpressed in CRC, and were considered as diagnostic biomarkers of CRC. In addition, circ_103809 has been shown to be down-expressed in CRC tissues and is considered as a potential novel biomarker for CRC diagnosis.
Both circ_0000338 and circZNF609 were associated with the diagnosis of CRC, but notably, their expression patterns were controversial in different reports. Down-regulation of circ_0000338 in CRC was detected by qRT-PCR by Hon et al. (Hon et al., 2019). However, Zhao et al. found that circ_0000338 expression was significantly higher in CRC tissues than in paracancerous tissue (Zhao et al., 2021). In addition, opposite descriptions of circZNF609 expression in CRC were also described in different papers (Oliveira et al., 1988; Zhang et al., 2019b). Further experiments are needed to verify the expression pattern of these controversial circRNAs in CRC tissues.
Among the 20 circRNAs repeatedly reported as therapeutic markers, circFBXW7 was down-regulated, 17 circRNAs were up-regulated, and two circRNAs (circ_0060745 and circZNF609) had inconsistent expression patterns in different reports. These 17 circRNAs were reported twice, except for circCSPP1, circZNF609 and circCCDC66, which were reported three times in duplicates.
circCSPP1 acts as an oncogene in three literature reports. Wang et al. found that circCSPP1 was significantly up-regulated in CRC tissues, and that circCSPP1 promotes the migration and invasion of colorectal carcinoma cells in vitro and in vivo. In addition, they found that circCSPP1 may act as a promising therapeutic target by regulating the EMT process in CRC via activation of the circCSPP1/miR-193a-5p/COL1A1 axis (Wang et al., 2020a). Xi et al. found that circCSPP1 knockdown inhibited the growth of Doxorubicin-resistant CRC cells and enhanced doxorubicin sensitivity through the miR-944/FZD7 axis, providing a potential target for CRC therapy (Xi et al., 2021). In addition, it was also found that knockdown of circCSPP1 promoted apoptosis and weaken tumour growth in vivo, providing evidence for circCSPP1 as a promising biomarker for CRC management (Li et al., 2021).
It has been reported that circCCDC66 facilitates the development of CRC cells under hypoxic conditions via regulation of miR-3140/autophagy pathway, and this finding may provide a novel therapeutic option for patients with CRC (Feng et al., 2020). Furthermore, Hsiao et al. found that circCCDC66 controlled multiple pathological processes, including cell proliferation, migration, invasion (Hsiao et al., 2017).
Whether circZNF609 functions either as an oncogene or as a tumour suppressor gene in CRC is controversial. Zhang et al., 2019b found that circZNF609 was down-regulated at the RNA and protein levels. Overexpression of circZNF609 could induce apoptosis, up-regulate the expression of the pro-apoptotic protein Bax, down-regulate the expression of the anti-apoptotic protein Bcl-2, and up-regulate the expression of p53. However, Wu et al. (2018) found that circZNF609 was upregulated at both the RNA level and protein level, regulating the expression of Gli1 through miR-150 and thus promoting CRC migration.
Besides circZNF609 is controversial, the expression of circ_0060745 and circFBXW7 in CRC is also controversial. Some results show that circ_0060745 is up-regulated in CRC and promotes CRC cell proliferation and metastasis by regulating miR-4736/CSE1L signaling pathway, which is considered a novel target for the treatment of CRC (Wang et al., 2020b). However, Zhang et al. found that the expression of circ_0060745 was down-regulated in CRC and inhibited the proliferation of CRC cells (Xu et al., 2020). Further experimental validation is needed to determine their expression pattern in CRC tissues and whether they function as tumour suppressors or oncogenes in humans.
circ_0005075 and circ_100876 were repeatedly reported as prognostic markers in different literatures. They have been found to be upregulated in CRC and are considered to be potential prognostic markers in CRC. Clinical assays indicated that overexpression of circ_0005075 was significantly associated with histology differentiation, depth of invasion, advanced TNM stage, shorter overall survival and disease-free survival of CRC patients (Jin et al., 2019). Zhong et al. found that circ_0005075 was overexpressed in CRC tissues, and its expression level was correlated with distant metastasis, invasion, tumour lymph node metastasis stage, and tumour diameter of CRC, and was negatively correlated with overall survival of CRC patients (Zhong et al., 2019). Circ_100,876 was reported to be up-regulated in CRC tissues and is closely associated with adverse clinical outcomes (Zhang et al., 2020b; Zhou et al., 2020).
In addition to circRNAs related to diagnosis, treatment and prognosis, some circRNAs are also correlated with metastasis, drug resistance, and radioresistance. Among the 217 circRNAs, 42 were metastasis-related circRNAs. Among them, 19 were related to lymph node metastasis, 10 were related to distant metastasis, seven were related to both lymph node metastasis and distant metastasis, and the remaining six were related to liver metastasis (Supplementary Table S7). There were 13 circRNAs associated with drug resistance, of which circDDX17 (Ren et al., 2020), circ_0007031 (He et al., 2020), circ_0032833 (Li and Zheng, 2020), circ_0000338 (Zhao et al., 2021), circNRIP1 (Liu et al., 2021) were associated with 5-fluorouracil resistance; circ_0005963 (Wang et al., 2020c), circFBXW7 (Xu et al., 2021) were associated with oxaliplatin resistance; circ_0000338 (Hon et al., 2019), circ_32883 (Abu et al., 2019) were associated with both 5-fluorouracil and oxaliplatin; circ_ 0020095 (Sun et al., 2020), circ_0071589 (Zhang et al., 2021b), and circ_0131,663 (Wang et al., 2021) were associated with cisplatin resistance; circCSPP1 (Xi et al., 2021), and circ_0006174 (Zhang et al., 2022) were associated with Adriamycin resistance. In addition, circ_0001313 was associated with radiotherapy resistance (Wang et al., 2019a) (Supplementary Table S7).
32 exosomal circRNAs were found from 217 circRNAs. Of these, 21 exosomal circRNAs were associated with diagnosis, 13 with treatment and 10 with prognosis. Some exosomal circRNAs have the potential to be multiple biomarkers. For example, circ_0005100 was a diagnostic, therapeutic, and prognostic marker for CRC. circ_0021977 and circ_0067835 are prognostic and therapeutic markers for CRC. circIFT80, circ_0000338 and circEPB41L2 may function as diagnostic markers and therapeutic markers in CRC. In addition, circMBOAT2, circFMN2, circLMNB1, circZNF609 and circ_0007534 have been considered as diagnostic and prognostic markers for CRC. Exosome circRNAs exist in different forms in vivo. CircEIF3K, circ_0007334, circRNF121 and circ_0006174 were detected in the cell culture medium, while the other 28 circRNAs were found in blood. Nine exosomes, including circ_0001649, were expressed down-regulated in CRC patients, and another 23 exosomal circRNAs were up-regulated (Table 2).
As a novel endogenous non-coding RNA (ncRNA) molecule, circRNA have shown great potential as biomarkers due to their high stability, abundance, evolutionary conservation, and wide distribution in various body fluids and exosomes (Jeck et al., 2013; Zhou et al., 2018). Although previous reviews and meta-analyses have reported some studies of circRNAs in CRC, they are small in size and the analyses are relatively fragmented, lacking a comprehensive systematic summary. Therefore, we performed a systematic analysis of the role of circRNAs as markers in CRC. From the 236 included papers, we identified 217 circRNAs associated with CRC. We comprehensively elucidated the relationship between circRNAs and CRC in terms of molecular mechanisms, functions and clinical applications.
Among the 217 circRNAs, the majority of biomarkers were associated with treatment (n = 154), 82 were associated with diagnosis, and 43 were associated with prognosis. Some circRNAs are considered to have multiple marker potential. For example, circ_0005075 is upregulated in CRC and its high expression is related to tumour proliferation, distant metastasis and poor prognosis of patients, and has been reported as three markers for the diagnosis, treatment and prognosis of CRC (Zhong et al., 2019). circEPB41L2 is down-expressed in CRC, promoting tumour cell proliferation, migration, and inhibiting apoptosis. It has also been detected in patient plasma samples, and is considered to be a marker for CRC diagnosis and treatment (Jiang et al., 2021). Notably, many circRNAs play a role in the development of CRC through the ceRNA mechanism, illustrating the importance of the ceRNA mechanism for circRNAs. In addition, the majority of these circRNAs markers are oncogenes, probably due to the relatively low cost of studying oncogenes, resulting in an artificial selection bias.
Relatively, circRNAs that were reported repeatedly may have a high discriminatory significance. Thirty circRNAs were reported repeatedly, of which 23 were up-expressed, two were down-expressed, and five were controversial. Among the 23 highly expressed circRNAs, the most studied were circ_0007142 (Zhu et al., 2019; Yin et al., 2020; Wen et al., 2021) and circCSPP1 (Wang et al., 2020a; Li et al., 2021; Xi et al., 2021), both of which act as oncogenes that promote proliferation, migration and invasion of tumour cells and inhibit apoptosis; in addition, circCSPP1 was found to promote liver metastasis and reduce Adriamycin sensitivity in CRC through different pathways. circFBXW7 (Lu et al., 2019; Xu et al., 2021) and circFNDC3B (Pan et al., 2020; Zeng et al., 2020) function as tumour suppressor genes and both inhibit cancer cell proliferation, migration and invasion. Moreover, circ-FBXW7 can improve chemoresistance to oxaliplatin in CRC by binding to miR-128-3p, and circFNDC3B can promote angiogenesis and liver metastasis.
Exosomes are nanoscale spherical lipid bilayer vesicles secreted by cells containing DNA, RNA, proteins, lipids, and other biologically active substances (Zhang et al., 2020c). Both normal and tumour cells can produce exosomes, which are widely distributed in various biological fluids, such as saliva, blood, urine, and ascites bile (Zhang et al., 2020c). Exosomes produced by tumour cells not only play an important role in tumour growth, metastasis and immunomodulation (Poggio et al., 2019; Sanderson et al., 2019), but also monitor disease progression and serve as diagnostic markers of the disease (Zhang et al., 2020c). Recent studies have shown that circRNAs are enriched and stable in exosomes, which confers the potential of exosomal circRNAs as biomarkers and new therapeutic targets for tumours (Wang et al., 2019b). In this study, we identified 32 exosomal circRNAs, most of which were found in serum or plasma. Maybe for the reason that blood samples from cancer patients are easier to be obtained. In contrast, saliva, urine and other body fluid samples are rarely collected in research. We suggest that future researchers should not only collect cancer tissues, but also try to collect serum, urine and saliva to help discover more exosomal circRNA forms in CRC.
In addition to the research on exosomal circRNAs to be improved, there are still some deficiencies in the research of circRNAs in colorectal cancer. For example, the expression levels of some circRNAs are inconsistent as reported in different articles. For controversial circRNAs, additional sample sizes for studies are needed to obtain reliable expression data. Alternatively, some articles do not go far enough into the study of circRNAs and lack exploration of specific mechanisms of action and pathways. Correspondingly, research on the mechanisms and pathways associated with circRNAs in tumours needs to be increased. Moreover, the current study just demonstrated that circRNAs proved to be highly stable in serum. For example, Li et al. found that human serum exosomes contain large amounts of intact and stable circRNA. They demonstrated that circRNA is resistant to digestion by RNase R exonuclease and incubation of serum at room temperature for up to 24 h showed little effect on circRNA levels (Li et al., 2015). When is diagnostic circRNA produced in the blood during cancer evolution, and how does the concentration of circRNA change after it is produced? How long does it last? These questions are worth exploring in the future.
In the last decade we have witnessed a revolution in single-cell transcriptomics (Aldridge and Teichmann, 2020). Single-cell sequencing is a technical means of separating cell populations in tissues or body fluids into individual cells and analyzing their genetic material by high-throughput sequencing (Lei et al., 2021). Unlike traditional sequencing methods, single-cell sequencing provides unbiased, high-throughput and high-resolution transcriptome analysis of individual cells, thereby revealing the gene structure and gene expression status of individual cells, reflecting the heterogeneity between cells and categorizing them for personalized analysis (Natarajan et al., 2019). As cellular heterogeneity is prevalent in tumors, understanding the heterogeneity of tumour cells has important implications for diagnosis, clarification of the nature of the tumour and targeted treatment (Chen et al., 2020b). Therefore, this technology can be applied to detect molecular phenotypic typing of circulating tumour cells, detect early tumour cells, monitor intra-tumour heterogeneity and guide targeted precision therapy (Rantalainen, 2018; Winterhoff et al., 2019). In 2022, Wu et al. explored the cellular landscape of circRNAs using full-length single-cell RNA sequencing to investigate the expression of circRNAs in neurons in the human brain and circRNAs in breast cancer cells, extending knowledge of circRNA expression to the single-cell level (Fan et al., 2015). However, single-cell analysis of circRNA in colorectal cancer is currently lacking, and we believe this is a highly promising area for future research.
To our knowledge, this study is the first systematic analysis focusing on CRC circRNA as a diagnostic, therapeutic and prognostic marker. Our literature analysis is relatively objective and comprehensive, recording the marker types, research methods, mechanism of action, function, expression, sample size and other indicators of these circRNAs. However, this study has some limitations. First, the literature search deadline was 4 December 2021, and subsequent studies were not included. Second, the search database was limited to PubMed. Third, only articles written in English were included in our analysis, which may introduce some linguistic bias. Fourth, we only recorded circRNAs associated with diagnostic, therapeutic, and prognostic markers. Finally, due to the large number of circRNAs, we only focused on the circRNAs that were repeatedly reported. Therefore, the circRNAs included in our analysis may not fully reflect all circRNA studies. However, our analysis covers the vast majority of circRNAs as biomarkers in CRC, which is enough to illustrate the basic situation of the research on circRNAs as CRC markers in recent years.
In this study, we summarized and analyzed the expression patterns, biological functions and clinical significance of circRNAs and related miRNAs, host and target genes in colorectal cancer. Exosomal circRNAs may be the most promising biomarkers in the future due to their specific mode of existence. In addition, we have summarised circRNAs associated with metastasis, drug resistance and radioresistance. Finally, we analyzed the current status and shortcomings of circRNA research on colorectal cancer according to the summary and make some suggestions.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
CQ and YX: study concept and design. MQ, CT, WH, SH, XJ, MG, CW, JL, YW, ML, and QL: acquisition of data. YX, MQ CT, WH, SH,and XJ: analysis and interpretation of data. YX, MQ, CT, WH, and SH: draft of the manuscript. CQ and QL: critical revision of the manuscript for intellectual content.
This study was supported by Science and Technology Foundation of Henan Province (172102310152), Natural Science Foundation of Henan Province (182300410359), Science and Technology Foundation of Henan Province (SBGJ202002097), Science and Technology Foundation of Henan Province (192102310099), Henan Provincial Education Fund (19A320020), Postgraduate Cultivating Innovation and Quality Improvement Action Plan of Henan University (SYL20060192, SYLYC2022141 and SYLJD2022009). Henan Province United Common Project Fund (LHGJ20220663).
We would like to thank Professor Longxiang Xie, Institute of Biomedical Informatics, School of Basic Medical Sciences, Henan University, for his informatics and statistical advice and support for this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.938672/full#supplementary-material
Abu, N., Hon, K. W., Jeyaraman, S., Yahaya, A., Abdullah, N. M., Mustangin, M., et al. (2019). Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics 11, 875–884. doi:10.2217/epi-2019-0042
Aldridge, S., and Teichmann, S. A. (2020). Single cell transcriptomics comes of age. Nat. Commun. 11, 4307. doi:10.1038/s41467-020-18158-5
Anastasiadou, E., Jacob, L. S., and Slack, F. J. (2018). Non-coding RNA networks in cancer. Nat. Rev. Cancer 18, 5–18. doi:10.1038/nrc.2017.99
Bian, J., Dannappel, M., Wan, C., and Firestein, R. (2020). Transcriptional regulation of wnt/β-catenin pathway in colorectal cancer. Cells 9, E2125. doi:10.3390/cells9092125
Bian, L., Zhi, X., Ma, L., Zhang, J., Chen, P., Sun, S., et al. (2018). Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem. Biophys. Res. Commun. 505, 346–352. doi:10.1016/j.bbrc.2018.09.073
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Chen, L. L., and Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12, 381–388. doi:10.1080/15476286.2015.1020271
Chen, W., Li, S., Kulkarni, A. S., Huang, L., Cao, J., Qian, K., et al. (2020). Single cell omics: From assay design to biomedical application. Biotechnol. J. 15, e1900262. doi:10.1002/biot.201900262
Chen, Z., Xiao, K., Chen, S., Huang, Z., Ye, Y., and Chen, T. (2020). Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci. 111, 713–726. doi:10.1111/cas.14261
Cortes-Lopez, M., and Miura, P. (2016). Emerging functions of circular RNAs. Yale J. Biol. Med. 89, 527–537.
Falagas, M. E., Pitsouni, E. I., Malietzis, G. A., and Pappas, G. (2008). Comparison of PubMed, scopus, web of science, and google scholar: Strengths and weaknesses. FASEB J. 22, 338–342. doi:10.1096/fj.07-9492LSF
Fan, X., Zhang, X., Wu, X., Guo, H., Hu, Y., Tang, F., et al. (2015). Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148. doi:10.1186/s13059-015-0706-1
Feng, J., Li, Z., Li, L., Xie, H., Lu, Q., and He, X. (2020). Hypoxiainduced circCCDC66 promotes the tumorigenesis of colorectal cancer via the miR3140/autophagy pathway. Int. J. Mol. Med. 46, 1973–1982. doi:10.3892/ijmm.2020.4747
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi:10.1038/nature11993
He, X., Ma, J., Zhang, M., Cui, J., and Yang, H. (2020). Circ_0007031 enhances tumor progression and promotes 5-fluorouracil resistance in colorectal cancer through regulating miR-133b/ABCC5 axis. Cancer Biomark. 29, 531–542. doi:10.3233/CBM-200023
Hon, K. W., Ab-Mutalib, N. S., Abdullah, N. M. A., Jamal, R., and Abu, N. (2019). Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Sci. Rep. 9, 16497. doi:10.1038/s41598-019-53063-y
Hsiao, K. Y., Lin, Y. C., Gupta, S. K., Chang, N., Yen, L., Sun, H. S., et al. (2017). Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 77, 2339–2350. doi:10.1158/0008-5472.CAN-16-1883
Huang, M., Zhong, Z., Lv, M., Shu, J., Tian, Q., and Chen, J. (2016). Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget 7, 47186–47200. doi:10.18632/oncotarget.9706
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi:10.1261/rna.035667.112
Jiang, Z., Hou, Z., Li, L., Liu, W., Yu, Z., and Chen, S. (2021). Exosomal circEPB41L2 serves as a sponge for miR-21-5p and miR-942-5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway. Eur. J. Clin. Invest. 51, e13581. doi:10.1111/eci.13581
Jin, X., Feng, C. Y., Xiang, Z., Chen, Y. P., and Li, Y. M. (2016). CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget 7, 66455–66467. doi:10.18632/oncotarget.12186
Jin, Y. D., Ren, Y. R., Gao, Y. X., Zhang, L., and Ding, Z. (2019). Hsa_circ_0005075 predicts a poor prognosis and acts as an oncogene in colorectal cancer via activating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 3311–3319. doi:10.26355/eurrev_201904_17693
Kern, F., Aparicio-Puerta, E., Li, Y., Fehlmann, T., Kehl, T., Wagner, V., et al. (2021). miRTargetLink 2.0-interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 49, W409–W416. doi:10.1093/nar/gkab297
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Lei, Y., Tang, R., Xu, J., Wang, W., Zhang, B., Liu, J., et al. (2021). Applications of single-cell sequencing in cancer research: Progress and perspectives. J. Hematol. Oncol. 14, 91. doi:10.1186/s13045-021-01105-2
Li, M., Zhuang, J., Kang, D., Chen, Y., and Song, W. (2021). Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis. Open Life Sci. 16, 523–536. doi:10.1515/biol-2021-0053
Li, S., and Zheng, S. (2020). Down-regulation of Circ_0032833 sensitizes colorectal cancer to 5-fluorouracil and oxaliplatin partly depending on the regulation of miR-125-5p and MSI1. Cancer Manag. Res. 12, 11257–11269. doi:10.2147/CMAR.S270123
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015). Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell. Res. 25, 981–984. doi:10.1038/cr.2015.82
Liu, F., Li, R., Zhang, R., He, M., and Zhang, Y. (2021). Knockdown of circNRIP1 sensitizes colorectal cancer to 5FU via sponging miR5323p. Oncol. Rep. 46, 218. doi:10.3892/or.2021.8169
Liu, J., Li, D., Luo, H., and Zhu, X. (2019). Circular RNAs: The star molecules in cancer. Mol. Asp. Med. 70, 141–152. doi:10.1016/j.mam.2019.10.006
Liu, J., Liu, T., Wang, X., and He, A. (2017). Circles reshaping the RNA world: From waste to treasure. Mol. Cancer 16, 58. doi:10.1186/s12943-017-0630-y
Liu, Q., Zhang, X., Hu, X., Dai, L., Fu, X., Zhang, J., et al. (2016). Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'sponge' in human cartilage degradation. Sci. Rep. 6, 22572. doi:10.1038/srep22572
Liu, W., Zhang, J., Zou, C., Xie, X., Wang, Y., Wang, B., et al. (2017). Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell. Physiol. biochem. 43, 969–985. doi:10.1159/000481650
Louis, C., Desoteux, M., and Coulouarn, C. (2019). Exosomal circRNAs: New players in the field of cholangiocarcinoma. Clin. Sci. 133, 2239–2244. doi:10.1042/CS20190940
Lu, H., Yao, B., Wen, X., and Jia, B. (2019). FBXW7 circular RNA regulates proliferation, migration and invasion of colorectal carcinoma through NEK2, mTOR, and PTEN signaling pathways in vitro and in vivo. BMC Cancer 19, 918. doi:10.1186/s12885-019-6028-z
Lv, C., Sun, L., Guo, Z., Li, H., Kong, D., Xu, B., et al. (2018). Circular RNA regulatory network reveals cell-cell crosstalk in acute myeloid leukemia extramedullary infiltration. J. Transl. Med. 16, 361. doi:10.1186/s12967-018-1726-x
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi:10.1038/nature11928
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P., et al. (2017). CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16, 94. doi:10.1186/s12943-017-0663-2
Millas, S. G., Alawadi, Z. M., Wray, C. J., Silberfein, E. J., Escamilla, R. J., Karanjawala, B. E., et al. (2015). Treatment delays of colon cancer in a safety-net hospital system. J. Surg. Res. 198, 311–316. doi:10.1016/j.jss.2015.03.078
Natarajan, K. N., Miao, Z., Jiang, M., Huang, X., Zhou, H., Xie, J., et al. (2019). Comparative analysis of sequencing technologies for single-cell transcriptomics. Genome Biol. 20, 70. doi:10.1186/s13059-019-1676-5
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled exons. Cell. 64, 607–613. doi:10.1016/0092-8674(91)90244-s
Nusse, R., and Clevers, H. (2017). Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell. 169, 985–999. doi:10.1016/j.cell.2017.05.016
Oliveira, H. C., Hirata, M. H., Redgrave, T. G., and Maranhao, R. C. (1988). Competition between chylomicrons and their remnants for plasma removal: A study with artificial emulsion models of chylomicrons. Biochim. Biophys. Acta 958, 211–217. doi:10.1016/0005-2760(88)90179-8
Pan, Z., Cai, J., Lin, J., Zhou, H., Peng, J., Liang, J., et al. (2020). A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol. Cancer 19, 71. doi:10.1186/s12943-020-01179-5
Panda, A. C. (2018). Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 1087, 67–79. doi:10.1007/978-981-13-1426-1_6
Poggio, M., Hu, T., Pai, C. C., Chu, B., Belair, C. D., Chang, A., et al. (2019). Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 177, 414–427. doi:10.1016/j.cell.2019.02.016
Rantalainen, M. (2018). Application of single-cell sequencing in human cancer. Brief. Funct. Genomics 17, 273–282. doi:10.1093/bfgp/elx036
Ren, T. J., Liu, C., Hou, J. F., and Shan, F. X. (2020). CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 1743–1754. doi:10.26355/eurrev_202002_20351
Sanderson, R. D., Bandari, S. K., and Vlodavsky, I. (2019). Proteases and glycosidases on the surface of exosomes: Newly discovered mechanisms for extracellular remodeling. Matrix Biol. 75-76, 160–169. doi:10.1016/j.matbio.2017.10.007
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73, 3852–3856. doi:10.1073/pnas.73.11.3852
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shen, L., Li, Q., Wang, W., Zhu, L., Zhao, Q., Nie, Y., et al. (2020). Treatment patterns and direct medical costs of metastatic colorectal cancer patients: A retrospective study of electronic medical records from urban China. J. Med. Econ. 23, 456–463. doi:10.1080/13696998.2020.1717500
Siegel, R. L., Miller, K. D., Fedewa, S. A., Ahnen, D. J., Meester, R. G. S., Barzi, A., et al. (2017). Colorectal cancer statistics, 2017. Ca. Cancer J. Clin. 67, 177–193. doi:10.3322/caac.21395
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer statistics, 2021. CA Cancer J. Clin. 71, 7–33. doi:10.3322/caac.21654
Su, S., Lu, W., Liu, J., Li, L., Liu, L., Li, X., et al. (2022). Circ_0007031 silencing inhibits cell proliferation and induces cell apoptosis via downregulating MELK at a miR-485-3p-dependent way in colorectal cancer. Biochem. Genet. 60, 576–597. doi:10.1007/s10528-021-10111-5
Sun, Y., Cao, Z., Shan, J., Gao, Y., Liu, X., Ma, D., et al. (2020). Hsa_circ_0020095 promotes oncogenesis and cisplatin resistance in colon cancer by sponging miR-487a-3p and modulating SOX9. Front. Cell. Dev. Biol. 8, 604869. doi:10.3389/fcell.2020.604869
Vallee, A., and Lecarpentier, Y. (2018). Crosstalk between peroxisome proliferator-activated receptor gamma and the canonical WNT/β-Catenin pathway in chronic inflammation and oxidative stress during carcinogenesis. Front. Immunol. 9, 745. doi:10.3389/fimmu.2018.00745
Wang, L., Peng, X., Lu, X., Wei, Q., Chen, M., and Liu, L. (2019). Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: Effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol. Res. Pract. 215, 689–696. doi:10.1016/j.prp.2018.12.032
Wang, L., Zhou, J., Zhang, C., Chen, R., Sun, Q., Yang, P., et al. (2021). A novel tumour suppressor protein encoded by circMAPK14 inhibits progression and metastasis of colorectal cancer by competitively binding to MKK6. Clin. Transl. Med. 11, e613. doi:10.1002/ctm2.613
Wang, Q., Shi, L., Shi, K., Yuan, B., Cao, G., Kong, C., et al. (2020). CircCSPP1 functions as a ceRNA to promote colorectal carcinoma cell EMT and liver metastasis by upregulating COL1A1. Front. Oncol. 10, 850. doi:10.3389/fonc.2020.00850
Wang, X., Ren, Y., Ma, S., and Wang, S. (2020). Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. Onco. Targets. Ther. 13, 1941–1951. doi:10.2147/OTT.S240642
Wang, X., Zhang, H., Yang, H., Bai, M., Ning, T., Deng, T., et al. (2020). Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 14, 539–555. doi:10.1002/1878-0261.12629
Wang, Y., Liu, J., Ma, J., Sun, T., Zhou, Q., Wang, W., et al. (2019). Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 18, 116. doi:10.1186/s12943-019-1041-z
Wen, T., Wu, H., Zhang, L., Li, K., Xiao, X., Zhang, L., et al. (2021). Circular RNA circ_0007142 regulates cell proliferation, apoptosis, migration and invasion via miR-455-5p/SGK1 axis in colorectal cancer. Anticancer. Drugs 32, 22–33. doi:10.1097/CAD.0000000000000992
Weng, W., Wei, Q., Toden, S., Yoshida, K., Nagasaka, T., Fujiwara, T., et al. (2017). Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 23, 3918–3928. doi:10.1158/1078-0432.CCR-16-2541
Winterhoff, B., Talukdar, S., Chang, Z., Wang, J., and Starr, T. K. (2019). Single-cell sequencing in ovarian cancer: A new frontier in precision medicine. Curr. Opin. Obstet. Gynecol. 31, 49–55. doi:10.1097/GCO.0000000000000516
Wu, L., Xia, J., Yang, J., Shi, Y., Xia, H., Xiang, X., et al. (2018). Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J. BUON 23, 1343–1349.
Xi, L., Liu, Q., Zhang, W., Luo, L., Song, J., Liu, R., et al. (2021). Circular RNA circCSPP1 knockdown attenuates doxorubicin resistance and suppresses tumor progression of colorectal cancer via miR-944/FZD7 axis. Cancer Cell. Int. 21, 153. doi:10.1186/s12935-021-01855-6
Xu, B., Yang, N., Liu, Y., Kong, P., Han, M., and Li, B. (2020). Circ_cse1l inhibits colorectal cancer proliferation by binding to eIF4A3. Med. Sci. Monit. 26, e923876. doi:10.12659/MSM.923876
Xu, Y., Qiu, A., Peng, F., Tan, X., Wang, J., and Gong, X. (2021). Exosomal transfer of circular RNA FBXW7 ameliorates the chemoresistance to oxaliplatin in colorectal cancer by sponging miR-18b-5p. Neoplasma 68, 108–118. doi:10.4149/neo_2020_200417N414
Yang, G., Zhang, Y., and Yang, J. (2019). Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in gastric carcinoma using bioinformatics analysis. Med. Sci. Monit. 25, 8777–8796. doi:10.12659/MSM.916902
Yin, W., Xu, J., Li, C., Dai, X., Wu, T., and Wen, J. (2020). Circular RNA circ_0007142 facilitates colorectal cancer progression by modulating CDC25A expression via miR-122-5p. Onco. Targets. Ther. 13, 3689–3701. doi:10.2147/OTT.S238338
Zeng, H., Chen, W., Zheng, R., Zhang, S., Ji, J. S., Zou, X., et al. (2018). Changing cancer survival in China during 2003-15: A pooled analysis of 17 population-based cancer registries. Lancet Glob. Health 6, e555–e567. doi:10.1016/S2214-109X(18)30127-X
Zeng, W., Liu, Y., Li, W. T., Li, Y., and Zhu, J. F. (2020). CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol. Oncol. 14, 2960–2984. doi:10.1002/1878-0261.12796
Zhang, J., Wang, H., Wu, K., Zhan, F., and Zeng, H. (2020). Dysregulated circRNA_100876 contributes to proliferation and metastasis of colorectal cancer by targeting microRNA-516b (miR-516b). Cancer Biol. Ther. 21, 733–740. doi:10.1080/15384047.2020.1776075
Zhang, L., Yu, R., Li, C., Dang, Y., Yi, X., and Wang, L. (2021). Circ_0026416 downregulation blocks the development of colorectal cancer through depleting MYO6 expression by enriching miR-545-3p. World J. Surg. Oncol. 19, 299. doi:10.1186/s12957-021-02407-y
Zhang, W., Wang, Z., Cai, G., and Huang, P. (2021). Downregulation of Circ_0071589 suppresses cisplatin resistance in colorectal cancer by regulating the MiR-526b-3p/KLF12 Axis. Cancer Manag. Res. 13, 2717–2731. doi:10.2147/CMAR.S294880
Zhang, X., Xu, Y., Yamaguchi, K., Hu, J., Zhang, L., Wang, J., et al. (2020). Circular RNA circVAPA knockdown suppresses colorectal cancer cell growth process by regulating miR-125a/CREB5 axis. Cancer Cell. Int. 20, 103. doi:10.1186/s12935-020-01178-y
Zhang, X., Zhao, Y., Kong, P., Han, M., and Li, B. (2019). Expression of circZNF609 is down-regulated in colorectal cancer tissue and promotes apoptosis in colorectal cancer cells by upregulating p53. Med. Sci. Monit. 25, 5977–5985. doi:10.12659/MSM.915926
Zhang, Y., Bi, J., Huang, J., Tang, Y., Du, S., and Li, P. (2020). Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int. J. Nanomedicine 15, 6917–6934. doi:10.2147/IJN.S264498
Zhang, Y., Ke, X., Liu, J., Ma, X., Liu, Y., Liang, D., et al. (2019). Characterization of circRNAassociated ceRNA networks in patients with nonvalvular persistent atrial fibrillation. Mol. Med. Rep. 19, 638–650. doi:10.3892/mmr.2018.9695
Zhang, Y., Liu, H., Li, W., Yu, J., Li, J., Shen, Z., et al. (2017). CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 9, 1585–1594. doi:10.18632/aging.101254
Zhang, Y., Sun, X., Icli, B., and Feinberg, M. W. (2017). Emerging roles for MicroRNAs in diabetic microvascular disease: Novel targets for therapy. Endocr. Rev. 38, 145–168. doi:10.1210/er.2016-1122
Zhang, Y., Tan, X., and Lu, Y. (2022). Exosomal transfer of circ_0006174 contributes to the chemoresistance of doxorubicin in colorectal cancer by depending on the miR-1205/CCND2 axis. J. Physiol. Biochem. 78, 39–50. doi:10.1007/s13105-021-00831-y
Zhao, K., Cheng, X., Ye, Z., Li, Y., Peng, W., Wu, Y., et al. (2021). Exosome-mediated transfer of circ_0000338 enhances 5-fluorouracil resistance in colorectal cancer through regulating MicroRNA 217 (miR-217) and miR-485-3p. Mol. Cell. Biol. 41, e00517–e00520. doi:10.1128/MCB.00517-20
Zhong, D., Li, P., and Gong, P. Y. (2019). Hsa_circ_0005075 promotes the proliferation and invasion of colorectal cancer cells. Int. J. Biol. Markers 34, 284–291. doi:10.1177/1724600819872765
Zhou, G. R., Huang, D. P., Sun, Z. F., and Zhang, X. F. (2020). Characteristics and prognostic significance of circRNA-100876 in patients with colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 24, 11587–11593. doi:10.26355/eurrev_202011_23801
Zhou, R., Wu, Y., Wang, W., Su, W., Liu, Y., Wang, Y., et al. (2018). Circular RNAs (circRNAs) in cancer. Cancer Lett. 425, 134–142. doi:10.1016/j.canlet.2018.03.035
Keywords: biomarkers, circRNA, colorectal cancer, miRNA, ceRNA, exosome
Citation: Xiao Y, Qiu M, Tan C, Huang W, Hu S, Jiang X, Guo M, Wang C, Liang J, Wu Y, Li M, Li Q and Qin C (2022) Systematic analysis of circRNA biomarkers for diagnosis, prognosis and therapy in colorectal cancer. Front. Genet. 13:938672. doi: 10.3389/fgene.2022.938672
Received: 16 May 2022; Accepted: 23 September 2022;
Published: 12 October 2022.
Edited by:
Parvin Mehdipour, Tehran University of Medical Sciences, IranReviewed by:
Pouria Samadi, Hamadan University of Medical Sciences, IranCopyright © 2022 Xiao, Qiu, Tan, Huang, Hu, Jiang, Guo, Wang, Liang, Wu, Li, Li and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjiang Qin, cWluY2o4ODhAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.