
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 25 July 2022
Sec. Livestock Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.938526
This article is part of the Research Topic Epigenetics and Epigenomics in Aquatic Organisms View all 7 articles
 Chenxi Zhu1†
Chenxi Zhu1† Guoxing Liu1,2†
Guoxing Liu1,2† Xiankun Gu1,3
Xiankun Gu1,3 Jiawen Yin1,3
Jiawen Yin1,3 Aijun Xia1,3
Aijun Xia1,3 Mingming Han4
Mingming Han4 Tongqing Zhang1,3
Tongqing Zhang1,3 Qichen Jiang1,3*
Qichen Jiang1,3*Quercetin is a flavanol beneficial in reducing fat, promoting muscle growth, and Anti-oxidation. To study its effects in freshwater fish, the full-length cDNA of the follistatin (FST) and myostatin (MSTN) genes of the dark sleeper Odontobutis potamophila were cloned for the first time. Juvenile individual O. potamophila was exposed to quercetin at one of four concentrations (0, 2.5, 5, and 10 mg/L) for 21 days. The expression level of MSTN which inhibits muscle growth in the quercetin solution was lower than in the unexposed control group. The genes that promote muscle growth are in TGF-β superfamily like FST, TGF-β1 (transforming growth factor-beta 1), and Myogenic regulatory factors (MRFs) like Myf5 (myogenic factor 5), MyoD (myogenic differentiation), MyoG (myogenin), were higher than in the control group. Apolipoprotein and growth hormone receptor transcription levels in the quercetin-treated fish were significantly lower than in the control group. The concentrations of triglyceride, low-density lipoprotein cholesterol, and high-density lipoprotein cholesterol in the muscle tissue decreased, and the lipid-lowering function of quercetin was also demonstrated at the biochemical level. In this study, we analyzed the mRNA levels of AKT, Keap1 (kelch-like ECH-associated protein 1), Nrf2 (NF-E2-related factor 2) oxidation-related genes in the Nrf2/ARE antioxidant pathway, and Malondialdehyde (MDA), catalase (CAT) activity and glutathione (GSH) content in the hepatopancreas of O. potamophila after quercetin treatment, the mRNA expression of AKT, Nrf2 and CAT activity and GSH content are higher than in the control group. Quercetin enhances antioxidant properties and positively affects muscle growth. The results showed that quercetin has no significant effects on the growth performance of O. potamophila, but is effective in increasing muscle growth rate and lowering muscle fat content.
Quercetin is a flavanol widely distributed in food and vegetables such as tea, apples, cocoa, onions, and red wine (Aebi, 1984). It has been shown to have beneficial biological effects on health, including antioxidant, anti-inflammatory, anti-tumor, and anti-bacterial effects, as well as pharmacological effects on cardiovascular system protection (da Silva et al., 1998; Shutenko et al., 1999; Prince and Sathya, 2010). Quercetin can inhibit dietary energy absorption, regulate body fat metabolism, and inhibit triglyceride deposition, showing significant hypolipidemic effects both in vivo and in vitro in animals (Cai-ke et al., 2012; Pallauf et al., 2017; Forney et al., 2018; Pourteymour Fard Tabrizi et al., 2020). It ameliorates lipemia, hypertension, and hyperinsulinemia in obese rats while reducing weight gain and prolonging life span (Rivera et al., 2008). However, information on its effects on aquatic animals is quite limited.
Follistatin (FST) is a secreted protein that binds to, and inhibits, the activity of many proteins, including bone morphogenetic proteins (BMPs) and myostatin (MSTN), and growth differentiation factor-9 (GDF-9) and growth differentiation factor 11 (GDF-11) in the transforming growth factor β (TGFβ) superfamily (Funkenstein and Jakowlew, 1997; Iemura et al., 1998; Gamer et al., 1999; Amthor et al., 2002). FST is widely located in organisms and plays a role in a variety of physiological activities. A higher level of FST occurs in groupers (Family Serranidae) after consuming hydrolyzed porcine mucosa (HPM) feed, promoting muscle growth and improving meat quality (Lee and McPherron, 2001). The binding of MSTN to the activin A complex (Act RIIB) can be blocked by the activin-binding protein follistatin, suggesting that propeptide, follistatin, or other molecules can inhibit signaling through this pathway. MSTN is a member of the TGFβ superfamily and is involved in the inhibition of muscle differentiation and growth (Ferrell et al., 1999). In Zebrafish Danio rerio (Dong et al., 2014; Gao et al., 2016), Japanese ricefish Oryzias latipes (Chiang et al., 2016), and Eurasian carp Cyprinus carpio (Zhong et al., 2016), MSTN leads to inhibition of muscle growth. Knocking out MSTN in the red seabream Pagrus major using CRISPR/Cas9 increased skeletal muscle growth and the gene mutation resulted in a loss of protein function, causing an increase in the number of muscle cells and the diameter of muscle fibers, and consequent muscle overgrowth (Argilés et al., 2012). Marcelos (Aoki et al., 2010) demonstrated that all homologs of FST, such as FST-288, FST-315, and FST-L3 promote muscle growth in mice. By using FST in transgenic zebrafish, MSTN in the muscles was suppressed, promoting muscles growth (Xu et al., 2003). The co-expression of FST and MSTN in the different somatic cell groups in the brain and muscles of bighead carp Hypophthalmichthys nobilis indicated that FST could suppress the expression of MSTN, and thus, promote their growth (Pang et al., 2018). A-I transports lipids and stabilizes the structure of plasma lipoproteins.
Upon binding to its receptor ghra, growth hormone (GH) affects the metabolism of carbohydrates, lipids, and proteins in animals. The ghra mediates a wide range of growth-related and metabolic functions, both directly and via insulin-like growth factor 1 (IGF-1) (Brooks and Waters, 2010; Waters, 2016). Quercetin has been used in animal experiments to treat oxidative damage caused by the triose phosphate/phosphate translocator (TPT) in zebrafish (Zhang et al., 2021). Oral administration of quercetin was found to lower blood glucose and normalize plasma lipid and protein profiles in rats suffering from diabetes (Ahmad et al., 2017). However, The effects of quercetin on muscle growth-related genes in freshwater fish have not been reported.
P2X7R is an ion path for ATP that can activate and induce ROS production through high levels of ATP(S) and alter the levels of oxidative stress markers (GSH, SOD, CAT, GPX, and GR) (Jiang et al., 2017), suggesting a correlation between P2X7R and oxidative stress. Quercetin alleviates oxidative stress through the P2X7R-mediated Nrf2/ARE antioxidant pathway, which further mediates the mRNA expression of PI3K, Keap1, and Nrf2 to prevent liver damage (Lee et al., 2019; Rubio-Ruiz et al., 2019; Zhao et al., 2021). In terms of the protective mechanisms, the excessive accumulation of reactive oxygen species (ROS) in fish can lead to tissue lipid peroxidation (POD), which can seriously damage cells (Valavanidis et al., 2006; Wu et al., 2017). Antioxidant enzyme systems, such as superoxide dismutase (SOD), catalase (CAT), peroxidase (GPX), and glutathione (GSH), can prevent excessive ROS (Lu et al., 2019) from potentially damaging tissues. High levels of ROS can interact with lipids and proteins and induce oxidative stress (Martínez-Álvarez et al., 2005). Importantly, fish muscle tissue is more sensitive to oxidative stress due to high levels of polyunsaturated fatty acids (Tokur and Korkmaz, 2007). The decrease in fish flesh quality may be related to the disruption of muscle structural integrity due to oxidative damage in fish (Buckley et al., 1995). In this study, we verified the antioxidant properties of quercetin by analyzing the mRNA levels of AKT, Keap1, and Nrf2 oxidation-related genes in the Nrf2/ARE antioxidant pathways and the changes of antioxidant enzymes MDA, CAT activity, and nonenzymatic substance GSH content after quercetin treatment.
The dark sleeper Odontobutis potamophila is a commercially valuable freshwater fish that is widely distributed in the river systems of China and Southeast Asian countries (Hou et al., 2014), and shows significant sexual dimorphism in growth, with males growing more than 30% faster than females over the same period (Zhao et al., 2017). Aquaculture of this species is of interest because of its high meat content, taste, nutritional value, and potentially high profitability (Wang et al., 2017; Jia et al., 2021). Therefore, O. potamophila was selected as the study species for this investigation of the molecular mechanism of quercetin effects in fish.
The aims of this study were: 1) to clone the full-length cDNA of the FST and MSTN genes; 2) to test the effect of quercetin on the growth of O. potamophila; 3) to examine the mRNA expression of the FST, MSTN, A-I, ghra, genes in different tissues, and the Myf5, MyoD, MyoG, AKT, Keap1, Nrf2 and TGF-β1 in muscle; and 4) to detect the effects of quercetin on growth-related and biochemical parameters and antioxidant enzyme activity in O. potamophila. The results of this study will provide insight into the mechanisms by which growth-related genes regulate muscle development in O. potamophila under various quercetin treatments at the molecular level, and extend the use of quercetin in fish culture.
This article does not contain any studies with human participants by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
The O. potamophila were obtained from the Freshwater Fisheries Research Institute of Jiangsu Province, Nanjing, China. The 144 O. potamophila used in this study had an individual weight of 1.1 ± 0.05 g. The temperature and pH of the water were maintained at 25 ± 1°C and 7.2 ± 0.2, respectively. The dissolved oxygen concentration in the water was maintained at about 5.0 mg L−1. During the acclimation period, all were fed a diet of Limnodrilus hoffmeisteri. The feeding rate was set at 5% of the fish body weight. Quercetin solution was completely changed every 2 days to ensure stable experimental concentration.
After acclimation, the fish were not fed for 24 h before being exposed to the different experimental quercetin concentrations (0, 2.5, 5, and 10 mg/L) for 21 d. Judging by preliminary experiments, 10 mg/L of quercetin was not harmful to O. potamophila. Quercetin (Sigma-Aldrich), purity >98%, was dissolved in dimethyl sulfoxide before use and stored at −20°C in the dark. There were six replicate aquariums for each treatment condition and each aquarium contained six O. potamophila, all living under the same cultural conditions.
During the experiments, all of the fish were fed L. hoffmeisteri twice daily (at 7:00 a.m. and 8:00 p.m.). The feeding rate was set at 5% of the fish body weight. Individuals were selected and treated with different concentrations of quercetin (0, 2.5, 5, and 10 mg/L) for 21 d. The quercetin solution was changed every day. Weight data were recorded at 7 d, 14 d, and 21 d during the experiment.
After 3 weeks, 144 sample fishes were anesthetized over ice, and samples of muscle tissue, gill tissue, intestinal tissue, and hepatopancreas tissue were collected using sterile scissors and forceps. The tissue samples were stored in liquid nitrogen. A control group without quercetin exposure was used for comparison and the gene cloning experiments.
TRIzol reagent (Aidlab Biotech Co., Beijing, China) was used to extract the total RNA for differential gene expression, according to the manufacturer’s instructions. FST and MSTN gene fragment data were obtained from the O. potamophila genome database or existing transcriptome data and analyzed by comparing their open reading frames (ORFs). Primers for the relevant genes were designed using Primer Premier 5.0. The names and sequences of the primers used are shown in Table 1. Full-length fluorescent quantitative primers FST F, FST R, MSTN F, MSTN R, ghra F, ghra R, A-1 F, A-1 R, AKT F, AKT R, Keap1 F, Keap1 R, Nrf2 F, Nrf2 R, Myf5 F, Myf5 R, MyoD F, MyoD R, MyoG F, MyoG R, TGF-β1 F, TGF-β1 R. (Table 1). and O. potamophila β-actin gene-specific upstream and downstream primers β-actin F, β-actin R were designed as internal reference genes. The RACE PCR primers used in this paper are also shown in Table 1.
First-strand synthesis of cDNA was performed using a RevertAid First Strand cDNA Synthesis Kit (Fermentas, Burlington, Canada). cDNA was synthesized for gene cloning using the PrimeScript RT reagent kit (Takara, Shinga, Japan). Full-length sequences of cDNAs for FST and MSTN were obtained according to the instructions for the SMARTer RACE5 '/3′ kit, and the expression products were negative. In order to check the sharpness of bands and fragment lengths, the PCR products of amplified FST and MSTN were examined using gel electrophoresis on an agarose matrix.
The MSTN and FST PCR products with clear, accurate bands were sent for sequencing (by Baibaxun Biotechnology Co., Shanghai, China). The validity of the cDNA sequences was checked by comparing the sequencing results with the amino acid sequences of the same genes in the NCBI database, using BLASTP. Following completion of the comparisons, the 5 and 3′ ends were obtained, and the full-length FST and MSTN sequences were obtained by splicing the ends and intermediate sequences using the DNAMan software.
In this study, ORF intervals were predicted using the NCBI ORF Finder. Homologous proteins were retrieved and analyzed using BLASTP, and protein hydrophobic regions were analyzed using ProtScale on the ExPASy server. ProtParam on the ExPASy server was used to calculate amino acid compositions, relative molecular weights, and isoelectric points. The signal peptides were predicted using the SignalP 5.0 Server from DTU Health Tech (Lyngby, Denmark). Multiple sequence alignment was performed using seeded guide trees and HMM profile-profile techniques using Clustal Omega from EMBL-EBI (Hinxton, United Kingdom). Protein structural domains were analyzed using SMART, and secondary and tertiary structures were analyzed using the PSIRED Protein Structure Prediction Server and the SWISS-MODEL Server, respectively. Sequences were aligned with MAFFT (Katoh and Standley, 2013) using “--auto” strategy and normal alignment mode. Gap sites were removed with trimAl (Katoh and Standley, 2013) using “-strictplus” command. ModelFinder (Katoh and Standley, 2013)was used to select the best-fit model using BIC criterion. Maximum likelihood phylogenies were inferred using IQ-TREE (Nguyen et al., 2015) under the JTTDCMut + G4 model for 20,000 ultrafast (Minh et al., 2013) bootstraps, as well as the Shimodaira–Hasegawa–like approximate likelihood-ratio test (Minh et al., 2013).
The RNA extracted from the hepatopancreas, muscle, gill, and intestinal tissues of O. potamophila was reverse transcribed into cDNA using a PrimeScript RT reagent Kit (Takara, Shinga, Japan) and stored at −80° for real-time fluorescence qPCR analysis. Total RNA from each tissue sample was analyzed by RT-qPCR using CFX96 RT-PCR (BioRad, Hercules, CA, United States) and TransStart Top Green qPCR SuperMix (TransGen, Beijing, China).
Triglycerides (TGs), low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) levels were measured using a kit supplied by the Nanjing Jiancheng Bioengineering Research Institute (Nanjing, China). The TG content was calculated by mixing 10 μl of tissue homogenate with 1 L of enzyme agent, incubating at 37°C for 10 min, and then measuring the color at 510 nm colorimetrically. The resulting H2O2 reacted with 4-AAP to produce a red-purple pigment, and the LDL-C and HDL-C contents were tested by absorbance at 546 nm. GSH in tissues can be measured colorimetrically at 405 nm. CAT activity was measured by monitoring the stable complex produced by H2O2 with ammonium molybdate, measured with ammonium molybdate at 405 nm optical diameter. MDA in peroxidized lipid degradation products was measured calorimetrically at 405 nm.
The experimental data were examined graphically using GraphPad Prism 8 (GraphPad Software, San Diego, CA, United States) and SPSS 20.0 software (IBM, Armonk, NY, United States). The relative mRNA levels of target genes were analyzed using the 2−ΔΔCt method (Livak and Schmittgen, 2001). One-way analysis of variance (ANOVA) was used to test the significance of differences in growth data and muscle growth-related gene expression between the various quercetin-treated groups and the control.
The datasets presented in this study can be found in online repositories. The complete mRNA sequences for FST and MSTN were submitted to GenBank with the accession numbers OK641659 and OK641660, respectively. The FST sequence encodes for 506 amino acids. The predicted molecular mass of the protein was 38.60 kDa with an estimated pI of 6.45. The MSTN sequence encodes 376 amino acids. The predicted molecular mass of the protein was 42.42 kDa and the estimated pI is 5.66.
Sequencing results and amino acid sequence analysis of transcription factor FST (Figure 1) and MSTN (Figure 2) of O. potamophila. The homology comparison results showed that the similarity of the FST sequences was very high, with the highest homology between the O. potamophila FST sequence and that of Sparus aurata, followed by Paralichthys olivaceus. The signal peptide region, TB (TGFβ-binding structural domain), three repeats in the FST of fish, mammals, amphibians, and birds, the follistatin structural domain, and three duplicated Kazal structural domains were very similar. The homology comparison results showed that the similarity between the corresponding MSTN sequences was very low in the signal peptide region, high in the pre-peptide structural domain, and very conserved in the C-terminal GDF8 structural domain (Figure 3).
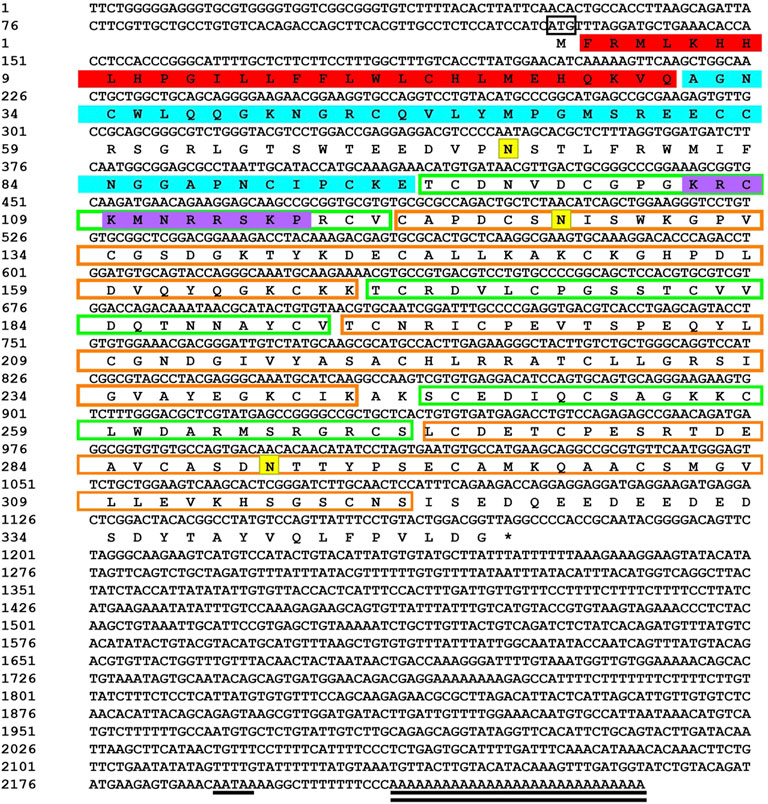
FIGURE 1. Sequencing results and amino acid sequence analysis of transcription factor FST of Odontobutis potamophila. The black box indicates the ATG start codon; * indicates the termination codon; underlines indicate the Poly(A) plus tail signal; double underlines indicate the poly(A) sequence; red shading indicates a possible signal peptide region; blue shading indicates a possible TGFβ binding structural domain; green boxes indicate three possible repetitive follistatin structural domains; orange boxes indicate three duplicated Kazal structural domains; purple shading indicates possible nuclear localization signals; and yellow shading indicates possible N-glycosylation sites.
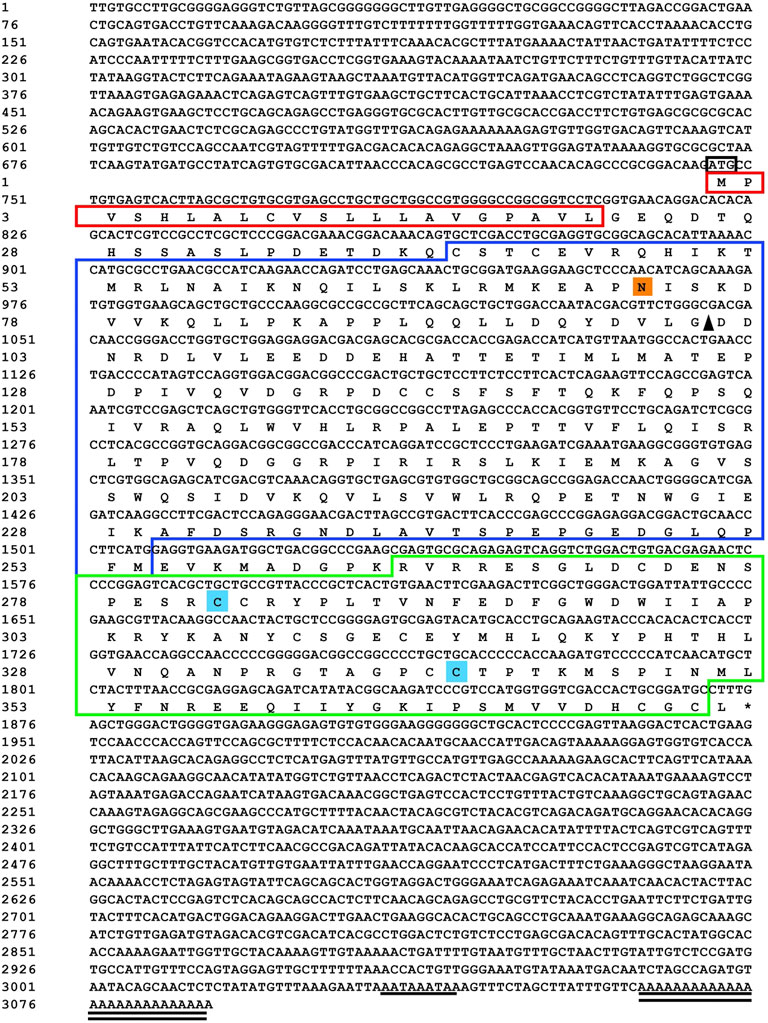
FIGURE 2. Sequencing results and amino acid sequence analysis of MSTN in Odontobutis potamophila. The black box indicates the ATG start codon; * indicates the termination codon; underlines indicate the Poly(A) plus tail signal; double underlines indicate the poly(A) sequence; red shading indicates possible signal peptide regions; blue shading indicates possible TGFβ binding structural domains; green boxes indicate three possible repetitive follistatin structural domains; orange boxes indicate three duplicated Kazal structural domains; purple shading indicates possible nuclear localization signals; and yellow shading indicates possible N-glycosylation sites.
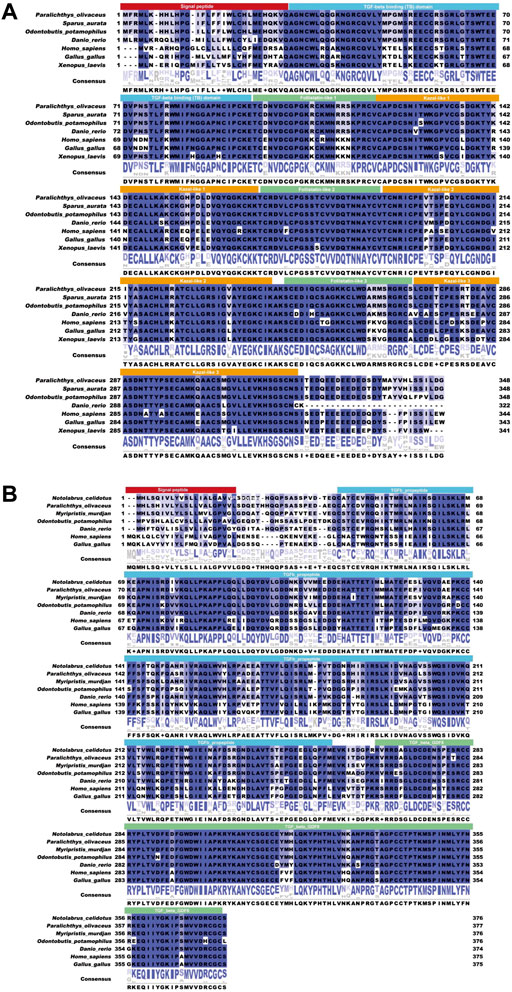
FIGURE 3. The results of the similarity analysis. (A) FST: the solid red box shows the signal peptide; the solid blue box shows the TGFβ binding domain; the solid yellow box shows the kazoo-like 2; the solid green box shows the FST-like 3. (B) MSTN: the solid red box shows the signal peptide; the solid blue box shows the TGFβ binding domain; and the solid green box shows the TGFβ GDF8.
The results of the FST evolutionary tree comparison are shown in Figure 4A. The total length of the evolutionary tree score was 0.60789282, and the FST sequences of each species were relatively close, indicating a very high homology in the sequences. FSTO. potamophilaFST. The MSTN evolutionary tree comparison showed that the total length of the evolutionary tree score was 1.08964954 and that the MSTN sequences of each species were not very homologous. Notolabrus celidotus. The MSTN and FST sequences of O. potamophila clustered together with those of other fishes and split into two large branches in the evolutionary tree, with a split between fishes, birds, and mammals (Figure 4AB).
FST is a multi-structural protein consisting of an N-terminal structural domain (labeled ND in Figure 5A for the TB structural domain) and three resultant FST structural domains (FSD1-3). FST is an antagonist, and FST-type molecules block all four MSTN receptor binding sites to suppress signaling (Figure 5A).
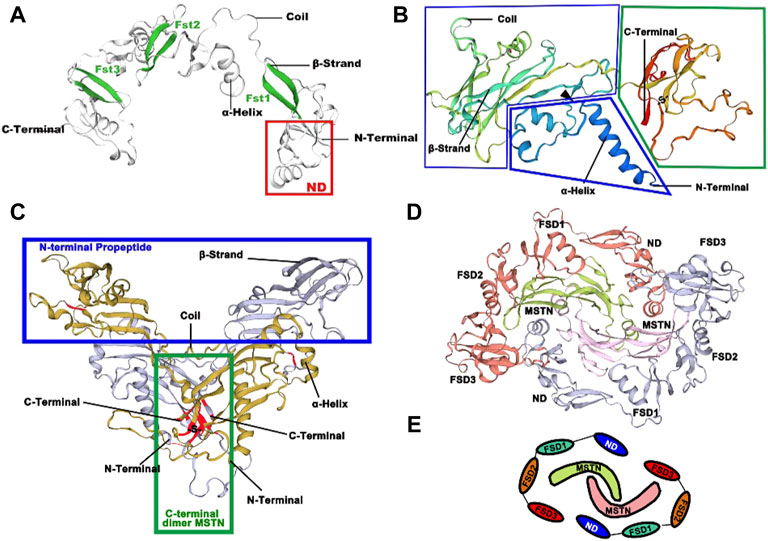
FIGURE 5. Three-dimension structures of FST and MSTN in O. potamophila. (A) Predicted 3D protein structure of transcription factor FST. (B) Predicted 3D protein structure of MSTN. (C) Predicted 3D protein structure of the MSTN homodimer. (D) Predicted 3D protein structure of the MSTN homodimer and the FST homodimer. (E) Schematic representation of the antagonistic relationship between MSTN and FST.
The possible tertiary structure of MSTN is shown in Figure 5B, where the protein is first synthesized as a large precursor molecule, which is then cleaved by protein hydrolysis to generate an N-terminal pro-peptide and a disulfide-linked C-terminal dimer. The cyclic form comprises a potential compound of the C-terminal dimer and other proteins (including its pre-peptide) that keep the C-terminal dimer in a potentially non-active state. Activation of the ligand requires further cleavage of the precursor protein by a tolylene-like metalloprotease that breaks at a pair of Asp residues (the black triangle in Figure 5B). The C-terminus forms a dimer through a disulfide bond, with the position of the Cys residue marked in red in Figure 5C. It is concentrated in the middle of the C-terminal mature peptide, both within the single subunit and between the two dimeric subunits, and has the potential to form disulfide bonds to create a stable dimer structure at the point where one of our predicted disulfide bonds occurs between Cys282-Cys341. In Figure 5D, red and gray colors represent the two FST homodimers, and the MSTN dimer is indicated in green and pink. Figure 5E shows a schematic representation of possible FST and MSTN antagonism (Cash et al., 2009).
No significant effect of quercetin on the growth and specific growth rate of O. potamophila was observed in this study (Du and Turchini, 2021) (Figure 6).
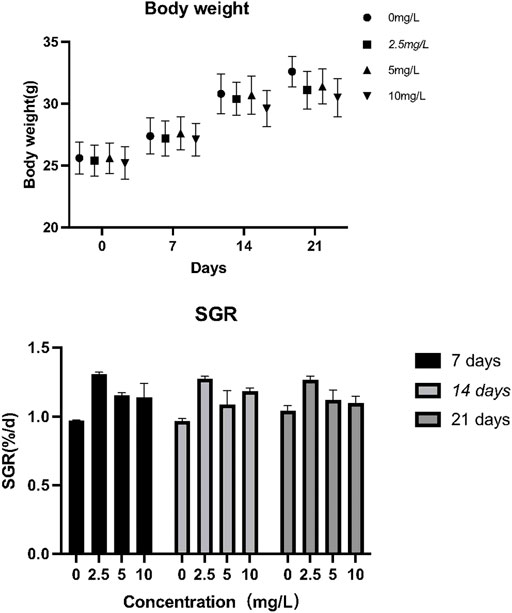
FIGURE 6. The effect of quercetin on the growth performance of Odontobutis potamophila. The bars indicate the mean weight ± SD (n = 3) of fish in the three treatment groups and the control. Statistical significance was taken as *(p < 0.05) and **(p < 0.01), compared with the control.
As can be seen from Figure 7, when O. potamophila were exposed to increasing concentrations of quercetin, a decreasing trend was observed in the muscles TG and LDL-C levels, and when the quercetin concentration was 10 mg/L, the muscle HDL-C and LDL-C levels were significantly lower than in the control group (p < 0.01). The activities of CAT and contents of GSH increased with increasing quercetin concentrations, with significant increases in GSH contents and CAT activities at 10 mg/L quercetin (p < 0.05). MDA content in the hepatopancreas of O. potamophila decreased as concentrations of quercetin levels increased, with significant decreases at 5 mg/L and 10 mg/L quercetin (p < 0.05).
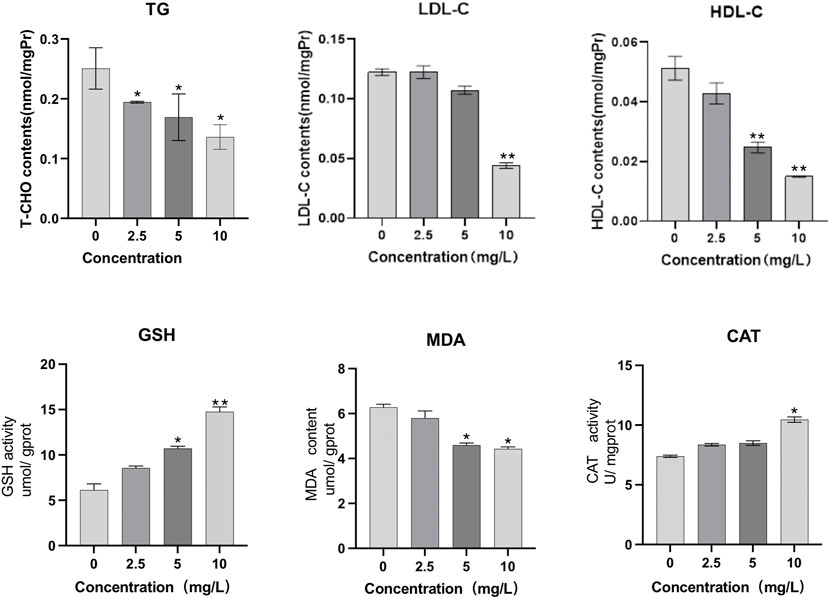
FIGURE 7. The effect of exposure to increasing concentrations of quercetin on triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), glutathione (GSH), Malondialdehyde (MDA) and catalase (CAT) in the muscles of O. potamophila. The bars indicate the Mean ± SD (n = 3). Statistical significance was taken as *(p < 0.05) and **(p < 0.01) compared with the control.
The distribution of FST and MSTN in different tissues was examined. MSTN and FST mRNA were detected in the gills, muscles, intestines, and hepatopancreas samples (Figure 9), with the highest levels found in muscle. An increasing trend of FST expression and a decreasing trend of MSTN expression in muscle and hepatopancreas tissues was found between the control and the 2.5 mg/L and 5 mg/L treatment groups. FST and MSTN were significantly expressed in muscle tissue (Figure 8). In Figure 9, the expression of the FST gene in the muscles of O. potamophila at quercetin exposure levels of 0, 2.5, and 5 mg/L increased in a stepwise manner with the highest expression in the 5 mg/L quercetin treatment group (p < 0.05). In contrast, the expression of MSTN in muscle tissue was lower in the quercetin-treated groups, and gene expression was significantly lower in the 2.5 mg/L quercetin-treated group (p < 0.01). Quercetin treatment reduced the expression of A-1 and ghra in all tissues. In muscle tissue, A-1 and ghra gene expressions were significantly reduced in the 10 mg/L treatment group (p < 0.01).
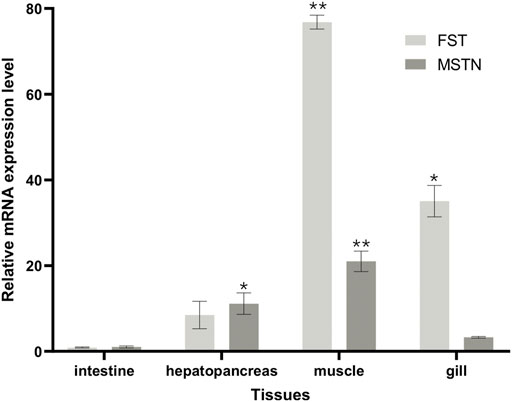
FIGURE 8. FST and MSTN gene expressions in various tissues of O. potamophila exposed to 0, 2.5, 5 and 10 mg/L of quercetin. The bars indicate the Mean ± SD (n ± 3). Statistical significance was taken as *(p < 0.05) and **(p < 0.01) compared with the intestine samples.
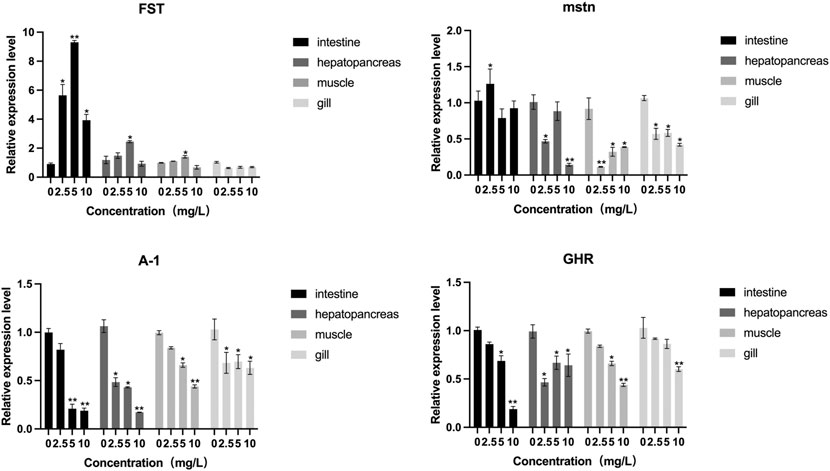
FIGURE 9. The effect of increasing concentrations of quercetin exposure on the relative expression of the FST, MSTN, A-I and GHR genes in the intestine, hepatopancreas, muscle and gill tissue of Odontobutis potamophila after 21 d. The bars indicate the Mean ± SD (n = 3). Statistical significance was taken as *(p < 0.05) and **(p < 0.01) compared with the control.
The relative mRNA expression of AKT and Nrf2 increased with increasing quercetin concentrations, with significant increases in AKT and Nrf2 activities at 10 mg/L quercetin (p < 0.01). The relative mRNA expression of Keap1 in the muscle of O. potamophila decreased as concentrations of quercetin levels increased, with significant decreases at 5 mg/L and 10 mg/L quercetin (p < 0.05). The relative mRNA expression of MyoD, MyoG and TGF-β1 increased significantly at 10 mg/L quercetin (p < 0.05). The relative mRNA expression of Myf5 increased significantly at 2.5 mg/L,5 mg/L (p < 0.05) and 10 mg/L quercetin (p < 0.01) (Figure 10).
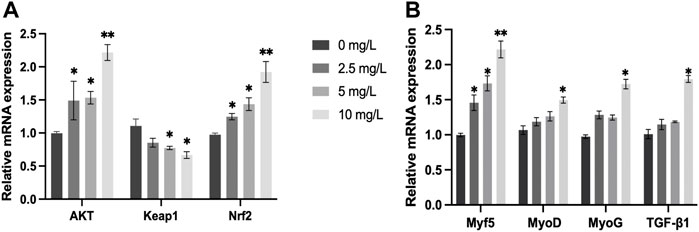
FIGURE 10. The effect of increasing concentrations of quercetin exposure on the relative expression of the (A): AKT, Keap1 and Nrf2 genes and (B): Myf5, MyoD, MyoG, and TGF-β1 genes in the muscle tissue of O. potamophila after 21 d. The bars indicate the Mean ± SD (n = 3). Statistical significance was taken as *(p < 0.05) and **(p < 0.01) compared with the control.
In this study, we demonstrated changes in muscle growth-related genes and biochemical parameters after treating O. potamophila with quercetin resulted in the expression of. It has been shown that quercetin supplemented feed can improve the quality of lamb (Andrés et al., 2013) and chicken (Goliomytis et al., 2014) meat. MSTN is a member of the TGF-β superfamily and is involved in inhibiting muscle differentiation and growth. TGF-β1 mediates the synthesis of collagen (Bradley et al., 2008). Due to the structure of MSTN, the FST complex appears to generate a complex TGF-β binding site where a TGF-β molecule can bind across a persistent electropositive gap between the two elements of FST. The gap, with a width of 60 A, allows sufficient space for the TGF-β molecules, with a length of 14–16 hexoses, analogous to that observed in the FGF growth factor acceptor heparin complex (Oelgeschläger et al., 2000). FSTMSTN. Two FST molecules surround the MSTN, blocking all four of its receptor binding sites, and possibly causing an antagonistic effect between FST and MSTN. the expression of FST in O. potamophila after exposure to 2.5 and 5 mg/L quercetin over 21 days increased in the intestine, muscle, and hepatopancreas and decreased in that of MSTN (a related gene responsible for controlling muscle growth) compared to the control group, further supporting the antagonistic effect of FST on MSTN. FSTFST The overexpression of FST in animals results in increased muscle mass, while its underexpression results in reduced muscle mass (Lee and Mcpherron, 1999). The FST in the brain and muscles with different somatic organizations suppressed MSTN expression and, as such, promoted growth in large-headed carp (Meixia et al., 2018). Myogenic regulatory factors (MRFs) related genes, as intrinsic factors affecting meat quality (Hernández-Hernández et al., 2017), have important regulatory roles in myogenic cell differentiation, muscle fiber development, and muscle tissue formation and growth (Lin et al., 2015). MRF is represented chronologically (Johnston et al., 2007). During somatic cell formation for carp, the first MRF family factor to be expressed is Myf5, followed by MyoD and MEF2C, and finally MEF2A (Watabe, 1999). Among them, Myf5 and MyoD mainly act as myogenic determinants and MyoG plays an important role in myoblast differentiation (Kassar-Duchossoy et al., 2004). (Rı́os et al., 2002) found experimentally that if MSTN overexpression occurs, reversible inhibition of myogenic functions can regulate myogenic fiber differentiation through downregulation of myogenin, MyoD and Myf5, and downstream creatine kinase activity. MSTN signaling specifically induces Smad 3 phosphorylation and increases Smad 3- MyoD association, suggesting that Smad 3 regulates myostatin signaling by inhibiting MyoD activity and expression (Langley et al., 2002). In the present experiment, the expression levels of Myf5, MyoD, MyoG, and TGF-β1 were increased when exposed to quercetin solution. This suggests that quercetin can improve muscle formation and differentiation in O. potamophila. Yang (Xya et al., 2022)also found that the expression of the TGF-β1 and FST genes involved in muscle growth was upregulated by ingestion of diets containing 3% HPM compared to those without, followed by improved muscle mass and increased meat firmness and chewiness. Quercetin feed supplements may therefore be able to increase muscle mass by increasing FST, Myf5, MyoD, MyoG, and TGF-β1 expression while decreasing MSTN expression.
Quercetin reduced the levels of triglycerides and total cholesterol. APOA1 is an important component of blood lipoproteins. The principal role of APOA1 is to transport lipids and stabilize the structure of lipoproteins, which can affect liver function when impaired (Bradbury et al., 2014). Growth hormones play a role in promoting lipolysis in the body (Holder et al., 2002; Hirano et al., 2011). The relative expressions of apolipoprotein APOA1 and the ghra gene in liver, muscle, and intestine tissues were lower than those in the control group, especially in the intestinal, which is due to the fact that the intestine is the primary site for lipid absorption and transport, as well as the fact that excessive quercetin concentrations are primarily concentrated in the intestine, which makes inhibition greater. Based on these findings, we conclude that quercetin functions effectively in lowering lipid levels, and maintains and promotes lipid metabolism in an organism by regulating lipid-related parameters, conserving liver function, and reducing the body-fat accumulation rate in O. potamophila. In general, muscle growth is usually associated with weight gain, but in this study, there was no significant weight gain over 21 days because quercetin strengthens the antioxidant capacity of the liver, strengthens genes related to lipid metabolism and affects growth, muscle growth is enhanced and lipid metabolism is also strengthened, which may be the reason for the non-gain in weight over 21 days.
To scavenge ROS, non-enzymatic and enzymatic antioxidant systems have been developed for fish (Valko et al., 2007).
GSH and CAT are important antioxidant enzymes in fish, which can scavenge hydroxyl radicals (Costantini and Verhulst, 2009), MDA is the end product of lipid peroxidation. In this study, we found that quercetin induced an increase in antioxidant enzyme activity and a decrease in MDA activity, This suggests that phosphorus can reduce lipid and protein oxidation in fish muscle. Lipid peroxidation is usually caused by ROS (Valko et al., 2007). Moreover, quercetin further mediated the mRNA expression of AKT and Nrf2 by activating the PI3K/Akt/Nrf2 pathway mediated by P2X7R to improve the antioxidant capacity of the body. Nrf2 induces the expression of protein genes that act as antioxidants and anti-inflammatory regulators and are important antioxidant genes, Nrf2 binds to its cytoplasmic inhibitor Keap1 and is present in the cytoplasm before degradation by the proteasome (Jiang et al., 2015). Downregulation of Keap1 by quercetin treatment allows Nrf2 to move from the cytoplasm to the nucleus to exert antioxidant effects. Upstream of Nrf2 TOR and ribosomal S6 protein kinase1 (S6K1) also promote Nrf2 expression through oxidative phosphorylation (Shay et al., 2012). PI3K/Akt signaling pathway activation promotes Nrf2 nuclear translocation, The upregulation of the AKT gene indicates that quercetin has an anti-apoptotic effect (Deng et al., 2013). Through the activation of Nrf2, Wang (Wang et al., 2015) found that antioxidant capacity could improve the quality of grass carp muscle and meat, water holding capacity, and tenderness. Additionally, phosphorus supplementation significantly enhances growth performance, meat and water retention in grass carp by enhancing SOD, CAT and GST activity and GSH content in grass carp (Wen et al., 2015). The enhanced antioxidant enzyme activity in fish muscle may be the result of improved transcription of antioxidant enzyme and antioxidant-related signaling molecule genes (Olsen et al., 2012; Wang et al., 2015). However, the relationship between the antioxidant mechanism of quercetin and muscle growth remains to be investigated in depth.
Feeding techniques can alter the quality of fish meat by affecting the condition of the fish and the structural and metabolic properties of the muscle tissues, resulting in changes in meat quality. Studies have shown that the juiciness and tenderness of the meat are related to its fat content and moisture, the lower the fat content the better the quality (Jeremiah et al., 1997; Rivero et al., 1999). The greater the muscle fiber density, the more tender the meat is, while muscle fiber diameter is positively correlated with tenderness. In mice fed on a quercetin supplemented diet, the reduction of muscle histopathology, the retention of muscle fiber number, and the reduction of fibrosis could result in both finer muscle fibers and greater muscle fiber density (Selsby et al., 2015). It was found that when the concentration of quercetin supplementation was higher than 5 mg/L, the muscle HDL-C and LDL-C contents were significantly lower than in the control group, that body fat was at its lowest, and body weight was relatively lower. In vivo studies on TG and cholesterol revealed that quercetin stimulated lipid oxidation and decreased muscle triglyceride levels (Zhang et al., 2012). The effects of quercetin supplementation discovered in this study, when taken collectively, may govern muscle growth through influencing gene expression.
In conclusion, full-length cDNA sequences of FST and MSTN in O. potamophila were obtained. The 3D structure of FST and MSTN showed that FST surrounded the MSTN ligand and blocked all four of its receptor binding sites. The biochemical parameters of muscle decreased, and quercetin was also effective in lowering lipid levels in the tissues examined. Quercetin-induced activation of Nrf2 can upregulate several antioxidant enzymes that play important roles in combating oxidative stress. Our study provides new insights into the potential effects of quercetin supplementation on the mechanism of muscle growth and anti-oxidation properties and offers new perspectives into the potential enhancement of fish meat quality by increasing muscle fiber diameter and muscle density.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was reviewed and approved by All applicable international, national and/or institutional guidelines for the care and use of animals were followed. (Freshwater Fisheries Research Institute of Jiangsu Provinceï). Written informed consent was obtained from the owners for the participation of their animals in this study.
Data curation and roles/writing—original draft: XW; funding acquisition and project admin-istration: QX; methodology: PW, JZ, and CZ; supervision: YZ; writing—re‐ view and editing: YL and QX.
This work was supported by the National Key R&D Program of China (2020YFD0900305), the Science Foundation of Jiangsu (NO. BK20191488) in China, the Major project of hydrobios resources in Jiangsu province (ZYHB16-3) and the Agricultural Major New Variety Creation Project in Jiangsu province (PZCZ201743).
The authors would like to thank Mengling Sun for her help in collecting tissue samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, M., Sultana, M., Raina, R., Pankaj, N. K., Verma, P. K., Prawez, S., et al. (2017). Hypoglycemic, hypolipidemic, and wound healing potential of quercetin in streptozotocin-induced diabetic rats. Pharmacogn. Mag. 13 (Suppl. 3), S633–S639. doi:10.4103/pm.pm_108_17
Amthor, H., Christ, B., Rashid-Doubell, F., Kemp, C. F., Lang, E., Patel, K., et al. (2002). Follistatin regulates bone morphogenetic protein-7 (BMP-7) activity to stimulate embryonic muscle growth. Dev. Biol. 243 (1), 115–127. doi:10.1006/dbio.2001.0555
Andrés, S., Tejido, M. L., Bodas, R., Morán, L., Prieto, N., Blanco, C., et al. (2013). Quercetin dietary supplementation of fattening lambs at 0.2% rate reduces discolouration and microbial growth in meat during refrigerated storage. Meat Sci. 93 (2), 207–212. doi:10.1016/j.meatsci.2012.08.023
Aoki, M. S., Soares, A. G., Miyabara, E. H., Baptista, I. L., and Moriscot, A. S. (2010). Expression of genes related to myostatin signaling during rat skeletal muscle longitudinal growth. Muscle Nerve 40 (6), 992–999. doi:10.1002/mus.21426
Argilés, J. M., Orpí, M., Busquets, S., and López-Soriano, F. J. (2012). Myostatin: More than just a regulator of muscle mass. Drug Discov. Today 17 (13-14), 702–709. doi:10.1016/j.drudis.2012.02.001
Bradbury, K. E., Crowe, F. L., Appleby, P. N., Schmidt, J. A., Travis, R. C., Key, T. J., et al. (2014). Serum concentrations of cholesterol, apolipoprotein A-I and apolipoprotein B in a total of 1694 meat-eaters, fish-eaters, vegetarians and vegans. Eur. J. Clin. Nutr. 68, 178–183. doi:10.1038/ejcn.2013.248
Bradley, L., Yaworsky, P., and Walsh, F. (2008). Myostatin as a therapeutic target for musculoskeletal disease. Cell. Mol. Life Sci. 65 (14), 2119–2124. doi:10.1007/s00018-008-8077-3
Brooks, A. J., and Waters, M. J. (2010). The growth hormone receptor: Mechanism of activation and clinical implications. Nat. Rev. Endocrinol. 6 (9), 515–525. doi:10.1038/nrendo.2010.123
Buckley, D., Morrissey, P., and Gray, J. (1995). Influence of dietary vitamin E on the oxidative stability and quality of pig meat. J. Anim. Sci. 73 (10), 3122–3130. doi:10.2527/1995.73103122x
Cai-ke, Z., Jing, B., Hui, Y., Zheng-ha, S., and Guo-wei, L. (2012). Effect of quercetin on serum lipid metabolism of mice and its antioxidant activity in vitro. Nat. Prod. Res. Dev. 24 (5).
Cash, J. N., Rejon, C. A., McPherron, A. C., Bernard, D. J., and Thompson, T. B. (2009). The structure of myostatin: Follistatin 288: Insights into receptor utilization and heparin binding. EMBO J. 28 (17), 2662–2676. doi:10.1038/emboj.2009.205
Chiang, Y.-A., Kinoshita, M., Maekawa, S., Kulkarni, A., Lo, C.-F., Yoshiura, Y., et al. (2016). TALENs-mediated gene disruption of myostatin produces a larger phenotype of medaka with an apparently compromised immune system. Fish. Shellfish Immunol. 48, 212–220. doi:10.1016/j.fsi.2015.11.016
Costantini, D., and Verhulst, S. (2009). Does high antioxidant capacity indicate low oxidative stress? Funct. Ecol. 23 (3), 506–509. doi:10.1111/j.1365-2435.2009.01546.x
da Silva, E. L., Tsushida, T., and Terao, J. (1998). Inhibition of mammalian 15-lipoxygenase-dependent lipid peroxidation in low-density lipoprotein by quercetin and quercetin monoglucosides. Arch. Biochem. Biophys. 349 (2), 313–320. doi:10.1006/abbi.1997.0455
Deng, C., Sun, Z., Tong, G., Yi, W., Ma, L., Zhao, B., et al. (2013). α-Lipoic acid reduces infarct size and preserves cardiac function in rat myocardial ischemia/reperfusion injury through activation of PI3K/Akt/Nrf2 pathway. PloS one 8 (3), e58371. doi:10.1371/journal.pone.0058371
Dong, Z., Ge, J., Xu, Z., Dong, X., Cao, S., Pan, J., et al. (2014). Generation of myostatin B knockout yellow catfish (Tachysurus fulvidraco) using transcription activator-like effector nucleases. Zebrafish 11 (3), 265–274. doi:10.1089/zeb.2014.0974
Du, Z.-Y., and Turchini, G. M. (2021). Are we actually measuring growth?-An appeal to use a more comprehensive growth index system for advancing aquaculture research. Rev. Aquac. 1.
Ferrell, R. E., Conte, V., Lawrence, E. C., Roth, S. M., Hagberg, J. M., Hurley, B. F., et al. (1999). Frequent sequence variation in the human myostatin (GDF8) gene as a marker for analysis of muscle-related phenotypes. Genomics 62 (2), 203–207. doi:10.1006/geno.1999.5984
Forney, L. A., Lenard, N. R., Stewart, L. K., and Henagan, T. M. (2018). Dietary quercetin attenuates adipose tissue expansion and inflammation and alters adipocyte morphology in a tissue-specific manner. Int. J. Mol. Sci. 19 (3), 895. doi:10.3390/ijms19030895
Funkenstein, B., and Jakowlew, S. (1997). Piscine (Sparus aurata) α subunit of the G-protein transducin is homologous to mammalian cone and rod transducin. Vis. Res. 37 (18), 2487–2493. doi:10.1016/s0042-6989(97)00062-x
Gamer, L. W., Wolfman, N. M., Celeste, A. J., Hattersley, G., Hewick, R., Rosen, V., et al. (1999). A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer inXenopusEmbryos. Dev. Biol. 208 (1), 222–232. doi:10.1006/dbio.1998.9191
Gao, Y., Dai, Z., Shi, C., Zhai, G., Jin, X., He, J., et al. (2016). Depletion of myostatin b promotes somatic growth and lipid metabolism in zebrafish. Front. Endocrinol. 7, 88. doi:10.3389/fendo.2016.00088
Goliomytis, M., Tsoureki, D., Simitzis, P., Charismiadou, M., Hager-Theodorides, A., Deligeorgis, S., et al. (2014). The effects of quercetin dietary supplementation on broiler growth performance, meat quality, and oxidative stability. Poult. Sci. 93 (8), 1957–1962. doi:10.3382/ps.2013-03585
Hernández-Hernández, J. M., García-González, E. G., Brun, C. E., and Rudnicki, M. A. (2012). “The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration,” in Seminars in cell & developmental biology (Elsevier), 10–18.
Hirano, Y., Kaneko, G., Koyama, H., Ushio, H., and Watabe, S. (2011). cDNA cloning of two types of growth hormone receptor in torafugu Takifugu rubripes: tissue distribution is possibly correlated to lipid accumulation patterns. Fish. Sci. 77 (5), 855–865. doi:10.1007/s12562-011-0377-0
Holder, J. R., Bauzo, R. M., Xiang, Z., and Haskell-Luevano, C. (2002). Structure−Activity relationships of the melanocortin tetrapeptide Ac-His-DPhe-Arg-Trp-NH2 at the mouse melanocortin receptors. 1. Modifications at the his position. J. Med. Chem. 45, 2801–2810. doi:10.1021/jm0104872
Hou, X., Zhu, F., Yin, S., Zhang, L., Hu, Y., Wang, Y., et al. (2014). Genetic diversity of Odontobutis potamophila from different geographic populations inferred from mtDNA control region. Mitochondrial DNA 25 (5), 400–406. doi:10.3109/19401736.2013.803084
Iemura, S.-i., Yamamoto, T. S., Takagi, C., Uchiyama, H., Natsume, T., Shimasaki, S., et al. (1998). Direct binding of follistatin to a complex of bone-morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proc. Natl. Acad. Sci. U. S. A. 95 (16), 9337–9342. doi:10.1073/pnas.95.16.9337
Jeremiah, L. E., Aalhus, J. L., Robertson, W. M., and Gibson, L. L. (1997). The effects of grade, gender, and postmortem treatment on beef. I. Composition, cutability, and meat quality. Can. J. Anim. Sci. 77 (1), 33–40. doi:10.4141/a94-136
Jia, Y., Zheng, J., Liu, S., Li, F., Chi, M., Cheng, S., et al. (2021). A chromosome-level genome assembly of the dark sleeper Odontobutis potamophila. Genome Biol. Evol. 13 (2), evaa271. doi:10.1093/gbe/evaa271
Jiang, S., Zhang, Y., Zheng, J.-H., Li, X., Yao, Y.-L., Wu, Y.-L., et al. (2017). Potentiation of hepatic stellate cell activation by extracellular ATP is dependent on P2X7R-mediated NLRP3 inflammasome activation. Pharmacol. Res. 117, 82–93. doi:10.1016/j.phrs.2016.11.040
Jiang, W.-D., Liu, Y., Jiang, J., Wu, P., Feng, L., Zhou, X.-Q., et al. (2015). Copper exposure induces toxicity to the antioxidant system via the destruction of Nrf2/ARE signaling and caspase-3-regulated DNA damage in fish muscle: Amelioration by myo-inositol. Aquat. Toxicol. 159, 245–255. doi:10.1016/j.aquatox.2014.12.020
Johnston, I. A., Bickerdike, R., Li, X., Dingwall, A., Nickell, D., Alderson, R., et al. (2007). Fast growth was not associated with an increased incidence of soft flesh and gaping in two strains of Atlantic salmon (Salmo salar) grown under different environmental conditions. Aquaculture 265 (1-4), 148–155. doi:10.1016/j.aquaculture.2007.01.045
Kassar-Duchossoy, L., Gayraud-Morel, B., Gomès, D., Rocancourt, D., Buckingham, M., Shinin, V., et al. (2004). Mrf4 determines skeletal muscle identity in Myf5: Myod double-mutant mice. Nature 431 (7007), 466–471. doi:10.1038/nature02876
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi:10.1093/molbev/mst010
Langley, B., Thomas, M., Bishop, A., Sharma, M., Gilmour, S., Kambadur, R., et al. (2002). Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J. Biol. Chem. 277 (51), 49831–49840. doi:10.1074/jbc.M204291200
Lee, S.-J., and McPherron, A. C. (2001). Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. U. S. A. 98 (16), 9306–9311. doi:10.1073/pnas.151270098
Lee, S. J., and Mcpherron, A. C. (1999). Methods for detection of mutations in myostatin variants. WO.
Lee, S., Lee, J., Lee, H., and Sung, J. (2019). Relative protective activities of quercetin, quercetin‐3‐glucoside, and rutin in alcohol‐induced liver injury. J. Food Biochem. 43 (11), e13002. doi:10.1111/jfbc.13002
Lin, Y., Zhou, J., Li, R., Zhao, Y., and Zheng, Y. (2015). Cloning and expression patterns of MRFs and effect of replacing dietary fish oil with vegetable oils on MRFs expression in grass carp (Ctenopharyngodon idellus). Turkish J. Fish. Aquatic Sci. 15 (2), 255–264.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lu, J., Qi, C., Limbu, S. M., Han, F., Yang, L., Wang, X., et al. (2019). Dietary mannan oligosaccharide (MOS) improves growth performance, antioxidant capacity, non-specific immunity and intestinal histology of juvenile Chinese mitten crabs (Eriocheir sinensis). Aquaculture 510, 337–346. doi:10.1016/j.aquaculture.2019.05.048
Martínez-Álvarez, R. M., Morales, A. E., and Sanz, A. (2005). Antioxidant defenses in fish: Biotic and abiotic factors. Rev. Fish. Biol. Fish. 15 (1), 75–88. doi:10.1007/s11160-005-7846-4
Meixia, L., Yudan, W., Yidian, L., Chen, Z., He, S., Zhang, H., et al. (2018). Basic fibroblast growth factor protects astrocytes against ischemia/reperfusion injury by upregulating the caveolin-1/VEGF signaling pathway. J. Mol. Neurosci. 64, 211–223. doi:10.1007/s12031-017-1023-9
Minh, B. Q., Nguyen, M. A. T., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30 (5), 1188–1195. doi:10.1093/molbev/mst024
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi:10.1093/molbev/msu300
Oelgeschläger, M., Larraín, J., Geissert, D., and De Robertis, E. M. (2000). The evolutionarily conserved BMP-binding protein Twisted gastrulation promotes BMP signalling. Nature 405 (6788), 757–763. doi:10.1038/35015500
Olsen, B. B., Svenstrup, T. H., and Guerra, B. (2012). Downregulation of protein kinase CK2 induces autophagic cell death through modulation of the mTOR and MAPK signaling pathways in human glioblastoma cells. Int. J. Oncol. 41 (6), 1967–1976. doi:10.3892/ijo.2012.1635
Pallauf, K., Duckstein, N., and Rimbach, G. (2017). A literature review of flavonoids and lifespan in model organisms. Proc. Nutr. Soc. 76 (2), 145–162. doi:10.1017/S0029665116000720
Pang, M., Tong, J., Yu, X., Fu, B., and Zhou, Y. (2018). Molecular cloning, expression pattern of follistatin gene and association analysis with growth traits in bighead carp (Hypophthalmichthys nobilis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 218, 44–53. doi:10.1016/j.cbpb.2018.02.003
Pourteymour Fard Tabrizi, F., Hajizadeh-Sharafabad, F., Vaezi, M., Jafari-Vayghan, H., Alizadeh, M., Maleki, V., et al. (2020). Quercetin and polycystic ovary syndrome, current evidence and future directions: A systematic review. J. Ovarian Res. 13 (1), 11. doi:10.1186/s13048-020-0616-z
Prince, P. S. M., and Sathya, B. (2010). Pretreatment with quercetin ameliorates lipids, lipoproteins and marker enzymes of lipid metabolism in isoproterenol treated cardiotoxic male Wistar rats. Eur. J. Pharmacol. 635 (1-3), 142–148. doi:10.1016/j.ejphar.2010.02.019
Rivera, L., Morón, R., Sánchez, M., Zarzuelo, A., and Galisteo, M. (2008). Quercetin ameliorates metabolic syndrome and improves the inflammatory status in obese Zucker rats. Obesity 16 (9), 2081–2087. doi:10.1038/oby.2008.315
Rivero, J., Talmadge, R. J., and Edgerton, V. R. (1999). Interrelationships of myofibrillar ATPase activity and metabolic properties of myosin heavy chain-based fibre types in rat skeletal muscle. Histochem. Cell Biol. 111 (4), 277–287. doi:10.1007/s004180050358
Rı́os, R., Carneiro, I., Arce, V. M., and Devesa, J. (2002). Myostatin is an inhibitor of myogenic differentiation. Am. J. Physiol. Cell Physiol. 282 (5), C993–C999. doi:10.1152/ajpcell.00372.2001
Rubio-Ruiz, M. E., Guarner-Lans, V., Cano-Martínez, A., Díaz-Díaz, E., Manzano-Pech, L., Gamas-Magaña, A., et al. (2019). Resveratrol and quercetin administration improves antioxidant DEFENSES and reduces fatty liver in metabolic syndrome rats. Molecules 24 (7), 1297. doi:10.3390/molecules24071297
Selsby, J., Ballmann, C., and Quindry, J. (2015). Long‐term dietary quercetin enrichment improves muscle function in dystrophic skeletal muscle. FASEB J. 29 (S1). doi:10.1096/fasebj.29.1_supplement.1039.6
Shay, K. P., Michels, A. J., Li, W., Kong, A.-N. T., and Hagen, T. M. (2012). Cap-independent Nrf2 translation is part of a lipoic acid-stimulated detoxification stress response. Biochim. Biophys. Acta 1823 (6), 1102–1109. doi:10.1016/j.bbamcr.2012.04.002
Shutenko, Z., Henry, Y., Pinard, E., Seylaz, J., Potier, P., Berthet, F., et al. (1999). Influence of the antioxidant quercetin in vivo on the level of nitric oxide determined by electron paramagnetic resonance in rat brain during global ischemia and reperfusion. Biochem. Pharmacol. 57 (2), 199–208. doi:10.1016/s0006-2952(98)00296-2
Tokur, B., and Korkmaz, K. (2007). The effects of an iron-catalyzed oxidation system on lipids and proteins of dark muscle fish. Food Chem. 104 (2), 754–760. doi:10.1016/j.foodchem.2006.12.033
Valavanidis, A., Vlahogianni, T., Dassenakis, M., and Scoullos, M. (2006). Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol. Environ. Saf. 64 (2), 178–189. doi:10.1016/j.ecoenv.2005.03.013
Valko, M., Leibfritz, D., Moncol, J., Cronin, M. T., Mazur, M., Telser, J., et al. (2007). Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39 (1), 44–84. doi:10.1016/j.biocel.2006.07.001
Wang, B., Liu, Y., Feng, L., Jiang, W.-D., Kuang, S.-Y., Jiang, J., et al. (2015). Effects of dietary arginine supplementation on growth performance, flesh quality, muscle antioxidant capacity and antioxidant-related signalling molecule expression in young grass carp (Ctenopharyngodon idella). Food Chem. 167, 91–99. doi:10.1016/j.foodchem.2014.06.091
Wang, P., Ren, M., Chen, S., Yin, S., Zhao, C., Zhang, H., et al. (20174238). Characterization and development of 56 EST-SSR markers derived from the transcriptome of Odontobutis potamophila. Genet. Mol. Res. 16 (10). doi:10.4238/gmr16029129
Watabe, S. (1999). Myogenic regulatory factors and muscle differentiation during ontogeny in fish. J. Fish Biol. 55, 1–18. doi:10.1111/j.1095-8649.1999.tb01042.x
Waters, M. J. (2016). The growth hormone receptor. Growth Horm. IGF Res. 28, 6–10. doi:10.1016/j.ghir.2015.06.001
Wen, J., Jiang, W., Feng, L., Kuang, S., Jiang, J., Tang, L., et al. (2015). The influence of graded levels of available phosphorus on growth performance, muscle antioxidant and flesh quality of young grass carp (Ctenopharyngodon idella). Anim. Nutr. 1 (2), 77–84. doi:10.1016/j.aninu.2015.05.004
Wu, Y.-S., Lee, M.-C., Huang, C.-T., Kung, T.-C., Huang, C.-Y., Nan, F.-H., et al. (2017). Effects of traditional medical herbs “minor bupleurum decoction” on the non-specific immune responses of white shrimp (Litopenaeus vannamei). Fish. Shellfish Immunol. 64, 218–225. doi:10.1016/j.fsi.2017.03.018
Xu, C., Wu, G., Zohar, Y., and Du, S.-J. (2003). Analysis of myostatin gene structure, expression and function in zebrafish. J. Exp. Biol. 206 (22), 4067–4079. doi:10.1242/jeb.00635
Xya, B., Xz, A., Zs, A., Gw, D., Xz, D., Scab, C., et al. (2022). Flesh quality of hybrid grouper ( Epinephelus fuscoguttatus ♀× Epinephelus lanceolatus ♂) fed with hydrolyzed porcine mucosa-supplemented low fishmeal diet. Anim. Nutr. 8, 114–124. doi:10.1016/j.aninu.2021.05.011
Zhang, C., Jiang, D., Wang, J., and Qi, Q. (2021). The effects of TPT and dietary quercetin on growth, hepatic oxidative damage and apoptosis in zebrafish. Ecotoxicol. Environ. Saf. 224, 112697. doi:10.1016/j.ecoenv.2021.112697
Zhang, C. K., Jing, B., Hui, Y., Shen, Z. H., and Le, G. W. (2012). Effect of quercetin on serum lipid metabolism of mice and its antioxidant activity in vitro. Natural Product Research & Development 24 (5), 663–667.
Zhao, C., Zhang, G., Yin, S., Li, Z., Wang, Q., Chen, S., et al. (2017). Integrated analysis of mRNA-seq and miRNA-seq reveals the potential roles of sex-biased miRNA-mRNA pairs in gonad tissue of dark sleeper (Odontobutis potamophila). BMC genomics 18 (1), 613. doi:10.1186/s12864-017-3995-9
Zhao, X., Gong, L., Wang, C., Liu, M., Hu, N., Dai, X., et al. (2021). Quercetin mitigates ethanol-induced hepatic steatosis in zebrafish via P2X7R-mediated PI3K/Keap1/Nrf2 signaling pathway. J. Ethnopharmacol. 268, 113569. doi:10.1016/j.jep.2020.113569
Keywords: muscle growth-related gene, quercetin, antioxidant, Odontobutis potamophila, flesh quality
Citation: Zhu C, Liu G, Gu X, Yin J, Xia A, Han M, Zhang T and Jiang Q (2022) Effect of quercetin on muscle growth and antioxidant status of the dark sleeper Odontobutis potamophila. Front. Genet. 13:938526. doi: 10.3389/fgene.2022.938526
Received: 07 May 2022; Accepted: 30 June 2022;
Published: 25 July 2022.
Edited by:
Feng He, Ocean University of China, ChinaReviewed by:
Hesham Eed Desouky, Nanjing Agricultural University, ChinaCopyright © 2022 Zhu, Liu, Gu, Yin, Xia, Han, Zhang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qichen Jiang, cWljaGVuamlhbmdAbGl2ZS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.