
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 04 August 2022
Sec. Neurogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.934154
This article is part of the Research TopicBioinformatics Applied to NeuroscienceView all 16 articles
Background: Numerous studies have suggested that programmed cell death (PCD) pathways play vital roles in cerebral ischemia/reperfusion (I/R) injury. However, the specific mechanisms underlying cell death during cerebral I/R injury have yet to be completely clarified. There is thus a need to identify the PCD-related gene signatures and the associated regulatory axes in cerebral I/R injury, which should provide novel therapeutic targets against cerebral I/R injury.
Methods: We analyzed transcriptome signatures of brain tissue samples from mice subjected to middle cerebral artery occlusion/reperfusion (MCAO/R) and matched controls, and identified differentially expressed genes related to the three types of PCD(apoptosis, pyroptosis, and necroptosis). We next performed functional enrichment analysis and constructed PCD-related competing endogenous RNA (ceRNA) regulatory networks. We also conducted hub gene analysis to identify hub nodes and key regulatory axes.
Results: Fifteen PCD-related genes were identified. Functional enrichment analysis showed that they were particularly associated with corresponding PCD-related biological processes, inflammatory response, and reactive oxygen species metabolic processes. The apoptosis-related ceRNA regulatory network was constructed, which included 24 long noncoding RNAs (lncRNAs), 41 microRNAs (miRNAs), and 4 messenger RNAs (mRNAs); the necroptosis-related ceRNA regulatory network included 16 lncRNAs, 20 miRNAs, and 6 mRNAs; and the pyroptosis-related ceRNA regulatory network included 15 lncRNAs, 18 miRNAs, and 6 mRNAs. Hub gene analysis identified hub nodes in each PCD-related ceRNA regulatory network and seven key regulatory axes in total, namely, lncRNA Malat1/miR-181a-5p/Mapt, lncRNA Malat1/miR-181b-5p/Mapt, lncRNA Neat1/miR-181a-5p/Mapt, and lncRNA Neat1/miR-181b-5p/Mapt for the apoptosis-related ceRNA regulatory network; lncRNA Neat1/miR-181a-5p/Tnf for the necroptosis-related ceRNA regulatory network; lncRNA Malat1/miR-181c-5p/Tnf for the pyroptosis-related ceRNA regulatory network; and lncRNAMalat1/miR-181a-5p for both necroptosis-related and pyroptosis-related ceRNA regulatory networks.
Conclusion: The results of this study supported the hypothesis that these PCD pathways (apoptosis, necroptosis, pyroptosis, and PANoptosis) and crosstalk among them might be involved in ischemic stroke and that the key nodes and regulatory axes identified in this study might play vital roles in regulating the above processes. This may offer new insights into the potential mechanisms underlying cell death during cerebral I/R injury and provide new therapeutic targets for neuroprotection.
Ischemic stroke is one of the leading causes of long-term severe disability and death worldwide, which is usually caused by a permanent or transient local reduction in blood supply to the brain (Campbell and Khatri, 2020; Mendelson and Prabhakaran, 2021). Currently, the most effective strategy for ischemic stroke patients is to restore cerebral blood flow in a timely manner through drugs and surgery (Herpich and Rincon, 2020). However, injury to brain tissue caused by ischemia and hypoxia is further aggravated following the short-term recovery of blood perfusion, which is known as cerebral ischemia/reperfusion (I/R) injury. The mechanism by which cerebral ischemia/reperfusion injury occurs has not been fully elucidated. Nonetheless, a growing body of evidence suggests that the overproduction of ROS and activation of inflammation and immune responses might be involved, which ultimately trigger cell death, including apoptosis, necroptosis, and pyroptosis (Eltzschig and Eckle, 2011; Jurcau and Simion, 2021). There is thus a need for a comprehensive understanding of the mechanisms underlying cell death during cerebral ischemia/reperfusion (I/R) injury to rescue injured cells, especially injured neurons in the brain, and seek new neuroprotective therapies.
Multiple cell death pathways are currently believed to be involved in cell death in ischemic stroke, among which apoptosis, pyroptosis, and necroptosis are three key programmed cell death (PCD) pathways (Tuo et al., 2022). Apoptosis can be triggered through the intrinsic and/or extrinsic pathway and may contribute to a significant proportion of neuron death following cerebral ischemia/reperfusion (Radak et al., 2017; Datta et al., 2020). Meanwhile, necroptosis is a newly discovered mechanism of cell death that is mainly regulated by receptor-interacting protein kinase 1 (RIPK1), receptor-interacting protein kinase 3 (RIPK3), and mixed-lineage kinase domain-like pseudokinase (MLKL) (Liao et al., 2020). Increasing studies have suggested that necroptosis participates in the pathogenesis of various diseases including ischemia stroke. Studies have also indicated that the inhibition of necroptosis can exert neuroprotective effects after cerebral I/R in mice by reducing cerebral infarct volume and improving motor and cognitive function (Deng et al., 2019; Yao et al., 2021). Pyroptosis is a kind of inflammatory programmed cell death that is characterized by rapid plasma-membrane rupture and the release of proinflammatory intracellular contents as well as cytokines (Yu et al., 2021). Pyroptosis was reported to be triggered by certain inflammasomes and activating caspases and executed by gasdermin family members (Dong et al., 2018). Accumulating evidence has shown that these three PCD pathways participate in the pathogenesis of ischemic stroke and that their inhibition could attenuate ischemic brain injury (Tuo et al., 2022). Recently, further evidence has also shown significant crosstalk among the three PCD pathways (Banoth et al., 2020; Zheng et al., 2020; Karki et al., 2021). Against this background, the concept of PANoptosis was proposed, which is defined as a proinflammatory PCD pathway with key features of pyroptosis, apoptosis, and/or necroptosis that cannot be accounted for by any of these PCD pathways alone (Malireddi et al., 2020; Wang and Kanneganti, 2021). PANoptosis is regulated by the cytoplasmic multimeric protein complex called the PANoptosome, which can participate in the three PCD pathways in parallel (Samir et al., 2020). PANoptosis has been implicated in various conditions, including infection, sterile inflammation, and cancer (Karki et al., 2020; Zheng et al., 2020; Place et al., 2021). A recent study that collected, integrated, and analyzed reports on research on cerebral I/R indicated that PANoptosis is observed in ischemic brain injury (Yan et al., 2022). Despite efforts to reveal the role of PCD pathways in cerebral I/R injury, the mechanisms underlying the involvement of the three PCD pathways in cerebral I/R injury are extremely complicated and remain largely unknown.
In this study, we collected PCD (apoptosis, pyroptosis, and necroptosis)-related genes based on previous literature and related databases, and analyzed transcriptome signatures of brain tissue samples from mice subjected to middle cerebral artery occlusion/reperfusion (MCAO/R) and matched controls to identify differentially expressed genes related to the three types of PCD. We then performed functional enrichment analysis of these differentially expressed PCD-related genes and their potential regulatory axes to explore their potential biological functions and regulatory mechanisms. This bioinformatic analysis might provide new insights into the potential mechanisms underlying cell death during cerebral I/R injury and new therapeutic targets for neuroprotection.
We searched the Gene Expression Omnibus (GEO) database (Barrett et al., 2013) (https://www.ncbi.nlm.nih.gov/geo) using the following terms: “cerebral ischemia–reperfusion OR cerebral ischemia OR ischemia stroke” AND “Mus musculus.” We included the gene expression profiling of adult mouse brain tissues after transient focal ischemia at 24 h of reperfusion and matched control samples. Then two datasets, GSE131193 and GSE58720, were downloaded for analysis. The dataset GSE131193 based on the GPL19057 platform is an mRNA high-throughput sequencing series that includes data on contralateral and ipsilateral brain tissues from mice subjected to transient middle cerebral artery occlusion (tMCAO) at different reperfusion timepoints (24 h and 7 days) and matched sham-operated mice. We selected a subset comprising three ipsilateral brain tissues after transient focal ischemia at 24 h of reperfusion and three matched sham-operated mice for analysis. The dataset GSE58720 based on the GPL10787 platform contains microarray gene expression data of brain tissue samples from three MCAO-operated mice at 24 h of reperfusion and three matched sham-operated mice.
For apoptosis-related genes (ARGs), 101 ARGs were downloaded from Reactome Pathway Database (https://reactome.org/) (Jassal et al., 2020) and two were extracted from the literature, thus 103 ARGs were collected (Supplementary Table S1); for necroptosis-related genes (NRGs), twenty-seven NRGs were downloaded from Reactome Pathway Database, eighty-two NRGs were extracted from the literature, after removing the overlapping genes, ninety-three NRGs were obtained (Supplementary Table S2); for pyroptosis-related genes (PRGs), twenty PRGs were downloaded from Reactome Pathway Database, sixty-seven PRGs were extracted from the literature, after removing the overlapping genes, seventy-eight PRGs were obtained for further study (Supplementary Table S3).
Differentially expressed genes (DEGs) of the microarray dataset GSE58720 were identified with NCBI’s GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) using the Limma package. For the sequencing dataset GSE131193, the processed count matrix data was downloaded and differential analysis between tMCAO-operated mice and their matched control was conducted using the “lmFit” and “eBayes” functions in the Limma package (Ritchie et al., 2015). A p-value < 0.05 and |log2 fold change (FC)| > 1 were regarded as cut-off criteria for significant DEGs. The common DEGs in common between the GSE58720 dataset and the GSE131193 dataset were intersected with PCD (apoptosis, necroptosis, and pyroptosis)-related genes, respectively, to obtain apoptosis-related DEGs (ARDEGs), necroptosis-related DEGs (NRDEGs), and pyroptosis-related DEGs (PRDEGs). The above results were visualized using the online tool Jvenn (http://jvenn.toulouse.inra.fr/app/index.html) (Bardou et al., 2014).
To obtain a better understanding of the biological mechanisms of the differentially expressed PCD-related genes, functional enrichment analysis, including Gene Ontology (GO) and pathway enrichment analysis, were performed using Metascape (http://metascape.org) (Zhou et al., 2019). The Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2017), Reactome (Jassal et al., 2020), and WikiPathways (Martens et al., 2021) databases were used for pathway annotations.
To explore the potential regulatory mechanisms of these differentially expressed PCD-related genes, we constructed intricate competing endogenous RNA (ceRNA) networks. First, target microRNAs (miRNAs) of the obtained differentially expressed PCD-related genes were predicted by four independent online databases: TargetScan (Agarwal et al., 2015) (v7.2, http://www.targetscan.org/vert_72/), miRTarBase (Huang et al., 2020) (v8.0, http://mirtarbase.mbc.nctu.edu.tw/php/index.php), StarBase (Li et al., 2014) (http://starbase.sysu.edu.cn/), and miRWalk (Sticht et al., 2018) (http://mirwalk.umm.uni-heidelberg.de/). Only the miRNAs that were shared by any three or all four databases were regarded as eligible. Next, target long noncoding RNAs (lncRNAs) of the above-obtained miRNAs were predicted by StarBase and the LncBase module of the DIANA tool (http://carolina.imis.athena-innovation.gr/) (Karagkouni et al., 2020). Only the lncRNAs that were shared between the two databases were regarded as eligible. Finally, we selected lncRNA–mRNA interactions and miRNA–mRNA interactions that shared the same miRNAs to construct the ceRNA network and visualized it using Cytoscape software (Shannon et al., 2003) (Version 3.8.0, http://cytoscape.org).
The cytoHubba plugin was applied to screen out the top ten genes of the above three ceRNA regulatory networks through seven different algorithms: MCC, Degree, Edge Percolated Component (EPC), EcCentricity, Closeness, Radiality, and Betweenness (Chin et al., 2014). UpSet R package was used to extract the overlapping genes obtained by the above seven different algorithms and visualize them (Conway et al., 2017). These overlapping genes were confirmed as the hub nodes.
A flow chart of this study is shown in Figure 1. We first analyzed the two datasets GSE58720 and GSE131193 to identify the common DEGs in MCAO/R-operated mice at 24 h of reperfusion compared with matched controls. Then, we identified the common DEGs that overlapped with PCD (apoptosis, necroptosis, and pyroptosis)-related genes to obtain differentially expressed PCD-related genes (DEARGs, DENRGs, DEPRGs). A total of six DEARGs (Figure 2A), nine DENRGs (Figure 2B), and ten DEPRGs (Figure 2C) were identified. The six DEARGs included Cd14, Zbp1, Tnfrsf10b, Bax, Mapt, and Pycard, among which Cd14, Zbp1, Tnfrsf10b, Bax, and Pycard were upregulated in the dataset GSE58720 but downregulated in the dataset GSE131193, while Mapt showed the opposite pattern (Table 1). The nine DENRGs included Cxcl1, Zbp1, Il1b, Tnf, Ripk3, Tnfrsf10b, Mlkl, Pycard, and Bax (Table 1) and the ten DEPRGs included Cd14, Zbp1, Il1b, Mefv, Tnf, Il1rn, Anxa2, Ccr5, Pycard, and Bax (Table 1). All of these DENRGs and DEPRGs were upregulated in the dataset GSE58720 but downregulated in the dataset GSE131193. We also attempted to identify the common genes among the above three kinds of differentially expressed PCD-related genes and found that three genes, namely, Zbp1, Bax, and Pycard, overlapped among the three sets of differentially expressed PCD-related genes. Cd14 was in common between DEARGs and DEPRGs, Tnfrsf10b was in common between DEARGs and DENRGs, while Il1b and Tnf were in common between DENRGs and DEPRGs (Figure 2D and Table 1).
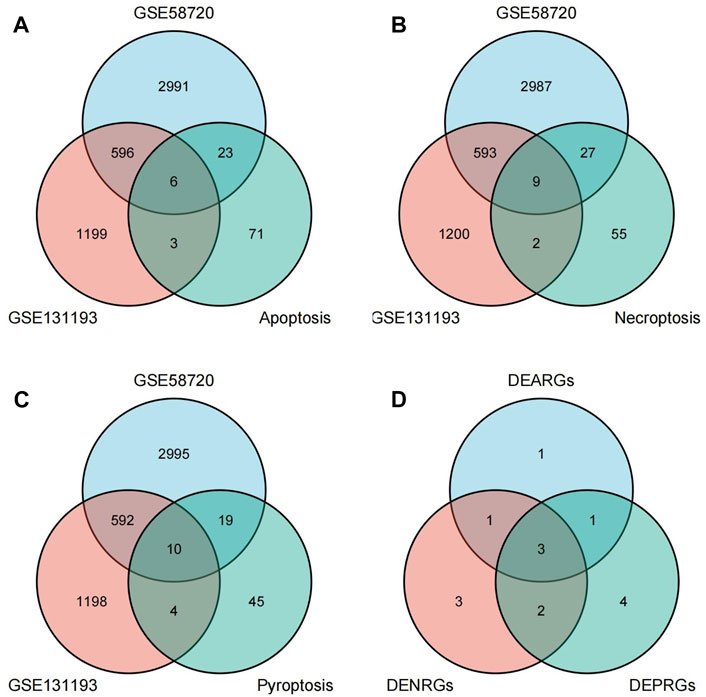
FIGURE 2. Identification of differentially expressed programmed cell death (PCD)-related genes. (A) Differentially expressed apoptosis-related genes (DEARGs) were identified by determining the overlap of datasets GSE58720 and GSE131193 with apoptosis-related genes. (B) Differentially expressed necroptosis-related genes (DENRGs) were identified by determining the overlap of datasets GSE58720 and GSE131193 with necroptosis-related genes. (C) Differentially expressed pyroptosis-related genes (DEPRGs) were identified by determining the overlap of datasets GSE58720 and GSE131193 with pyroptosis-related genes. (D) The overlapping genes were identified among DEARGs, DENRGs, and DEPRGs.
To further explore the potential functions of DEARGs, DENRGs, and DEPRGs, functional enrichment analysis was performed using the online database Metascape. The results of GO analysis revealed that the DEARGs were particularly associated with the positive regulation of cell death, apoptotic signaling pathway, negative regulation of mitochondrial membrane potential, extrinsic apoptotic signaling pathway via death domain receptors, positive regulation of interleukin-8 production, membrane rafts, and left-handed Z-DNA binding (Figure 3A and Table 2). The DENRGs were mainly associated with programmed necrotic cell death, response to virus, positive regulation of apoptotic process, necroptotic process, defense response, release of cytochrome c from mitochondria, cytosolic calcium ion concentration, reactive oxygen species metabolic process, and regulation of interferon-gamma production (Figure 3B and Table 3). The DEPRGs were particularly involved in the positive regulation of inflammatory response, tumor necrosis factor production, fever generation, interleukin-8 production, regulation of interleukin-1-mediated signaling pathway, regulation of neurogenesis, negative regulation of membrane potential and neural precursor cell proliferation, and inflammatory response to antigenic stimulus (Figure 3C and Table 4).
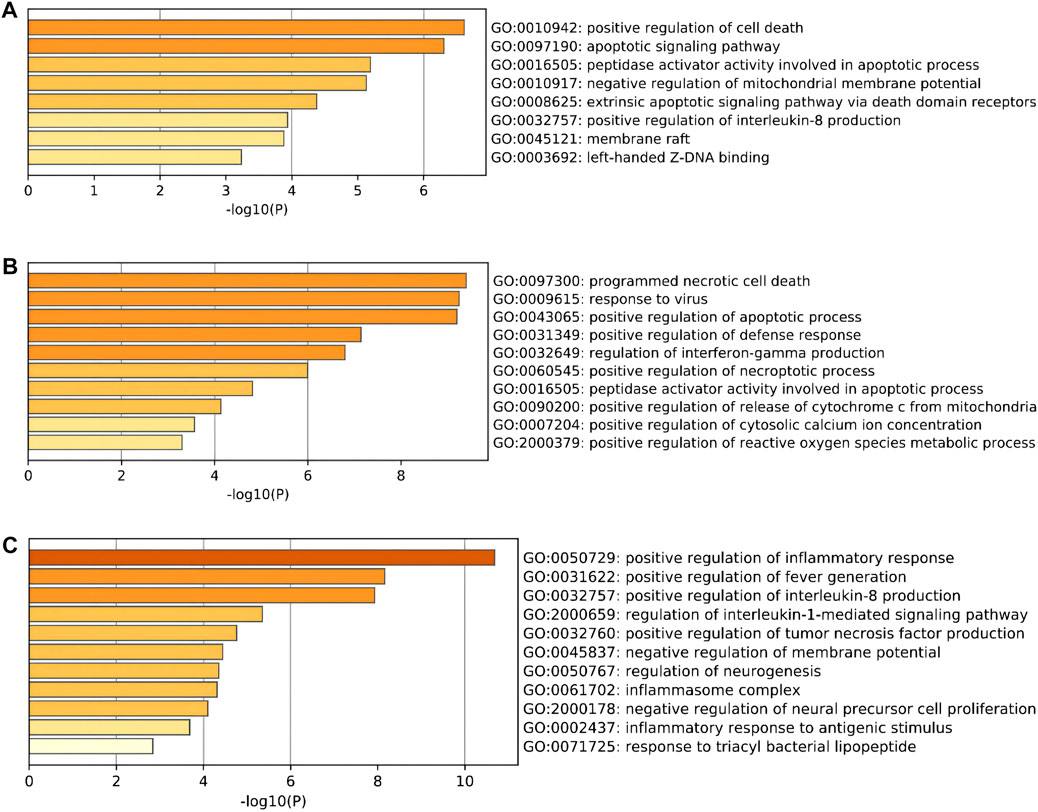
FIGURE 3. Gene Ontology (GO) enrichment analysis of these differentially expressed programmed cell death (PCD)-related genes. (A) Significantly enriched GO terms of differentially expressed apoptosis-related genes (DEARGs). (B) Significantly enriched GO terms of differentially expressed necroptosis-related genes (DENRGs). (C) Significantly enriched GO terms of differentially expressed pyroptosis-related genes (DEPRGs).
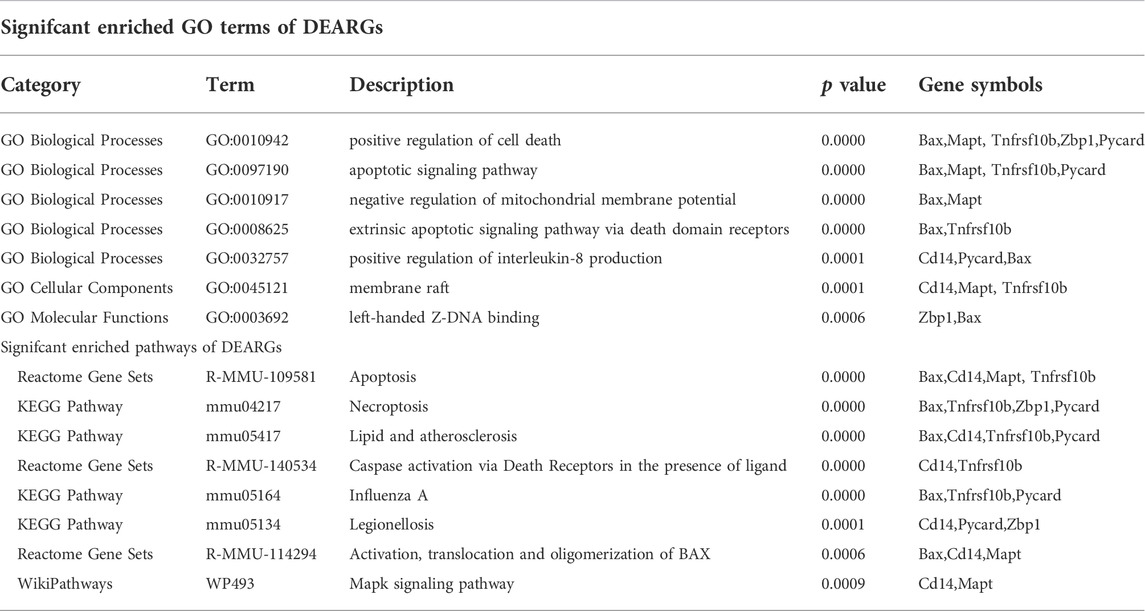
TABLE 2. Functional enrichment analysis of differentially expressed apoptosis-related genes (DEARGs).
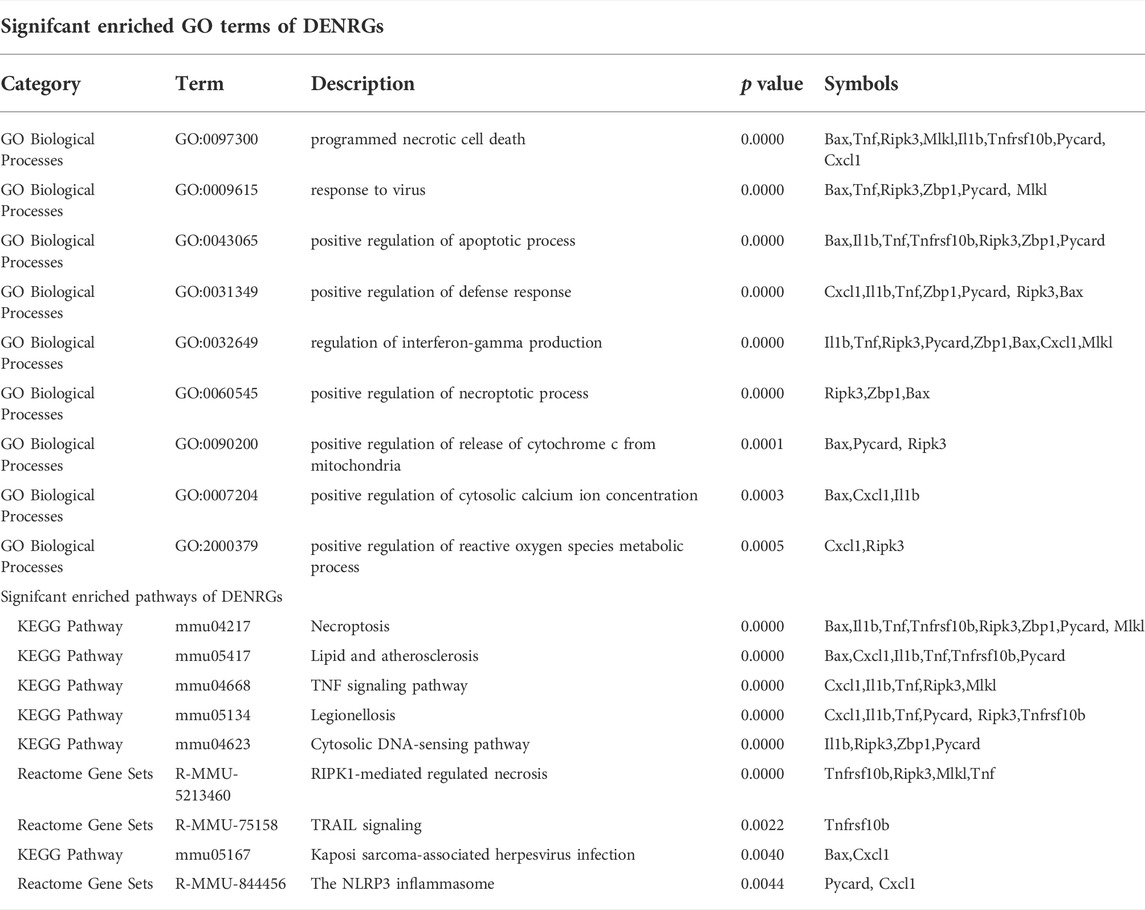
TABLE 3. Functional enrichment analysis of differentially expressed necroptosis-related genes (DENRGs).
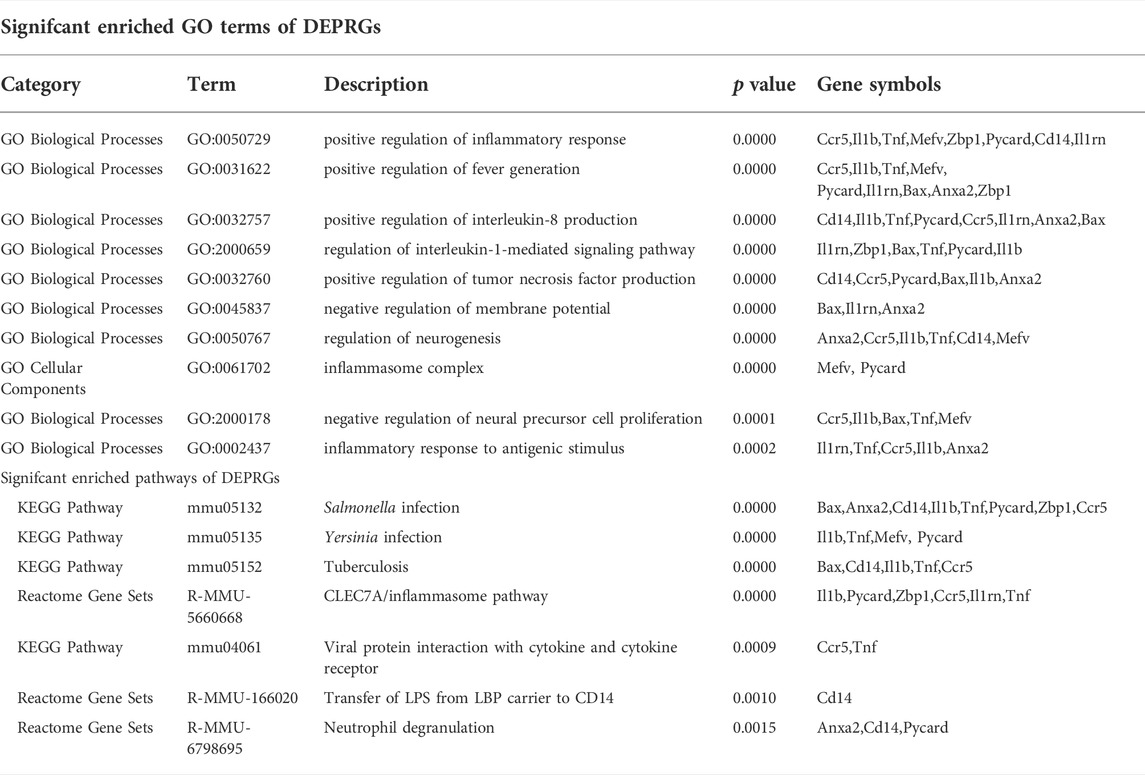
TABLE 4. Functional enrichment analysis of differentially expressed pyroptosis-related genes (DEPRGs).
Moreover, regarding the results of pathway analysis, these revealed that the DEARGs were mainly associated with apoptosis, necroptosis, lipids and atherosclerosis, caspase activation via death receptors in the presence of ligand influenza A, legionellosis, activation, translocation and oligomerization of BAX, and the Mapk signaling pathway (Figure 4A and Table 2). The DENRGs were particularly involved in necroptosis, lipids and atherosclerosis, TNF signaling pathway, legionellosis, cytosolic DNA-sensing pathway, RIPK1-mediated regulated necrosis, TRAIL signaling, Kaposi sarcoma-associated herpesvirus infection, and the NLRP3 inflammasome (Figure 4B and Table 3). The DEPRGs were particularly associated with Salmonella infection, Yersinia infection, tuberculosis, CLEC7A/inflammasome pathway, viral protein interaction with cytokine and cytokine receptor, transfer of LPS from LBP carrier to CD14, and neutrophil degranulation (Figure 4C and Table 4).
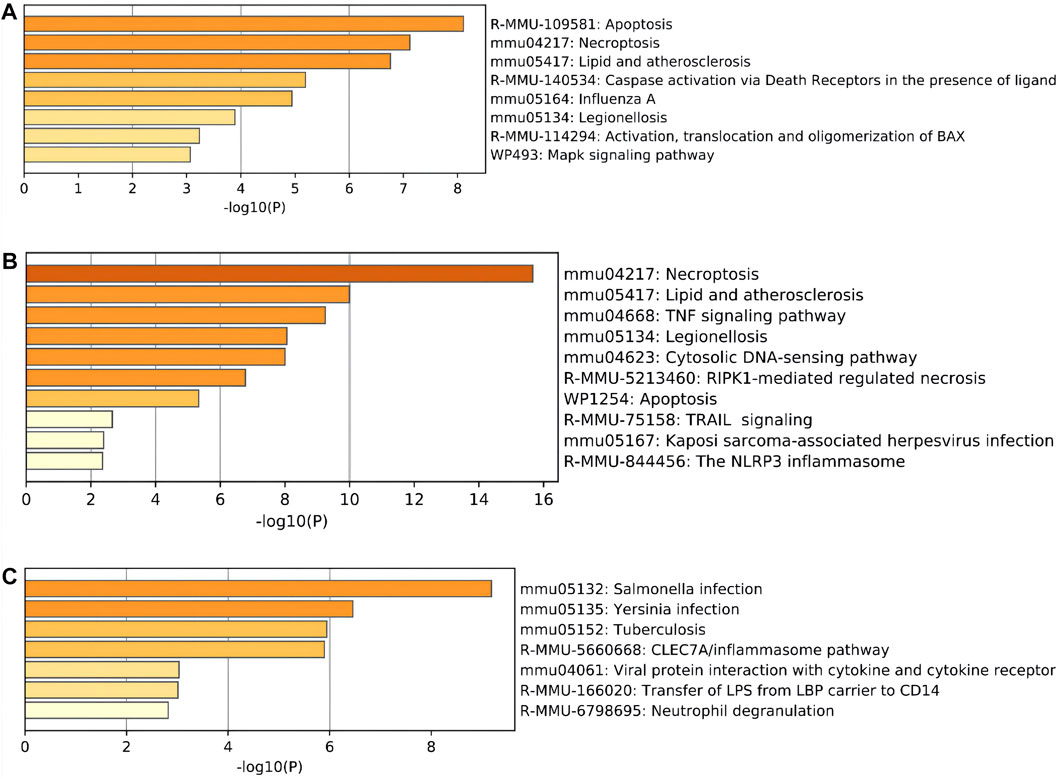
FIGURE 4. Pathway enrichment analysis of these differentially expressed programmed cell death (PCD)-related genes. (A) Significantly enriched pathways of differentially expressed apoptosis-related genes (DEARGs). (B) Significantly enriched pathways of differentially expressed necroptosis-related genes (DENRGs). (C) Significantly enriched pathways of differentially expressed pyroptosis-related genes (DEPRGs).
To clarify the potential molecular regulatory mechanisms of these differentially expressed PCD-related genes, we then constructed PCD-related ceRNA regulatory networks of lncRNA–miRNA–mRNA. First, four independent online databases, namely, TargetScan, miRTarBase, StarBase, and miRWalk, were used to predict the interactions between miRNAs and mRNAs. Only the miRNAs that were shared by any three or all four of the databases were regarded as eligible. A total of 133 miRNA–mRNA interactions for apoptosis-related ceRNA regulatory networks, 91 miRNA–mRNA interactions for necroptosis-related ceRNA regulatory networks, and 70 miRNA–mRNA interactions for pyroptosis-related ceRNA regulatory networks were obtained based on the above methods. Next, target lncRNAs of the above-obtained miRNAs were predicted by StarBase and the LncBase module of the DIANA tool; only the lncRNAs that were shared between the two databases were regarded as eligible. A total of 107 lncRNA–miRNA pairs for apoptosis-related ceRNA regulatory networks, 58 lncRNA–miRNA pairs for necroptosis-related ceRNA regulatory networks, and 49 lncRNA–miRNA pairs for pyroptosis-related ceRNA regulatory networks were identified. Then, the ceRNA networks were constructed using the miRNAs shared between the lncRNAs and mRNAs according to the ceRNA hypothesis. The apoptosis-related ceRNA regulatory network included 152 edges and 69 nodes (including 24 lncRNAs, 41 miRNAs, and 4 mRNAs) (Figure 5A). The necroptosis-related ceRNA regulatory network included 82 edges and 42 nodes (including 16 lncRNAs, 20 miRNAs, and 6 mRNAs) (Figure 5B). Finally, the pyroptosis-related ceRNA regulatory network included 69 edges and 39 nodes (including 15 lncRNAs, 18 miRNAs, and 6 mRNAs) (Figure 5C).
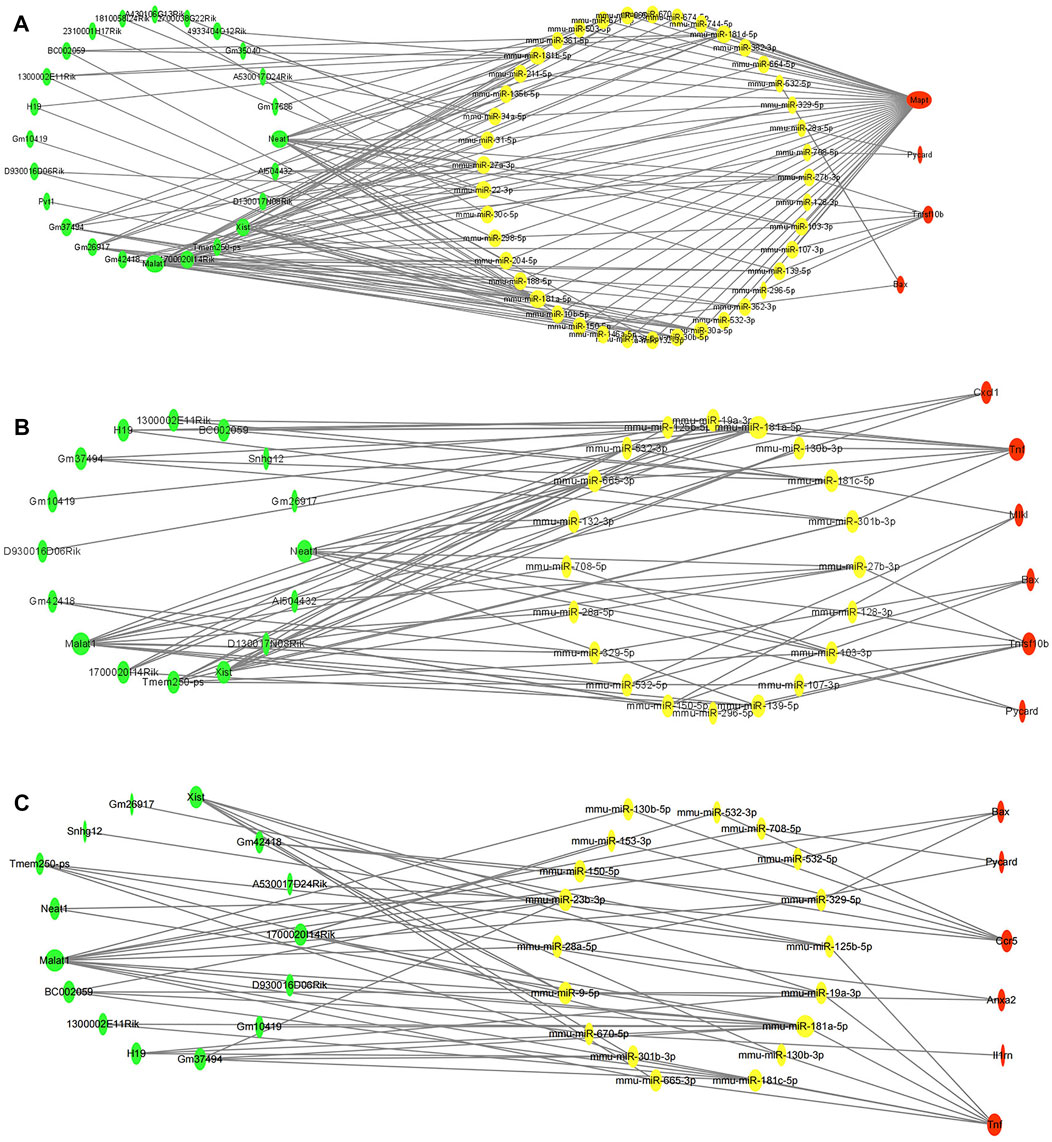
FIGURE 5. Construction of programmed cell death (PCD)-related ceRNA regulatory networks. (A) Apoptosis-related ceRNA regulatory networks. (B). Necroptosis-related ceRNA regulatory networks. (C) Pyroptosis-related ceRNA regulatory networks. Red color denotes differentially expressed programmed cell death (PCD)-related genes, yellow color denotes eligible target miRNAs of these PCD-related genes, and green color denotes eligible target lncRNAs of miRNAs. The size of nodes represented closeness score calculated by cytoHubba plugin.
The cytoHubba plugin was used to identify hub nodes of each of these PCD-related ceRNA regulatory networks based on the above methods. In the hub gene analysis, five hub nodes, namely, the mRNA Mapt, miR-181a-5p, miR-181b-5p, and the lncRNAs Malat1 and Neat1, were identified in the apoptosis-related ceRNA regulatory network (Figure 6A) and these hub nodes formed four ceRNA regulatory pathways, namely, lncRNA Malat1/miR-181a-5p/Mapt, lncRNA Malat1/miR-181b-5p/Mapt, lncRNA Neat1/miR-181a-5p/Mapt, and lncRNA Neat1/miR-181b-5p/Mapt (Figure 6B). Hub nodes in the necroptosis-related ceRNA regulatory network included mRNA Tnf, miR-181a-5p, lncRNA Malat1, lncRNA Xist, and lncRNA Neat1 (Figure 6C). These hub nodes formed two ceRNA regulatory pathways, namely, lncRNA Malat1/miR-181a-5p/Tnf and lncRNA Neat1/miR-181a-5p/Tnf (Figure 6D). Hub nodes in the pyroptosis-related ceRNA regulatory network included the mRNA Tnf, miR-181a-5p, miR-181c-5p, lncRNA Malat1, and lncRNA Xist (Figure 6E); these hub nodes formed two ceRNA regulatory pathways, namely, lncRNA Malat1/miR-181a-5p/Tnf and lncRNA Malat1/miR-181c-5p/Tnf (Figure 6F).
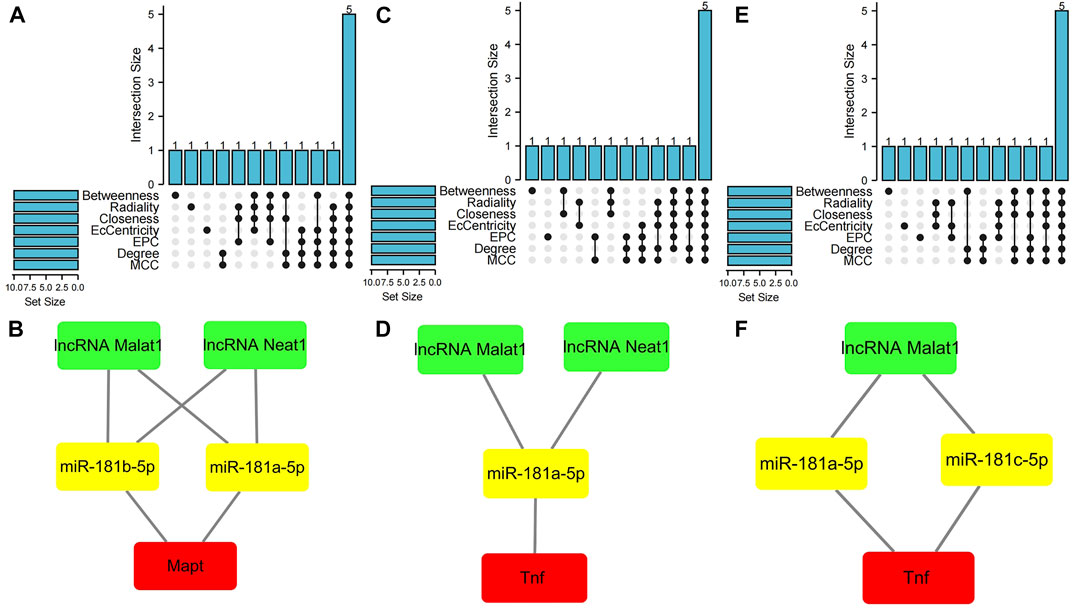
FIGURE 6. Hub gene analysis of three programmed cell death (PCD)-related ceRNA regulatory networks. (A) Hub nodes in the apoptosis-related ceRNA regulatory network that were shared by seven different algorithms. (B) Key ceRNA regulatory pathways for apoptosis-related ceRNA regulatory network. (C) Hub nodes in the necroptosis-related ceRNA regulatory network that were shared by seven different algorithms. (D) Key ceRNA regulatory pathways for necroptosis-related ceRNA regulatory network. (E) Hub nodes in the pyroptosis-related ceRNA regulatory network that were shared by seven different algorithms. (F) Key ceRNA regulatory pathways for pyroptosis-related ceRNA regulatory network. Red color denotes differentially expressed programmed cell death (PCD)-related genes, yellow color denotes miRNAs, and green color denotes lncRNAs.
To validate the hub nodes in these PCD-related ceRNA regulatory networks, we searched the literature and found that they were abnormally expressed in ischemic stroke. Shi et al. (Shi et al., 2021) found that the levels of acetylated tau (ac-MAPT) and phosphorylated tau (p-MAPT) increased in rats subjected to MCAO/R compared with that in the sham group. The protein and mRNA levels of total-tau (T-MAPT) showed no significant differences between the sham and MCAO/R groups. Basurto-Islas et al. (Basurto-Islas et al., 2018) observed higher phosphorylation of tau and total tau in MCAO/R mice. Other studies also reported that the hyperphosphorylation of tau increases during MCAO/R in animal models (Dong et al., 2014; Fujii et al., 2017). Tnf was also reported to be significantly upregulated in MCAO/R animal models and OGD/R cell models (Li et al., 2019; Zhang et al., 2021a; Zhou et al., 2021). Moreover, it was reported that miR-181a-5p was highly expressed in serum of ischemic stroke patients, brain tissues of MCAO/R mice, and an oxygen-glucose-deprivation/reoxygenation (OGD/R) N2a cell model (Ouyang et al., 2012; Wu et al., 2017; Song et al., 2021). Studies also reported that miR-181b-5p and miR-181c-5p expression was significantly decreased in cerebral ischemia in vivo and in vitro (Deng et al., 2016; Ma et al., 2016; Zhang et al., 2018; Meng et al., 2020). Accumulating evidence has also revealed that expression of the lncRNA Malat1 was upregulated after MCAO/R in rats and mice and OGD/R in different cells including primary neuronal cells, HT-22 cells, mouse astrocyte cells, and brain vascular endothelial cells (Xin and Jiang, 2017; Zhang et al., 2021b; Jia et al., 2021; Tan et al., 2021). Moreover, a recent study reported that the lncRNA Malat1 significantly increased in the blood of ischemic stroke patients compared with the level in normal controls (Tan et al., 2021). Furthermore, several studies reported that the lncRNA Neat1 was increased in an MCAO/R animal model, an OGD/R-induced cell model, and ischemic stroke patients (Ni et al., 2020; Zhang et al., 2021c; Jin et al., 2021). Another recent study reported that expression of the lncRNA Neat1 was significantly decreased in OGD/R-induced BV-2 and N2a cells compared with that in control cells (Zhou et al., 2022). The lncRNA XIST was also reported to be highly expressed in an MCAO/R-treated animal model and an OGD/R-treated cell model (Zhang et al., 2021d; Wang et al., 2021; Xiong et al., 2021). Wang et al. also reported that the lncRNA XIST was upregulated in brain tissues under MCAO/R treatment and in OGD/R-treated PC12 cells (Wang et al., 2021). Furthermore, Xiong et al. found that XIST was significantly highly expressed in SH-SY5Y cells after OGD/R treatment (Xiong et al., 2021). Finally, another study identified that XIST expression was upregulated in the brain tissues of an I/R mouse model and OGD/R-induced N2a cells (Zhang et al., 2021d). The findings of these previous studies are in accordance with our results, indicating the robustness of our analysis.
Accumulating evidence supports the involvement of PCD pathways in the pathogenesis of ischemic stroke and highlights the importance of each form of cell death. However, the specific mechanisms underlying them remain incompletely clarified. There is also currently a lack of specific neuroprotective drugs in clinical practice. Nonetheless, increasing studies have indicated significant crosstalk among these PCD pathways. Therefore, we applied bioinformatic analysis to identify differentially expressed PCD-related genes during cerebral I/R injury and investigated their potential regulatory axes by constructing ceRNA networks. This may contribute to elucidating the molecular mechanisms behind these PCD pathways and provide a basis for developing novel therapeutic targets against cerebral I/R injury.
A total of six DEARGs, nine DENRGs, and ten DEPRGs were identified in this study. Among them, three genes, namely, Bax, Zbp1, and Pycard, overlapped among these three sets of genes, indicating that they may play key roles in the crosstalk among these PCD pathways. The protein encoded by the Bax gene belongs to the BCL2 protein family and is regarded as the fundamental effector of the intrinsic apoptotic pathway (Spitz and Gavathiotis, 2022). Numerous studies have indicated that Bax-dependent initiation and activation of subsequent apoptotic pathways play critical roles in ischemic brain injury (Li et al., 2021; Tu and Hu, 2021). In addition, it has been suggested that inhibition of Bax function may provide a new strategy for neuroprotection and functional improvement against cerebral ischemia (Han et al., 2011). Recently, some studies also reported that Bax is a key regulator of caspase-independent necroptosis and pyroptosis (Cabon et al., 2012; Hu et al., 2020). However, whether Bax is involved in necroptosis and pyroptosis in cerebral ischemia–reperfusion injury has remained unclear, so further research on this issue is needed. The gene Pycard encodes the adaptor protein ASC, which comprises two protein–protein interaction domains: an N-terminal PYRIN-PAAD-DAPIN domain (PYD) and a C-terminal caspase-recruitment domain (CARD) (Hoss et al., 2017). Previous studies demonstrated that ASC was upregulated in an ischemic stroke model and played a key role in cerebral ischemia–reperfusion injury by participating in the inflammatory response and cell death, including apoptosis, necroptosis, and pyroptosis (Meng et al., 2019; Liang et al., 2020; Xu et al., 2021a). The gene Zbp1 encodes Z-DNA binding protein 1 with two Zα domains, which is a critical innate immune sensor of not only viral RNA products but also endogenous nucleic acid ligands (Zhang et al., 2020). Previous studies showed that Zbp1 plays a role in the innate immune response by binding to foreign DNA and inducing type I interferon production (Jiao et al., 2020). In addition, in response to influenza virus infection, it could induce cell death in the form of pyroptosis, apoptosis, and necroptosis, that is, PANoptosis (Zheng and Kanneganti, 2020). However, the role of Zbp1 in cerebral I/R injury remains unknown. Recent studies demonstrated that Zbp1 and ASC are components of the PANoptosome (Zheng and Kanneganti, 2020; Lee et al., 2021). Yan et al. (Yan et al., 2022) also indicated that PANoptosis is observed in ischemic brain injury. Based on previous related studies, we speculated that these three genes might be components of the PANoptosome and be involved in PANoptosis in cerebral I/R injury. Our study further confirmed previous findings supporting the hypothesis that these three genes might be involved in PANoptosis and crosstalk among apoptosis, necroptosis, and pyroptosis in cerebral I/R injury, thus providing new targets for neuroprotection.
Functional enrichment analyses were performed to obtain a more in-depth understanding of the differentially expressed PCD-related genes. The results showed that these genes were not only particularly associated with corresponding PCD-related biological processes and pathways, but also involved in other biological processes and pathways, such as inflammatory response and reactive oxygen species metabolic process. This indicates that these genes have different functions under particular circumstances and that there might be crosstalk among these biological processes. These results are in line with previous studies that revealed significant crosstalk between PCD and inflammatory response (Jayaraj et al., 2019).
In recent years, increasing studies have suggested that the regulatory network composed of lncRNAs, miRNAs, and mRNAs plays a critical role in the mechanisms underlying cerebral ischemia–reperfusion injury (Xu et al., 2021b). To better understand the molecular regulatory mechanisms of these differentially expressed PCD-related genes, we constructed PCD-related ceRNA regulatory networks and performed hub gene analysis to identify key nodes in these networks. The gene Mapt was found to be a hub node of the apoptosis-related ceRNA regulatory network. It can encode several isoforms of tau protein as a result of complex, regulated alternative splicing of its messenger RNA (Zhang et al., 2009). Mapt transcripts are differentially expressed in the nervous system, depending on the stage of neuronal maturation and neuron type. Mapt gene mutations have been shown to be associated with several neurodegenerative disorders (Michalicova et al., 2020). In recent years, increasing evidence has demonstrated that Mapt plays a role in ischemic stroke. In addition, Basurto-Islas et al. (Basurto-Islas et al., 2018) found that a large amount of hyperphosphorylated MAPT (Ser262/356) was colocalized with apoptotic cells in MCAO/R-treated mice. Moreover, Fujii et al. (Fujii et al., 2017) illustrated that the knockout of MAPT reduced infarct area and alleviated symptoms of neurological deficit. A recent study also showed that astragaloside IV exerted neuroprotective effects in rats with cerebral ischemia/reperfusion (CIR) injury, probably through the Sirt1/Mapt pathway (Shi et al., 2021). In this study, the gene Tnf was identified as a hub node in both the necroptosis-related ceRNA regulatory network and the pyroptosis-related ceRNA regulatory network. This gene encodes the multifunctional proinflammatory cytokine TNF-α, which belongs to the tumor necrosis factor (TNF) superfamily (Watters and O'Connor, 2011). It can bind to its surface receptors and functions through their activation. Generally, TNF is a classical activator of necroptosis that binds to its receptor to recruit RIPK1, which interacts with RIPK3 to form necrosome and phosphorylate MLKL to mediate necroptosis in the absence of caspases-8 (Green, 2019). Studies have shown that the level of TNF-α was elevated in ischemic stroke and it has been implicated in cerebral I/R injury, exerting effects by regulating the inflammatory response and PCD pathways including apoptosis, necroptosis, and pyroptosis (Hallenbeck, 2002; Maddahi et al., 2011), which is consistent with our results. Many studies have demonstrated that the inhibition of TNF signaling pathways may have neuroprotective effects against cerebral I/R injury. For example, Zhang et al. showed that preconditioning with Carbonisatus significantly decreased the levels of TNF-α and IL-6, reduced ischemic lesion volume, and improved neurological deficits in MCAO/R rats (Zhang et al., 2021a).
MicroRNAs (miRNAs) are conserved small regulatory noncoding RNAs of about 20–22 bp in length. They can regulate protein expression by binding to the 3′ untranslated region (3′UTR) of their target genes, degrading or inhibiting their expression (Krol et al., 2010). Numerous studies have shown that miRNAs are involved in the regulation of PCD pathways in many diseases, including ischemic stroke (Ghafouri-Fard et al., 2020). The hub gene analysis in this study demonstrated that mir-181a-5p was a hub node in all of the above-mentioned three PCD-related ceRNA networks, and that mir-181b-5p was a hub node of the apoptosis-related ceRNA regulatory network and mir-181c-5p was a hub node of the pyroptosis-related ceRNA regulatory network. miR-181a-5p, miR-181b-5p, and miR-181c-5p all belong to the miR-181 family and their aberrant expression has been associated with various diseases including stroke, neurodegeneration, and cancer (Indrieri et al., 2020). Previous studies suggested that the miR-181 family participates in the regulation of a range of biological processes including cell proliferation (Huo et al., 2016), apoptosis (Zhang et al., 2018), autophagy (Guo et al., 2019), and immune and inflammatory responses (Hutchison et al., 2013; Lu et al., 2019). Moreover, several studies have demonstrated that the inhibition of miR-181a-5p played a neuroprotective role in cerebral ischemic injury, as evidenced by reductions in cell apoptosis, pyroptosis, and cerebral infarction area (Moon et al., 2013; Stary et al., 2017; Yan et al., 2020; Song et al., 2021). However, the roles of miR-181b-5p and miR-181c-5p in cerebral ischemia have remained controversial. Peng et al. showed that downregulated miR-181b played a neuroprotective role against ischemic injury through negatively regulating HSPA5 and UCHL1 protein levels (Peng et al., 2013). In addition, Zhang et al. suggested that the downregulation of miRNA-181b protects against cerebral ischemic injury via the inhibition of NF-κB-mediated inflammatory and apoptotic responses (Zhang et al., 2018). In contrast, another two reports demonstrated the possible neuroprotective effects of increased miR-181b in ischemia-caused neuronal cell apoptosis and mechanical repair of brain tissue (Deng et al., 2016; Liu et al., 2016). Most studies supported the assertion that miR-181c-5p plays a positive role in brain injury caused by cerebral ischemia–reperfusion and that its overexpression can inhibit brain injury caused by ischemic stroke through regulating proliferation, inflammatory response, and apoptosis of neuronal cells (Zhang et al., 2019; Cao et al., 2020; Bu et al., 2021). However, in two other studies, the opposite conclusions were drawn. Specifically, Ma et al. (Ma et al., 2016) found a positive correlation between the NIHSS score and miR-181c level, and showed that plasma miR-181c concentration was positively correlated with the number of neutrophils and blood platelet count and negatively correlated with the number of lymphocytes. They also found that miR-181c promoted the apoptosis of BV2 and Neuro-2a cells and aggravated brain ischemia–reperfusion injury in a mouse model of stroke via the modulation of pro- and anti-apoptotic proteins. Moreover, a recent study showed that downregulated miR-181c ameliorated cerebral ischemic injury via increasing the expression of c-Fos and its downstream genes (Meng et al., 2020). Taken together, these findings indicated that mir-181a-5p, miR-181b-5p, and miR-181c-5p are all involved in the mechanism of cerebral I/R injury, but might play different roles depending on the specific target gene to which they bind.
lncRNAs are the most abundant noncoding RNAs (ncRNAs). They are greater than 200 bp in length, lack protein-coding function, and are associated with a variety of neurological diseases including ischemic stroke (Wu et al., 2013; Bao et al., 2018). Our hub analysis identified the lncRNA Malat1 as a hub node in all three PCD-related ceRNA regulatory networks, which is consistent with previous studies, indicating its critical role in regulating PCD pathways. Malat1 is known as a long intergenic noncoding RNA and is highly abundant in the nervous system. Accumulating evidence has indicated that this lncRNA plays vital roles in regulating various physiological processes, including apoptosis, autophagy, immune and inflammatory responses, and endothelial dysfunction of ischemic stroke (Wang et al., 2022). The expression of Malat1 was also found to be upregulated in ischemic stroke, while its downregulation was shown to improve the neurological deficit score and reduce neuronal apoptosis and the size of cerebral infarction by regulating miR-211-5p to in turn regulate the expression of COX-2 (Tan et al., 2021). Other studies also demonstrated that the inhibition of Malat1 expression could protect against cerebral I/R injury by alleviating neuronal apoptosis, endoplasmic reticulum stress, and inflammation (Shi et al., 2019; Cao et al., 2020; Jia et al., 2021). Moreover, it was reported that Malat1 was highly expressed in OGD/R-induced astrocyte injury models, and that its silencing protected against cerebral ischemia–reperfusion injury by downregulating AQP4 levels via miR-145 (Wang et al., 2020). In contrast, some studies supported the neuroprotective role of Malat1 in cerebral ischemia–reperfusion injury. For example, Xin et al. (Xin and Jiang, 2017) found that Malat1 could protect human brain vascular endothelial cells from OGD/R-induced apoptosis via a PI3K-dependent mechanism. Another study showed that mice with lncRNA Malat1 KO presented larger brain infarct size and worse neurological scores, indicating that Malat1 plays critical protective roles in ischemic stroke via anti-apoptotic and anti-inflammatory effects in the brain microvasculature (Zhang et al., 2017). Accumulating evidence has also indicated that Malat1 is an important regulator of pyroptosis in various diseases (Song et al., 2019; Shu et al., 2021). However, the specific roles and mechanisms by which Malat1 regulates pyroptosis and necroptosis in cerebral ischemia–reperfusion injury have remained unclear. In addition, the lncRNAMalat1/miR-181a-5p/Mapt regulatory axis, lncRNAMalat1/miR-181b-5p/Mapt regulatory axis, and lncRNA Malat1/miR-181a-5p/Tnf regulatory axis were not previously reported to be involved in cerebral I/R injury, so they need further investigation. In this study, the lncRNA Neat1 was also identified as a hub node in both apoptosis-related and necroptosis-related ceRNA regulatory networks. Recently, increasing evidence has shown that this lncRNA plays an essential role in physiological and pathological responses in ischemic stroke (Ni et al., 2020; Jin et al., 2021). Li et al. (Li et al., 2020) found that the expression of Neat1 was elevated in patients with ischemic stroke compared with that in controls, and that lncRNA Neat1 expression positively correlated with NIHSS score and inflammatory factors and could predict an increased risk of recurrence/death. Ni et al. (Ni et al., 2020) also showed that Neat1 knockdown alleviated OGD/R-induced apoptosis and increased neuronal viability. Another study demonstrated that Gastrodin significantly alleviated cerebral I/R injury by regulating the lncRNA Neat1/miR-22-3p axis; it also showed that the overexpression of Neat1 promoted neuronal pyroptosis (Zhang et al., 2021c). Previous studies also reported that the downregulation of Neat1 could exert anti-inflammatory effects in cerebral I/R injury (Han and Zhou, 2019; Jin et al., 2021). Taken together, these findings indicate that the lncRNA Neat1 might play crucial roles in PCD pathways and inflammation in cerebral I/R injury and is a potential therapeutic target. Another lncRNA identified as a hub node in both necroptosis-related ceRNA and pyroptosis-related regulatory networks is Xist. Previous studies confirmed that Xist contributes to cerebral I/R injury through modulating cell apoptosis, ROS production, and inflammation. Wang et al. also demonstrated that the silencing of XIST protected against cerebral I/R injury by inhibiting neuronal deficit and inflammation via the miR-362/ROCK2 axis (Wang et al., 2021). In addition, Xiong et al. reported that XIST reduced cell viability and induced cell apoptosis via modulating miR-486-5p and GAB2, which promoted cerebral I/R injury (Xiong et al., 2021). Another two studies also indicated that knockdown of XIST inhibited brain injury by suppressing apoptosis and ROS production (Zhang et al., 2021d; Weng et al., 2021). Moreover, a recent study illustrated that Xist was involved in the regulation of pyroptosis in MCAO/R-treated rats and OGD/R-treated rat brain microvascular endothelial cells (Guo et al., 2022). Nevertheless, the molecular roles and regulatory mechanisms of XIST in necroptosis and pyroptosis in cerebral I/R injury have not been fully elucidated and require further research.
In summary, we analyzed datasets GSE131193 and GSE58720 to identify PCD-related genes signature and potential regulatory axes in cerebral I/R injury and the results were validated through previous work. To our knowledge, this was the first study to focused on PCD (apoptosis, necroptosis, and pyroptosis)-related genes and potential regulatory axes in cerebral I/R injury, which might have profound significance for ischemia stroke. We identified hub nodes and seven key ceRNA regulatory axes that has never been reported before in ischemia stroke, which may contribute to elucidating the molecular mechanisms and provide a basis for developing novel therapeutic targets against cerebral I/R injury. Further in vivo and in vitro studies should be conducted to verify these regulatory axes.
There were several limitations to this study that should be acknowledged. First, the PCD (apoptosis, necroptosis, and pyroptosis)-related genes included in this study were mainly identified in previous studies, so some unreported related genes may have been ignored or excluded. Second, both lncRNAs and miRNAs were obtained by online database prediction because neither lncRNA nor miRNA datasets on adult mouse brain tissues after transient focal ischemia at 24 h of reperfusion and matched control samples were available. In future studies, if available, lncRNA and miRNA datasets should be analyzed simultaneously to increase the reliability of the results. Third, the selected datasets were performed in different laboratories and therefore, the differences in sample preparation, sample collection methods, and microarray platforms might influence the results. Finally, our hypothesized potential binding affinity among lncRNAs, miRNAs, and mRNAs should be subjected to further experimental investigation.
Taken together, our findings indicated that these PCD pathways (apoptosis, necroptosis, pyroptosis, and PANoptosis) and crosstalk among them might be involved in ischemic stroke. And the key nodes and regulatory axes identified in this study might play vital roles in regulating the above processes, which may offer new insights into the potential mechanisms underlying cell death during cerebral I/R injury and provide new therapeutic targets for neuroprotection.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
JS, LY, WW, and LZ conceived and designed the study, analyzed the data, wrote the manuscript, and contributed to the designs of the methods used in this study. All authors have read and agreed to publication of this version of the manuscript.
This work was funded by Shanghai Municipal Key Clinical Specialty, grant number shslczdzk02801, the Shanghai Municipal Health Commission, grant number 2020YJZX0109, the Shanghai Pujiang Program, grant number 2019PJD013, and the National Natural Science Foundation of China (NSFC), grant number 81471103.
We thank Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the language of a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.934154/full#supplementary-material
Agarwal, V., Bell, G. W., and Nam, J. W., (2015). Predicting effective microRNA target sites in mammalian mRNAs[J]. United Kingdom: Elife, 4.
Banoth, B., Tuladhar, S., Karki, R., Sharma, B. R., Briard, B., Kesavardhana, S., et al. (2020). ZBP1 promotes fungi-induced inflammasome activation and pyroptosis, apoptosis, and necroptosis (PANoptosis). J. Biol. Chem. 295 (52), 18276–18283. doi:10.1074/jbc.RA120.015924
Bao, M., Szeto, V., Yang, B. B., Zhu, S. Z., Sun, H. S., and Feng, Z. P. (2018). Long non-coding RNAs in ischemic stroke. Cell Death Dis. 9 (3), 281. doi:10.1038/s41419-018-0282-x
Bardou, P., Mariette, J., Escudie, F., Djemiel, C., and Klopp, C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinforma. 15, 293. doi:10.1186/1471-2105-15-293
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI geo: Archive for functional genomics data sets--update. Nucleic Acids Res. 41 (Database issue), D991–D995. doi:10.1093/nar/gks1193
Basurto-Islas, G., Gu, J., Tung, Y. C., Liu, F., and Iqbal, K. (2018). Mechanism of tau hyperphosphorylation involving lysosomal enzyme asparagine endopeptidase in a mouse model of brain ischemia. J. Alzheimer's Dis. 63 (2), 821–833. doi:10.3233/jad-170715
Bu, X., Zhao, Y., Chang, M., and Ge, X. (2021). Downregulation of lncRNA SNHG14 alleviates neurons injury by modulating the miR-181c-5p/BMF axis in ischemic stroke. Brain Res. Bull. 174, 379–388. doi:10.1016/j.brainresbull.2021.06.026
Cabon, L., Galan-Malo, P., Bouharrour, A., Brunelle-Navas, M. N., Lorenzo, H. K., Gross, A., et al. (2012). BID regulates AIF-mediated caspase-independent necroptosis by promoting BAX activation. Cell Death Differ. 19 (2), 245–256. doi:10.1038/cdd.2011.91
Campbell, B., and Khatri, P. (2020). Stroke. Lancet 396 (10244), 129–142. doi:10.1016/S0140-6736(20)31179-X
Cao, D. W., Liu, M. M., Duan, R., Tao, Y. F., Zhou, J. S., Fang, W. R., et al. (2020). The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol. Sin. 41 (1), 22–33. doi:10.1038/s41401-019-0284-y
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8 (Suppl. 4), S11. doi:10.1186/1752-0509-8-S4-S11
Conway, J. R., Lex, A., and Gehlenborg, N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33 (18), 2938–2940. doi:10.1093/bioinformatics/btx364
Datta, A., Sarmah, D., Mounica, L., Kaur, H., Kesharwani, R., Verma, G., et al. (2020). Cell death pathways in ischemic stroke and targeted pharmacotherapy. Transl. Stroke Res. 11 (6), 1185–1202. doi:10.1007/s12975-020-00806-z
Deng, B., Bai, F., Zhou, H., Zhou, D., Ma, Z., Xiong, L., et al. (2016). Electroacupuncture enhances rehabilitation through miR-181b targeting PirB after ischemic stroke. Sci. Rep. 6, 38997. doi:10.1038/srep38997
Deng, X. X., Li, S. S., and Sun, F. Y. (2019). Necrostatin-1 prevents necroptosis in brains after ischemic stroke via inhibition of RIPK1-mediated RIPK3/MLKL signaling. Aging Dis. 10 (4), 807–817. doi:10.14336/AD.2018.0728
Dong, D. W., Zhang, Y. S., Yang, W. Y., Wang-Qin, R. Q., Xu, A. D., and Ruan, Y. W. (2014). Hyperphosphorylation of tau protein in the ipsilateral thalamus after focal cortical infarction in rats. Brain Res. 1543, 280–289. doi:10.1016/j.brainres.2013.11.004
Dong, Z., Pan, K., Pan, J., Peng, Q., and Wang, Y. (2018). The possibility and molecular mechanisms of cell pyroptosis after cerebral ischemia. Neurosci. Bull. 34 (6), 1131–1136. doi:10.1007/s12264-018-0294-7
Eltzschig, H. K., and Eckle, T. (2011). Ischemia and reperfusion--from mechanism to translation. Nat. Med. 17 (11), 1391–1401. doi:10.1038/nm.2507
Fujii, H., Takahashi, T., Mukai, T., Tanaka, S., Hosomi, N., Maruyama, H., et al. (2017). Modifications of tau protein after cerebral ischemia and reperfusion in rats are similar to those occurring in Alzheimer's disease - hyperphosphorylation and cleavage of 4- and 3-repeat tau. J. Cereb. Blood Flow. Metab. 37 (7), 2441–2457. doi:10.1177/0271678X16668889
Ghafouri-Fard, S., Shoorei, H., and Taheri, M. (2020). Non-coding RNAs participate in the ischemia-reperfusion injury. Biomed. Pharmacother. 129, 110419. doi:10.1016/j.biopha.2020.110419
Green, D. R. (2019). The coming decade of cell death research: Five riddles. Cell 177 (5), 1094–1107. doi:10.1016/j.cell.2019.04.024
Guo, J., Ma, Y., Peng, X., Jin, H., and Liu, J. (2019). LncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging miR-181 in hepatocellular carcinoma. J. Cell. Biochem. 120 (10), 17975–17983. doi:10.1002/jcb.29064
Guo, Y., Yang, J. H., He, Y., Zhou, H. F., Wang, Y., Ding, Z. S., et al. (2022). Protocatechuic aldehyde prevents ischemic injury by attenuating brain microvascular endothelial cell pyroptosis via lncRNA Xist. Phytomedicine. 94, 153849. doi:10.1016/j.phymed.2021.153849
Hallenbeck, J. M. (2002). The many faces of tumor necrosis factor in stroke. Nat. Med. 8 (12), 1363–1368. doi:10.1038/nm1202-1363
Han, B., Wang, Q., Cui, G., Shen, X., and Zhu, Z. (2011). Post-treatment of Bax-inhibiting peptide reduces neuronal death and behavioral deficits following global cerebral ischemia. Neurochem. Int. 58 (2), 224–233. doi:10.1016/j.neuint.2010.12.008
Han, D., and Zhou, Y. (2019). YY1-induced upregulation of lncRNA NEAT1 contributes to OGD/R injury-induced inflammatory response in cerebral microglial cells via Wnt/β-catenin signaling pathway. Vitro Cell. Dev. Biol. Anim. 55 (7), 501–511. doi:10.1007/s11626-019-00375-y
Herpich, F., and Rincon, F. (2020). Management of acute ischemic stroke. Crit. Care Med. 48 (11), 1654–1663. doi:10.1097/CCM.0000000000004597
Hoss, F., Rodriguez-Alcazar, J. F., and Latz, E. (2017). Assembly and regulation of ASC specks. Cell. Mol. Life Sci. 74 (7), 1211–1229. doi:10.1007/s00018-016-2396-6
Hu, L., Chen, M., Chen, X., Zhao, C., Fang, Z., Wang, H., et al. (2020). Chemotherapy-induced pyroptosis is mediated by BAK/BAX-caspase-3-GSDME pathway and inhibited by 2-bromopalmitate. Cell Death Dis. 11 (4), 281. doi:10.1038/s41419-020-2476-2
Huang, H. Y., Lin, Y. C., Li, J., Huang, K. Y., Shrestha, S., Hong, H. C., et al. (2020). miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 48 (D1), D148–D154. doi:10.1093/nar/gkz896
Huo, X., Zhang, K., Yi, L., Mo, Y., Liang, Y., Zhao, J., et al. (2016). Decreased epithelial and plasma miR-181b-5p expression associates with airway eosinophilic inflammation in asthma. Clin. Exp. Allergy 46 (10), 1281–1290. doi:10.1111/cea.12754
Hutchison, E. R., Kawamoto, E. M., Taub, D. D., Lal, A., Abdelmohsen, K., Zhang, Y., et al. (2013). Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 61 (7), 1018–1028. doi:10.1002/glia.22483
Indrieri, A., Carrella, S., Carotenuto, P., Banfi, S., and Franco, B. (2020). The pervasive role of the miR-181 family in development, neurodegeneration, and cancer. Int. J. Mol. Sci. 21 (6), 2092. doi:10.3390/ijms21062092
Jassal, B., Matthews, L., Viteri, G., Gong, C., Lorente, P., Fabregat, A., et al. (2020). The reactome pathway knowledgebase. Nucleic Acids Res. 48 (D1), D498–D503. doi:10.1093/nar/gkz1031
Jayaraj, R. L., Azimullah, S., Beiram, R., Jalal, F. Y., and Rosenberg, G. A. (2019). Neuroinflammation: Friend and foe for ischemic stroke. J. Neuroinflammation 16 (1), 142. doi:10.1186/s12974-019-1516-2
Jia, Y., Yi, L., Li, Q., Liu, T., and Yang, S. (2021). LncRNA MALAT1 aggravates oxygen-glucose deprivation/reoxygenation-induced neuronal endoplasmic reticulum stress and apoptosis via the miR-195a-5p/HMGA1 axis. Biol. Res. 54 (1), 8. doi:10.1186/s40659-021-00331-9
Jiao, H., Wachsmuth, L., Kumari, S., Schwarzer, R., Lin, J., Eren, R. O., et al. (2020). Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 580 (7803), 391–395. doi:10.1038/s41586-020-2129-8
Jin, F., Ou, W., Wei, B., Fan, H., Wei, C., Fang, D., et al. (2021). Transcriptome-wide analysis to identify the inflammatory role of lncRNA Neat1 in experimental ischemic stroke. J. Inflamm. Res. 14, 2667–2680. doi:10.2147/JIR.S315281
Jurcau, A., and Simion, A. (2021). Neuroinflammation in cerebral ischemia and ischemia/reperfusion injuries: From pathophysiology to therapeutic strategies. Int. J. Mol. Sci. 23 (1), 14. doi:10.3390/ijms23010014
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). Kegg: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45 (D1), D353–D361. doi:10.1093/nar/gkw1092
Karagkouni, D., Paraskevopoulou, M. D., Tastsoglou, S., Skoufos, G., Karavangeli, A., Pierros, V., et al. (2020). DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 48 (D1), D101–D110. doi:10.1093/nar/gkz1036
Karki, R., Sharma, B. R., Lee, E., Banoth, B., Malireddi, R. K. S., Samir, P., et al. (2020). Interferon regulatory factor 1 regulates PANoptosis to prevent colorectal cancer. JCI Insight 5 (12), 136720. doi:10.1172/jci.insight.136720
Karki, R., Sharma, B. R., Tuladhar, S., Williams, E. P., Zalduondo, L., Samir, P., et al. (2021). Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184 (1), 149–168.e17. doi:10.1016/j.cell.2020.11.025
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11 (9), 597–610. doi:10.1038/nrg2843
Lee, S., Karki, R., Wang, Y., Nguyen, L. N., Kalathur, R. C., and Kanneganti, T. D. (2021). AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature 597 (7876), 415–419. doi:10.1038/s41586-021-03875-8
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42 (Database issue), D92–D97. doi:10.1093/nar/gkt1248
Li, J., Zhang, J., Zhang, Y., Wang, Z., Song, Y., Wei, S., et al. (2019). TRAF2 protects against cerebral ischemia-induced brain injury by suppressing necroptosis. Cell Death Dis. 10 (5), 328. doi:10.1038/s41419-019-1558-5
Li, P., Duan, S., and Fu, A. (2020). Long noncoding RNA NEAT1 correlates with higher disease risk, worse disease condition, decreased miR-124 and miR-125a and predicts poor recurrence-free survival of acute ischemic stroke. J. Clin. Lab. Anal. 34 (2), e23056. doi:10.1002/jcla.23056
Li, Y., Xiang, L., Wang, C., Song, Y., Miao, J., and Miao, M. (2021). Protection against acute cerebral ischemia/reperfusion injury by Leonuri Herba Total Alkali via modulation of BDNF-TrKB-PI3K/Akt signaling pathway in rats. Biomed. Pharmacother. 133, 111021. doi:10.1016/j.biopha.2020.111021
Liang, Y., Song, P., Zhu, Y., Xu, J., Zhu, P., Liu, R., et al. (2020). TREM-1-targeting LP17 attenuates cerebral ischemia-induced neuronal injury by inhibiting oxidative stress and pyroptosis. Biochem. Biophys. Res. Commun. 529 (3), 554–561. doi:10.1016/j.bbrc.2020.05.056
Liao, S., Apaijai, N., Chattipakorn, N., and Chattipakorn, S. C. (2020). The possible roles of necroptosis during cerebral ischemia and ischemia/reperfusion injury. Arch. Biochem. Biophys. 695, 108629. doi:10.1016/j.abb.2020.108629
Liu, X., Hou, L., Huang, W., Gao, Y., Lv, X., and Tang, J. (2016). The mechanism of long non-coding RNA MEG3 for neurons apoptosis caused by hypoxia: Mediated by miR-181b-12/15-LOX signaling pathway. Front. Cell. Neurosci. 10, 201. doi:10.3389/fncel.2016.00201
Lu, Y., Xu, X., Dong, R., Sun, L., Chen, L., Zhang, Z., et al. (2019). MicroRNA-181b-5p attenuates early postoperative cognitive dysfunction by suppressing hippocampal neuroinflammation in mice. Cytokine 120, 41–53. doi:10.1016/j.cyto.2019.04.005
Ma, Q., Zhao, H., Tao, Z., Wang, R., Liu, P., Han, Z., et al. (2016). MicroRNA-181c exacerbates brain injury in acute ischemic stroke. Aging Dis. 7 (6), 705–714. doi:10.14336/AD.2016.0320
Maddahi, A., Kruse, L. S., Chen, Q. W., and Edvinsson, L. (2011). The role of tumor necrosis factor-α and TNF-α receptors in cerebral arteries following cerebral ischemia in rat. J. Neuroinflammation 8, 107. doi:10.1186/1742-2094-8-107
Malireddi, R., Tweedell, R. E., and Kanneganti, T. D. (2020). PANoptosis components, regulation, and implications. Aging (Albany NY) 12 (12), 11163–11164. doi:10.18632/aging.103528
Martens, M., Ammar, A., Riutta, A., Waagmeester, A., Slenter, D. N., Hanspers, K., et al. (2021). WikiPathways: Connecting communities. Nucleic Acids Res. 49 (D1), D613–D621. doi:10.1093/nar/gkaa1024
Mendelson, S. J., and Prabhakaran, S. (2021). Diagnosis and management of transient ischemic attack and acute ischemic stroke: A review. JAMA 325 (11), 1088–1098. doi:10.1001/jama.2020.26867
Meng, C., Zhang, J., Zhang, L., Wang, Y., Li, Z., and Zhao, J. (2019). Effects of NLRP6 in cerebral ischemia/reperfusion (I/R) injury in rats. J. Mol. Neurosci. 69 (3), 411–418. doi:10.1007/s12031-019-01370-4
Meng, Q., Ye, C., and Lu, Y. (2020). miR-181c regulates ischemia/reperfusion injury-induced neuronal cell death by regulating c-Fos signaling. Pharmazie 75 (2), 90–93. doi:10.1691/ph.2020.9856
Michalicova, A., Majerova, P., and Kovac, A. (2020). Tau protein and its role in blood–brain barrier dysfunction[J]. Front. Mol. Neurosci., 13.
Moon, J. M., Xu, L., and Giffard, R. G. (2013). Inhibition of microRNA-181 reduces forebrain ischemia-induced neuronal loss. J. Cereb. Blood Flow. Metab. 33 (12), 1976–1982. doi:10.1038/jcbfm.2013.157
Ni, X., Su, Q., Xia, W., Zhang, Y., Jia, K., Su, Z., et al. (2020). Knockdown lncRNA NEAT1 regulates the activation of microglia and reduces AKT signaling and neuronal apoptosis after cerebral ischemic reperfusion. Sci. Rep. 10 (1), 19658. doi:10.1038/s41598-020-71411-1
Ouyang, Y. B., Lu, Y., Yue, S., Xu, L. J., Xiong, X. X., White, R. E., et al. (2012). miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol. Dis. 45 (1), 555–563. doi:10.1016/j.nbd.2011.09.012
Peng, Z., Li, J., Li, Y., Yang, X., Feng, S., Han, S., et al. (2013). Downregulation of miR-181b in mouse brain following ischemic stroke induces neuroprotection against ischemic injury through targeting heat shock protein A5 and ubiquitin carboxyl-terminal hydrolase isozyme L1. J. Neurosci. Res. 91 (10), 1349–1362. doi:10.1002/jnr.23255
Place, D. E., Lee, S., and Kanneganti, T. D. (2021). PANoptosis in microbial infection. Curr. Opin. Microbiol. 59, 42–49. doi:10.1016/j.mib.2020.07.012
Radak, D., Katsiki, N., Resanovic, I., Jovanovic, A., Sudar-Milovanovic, E., Zafirovic, S., et al. (2017). Apoptosis and acute brain ischemia in ischemic stroke. Curr. Vasc. Pharmacol. 15 (2), 115–122. doi:10.2174/1570161115666161104095522
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. doi:10.1093/nar/gkv007
Samir, P., Malireddi, R. K. S., and Kanneganti, T. (2020). The PANoptosome: A deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 238. doi:10.3389/fcimb.2020.00238
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shi, Y. L., Wang, Q., and Wei, J. C. (2019). Influence of lncRNA-MALAT1 on neuronal apoptosis in rats with cerebral infarction through regulating the ERK/MAPK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (18), 8039–8048. doi:10.26355/eurrev_201909_19020
Shi, Y., Zhang, X., and Ying, P., (2021). Neuroprotective effect of astragaloside IV on cerebral ischemia/reperfusion injury rats through sirt1/mapt pathway[J]. Front. Pharmacol., 12.
Shu, B., Zhou, Y. X., Li, H., Zhang, R. Z., He, C., and Yang, X. (2021). The METTL3/MALAT1/PTBP1/USP8/TAK1 axis promotes pyroptosis and M1 polarization of macrophages and contributes to liver fibrosis. Cell Death Discov. 7 (1), 368. doi:10.1038/s41420-021-00756-x
Song, X., Xue, Y., and Cai, H. (2021). Down-regulation of miR-181a-5p prevents cerebral ischemic injury by upregulating En2 and activating wnt/β-catenin pathway. J. Stroke Cerebrovasc. Dis. 30 (3), 105485. doi:10.1016/j.jstrokecerebrovasdis.2020.105485
Song, Y., Yang, L., Guo, R., Lu, N., Shi, Y., and Wang, X. (2019). Long noncoding RNA MALAT1 promotes high glucose-induced human endothelial cells pyroptosis by affecting NLRP3 expression through competitively binding miR-22. Biochem. Biophys. Res. Commun. 509 (2), 359–366. doi:10.1016/j.bbrc.2018.12.139
Spitz, A. Z., and Gavathiotis, E. (2022). Physiological and pharmacological modulation of BAX. Trends Pharmacol. Sci. 43 (3), 206–220. doi:10.1016/j.tips.2021.11.001
Stary, C. M., Xu, L., Li, L., Sun, X., Ouyang, Y. B., Xiong, X., et al. (2017). Inhibition of miR-181a protects female mice from transient focal cerebral ischemia by targeting astrocyte estrogen receptor-α. Mol. Cell. Neurosci. 82, 118–125. doi:10.1016/j.mcn.2017.05.004
Sticht, C., De La Torre, C., Parveen, A., and Gretz, N. (2018). miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 13 (10), e0206239. doi:10.1371/journal.pone.0206239
Tan, X., Guo, W., Peng, Z., Gu, C., Xiang, P., Tu, Y., et al. (2021). LncRNA-Malat1 down-regulates miR-211-5p expression to promote neuronal damage from cerebral ischemia reperfusion injury. Biochem. Pharmacol. 192, 114694. doi:10.1016/j.bcp.2021.114694
Tu, Y., and Hu, Y. (2021). MiRNA-34c-5p protects against cerebral ischemia/reperfusion injury: Involvement of anti-apoptotic and anti-inflammatory activities. Metab. Brain Dis. 36 (6), 1341–1351. doi:10.1007/s11011-021-00724-5
Tuo, Q. Z., Zhang, S. T., and Lei, P. (2022). Mechanisms of neuronal cell death in ischemic stroke and their therapeutic implications. Med. Res. Rev. 42 (1), 259–305. doi:10.1002/med.21817
Wang, H., Zheng, X., Jin, J., Zheng, L., Guan, T., Huo, Y., et al. (2020). LncRNA MALAT1 silencing protects against cerebral ischemia-reperfusion injury through miR-145 to regulate AQP4. J. Biomed. Sci. 27 (1), 40. doi:10.1186/s12929-020-00635-0
Wang, J., Fu, Z., Wang, M., Lu, J., Yang, H., and Lu, H. (2021). Knockdown of XIST attenuates cerebral ischemia/reperfusion injury through regulation of miR-362/ROCK2 Axis. Neurochem. Res. 46 (8), 2167–2180. doi:10.1007/s11064-021-03354-6
Wang, L., Li, S., Stone, S. S., Liu, N., Gong, K., Ren, C., et al. (2022). The role of the lncRNA MALAT1 in neuroprotection against hypoxic/ischemic injury. Biomolecules 12 (1), 146. doi:10.3390/biom12010146
Wang, Y., and Kanneganti, T. (2021). From pyroptosis, apoptosis and necroptosis to PANoptosis: A mechanistic compendium of programmed cell death pathways. Comput. Struct. Biotechnol. J. 19, 4641–4657. doi:10.1016/j.csbj.2021.07.038
Watters, O., and O'Connor, J. J. (2011). A role for tumor necrosis factor-α in ischemia and ischemic preconditioning. J. Neuroinflammation 8, 87. doi:10.1186/1742-2094-8-87
Weng, S., Wang, S., and Jiang, J. (2021). Long noncoding RNA X-inactive specific transcript regulates neuronal cell apoptosis in ischemic stroke through miR-98/BACH1 Axis. DNA Cell Biol. 40 (7), 979–987. doi:10.1089/dna.2020.6354
Wu, J., Fan, C. L., Ma, L. J., Liu, T., Wang, C., Song, J. X., et al. (2017). Distinctive expression signatures of serum microRNAs in ischaemic stroke and transient ischaemic attack patients. Thromb. Haemost. 117 (5), 992–1001. doi:10.1160/TH16-08-0606
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 97, 69–80. doi:10.1016/j.brainresbull.2013.06.001
Xin, J. W., and Jiang, Y. G. (2017). Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp. Ther. Med. 13 (4), 1225–1234. doi:10.3892/etm.2017.4095
Xiong, F., Wei, W. P., Liu, Y. B., Wang, Y., Zhang, H. Y., and Liu, R. (2021). Long noncoding RNA XIST enhances cerebral ischemia-reperfusion injury by regulating miR-486-5p and GAB2. Eur. Rev. Med. Pharmacol. Sci. 25 (4), 2013–2020. doi:10.26355/eurrev_202102_25103
Xu, Q., Guohui, M., Li, D., Bai, F., Fang, J., Zhang, G., et al. (2021). lncRNA C2dat2 facilitates autophagy and apoptosis via the miR-30d-5p/DDIT4/mTOR axis in cerebral ischemia-reperfusion injury. Aging (Albany NY) 13 (8), 11315–11335. doi:10.18632/aging.202824
Xu, Q., Zhao, B., Ye, Y., Li, Y., Zhang, Y., Xiong, X., et al. (2021). Relevant mediators involved in and therapies targeting the inflammatory response induced by activation of the NLRP3 inflammasome in ischemic stroke. J. Neuroinflammation 18 (1), 123. doi:10.1186/s12974-021-02137-8
Yan, W., Yang, Y., Hu, X., Ning, W. Y., Liao, L. S., Lu, S., et al. (2022). Do pyroptosis, apoptosis, and necroptosis (PANoptosis) exist in cerebral ischemia? Evidence from cell and rodent studies[J]. Neural Regen. Res. 17 (8), 1761–1768. doi:10.4103/1673-5374.331539
Yan, Y., Chen, L., Zhou, J., and Xie, L. (2020). SNHG12 inhibits oxygen‑glucose deprivation‑induced neuronal apoptosis via the miR‑181a‑5p/NEGR1 axis. Mol. Med. Rep. 22 (5), 3886–3894. doi:10.3892/mmr.2020.11459
Yao, D., Zhang, S., Hu, Z., Luo, H., Mao, C., Fan, Y., et al. (2021). CHIP ameliorates cerebral ischemia-reperfusion injury by attenuating necroptosis and inflammation. Aging (Albany NY) 13 (23), 25564–25577. doi:10.18632/aging.203774
Yu, P., Zhang, X., Liu, N., Tang, L., Peng, C., and Chen, X. (2021). Pyroptosis: Mechanisms and diseases. Signal Transduct. Target. Ther. 6 (1), 128. doi:10.1038/s41392-021-00507-5
Zhang, G., Wang, Q., and Su, D., (2021). Long non-coding RNAMALAT1 knockdown alleviates cerebral ischemia/reperfusion injury of rats through regulating the miR-375/pde4d Axis[J]. Front. Neurology, 11.
Zhang, H., Ouyang, B., Ji, X., and Liu, M. F. (2021). Gastrodin alleviates cerebral ischaemia/reperfusion injury by inhibiting pyroptosis by regulating the lncRNA NEAT1/miR-22-3p Axis. Neurochem. Res. 46 (7), 1747–1758. doi:10.1007/s11064-021-03285-2
Zhang, H., Xia, J., Hu, Q., Xu, L., Cao, H., Wang, X., et al. (2021). Long non‑coding RNA XIST promotes cerebral ischemia/reperfusion injury by modulating miR‑27a‑3p/FOXO3 signaling. Mol. Med. Rep. 24 (2), 566. doi:10.3892/mmr.2021.12205
Zhang, H., Zhong, K., Lu, M., Mei, Y., Tan, E., Sun, X., et al. (2018). Neuroprotective effects of isosteviol sodium through increasing CYLD by the downregulation of miRNA-181b. Brain Res. Bull. 140, 392–401. doi:10.1016/j.brainresbull.2018.05.015
Zhang, T., Yin, C., Boyd, D. F., Quarato, G., Ingram, J. P., Shubina, M., et al. (2020). Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180 (6), 1115–1129. doi:10.1016/j.cell.2020.02.050
Zhang, X., Liu, Z., Shu, Q., Yuan, S., Xing, Z., and Song, J. (2019). LncRNA SNHG6 functions as a ceRNA to regulate neuronal cell apoptosis by modulating miR-181c-5p/BIM signalling in ischaemic stroke. J. Cell. Mol. Med. 23 (9), 6120–6130. doi:10.1111/jcmm.14480
Zhang, X., Tang, X., Liu, K., Hamblin, M. H., and Yin, K. J. (2017). Long noncoding RNA Malat1 regulates cerebrovascular pathologies in ischemic stroke. J. Neurosci. 37 (7), 1797–1806. doi:10.1523/JNEUROSCI.3389-16.2017
Zhang, Y., Tian, Q., Zhang, Q., Zhou, X., Liu, S., and Wang, J. Z. (2009). Hyperphosphorylation of microtubule-associated tau protein plays dual role in neurodegeneration and neuroprotection. Pathophysiology 16 (4), 311–316. doi:10.1016/j.pathophys.2009.02.003
Zhang, Y., Wang, S., Lu, F., Zhang, M., Kong, H., Cheng, J., et al. (2021). The neuroprotective effect of pretreatment with carbon dots from Crinis Carbonisatus (carbonized human hair) against cerebral ischemia reperfusion injury. J. Nanobiotechnology 19 (1), 257. doi:10.1186/s12951-021-00908-2
Zheng, M., and Kanneganti, T. D. (2020). The regulation of the ZBP1-NLRP3 inflammasome and its implications in pyroptosis, apoptosis, and necroptosis (PANoptosis). Immunol. Rev. 297 (1), 26–38. doi:10.1111/imr.12909
Zheng, M., Karki, R., Vogel, P., and Kanneganti, T. D. (2020). Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell 181 (3), 674–687. doi:10.1016/j.cell.2020.03.040
Zhou, F., Wang, Y. K., Zhang, C. G., and Wu, B. Y. (2021). miR-19a/b-3p promotes inflammation during cerebral ischemia/reperfusion injury via SIRT1/FoxO3/SPHK1 pathway. J. Neuroinflammation 18 (1), 122. doi:10.1186/s12974-021-02172-5
Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A. H., Tanaseichuk, O., et al. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10 (1), 1523. doi:10.1038/s41467-019-09234-6
Keywords: apoptosis, pyroptosis, necroptosis, cerebral ischemia/reperfusion (I/R) injury, competing endogenous RNA (ceRNA) network
Citation: Shu J, Yang L, Wei W and Zhang L (2022) Identification of programmed cell death-related gene signature and associated regulatory axis in cerebral ischemia/reperfusion injury. Front. Genet. 13:934154. doi: 10.3389/fgene.2022.934154
Received: 02 May 2022; Accepted: 06 July 2022;
Published: 04 August 2022.
Edited by:
Mariana Recamonde-Mendoza, Federal University of Rio Grande do Sul, BrazilReviewed by:
Qiuhuan Yuan, Shandong University, ChinaCopyright © 2022 Shu, Yang, Wei and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenshi Wei, d2Vuc2hpd2VpMTk5OUAxNjMuY29t; Li Zhang, bGl6aGFuZ19odWFkb25nQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.