- 1Department of Biochemistry, University of Lucknow, Lucknow, India
- 2Department of Microbiology, College of Basic Science and Humanities, Sardarkrushinagar Dantiwada Agriculture University, Banaskantha, India
- 3Department of Biochemistry, Era Medical University and Hospital, Lucknow, India
The global malnutrition burden imparts long-term developmental, economic, social, and medical consequences to individuals, communities, and countries. The current developments in biotechnology have infused biofortification in several food crops to fight malnutrition. However, these methods are not sustainable and suffer from several limitations, which are being solved by the CRISPR-Cas-based system of genome editing. The pin-pointed approach of CRISPR-based genome editing has made it a top-notch method due to targeted gene editing, thus making it free from ethical issues faced by transgenic crops. The CRISPR-Cas genome-editing tool has been extensively used in crop improvement programs due to its more straightforward design, low methodology cost, high efficiency, good reproducibility, and quick cycle. The system is now being utilized in the biofortification of cereal crops such as rice, wheat, barley, and maize, including vegetable crops such as potato and tomato. The CRISPR-Cas-based crop genome editing has been utilized in imparting/producing qualitative enhancement in aroma, shelf life, sweetness, and quantitative improvement in starch, protein, gamma-aminobutyric acid (GABA), oleic acid, anthocyanin, phytic acid, gluten, and steroidal glycoalkaloid contents. Some varieties have even been modified to become disease and stress-resistant. Thus, the present review critically discusses CRISPR-Cas genome editing-based biofortification of crops for imparting nutraceutical properties.
Introduction
Malnutrition is becoming a rapidly increasing and serious problem as the world’s population grows. The world’s population will reach 8.3 billion in 2030, up from 7.8 billion at present. According to estimates, almost 800 million people are malnourished worldwide, with 98 percent living in underdeveloped countries (Sinha et al., 2019). Undernutrition (wasting, stunting, and being underweight), insufficient vitamins and minerals, obesity, and the consequent diet-related non-communicable disorders are all examples of malnutrition. Furthermore, more than 340 million people suffer from one or more micronutrient deficiencies, including deficiencies in vitamin A, iron, iodine, and zinc (UNICEF, 2021).
Fortification and biofortification are food enrichment technologies that differ in their approach. In the former method, fortificants are added directly to the food during processing, but the latter involves fortification at the crop production level. In comparison to fortification, biofortification is cost-effective as it is a one-time investment to develop a biofortified crop and recurrent costs are low (Shoab and Hefferon, 2022). Biofortified crops hold a brighter future to address nutritional challenges (Yadav et al., 2020; Buturi et al., 2021; Mushtaq and Nazir, 2021; Ziarati et al., 2021). Biofortification is considered a sustainable and long-term solution to provide micronutrient-rich crops to people (Bouis and Welch, 2010; Bouis and Saltzman, 2017; Garg et al., 2018; Heck et al., 2020; Van Der Straeten et al., 2020).
The crops are biofortified for desired nutrients through nutrient treatments as well as breeding (Garg et al., 2018). Agronomic biofortification involves the deliberate use of mineral fertilizers to increase the concentration of a target mineral in edible portions of crops (Adu et al., 2018). Advanced agronomic biofortification includes engineered nanoparticles attached with fertilizers (nano fertilizers) and PGPR (plant growth–promoting rhizobacteria) (Nayana et al., 2020). Moreover, crucial quantitative trait loci (QTLs) are also utilized in crop breeding programs to improve crop nutrient profiles (Gangashetty et al., 2016). Nevertheless, plant breeding, especially polyploid crop breeding, is a time-consuming and laborious method for improving crop productivity (Parry et al., 2009; Nagamine and Ezura, 2022). Some crops have also been biofortified for desired nutrients through transgenic technology-based genetic alterations (Pérez-Massot et al., 2013). Disadvantages of genetically modified (GM) crops include allergic reactions in humans and reduced nutrition. Also, they cause environmental impact by releasing toxins in the soil, induce pest resistance, and disruption of crop biodiversity. Several ethical concerns are associated with GM crops.
In view of the disadvantages of GM crops, genome editing (GE) technology offers distinct advantages (Gaj et al., 2013; Xiong et al., 2015; Aglawe et al., 2018; Fiaz et al., 2021; Nagamine and Ezura, 2022). Thus, genome editing produces predictable and inheritable mutations in specified regions of the genome, with minimal off-target effects and no external gene sequence integration (Bhattacharya et al., 2021). Deletions, insertions, single-nucleotide substitutions, and extensive fragment substitutions are used for GE-mediated DNA alterations. Systems such as homing endonucleases or meganucleases (HEs) (Daboussi et al., 2015), Zinc-Finger Nucleases (ZFNs) (Urnov et al., 2010), and transcription activator-like effector nucleases (TALENs) (Joung and Sander, 2013) were engaged as genome editing tools before the discovery of CRISPR-associated protein (Cas) (Wang et al., 2016). The brief details of previously used genome editing methods are described below.
Meganucleases, also known as homing endonucleases, are rare-cutting enzymes found in all microbial genomes. These enzymes identify and cleave lengthy DNA sequences (usually 18–30 base pairs), resulting in double-strand DNA breaks (DSBs). Various designed meganuclease variants are available to cleave unique DNA targets for genomic changes for creating important characteristics in crop species (Daboussi et al., 2015). Homing endonucleases technology has suffered from technical problems in the manufacture of these nucleases and designing vectors for their entrance into cells and off-targeting consequences (Jin et al., 2016; Rey-Rico and Cucchiarini, 2016).
Zinc-finger nucleases (ZFNs) are “nucleases” consisting of engineered zinc-finger DNA-binding domains paired with a nuclease, most often the FokI nuclease. ZFN-induced double-strand breaks are exposed to cellular DNA repair processes, resulting in remarkably targeted mutagenesis and targeted gene replacement (Carroll, 2011). The zinc-finger domains consist of four to six 30 amino acid domains that may bind to trinucleotide sequences, limiting total DNA-binding domain specificity to 12–18 nucleotide sequences (Davies et al., 2017). However, ZFN technology has disadvantages such as complex design (which requires customized protein for each DNA sequence), low engineering feasibility, low specificity, normal efficacy, and inability to gene knockout and RNA editing (Zhang J.-H. et al., 2016; Zhang B. et al., 2016; Callaway, 2016; Wang et al., 2016; Salsman and Dellaire, 2017; Sun et al., 2018).
Transcription activator-like effector nucleases (TALENs) are a nonspecific DNA-cleaving nuclease linked to a DNA-binding domain that could be tailored to target specific sequences. TALENs are made up of an engineered array of TALE repeats fused to the FokI nuclease domain and are used for editing the genome (Reyon et al., 2013). However, TALENs also have disadvantages such as complex design, lower engineering feasibility, lower specificity, low efficacy, and inability to gene knockout and RNA editing (Zhang J.-H. et al., 2016; Zhang B. et al., 2016; Callaway, 2016; Wang et al., 2016; Salsman and Dellaire, 2017; Sun et al., 2018).
Thus, CRISPR-Cas technology has emerged as a promising genome editing tool overcoming many of the abovementioned disadvantages of HEs, ZFNs, and TALENs. Therefore, it is currently the most extensively used genome editing technique worldwide because of its simple design, cost-effectiveness, high efficiency, good reproducibility, high engineering feasibility, ability to create gene knockout, RNA editing, and quick cycle (Asmamaw and Zawdie, 2021).
Thus, the present review critically discusses CRISPR-Cas genome editing-based biofortification of crops with respect to enhancement of carbohydrate, protein, fatty acids, secondary metabolite contents, as well as imparting disease and stress-resistance.
Overview of the CRISPR-Cas Systems
Most bacteria and Archaea have an adaptive defense system called CRISPR-Cas that protects them from phages, viruses, and other foreign genetic material (Marraffini and Sontheimer, 2010). The main components of the CRISPR-Cas9 system are an RNA-guided Cas9 endonuclease and a single-guide RNA (sgRNA). Type II CRISPR-Cas9 is one of the best-defined and most commonly utilized categories in multiple CRISPR-Cas systems (Jiang and Doudna, 2017). Cas9 endonucleases HNH domain cuts one strand of sgRNA, while the RuvC-like domain cuts the opposite strand of dsDNA, resulting in double-strand breaks (DSBs). As a result, the plant endogenous repair system automatically repairs DSBs in vivo, utilizing error-prone non-homologous end-joining (NHEJ) or homology-directed repair (HDR), resulting in massive insertion or fragment replacement (Liu X. et al., 2017).
The schematic representation of the CRISPR-Cas genome editing methodology for developing genetically edited biofortified plants is shown in Figure 1.
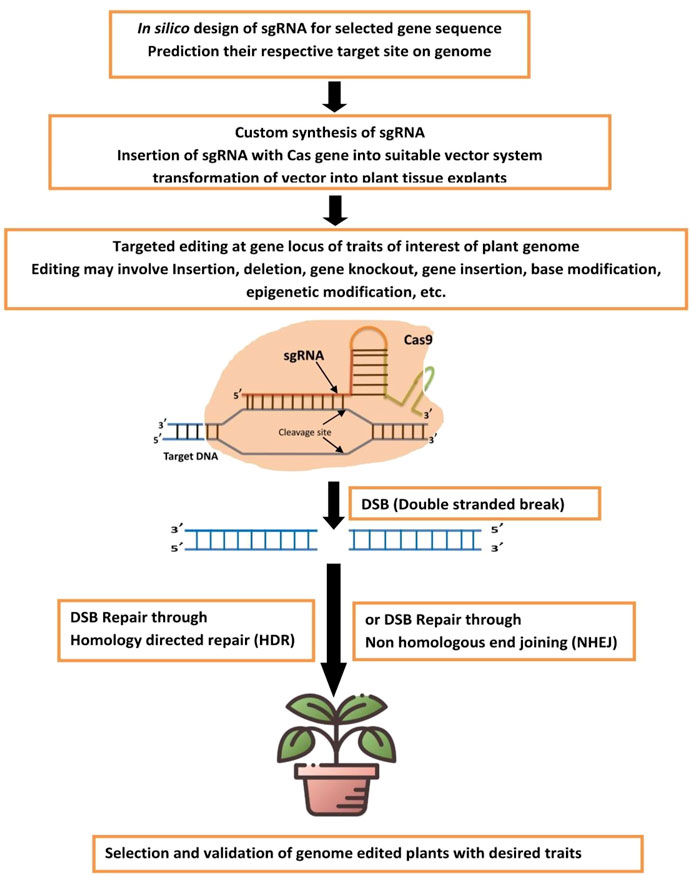
FIGURE 1. Schematic representation for CRISPR-Cas genome editing methodology for developing genetically edited biofortified plants.
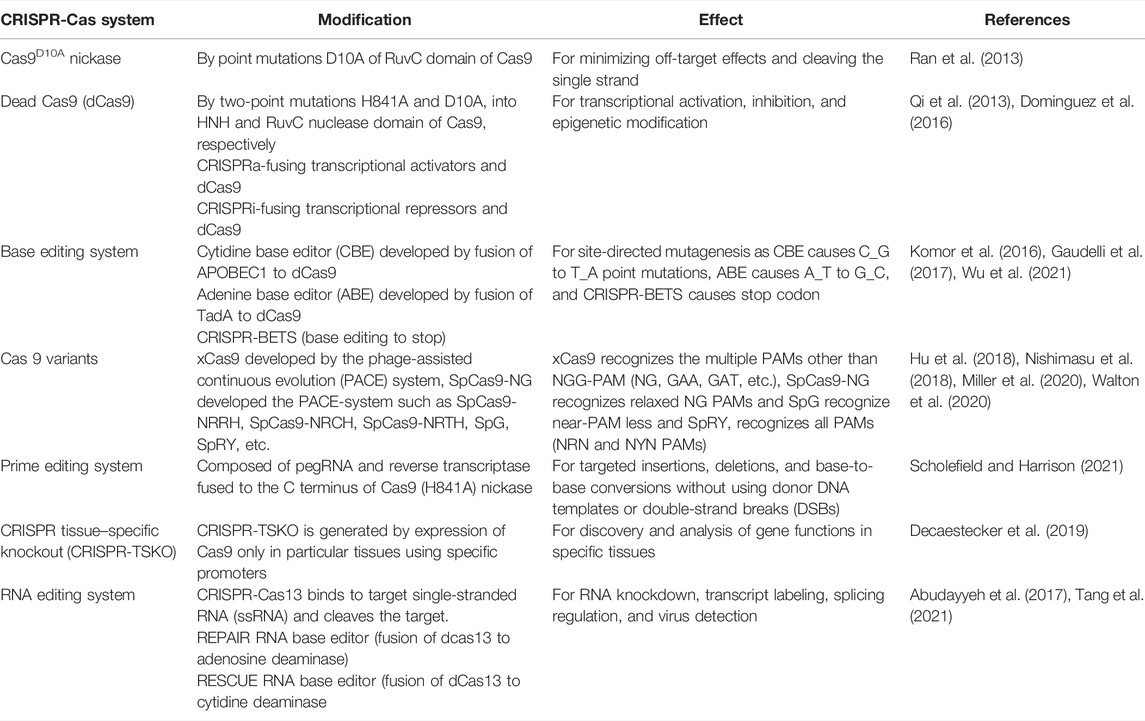
Efforts have been made by researchers to modify the CRISPR-Cas system to make its application more efficient and specific as depicted in Table 1.
Development of Biofortified Crops Through CRISPR-Cas Genome Editing Approach
The CRISPR-Cas system allows rapid, site-specific genome modification in a single cell or a whole organism. It regulates transcription, changes epigenomes, runs genome-wide screens, and imaging chromosomes. The CRISPR-Cas system is now increasingly used for developing edited crops due to its diverse applications in genome editing. Using the CRISPR-Cas, research groups have engineered several crop systems for disease resistance (Schenke and Cai, 2020), drought, salinity, and thermotolerance (Chennakesavulu et al., 2021). CRISPR-Cas is aiding in developing climate-ready crops (Razzaq et al., 2021) and improving crop quality parameters such as appearance, palatability, nutritional components, and other preferred traits (Liu Q et al., 2021). This study has reviewed the nutritional enrichment of important crops using the CRISPR-Cas genome editing method.
Vitamin A Enriched Crops
Carotenoids are widely distributed isoprenoid pigments essential for photosynthetic organisms. Humans do not produce carotenoids de novo, but they require them in their food, notably as β-carotene and vitamin A precursors (Maoka, 2020). Vitamin A is necessary for biological functions that include light transduction in vision, embryonic development, immunological operation, and overall health maintenance in human beings (Timoneda et al., 2018). Vitamin A deficiency (VAD) is one of the most severe worldwide health issues, resulting in a variety of symptoms such as xerophthalmia, night blindness, pediatric blindness, and an increased risk of morbidity and mortality, particularly in young children (Sommer, 2008; Reddy et al., 2022). Given this, carotenoid-enriched staple crops (golden crops), either through traditional breeding or genetic engineering, were initiated to combat VAD (Zheng et al., 2020). Among all the genetic engineering techniques, the CRISPR-based genome-editing technique seems the most efficient and widespread method, making rapid, DNA/transgene-free, and targeted multiplex genetic modification of organisms: a reality for developing “golden” staple crops (Zheng X. et al., 2021). Through CRISPR-Cas-based systems, various genome-edited golden crops, that is, carotenoid biofortified crops have been produced to combat VAD (Table 2). For instance, the Golden rice cultivar Kitaake has been developed by Knock-in, a 5.2-kb carotenogenesis cassette consisting of CrtI and maize PSY genes. The variety contains 7.9 μg/g dry weight (DW) β-carotene in the endosperm (Dong et al., 2020).
Vitamin E–Enriched Crops
Vitamin E (tocopherol) is a potent lipid-soluble antioxidant and an essential component of the human diet. Many human diseases, such as cardiovascular disease and certain cancers, are associated with insufficient vitamin E intake (Rizvi et al., 2014). The daily requirement of vitamin E for humans lies between 15 and 30 mg (Ungurianu et al., 2021).
Through CRISPR-Cas9 technology, significant increase in tocopherols and tocotrienol content was achieved by targeted overexpression of Hordeum vulgare homogentisate phytyltransferase (HvHPT) and Hordeum vulgare homogentisate geranylgeranyltransferase (HvHGGT) (Zeng et al., 2020). These genes can be utilized for enhancing vitamin E content in other crops.
Iron-Enriched Crops
Iron is involved in various metabolic activities such as oxygen transport and electron transport chain. Iron metabolism disorders cause the most frequent diseases in humans, encompassing multiple conditions with various clinical presentations, ranging from anemia to neurodegenerative diseases (Abbaspour et al., 2014). The iron need in humans can be fulfilled through dietary and crop biofortification (Liberal et al., 2020). CRISPR-Cas9 is responsible for the latter type of biofortification. This system has been reported to disrupt the Inositol pentakisphosphate 2- kinase 1 (IPK1) gene causing iron biofortification in wheat (Ibrahim et al., 2021).
Zn-Enriched Crops
The mineral Zn is involved in numerous cellular metabolic processes and catalytic activity of approximately 100 enzymes (Sandstead, 1994). It plays a role in immune functions (Prasad, 1995; Solomons, 1998), protein synthesis (Prasad, 1995; Heyneman, 1996), wound healing (Heyneman, 1996), DNA synthesis, and cell division (Prasad, 1995). Zinc keeps our immune system healthy (Maggini et al., 2010). Zinc maintains cell normal development and activation for innate and adaptive immune responses. It facilitates the integrity of epithelial barriers, which are essential for protecting organisms and preventing pathogen entry (Sturniolo et al., 2002; Finamore et al., 2008; Maares and Haase, 2016). Moreover, Zn can modulate the development and activity of T cells and hence be used as an immunomodulatory candidate. There is only one report on Zn enhancement in wheat, to the best of our knowledge. In this crop, the CRISPR-Cas system disrupts Triticum aestivum Inositol Pentakisphosphate 2-kinase 1 (TaIPK1) that reduces phytic acid to cause improvement in zinc accumulation in wheat grains (Ibrahim et al., 2021). More crops are needed to be biofortified with Zn through the CRISPR-Cas system to make available the products worldwide.
Biofortification Through Targeting Cytokinin Metabolism
Plants absorb a range of mineral elements essential for growth, including C, H, O, N, Fe, Zn, K, Na, and others, in various forms. C, H, and O are acquired from gases or water, and their uptake pathways are straightforward and well-known (Reid and Hayes, 2003). In contrast, other elements are primarily classified as mineral elements and are mainly taken from soil in terrestrial plants or water in aquatic plants (Reid and Hayes, 2003). Root system architecture (RSA), which constitutes the structure of root length, spread, number, and length of lateral roots, is a critical developmental and agronomic characteristic that affects overall plant architecture, growth rate and yield, abiotic stress resistance, nutrient absorption, and developmental flexibility in response to environmental changes (Jung and McCouch, 2013). Phytohormones are communicators between soil and RSA, regulating root development processes, extending from organogenesis to creating higher-order lateral roots (LRs) via various mechanisms (Sharma et al., 2021). The hormones cytokinin (CK) and auxin (IAA), along with ethylene, are essential regulators of root growth, vascular differentiation, and root gravitropism (Aloni et al., 2006). Cytokinin negatively regulates root elongation and branching and crucially shapes RSA (Ramireddy et al., 2014). The enzymes isopentenyl transferase (IPT) present in plants regulates cytokinin levels, which are later destroyed by cytokinin oxidase/dehydrogenase (CKX) and inactivated by glucosylation through cytokinin glucosyl transferases (CGTs) (Chen et al., 2021). Since cytokinin level in roots negatively correlates with crop yields. The reduced cytokine level induced by the enzymes mentioned above increases root growth and uptake of mineral nutrients, particularly Zn and Fe (Chen et al., 2020). Therefore, overexpression of CKX and CGTs in the root zone increases the crop yield.
Genetically edited plants have been developed with indigenously lowered cytokinin levels that favor enrichment of P, Ca, S, Cu, Mn, Fe, and Zn in plant biomass (Ramireddy et al., 2018a). Ramireddy et al. (2018b) developed Zn-fortified field-grown barley which breaks down plant cytokinin through transgenics. In the additional study carried out by the author, the grain yield of barley was increased by knocking out CKX genes through an RNA-guided Cas9 system to generate ckx1 and ckx3 mutant lines with knockout mutations in the HvCKX1 and HvCKX3 genes, respectively. Reduced CKX activity in the ckx1 lines induced longer roots, increased surface area, and a higher number of root hairs. In contrast, enhanced CKX activity in the ckx3 mutants had opposite results. The authors’ findings show that the control of cytokinin activity is complicated, where alterations in just a single component might have unexpected consequences (Gasparis et al., 2019). In another study, silencing the OsCKX4 gene or knockout of the homologous gene OsCKX2 resulted in decreased Zn concentrations in brown rice. However, CRISPR-Cas9 mediated knockout of CYP735A involved in the formation of trans-zeatin (tZ-type) cytokinins elevates Zn concentrations (Gao et al., 2019). In yet another study, Karunarathne et al. (2022) developed the barley abnormal cytokinin response 1 repressor (HvARE1) mutants with high nitrogen content in shoots in nitrogen-deficient soil through Agrobacterium-mediated genetic transformation of immature embryos (cv. Golden Promise) with sgRNAs targeting HvARE1. Such crop types possess nitrogen use efficiency (NUE) and can reduce fertilizer input in soils, rendering them a cost-effective alternative that can prevent environmental pollution due to excessive fertilizer applications.
Quality Improved Crops
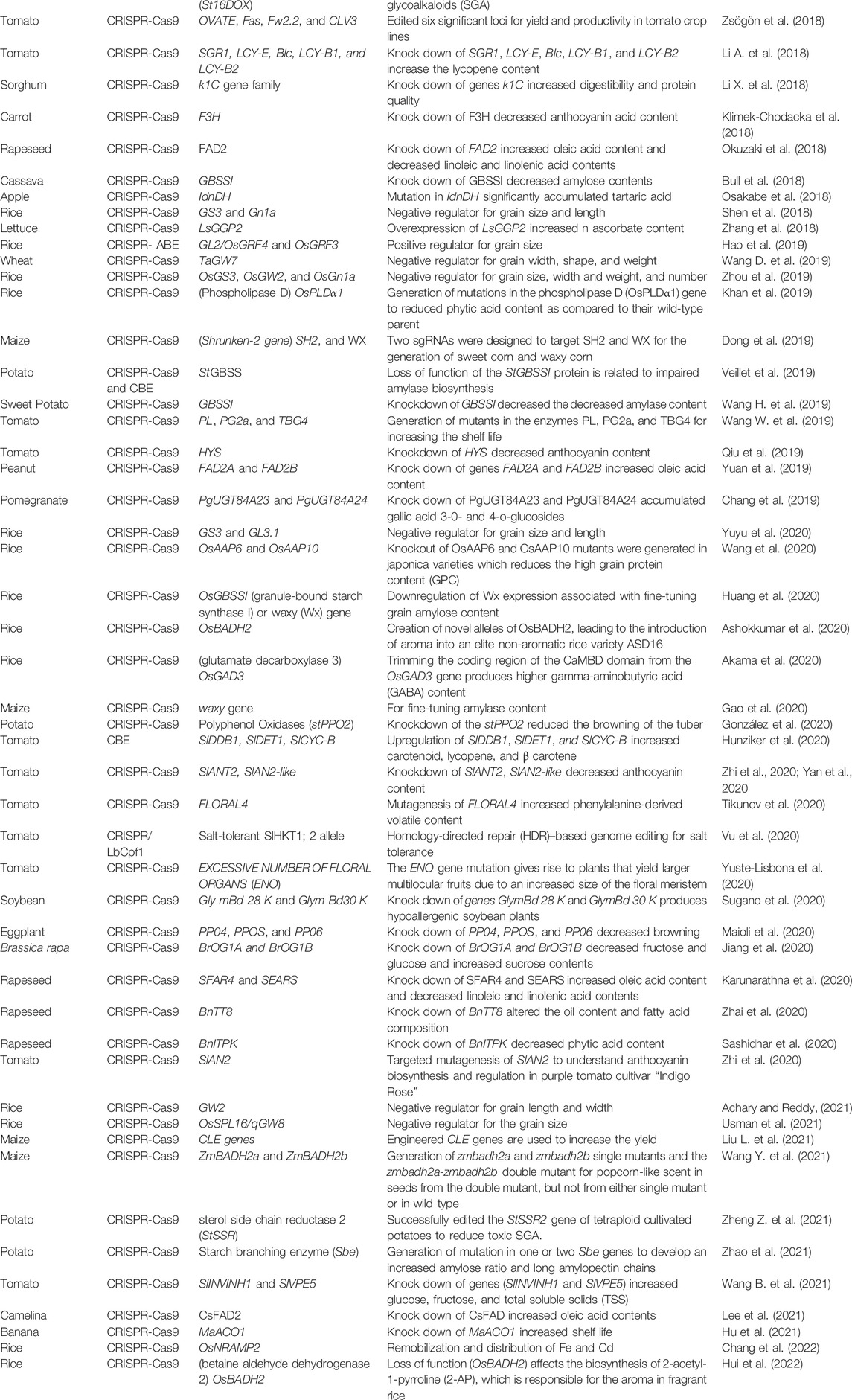
In addition to developing nutrient-enriched crops, many crops are improved to boost production, biotic and abiotic stress resistance, and quality and nutritional value. Over several decades, innovative agricultural technology has considerably enhanced crop productivity. Consumers are more concerned about crop quality because it is linked to human health by delivering nutrients such as proteins, fiber, vitamins, minerals, and bioactive substances (Slavin and Lloyd, 2012). Compared to conventional breeding, CRISPR-based systems have increased the quality of staple, oilseed, and horticultural crops with significant accuracy and efficiency (Ku and Ha, 2020). To the best of the literature review, through CRISPR-Cas mediated genome editing, various crops have been reported for improvement in their diverse categories of quality (Table 3).
Conclusion and Future Prospects
The CRISPR-Cas system is an efficient, convenient, and cost-effective genome editing tool through which major crops can be biofortified for deficient vitamins or minerals. Furthermore, due to the non-insertion of foreign DNA and lesser regulatory restrictions, the products of this technology are easily acceptable by people in society. This genome editing tool has immense potential to eliminate the nutrient deficiency of crops and provide food security for the ever-increasing population. So far, CRISPR technology has been specifically utilized to modify a single gene for crop improvement. However, it holds the potential to manipulate several genes simultaneously, either by assembling multiple sgRNA expression cassettes in a single CRISPR vector or by producing more RNA through an endogenous RNA processing system.
There are several challenges to the widespread CRISPR-based agriculture revolution which include varying legislation and regulatory frameworks for gene-edited crops, delivery of CRISPR-Cas payloads, and off-target activity in CRISPR-Cas systems. In this direction, the United States, Argentina, Brazil, Chile, and Colombia have established product-based regulations by illustrating that gene-edited crops are free from GMO monitoring, provided the end products contain no exogenous DNA. Furthermore, 13 World Trade Organization countries have issued a declaration favoring gene editing for agricultural innovation, marking the first step toward drafting a worldwide regulatory framework.
Recently in India, the Department of Biotechnology, Ministry of Science and Technology has come up with “Guidelines for Safety Assessment of Genome Edited Plants, 2022” (https://ibkp.dbtindia.gov.in/Page Content/ShowBrowsedFile? FileName=20220521202 445079_Final11052022Annexure%20I,%20Genome_Edited_Plants_2022_Hyperlink.pdf&FPath=E:\\DBT_Content_Test\CMS\Guidelines/20220521202445079_Final11052022Annexure%20I,%20Genome_Edited_Plants_2022_Hyperlink.pdf), which provides a road map for the development and sustainable use of genome editing technologies in India, specifying the biosafety and/or environmental safety concerns and describing the regulatory pathways to be adopted while undertaking the genome editing of plants.
Another limitation of CRISPR technology is off-target changes in the host genome, which is needed to be rectified through alteration in the current CRISPR-Cas methodology to minimize off-target binding for optimizing sgRNA, Cas proteins, and delivery methods. Inclusion of highly specific sgRNAs may lower off-target rates. Other strategies, including the extension of the sgRNA sequence and 3′-terminal cleavage of the sgRNA, may reduce the off-targeting effect1.
Despite the persisting challenges, several essential crops have been altered using CRISPR-Cas for nutrition, quality, and productivity enhancement. Nonetheless, additional study is needed to investigate more crop diversity in terms of nutrition, quality, and productivity enrichment to identify effective biofortification targets and optimize CRISPR delivery methods. The CRISPR-based agricultural genome editing is the future of crop fortification as it can design genes for increased vitamin synthesis, crop quality features, and crop production qualities. CRISPR-based genome editing has a great potential to achieve the 2030 goal of eradicating hunger, food insecurity, and all forms of human malnutrition.
Author Contributions
KY and UND conceptualized the manuscript. DK and KY drafted the manuscript. UND, AY, and RA proof-read the manuscript. All authors agreed to the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
DK acknowledges Junior and Senior Research Fellowships by the University Grants Commission (UGC), Government of India, New Delhi. KY acknowledges financial assistance under the Department of Science and Technology (DST-SERB), the Council of Science and Technology (CST), UP, and the Center of Excellence and Research and Development projects from the Department of Higher Education, Government of Uttar Pradesh, Lucknow, India.
Footnotes
1https://www.who.int/news-room/fact-sheets/detail/malnutrition
References
Abbaspour, N., Hurrell, R., and Kelishadi, R. (2014). Review on Iron and its Importance for Human Health. J. Res. Med. Sci. 19 (2), 164–174.
Abe, K., Araki, E., Suzuki, Y., Toki, S., and Saika, H. (2018). Production of High Oleic/low Linoleic Rice by Genome Editing. Plant Physiology Biochem. 131, 58–62. doi:10.1016/j.plaphy.2018.04.033
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA Targeting with CRISPR-Cas13. Nature 550 (7675), 280–284. doi:10.1038/nature24049
Achary, V. M. M., and Reddy, M. K. (2021). CRISPR-Cas9 Mediated Mutation in GRAIN WIDTH and WEIGHT2 (GW2) Locus Improves Aleurone Layer and Grain Nutritional Quality in Rice. Sci. Rep. 11 (1), 21941. doi:10.1038/s41598-021-00828-z
Adu, M. O., Asare, P. A., Yawson, D. O., Nyarko, M. A., and Osei-Agyeman, K. (2018). Agronomic Biofortification of Selected Underutilised Solanaceae Vegetables for Improved Dietary Intake of Potassium (K) in Ghana. Heliyon 4 (8), e00750. doi:10.1016/j.heliyon.2018.e00750
Aglawe, S. B., Barbadikar, K. M., Mangrauthia, S. K., and Madhav, M. S. (2018). New Breeding Technique "genome Editing" for Crop Improvement: Applications, Potentials and Challenges. 3 Biotech. 8 (8), 336. doi:10.1007/s13205-018-1355-3
Akama, K., Akter, N., Endo, H., Kanesaki, M., Endo, M., and Toki, S. (2020). An In Vivo Targeted Deletion of the Calmodulin-Binding Domain from Rice Glutamate Decarboxylase 3 (OsGAD3) Increases γ-Aminobutyric Acid Content in Grains. Rice (N Y) 13 (1), 20–12. doi:10.1186/s12284-020-00380-w
Aloni, R., Aloni, E., Langhans, M., and Ullrich, C. I. (2006). Role of Cytokinin and Auxin in Shaping Root Architecture: Regulating Vascular Differentiation, Lateral Root Initiation, Root Apical Dominance and Root Gravitropism. Ann. Bot. 97 (5), 883–893. doi:10.1093/aob/mcl027
Ashokkumar, S., Jaganathan, D., Ramanathan, V., Rahman, H., Palaniswamy, R., Kambale, R., et al. (2020). Creation of Novel Alleles of Fragrance Gene OsBADH2 in Rice through CRISPR/Cas9 Mediated Gene Editing. PloS one 15 (8), e0237018. doi:10.1371/journal.pone.0237018
Asmamaw, M., and Zawdie, B. (2021). Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biol. targets & Ther. 15, 353–361. doi:10.2147/BTT.S326422
Bhattacharya, A., Parkhi, V., and Char, B. (2021). Genome Editing for Crop Improvement: A Perspective from India. Vitro Cell.Dev.Biol.-Plant 57, 565–573. Advance online publication. doi:10.1007/s11627-021-10184-2
Bouis, H. E., and Saltzman, A. (2017). Improving Nutrition through Biofortification: a Review of Evidence from HarvestPlus, 2003 through 2016. Glob. food Secur. 12, 49–58. doi:10.1016/j.gfs.2017.01.009
Bouis, H. E., and Welch, R. M. (2010). Biofortification—a Sustainable Agricultural Strategy for Reducing Micronutrient Malnutrition in the Global South. Crop Sci. 50–S-20-S-32. doi:10.2135/cropsci2009.09.0531
Bull, S. E., Seung, D., Chanez, C., Mehta, D., Kuon, J. E., Truernit, E., et al. (2018). Accelerated Ex Situ Breeding of GBSS- and PTST1-Edited Cassava for Modified Starch. Sci. Adv. 4 (9), eaat6086. doi:10.1126/sciadv.aat6086
Buturi, C. V., Mauro, R. P., Fogliano, V., Leonardi, C., and Giuffrida, F. (2021). Mineral Biofortification of Vegetables as a Tool to Improve Human Diet. Foods 10 (2), 223. doi:10.3390/foods10020223
Callaway, E. (2016). Gene-editing Research in Human Embryos Gains Momentum. Nature 532 (7599), 289–290. doi:10.1038/532289a
Carroll, D. (2011). Genome Engineering with Zinc-Finger Nucleases. Genetics 188 (4), 773–782. doi:10.1534/genetics.111.131433
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, Precise Modification of the Tomato Genome. Genome Biol. 16, 232. doi:10.1186/s13059-015-0796-9
Chang, J.-D., Xie, Y., Zhang, H., Zhang, S., and Zhao, F.-J. (2022). The Vacuolar Transporter OsNRAMP2 Mediates Fe Remobilization during Germination and Affects Cd Distribution to Rice Grain. Plant Soil, 1–17. doi:10.21203/rs.3.rs-1206604/v1
Chang, L., Wu, S., and Tian, L. (2019). Effective Genome Editing and Identification of a Regiospecific Gallic Acid 4-O-Glycosyltransferase in Pomegranate (Punica Granatum L.). Hortic. Res. 6, 123. doi:10.1038/s41438-019-0206-7
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2020). Cytokinin Dehydrogenase: a Genetic Target for Yield Improvement in Wheat. Plant Biotechnol. J. 18 (3), 614–630. doi:10.1111/pbi.13305
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2021). Cytokinin Glucosyl Transferases, Key Regulators of Cytokinin Homeostasis, Have Potential Value for Wheat Improvement. Plant Biotechnol. J. 19 (5), 878–896. doi:10.1111/pbi.13595
Chennakesavulu, K., Singh, H., Trivedi, P. K., Jain, M., and Yadav, S. R. (2021). State-of-the-Art in CRISPR Technology and Engineering Drought, Salinity, and Thermo-Tolerant Crop Plants. Plant Cell. Rep. 41, 815–831. Advance online publication. doi:10.1007/s00299-021-02681-w
Daboussi, F., Stoddard, T. J., and Zhang, F. (2015). “Engineering Meganuclease for Precise Plant Genome Modification,” in Advances in New Technology for Targeted Modification of Plant Genomes (New York, NY: Springer), 21–38. doi:10.1007/978-1-4939-2556-8_2
Davies, J. P., Kumar, S., and Sastry-Dent, L. (2017). Use of Zinc-Finger Nucleases for Crop Improvement. Prog. Mol. Biol. Transl. Sci. 149, 47–63. doi:10.1016/bs.pmbts.2017.03.006
Decaestecker, W., Buono, R. A., Pfeiffer, M. L., Vangheluwe, N., Jourquin, J., Karimi, M., et al. (2019). CRISPR-TSKO: a Technique for Efficient Mutagenesis in Specific Cell Types, Tissues, or Organs in Arabidopsis. Plant Cell. 31 (12), 2868–2887. doi:10.1105/tpc.19.00454
Demorest, Z. L., Coffman, A., Baltes, N. J., Stoddard, T. J., Clasen, B. M., Luo, S., et al. (2016). Direct Stacking of Sequence-specific Nuclease-Induced Mutations to Produce High Oleic and Low Linolenic Soybean Oil. BMC Plant Biol. 16 (1), 225. doi:10.1186/s12870-016-0906-1
Dominguez, A. A., Lim, W. A., and Qi, L. S. (2016). Beyond Editing: Repurposing CRISPR-Cas9 for Precision Genome Regulation and Interrogation. Nat. Rev. Mol. Cell. Biol. 17 (1), 5–15. doi:10.1038/nrm.2015.2
Dong, L., Qi, X., Zhu, J., Liu, C., Zhang, X., Cheng, B., et al. (2019). Supersweet and Waxy: Meeting the Diverse Demands for Specialty Maize by Genome Editing. Plant Biotechnol. J. 17 (10), 1853–1855. doi:10.1111/pbi.13144
Dong, O. X., Yu, S., Jain, R., Zhang, N., Duong, P. Q., Butler, C., et al. (2020). Marker-free Carotenoid-Enriched Rice Generated through Targeted Gene Insertion Using CRISPR-Cas9. Nat. Commun. 11 (1), 1178. doi:10.1038/s41467-020-14981-y
Fiaz, S., Ahmar, S., Saeed, S., Riaz, A., Mora-Poblete, F., and Jung, K.-H. (2021). Evolution and Application of Genome Editing Techniques for Achieving Food and Nutritional Security. Ijms 22 (11), 5585. doi:10.3390/ijms22115585
Finamore, A., Massimi, M., Conti Devirgiliis, L., and Mengheri, E. (2008). Zinc Deficiency Induces Membrane Barrier Damage and Increases Neutrophil Transmigration in Caco-2 Cells. J. Nutr. 138 (9), 1664–1670. doi:10.1093/jn/138.9.1664
Gaj, T., Gersbach, C. A., and Barbas, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based Methods for Genome Engineering. Trends Biotechnol. 31 (7), 397–405. doi:10.1016/j.tibtech.2013.04.004
Gangashetty, P. I., Motagi, B. N., Pavan, R., and Roodagi, M. B. (2016). “Breeding Crop Plants for Improved Human Nutrition through Biofortification: Progress and Prospects,” in Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits (Cham: Springer), 35–76. doi:10.1007/978-3-319-22518-0_2
Gao, H., Gadlage, M. J., Lafitte, H. R., Lenderts, B., Yang, M., Schroder, M., et al. (2020). Superior Field Performance of Waxy Corn Engineered Using CRISPR-Cas9. Nat. Biotechnol. 38 (5), 579–581. doi:10.1038/s41587-020-0444-0
Gao, S., Xiao, Y., Xu, F., Gao, X., Cao, S., Zhang, F., et al. (2019). Cytokinin‐dependent Regulatory Module Underlies the Maintenance of Zinc Nutrition in Rice. New Phytol. 224 (1), 202–215. doi:10.1111/nph.15962
Garg, M., Sharma, N., Sharma, S., Kapoor, P., Kumar, A., Chunduri, V., et al. (2018). Biofortified Crops Generated by Breeding, Agronomy, and Transgenic Approaches Are Improving Lives of Millions of People Around the World. Front. Nutr. 5, 12. doi:10.3389/fnut.2018.00012
Gasparis, S., Przyborowski, M., Kała, M., and Nadolska-Orczyk, A. (2019). Knockout of the HvCKX1 or HvCKX3 Gene in Barley (Hordeum Vulgare L.) by RNA-Guided Cas9 Nuclease Affects the Regulation of Cytokinin Metabolism and Root Morphology. Cells 8 (8), 782. doi:10.3390/cells8080782
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable Base Editing of at to GC in Genomic DNA without DNA Cleavage. Nature 551 (7681), 464–471. doi:10.1038/nature24644
González, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Fält, A. S., et al. (2020). Reduced Enzymatic Browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 10, 1649. doi:10.3389/fpls.2019.01649
Hao, L., Ruiying, Q., Xiaoshuang, L., Shengxiang, L., Rongfang, X., Jianbo, Y., et al. (2019). CRISPR/Cas9-mediated Adenine Base Editing in Rice Genome. Rice Sci. 26 (2), 125–128. doi:10.1016/j.rsci.2018.07.002
Haun, W., Coffman, A., Clasen, B. M., Demorest, Z. L., Lowy, A., Ray, E., et al. (2014). Improved Soybean Oil Quality by Targeted Mutagenesis of the Fatty Acid Desaturase 2 Gene Family. Plant Biotechnol. J. 12 (7), 934–940. doi:10.1111/pbi.12201
Heck, S., Campos, H., Barker, I., Okello, J. J., Baral, A., Boy, E., et al. (2020). Resilient Agri-Food Systems for Nutrition amidst COVID-19: Evidence and Lessons from Food-Based Approaches to Overcome Micronutrient Deficiency and Rebuild Livelihoods after Crises. Food Sec. 12 (4), 823–830. doi:10.1007/s12571-020-01067-2
Heyneman, C. A. (1996). Zinc Deficiency and Taste Disorders. Ann. Pharmacother. 30 (2), 186–187. doi:10.1177/106002809603000215
Hu, C., Sheng, O., Deng, G., He, W., Dong, T., Yang, Q., et al. (2021). CRISPR/Cas9‐mediated Genome Editing of MaACO1 (Aminocyclopropane‐1‐carboxylate Oxidase 1) Promotes the Shelf Life of Banana Fruit. Plant Biotechnol. J. 19 (4), 654–656. doi:10.1111/pbi.13534
Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., et al. (2018). Evolved Cas9 Variants with Broad PAM Compatibility and High DNA Specificity. Nature 556 (7699), 57–63. doi:10.1038/nature26155
Huang, L., Li, Q., Zhang, C., Chu, R., Gu, Z., Tan, H., et al. (2020). Creating Novel Wx Alleles with Fine‐tuned Amylose Levels and Improved Grain Quality in Rice by Promoter Editing Using CRISPR/Cas9 System. Plant Biotechnol. J. 18 (11), 2164–2166. doi:10.1111/pbi.13391
Hui, S., Li, H., Mawia, A. M., Zhou, L., Cai, J., Ahmad, S., et al. (2022). Production of Aromatic Three‐line Hybrid Rice Using Novel Alleles of BADH2. Plant Biotechnol. J. 20 (1), 59–74. doi:10.1111/pbi.13695
Hunziker, J., Nishida, K., Kondo, A., Kishimoto, S., Ariizumi, T., and Ezura, H. (2020). Multiple Gene Substitution by Target-AID Base-Editing Technology in Tomato. Sci. Rep. 10 (1), 20471. doi:10.1038/s41598-020-77379-2
Ibrahim, S., Saleem, B., Rehman, N., Zafar, S. A., Naeem, M. K., and Khan, M. R. (2021). CRISPR/Cas9 Mediated Disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) Reduces Phytic Acid and Improves Iron and Zinc Accumulation in Wheat Grains. J. Adv. Res. 37, 33–41. doi:10.1016/j.jare.2021.07.006
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., Shima, Y., Nakamura, N., et al. (2017). Re-evaluation of the Rin Mutation and the Role of RIN in the Induction of Tomato Ripening. Nat. plants 3 (11), 866–874. doi:10.1038/s41477-017-0041-5
Jiang, F., and Doudna, J. A. (2017). CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 46, 505–529. doi:10.1146/annurev-biophys-062215-010822
Jiang, M., Zhan, Z., Li, H., Dong, X., Cheng, F., and Piao, Z. (2020). Brassica Rapa Orphan Genes Largely Affect Soluble Sugar Metabolism. Hortic. Res. 7, 181. doi:10.1038/s41438-020-00403-z
Jin, L., Lange, W., Kempmann, A., Maybeck, V., Günther, A., Gruteser, N., et al. (2016). High-efficiency Transduction and Specific Expression of ChR2opt for Optogenetic Manipulation of Primary Cortical Neurons Mediated by Recombinant Adeno-Associated Viruses. J. Biotechnol. 233, 171–180. doi:10.1016/j.jbiotec.2016.07.001
Joung, J. K., and Sander, J. D. (2013). TALENs: a Widely Applicable Technology for Targeted Genome Editing. Nat. Rev. Mol. Cell. Biol. 14 (1), 49–55. doi:10.1038/nrm3486
Jung, J. K. H., and McCouch, S. (2013). Getting to the Roots of it: Genetic and Hormonal Control of Root Architecture. Front. Plant Sci. 4, 186. doi:10.3389/fpls.2013.00186
Karunarathna, N. L., Wang, H., Harloff, H. J., Jiang, L., and Jung, C. (2020). Elevating Seed Oil Content in a Polyploid Crop by Induced Mutations in SEED FATTY ACID REDUCER Genes. Plant Biotechnol. J. 18 (11), 2251–2266. doi:10.1111/pbi.13381
Karunarathne, S. D., Han, Y., Zhang, X. Q., and Li, C. (2022). CRISPR/Cas9 Gene Editing and Natural Variation Analysis Demonstrate the Potential for HvARE1 in Improvement of Nitrogen Use Efficiency in Barley. J. Integr. Plant Biol. 64 (3), 756–770. doi:10.1111/jipb.13214
Kaur, N., Alok, A., ShivaniKumar, P., Kumar, P., Kaur, N., Awasthi, P., et al. (2020). CRISPR/Cas9 Directed Editing of Lycopene Epsilon-Cyclase Modulates Metabolic Flux for β-carotene Biosynthesis in Banana Fruit. Metab. Eng. 59, 76–86. doi:10.1016/j.ymben.2020.01.008
Khan, M. S. S., Basnet, R., Islam, S. A., and Shu, Q. (2019). Mutational Analysis of OsPLDα1 Reveals its Involvement in Phytic Acid Biosynthesis in Rice Grains. J. Agric. Food Chem. 67 (41), 11436–11443. doi:10.1021/acs.jafc.9b05052
Klimek-Chodacka, M., Oleszkiewicz, T., Lowder, L. G., Qi, Y., and Baranski, R. (2018). Efficient CRISPR/Cas9-based Genome Editing in Carrot Cells. Plant Cell. Rep. 37 (4), 575–586. doi:10.1007/s00299-018-2252-2
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 533 (7603), 420–424. doi:10.1038/nature17946
Ku, H.-K., and Ha, S.-H. (2020). Improving Nutritional and Functional Quality by Genome Editing of Crops: Status and Perspectives. Front. Plant Sci. 11, 577313. doi:10.3389/fpls.2020.577313
Lee, K.-R., Jeon, I., Yu, H., Kim, S.-G., Kim, H.-S., Ahn, S.-J., et al. (2021). Increasing Monounsaturated Fatty Acid Contents in Hexaploid Camelina Sativa Seed Oil by FAD2 Gene Knockout Using CRISPR-Cas9. Front. Plant Sci. 12, 702930. doi:10.3389/fpls.2021.702930
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S. J., Zhang, C., et al. (2018). Editing of an Alpha-Kafirin Gene Family Increases, Digestibility and Protein Quality in Sorghum. Plant Physiol. 177 (4), 1425–1438. Epub 2018 Jun 20. PMID: 29925584; PMCID: PMC6084649. doi:10.1104/pp.18.00200
Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., Pan, X., et al. (2016). Reassessment of the Four Yield-Related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 7, 377. doi:10.3389/fpls.2016.00377
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018). Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-mediated Multiplex Genome Editing. Front. Plant Sci. 9, 559. doi:10.3389/fpls.2018.00559
Liberal, Â., Pinela, J., Vívar-Quintana, A. M., Ferreira, I. C. F. R., and Barros, L. (2020). Fighting Iron-Deficiency Anemia: Innovations in Food Fortificants and Biofortification Strategies. Foods 9 (12), 1871. doi:10.3390/foods9121871
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 Acts in the Brassinosteroid Signalling Pathway to Regulate Grain Width and Weight in Rice. Nat. plants 3, 17043. doi:10.1038/nplants.2017.43
Liu, L., Gallagher, J., Arevalo, E. D., Chen, R., Skopelitis, T., Wu, Q., et al. (2021). Enhancing Grain-Yield-Related Traits by CRISPR-Cas9 Promoter Editing of Maize CLE Genes. Nat. Plants 7 (3), 287–294. doi:10.1038/s41477-021-00858-5
Liu, Q., Yang, F., Zhang, J., Liu, H., Rahman, S., Islam, S., et al. (2021). Application of CRISPR/Cas9 in Crop Quality Improvement. Ijms 22 (8), 4206. doi:10.3390/ijms22084206
Liu, X., Wu, S., Xu, J., Sui, C., and Wei, J. (2017). Application of CRISPR/Cas9 in Plant Biology. Acta Pharm. sin. B 7 (3), 292–302. doi:10.1016/j.apsb.2017.01.002
Maares, M., and Haase, H. (2016). Zinc and Immunity: An Essential Interrelation. Archives Biochem. biophysics 611, 58–65. doi:10.1016/j.abb.2016.03.022
Maggini, S., Wenzlaff, S., and Hornig, D. (2010). Essential Role of Vitamin C and Zinc in Child Immunity and Health. J. Int. Med. Res. 38 (2), 386–414. doi:10.1177/147323001003800203
Maioli, A., Gianoglio, S., Moglia, A., Acquadro, A., Valentino, D., Milani, A. M., et al. (2020). Simultaneous CRISPR/Cas9 Editing of Three PPO Genes Reduces Fruit Flesh Browning in Solanum Melongena L. Front. Plant Sci. 11, 607161. doi:10.3389/fpls.2020.607161
Maoka, T. (2020). Carotenoids as Natural Functional Pigments. J. Nat. Med. 74 (1), 1–16. doi:10.1007/s11418-019-01364-x
Marraffini, L. A., and Sontheimer, E. J. (2010). CRISPR Interference: RNA-Directed Adaptive Immunity in Bacteria and Archaea. Nat. Rev. Genet. 11 (3), 181–190. doi:10.1038/nrg2749
Miller, S. M., Wang, T., Randolph, P. B., Arbab, M., Shen, M. W., Huang, T. P., et al. (2020). Continuous Evolution of SpCas9 Variants Compatible with Non-G PAMs. Nat. Biotechnol. 38 (4), 471–481. doi:10.1038/s41587-020-0412-8
Mushtaq, Z., and Nazir, A. (2021). Biofortification: Way Forward toward Micronutrient Deficiency: Biofortification a Sustainable Approach. EQA-International J. Environ. Qual. 42, 36–41.
Nagamine, A., and Ezura, H. (2022). Genome Editing for Improving Crop Nutrition. Front. genome Ed. 4. doi:10.3389/fgeed.2022.850104
Nakayasu, M., Akiyama, R., Lee, H. J., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of α-solanine-free Hairy Roots of Potato by CRISPR/Cas9 Mediated Genome Editing of the St16DOX Gene. Plant Physiology Biochem. 131, 70–77. doi:10.1016/j.plaphy.2018.04.026
Nayana, A. R., Joseph, B. J., Jose, A., and Radhakrishnan, E. K. (2020). “Nanotechnological Advances with PGPR Applications,” in Sustainable Agriculture Reviews (Cham: Springer), 41, 163–180. doi:10.1007/978-3-030-33996-8_9
Nieves-Cordones, M., Mohamed, S., Tanoi, K., Kobayashi, N. I., Takagi, K., Vernet, A., et al. (2017). Production of Low-Cs+ Rice Plants by Inactivation of the K+ Transporter OsHAK1 with the CRISPR-Cas System. Plant J. 92 (1), 43–56. doi:10.1111/tpj.13632
Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., et al. (2018). Engineered CRISPR-Cas9 Nuclease with Expanded Targeting Space. Science 361 (6408), 1259–1262. doi:10.1126/science.aas9129
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., and Ezura, H. (2017). Efficient Increase of Ɣ-Aminobutyric Acid (GABA) Content in Tomato Fruits by Targeted Mutagenesis. Sci. Rep. 7 (1), 7057. doi:10.1038/s41598-017-06400-y
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated Genome Editing of the Fatty Acid Desaturase 2 Gene in Brassica Napus. Plant Physiology Biochem. 131, 63–69. doi:10.1016/j.plaphy.2018.04.025
Osakabe, Y., Liang, Z., Ren, C., Nishitani, C., Osakabe, K., Wada, M., et al. (2018). CRISPR-Cas9-mediated Genome Editing in Apple and Grapevine. Nat. Protoc. 13 (12), 2844–2863. doi:10.1038/s41596-018-0067-9
Parry, M. A. J., Madgwick, P. J., Bayon, C., Tearall, K., Hernandez-Lopez, A., Baudo, M., et al. (2009). Mutation Discovery for Crop Improvement. J. Exp. Bot. 60 (10), 2817–2825. doi:10.1093/jxb/erp189
Pérez-Massot, E., Banakar, R., Gómez-Galera, S., Zorrilla-López, U., Sanahuja, G., Arjó, G., et al. (2013). The Contribution of Transgenic Plants to Better Health Through Improved Nutrition: Opportunities and Constraints. Genes Nutr. 8 (1), 29–41.
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-Guided Platform for Sequence-specific Control of Gene Expression. Cell. 152 (5), 1173–1183. doi:10.1016/j.cell.2013.02.022
Qiu, Z., Wang, H., Li, D., Yu, B., Hui, Q., Yan, S., et al. (2019). Identification of Candidate HY5-dependent and -Independent Regulators of Anthocyanin Biosynthesis in Tomato. Plant Cell. Physiology 60 (3), 643–656. doi:10.1093/pcp/pcy236
Ramireddy, E., Chang, L., and Schmülling, T. (2014). Cytokinin as a Mediator for Regulating Root System Architecture in Response to Environmental Cues. Plant Signal. Behav. 9 (1), e27771. doi:10.4161/psb.27771
Ramireddy, E., Galuszka, P., and Schmülling, T. (2018a). Zn-fortified Cereal Grains in Field-Grown Barley by Enhanced Root Cytokinin Breakdown. Plant Signal. Behav. 13 (11), e1530023. doi:10.1080/15592324.2018.1530023
Ramireddy, E., Hosseini, S. A., Eggert, K., Gillandt, S., Gnad, H., von Wirén, N., et al. (2018b). Root Engineering in Barley: Increasing Cytokinin Degradation Produces a Larger Root System, Mineral Enrichment in the Shoot and Improved Drought Tolerance. Plant Physiol. 177 (3), 1078–1095. doi:10.1104/pp.18.00199
Ran, F. A., Hsu, P. D., Lin, C.-Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., et al. (2013). Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 154 (6), 1380–1389. doi:10.1016/j.cell.2013.08.021
Razzaq, A., Kaur, P., Akhter, N., Wani, S. H., and Saleem, F. (2021). Next-Generation Breeding Strategies for Climate-Ready Crops. Front. Plant Sci. 12, 620420. doi:10.3389/fpls.2021.620420
Reddy, G. B., Shalini, T., Ghosh, S., Pullakhandam, R., Kumar, B. N., Kulkarni, B., et al. (2022). Prevalence of Vitamin A Deficiency and Dietary Inadequacy in Indian School-Age Children and Adolescents. Eur. J. Nutr. 61 (1), 197–209. doi:10.1007/s00394-021-02636-7
Reid, R., and Hayes, J. (2003). Mechanisms and Control of Nutrient Uptake in Plants. Int. Rev. Cytol. 229, 73–114. doi:10.1016/s0074-7696(03)29003-3
Ren, C., Liu, X., Zhang, Z., Wang, Y., Duan, W., Li, S., et al. (2016). CRISPR/Cas9-mediated Efficient Targeted Mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 6 (1), 32289–9. doi:10.1038/srep32289
Rey-Rico, A., and Cucchiarini, M. (2016). Controlled Release Strategies for rAAV-Mediated Gene Delivery. Acta biomater. 29, 1–10. doi:10.1016/j.actbio.2015.10.015
Reyon, D., Maeder, M. L., Khayter, C., Tsai, S. Q., Foley, J. E., Sander, J. D., et al. (2013). Engineering Customized TALE Nucleases (TALENs) and TALE Transcription Factors by Fast Ligation-Based Automatable Solid-phase High-Throughput (FLASH) Assembly. Curr. Protoc. Mol. Biol. Chapter 12 (1), Unit–16. doi:10.1002/0471142727.mb1216s103
Rizvi, S., Raza, S. T., Ahmed, F., Ahmad, A., Abbas, S., and Mahdi, F. (2014). The role of vitamin e in human health and some diseases. Sultan Qaboos Univ. Med. J. 14 (2), e157–65.
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell. 171 (2), 470–e8. doi:10.1016/j.cell.2017.08.030
Salsman, J., and Dellaire, G. (2017). Precision Genome Editing in the CRISPR Era. Biochem. Cell. Biol. 95 (2), 187–201. doi:10.1139/bcb-2016-0137
Sánchez-León, S., Gil-Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, Nontransgenic Wheat Engineered with CRISPR/Cas9. Plant Biotechnol. J. 16 (4), 902–910. doi:10.1111/pbi.12837
Sandstead, H. H. (1994). Understanding Zinc: Recent Observations and Interpretations. J. Lab. Clin. Med. 124 (3), 322–327.
Sashidhar, N., Harloff, H. J., Potgieter, L., and Jung, C. (2020). Gene Editing of Three BnITPK Genes in Tetraploid Oilseed Rape Leads to Significant Reduction of Phytic Acid in Seeds. Plant Biotechnol. J. 18 (11), 2241–2250. doi:10.1111/pbi.13380
Schenke, D., and Cai, D. (2020). Applications of CRISPR/Cas to Improve Crop Disease Resistance: Beyond Inactivation of Susceptibility Factors. iScience 23 (9), 101478. Advance online publication. doi:10.1016/j.isci.2020.101478
Scholefield, J., and Harrison, P. T. (2021). Racism in Science: a Perspective from Gene Therapy. Gene Ther. 28, 1–2. doi:10.1038/s41434-020-00188-9
Sharma, M., Singh, D., Saksena, H. B., Sharma, M., Tiwari, A., Awasthi, P., et al. (2021). Understanding the Intricate Web of Phytohormone Signalling in Modulating Root System Architecture. Ijms 22 (11), 5508. doi:10.3390/ijms22115508
Shen, L., Wang, C., Fu, Y., Wang, J., Liu, Q., Zhang, X., et al. (2018). QTL Editing Confers Opposing Yield Performance in Different Rice Varieties. J. Integr. Plant Biol. 60 (2), 89–93. doi:10.1111/jipb.12501
Shoab, E., and Hefferon, K. (2022). “Crop Biofortification and Food Security,” in Plant Nutrition and Food Security in the Era of Climate Change (Cambridge, MA, United States: Academic Press), 423–436.
Sinha, P., Davis, J., Saag, L., Wanke, C., Salgame, P., Mesick, J., et al. (2019). Undernutrition and Tuberculosis: Public Health Implications. J. Infect. Dis. 219 (9), 1356–1363. doi:10.1093/infdis/jiy675
Slavin, J. L., and Lloyd, B. (2012). Health Benefits of Fruits and Vegetables. Adv. Nutr. 3 (4), 506–516. doi:10.3945/an.112.002154
Solomons, N. W. (1998). Mild Human Zinc Deficiency Produces an Imbalance between Cell-Mediated and Humoral Immunity. Nutr. Rev. 56 (1 Pt 1), 27–28. doi:10.1111/j.1753-4887.1998.tb01656.x
Sommer, A. (2008). Vitamin a Deficiency and Clinical Disease: an Historical Overview. J. Nutr. 138 (10), 1835–1839. doi:10.1093/jn/138.10.1835
Sturniolo, G. C., Fries, W., Mazzon, E., Di Leo, V., Barollo, M., and D'inca, R. (2002). Effect of Zinc Supplementation on Intestinal Permeability in Experimental Colitis. J. Laboratory Clin. Med. 139 (5), 311–315. doi:10.1067/mlc.2002.123624
Sugano, S., Hirose, A., Kanazashi, Y., Adachi, K., Hibara, M., Itoh, T., et al. (2020). Simultaneous Induction of Mutant Alleles of Two Allergenic Genes in Soybean by Using Site-Directed Mutagenesis. BMC Plant Biol. 20 (1), 513–515. doi:10.1186/s12870-020-02708-6
Sun, B., Yang, J., Yang, S., Ye, R. D., Chen, D., and Jiang, Y. (2018). A CRISPR-Cpf1-Assisted Non-Homologous End Joining Genome Editing System of Mycobacterium Smegmatis. Biotechnol. J. 13 (9), e1700588. doi:10.1002/biot.201700588
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of High-Amylose Rice through CRISPR/Cas9-mediated Targeted Mutagenesis of Starch Branching Enzymes. Front. Plant Sci. 8, 298. doi:10.3389/fpls.2017.00298
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 Using the CRISPR/Cas9 System Produces Low Cd-Accumulating Indica Rice without Compromising Yield. Sci. Rep. 7 (1), 14438. doi:10.1038/s41598-017-14832-9
Tang, T., Han, Y., Wang, Y., Huang, H., and Qian, P. (2021). Programmable System of Cas13-Mediated RNA Modification and its Biological and Biomedical Applications. Front. Cell. Dev. Biol. 9, 677587. doi:10.3389/fcell.2021.677587
Tikunov, Y. M., Roohanitaziani, R., Meijer‐Dekens, F., Molthoff, J., Paulo, J., Finkers, R., et al. (2020). The Genetic and Functional Analysis of Flavor in Commercial Tomato: the FLORAL4 Gene Underlies a QTL for Floral Aroma Volatiles in Tomato Fruit. Plant J. 103 (3), 1189–1204. doi:10.1111/tpj.14795
Timoneda, J., Rodríguez-Fernández, L., Zaragozá, R., Marín, M., Cabezuelo, M., Torres, L., et al. (2018). Vitamin A Deficiency and the Lung. Nutrients 10 (9), 1132. doi:10.3390/nu10091132
Tzuri, G., Zhou, X., Chayut, N., Yuan, H., Portnoy, V., Meir, A., et al. (2015). A 'golden' SNP inCmOrgoverns the Fruit Flesh Color of Melon (Cucumis Melo). Plant J. 82 (2), 267–279. doi:10.1111/tpj.12814
Ungurianu, A., Zanfirescu, A., Nițulescu, G., and Margină, D. (2021). Vitamin E beyond its Antioxidant Label. Antioxidants 10 (5), 634. doi:10.3390/antiox10050634
UNICEF (2021). 2021: A Year Marked by Conflict, COVID and Climate Change. Available at: https://www.unicef.org/reports/unicef-annual-report 2021.
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., and Gregory, P. D. (2010). Genome Editing with Engineered Zinc Finger Nucleases. Nat. Rev. Genet. 11 (9), 636–646. doi:10.1038/nrg2842
Usman, B., Nawaz, G., Zhao, N., Liao, S., Qin, B., Liu, F., et al. (2021). Programmed Editing of Rice (Oryza Sativa L.) OsSPL16 Gene Using CRISPR/Cas9 Improves Grain Yield by Modulating the Expression of Pyruvate Enzymes and Cell Cycle Proteins. Int. J. Mol. Sci. 22 (1), 249. doi:10.3390/ijms22063225
Van Der Straeten, D., Bhullar, N. K., De Steur, H., Gruissem, W., MacKenzie, D., Pfeiffer, W., et al. (2020). Multiplying the Efficiency and Impact of Biofortification through Metabolic Engineering. Nat. Commun. 11 (1), 5203–5210. doi:10.1038/s41467-020-19020-4
Veillet, F., Chauvin, L., Kermarrec, M.-P., Sevestre, F., Merrer, M., Terret, Z., et al. (2019). The Solanum tuberosum GBSSI Gene: a Target for Assessing Gene and Base Editing in Tetraploid Potato. Plant Cell. Rep. 38 (9), 1065–1080. doi:10.1007/s00299-019-02426-w
Vu, T. V., Sivankalyani, V., Kim, E. J., Doan, D. T. H., Tran, M. T., Kim, J., et al. (2020). Highly Efficient Homology‐directed Repair Using CRISPR/Cpf1‐geminiviral Replicon in Tomato. Plant Biotechnol. J. 18 (10), 2133–2143. Advance online publication. doi:10.1111/pbi.13373
Walton, R. T., Christie, K. A., Whittaker, M. N., and Kleinstiver, B. P. (2020). Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science 368 (6488), 290–296. doi:10.1126/science.aba8853
Wang, B., Li, N., Huang, S., Hu, J., Wang, Q., Tang, Y., et al. (2021). Enhanced Soluble Sugar Content in Tomato Fruit Using CRISPR/Cas9-mediated SlINVINH1 and SlVPE5 Gene Editing. PeerJ 9, e12478. doi:10.7717/peerj.12478
Wang, D., Samsulrizal, N., Yan, C., Allcock, N. S., Craigon, J., Blanco-Ulate, B., et al. (2019). Characterisation of CRISPR Mutants Targeting Genes Modulating Pectin Degradation in Ripening Tomato. Plant Physiol. 179 (2), 544–557. doi:10.1104/pp.18.01187
Wang, H., La Russa, M., and Qi, L. S. (2016). CRISPR/Cas9 in Genome Editing and beyond. Annu. Rev. Biochem. 85, 227–264. doi:10.1146/annurev-biochem-060815-014607
Wang, H., Wu, Y., Zhang, Y., Yang, J., Fan, W., Zhang, H., et al. (2019). CRISPR/Cas9-based Mutagenesis of Starch Biosynthetic Genes in Sweet Potato (Ipomoea Batatas) for the Improvement of Starch Quality. Ijms 20 (19), 4702. doi:10.3390/ijms20194702
Wang, S., Yang, Y., Guo, M., Zhong, C., Yan, C., and Sun, S. (2020). Targeted Mutagenesis of Amino Acid Transporter Genes for Rice Quality Improvement Using the CRISPR/Cas9 System. Crop J. 8 (3), 457–464. doi:10.1016/j.cj.2020.02.005
Wang, W., Pan, Q., He, F., Akhunova, A., Chao, S., Trick, H., et al. (2018). Transgenerational CRISPR-Cas9 Activity Facilitates Multiplex Gene Editing in Allopolyploid Wheat. CRISPR J. 1 (1), 65–74. doi:10.1089/crispr.2017.0010
Wang, W., Pan, Q., Tian, B., He, F., Chen, Y., Bai, G., et al. (2019). Gene Editing of the Wheat Homologs of TONNEAU 1‐recruiting Motif Encoding Gene Affects Grain Shape and Weight in Wheat. Plant J. 100 (2), 251–264. doi:10.1111/tpj.14440
Wang, Y., Liu, X., Zheng, X., Wang, W., Yin, X., Liu, H., et al. (2021). Creation of Aromatic Maize by CRISPR/Cas. J. Integr. Plant Biol. 63 (9), 1664–1670. doi:10.1111/jipb.13105
Wu, Y., He, Y., Sretenovic, S., Liu, S., Cheng, Y., Han, Y., et al. (2021). CRISPR‐BETS: a Base‐editing Design Tool for Generating Stop Codons. Plant Biotechnol. J. 20 (3), 499–510. doi:10.1111/pbi.13732
Xiong, J. S., Ding, J., and Li, Y. (2015). Genome-editing Technologies and Their Potential Application in Horticultural Crop Breeding. Hortic. Res. 2 (1), 15019. doi:10.1038/hortres.2015.19
Xu, R., Yang, Y., Qin, R., Li, H., Qiu, C., Li, L., et al. (2016). Rapid Improvement of Grain Weight via Highly Efficient CRISPR/Cas9-mediated Multiplex Genome Editing in Rice. J. Genet. Genomics 43 (8), 529–532. doi:10.1016/j.jgg.2016.07.003
Yadav, D. N., Bansal, S., Tushir, S., Kaur, J., and Sharma, K. (2020). “Advantage of Biofortification over Fortification Technologies,” in Wheat and Barley Grain Biofortification (Sawston, United Kingdom: Woodhead Publishing), 257–273.
Yan, S., Chen, N., Huang, Z., Li, D., Zhi, J., Yu, B., et al. (2020). Anthocyanin Fruit Encodes an R2R3‐MYB Transcription Factor, SlAN2‐like, Activating the Transcription of SlMYBATV to Fine‐tune Anthocyanin Content in Tomato Fruit. New Phytol. 225 (5), 2048–2063. doi:10.1111/nph.16272
Yu, Q.-h., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced Targeted Mutagenesis and Gene Replacement to Generate Long-Shelf Life Tomato Lines. Sci. Rep. 7 (1), 11874. doi:10.1038/s41598-017-12262-1
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 Genes in Peanut with CRISPR/Cas9 Based Gene Editing. BMC Biotechnol. 19 (1), 24. doi:10.1186/s12896-019-0516-8
Yuste-Lisbona, F. J., Fernández-Lozano, A., Pineda, B., Bretones, S., Ortíz-Atienza, A., García-Sogo, B., et al. (2020). ENO Regulates Tomato Fruit Size through the Floral Meristem Development Network. Proc. Natl. Acad. Sci. U.S.A. 117 (14), 8187–8195. doi:10.1073/pnas.1913688117
Yuyu, C., Aike, Z., Pao, X., Xiaoxia, W., Yongrun, C., Beifang, W., et al. (2020). Effects of GS3 and GL3.1 for Grain Size Editing by CRISPR/Cas9 in Rice. Rice Sci. 27 (5), 405–413. doi:10.1016/j.rsci.2019.12.010
Zeng, Z., Han, N., Liu, C., Buerte, B., Zhou, C., Chen, J., et al. (2020). Functional Dissection of HGGT and HPT in Barley Vitamin E Biosynthesis via CRISPR/Cas9-enabled Genome Editing. Ann. Bot. 126 (5), 929–942. doi:10.1093/aob/mcaa115
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted Mutagenesis of BnTT8 Homologs Controls Yellow Seed Coat Development for Effective Oil Production in Brassica Napus L. Plant Biotechnol. J. 18 (5), 1153–1168. doi:10.1111/pbi.13281
Zhang, B., Yang, Q., Chen, J., Wu, L., Yao, T., Wu, Y., et al. (2016b). CRISPRi-Manipulation of Genetic Code Expansion via RF1 for Reassignment of Amber Codon in Bacteria. Sci. Rep. 6, 20000. doi:10.1038/srep20000
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome Editing of Upstream Open Reading Frames Enables Translational Control in Plants. Nat. Biotechnol. 36 (9), 894–898. doi:10.1038/nbt.4202
Zhang, J.-H., Adikaram, P., Pandey, M., Genis, A., and Simonds, W. F. (2016a). Optimization of Genome Editing through CRISPR-Cas9 Engineering. Bioengineered 7 (3), 166–174. doi:10.1080/21655979.2016.1189039
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 Acts as a Transcriptional Activator to Regulate Rice Grain Shape and Appearance Quality. Nat. Commun. 9 (1), 1240. doi:10.1038/s41467-018-03616-y
Zhao, X., Jayarathna, S., Turesson, H., Fält, A.-S., Nestor, G., González, M. N., et al. (2021). Amylose Starch with No Detectable Branching Developed through DNA-free CRISPR-Cas9 Mediated Mutagenesis of Two Starch Branching Enzymes in Potato. Sci. Rep. 11 (1), 4311. doi:10.1038/s41598-021-83462-z
Zheng, X., Giuliano, G., and Al-Babili, S. (2020). Carotenoid Biofortification in Crop Plants: Citius, Altius, Fortius. Biochimica Biophysica Acta (BBA) - Mol. Cell. Biol. Lipids 1865 (11), 158664. doi:10.1016/j.bbalip.2020.158664
Zheng, X., Kuijer, H. N. J., and Al-Babili, S. (2021). Carotenoid Biofortification of Crops in the CRISPR Era. Trends Biotechnol. 39 (9), 857–860. doi:10.1016/j.tibtech.2020.12.003
Zheng, Z., Ye, G., Zhou, Y., Pu, X., Su, W., and Wang, J. (2021). Editing Sterol Side Chain Reductase 2 Gene (StSSR2) via CRISPR/Cas9 Reduces the Total Steroidal Glycoalkaloids in Potato. All Life 14 (1), 401–413. doi:10.1080/26895293.2021.1925358
Zhi, J., Liu, X., Li, D., Huang, Y., Yan, S., Cao, B., et al. (2020). CRISPR/Cas9-mediated SlAN2 Mutants Reveal Various Regulatory Models of Anthocyanin Biosynthesis in Tomato Plant. Plant Cell. Rep. 39 (6), 799–809. doi:10.1007/s00299-020-02531-1
Zhong, Y., Blennow, A., Kofoed-Enevoldsen, O., Jiang, D., and Hebelstrup, K. H. (2019). Protein Targeting to Starch 1 Is Essential for Starchy Endosperm Development in Barley. J. Exp. Bot. 70 (2), 485–496. doi:10.1093/jxb/ery398
Zhou, J., Xin, X., He, Y., Chen, H., Li, Q., Tang, X., et al. (2019). Multiplex QTL Editing of Grain-Related Genes Improves Yield in Elite Rice Varieties. Plant Cell. Rep. 38 (4), 475–485. doi:10.1007/s00299-018-2340-3
Ziarati, P., Rodriguez, L. C., Hochwimmer, B., Vambol, V., and Vambol, S. (2021). Biofortification of Oryza Sativa L. With Agri-Food Waste to Improve the Dietary Value of Crops. Ecol. Quest. 33 (1), 47–54. doi:10.12775/EQ.2022.004
Keywords: biofortification, biofortified crops, CRISPR-cas system, genome editing, hidden hunger, malnutrition, micronutrients
Citation: Kumar D, Yadav A, Ahmad R, Dwivedi UN and Yadav K (2022) CRISPR-Based Genome Editing for Nutrient Enrichment in Crops: A Promising Approach Toward Global Food Security. Front. Genet. 13:932859. doi: 10.3389/fgene.2022.932859
Received: 30 April 2022; Accepted: 08 June 2022;
Published: 14 July 2022.
Edited by:
Santosh Kumar Gupta, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Jaindra Nath Tripathi, International Institute of Tropical Agriculture (IITA), KenyaPriti Sharma, Punjab Agricultural University, India
Copyright © 2022 Kumar, Yadav, Ahmad, Dwivedi and Yadav. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kusum Yadav, YW51a3VzdW1AZ21haWwuY29t
 Dileep Kumar1
Dileep Kumar1 Kusum Yadav
Kusum Yadav