
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 02 August 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.932166
This article is part of the Research TopicAccelerating Genetic Gain for Key Traits Using Genome-Wide Association Studies and Genomic Selection: Promising Breeding Tools for Sustainable AgricultureView all 11 articles
 Ankit Malik1,2
Ankit Malik1,2 Aruna Kumar2
Aruna Kumar2 Ranjith Kumar Ellur1
Ranjith Kumar Ellur1 Gopala Krishnan S1
Gopala Krishnan S1 Deepshikha Dixit1
Deepshikha Dixit1 Haritha Bollinedi1
Haritha Bollinedi1 KK Vinod1
KK Vinod1 M Nagarajan3
M Nagarajan3 PK Bhowmick1
PK Bhowmick1 NK Singh4
NK Singh4 AK Singh1*
AK Singh1*Basmati rice is known for its extra-long slender grains, exceptional kernel dimensions after cooking, high volume expansion, and strong aroma. Developing high yielding Basmati rice varieties with good cooking quality is a gigantic task. Therefore, identifying the genomic regions governing the grain and cooked kernel dimension traits is of utmost importance for its use in marker-assisted breeding. Although several QTLs governing grain dimension traits have been reported, limited attempts have been made to map QTLs for grain and cooked kernel dimension traits of Basmati rice. In the current study, a population of recombinant inbred lines (RIL) was generated from a cross of Sonasal and Pusa Basmati 1121 (PB1121). In the RIL population, there was a significant positive correlation among the length (RRL: rough rice length, MRL: milled rice length, CKL: cooked kernel length) and breadth (RRB: rough rice breadth, MRB: milled rice breadth and CKB: cooked kernel breadth) of the related traits, while there was significant negative correlation between them. QTL mapping has led to the identification of four major genomic regions governing MRL and CKL. Two QTLs co-localize with the earlier reported major gene GS3 and a QTL qGRL7.1, while the remaining two QTLs viz., qCKL3.2 (qMRL3.2) and qCKL4.1 (qMRL4.1) were novel. The QTL qCKL3.2 has been bracketed to a genomic region of 0.78 Mb between the markers RM15247 and RM15281. Annotation of this region identified 18 gene models, of which the genes predicted to encode pentatricopeptides and brassinosteroid insensitive 1-associated receptor kinase 1 precursor may be the putative candidate genes. Furthermore, we identified a novel QTL qKER2.1 governing kernel elongation ratio (KER) in Basmati rice.
Basmati rice attracts consumers worldwide due to its pleasant aroma and unique grain and cooking quality parameters. Among the Basmati rice varieties developed, PB1121 stands out due to its exclusive grain qualities, making it one of the most traded crop varieties in the world. It possesses extra-long slender grains, very long cooked kernel (upto 22 mm), high kernel elongation ratio, volume expansion of more than four times upon cooking, and pleasant aroma. The grain dimension traits that determine rice grain quality are the length and breadth of rough, milled, and cooked rice grain as well as the kernel elongation ratio (KER). The genetics of rice grain size and weight has been extensively studied and >300 QTLs have been mapped (www.gramene.org) (Bhatia et al., 2018). However, only few of them viz., GS3 (Fan et al., 2006), GS5 (Li et al., 2011), GS7 (Shao et al., 2012), qGL3 (Zhang et al., 2012b), GW2 (Song et al., 2007), GW8 (Wang et al., 2012), TGW6 (Ishimaru et al., 2013), GW5 (Duan et al., 2017), qTGW3 (Zejun et al., 2018), and qGS10 (Zhu et al., 2019) have been cloned and functionally validated. Among these, GS3 has been identified as a major gene governing the rice grain length and weight (Mao et al., 2010). Several polymorphic loci of GS3 viz, SR17 (2nd intron), RGS1 (4th intron), RGS2 (5th exon), aksGS3-12 (5th exon) etc., have been reported (Wang et al., 2011; Anand et al., 2013a). The gene qGL3 has been identified to encode a putative serine/threonine protein phosphatase that contains a Kelch-like repeat domain and acts on its cell cycle-related substrate, Cyclin-T1; 3, to increase the rice grain length (Zhang et al., 2012b).
Several independent studies have identified QTLs for the grain width and weight. In a large-grained genotype WY3, a major gene GW2 encoding a RING-type E3 ubiquitin ligase was identified wherein, a single base pair deletion results into premature stop codon, thereby reducing its expression while increasing the grain width and weight (Song et al., 2007). GW5 was mapped on chromosome 5 which encodes a putative serine carboxypeptidase, wherein a deletion increases the cell number in the outer glume of the rice flower thereby regulating the grain width, filling, and weight (Li et al., 2011). TGW6 was mapped on chromosome 6 which encodes a novel protein with an indole-3-acetic acid (IAA)-glucose hydrolase activity and its loss of function enhances the rice grain weight (Ishimaru et al., 2013).
Identifying genomic regions governing grain dimension traits in Basmati rice genotypes helps in their precise manifestation in Basmati improvement programs. Earlier, in a RIL population generated from a cross Pusa 1342/PB1121, several novel QTLs governing grain length, elongation ratio, and aroma in Basmati rice were identified (Amarawathi et al., 2008). Subsequently, a major gene GW8 promoting cell division and proliferation thereby enhancing the grain width and yield was identified in a traditional Basmati rice variety, Basmati 385 (Wang et al., 2012). Furthermore, three QTLs for grain elongation on chromosomes 4 (qGE4.1) and 6 (qGE6.1 and qGE6.2) were identified in a Basmati rice genotype (Arikit et al., 2019).
Although, Basmati rice possesses unique grain dimension traits such as extra-long slender grains and exceptional cooked kernel dimensions, there are limited efforts made toward identifying the genomic regions governing these traits. In the present study, we attempted to map QTLs governing the grain and cooked kernel dimension traits using a RIL population generated from the cross of Sonasal and PB1121.
A RIL population comprising of 173 lines was developed by crossing an aromatic short grain genotype Sonasal with an extra-long grained Basmati rice variety PB1121. Sonasal has extremely shorter grains, which are at least 50% shorter than that of PB1121, with slightly wider grains. Both parents were aromatic. The population was advanced till the F13 generation through the single seed descent method (Figure 1).
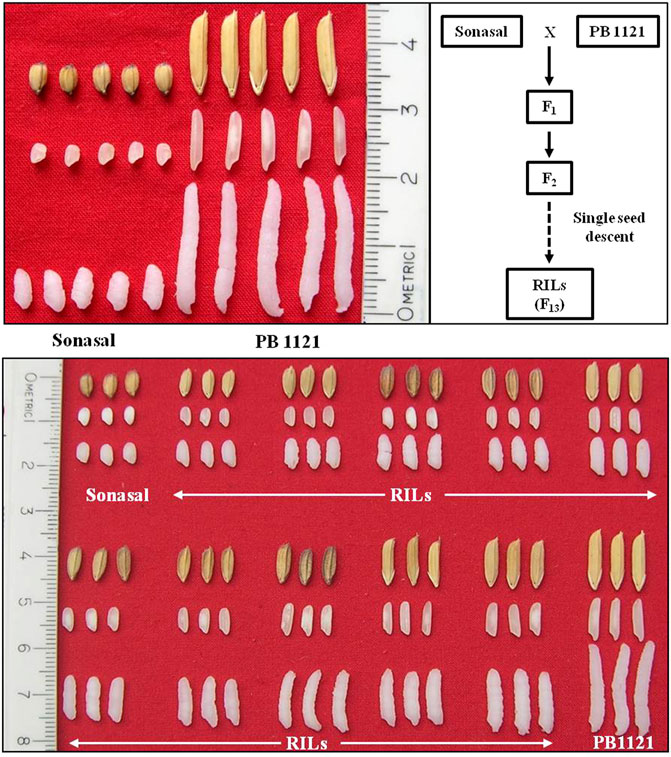
FIGURE 1. Variation for grain dimension traits between parental lines, Sonasal, and PB1121; and RIL population.
The RIL population along with the parental lines and checks were grown in an augmented randomized block design following all the standard agronomic practices. The seed harvested from each of the lines was shade-dried to a uniform moisture content of 14% and stored at room temperature. Observations on rough, milled, and cooked grain dimension traits were recorded after six months. The grain dimension traits namely, RRL, RRB, MRL, MRB, CKL, and CKB were recorded in three replications with ten grains in each replication. For CKL and CKB, ten milled rice grains were soaked in 20 ml of distilled water for 20 min in a test tube. The samples were placed in a water bath at 100°C for 7 min followed by cooling them to room temperature. A photo analyzer was used (C-DAC) to record the data on the length and breadth of the grains.
DNA was extracted using a standard CTAB (Cetyl- Tri Methyl Ammonium Bromide) method (Murray and Thompson, 1980). The PCR reaction was carried out using 25 ng of genomic DNA, 1X PCR assay buffer with 1.5 mM MgCl2, 5 pmol of forward and reverse primers, 0.05 mM dNTPs, and 1U of Taq polymerase. The final reaction volume was made up to 10 μl using nuclease-free water. Amplified PCR products were resolved by 3.5% agarose gel electrophoresis and documented on a gel documentation system (BIO-RAD Gel Doc XR+, California, United States). A total of 1,054 markers comprising of sequence-tagged microsatellite site (STMS) and gene-based markers were used in a parental polymorphism survey. The polymorphic markers were used to genotype the RIL population. The linkage map was constructed using the QTL IciMapping software (Li et al., 2007; Meng et al., 2015). Grouping and ordering of polymorphic markers were carried out using the regression mapping algorithm using recombination frequency (REC) with a default value set to 0.3. Furthermore, rippling was carried out for fine-tuning the ordered markers on their respective chromosomes using the REC algorithm with a default window size of 5. QTLs were identified using the inclusive composite interval mapping (ICIM) algorithm for additive gene effects.
The RIL population comprising of 173 lines were evaluated during three subsequent crop growing seasons, viz., Kharif 2019, 2020, and 2021 (Table 1). The population showed significant variations for all the grain dimension-related traits. The RRL ranged from 4.67 to 12.33 mm while, RRB ranged from 1.40 to 3.00 mm. The corresponding measures for RRL in Pusa Basmati 1121 were 12.43 and 6.37 mm in Sonasal. Similarly, for RRB, the parents showed average values of 2.30 and 2.60 mm for Pusa Basmati 1121 and Sonasal, respectively. The MRL among the RILs varied from 3.37 to 8.07 mm, while Pusa Basmati 1121 measured 8.07 mm and Sonasal 4.07 mm. For MRB, RILs ranged between 1.33 and 2.47 mm, while the parents recorded values of 1.80 and 2.20 mm, respectively for Pusa Basmati 1121 and Sonasal. There was a huge variation for CKL which ranged from 6.27 to 17.13 mm among the RILs, while CKB ranged between 1.78 and 3.10 mm. CKL and CKB for Pusa Basmati 1121 were 16.60 and 2.33 mm, respectively, while that of Sonasal was 6.25 and 2.47 mm, in that order. Among the RILs, KER varied from 1.23 to 2.59, compared to 2.06 and 1.54 recorded in Pusa Basmati 1121 and Sonasal, respectively. The phenotypic coefficient of variation (PCV) among the traits ranged from a minimum of 7.12% for CKB to a maximum of 25.86% for MRLB during Kharif 2019, while during Kharif 2020, it ranged from 6.40% for CKB to 24.83% in RRLB. During Kharif 2021, PCV was minimum for MRB (5.14%) and maximum for RRLB (23.67%). The PCV for KER ratio was 9.33%, 10.83%, and 10.84% during Kharif 2019, Kharif 2020, and Kharif 2021, respectively (Table 1; Figure 1). The transgressive segregation was observed for the traits RRL, MRL, RRB, MRB, CKB, and KER (Figure 2).
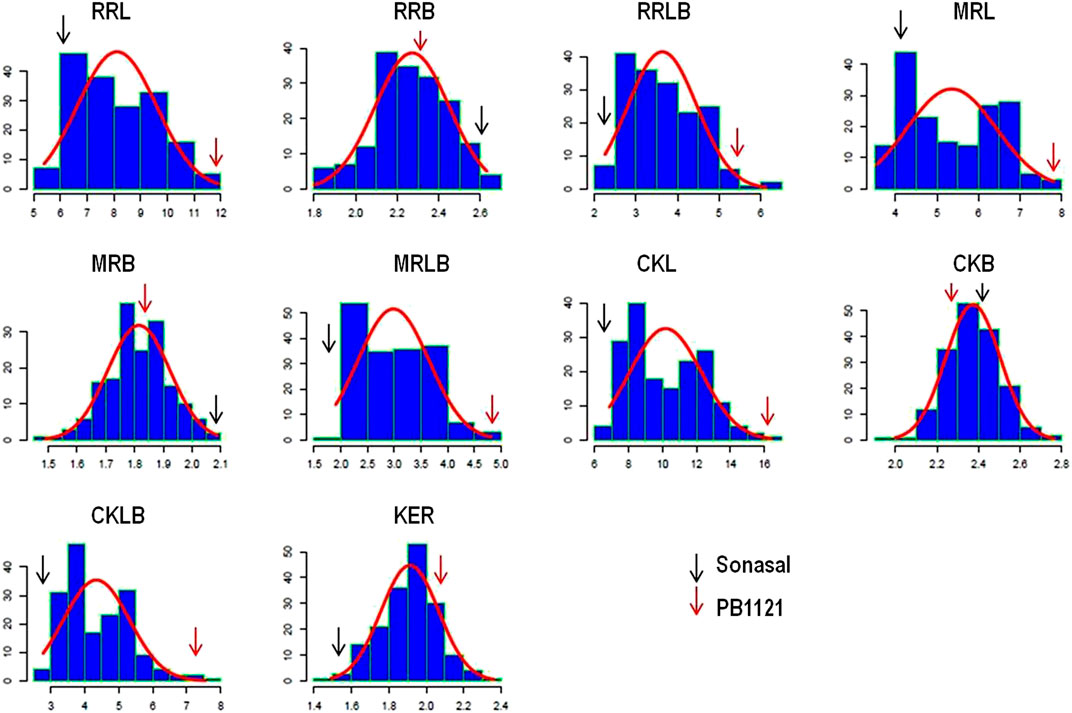
FIGURE 2. Distribution pattern of grain dimension traits in an RIL population. rough rice length (RRL), rough rice breadth (RRB), rough rice length to breadth ratio (RRLB), milled rice length (MRL), milled rice breadth (MRB), milled rice length to breadth ratio (MRLB), cooked kernel length (CKL), cooked kernel breadth (CKB), cooked kernel length to breadth ratio (CKLB), and kernel elongation ratio (KER).
The significant positive correlation was observed among the grain length and length/breadth (rough rice, milled, and cooked) traits. However, there was a significant negative correlation between the grain length and breadth traits. Furthermore, there was a significant positive correlation between CKL and KER; while negative correlation was observed between CKB and CKL, MRLB, and KER; and no correlation between KER and rough rice dimension traits (Figure 3).
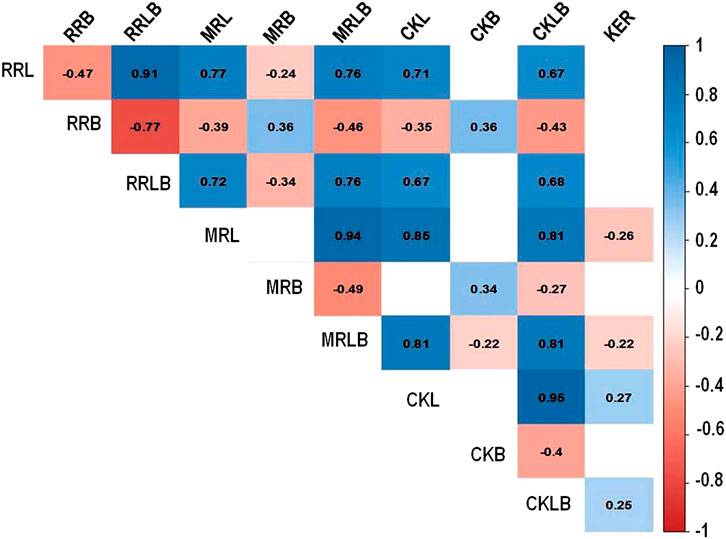
FIGURE 3. Correlation among grain dimension traits in rice. Rough rice length (RRL), rough rice breadth (RRB), rough rice length to breadth ratio (RRLB), milled rice length (MRL), milled rice breadth (MRB), milled rice length to breadth ratio (MRLB), cooked kernel length (CKL), cooked kernel breadth (CKB), cooked kernel length to breadth ratio (CKLB), and kernel elongation ratio (KER).
A total of 116 markers polymorphic between Sonasal and PB1121 were used to construct the linkage map. A total of 12 linkage groups were constructed with a cumulative map distance of 2,469.38 cM. The number of markers on different chromosomes ranged from 4 on chromosome 10 to 16 on chromosome 3 (Table 2). The average genetic distance between two adjacent markers was 21.29 cM.
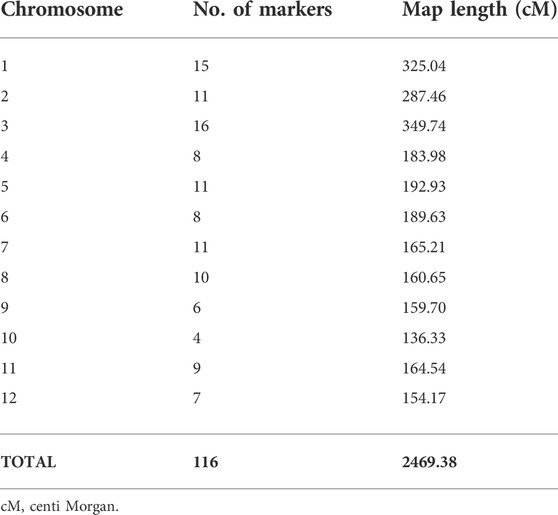
TABLE 2. Chromosome-wise number of the markers used for construction of linkage map, map length, and average marker interval.
QTLs were identified for ten grain dimension traits using ICIM. A total of twenty-three QTLs were detected on six different chromosomes (chromosomes 2, 3, 4, 6, 7, and 8) for grain dimension-related traits (Figure 4). For each of these grain dimension traits, the proportion of phenotypic variance explained (PVE), its additive effect, nearest left marker, and nearest right marker were determined (Table 3).
A total of eight QTLs associated with rough rice dimension traits were identified on chromosomes 3, 4, and 7 across 3 years of testing. A novel QTL qRRL4.1 flanking the markers RM551 and HvSSR04-15 was identified on chromosome 4 with PVE of 10.86% and 18.11% during the years 2019 and 2021, respectively. The PVE of qRRL3.1 ranged from 51.37% to 65.92% and the PVE of qRRL7.1 ranged from 19.19% to 21.95% during 3 years of testing. The QTLs qRRL3.1 and qRRL7.1 correspond to already cloned genes GS3 and qGRL7.1, respectively.
The trait RRB was found to mainly governed by two major QTLs qRRB3.1 (PVE: 3.37%) and qRRB7.1 (PVE: 12.49% and 23.48%) and for the trait RRLB, two robust QTLs qRRLB3.1 (PVE: 30.11–46.15%) and qRRLB7.1 (PVE: 22.41–29.21%) were mapped on chromosome 3 and chromosome 7 which correspond to the major genes GS3 and qGRL7.1, respectively (Table 3). A major novel QTL qRRLB4.1 flanking the region RM551 and HvSSR04-15 was identified for the trait RRLB on chromosome 4 during Kharif 2020 and Kharif 2021 which explained the phenotypic variance of 10.03%–20.51%, respectively.
A total of four QTLs were found to govern the trait MRL. The major QTL qMRL3.1 was identified on chromosome 3 with PVE of 64.59%–76.14% during 3 years of testing. The QTL qMRL7.1 was mapped on chromosome 7 which accounted for the PVE of 7.36 and 8.72% during Kharif 2019 and Kharif 2020, respectively. Two novel QTLs, namely, qMRL3.2 flanked by the markers RM15247 and RM15281; and qMRL4.1 flanked by the markers RM551 and HvSSR04-15 located on chromosomes 3 and 4, respectively, were mapped. The PVE of QTL qMRL3.1 was 12.92% during Kharif 2020 while, the PVE of qMRL4.1 was 14.34% and 13.65% during Kharif 2019 and Kharif 2021, respectively.
For the trait MRB, a robust QTL qMRB7.1 was identified on chromosome 7 with PVE of 15.54% and 10.86% during Kharif 2019 and Kharif 2020, respectively. For the trait MRLB, two robust QTLs, namely, qMRLB3.1 and qMRLB7.1 were mapped on chromosomes 3 and 7, respectively. Another novel QTL qMRLB4.1 flanked by the markers RM551 and HvSSR04-15 with PVEs of 9.83% and 14.30% during kharif 2019 and kharif 2020, respectively.
CKL is one of the key Basmati traits which determine the cooking quality. In the current population, four QTLs governing the trait CKL were identified of which three QTLs namely; qCKL3.2, qCKL4.1, and qCKL6.1 were identified to be novel. A major QTL qCKL3.1 with PVE ranging from 29.81% to 58.26% and an additive effect ranging from 1.12 to 1.62 mm was identified during all the 3 years of testing. The QTL qCKL3.2 (PVE: 13.01%–13.96%) flanked by RM15247 and RM15281 and the QTL qCKL4.1 (PVE: 11.26%–11.89%) flanked by the markers RM551 and HvSSR04-15 were identified during Kharif 2019 and Kharif 2020. Another novel QTL, qCKL6.1 was mapped on chromosome 6 with PVE of 5.60% during Kharif 2020. The promising QTL qCKL3.2 was bracketed between the markers RM15247 and RM15281 which spans a region of 0.78 Mb (17.90 Mb–18.68 Mb) on chromosome 3. The region comprises of 114 annotated gene models, of which 18 genes were putative candidates which may play a role in determining the grain length (Table 4).
KER is one of the key traits of Basmati rice which determines the extent of the linear elongation of the grain upon cooking. A novel QTL qKER2.1 was mapped on chromosome 2 flanked by markers RM424 and RM452 with PVE of 9.56% during Kharif 2020.
Rice grain shape is mainly determined by the grain length and breadth. These traits are under the influence of a few large effects and multiple small effect genes, thus possessing complex patterns of inheritance. All the grain dimension traits showed a high degree of phenotypic variation in the RIL population during all the 3 years of testing. Furthermore, the grain dimension traits showed a near-normal distribution with slight skewness toward either of the parental lines which indicated the influence of the combination of few oligo-genes and many minor genes in determining these traits. The significant positive correlation was observed among length-related traits while a significant negative correlation was observed between the length- and breadth-related traits. The observed correlation among the traits in the present study was consistent with the previous studies (Amarawathi et al., 2008; Bai et al., 2010).
The linkage map comprising of 12 linkage groups with a cumulative map distance of 2,469.38 cM was constructed using 116 polymorphic markers with an average marker interval distance of 21.29 cM. Although the average marker interval distance was 21.29 cM, the genetic distances between neighboring markers varied across the linkage groups due to the spatial variation in recombination frequencies. Sonasal is a short grain aromatic rice variety and PB1121 is an extra-long grain Basmati rice variety with sufficient genomic and phenotypic diversity among the parental lines. It was demonstrated that there exists tremendous diversity between the aromatic short grain and Basmati varieties (Garris et al., 2005), indicating the existence of tremendous structural changes, which may lead to the occurrence of segregation distortion at a certain loci in the mapping population generated. There are several possible reasons for segregation distortion, such as gametic or zygotic selection, genetic incompatibility, chromosome rearrangement, pollen competition, preferential fertilization etc., (Xu et al., 1997).
The genetic and molecular dissection of grain dimension parameters in rice have received a lot of attention, and until now, several QTLs have been identified and few genes have been functionally characterized (www.gramene.org). The major focus has been to understand the genetics and the identification of genomic regions governing the grain length and grain width in non-Basmati rice genotypes. This has resulted in the identification of major effect QTLs/genes such as GS3, a major gene governing the grain length and weight which was identified in a population generated from a cross Minghui 63/Chaun 7; GW2, a major gene for the grain width and weight mapped in a population generated from a cross WY3/Fengaizhan 1; GS7, associated with grain length was mapped using a population generated from a cross D50/HB277; qSW5, associated with the grain breadth was mapped in a population Nipponbare/Kasalath etc.,
Basmati is a unique combination of grain and cooking quality parameters with a strong and pleasing aroma. However, limited attempts were made to understand the grain and cooked rice dimension-related traits in Basmati genotypes. Earlier, in a population generated from the cross Basmati 385 and HJX74, a major gene GW8 was identified. In the present study, a RIL population was generated using highly contrasting parental lines for the target traits. The major effect QTLs viz., qRRL3.1, qRRLB3.1, qMRL3.1, qMRLB3.1, and qCKL3.1 were found to be co-mapped to a genomic region on chromosome 3 which is known to possess the GS3 gene. The functional allele of GS3 reduces the grain size and weight (Fan et al., 2009). In GS3 a C to A mutation in the exon 2 introduces a stop codon leading to long grains. Based on this mutation, a CAPS marker SF28 was developed for its use in molecular breeding (Fan et al., 2009). Furthermore, previously we identified a novel indel aksGS3-12 in the short grain genotype Sonasal, which was predominant in short grain landraces (Anand et al., 2013b). The functional allele at SF28 reduces the grain size, and if coupled with the 342bp deletion at aksGS3-12, it may further reduce the grain size, as in Sonasal. In the current study, it has been demonstrated that GS3 has a key role in governing rough, milled, and cooked rice length as well as MRLB. The other major effect QTLs identified were qRRL7.1, qRRB7.1, qMRL7.1, qMRB7.1, and qMRLB7.1 which were co-mapped to the genomic region on chromosome 7. This region co-localizes with the earlier reported QTL qGRL7.1 (Amarawathi et al., 2008).
The novel QTLs viz., qMRL3.2, qCKL3.2, and qCKLB3.2 are co-mapped between the markers RM15247 and RM15281 on chromosome 3. The annotation of the QTL-bracketed region of 0.78 Mb identified 18 putative candidate gene models which may have a significant role in determining the kernel length. This genomic region possessed gene models which code for pentatricopeptides, brassinosteroid insensitive 1-associated receptor kinase 1 precursor and transcription factors. A maize QTL qKW9 has been reported to encode a pentatricopeptide repeat protein which affects the maternal photosynthates available during grain filling (Huang et al., 2020), while, brassinosteroids are known to play a key role in determining grain size and plant architecture (Zhang et al., 2021). Other gene models code for proteins such as expansin, aldehyde oxidase, N-acetyltransferase, ploygalacturonase, allene oxide cyclase, etc., which are known to play important roles in cell wall loosening, elongation, and cell division (Agrawal et al., 2003; Riemann et al., 2013; Byeon and Back, 2014; Marowa et al., 2016; Lee and Back, 2017; Cao et al., 2021; Shi et al., 2021), which may therefore be associated with an increased grain length. Furthermore, fine-mapping and functional validation is required to pin-point the gene underlying this important QTL.
The KER is one of the important traits of Basmati rice which determines the degree of linear elongation after cooking. This trait is proportional to other economically important cooking quality traits such as volume expansion and water uptake. In the current study, we have identified a novel QTL viz., qKER2.1 which has a large effect on KER in Basmati rice.
In all, the Basmati rice grain length is mainly governed by three loci, namely GS3, qGL7.1, and qMRL3.2. However, the cooked grain length is mainly governed by four loci, viz., GS3, qGL7.1, qCKL3.2, and qCKL4.1. Therefore, targeting these loci through marker-assisted breeding can help in developing Basmati rice genotypes with extra-long slender grains and exceptional cooking quality traits.
The present study identified novel QTLs governing grain dimension traits in a RIL population generated by crossing a short grain aromatic rice landrace, Sonasal and Pusa Basmati 1121, a Basmati rice variety possessing extra-long slender grains. Two previously reported QTLs and two novel QTLs identified in the current study were identified to influence the exceptional milled rice length and cooked kernel length in Basmati rice. One of the novel QTLs was bracketed to a genomic region of 0.78 Mb which possesses putative candidate genes. However, further fine mapping and map-based cloning is required to precisely identify the candidate genes governing the trait. Furthermore, the markers flanking the QTLs identified can be utilized in the marker-assisted breeding to develop Basmati rice varieties with extra-long slender grains and high kernel elongation upon cooking
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
AS: conceptualization. GK, RE, NS, and AS: methodology. AM: genotyping and phenotyping. RE and AM: data curation, statistical analysis. AM and RE: original draft preparation. RE, KV, and DD: editing. RE, GS, NM, PB, HB, AS, AK: supervision. All authors have read and agreed to the final version of the manuscript.
This research was funded by the ICAR- “Network Project on Functional Genomics and Genetic Modification in Crops (Rice).”
The study is part of the Ph.D. thesis research of AM and AM wishes to acknowledge ICAR-Indian Agricultural Research Institute, New Delhi, and Indian Council of Agriculture Research (ICAR) for providing all the necessary facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agrawal, G. K., Jwa, N. S., Shibato, J., Han, O., Iwahashi, H., Rakwal, R., et al. (2003). Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochem. Biophys. Res. Commun. 310, 1073–1082. doi:10.1016/j.bbrc.2003.09.123
Amarawathi, Y., Singh, R., Singh, A. K., Singh, V. P., Mohapatra, T., Sharma, T. R., et al. (2008). Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol. Breed. 21, 49–65. doi:10.1007/s11032-007-9108-8
Anand, D., Baunthiyal, M., Gopala Krishnan, S., Singh, N. K., Prabhu, K. V., Singh, A. K., et al. (2013a). Novel InDel variation in GS3 locus and development of InDel based marker for marker assisted breeding of short grain aromatic rices. J. Plant Biochem. Biotechnol. 24, 120–127. doi:10.1007/s13562-013-0243-5
Anand, D., Baunthiyal, M., Singh, A., Gopala Krishnan, S., Singh, N. K., Prabhu, K. V., et al. (2013b). Validation of gene based marker-QTL association for grain dimension traits in rice. J. Plant Biochem. Biotechnol. 22, 467–473. doi:10.1007/s13562-012-0176-4
Arikit, S., Wanchana, S., Khanthong, S., Saensuk, C., Thianthavon, T., Vanavichit, A., et al. (2019). QTL-seq identifies cooked grain elongation QTLs near soluble starch synthase and starch branching enzymes in rice (Oryza sativa L.). Sci. Rep. 9, 8328. doi:10.1038/s41598-019-44856-2
Bai, X., Luo, L., Yan, W., Kovi, M. R., Zhan, W., Xing, Y., et al. (2010). Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11, 16. doi:10.1186/1471-2156-11-16
Balram, M., Smriti, G., Trilochan, M., Rajender, P., Nagarajan, M., Vinod, K. K., et al. (2012). QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L.). BMC Plant Biol. 12, 137. doi:10.1186/1471-2229-12-137
Bhatia, D., Wing, R. A., Yu, Y., Chougule, K., Kudrna, D., Lee, S., et al. (2018). Genotyping by sequencing of rice interspecific backcross inbred lines identifies QTLs for grain weight and grain length. Euphytica 214, 41. doi:10.1007/s10681-018-2119-1
Byeon, Y., and Back, K. (2014). An increase in melatonin in transgenic rice causes pleiotropic phenotypes, including enhanced seedling growth, delayed flowering, and low grain yield. J. pineal Res. 56, 408–414. doi:10.1111/jpi.12129
Cao, Y., Zhang, Y., Chen, Y., Yu, N., Liaqat, S., Wu, W., et al. (2021). OsPG1 encodes a polygalacturonase that determines cell wall architecture and affects resistance to bacterial blight pathogen in rice. Rice 14, 36. doi:10.1186/s12284-021-00478-9
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10, 685–694. doi:10.1016/j.molp.2017.03.009
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 12, 1164–1171. doi:10.1007/s00122-006-0218-1
Fan, C., Yu, S., Wang, C., and Xin, Y. (2009). A causal C–A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 118, 465–472. doi:10.1007/s00122-008-0913-1
Garris, A. J., Tai, T. H., Coburn, J., Kresovich, S., and McCouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169, 1631–1638. doi:10.1534/genetics.104.035642
Hu, Z., Lu, S. J., Wang, M. J., He, H., Sun, L., Wang, H., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. doi:10.1016/j.molp.2018.03.005
Huang, J., Lu, G., Liu, L., Raihan, M. S., Xu, J., Jian, L., et al. (2020). The kernel size-related quantitative trait locus qkw9 encodes a pentatricopeptide repeat protein that affects photosynthesis and grain filling. Plant Physiol. 183, 1696–1709. doi:10.1104/pp.20.00374
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. doi:10.1038/ng.2612
Jiang, G. H., Hong, X. Y., Xu, C. G., Li, X. H., and He, Y. Q. (2005). Identification of quantitative trait loci for grain appearance and milling quality using a doubled- haploid rice population. J. Integr. Plant Biol. 47, 1391–1403. doi:10.1111/j.1744-7909.2005.00089.x
Kosambi, D. D. (1944). The estimation of map distance from recombination values. Ann. Eugen. 12, 172–175. doi:10.1111/j.1469-1809.1943.tb02321.x
Lander, E. S., and Botstein, D. (1989). Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121, 185–199. doi:10.1093/genetics/121.1.185
Lee, K., and Back, K. (2017). Overexpression of rice serotonin N‐acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. pineal Res. 62, e12392. doi:10.1111/jpi.12392
Li, H., Ye, G., and Wang, J. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361–374. doi:10.1534/genetics.106.066811
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi:10.1038/ng.977
Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U. S. A. 107, 19579–19584. doi:10.1073/pnas.1014419107
Marowa, P., Ding, A., and Kong, Y. (2016). Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell. Rep. 35, 949–965. doi:10.1007/s00299-016-1948-4
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi:10.1016/j.cj.2015.01.001
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi:10.1093/nar/8.19.4321
Riemann, M., Haga, K., Shimizu, T., Okada, K., Ando, S., Mochizuki, S., et al. (2013). Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74, 226–238. doi:10.1111/tpj.12115
Shao, G., Wei, X., Chen, M., Tang, S., Luo, J., Jiao, G., et al. (2012). Allelic variation for a candidate gene for GS7, responsible for grain shape in rice. Theor. Appl. Genet. 125, 1303–1312. doi:10.1007/s00122-012-1914-7
Shi, X., Tian, Q., Deng, P., Zhang, W., and Jing, W. (2021). The rice aldehyde oxidase OsAO3 gene regulates plant growth, grain yield, and drought tolerance by participating in ABA biosynthesis. Biochem. Biophys. Res. Commun. 548, 189–195. doi:10.1016/j.bbrc.2021.02.047
Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. doi:10.1038/ng.169
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi:10.1038/ng2014
Wang, C., Chen, S., and Yu, S. (2011). Functional markers developed from multiple loci in GS3 for fine marker assisted selection of grain length in rice. Theor. Appl. Genet. 122, 905–913. doi:10.1007/s00122-0101497-0
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi:10.1038/ng.2327
Xu, Y., Zhu, L., Xiao, J., Huang, N., and McCouch, S. R. (1997). Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, double haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol. Gen. Genet. 253, 535–545. doi:10.1007/s004380050355
Zhang, J., Gao, X., Cai, G., Wang, Y., Li, J., Du, H., et al. (2021). An adenylate kinase OsAK3 involves brassinosteroid signaling and grain length in rice (Oryza sativa L.). Rice 14, 105. doi:10.1186/s12284-021-00546-0
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012b). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U. S. A. 109, 21534–21539. doi:10.1073/pnas.1219776110
Keywords: quantitative trait loci, basmati rice, grain quality, SSR markers, mapping
Citation: Malik A, Kumar A, Ellur RK, Krishnan S G, Dixit D, Bollinedi H, Vinod K, Nagarajan M, Bhowmick P, Singh N and Singh A (2022) Molecular mapping of QTLs for grain dimension traits in Basmati rice. Front. Genet. 13:932166. doi: 10.3389/fgene.2022.932166
Received: 29 April 2022; Accepted: 28 June 2022;
Published: 02 August 2022.
Edited by:
Sundeep Kumar, National Bureau of Plant Genetic Resources (ICAR), IndiaReviewed by:
Pallavi Sinha, IRRI South Asia Hub, IndiaCopyright © 2022 Malik, Kumar, Ellur, Krishnan S, Dixit, Bollinedi, Vinod, Nagarajan, Bhowmick, Singh and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: AK Singh, YWtzX2dlbmVAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.