- 1Centre for Agricultural Bioinformatics, ICAR-Indian Agricultural Statistics Research Institute, New Delhi, India
- 2ICAR-National Bureau of Plant Genetic Resources, New Delhi, India
- 3Department of Biotechnology, School of Interdisciplinary and Allied Sciences (SIAS), Central University of Haryana, Haryana, India
Cytokinin dehydrogenase (CKX; EC.1.5.99.12) regulates the level of cytokinin (CK) in plants and is involved in CK regulatory activities. In different plants, a small gene family encodes CKX proteins with varied numbers of members. These genes are expanded in the genome mainly due to segmental duplication events. Despite their biological importance, CKX genes in Triticum aestivum have yet to be studied in depth. A total of 11 CKX subfamilies were identified with similar gene structures, motifs, domains, cis-acting elements, and an average signal peptide of 25 amino acid length was found. Introns, ranging from one to four, were present in the coding regions at a similar interval in major CKX genes. Putative cis-elements such as abscisic acid, auxin, salicylic acid, and low-temperature-, drought-, and light-responsive cis-regulatory elements were found in the promoter region of majority CKX genes. Variation in the expression pattern of CKX genes were identified across different tissues in Triticum. Phylogenetic analysis shows that the same subfamily of CKX clustered into a similar clade that reflects their evolutionary relationship. We performed a genome-wide identification of CKX family members in the Triticum aestivum genome to get their chromosomal location, gene structure, cis-element, phylogeny, synteny, and tissue- and stage-specific expression along with gene ontology. This study has also elaborately described the tissue- and stage-specific expression and is the resource for further analysis of CKX in the regulation of biotic and abiotic stress resistance, growth, and development in Triticum and other cereals to endeavor for higher production and proper management.
Introduction
Wheat (Triticum aestivum L.) is a widely cultivated cereal grain that provides approximately 20% of the protein in human diet. It ranks second to corn and rice as the most important staple food, with global production of around 778.6 million tonnes of grain (USDA, 2022). Looking at the increasing human population, its consumption is estimated to rise to 900 million tonnes by 2050 (FAO, 2022). Cytokinins, the plant hormone family, affect crop yield in terms of grain size and number, and are involved in the regulation of biotic and abiotic stress (Cortleven et al., 2019). They are reported to control both shoot and root architecture as well as crown formation (Chen L. et al., 2020). Cytokinin dehydrogenase (CKX) gene family members encode the enzyme CKX, and play an important role in the cell cycle, developmental processes, and senescence process of plants such as seed germination, shoot/root growth, photosynthesis, chloroplast formation, and crop productivity (Mok and Mok 2001; Takei et al., 2001; Werner et al., 2001). Recent research suggests that a majority of plants had cytokinin (CK) metabolic genes closely linked to them and, therefore, strongly regulate cytokinin content in different organs of plants. (Ashikari et al., 2005; Bartrina et al., 2011; Yuldashev et al., 2012). Further, CKs are adenine derivatives with a side chain at the N6 position, which causes the N6 substituent to be classified as an isoprenoid or aromatic side chain (Galuszka et al., 2000; Frebort et al., 2011). Since numerous changes have been observed in the CK content during grain development in crops like wheat and barley, genetic manipulation of the genes involved in CK homeostasis is frequently utilized for yield improvement in these crops (Schmuelling et al., 2003).
CKX (EC: 1.5.99.12) is the only known enzyme that permanently degrades CKs by cleaving the N6-unsaturated side chain of the CK to adenine and adenosine in a single step. The catalytic activity of CKX is catalyzed by two conserved domains, namely, a FAD-binding domain at the N terminus and a CK-binding domain at the C terminus of the protein. CKX activity was first discovered in crude extracts from tobacco plants (Paces et al., 1971). Subsequently, CKX was recognized as a member of a small gene family and several CKX genes were duplicated and identified in different plants. Cytokinin dehydrogenase has been identified as a significant enzyme present in different cereal crops. It has been reported in 13 gene families in Zea mays (Gu et al., 2010; Houba-Hérin et al., 1999; Brugière et al., 2003), 11 gene families in rice (Ashikari et al., 2005), 9 gene families in Arabidopsis (Galuszka et al., 2007), 13 gene families in Brachypodium distachyon (Mameaux et al., 2012), four gene families in Hordeum vulgare (Mameaux et al., 2012), and 11–14 gene families in wheat (Ogonowska et al., 2019; Shoaib et al., 2019; Song et al., 2012). In rice, an increase in CK is a product of low OsCKX2 expression which promotes the total yield by increasing the number of reproductive organs using short hairpin RNA-mediated silencing technology. This hinders the expression of OsCKX2 in rice, resulting in an increased number of tillers, thus increasing the grain weight (Ashikari et al., 2005; Yeh et al., 2015). Similarly, Arabidopsis CKX genes showed diverse countenance, which supports that differential expression of CKX genes can play a vital role in regulating CK levels (Bartrina et al., 2011).
The CKX gene family has been reported to show natural variations in the form of SNPs which were used for the potential marker-assisted selection for higher yield as in soybean (Nguyen et al., 2021). Besides, CKs are widely used for crop yield improvement/management because of their broader effects on plant growth/development and physiology. Applications of CRISPR-based gene-editing tools in manipulating cytokinin metabolism at the genetic level for yield improvement has been reported in literature (Mandal et al., 2022). Genome editing occurs via natural process of random mutagenesis, mostly through SNPs to improve yield (Songstad et al., 2017).
Although > 20 different cytokinins are reported in wheat (Sýkorová et al., 2008), the complete analysis on the early development time frame has not yet been reported for wheat. The hexaploidy, large genome size (∼17 GB), and complicated interactions between the three genomes of wheat (Triticum aestivum L.) have led to limited information on the CKX gene families in this cereal crop.
This study aims at studying the genome-wide identification and distribution of CKX genes in wheat, motif characteristics, distribution and evolutionary relationships along with the presence of cis-elements in the promoter regions across 11 subfamily members of CKX. This study also aims at comparing motifs and domains present among the CKX family members at the protein level along with tissue- and stage-specific expression. This analysis will lead to useful information for further functional dissection of CKX genes in plants.
Materials and Methods
Identification of CKX Family Members in Triticum
The CKX genes were identified from the Triticum aestivum (bread wheat) genome (IWGSC RefSeq), available at Ensemble plant database (https://plants.ensembl.org/index.html). The cytokinin dehydrogenase (CKX), FAD- and cytokinin-binding (cytokinin-bind PF09265) protein domain, family was obtained from the PFAM database (https://pfam.xfam.org/). HMMER 3.0 program was used to identify the CKX protein domain-containing putative protein in bread wheat. Blastp was performed on these proteins with parameters e < 1e− 5 and 50% identity, keeping the query as Oryza sativa, Arabidopsis thaliana, Hordeum vulgare, and Zea mays to find the putative CKX protein from the bread wheat protein sequence. The NCBI-CDD web server (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) was used to confirm the candidate CKX genes in Triticum aestivum. ProtComp was used to predict the subcellular localization (Kamper et al., 2006) followed by the identification of the signal peptides and the location of their cleavage sites in CKX protein using SignalP (Armenteros et al., 2019). TargetP was used to identify the presence of N-terminal presequences: signal peptide (SP), mitochondrial transit peptide (MTP), chloroplast transit peptide (cTP), or thylakoid luminal transit peptide (lTP) in CKX proteins. Other protein parameters like molecular weight (MW) and theoretical isoelectric point (PI) of CKX proteins was found using the ExPASy server (Gasteiger et al., 2003). The subcellular localization predictions of the analyzed proteins were also performed using WoLF PSORT (https://wolfpsort.hgc.jp/).
Chromosomal Distribution, Gene Structure, and Conserved Motif of CKX Gene Family
The distribution of CKX genes was observed with its chromosomal location using Plant ensemble database (Bolser et al., 2016). Also, their gene structures were displayed in the Gene Structure Display Server (GSDS: http://gsds.gao-lab.org/; Guo et al., 2007). Motif identification was performed using the MEME suite. The multiple expectation maximization for the motif elicitation with MEME v4.9. (Bailey and Elkan, 1994) utility program (http://meme-suite.org/tools/meme) was used to display motifs in CKX proteins.
Phylogenetic and Synteny Analysis of the CKX Gene Family
Multiple sequence alignment was performed through ClustalW (Thompson et al., 1994) based on the full sequence of the proteins with default parameters from Triticum aestivum, Oryza sativa, Arabidopsis thaliana, Hordeum vulgare, and Zea mays. The phylogenetic tree was constructed by maximum likelihood method using the MEGA11 (Tamura et al., 2021) software with 1,000 replicates as bootstrap values. iTOL (https://itol.embl.de/) was used to visualize the phylogenetic tree and motifs in the CKX proteins (Letunic and Bork, 2019).
TBtool (Chen et al., 2020b) (https://github.com/CJ-Chen/TBtools/releases) was used for synteny analysis of CKX genes in Triticum aestivum with Aegilops tauschii via the MCScanX function. MCScanX is a toolkit for the detection and evolutionary analysis of gene collinearity (Wang et al., 2012).
Promoter and Gene Ontology Analysis of CKX Gene Family With Expression Across Different Tissue and Stages
A 1.5-kb region upstream from the start of the gene was retrieved for the promoter analysis from the CKX gene. The PlantCARE (Lescot et al., 2002) database was used for the identification of cis-regulatory elements in the promoter region. CKX genes in developmental and tissue time course expression in Triticum varieties were obtained from the wheat expression browser (http://www.wheat-expression.com/download) to detect the expression profile of the CKX gene. Gene ontology analysis was performed using the PlantRegMap tool (Tian et al., 2020).
Results and Discussion
Identification of the CKX Gene Family in Triticum aestivum
CKX proteins were identified from the whole genome of Triticum aestivum using blastp and HMM. The putative CKX proteins were confirmed using Pfam and a conserved domain database. In earlier published literature, 11–14 gene family members have been reported in wheat (Ogonowska et al., 2019; Shoaib et al., 2019). In rice, 11 CKX homologs have been identified and in Arabidopsis thaliana, 7 homologs of the CKX gene, in Hordeum vulgare 15, and in Zea mays 34 homologs of CKX were identified.
Earlier studies on CKX have given different nomenclatures to the different gene families of CKX. We have renamed CKX based on recent nomenclature in IWGSC taking into consideration the earlier reports and true orthologs in rice and maize. Finally, 11 CKX families were identified and their genomic location was extracted from Plant Ensemble. The average molecular weight (MW) and theoretical isoelectric point (PI) of CKX proteins ranged from 50,000 to 60,000 Da and from 5‐8, respectively (Table 1).
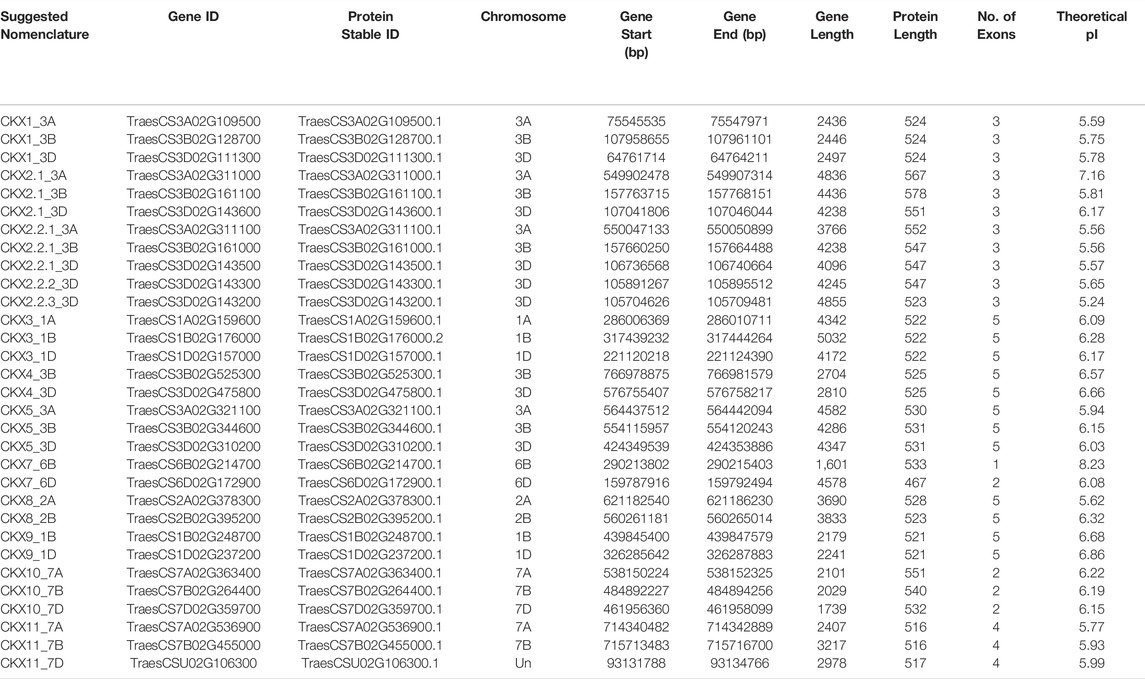
TABLE 1. Detailed information of the cytokinin dehydrogenase gene family (Triticum aestivum) with respect to their chromosomal location and general protein parameters.
To determine whether the analyzed proteins are directed to the secretory pathway and secreted out of the cell, predictions of the presence of signal peptides at the N terminus of the sequence were performed. For this purpose, program SignalP-6.0 and TargetP were used. Using the SignalP, the signal peptides were found in 21 of the analyzed proteins. The average length of sequences provided as a signal peptide was 25 aa. Similar results were obtained using the TargetP tool—the signal peptides were found in 25 of the investigated sequences (Figure 1A; Supplementary Tables S1A,B). The most likely location of the CKX protein was predicted as extracellular using the ProtComp tool. The subcellular localization prediction program, WoLF PSORT helps to trace the protein in the intracellular compartments. Out of the total analyzed proteins, 13 sequences were classified as chloroplast, 7 as the vacuole membrane protein, 4 as extracellular, 3 as endoplasmic reticulum, 2 as cytoplasmic, 1 as the mitochondria, and 1 as the nuclear pathway (Figure 1B).
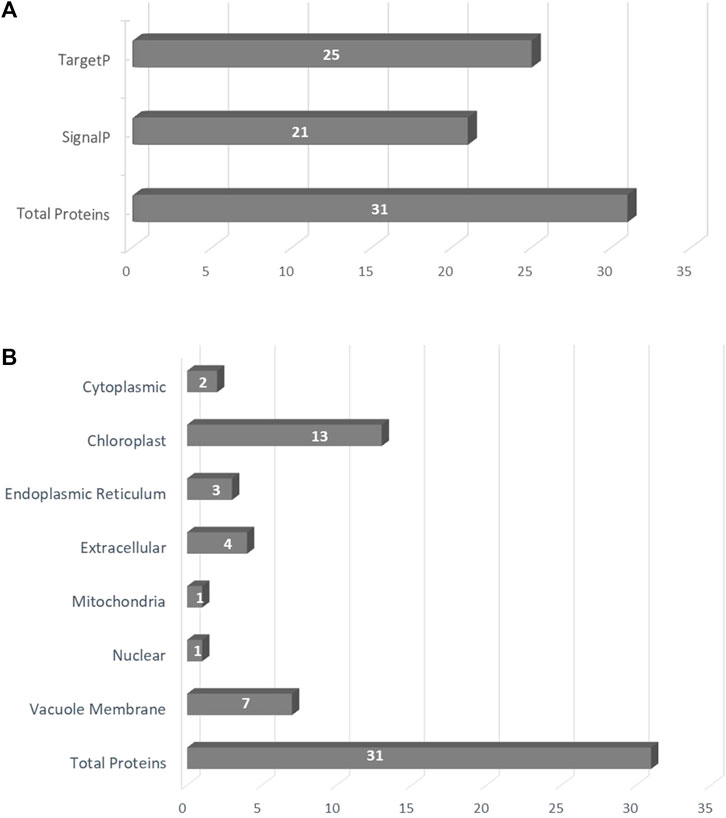
FIGURE 1. (A) The subcellular localization of predicted proteins; (B) The presence of signal peptides in the analyzed sequences using SignalP and TargetP.
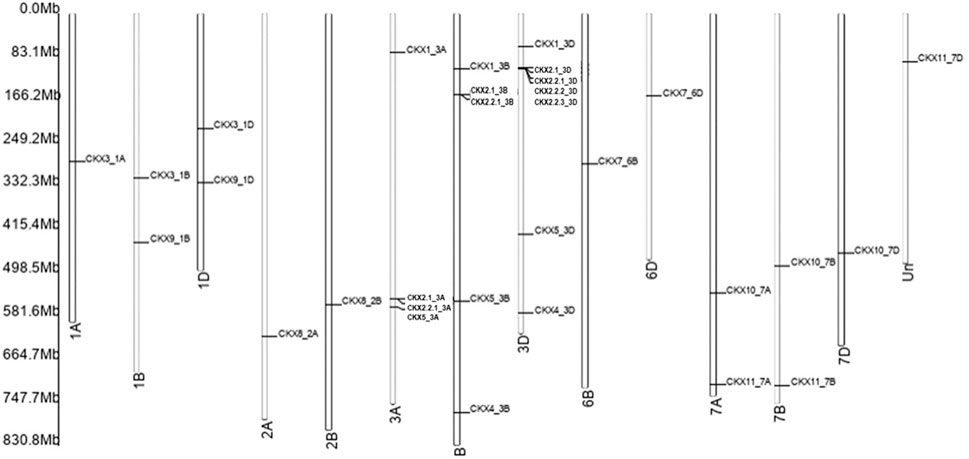
Chromosomal Distribution, Gene Structure, and Conserved Motif of CKX Genes
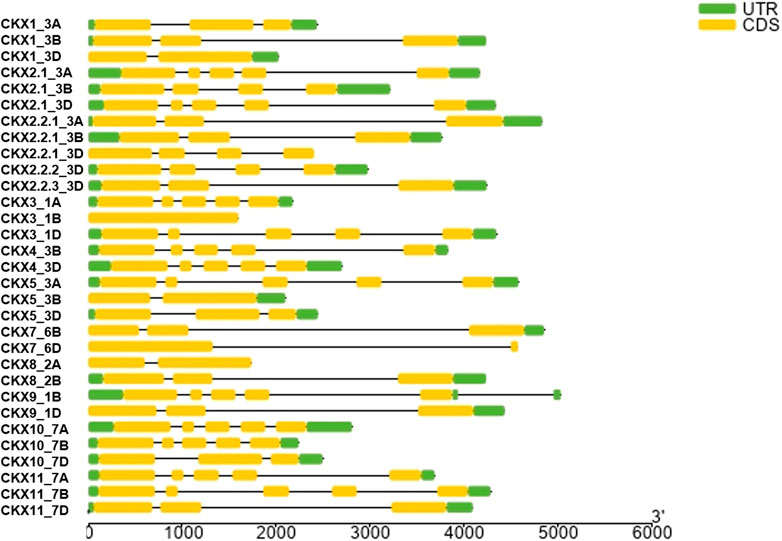
Motif diversity and gene structure promote evolution in gene families (Zhu et al., 2020). We identified 31 TaCKX gene family members that were present on chromosomes, linked with the A, B, and D subgenomes of Triticum. Among the 11 gene families, TaCKX2 has gene duplication on chromosome 3D, that is further subdivided, based on homology, into different groups (Mameaux et al., 2012; Lu et al., 2015). CKX gene families identified different homologs in the subgenome A, B, and D. All CKX gene family members were found on chromosomes 1 to 7, except chromosome 6 (Figure 2). One of the gene families, i.e., the CKX11 subfamily was not present on any of the 21 chromosomes (of A, B, and D subgenomes) in IWGSC Refseq. In the CKX genes, a number of exons varied from 1 to 5. Only CKX7_6B identified one exon and no intron while 11 CKX genes and 13 CKX genes had 3 exons and 5 exons in their gene structures, respectively (Figure 3). The gene transfer format (gff) was used to extract the 5′ UTR, 3′ UTR, exonic, intronic, start, and end position of the gene. It was found that the 5’ UTR position was absent for CKX2.1_3B, CKX2.2.3_3D, CKX7_6B, CKX7_6D, CKX10_7A, CKX11_7A, CKX10_7B, and CKX10_7D gene family members in the gff file. Intronic region regulated region expression via small and long noncoding RNA in genome (Jain et al., 2021). CKX gene structures showed that a majority of the genes had the presence of more than one intron. However, the length of the intron kept varying between the subgroups. The longest intron was found in CKX7_6D. Previous studies show that introns present in β-tubulin might regulate the expression and evolution of genes in Fusarium graminearum (Li et al., 2019).
Comparison of 31 CKX family members showed the presence of the PLN02441 domain family. The predicted CKX proteins had typical FAD- and CK-binding domains, which were specific to CKX family members as shown in Supplementary Figure S1. Similar results have been obtained in genome-wide analysis and identification of cytokinin oxidase/dehydrogenase (CKX) gene family in foxtail millet (Liu et al., 2018).
Phylogenetic and Synteny Analysis of the CKX Gene Family
Highly conserved motif regions in CKX proteins were identified using MEGA 11 in classic mode with the maximum number of motifs set at ten. Ten-motif consensus sequences were identified in all 31 CKX proteins as shown in Figure 4A. To uncover the evolutionary relationships among Triticum aestivum, Oryza sativa, Arabidopsis thaliana, Hordeum vulgare, and Zea mays CKXs, the amino acid sequences of the CKX genes were aligned using ClustalW. This was followed by the maximum likelihood analysis method to construct a phylogenetic tree (Figures 4A,B). All the proteins fell into four major clusters (I, II, III, and IV). Cluster I contained 39 members (with 16 of Triticum aestivum, 1 Oryza sativa,1 Hordeum vulgare, 3 Zea mays, and 3 Arabidopsis thaliana, respectively). This could be further divided into the following subclusters: IA,IB,1C,1D, 1E, and a diverge member (CKX). Cluster II included 18 members (with five of Triticum aestivum, two Oryza sativa, two Hordeum vulgare, and nine Zea mays) and could be separated into subclusters IIA and IIB. Cluster III contained 11 members (three Triticum aestivum, one Oryza sativa, one Hordeum vulgare, and five Zea mays) that could be separated into subclusters IIIA and IIIB. Cluster IV had 30 members (with seven Triticum aestivum, three Oryza sativa, five Hordeum vulgare, six Zea mays, and two Arabidopsis thaliana, respectively) which could be further divided into subclusters IVA and IVB.
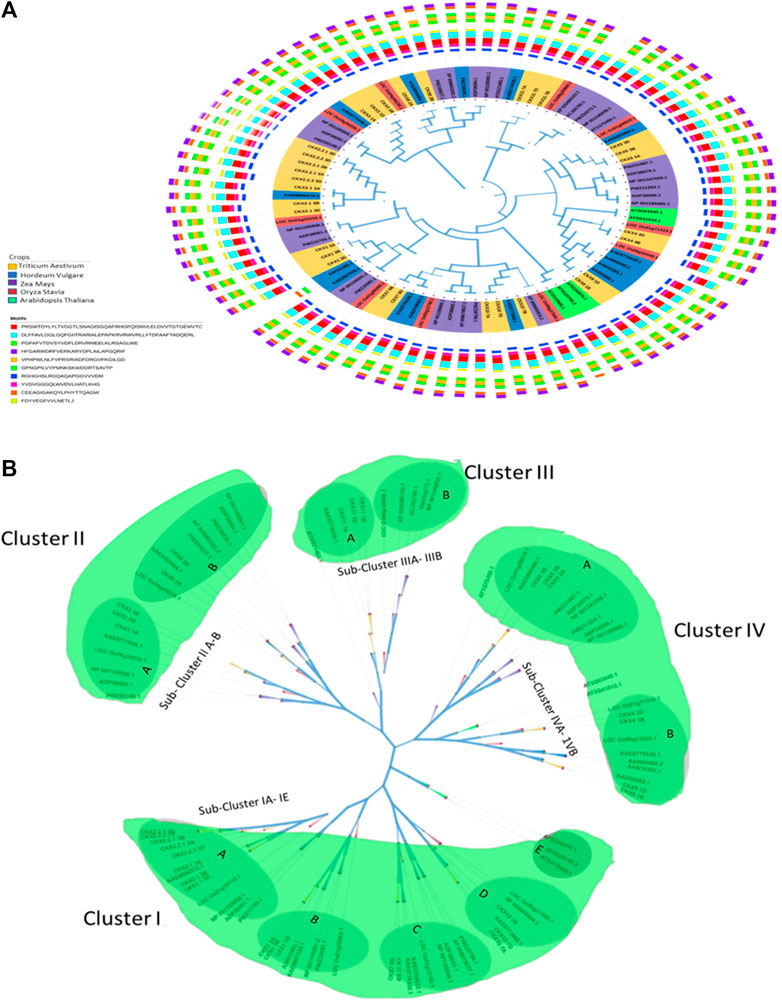
FIGURE 4. (A) Phylogenetic tree for CKX proteins of Triticum aestivum, Oryza sativa, Arabidopsis thaliana, Hordeum vulgare, and Zea mays. (B): Four major clusters of CKX proteins of Triticum aestivum, Oryza sativa, Arabidopsis thaliana, Hordeum vulgare, and Zea mays using the maximum likelihood method.
The paralogs have been separated into two subgene families (CKX2.1 and CKX2.2) as shown in the phylogenetic tree (Figure 4B). Notably, KAE8806376.1 and LOC_Os01g10110.1 from Hordeum vulgare and Oryza sativa, respectively grouped with the CKX2.1 and CKX2.2, CKX1 was grouped with AAK51495.1 and KAE8807039.1 (Hordeum vulgare) and CKX7 with KAE8776326.1 and KAE8768522.1 (Hordeum vulgare) subclusters, respectively. Similarly, Cluster II CKX3 1A was grouped with KAE8771049.1 and LOC_Os10g34230.1 (Oryza sativa) and CKX8 2A were grouped with KAE8805484.1 (Hordeum vulgare) while CKX8 2B was grouped with LOC_Os04g44230.1 (Oryza sativa). In Cluster III, CKX11 was clustered with KAE8774039.1 (Hordeum vulgare) and in Cluster IV, CKX5 3D was clustered with KAE8804960.1 and CKX7 3D LOC_Os01g71310.1. Functional investigation of these new members is critical as they could be of considerable importance for genetic improvement.
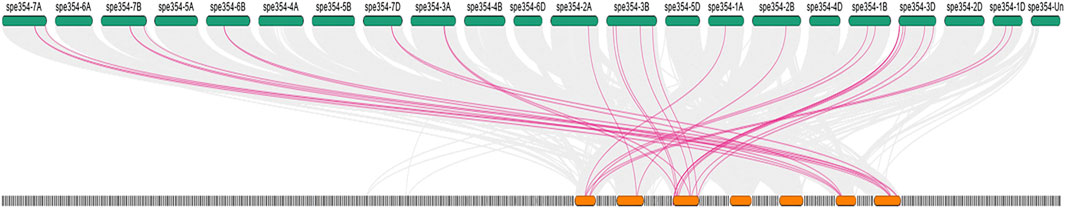
Gene duplication, an indispensable mechanism, can expand new genes that share similar or different functions (Ohno., 1970). Therefore, we analyzed the duplication events that occurred in the gene family through synteny and collinearity of the CKXS gene. Three CKX2 have undergone gene duplication suggesting that the CKX2s on chromosome 3D could be subdivided into two groups based on their homology. Collinearity is identified between Triticum aestivum and Aegilops tauschii as shown in (Figure 5; Supplementary Table S2). Similar results for synteny has been identified in Brassica oleracea and Arabidopsis thaliana CKX genes (Zhu et al., 2022)
Promoter and Gene Ontology Analysis of CKX Gene Family
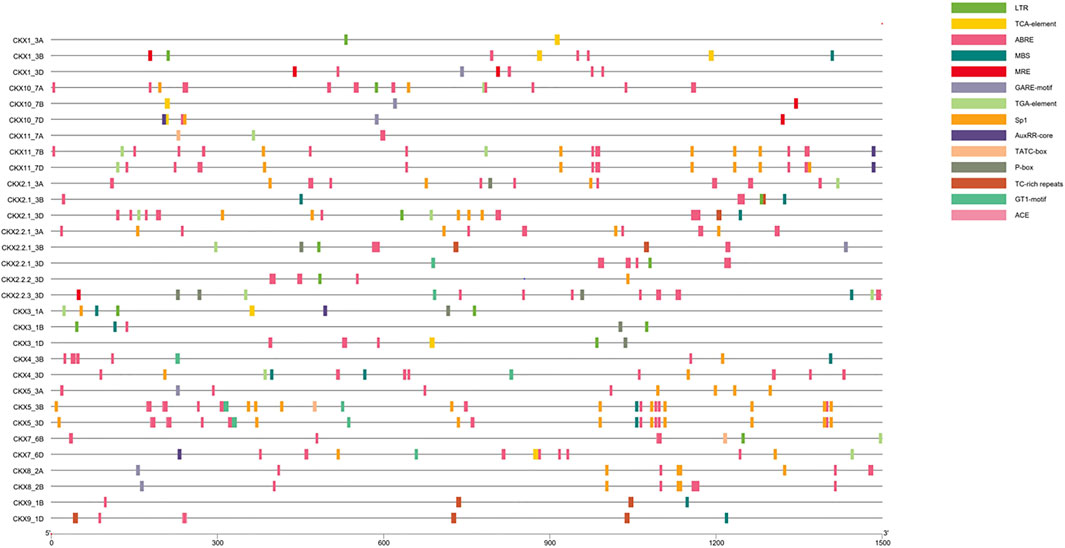
Cis-regulatory elements present in the promoter region play an important role in regulating gene expression of metabolic pathway-related genes (Lescot et al., 2002). In a 1.5-kb upstream region of CKX family members, the cis-regulatory elements predicted were mainly phytohormone responsive (salicylic acid, abscisic acid, gibberellin, auxin-responsive element), drought, light, defense, and stress responsive (Figure 6; Supplementary Table S3). Cis-acting element involved in low-temperature responsiveness (LTR) was found in CKX1_3A, CKX1_3B, CKX2.1_3B, CKX2.1_3D, CKX2.2.1_3B, CKX2.2.1_3D, CKX2.2.2_3D, CKX3_1A,CKX3_1B,CKX3_1D, CKX7_6B, and CKX1-_7A. Transcription factor MYB-binding site involved in drought inducibility (MBS) was found in CKX1_3B, CKX2.1_3B, CKX2.1_3D, CKX2.2.3_3D, CKX3_1A, CKX3_1B, CKX4_3B, CKX4_3D, CKX5_3B, CKX5_3D, CKX9_1B, and CKX9_1D. The MYB-binding site involved in light responsiveness (MRE) was found in CKX1_3B, CKX1_3D, CKX2.2.3_3D, CKX1-_7B, and CKX1-_7D. Light responsive element (Sp1) was found in CKX2.1_3A, CKX2.1_3D, CKX2.2.1_3A, CKX2.2.2_3D, CKX3_1A, CKX4_3B, CKX4_3D, CKX5_3A, CKX5_3B, CKX5_3D, CKX7_6D, CKX8_2A, CKX8_2B, CKX1-_7A, CKX1-_7D, CKX11_7B, and CKX11_7D. Light responsive element (GT1-motif) was found in CKX2.2.1_3D, CKX2.2.3_3D, CKX4_3B, CKX4_3D, CKX5_3B, CKX5_3D, and CKX7_6D. Cis-acting element involved in light responsiveness (ACE) was observed in CKX5_3B, CKX5_3D, CKX7_6B, and CKX9_1D. Cis-acting element involved in defense and stress responsiveness (TC-rich repeat) was found in CKX2.1_3B, CKX2.1_3D, CKX2.2.1_3B, CKX9_1B, and CKX9_1D. In tomato TC-rich repeats were found in seven promoters of protein disulfide isomerases (PDI) and high expression of PDI was found in response to abiotic stress in tomato (Wai et al., 2021). Cis-acting element involved in salicylic acid responsiveness (TCA element) was found in CKX1_3A, CKX1_3B, CKX2.1_3A, CKX3_1A, CKX3_1D, CKX7_6D, CKX1_7B, and CKX1-_7D. Cis-acting element involved in the abscisic acid responsiveness (ABRE) was found in CKX1_3B, CKX1_3D, CKX2.1_3A, CKX2.1_3B, CKX2.1_3D, CKX2.2.1_3A, CKX2.2.1_3B, CKX2.2.1_3D, CKX2.2.2_3D, CKX2.2.3_3D, CKX3_1B, CKX3_1D, CKX4_3B, CKX4_3D, CKX5_3A, CKX5_3B, CKX5_3D, CKX7_6B, CKX7_6D, CKX8_2A, CKX8_2B, CKX9_1B, CKX9_1D, CKX1-_7A, CKX1-_7D, CKX11_7B, and CKX11_7D. Gibberellin-responsive element (GARE-motif) was found in CKX1_3D, CKX2.2.1_3B, CKX5_3A, CKX8_2A, CKX8_2B, CKX1-_7B, and CKX1-_7D. Auxin-responsive element (TGA-element) was found in CKX2.1_3D, CKX2.2.1_3B, CKX2.2.3_3D, CKX3_1A, CKX4_3D, CKX7_6B, CKX7_6D, CKX1-_7A, CKX11_7B, and CKX11_7D. Cis-acting regulatory element involved in auxin responsiveness (AuxRR-core) was found in CKX3_1A, CKX7_6D, CKX1-_7D, CKX11_7B, and CKX11_7D. Cis-acting element involved in gibberellin responsiveness (TATC-box) was found in CKX5_3B and CKX7_6B. Gibberellin-responsive element (P-box) was found in CKX2.1_3A, CKX2.2.1_3B, CKX2.2.3_3D, CKX3_1A, CKX3_1B, and CKX3_1D. In Brassica napus, the CKX gene family also has abscisic acid-responsive elements (ABRE), auxin-responsive elements (TGA-element), gibberellin-responsive elements (GARE-motif, P-box, and TATC-box), and salicylic acid-responsive elements (TCA-element)(Liu et al., 2018). Phytohormone-responsive cis-elements were found in majority of the promoter region of CKX genes, reflecting that cytokinins are involved in the development and growth of the plant. Cis-acting element involved in the abscisic acid responsiveness (ABRE) was found in the highest number (27) among all responsive elements. Literature supports the cross-talk between cytokinin and abscisic acid (El-Showk et al., 2013).
Validation of CKX Gene Family’s Expression Across Different Tissues of Wheat
Cytokinin receptor kinases (AHK2 and AHK3) inhibit ABA signaling for regulating cold stress response (Jeon et al., 2010). Hence, CKX and ABA crosstalk to regulate the stress responses. It has been validated earlier using qRT-PCR that ABA downregulates CKX genes (Werner et al., 2006). CKX5_3B, a family member of CKX, has the highest number of cis-elements.
The different regulatory elements predicted responsive to abiotic and biotic stress show that the CKX family is stabilizing CK content under both abiotic and biotic stresses. Gene expression pattern helps to identify the key genes playing a role in an important biological function (Jain et al., 2017; Jain et al., 2019). CKX gene family members in our study show differential tissue-specific expression (Figure 7). Expression of GmCKX03, 05, 10, and 11 was reported to be low while GmCKX13, 15, and 16 were highly expressed in soybean (Le et al., 2012). From the comparative expression analysis of PtCKXs, OsCKXs, ZmCKXs, and AtCKXs, it was found that tissue-specific gene expression pattern was conserved in each phylogenetic group (Gu et al., 2010). The expression data for CKX genes for different studies in Triticum were studied from a wheat expression browser. In reproductive stages grain tissue shows expression of CKX1, CKX2, CKX3, CKX4, CKX5, CKX8, CKX9, and CKX11 while CKX7 And CKX10 show very low-level expression in the majority of the studies. In leaf and shoot tissue under all three stages, namely, reproductive, seedling, and vegetative, we found a good level of expression of CKX3, CKX4, CKX5, CKX8, CKX9, and CKX11 while CKX1 and CKX2 had a lower level of expression. CKX7 and CKX 10 were found to have the least expression in shoot and leaf tissues in all three developmental stages for the majority of the studies. In root tissues, CKX1, CKX3, CKX4, CKX5, CKX8, and CKX11 showed a good level of expression while CKX9 and CKX7 had a low level of expression while CKX2 and CKX10 have the lowest level of expression in reproductive, seedling, and vegetative stages for the majority of the samples for studies reported in wheat expression data. In spike tissue at the reproductive stage, CKX7 and CKX10 have very low expression while the rest of CKX have a good level of expression. Chen et al., 2020a reported the higher expression of CKX1 and CKX2 during grain development similar to our results. Overall, CKX7 expression was at lower levels in the leaf, inflorescence, and spike tissue. CKX4 and CKX5 had a better level of expression in the leaf tissue. In all the tissues studied, CKX3 and CKX11 were expressed at various levels while the expression of CKX10 was found at lower level in all tissues except leaves. These expression trends across tissues found in our study are in concordance with the qRT-PCR results of wheat (Ogonowska et al., 2019). CKX genes functionally diverged and expanded after duplication in angiosperm having tissue/organ-specific patterns that show abiotic stimulus response of expression (Wang et al., 2020).
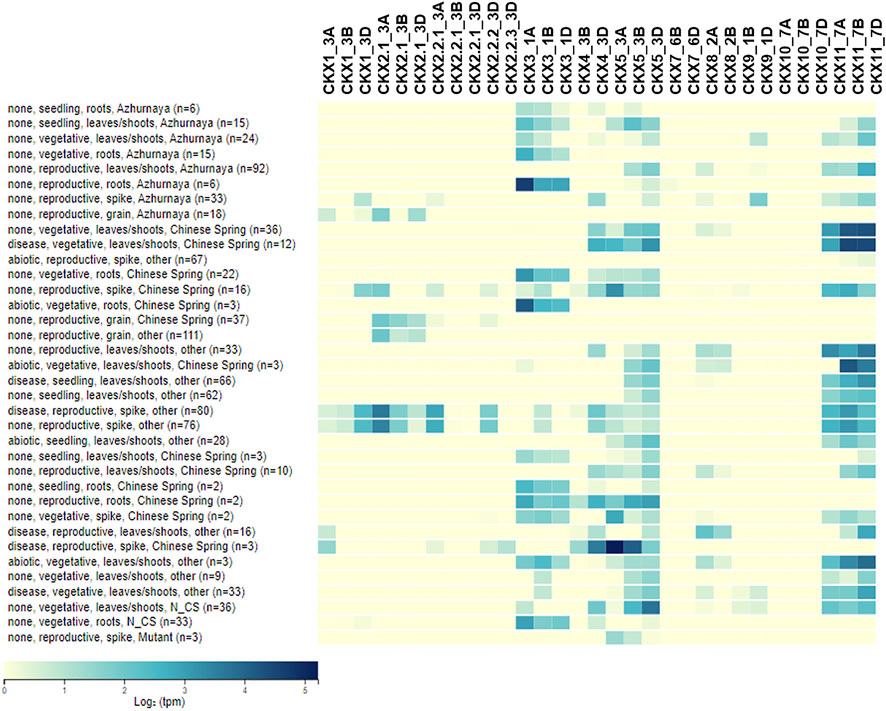
FIGURE 7. Heatmap for gene expression trend of CKX gene family in different tissue and stages of Triticum aestivum.
Gene ontology analysis shows the enrichment of the biological processes, cytokinin metabolic process, cellular hormone metabolic process, hormone metabolic process, and regulation of hormone levels (Supplementary Figure S2). The molecular functions enriched were cytokinin dehydrogenase activity, oxidoreductase activity, acting on the CH-NH group of donors, and flavin adenine dinucleotide binding (Supplementary Figure S3). Gene ontology analysis shows the enrichment of cellular hormone metabolic process, regulation of hormone levels, and molecular functions like oxidoreductase activity and flavin adenine dinucleotide binding were enriched which shows the role of CKX in regulating developmental events, enhancing grain yield in rice and enhancing drought and heat stress tolerance in tobacco (Mackova et al.,2013; Werner et al., 2010).
Conclusion
In this study, we identified 31 members of the CKX family in Triticum and analyzed their distribution on chromosome, subcellular localization, molecular weight, theoretical isoelectric point, signal peptide, motifs, domains, gene structure, cis-elements, and phylogenetic relationship among CKX members. The highest number of phytohormone-responsive cis-elements was found in the CKX gene, so, the protomer region has elements involved in the development and growth of Triticum. CKX7 and CKX10 were found to have very low gene expression compared to other CKX subgene family members in both reproductive and vegetative stages of Triticum. The CKX expression from the wheat express database and the analysis at vegetative and reproductive stages in different tissues showed that CKX has tissue-specific expression. Gene ontology analysis showed the enrichment of cellular hormone metabolic process, regulation of hormone levels, and molecular functions like oxidoreductase activity and flavin adenine dinucleotide binding that obviates the role of CKX in regulating developmental events, and enhancing grain yield and stress tolerance. This study gives a platform for the further identification of the CKX family and increases our understanding about the role of CKX in the regulation of biotic and abiotic stress resistance, growth, and development in Triticum to endeavor for higher production and proper management.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
MI and SJ conceived the theme of the study. PJ and AS performed the computational analysis and drafted the manuscript. MI, SJ, SK, and DK, and AR edited the manuscript. All co-authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are thankful to the Department of Biotechnology, Government of India (BT/Ag/Network/Wheat/2019-20), and Indian Council of Agricultural Research (ICAR), Ministry of Agriculture and Farmers’ Welfare, Government of India, for the use of the Advanced Super Computing Hub for Omics Knowledge in Agriculture (ASHOKA) facility at ICAR-IASRI, New Delhi, India, created under the National Agricultural Innovation Project. The authors further acknowledge the supportive role of the Director of ICAR-IASRI, New Delhi, and Director, ICAR-NBPGR, New Delhi, India. The authors are thankful to the Department of Biotechnology, Government of India, for financial support in the form of research project “Germplasm Characterization and Trait Discovery in Wheat using Genomics Approaches and its Integration for Improving Climate Resilience, Productivity and Nutritional quality” (BT/Ag/Network/Wheat/2019-20).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.931659/full#supplementary-material
References
Armenteros, J. J. A., Tsirigos, K. D., and Sønderby, C. K. (2019). SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 37, 420. doi:10.3390/genes12010023
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin Oxidase Regulates Rice Grain Production. Science 309, 741–745. doi:10.1126/science.1113373
Bailey, T. L., and Elkan, C. (1994). Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bartrina, I., Otto., E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield inArabidopsis Thaliana. Plant Cell. 23, 69–80. doi:10.1105/tpc.110.079079
Bolser, D., Staines, D. M., Pritchard, E., and Kersey, P. (2016). Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data. Methods Mol. Biol. 1374, 115–140. doi:10.1007/978-1-4939-3167-5_6
Brugière, N., Jiao, S., Hantke, S., Zinselmeier, C., Roessler, J. A., Niu, X., et al. (2003). Cytokinin Oxidase Gene Expression in Maize is Localized to the Vasculature, and is Induced by Cytokinins, Abscisic Acid, and Abiotic Stress. Plant Physiol. 132 (3), 1228–1240. doi:10.1104/pp.102.017707
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020b). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2020a). Cytokinin Dehydrogenase: A Genetic Target for Yield Improvement in Wheat. Plant Biotechnol. J. 18 (7), 614–630. doi:10.1111/pbi.13305
Cortleven, A., Leuendorf, J. E., Frank, M., Pezzetta, D., Bolt, S., and Schmülling, T. (2019). Cytokinin Action in Response to Abiotic and Biotic Stresses in Plants. Plant Cell. Environ. 42 (3), 998–1018. doi:10.1111/pce.13494
El-Showk, S., Ruonala, R., and Helariutta, Y. (2013). Crossing Paths: Cytokinin Signalling and Crosstalk. Development 140, 1373–1383. doi:10.1242/dev.086371
FAO (2022). FAOSTAT Statistical Database. Available at: http//www.fao.org/faostat/en/.Food and Agricultural Organization of the United Nations (FAO). 2016.
Frebort, I., Kowalska, M., Hluska, T., Frebortova, J., and Galuszka, P. (2011). Evolution of Cytokinin Biosynthesis and Degradation. J. Exp. Bot. 62, 2431–2452. doi:10.1093/jxb/err004
Galuszka, P., Frébort, I., Šebela, M., and Peč, P. (2000). Degradation of Cytokinins by Cytokinin Oxidases in Plants. Plant Growth Regul. 32, 315–327. doi:10.1023/a:1010735329297
Galuszka, P., Popelková, H., Werner, T., Frébortová, J., Pospíšilová, H., Mik, V., et al. (2007). Biochemical Characterization of Cytokinin Oxidases/Dehydrogenases from Arabidopsis thaliana Expressed in Nicotiana Tabacum L. J. Plant Growth Regul. 26, 255–267. doi:10.1007/s00344-007-9008-5
Gasteiger, E., Gattiker, A., Hoogland, C., and Ivanyi, I. (2003). ExPASy: The Proteomics Server for In-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 31 (13), 3784–3788. doi:10.1093/nar/gkg563
Gu, R., Fu, J., Guo, S., Duan, F., Wang, Z., Mi, G., et al. (2010). Comparative Expression and Phylogenetic Analysis of Maize Cytokinin Dehydrogenase/Oxidase (CKX) Gene Family. J. Plant Growth Regul. 29, 428–440. doi:10.1007/s00344-010-9155-y
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: a Gene Structure Display Server. Yi Chuan 29 (8), 1023–1026. doi:10.1360/yc-007-1023
Houba-Hérin, N., Pethe, C., d'Alayer, J., and Laloue, M. (1999). Cytokinin Oxidase from Zea mays: Purification, cDNA Cloning and Expression in Moss Protoplasts. Plant J. 17, 615–626. doi:10.1046/j.1365-313x.1999.00408.x
Jain, P., Singh, P. K., Kapoor, R., Khanna, A., Solanke, A. U., Krishnan, S. G., et al. (2017). Understanding Host-Pathogen Interactions with Expression Profiling of NILs Carrying Rice-Blast Resistance Pi9 Gene. Front. Plant Sci. 8, 93–20. doi:10.3389/fpls.2017.00093
Jain, P., Dubey, H., Singh, P. K., Solanke, A. U., Singh, A. K., and Sharma, T. R. (2019). Deciphering Signalling Network in Broad Spectrum Near Isogenic Lines of Rice Resistant to Magnaporthe Oryzae. Sci. Rep. 9, 16939. doi:10.1038/s41598-019-50990-8
Jain, P., Hussian, S., Nishad, J., Dubey, H., Bisht, D. S., Sharma, T. R., et al. (2021). Identification and Functional Prediction of Long Non-coding RNAs of Rice (Oryza Sativa L.) at Reproductive Stage under Salinity Stress. Mol. Biol. Rep. 48 (3), 2261–2271. doi:10.1007/s11033-021-06246-8
Jeon, J., Kim, N. Y., Kim, S., Kang, N. Y., Novák, O., Ku, S.-J., et al. (2010). A Subset of Cytokinin Two-Component Signaling System Plays a Role in Cold Temperature Stress Response in Arabidopsis. J. Biol. Chem. 285, 23371–23386. doi:10.1074/jbc.m109.096644
Kämper, J., Kahmann, R., Bölker, M., Ma, L. J., Brefort, T., Saville, B. J., et al. (2006). Insights from the Genome of the Biotrophic Fungal Plant Pathogen Ustilago Maydis. Nature 444, 97–101. doi:10.1038/nature05248
Le, D. T., Nishiyama, R., Watanabe, Y., Vankova, R., Tanaka, M., Seki, M., et al. (2012). Identification and Expression Analysis of Cytokinin Metabolic Genes in Soybean under Normal and Drought Conditions in Relation to Cytokinin Levels. PLoS ONE 7 (8), e42411. doi:10.1371/journal.pone.0042411
Lescot, M., Dhais, P., Thijs, G., Marchal, K., Moreau, Y., PeerVan, Y,de., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) V4: Recent Updates and New Developments. Nucleic Acids Res. 47 (W1), W256–W259. doi:10.1093/nar/gkz239
Li, Y., Chen, D., Luo, S., Zhu, Y., Jia, X., Duan, Y., et al. (2019). Intron-mediated Regulation of β-tubulin Genes Expression Affects the Sensitivity to Carbendazim in Fusarium Graminearum. Curr. Genet. 65, 1057–1069. doi:10.1007/s00294-019-00960-4
Liu, P., Zhang, C., Ma, J.-Q., Zhang, L.-Y., Yang, B., Tang, X.-Y., et al. (2018). Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica Napus L.) Genes. 9 (3), 168. doi:10.3390/genes9030168
Lu, J., Chang, C., Zhang, H.-P., Wang, S.-X., Sun, G., Xiao, S.-H., et al. (2015). Identification of a Novel Allele of TaCKX6a02 Associated with Grain Size, Filling Rate and Weight of Common Wheat. PLoS ONE 10, e0144765. doi:10.1371/journal.pone.0144765
Macková, H., Hronková, M., Dobrá, J., Turečková, V., Novák, O., Lubovská, Z., et al. (2013). Enhanced Drought and Heat Stress Tolerance of Tobacco Plants with Ectopically Enhanced Cytokinin Oxidase/dehydrogenase Gene Expression. J. Exp. Bot. 64, 2805–2815. doi:10.1093/jxb/ert131
Mameaux, S., Cockram, J., Thiel, T., Steuernagel, B., Stein, N., Taudien, S., et al. (2012). Molecular, Phylogenetic and Comparative Genomic Analysis of the Cytokinin Oxidase/dehydrogenase Gene Family in the Poaceae. Plant Biotech. J. 10, 67–82. doi:10.1111/j.1467-7652.2011.00645.x
Mandal, S., Ghorai, M., Anand, U., Roy, D., Kant, N., Mishra, T., et al. (2022). Cytokinins: A Genetic Target for Increasing Yield Potential in the CRISPR Era. Front. Genet. 13, 883930. doi:10.3389/fgene.2022.883930
Mok, D. W., and Mok, M. C. (2001). Cytokinin Metabolism and Action. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 52, 89–118. doi:10.1146/annurev.arplant.52.1.89
Nguyen, H. N., Kambhampati, S., Kisiala, A., Seegobin, M., and Emery, R. J. N. (2021). The Soybean (Glycine Max L.) Cytokinin Oxidase/dehydrogenase Multigene Family; Identification of Natural Variations for Altered Cytokinin Content and Seed Yield. Plant Direct 5 (2), e00308. doi:10.1002/pld3.308
Ogonowska, H., Barchacka, K., Gasparis, S., Jablonski, B., Orczyk, W., Dmochowska-Boguta, M., et al. (2019). Specificity of Expression of TaCKX Family Genes in Developing Plants of Wheat and Their Co-operation within and Among Organs. PLoS ONE 14, e0214239. doi:10.1371/journal.pone.0214239
Paces, V., Werstiuk, E., and Hall, R. H. (1971). Conversion of N-(Delta-Isopentenyl)adenosine to Adenosine by Enzyme Activity in Tobacco Tissue. Plant Physiol. 48 (6), 775–778. doi:10.1104/pp.48.6.775
Schmülling, T., Werner, T., Riefler, M., Krupková, E., and Bartrina y MannsManns, I. I. (2003). Structure and Function of Cytokinin Oxidase/dehydrogenase Genes of Maize, Rice, Arabidopsis and Other Species. J. Plant Res. 116 (3), 241–252. doi:10.1007/s10265-003-0096-4
Songstad, D. D., Petolino, J. F., Voytas, D. F., and Reichert, N. A. (2017). Genome Editing of Plants. Crit. Rev. Plant Sci. 36 (1), 1–23. doi:10.1080/07352689.2017.1281663
Sýkorová, B., Kurešová, G., Daskalova, S., Trckova, M., Hoyerová, K., Raimanová, I., et al. (2008). Senescence-induced Ectopic Expression of the A. Tumefaciens Ipt Gene in Wheat Delays Leaf Senescence, Increases Cytokinin Content, Nitrate Influx, and Nitrate Reductase Activity, but Does Not Affect Grain Yield. J. Exp. Bot. 59, 377–387. doi:10.1093/jxb/erm319
Takei, K., Sakakibara, H., Taniguchi, M., and Sugiyama, T. (2001). Nitrogen-Dependent Accumulation of Cytokinins in Root and theTranslocation to Leaf: Implication of Cytokinin Species that Induces GeneExpression of Maize ResponseRegulator. Plant Cell. Physiol. 42, 85–93. doi:10.1093/pcp/pce009
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-specific Gap Penalties and Weight Matrix Choice. Nucl. Acids Res. 22, 4673–4680. doi:10.1093/nar/22.22.4673
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., and Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104. doi:10.1093/nar/gkz1020
USDA (2022). USDA World Agricultural Production. Available at: https,//apps.fas.usda.gov/psdonline/circulars/production.pdf.
Wai, A. H., Waseem, M., and Khan, A. B. M. M. M. (2021). Genome-Wide Identification and Expression Profiling of the PDI Gene Family Reveals Their Probable Involvement in Abiotic Stress Tolerance in Tomato (Solanum lycopersicum L.) Genes. 12, 23. doi:10.1038/s41587-019-0036-z
Wang, X., Ding, J., Lin, S., Liu, D., Gu, T., Wu, H., et al. (2020). Evolution and Roles of Cytokinin Genes in Angiosperms 2: Do Ancient CKXs Play Housekeeping Roles while Non-ancient CKXs Play Regulatory Roles? Hortic. Res. 7, 29. doi:10.1038/s41438-020-0246-z
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Werner, T., Köllmer, I., Bartrina, I., Holst, K., and Schmülling, T. (2006). New Insights into the Biology of Cytokinin Degradation. Plant Biol. 8, 371–381. doi:10.1055/s-2006-923928
Werner, T., Motyka, V., Strnad, M., and Schmülling, T. (2001). Regulation of Plant Growth by Cytokinin. Proc. Natl. Acad. Sci. U.S.A. 98, 10487–10492. doi:10.1073/pnas.171304098
Werner, T., Nehnevajova, E., Köllmer, I., Novák, O., Strnad, M., Krämer, U., et al. (2010). Root-Specific Reduction of Cytokinin Causes Enhanced Root Growth, Drought Tolerance, and Leaf Mineral Enrichment in Arabidopsis and Tobacco. Plant Cell. 22, 3905–3920. doi:10.1105/tpc.109.072694
Yeh, S. Y., Chen, H. W., Ng, C. Y., Lin, C. Y., Tseng, T. H., Li, W. H., et al. (2015). Down-Regulation of Cytokinin Oxidase 2 Expression Increases Tiller Number and Improves Rice Yield. Rice 8 (1), 36. doi:10.1186/s12284-015-0070-5
Yuldashev, R., Avalbaev, A., Bezrukova, M., Vysotskaya, L., Khripach, V., and Shakirova, F. (2012). Cytokinin Oxidase is Involved in the Regulation of Cytokinin Content by 24-Epibrassinolide in Wheat Seedlings. Plant Physiol. Biochem. 55, 1–6. doi:10.1016/j.plaphy.2012.03.004
Zhu, M., Wang, Y., Lu, S., Yang, L., Zhuang, M., Zhang, Y., et al. (2022). Genome-wide Identification and Analysis of Cytokinin Dehydrogenase/oxidase (CKX) Family Genes in Brassica oleracea L. Reveals Their Involvement in Response to Plasmodiophora Brassicae Infections. Hortic. Plant J. 8 (1), 68–80. doi:10.1016/j.hpj.2021.05.003
Keywords: cytokinin, gene ontology, phylogenetics, synteny, wheat
Citation: Jain P, Singh A, Iquebal MA, Jaiswal S, Kumar S, Kumar D and Rai A (2022) Genome-Wide Analysis and Evolutionary Perspective of the Cytokinin Dehydrogenase Gene Family in Wheat (Triticum aestivum L.). Front. Genet. 13:931659. doi: 10.3389/fgene.2022.931659
Received: 29 April 2022; Accepted: 21 June 2022;
Published: 19 August 2022.
Edited by:
Jitender Singh, Sardar Vallabhbhai Patel University of Agriculture and Technology, IndiaReviewed by:
Sanjay Jha, Navsari Agricultural University, IndiaMeenakshi Tetorya, Donald Danforth Plant Science Center, United States
Copyright © 2022 Jain, Singh, Iquebal, Jaiswal, Kumar, Kumar and Rai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarika Jaiswal, c2FyaWthQGljYXIuZ292Lmlu
†These authors have contributed equally to this work and share first authorship
 Priyanka Jain
Priyanka Jain Ankita Singh
Ankita Singh Mir Asif Iquebal
Mir Asif Iquebal Sarika Jaiswal
Sarika Jaiswal Sundeep Kumar
Sundeep Kumar Dinesh Kumar
Dinesh Kumar Anil Rai
Anil Rai