
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 22 September 2022
Sec. Livestock Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.927670
This article is part of the Research TopicGenetic Engineering in Farm AnimalsView all 7 articles
 Yan Wu1,2
Yan Wu1,2 Yanyan Sun3
Yanyan Sun3 Hao Zhang1
Hao Zhang1 Hongwei Xiao1
Hongwei Xiao1 Ailuan Pan1
Ailuan Pan1 Jie Shen1
Jie Shen1 Yuejin Pu1
Yuejin Pu1 Zhenhua Liang1
Zhenhua Liang1 Jinping Du1
Jinping Du1 Jinsong Pi1*
Jinsong Pi1*Background: Eggshell strength and thickness are critical factors in reducing the egg breaking rate and preventing economic losses. The calcite biomineralization process is very important for eggshell quality. Therefore, we employed transcriptional sequencing and proteomics to investigate the differences between the uteruses of laying hens with high- and low-breaking-strength shells.
Results: A total of 1,028 differentially expressed genes (DEGs) and 270 differentially expressed proteins (DEPs) were identified. The analysis results of GO terms and KEGG pathways showed that most of the DEGs and DEPs were enriched in vital pathways related to processes such as calcium metabolism, hormone and amino acid biosynthesis, and cell proliferation and apoptosis. Several DEGs and DEPs that were coexpressed at mRNA and protein levels were verified. KRT14 (keratin-14) is a candidate gene (protein) obtained by multiple omics analysis due to the fold difference of KRT14 being the largest. After the overexpression of KRT14 in uterine epithelial cells, the expressions of OC116 (ovocleididin-116), CALB1 (calbindin 1), and BST1 (ADP-ribosyl cyclase 2) were found to be increased significantly, while the expression of OC17 (ovocleididin-17) was found to be decreased significantly.
Conclusion: In summary, this study confirms that during normal calcification, there are differences in ion transport between the uterus of hens producing high-breaking-strength eggshells and those producing low-breaking-strength eggshells, which may help elucidate the eggshell calcification process. The KRT14 gene may promote calcium metabolism and deposition of calcium carbonate in eggshells.
The ability of chickens to produce large numbers of eggs over an extended laying cycle is very essential for the poultry industry. The eggshells of birds play an important role in determining the physical and antimicrobial defenses of eggs and regulating the exchange of metabolic gases and water (Hincke et al., 2012). Eggshell quality is very important for hatchability (Hrnčár et al., 2012), food safety, and egg marketing (Park and Sohn, 2018). Therefore, during the entire laying period, improving the sustainability of egg quality (especially eggshell strength) is very important for the poultry industry. Many factors, including health status, age, nutrition, and housing system, affect the productive performance and egg quality of laying hens (Vlčková et al., 2019). Eggs with low eggshell strength are easily damaged during collection, storage, and transportation. On average, broken eggs account for 7.5% of eggs produced, which can lead to significant economic losses (Roland, 1988). The eggshell structure is complex and has important biological functions for protection of the inner content of eggs after deposition, as well as for chicken embryo development (Hincke et al., 1999). The eggshell contains 95% calcium carbonate and 3.5% organic matrix proteins (Marie et al., 2014). The eggshell is formed in the uterus, which is a segment of the oviduct of laying hens. The process of eggshell formation can be divided into three crucial periods: initiation of crystal growth, linear crystal growth, and termination of mineralization (Marie et al., 2015). The eggshell is composed of a bilayered membrane, mammillary layer, palisade layer, vertical crystal layer, and cuticle layer (Solomon, 1991).
Previous studies have shown that there are some differences in the ultrastructure between high-strength eggshells and low-strength eggshells (Ahmed et al., 2005; Stefanello et al., 2014; Zhang et al., 2017). The ultrastructure of eggshells is related to matrix proteins which regulate the morphology, growth kinetics, and crystallographic orientation of calcite crystals (Nys et al., 2004; Nys et al., 2010); these proteins include ovocleidin-116, ovocleidin-17, ovocalyxin-32, and ovocalyxin-36 (Hincke et al., 1995; Hincke et al., 1999; Gautron et al., 2001; Gautron et al., 2007). Matrix proteins have been shown to participate in the biomineralization of the eggshell (Mann, 2015). However, most of these discoveries only provide genes expressed in certain segments of the chicken oviduct (which is the principal organ for egg development); the overall mechanisms of eggshell development have not been determined yet. Similar to other economically important traits, eggshell strengthening remains relevant during the whole period of productive life for laying hens and generally deteriorates with age (Sirri et al., 2018; Crosara et al., 2019). The decline in eggshell quality is still one of the main reasons for replacing commercial flocks (Bain et al., 2016). Thus, clarifying the genetic basis and molecular mechanisms underlying eggshell strengthening at different chicken breeds and laying stages is very important for breeders. Therefore, we studied the differences in the gene expression profiles of the uteri of laying hens producing high- and low-breaking-strength eggshells during peak laying periods in this study.
Keratin 14 (KRT14) is a predominant cytoplasmic intermediate filament protein, which belongs to the type I keratin family and forms a heterodimer with type II keratin KRT5 (keratin 5). It is a prototypic marker of dividing basal keratinocytes where the KRT14 gene is transcribed at high levels (Rose-Anne et al., 2007). Previous studies have shown that the mutation of KRT14 and KRT5 causes epidermolysis bullosa simplex (Bolling et al., 2011; Jankowski et al., 2014); KRT14 may regulate the keratin turnover required for keratinocyte differentiation by interacting with receptor-interacting protein kinase 4 (RIPK4) (Smer et al., 2019). So far, the research on the KRT14 gene in poultry has not been reported. KRT14 is one of the genes with a large difference between the hard-eggshell group and the weak-eggshell group. In view of the important effect of KRT14 in the process of keratinization, we preliminarily explored the role of KRT14 in eggshell formation in this study.
All animal experiments were conducted according to the guidelines established by the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, 2004). The tissues were collected from 43-week-old hens. The hens were raised under free food intake and were humanely killed in the laboratory. All experimental protocols were approved by the Institute of Animal Husbandry and Veterinary of Hubei Academy of Agricultural Science.
Three eggs were collected from each of 1,820 hens from an experimental farm in Wuhan, China (Jianghan chicken, which is 43 weeks old and is a local chicken in China). Eggshell breaking strength, eggshell thickness, and egg weight were assessed. The eggshell thicknesses of eggs were tested using an electronic digital micrometer (Deli, Deli Group Co., Ltd, China). Eggshell breaking strength was assessed using an eggshell strength meter (NFN388, FHK Fujipin Co., Ltd, Japan). Eggs were weighed using an electronic balance (ES-E, Tianjin de ante Sensing Technology Co., Ltd, Tianjin, China).
The uteri of hens producing eggs with extreme eggshell thickness and eggshell breaking strength were collected immediately after oviposition for 6 h from six hens after being slaughtered (the hens were manually killed by the cervical dislocation method) in the laboratory. Three uterus samples were collected from the hard-eggshell group (HE group or thick group, exhibiting eggshells were strong and thick) and three uterus samples from the weak-eggshell group (LE group or thin group, exhibiting eggshells were weak and thin). The egg parameters of the six hens are listed in Table 1. Then, the uteri were frozen in liquid nitrogen for RNA-seq analysis and experimental validation.
Total RNA was isolated with TRIzol reagent (Invitrogen, United States). After total RNA was isolated from each individual sample and assessed, the concentration, quality, and integrity were determined by a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). A total of 5 μg RNA from each sample was employed for sequencing and library construction, which were performed at Novogene Biotech Co., Ltd. (Beijing, China).
First-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase, and then the RNaseH was used to degrade the RNA. Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and dNTP. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, adapters with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 370–420 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, United States). Then, after PCR amplification, the PCR product was purified by AMPure XP beads, and the library was finally obtained. After the construction of the library, the library was initially quantified by Qubit2.0 Fluorometer, then diluted to 1.5ng/ul, and the insert size of the library was detected by Agilent 2,100 bioanalyzer (Agilent Technologies, Palo Alto, CA, United States), and Also, being sequenced using the Illumina PE150 platform (Illumina, San Diego, CA) with paired-end 150 bp reads. Sequences were mapped to the chicken reference genome database from the NCBI database (ftp.ncbi.nlm.nih.gov/genomes/genbank/vertebrate_other/Gallus_gallus/latest_assembly_versions/GCA_000002315.5_GRCg6a/) with TopHat2 (Dou et al., 2016).
Differential expression analysis of two groups (three biological replicates per condition) was performed using the DESeq2 R package (Version 1.20.0). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. p-value <0.05 and |log2 (foldchange)| >0 were set as the thresholds for significantly differential expression.
Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package (version 3.8.1), in which gene length bias was corrected. GO terms with corrected p-value less than 0.05 were considered significantly enriched by differentially expressed genes. KEGG is a database resource for understanding high-level functions and utilities of the biological system such as the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). We used the clusterProfiler R package (version 3.8.1) to test the statistical enrichment of differential expression genes in KEGG pathways.
All RNA-seq data were uploaded to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA660090; SRA accession number: PRJNA660090).
Total proteins (the sample is the same as the RNA-seq) were extracted by the cold acetone method. The sample was ground individually in liquid nitrogen and lysed with lysis buffer (which contained 100 mM NH4HCO3 (pH 8), 8 M urea, and 0.2% SDS) and then ultrasonicated on ice for 5 min. After centrifuging at 12,000 g for 15 min at 4°C, the supernatant was transferred to a clean tube. Extracts from each sample were reduced with 10 mM DTT for 1 h at 56°C and subsequently alkylated with sufficient iodoacetamide for 1 h at room temperature in the dark. Then, samples were completely mixed with four times the volume of precooled acetone by vortexing and incubated at 20°C for at least 2 h. After being centrifuged, the precipitate was collected. After washing twice with cold acetone, the pellet was dissolved by dissolution buffer, which contained 0.1 M triethylammonium bicarbonate (TEAB, pH 8.5) and 6 M Urea.
A total of 120 μg of each protein sample was taken. The volume was made up to 100 μL with dissolution buffer. 1.5 μg trypsin and 500 μL of 100 mM TEAB buffer were added to each sample. Then, the sample was mixed and digested at 37°C for 4 h. After that, 1.5 μg trypsin and CaCl2 were added to each sample, and they were digested overnight. Formic acid was mixed with the digested sample, adjusted pH under 3, and then centrifuged at 12,000 g for 5 min at room temperature. The supernatant was slowly loaded to the C18 desalting column and washed three times with washing buffer (0.1% formic acid; 3% acetonitrile) and then eluted with some elution buffer (0.1% formic acid and 70% acetonitrile). The eluents of each sample were collected and lyophilized. 100 μL of 0.1 M TEAB buffer was added to reconstitute, and 41 μL of the acetonitrile-dissolved TMT labeling reagent was added, and then the sample was mixed with shaking for 2 h at room temperature. Then, the reaction was stopped by adding 8% ammonia. All labeling samples were mixed with equal volume, desalted, and lyophilized.
Mobile phase A (2% acetonitrile, adjusted pH to 10.0 using ammonium hydroxide) and B (98% acetonitrile) were used to develop a gradient elution. The lyophilized powder was dissolved in solution A and centrifuged at 12,000 g for 10 min at room temperature. The sample was fractionated using a C18 column (Waters BEH C18 4.6 × 250 mm, 5 μm) on a Rigol L3000 HPLC operating at 1 ml/min, and the column oven was set to 50°C. The elution gradient is listed in Table 2. The eluates were monitored at UV 214 nm, collected for a tube per minute, and combined into 10 fractions at last. All fractions were dried under vacuum and then reconstituted in 0.1% (v/v) formic acid (FA) in water.
An EASY-nLCTM 1200 UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher) operated in the data-dependent acquisition (DDA) mode was used to perform Shotgun proteomics analysis, which was performed at Novogene Biotech Co., Ltd. (Beijing, China).
Proteins with fold change in a comparison ≥ 1.2 or ≤ 0.83 and unadjusted significance level p < 0.05 were considered to be differentially expressed. Gene Ontology (GO) and InterPro (IPR) analyses were conducted using the InterProscan-5 program against the nonredundant protein database (including Pfam, PRINTS, ProDom, SMART, ProSiteProfiles, and PANTHER) (Jones et al., 2014). The protein family and pathway were analyzed by the KEGG database (Kyoto Encyclopedia of Genes and Genomes). The STRING-db server (http://string-db.org/) based on the related species was used to predict the probable interacting partners. The probable protein–protein interactions were predicted using the STRING-db server (Franceschini et al., 2013) (http://string.embl.de/). The enrichment analysis of GO and KEGG was performed by the enrichment pipeline (Huang et al., 2009).
The differentially expressed genes (DEGs) and the differentially expressed proteins (DEPs) were identified separately, and Venn diagrams were plotted according to the counted results. Correlation analysis was performed by R (version 3.5.1), and the maps were drawn based on changes in the transcriptome and proteome analysis.
Chicken KRT14 (NM_001001311) was amplified and then digested with KpnI and XhoI (NEB, Ipswich, MA, United States) and cloned into the pcDNA3.1 (+) vector (Promega).
The blood vessels, connective tissue, and fascia on the surface of the uterus were removed. The endometrial epithelial cells were gently scraped with a surgical blade. The scraped cells were collected and subjected to enzymatic digestion using collagenase type I with a final concentration of 1 mg/ml. The cells were filtered through a 74-μM mesh screen after enzymatic digestion for 60 min at 37°C and then washed and resuspended in culture media (DMEM/F12, 10% fetal bovine serum, and 1% penicillin and streptomycin). At last, the cell number was detected by the Trypan Blue method. The cells were seeded in 6-well culture plates with 1 × 106 cells/well. The cells were cultured at 37°C with 5% CO2. The transfection assays were conducted when cells reached 80–90% confluence. The overexpression vector of KRT14 and the pcDNA3.1 (+) vector were transfected into the cells by Lipofectamine 3,000 transfection reagent (Invitrogen, Carlsbad, California, United States). For each construct, experiments were carried out in triplicate.
RNA samples from the three chickens used for the RNA-seq experiment were analyzed by Q-PCR. The concentrations of RNA samples were measured using a NanoQuant Plate (TECAN, Infinite M200PRO). Total cDNA was synthesized using a PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Dalian, China). qRT-PCR was performed with a LightCycle® 480 II (Roche) in a final volume of 20 μL using THUNDERBIRD SYBR Q-PCR Mix (TOYOBO, Osaka, Japan). Each 20-μL reaction volume contained 10 μL of THUNDERBIRD SYBR Q-PCR Mix, 0.3 μL of each divergent primer, 1 μL of cDNA, and 8.4 μL of double-distilled H2O (ddH2O). The cycling conditions included an initial single cycle (98°C for 10 s) followed by 40 cycles (94°C for 15 s and 60°C for 30 s). β-actin was used as an internal control for expression of mRNA. The relative expression level was normalized and calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). All primers were designed using Primer 5 (listed in Supplementary Table S1).
The uterine protein lysates were generated by RIPA lysis buffer (Beyotime, Beijing, China). The SDS-PAGE was used to extract and separate uterine proteins. Then, the uterine protein was transferred to PVDF membranes which were blocked with skim milk. After that, the antibodies for β-actin (1:1,000, Abcam, Cambridge, MA, United States) and differential proteins (1:2000–1:1,000, Abcam, Cambridge, MA, United States) were used for immunoblotting. The protein expression levels were compared to β-actin expression using ImageJ (Wayne Rasband, National Institutes of Health, United States; Version 1.42q).
All data analyses was performed by GraphPad Prism 5 (San Diego, CA; Version 5). The experimental values are presented as the means ± SDs. The means of the groups were compared by Student’s t-test. p-values < 0.05 were considered to be statistically significant.
A total of 353 million raw reads were obtained from the RNA-seq data. After the adapter and low-quality sequences were removed, clean data were obtained. The total number of reads generated from the six samples is shown in Supplementary Table S2. Each sample yielded more than 45 million raw reads. A total of 19,128 expressed genes were detected between HE and LE groups. DEGs were identified by log2 (foldchange) >0 and p-values < 0.05 (Supplementary Table S3). There were 1,028 annotated genes that were significantly different (p < 0.05) in the uterus between hard and weak groups (Supplementary Table S3). Of these genes, 437 genes were upregulated, and 591 genes were downregulated (LE vs. HE) (Figure 1A).
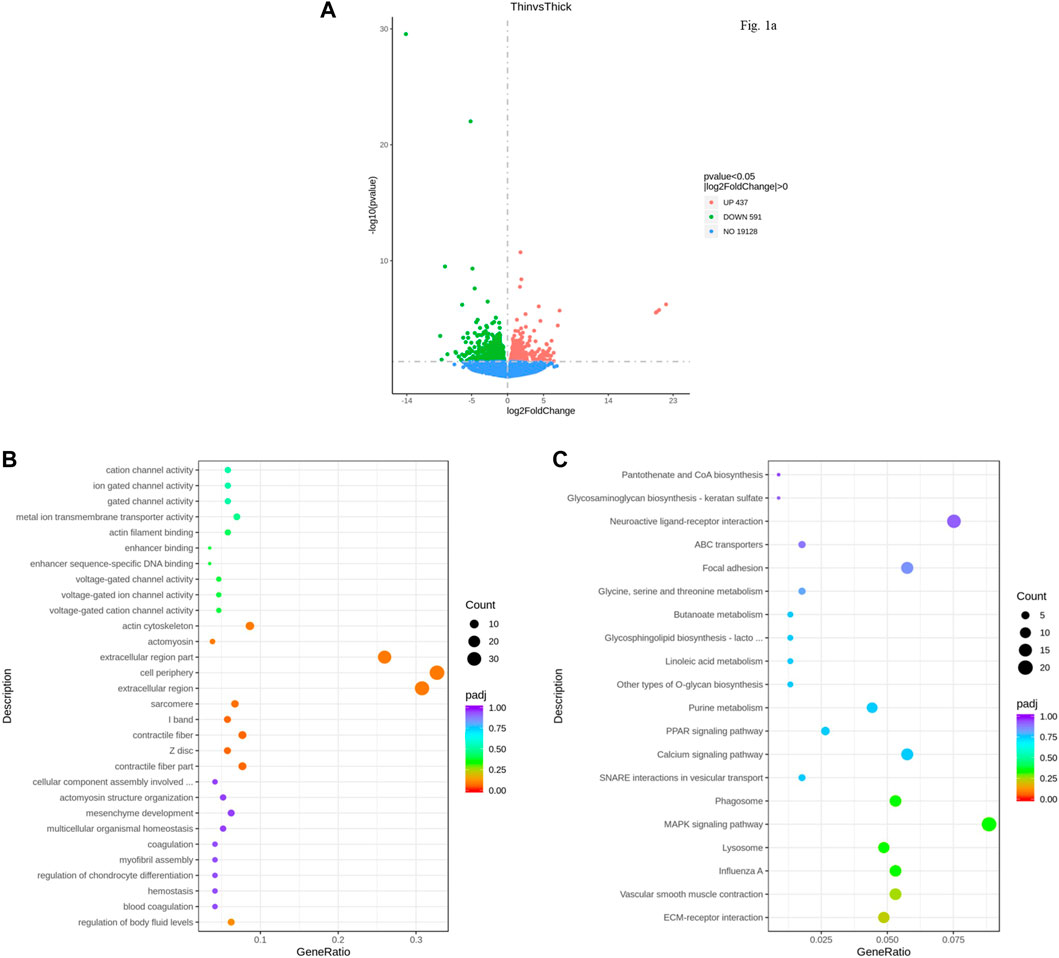
FIGURE 1. Differentially expressed genes (DEGs) identified for the uterus between HE and LE hens. (A). Volcano plot of DEGs between HE and LE hens; (B). GO analysis result of DEGs between HE and LE hens; (C). KEGG analysis result of DEGs between HE and LE hens.
GO analysis of the DEGs of the six uterus samples indicated that they were enriched in cellular component, biological process, and molecular function for terms such as cell periphery, extracellular region, and actin cytoskeleton (Figure 1B). The top 20 results from KEGG analysis showed that some vital pathways related to calcium metabolism, cell proliferation, and apoptosis could also be influenced by the pathway in which these DEGs in the uterus were enriched, including the calcium signaling pathway, MAPK signaling pathway, and PPAR signaling pathway (Figure 1C).
To elucidate the mechanism underlying differences in eggshell strength, the differentially expressed proteins of the HE and LE groups were detected by TMT proteomics. Totally, 6,786 proteins were identified. There were 270 differentially expressed proteins between the LE and HE groups that were significantly different (Supplementary Table S4), including 161 upregulated proteins and 109 downregulated proteins (LE vs. HE).
GO analysis showed that the DEPs were primarily enriched in biological processes, molecular functions, and cellular components, including calcium ion binding, molecular function regulators, and extracellular regions (Figure 2A). The top 20 pathways of KEGG enrichment analysis further indicated that these DEPs were most enriched in some vital pathways related to processes such as calcium metabolism, hormone and amino acid biosynthesis, and cell proliferation and apoptosis; these proteins were involved in the calcium signaling pathway, cell adhesion molecules (CAMs), ECM–receptor interactions, the p53 signaling pathway, biosynthesis of amino acids, and steroid hormone biosynthesis (Figure 2B).
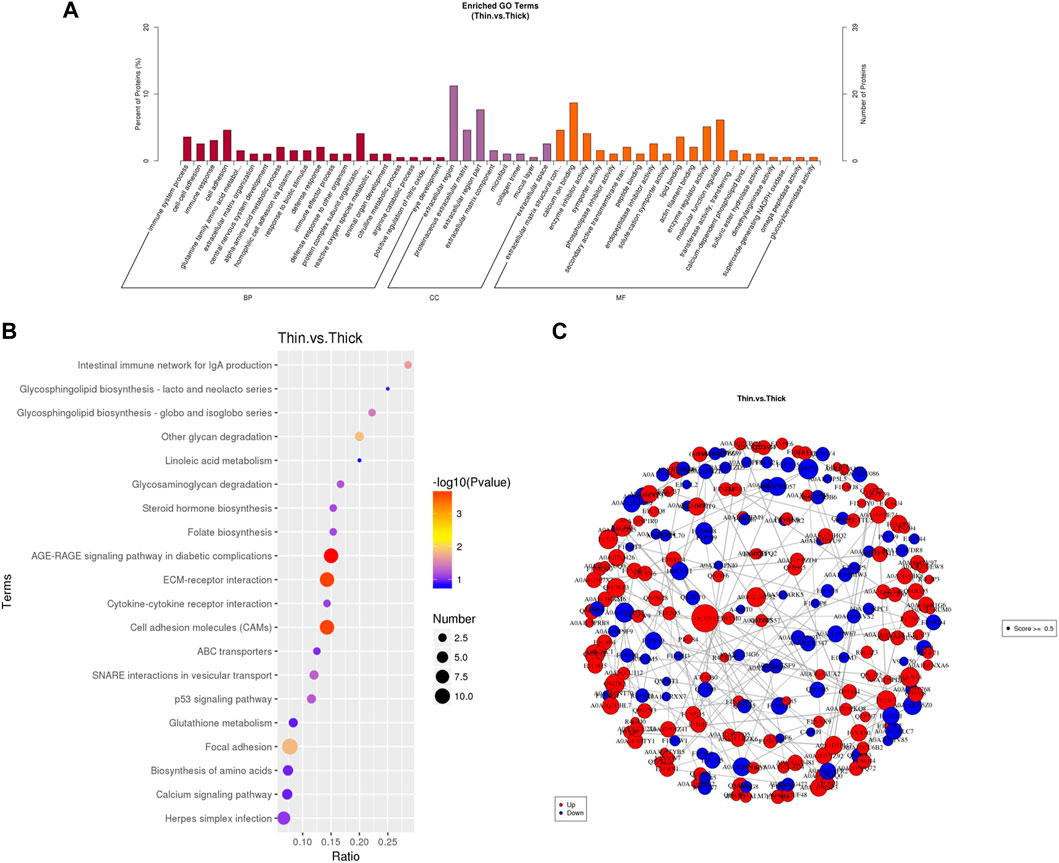
FIGURE 2. Differentially expressed proteins (DEGs) identified for the uterus between HE and LE hens. (A) GO analysis result of DEPs between HE and LE hens; (B) top 20 results of KEGG analysis of DEPs between HE and LE hens; (C) protein–protein interaction network of the DEPs.
The protein–protein interaction (PPI) relationship was also revealed by the STRING database. To further prove the function of DEPs, we constructed a protein–protein interaction network of the DEPs (Figure 2C). Most of those genes were associated with each other.
To elucidate the relationship between the transcriptome and proteome in the HE and LE groups, the two different omics datasets were integrated and analyzed. The results indicated that the number of DEGs and DEPs varied considerably in different eggshell-strength groups (Figures 3A–C), and some genes (or proteins) were differentially expressed at the gene level but not at the protein level, possibly because they were late-response genes. There were 20 genes and proteins detected at both the gene and protein levels (Table 2), and the regulation trend in mRNA and protein levels was matched. The GO enrichment analysis results showed that the correlated-expression DEGs/DEPs were primarily enriched in processes such as transporter activity, molecular function regulation, single-organism processes, membrane, and structural molecule activity (Figure 3D); meanwhile, KEGG enrichment analysis results showed that the correlated-expression DEGs/DEPs were primarily enriched in such processes as metabolic pathways, the p53 signaling pathway, focal adhesion, the calcium signaling pathway, and SNARE interactions in vesicular transport (Figure 3E).
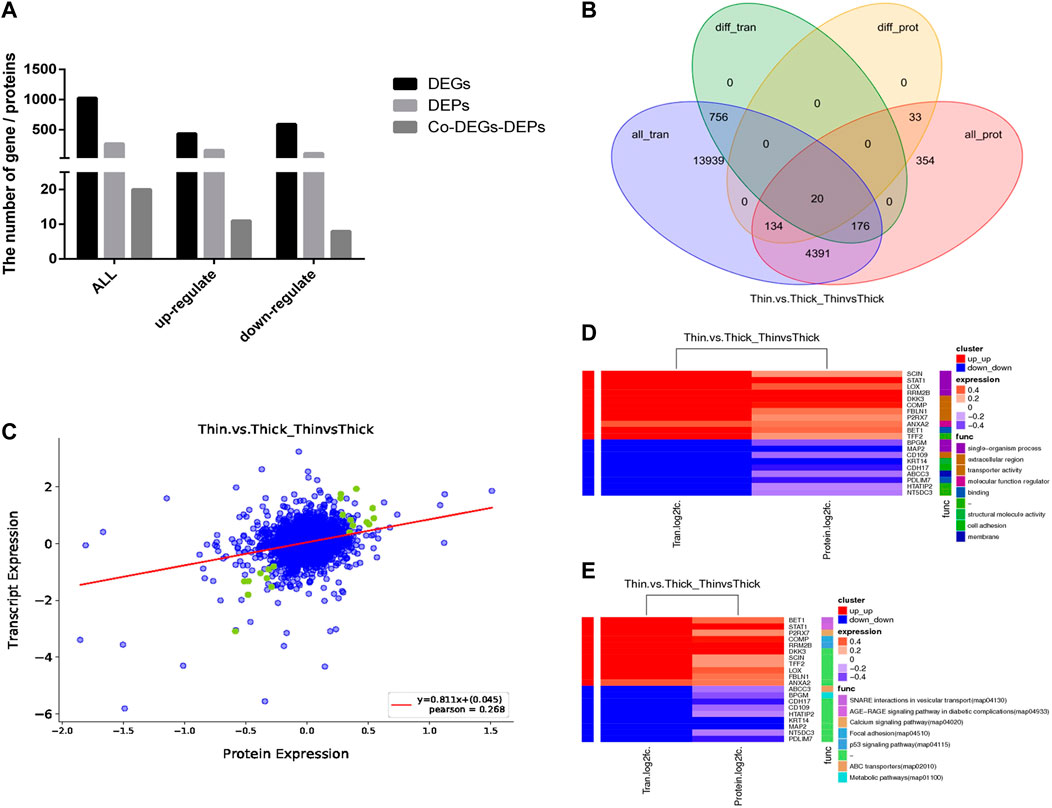
FIGURE 3. Integration and analysis of transcriptome and proteome between HE and LE groups. (A). DEGs, DEPs, and Co-DEGs-DEPs between HE and LE groups; (B). Venn diagram for DEGs and DEPs between HE and LE groups; (C). Corr_plot of RNA-seq and TMT between HE and LE groups; (D). Share GO analysis of RNA-seq and TMT between HE and LE groups; (E). Share KEGG analysis of RNA-seq and TMT between HE and LE groups.
The differentially expressed genes (DEGs) and differentially expressed proteins (DEPs) between LE and HE hens were identified. Seven upregulated and seven downregulated genes were randomly selected for verification via Q-PCR and Western blot, and eight proteins that were co-expressed at the mRNA and protein levels were randomly selected for verification by Western blotting. The results are shown in Figure 4A,B and supplement Figure 1. The log2 fold changes (LE/HE) were calculated based on the RNA-seq and Q-PCR results for DEGs and the TMT and Western blotting results for DEPs. The expression trends indicated that the two methods produced consistent results. Additionally, we investigated the relative expression levels of KRT14, ANXA2, and DKK3 in different chicken tissues (i.e., heart, liver, spleen, lung, kidney, muscle, and uterus) (Figures 4C–E). In particular, the KRT14, ANXA2, and DKK3 genes exhibited higher expression levels in the uterus than in other tissues, and these genes were expressed at the lowest levels in the spleens and livers of hens. Thus, the tissue expression profile of the KRT14, ANXA2, and DKK3 genes demonstrated tissue specificity.
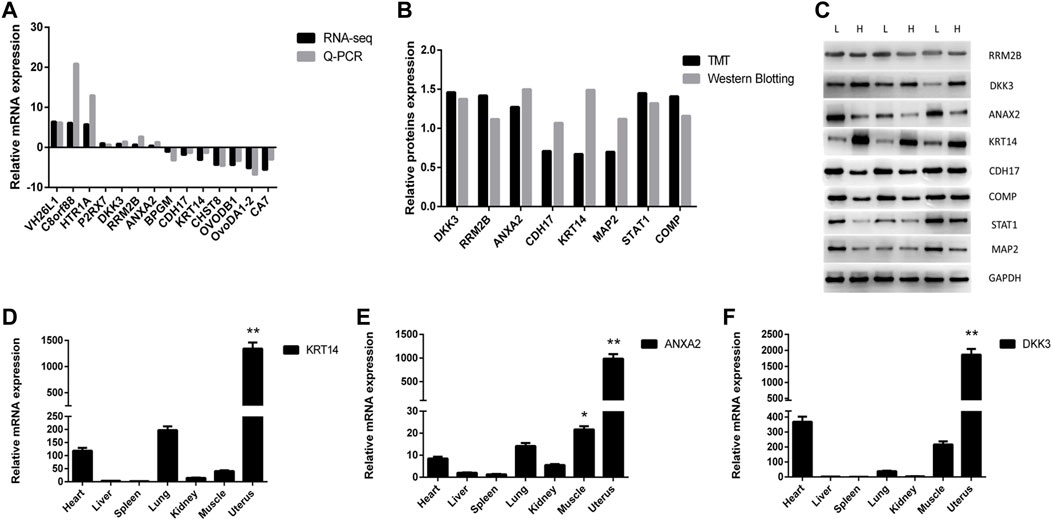
FIGURE 4. Identification and verification of differentially expressed gene and protein. (A). Changes in DEG expression validated by qRT-PCR. Comparison results of the 14 mRNAs using the qRT-PCR and RNA-seq; (B) changes of DEP expression validated by Western Blot. Comparison results of the eight proteins using the Western Blotting and TMT; (C). Western Blot result of part DEPs; (D) relative expression of KRT14 in different tissues; (E) relative expression of ANXA2 in different tissues; (F) relative expression of DKK3 in different tissues.
KRT14 is one of the genes with a large difference between the hard-eggshell group and the weak-eggshell group. It was differently expressed both in transcription and protein level. Also, the fold difference of KRT14 was the largest. So, KRT14 was selected for further study.
To verify the effect of the KRT14 gene on eggshell quality, the gene expressions of eggshell matrix proteins, calcium transport, and calcium metabolism-related proteins in uterine epithelial cells were detected by Q-PCR. The results showed that the KRT14 expression levels in the uterine epithelial cells were significantly increased after transfection with KRT14-pcDNA3.1 (KRT14) (Figures 5A–E). After the overexpression of the KRT14 gene, the mRNA expressions of KRT14, ovocleididin-116 (OC116) (the matrix protein), calbindin 1 (CALB1) (the calcium transport protein), and ADP-ribosyl cyclase 2 (also known as BST1, CD157) (calcium metabolism-related protein) were significantly increased (p < 0.01); the mRNA expressions of ovocleididin-17 (OC17) were significantly decreased (p < 0.01).
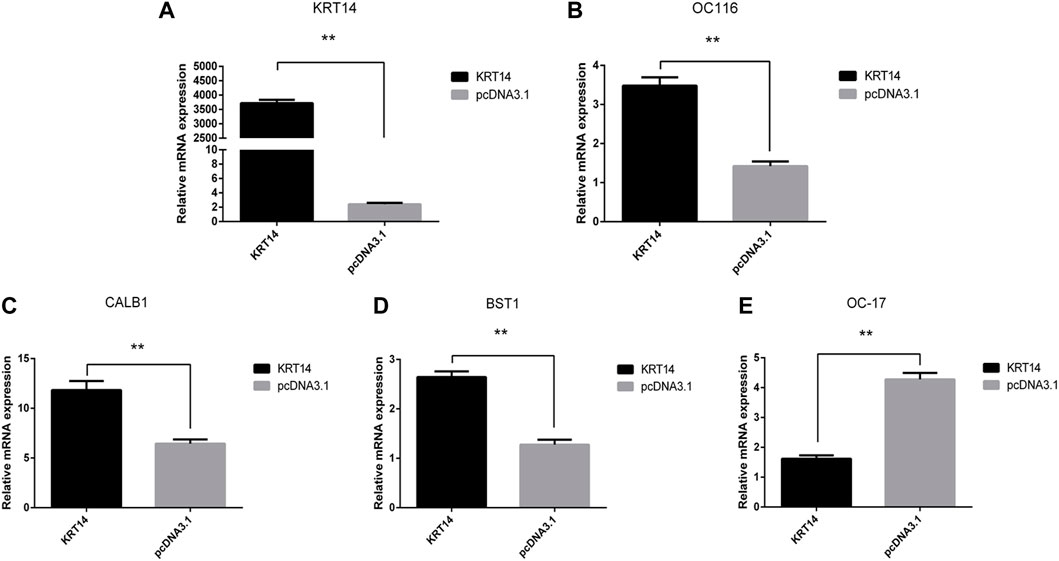
FIGURE 5. Effect of KRT14 on calcium metabolism and deposition of calcium carbonate in eggshell. (A–E): Expression levels of KRT14, OC116, CALB1, BST1, and OC17 mRNA.
Eggshell formation can be divided into several stages (Li et al., 2018). The mammillary knob layer is the first calcified layer, which is formatted with the formation of the outer surface of the outer shell membrane, and its tips are embedded in the outer shell membrane (Carnarius et al., 1996). The palisade layer formed with the mammillary knobs fuse. The vertical crystalline layer consists of short crystals running perpendicular to the shell membrane which is the last calcified layer to be deposited (Solomon, 1991; Hunton, 2005). Previous research studies have shown that shell quality is dependent on the mammillary layer (Bain, 1992), and column organization of the palisade layer is one of the major determinants of rigidity of the shell, strength, and shell resistance of the eggs (Fathi et al., 2007). Large amounts of calcium ions and matrix proteins are required when eggshells are calcified in the uterus. During the calcification periods (including initiation, growth, and termination), calcium metabolism and uterine proteins exhibit changes. The quality of eggshell is determined by its ultrastructure (Rodriguez-Navarro et al., 2002), especially the palisade layer (Meyer et al., 1973). Therefore, previous studies mainly focused on the growth period when the palisade layer is formed (Jonchère et al., 2010; Brionne et al., 2014).
It is reported that the initiation stage of calcification determines the strength of the eggshell (Zhang et al., 2019). The slit guidance ligand 3 (SLIT3) was validated as a candidate gene for eggshell strength (Gao et al., 2022). However, during synchronous calcification periods, few differences in the uterine between hens with high eggshell breaking strength and those with low eggshell breaking strength have been reported, and the genetic regulation of calcification and eggshell strength are still poorly understood. In this study, the differentially expressed gene and differentially expressed protein were identified in the uteri of LE and HE hens. The results showed that the numbers of DEPs were significantly lower than those of DEGs. Fewer proteins were detected than genes in the Venn diagram analysis, probably due to modification and activation at the protein level, the short length of the sample treatment period or the limitations of protein detection technologies (Ma et al., 2020).
The KEGG had been used to classify DEGs and DEPs. The results showed that most DEGs and DEPs were enriched in vital pathways related to calcium metabolism, hormone, amino acid biosynthesis, cell proliferation, or apoptosis. Thus, we hypothesized that quality of eggshells is closely related to deposition of calcium ions. The GO analysis showed that many DEGs and DEPs were enriched in ion transport functions or cytoskeletal functions associated with eggshell calcification. These results implied that there were differences in ion transport between the uteri of hens producing high- or low-breaking-strength eggshells during normal calcification, which helped clarify the eggshell calcification process. In addition to the common ion transport genes found in previous reports, some novel genes were also described in this study (e.g., ANXA2, DKK3, and KRT14).
Annexin A2 (ANXA2) is a multifunctional calcium (Ca) and phospholipid-binding protein that is expressed in a wide spectrum of cells, including those participating in the inflammatory response (Dallacasagrande and Hajjar, 2020). Dickkopf 3 (DKK3) is a secreted protein that belongs to the Dkk family, and it is encoded by the orthologous gene REIC. It was reported that DKK3 is a physiological ER stressor in the mouse adrenal gland (Fujita et al., 2020). Keratin-14 (KRT14) is a key regulator of spheroid integrity, mesothelial attachment, and invasion into the submesothelial matrix. Keratin-14 (KRT14)-positive leader cells mediate mesothelial clearance and invasion by ovarian cancer cells (Bilandzic et al., 2019). In this study, ANXA2, KRT14, and DKK3 were determined to be significantly upregulated in LE hens compared to HE hens. In addition, the mRNA expression levels of the ANXA2, KRT14, and DKK3 genes were significantly elevated in the uterus. So, it was presumed that ANXA2, KRT14, and DKK3 may be related to the quality of eggshells.
Eggshell biomineralization is an extremely complex biological process, which is regulated by multiple genes and signal pathways (Sah et al., 2018; Khan et al., 2019). Ovocleididin-17 is the most vital protein regulating the deposition of calcium carbonate in eggshells. It plays an important role in the formation of eggshells and regulates the formation process of calcite crystals in the eggshell matrix (Maxwell and Hincke, 2013). Ovocleididin-116 is a major protein in chicken eggshell matrices (Mann et al., 2006), which is localized throughout the palisade layer and the mammillary layer (Hincke et al., 1999). In vitro crystallization experiments indicated a direct interaction of OC116 with calcium carbonate (Hernández-Hernández et al., 2008). Calbindin 1 (CALB1) is the carrier of calcium ions in uterine epithelial cells (Sun et al., 2016). During the calcification process, the expression of the CALB1 gene in the uterus was increased significantly in order to meet the mineralized calcium requirement of the eggshell (Jonchère et al., 2012). ADP-ribosyl cyclase 2, also known as BST1 or CD157, could produce cyclic ADP‐ribose involved in regulating calcium homeostasis and plays an important regulatory role in calcium metabolism (Lee, 2001; Yamamoto‐Katayama et al., 2002). In this study, after the overexpression of the KRT14 gene, the expression of OC116, CALB1, and BST1 was significantly increased, while the expression of OC17 was significantly decreased. The abovementioned results suggested that KRT14 may promote calcium metabolism and deposition of calcium carbonate in eggshells.
Based on the transcriptome and proteome analysis, our results were consistent with those of previous studies, which indicated that the initiation period of calcification determines the eggshell strength in the uterus. During normal calcification, there are differences in ion transport between the uterus of hens producing high-breaking-strength eggshells and those producing low-breaking-strength eggshells, which may help elucidate the eggshell calcification process. KRT14 may promote calcium metabolism and the deposition of calcium carbonate in eggshells.
The data that support the findings of this study are openly available in NCBI Sequence Read Archive at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA660090, SRA reference number PRJNA660090. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD021449.
The animal study was reviewed and approved by The Institute of Animal Husbandry and Veterinary of Hubei Academy of Agricultural Science.
YW and JP conceived and designed the study; YS, HZ, AP, and HX performed the experiments; YP, JS, and ZL collected the tissue samples and tested the egg quality; YW analyzed the data and wrote the manuscript; and JP and JD revised the manuscript.
This research was supported by the National Natural Science Foundation of China (32072709), Hubei Science and Technology Major Project (2020ABA016), China Agriculture Research System of MOF and MARA (CARS-40-S15), Leading Talents Program of Hubei Academy of Agricultural Sciences (L2018017), and the Major R & D projects of Hubei Agricultural Innovation Center (2020-620-000-002-01).
We thank the reviewers for their critical reading and discussion of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.927670/full#supplementary-material
HE, hard-eggshell; LE, week-eggshell; DEGs, differentially expressed genes; DEPs, differentially expressed proteins; ANXA2, annexin A2; DKK3, Dickkopf 3; KRT14, keratin-14
Ahmed, A. M., Rodriguez-Navarro, A. B., Vidal, M. L., Gautron, J., García-Ruiz, J. M., and Nys, Y. (2005). Changes in eggshell mechanical properties, crystallographic texture and in matrix proteins induced by moult in hens. Br. Poult. Sci. 46, 268–279. doi:10.1080/00071660500065425
Bain, M. M. (1992). Eggshell strength: A relationship between the mechanism of failure and the ultrastructural organisation of the mammillary layer. Br. Poult. Sci. 33 (2), 303–319. doi:10.1080/00071669208417469
Bain, M. M., Nys, Y., and Dunn, I. C. (2016). Increasing persistency in lay and stabilising egg quality in longer laying cycles. What are the challenges? Br. Poult. Sci. 57, 330–338. doi:10.1080/00071668.2016.1161727
Bilandzic, M., Rainczuk, A., Green, E., Fairweather, N., Jobling, T. W., Plebanski, M., et al. (2019). Keratin-14 (KRT14) positive leader cells mediate mesothelial clearance and invasion by ovarian cancer cells. Cancers (Basel) 11 (9), 1228. doi:10.3390/cancers11091228
Bolling, M. C., Lemmink, H. H., Jansen, G., and Jonkman, M. F. (2011). Mutations in KRT5 and KRT14 cause epidermolysis bullosa simplex in 75 % of the patients. Br. J. Dermatol. 164 (3), 637–644. doi:10.1111/j.1365-2133.2010.10146.x
Brionne, A., Nys, Y., Hennequet-Antier, C., and Gautron, J. (2014). Hen uterine gene expression profiling during eggshell formation reveals putative proteins involved in the supply of minerals or in the shell mineralization process. BMC Genomics 15, 220. doi:10.1186/1471-2164-15-220
Carnarius, K. M., Conrad, K. M., Mast, M. G., and Macneil, J. H. (1996). Relationship of eggshell ultrastructure and shell strength to the soundness of shell eggs. Poult. Sci. 75, 656–663. doi:10.3382/ps.0750656
Crosara, F., Pereira, V. J., Lellis, C. G., Barra, K. C., Fernandes, E. A., Souza, L., et al. (2019). Is the eggshell quality influenced by the egg weight or the breeder age? Braz. J. Poult. Sci. 21 (2), 001–008. doi:10.1590/1806-9061-2018-0896
Dallacasagrande, V., and Hajjar, K. A. (2020). Annexin A2 in inflammation and host defense. Cells 9 (6), 1499. doi:10.3390/cells9061499
Dou, Y., Cha, D. J., Franklin, J. L., Higginbotham, J. N., Jeppesen, D. K., Weaver, A. M., et al. (2016). Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 6, 37982. doi:10.1038/srep37982
Fathi, M. M., El-Dein, A. Z., El-Safty, S. A., and Radwan, L. M. (2007). Using scanning electron microscopy to detect the ultrastructural variations in eggshell quality of Fayoumi and Dandarawi chicken breeds. Int. J. Poult. Sci. 6 (4), 236–241. doi:10.3923/ijps.2007.236.241
Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., et al. (2013). STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41 (D1), 808–815. doi:10.1093/nar/gks1094
Fujita, H., Bando, T., Oyadomari, S., Ochiai, K., Watanabe, M., Kumon, H., et al. (2020). Dkk3/REIC, an N-glycosylated protein, is a physiological endoplasmic reticulum stress inducer in the mouse adrenal gland. Acta Med. Okayama 74 (3), 199–208. doi:10.18926/AMO/59950
Gao, J., Xu, W., Zeng, T., Tian, Y., Wu, C., Liu, S., et al. (2022). Genome-wide association study of egg-laying traits and egg quality in LingKun chickens. Front. Vet. Sci. 9, 877739. doi:10.3389/fvets.2022.877739
Gautron, J., Hincke, M. T., Mann, K., PanheleuxM., , BainM., , McKee, M. D., et al. (2001). Ovocalyxin-32, a novel chicken eggshell matrix protein - isolation, amino acid sequencing, cloning, andimmunocytochemical localization. J. Biol. Chem. 276 (42), 39243–39252. doi:10.1074/jbc.M104543200
Gautron, J., Murayama, E., Vignal, A., Morisson, M., McKee, M. D., Réhault, S., et al. (2007). Cloning of Ovocalyxin-36, a novel chicken eggshell protein related to lipopolysaccharide-binding proteins, bactericidal permeability-increasing proteins, and plunc family proteins. J. Biol. Chem. 282 (8), 5273–5286. doi:10.1074/jbc.M610294200
Hernández-Hernández, A., Gómez-Morales, J., Rodríguez-Navarro, A. B., Gautron, J., Nys, Y., and García-Ruiz, J. M. (2008). Identification of some active proteins in the process of hen eggshell formation. Cryst. Growth Des. 8, 4330–4339. doi:10.1021/cg800786s
Hincke, M. T., Gautron, J. L., Charles, P. W. T., McKee, M. D., and Nys, Y. (1999). Molecular cloning and ultrastructural localization of the core protein of an eggshell matrix proteoglycan, Ovocleidin-116. J. Biol. Chem. 274 (46), 32915–32923. doi:10.1074/jbc.274.46.32915
Hincke, M. T., Nys, Y., Gautron, J., Mann, K., Rodriguez-Navarro, A. B., and McKee, M. D. (2012). The eggshell: Structure, composition and mineralization. Front. Biosci. 17 (4), 1266–1280. doi:10.2741/3985
Hincke, M. T., Tsang, C., Courtney, M., Hill, V., and Narbaitz, R. (1995). Purification and immunochemistry of a soluble matrix protein of the chicken eggshell (ovocleidin 17). Calcif. Tissue Int. 56 (6), 578–583. doi:10.1007/BF00298593
Hrnčár, C., Gič, D., and Bujko, J. (2012). The effect of eggshell quality on hatchability of pure chicken breeds. Sci. Pap. Animal Sci. Biotechnol. 45 (2), 415–418.
Huang, da W., Sherman, B. T., and Lempicki, R. A. (2009). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 (1), 1–13. doi:10.1093/nar/gkn923
Hunton, P. (2005). Research on eggshell structure and quality: An historical overview. Rev. Bras. Cienc. Avic. 7, 67–71. doi:10.1590/s1516-635x2005000200001
Jankowski, M., Wertheim-Tysarowska, K., Jakubowski, R., Sota, J., and Czajkowski, R. (2014). Novel KRT14 mutation causing epidermolysis bullosa simplex with variable phenotype. Exp. Dermatol. 23 (9), 684–687. doi:10.1111/exd.12478
Jonchère, V., Brionne, A., Gautron, J., and Nys, Y. (2012). Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC Physiol. 12 (1), 10. doi:10.1186/1472-6793-12-10
Jonchère, V., Réhault-Godbert, S., Hennequet-Antier, C., Cabau, C., Sibut, V., Cogburn, L. A., et al. (2010). Gene expression profiling to identify eggshell proteins involved in physical defense of the chicken egg. BMC Genomics 11 (1), 57. doi:10.1186/1471-2164-11-57
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi:10.1093/bioinformatics/btu031
Khan, S., Wu, S. B., and Roberts, J. (2019). RNA-sequencing analysis of shell gland shows differences in gene expression profile at two time-points of eggshell formation in laying chickens. BMC Genomics 20 (1), 89. doi:10.1186/s12864-019-5460-4
Lee, H. C. (2001). Physiological functions of cyclic ADP‐ribose and NAADP as calcium messengers. Annu. Rev. Pharmacol. Toxicol. 41, 317–345. doi:10.1146/annurev.pharmtox.41.1.317
Li, L. L., Zhang, N. N., Gong, Y. J., Zhou, M. Y., Zhan, H. Q., and Zou, X. T. (2018). Effects of dietary Mn-methionine supplementation on the egg quality of laying hens. Poult. Sci. 97 (1), 247–254. doi:10.3382/ps/pex301
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2− Δ Δ CT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Ma, Q., Shi, C., Su, C., and Liu, Y. (2020). Complementary analyses of the transcriptome and iTRAQ proteome revealed mechanism of ethylene dependent salt response in bread wheat (Triticum aestivum L.). Food Chem. 325, 126866. doi:10.1016/j.foodchem.2020.126866
Mann, K., Maček, B., and Olsen, J. V. (2006). Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics 6, 3801–3810. doi:10.1002/pmic.200600120
Mann, K. (2015). The calcified eggshell matrix proteome of a songbird, the zebra finch (Taeniopygia guttata). Proteome Sci. 13, 29. doi:10.1186/s12953-015-0086-1
Marie, P., Labas, V., Brionne, A., Harichaux, G., Hennequet-Antier, C., Nys, Y., et al. (2014). Data set for the proteomic inventory and quantitative analysis of chicken uterine fluid during eggshell biomineralization. Data Brief. 1 (C), 65–69. doi:10.1016/j.dib.2014.09.006
Marie, P., Labas, V., Brionne, A., Harichaux, G., Hennequet-Antier, C., Nys, Y., et al. (2015). Quantitative proteomics and bioinformatic analysis provide new insight into protein function during avian eggshell biomineralization. J. Proteomics 113, 178–193. doi:10.1016/j.jprot.2014.09.024
Maxwell, T., and Hincke, MT (2013). Understanding control of calcitic biomineralization proteomics to the rescue. Proteomics 13 (23-24), 3369–3370. doi:10.1002/pmic.201300471
Meyer, R., Baker, R. C., and Scott, M. L. (1973). Effects of hen egg-shell and other calcium sources upon egg-shell strength and ultrastructure. Poult. Sci. 52 (3), 949–955. doi:10.3382/ps.0520949
Nys, Y., Gautron, J., Garcia-Ruiz, J. M., and Hincke, M. T. (2004). Avian eggshell mineralization: Biochemical and functional characterization of matrix proteins. Comptes Rendus Palevol 3, 549–562. doi:10.1016/j.crpv.2004.08.002
Nys, Y., Hincke, M., Hernandez-Hernandez, A., Rodriguez-Navarro, A., Gomez-Morales, J., Jonchere, V., et al. (2010). Eggshell ultrastructure, properties and the process of mineralization: Involvement of organic matrix in the eggshell fabric. Prod. Animales-Paris- Inst. Natl. Rech. Agronomique- 23, 143–154.
Park, J. A., and Sohn, S. H. (2018). The influence of hen aging on eggshell ultrastructure and shell mineral components. Korean J. Food Sci. Anim. Resour. 38 (5), 1080–1091. doi:10.5851/kosfa.2018.e41
Rodriguez-Navarro, A., Kalin, O., Nys, Y., and Garcia-Ruiz, J. M. (2002). Influence of the microstructure on the shell strength of eggs laid by hens of different ages. Br. Poult. Sci. 433 (3), 395–403. doi:10.1080/00071660120103675
Roland, D. A. (1988). Research note: Egg shell problems: Estimates of incidence and economic impact. Poult. Sci. 67, 1801–1803. doi:10.3382/ps.0671801
Rose-Anne, Romano, Barbara, Birkaya, and Sinha, Satrajit (2007). A functional enhancer of keratin14 is a direct transcriptional target of deltaNp63. J. Invest. Dermatol. 127, 1175–1186. doi:10.1038/sj.jid.5700652
Sah, N., Kuehu, D. L., Khadka, V. S., Deng, Y., Peplowska, K., Jha, R., et al. (2018). RNA sequencing-based analysis of the laying hen uterus revealed the novel genes and biological pathways involved in the eggshell biomineralization. Sci. Rep. 8 (1), 16853. doi:10.1038/s41598-018-35203-y
Sirri, F., Zampiga, M., Berardinelli, A., and Meluzzi, A. (2018). Variability and interaction of some egg physical and eggshell quality attributes during the entire laying hen cycle. Poult. Sci. 97, 1818–1823. doi:10.3382/ps/pex456
Smer, C., Asiye, E. R., and Dner, T. (2019). Keratin 14 is a novel interaction partner of keratinocyte differentiation regulator: Receptor-interacting protein kinase 4. Turkish J. Biol. 43 (4), 225–234. doi:10.3906/biy-1904-37
Stefanello, C., Santos, T. C., Murakami, A. E., Martins, E. N., and Carneiro, T. C. (2014). Productive performance, eggshell quality, and eggshell ultrastructure of laying hens fed diets supplemented with organic trace minerals. Poult. Sci. 93, 104–113. doi:10.3382/ps.2013-03190
Sun, C. J., Duan, Z. Y., Lu-Jiang, Q. U., Zheng, J. X., Yang, N., and Gui-Yun, X. U. (2016). Expression analysis for candidate genes associated with eggshell mechanical property. J. Integr. Agric. 2, 397–402. doi:10.1016/s2095-3119(14)60969-2
Vlčková, J., Tůmová, E., Míková, K., Englmaierová, M., Okrouhlá, M., and Chodová, D. (2019). Changes in the quality of eggs during storage depending on the housing system and the age of hens. Poult. Sci. 98 (11), 6187–6193. doi:10.3382/ps/pez401
Yamamoto‐Katayama, S., Ariyoshi, M., Ishihara, K., Hirano, T., Jingami, H., and Morikawa, K. (2002). Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities. J. Mol. Biol. 316, 711–723. doi:10.1006/jmbi.2001.5386
Zhang, J., Wang, Y., Zhang, C., Xiong, M., Rajput, S. A., Liu, Y., et al. (2019). The differences of gonadal hormones and uterine transcriptome during shell calcification of hens laying hard or weak-shelled eggs. BMC Genomics 20 (1), 707. doi:10.1186/s12864-019-6017-2
Keywords: laying hen, eggshell, transcriptome, proteome, KRT14 gene
Citation: Wu Y, Sun Y, Zhang H, Xiao H, Pan A, Shen J, Pu Y, Liang Z, Du J and Pi J (2022) Multiomic analysis revealed the regulatory role of the KRT14 gene in eggshell quality. Front. Genet. 13:927670. doi: 10.3389/fgene.2022.927670
Received: 24 April 2022; Accepted: 29 August 2022;
Published: 22 September 2022.
Edited by:
Mike McGrew, University of Edinburgh, United KingdomReviewed by:
Hong Li, Henan Agricultural University, ChinaCopyright © 2022 Wu, Sun, Zhang, Xiao, Pan, Shen, Pu, Liang, Du and Pi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinsong Pi, cGlqaW5zb25nMDUyM0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.