
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 15 July 2022
Sec. RNA
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.927541
This article is part of the Research Topic Role of Non-coding RNAs, Metabolites and Extracellular Vesicles in Disease Regulation and Health View all 16 articles
CircRNA E3 ubiquitin protein ligase (ITCH) (circRNA ITCH, circ-ITCH), a stable closed-loop RNA derived from the 20q11.22 region of chromosome 20, is a new circRNA discovered in the cytoplasm in recent decades. Studies have shown that it does not encode proteins, but regulates proteins expression at different levels. It is down-regulated in tumor diseases and is involved in a number of biological activities, including inhibiting cell proliferation, migration, invasion, and promoting apoptosis. It can also alter disease progression in non-tumor disease by affecting the cell cycle, inflammatory response, and critical proteins. Circ-ITCH also holds a lot of promise in terms of tumor and non-tumor clinical diagnosis, prognosis, and targeted therapy. As a result, in order to aid clinical research in the hunt for a new strategy for diagnosing and treating human diseases, this study describes the mechanism of circ-ITCH as well as its clinical implications.
In brief, diseases were categorized into two groups: tumors and non-tumor disorders. Malignant tumors, for example, are a type of incurable polygenic disease that has claimed the lives of millions of individuals worldwide. According to GLOBOCAN 2020, there were 19.3 million new malignant tumor cases and 9.9 million deaths globally in 2020 (Sung et al., 2021). Although surgical resection and advanced therapeutic interventions have improved the 5-year survival rate in patients with early-stage GC, the prognosis for late-stage GC patients remains poor due to uncontrolled tumor cell growth and migration (Lasithiotakis et al., 2014). Non-tumor disorders, such as degenerative, metabolic, congenital, and inflammatory pathologies, make up the great bulk of all pathologies, aside from tumors. As a result, finding effective diagnostic biomarkers and treatment targets is crucial for disease fundamental research.
Benefiting from high-throughput sequencing, researchers can take a nuanced and complete picture of the transcriptome and genome of a species. RNA sequencing (RNA-seq) technology has become one of the important means of transcriptomic studies of high-throughput sequencing, which can discover all RNAs that a particular cell can transcribe in a certain functional state, mainly including mRNAs and non-coding RNAs, while avoiding detection using standard molecular techniques. CircRNAs, which were previously thought to be misspliced products, have lately been shown to have a range of biological regulatory activities owing to the development of RNA-seq (Memczak et al., 2013). Most circular RNAs (circRNAs) are mainly composed of one or more exons encoding known proteins. The 3' and 5' terminals of circRNAs are covalently bonded to form a closed-loop structure, unlike typical linear RNAs. It has no free terminal and is unaffected by RNA exonuclease, resulting in a more stable and difficult-to-degrade copy. CircRNAs is primarily involved in the following four processes: 1) sponging microRNAs (miRNAs) or long noncoding RNAs (lncRNAs) as a competing endogenous RNA (ceRNA); 2) binding RNA binding proteins (RBPs); 3) interfering with gene transcription and splicing regulation; and 4) translating protein/polypeptide (Kristensen et al., 2019). Maass et al. (2017) identified 71 differentially expressed circRNAs in 20 human clinical samples including tissues, blood, proving that circRNAs can express stably in vitro and are associated with multiple diseases, which has good potential for biomarker development.
As a member of the E3 ubiquitin protein ligase (ITCH) HECT family, ITCH can ubiquitinate phosphorylated disheveled-2 (Dvl2), promoting its degradation (Bernassola et al., 2008). Phosphorylated Dvl2 is the upstream target of activating β-catenin in the canonical Wnt pathway, therefore ITCH can block the canonical Wnt pathway, regulating cell cycle (Li et al., 2015). In addition, ITCH can regulate immune responses, epidermal keratinocyte differentiation and receptor trafficking/signaling (Melino et al., 2008). Previous studies have shown that in ITCH−/− mice, some signal proteins (such as Jun family members and Notch) are abnormally accumulated, seriously affecting the autoimmune phenotype (Parravicini et al., 2008). Jun and Notch are also transcription factors that control the maintenance of epidermal stem cells and the regulation of keratinocytes. The degradation of these proteins mediated by ITCH may play a regulatory role in skin biology (Candi et al., 2005), indicating that it has certain potential in radiotherapy protection. Moreover, Sundvall et al. (2008) emphasized the role of pruritus in regulating the endocytosis and protein stability of erbb-4, a receptor belonging to the epidermal growth factor receptor (EGFR)/ErbB family. Therefore, ITCH involves a variety of physiological and pathological regulation through different mechanisms, including regulating Wnt, Jun, Notch, MAPK signaling, immune cells differentiation and EGFR family. CircRNA E3 ubiquitin protein ligase (circRNA ITCH, hereinafter referred to as circ-ITCH), a stable closed-loop RNA with no protein coding ability and derived from the 20q11.22 region of chromosome 20, is a new circRNA discovered in recent decades (Bernassola et al., 2008; Memczak et al., 2013). The initial study reported that circ-ITCH came from exon 6–13 of E3 ubiquitin protein ligase (ITCH) encoding gene (Li et al., 2015), but then it became exon 7–14 in related studies without any explanation (Han et al., 2020), which is a point that needs to be clarified in subsequent experiments. Combining the database (Circular RNA Interactome and circBase) and related literature search, we considered it derived from exons 6–13 (Figure 1), while 7–14 was a writing error in correlative papers. Recently, studies have revealed that circ-ITCH is down-regulated in multiple tumor tissues, and regulates cell proliferation, migration, invasion and apoptosis of malignant tumor, indicating it might be an important tumor suppressor (Li Y. et al., 2020; Ghafouri-Fard et al., 2021; Su et al., 2022). Additionally, the down-regulation can also be seen in the peripheral blood and exosomes of patients. Its low expression has some diagnostic relevance and is linked to negative clinical outcomes (such as tumor sizes, lymph node metastasis and distant metastasis). Furthermore, circ-ITCH plays an essential function in non-tumor illnesses. Understanding its function and mechanism could help clinical researchers discover novel strategies to diagnose and treat a variety of diseases earlier.
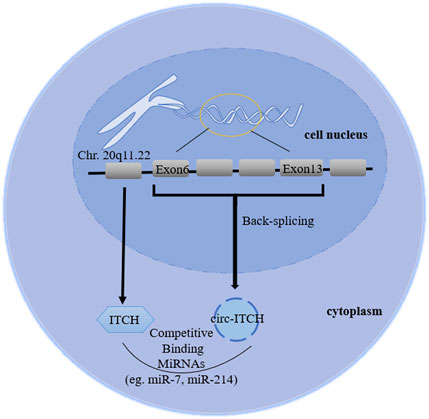
FIGURE 1. Biogenesis diagram of circ-ITCH. The ID number of circ-ITCH is hsa_circ_0001141, whose gene is located in chr 20q11.22, derived from exon 6–13 of ITCH coding gene, formed by back-splicing, and its mature sequence is 873bp.
MiRNA, as a part of the ceRNA network, causes polyadenylation by binding with target sites in the 3'-UTR of mRNA, lowering mRNA stability and interfering with translation to adversely control gene expression (Nawaz et al., 2016). CircRNAs may have biological impacts on tumors by sponging target miRNAs and limiting their function, according to previous studies (Li Y. et al., 2020; Ghafouri-Fard et al., 2021; Su et al., 2022). There is no exception for circ-ITCH. MiRNAs sponge locations of circ-ITCH downstream in malignant tumors now being researched, including miR-7, miR-10, miR-17, miR-20, miR-22, miR-93, miR-106, miR-145, miR-199, miR-214, miR-216, miR-224, miR-421, miR-524, and miR-615 (Huang et al., 2015; Li et al., 2015; Luo et al., 2018b; Hu et al., 2018; Wang et al., 2018; Yang et al., 2018; Lin et al., 2020; Liu et al., 2020; Wang et al., 2021). The specific regulation mechanism of circ-ITCH in a range of malignant tumors is also variable due to sponging distinct miRNAs (Table 1).
The Wnt/β-catenin signaling pathway is highly conserved and important for cell motility, invasion, polarity formation, organogenesis, and cell stemness maintenance (Wei et al., 2012). The Wnt/β-catenin signaling pathway has two pathways: the canonical pathway and the non-canonical pathway, with the conventional pathway being the more common (Wei et al., 2012; Muralidhar et al., 2019). Canonical signal pathways consist of the Wnt protein, Wnt receptor [frizzled family protein (FZD) and low density lipoprotein receptor associated protein-5/6 (LRP-5/6)], Dvl2, β-catenin protein and et al. (Muralidhar et al., 2019) Wnt protein interacts with the FZD receptor on the cell membrane surface in an autocrine or paracrine manner, then recruits LRP-5/6 to create a complex that activates intracellular Dvl2 protein via a phosphorylation cascade. Through its PDZ domain (one of Dvl2's domains), phosphorylated Dvl2 favorably regulates β-catenin protein, facilitating its entry into the nucleus as a transcriptional regulator and activating the expression of downstream target genes including cyclinD1 and c-Myc (Muralidhar et al., 2019).
Through targeting certain miRNAs, Circ-ITCH inhibits the function of matching miRNAs, particularly miRNAs that inhibit linear ITCH. Specifically, researchers showed that reporter gene assays in the presence of circ-ITCH demonstrated that the inhibitory effects of different miRNAs (including miR-7, miR-17, miR-20a, miR-22-3p, and miR-214) were dampened by the co-expression of circ-ITCH, consistent with the “sponge” hypothesis (Huang et al., 2015; Li et al., 2015; Wang et al., 2018). Among these miRNAs, the miR-7 and miR-214 are most important because they can share the binding sites with the 3′-UTR of circ-ITCH and its parental gene ITCH (Verduci et al., 2021). It is worth mentioning that, as one of the most conservative and oldest miRNAs, miR-7 plays different roles in different cancers and participates in many signal pathways involving differentiation, proliferation regulation, apoptosis and migration. In most tumors, its expression is down-regulated because its main activity is to inhibit tumor by inhibiting cell proliferation and survival. However, in lung cancer and oral cancer, its expression is up-regulated as a carcinogen, which is consistent with the research on circ-ITCH (Kora´c et al., 2021). Since circ-ITCH and ITCH share the 3'-UTR of miR-7, they will produce competitive inhibition. When circ-ITCH is up-regulated, the remaining ITCH content in vivo will be up-regulated (Verduci et al., 2021). ITCH can identify and ubiquitinate a range of proteins, the most important of which is phosphorylated Dvl2 (Wei et al., 2012). It is well known that Dvl family proteins are mostly made up of Dvl1-3, with Dvl2 serving as a key scaffold in the canonical Wnt pathway, connecting upstream Wnt protein with downstream β-catenin protein (Wei et al., 2012). As a result, below is the whole regulator mechanism: via sponging miR-7, miR-17, miR-20a, miR-22-3p, and miR-214, circ-ITCH increases ITCH levels. While phosphorylated Dvl2 labeled by ITCH ubiquitin promoted its degradation and inhibited the canonical Wnt pathway. A summary of recent studies has found that in esophageal squamous cell carcinoma (ESCC), colorectal cancer (CRC), lung cancer (LC), three negative breast cancer (TNBC), prostate cancer (PCa), hepatocellular carcinoma (HCC) and gastric cancer (GC), circ-ITCH could up-regulate the expression of linear ITCH via sponging miR-7, miR-17 and miR-20a, thereby inhibiting the canonical Wnt pathway and further suppressing the activation of c-Myc and cyclinD1 (Wan et al., 2016; Wang S. et al., 2019; Li S. et al., 2020; Peng and Wang, 2020; Yang et al., 2020). As widely reported, c-Myc is an oncogene. Its aberrant activation frequently results in unrestricted cell proliferation and immortalization, promoting cell malignancy and tumorigenesis (Glöckner et al., 2002). CyclinD1 (also known as G1/S-specific cyclin D1), on the other hand, regulates the cell cycle and promotes cell proliferation, and is up-regulated in a number of malignancies (Glöckner et al., 2002). In addition, Wang et al. (2018) also confirmed that circ-ITCH can sponge miR-22-3p and increase the expression of CBL in papillary thyroid cancer (PTC), inhibiting cell proliferation and invasion, increasing apoptosis, and repressing PTC progression. CBL is also a member of the E3 ubiquitin ligase family, which can ubiquitinate as well as label β-catenin to promote its destruction and so block the Wnt pathway (Shashar et al., 2016). In most tumors, the glucose transporter 1 (GLUT1) gene is overexpressed. By mediating glucose via the plasma membrane and increasing glucose absorption, it plays a vital function in the early stages of intracellular glucose metabolism and promotes tumor growth (Koch et al., 2015). ITCH can down-regulate the expression of GLUT1 in melanoma, reducing glucose uptake and tumor cell growth, according to Lin et al. (2021), but whether this regulation also involves the Wnt pathway needs to be confirmed in further experiments.
In the progression of many malignancies, activation of the PI3K/Akt pathway and MEK/Erk cascade have been confirmed. Following activation, PI3K activates the Akt protein, which subsequently enters the nucleus and regulates cell proliferation, invasion, migration, metabolic reprogramming, autophagy, and aging, potentially causing malignant tumors (Hermida et al., 2017). The MEK/Erk cascade interacts closely with the PI3K/Akt cascade and is involved in tumor development. After activating signaling pathways, many phosphorylated Erk substrates have been shown to contribute to cell proliferation and invasion (Burotto et al., 2014). Phosphatase and tensin homolog deleted on chromosome ten (PTEN) is a miR-7, miR-22, and miR-224 target that inhibits the PI3K/Akt cascade (Sadri Nahand et al., 2021). P21 is the downstream target of the PI3K/Akt cascade. To promote cell proliferation, activated Akt can phosphorylate p21, blocking its cell cycle arrest function (Cheng et al., 2019). Circ-ITCH sponges these miRNAs to up-regulate PTEN expression in bladder cancer (BCa) and OS, blocking the PI3K/Akt cascade, and up-regulating p21 protein to prevent tumor cell proliferation, migration, invasion and promote apoptosis (Yang et al., 2018; Ren et al., 2019). A recent vitro experimental investigation found that circ-ITCH might further up-regulate PTEN in nasopharyngeal cancer (NPC) via sponging miR-214 (Wang et al., 2022), indicating that it can prevent NPC progression by blocking the PI3K/Akt pathway. Ras p21 protein activator 1 (RASA1) is a regulator of Ras-GDP and GTP, which promotes apoptosis and inhibits angiogenesis, cell proliferation by inhibiting Ras/Raf/MEK/Erk signals cascade (Zhang et al., 2020). RASA1 has been shown to be low expressed in a number of tumors, and miR-14 has been identified to mute it (Zhang et al., 2020). Additionally, Hu et al. (2018) found that in ovarian cancer (OC), circ-ITCH up-regulated RASA1 by sponging miR-145, blocking the PI3K/Akt pathway and MEK/Erk cascade, therefore decreasing tumor cell malignancy. Yan et al. (2020) found a negative connection between circ-ITCH expression and lncRNA HULC expression in OC. Previous research has demonstrated that through down-regulating the miR-125a-3p level, lncRNA HULC can activate the PI3K/Akt/mTOR pathway, promoting the proliferation, migration, and invasion of OC cells (Chu et al., 2019). Therefore, circ-ITCH might compete with lncRNA HULC for miR-125a-3p binding, blocking the PI3K/Akt/mTOR pathway and so acting as an anti-tumor agent. However, the hypothesis needs to be confirmed by further experiments. Published reports by two independent groups of researchers in 2019 and 2021 suggested that circ-ITCH expression decreased in patients’ OS sample tissue, and that circ-ITCH hindered the proliferation, migration, and invasion of OS cells via sponging miR-22 and miR-524 (Ren et al., 2019; Zhou W. et al., 2021). But interestingly, in 2020, Li et al. (Li H. et al., 2020) discovered that the expression of circ-ITCH was up-regulated in U2OS and SJSA-1 cell lines, and enhanced the expression of epidermal growth factor receptor (EGFR) by reducing the level of tumor suppressor miR-7 in OS. Then, when EGFR is overexpressed, it activates the PI3K/Akt and MEK/Erk pathways, promoting OS development. This finding contradicts the findings of two previous investigations. However, there is a flaw in the experiment: it did not verify the degree of circ-ITCH expression in the OS tissue sample. It's possible that this is due to the heterogeneity of OS cell lines or the complexity of studying the regulatory network. Consequently, in the research of circ-ITCH in OS, more parameters should be explored.
Programed cell death receptor 4 (PDCD4) is described as a tumor suppressor, which is often down-regulated in tumors, promoting tumor cell apoptosis and inhibiting its proliferation, invasion and metastasis (Yang et al., 2021). MiR-106b-5p and miR-421 are common upstream targeting miRNAs of PDCD4 and can inhibit its expression (Wang Y. et al., 2019; Yang et al., 2021). Circ-ITCH specifically targets miR-106b-5p and miR-421 in clear cell renal cell carcinoma (ccRCC) and oral squamous cell carcinoma (OSCC) to up-regulate PDCD4 expression and prevent tumor progression, respectively (Hao et al., 2020; Gao et al., 2021). RAS association domain family member 6 (RASSF6) inhibits cell growth and promotes apoptosis in a variety of tumors by interrupting the cell cycle (van der Weyden and Adams, 2007). In OS, circ-ITCH sponges miR-524 to up-regulate RASSF6, inducing OS cell death and limiting its proliferation, according to Zhou et al. (Zhou W. et al., 2021). SAM and SH3 domain containing protein 1 (SASH1) is a tumor-suppressive protein that can regulate cell apoptosis and proliferation (Burgess et al., 2020). Circ-ITCH can suppress glioma growth and invasion by up-regulating SASH1 by targeting miR-106a-5p (Chen et al., 2021). Cytoplasmic polyadenylation element binding protein 3 (CPEB3), a RNA binding protein, plays a tumor-suppressive role though regulating the expression of malignant transformation-related genes through post-transcriptional control (Pichon et al., 2012). In HCC, circ-ITCH binds to miR-421 to prevent CPEB3 down-regulation and tumor growth (Zhao et al., 2021). MafF belonging to the Maf family, a basic leucine zipper (bZIP) transcription factor, has been found to have anti-tumor properties in HCC (Tsuchiya and Oura, 2018). By modulating the miR-224-5p/MAFF axis, Circ-ITCH can also decrease cell growth and increase apoptosis (Wu et al., 2020). Li et al. (2020b) found that circ-ITCH sponges miR-93-5p to up-regulate forkhead box K2 (FoxK2) and block tumor growth in cervical cancer (CC). Simultaneously, FoxK2 expression was dramatically reduced in CC tissues, and miR-93-5p mimic transfection further decreased FoxK2 expression in CC cell lines. FoxK2 deletion improved the capacity of cells transfected with miR-93-5p mimic to invade. Homeobox B13 (HOXB13), according to earlier research, inhibits the cell cycle by promoting the ubiquitination and degradation of cyclinD1 in a variety of malignancies (Hamid et al., 2014). Circ-ITCH restrains PCa cell proliferation, invasion, and migration via sponging miR-17-5p and boosting HOXB13 up-regulation, as well as promoting apoptosis (Wang X. et al., 2019). Furthermore, via targeting miR-197, circ-ITCH can attenuate PCa cell proliferation and increase apoptosis, however the underlying mechanism is uncertain (Yuan et al., 2019). In melanoma, circ-ITCH suppresses cell proliferation and metastasis through sponging miR-660, a previously reported tumor-promoting miRNA, further up-regulating transcription factor cellular promoter 2 (TFCP2) (Zhang et al., 2022). TFCP2, as a cell cycle regulating molecule, mainly plays a tumor suppressor role in melanoma. Its role is mainly to positively regulate the DAPK transcription by binding to the promoter of the death associated protein kinase (DAPK) gene, a tumor suppressor that is silenced in many cancers (Kotarba et al., 2018). Besides, TFCP2 can positively regulate the transcription of p21CIP1, a well-known cell cycle inhibitor protein (Goto et al., 2016; Kotarba et al., 2018). In addition, previous studies have shown that Klotho can inhibit the IGF-1/insulin pathway and regulate the expression of Bax/Bcl-2, thereby inhibiting cell proliferation and promoting apoptosis in A549 cells (Chen et al., 2010). While recently Wang et al. (Wang et al., 2021) found that circ-ITCH boosted Klotho expression though acting as a miR-199a-5p sponge, thus, suggesting that the function of circ-ITCH in GC may involve cell cycle-related regulatory proteins. In short, recent studies have discovered that circ-ITCH regulates the expression level of a number of cell cycle-related proteins, as a ceRNA, to induce tumor cell apoptosis and limit tumor cell growth, thereby acting as an anti-tumor agent.
Epithelial mesenchymal transition (EMT) is a process that occurs in almost all forms of tumors and is linked to tumor incidence, invasion, metastasis, recurrence, and medication resistance (Iwatsuki et al., 2010). E-cadherin and vimentin are two crucial proteins that are frequently used as EMT indicators (Iwatsuki et al., 2010). E-cadherin, which is encoded by CDH1, is involved in EMT and is linked to tumor invasion and diffusion (Ye et al., 2012). Vimentin, in particular, promotes EMT, whereas E-cadherin opposes it. Lin et al. (2020) discovered that circ-ITCH inhibits EMT in OC by increasing CDH1 expression via sponging miR-106a. Guo et al. (2022) revealed that the tumor suppressor role of circ-ITCH in HCC is associated with regulating EMT progression through KEGG enrichment analysis, and its regulation function is associated with sponging miR-184. It is well known that Klotho-mediated regulation of cellular EMT is a way to regulate tumor progression (Chen et al., 2019). Therefore, by modulating the miR-199a-5p/Klotho axis, circ-ITCH can block EMT and delay tumor growth in GC (Wang et al., 2021). In addition, earlier research has demonstrated that the Wnt pathway is important for regulating EMT (Kumari et al., 2021). As a result, circ-ITCH's modulation of the Wnt pathway might have an impact on the downstream EMT process, but more research is needed to confirm this.
In summary, circ-ITCH modulates downstream targets including the Wnt pathway, the PI3K/Akt cascade, the MEK/Erk cascade, cell cycle-related proteins, and EMT process via sponging different miRNAs, performing an anti-tumor effect in a range of malignant tumors (Figure 2).
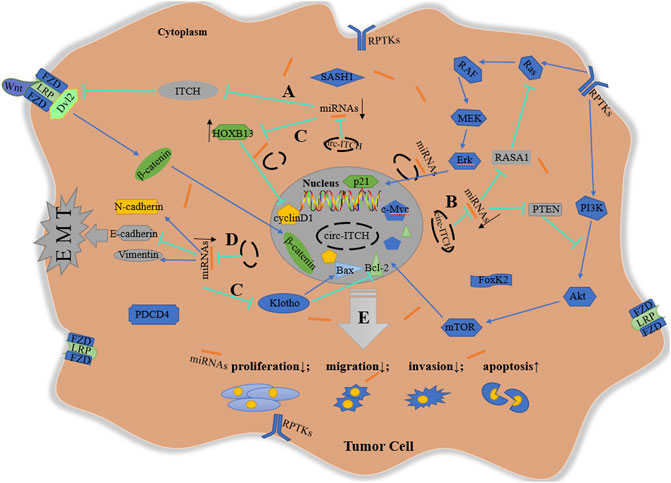
FIGURE 2. Schematic diagram of anti-tumor mechanism of circ-ITCH. (A) via sponging miR-7, miR-17, miR-20a, miR-22-3p, and miR-214, circ-ITCH increases ITCH levels to inhibit Wnt signaling pathway in ESCC, CRC, LC, TNBC, PCa, HCC and GC; (B) via sponging miR-7, miR-14, miR-22, miR-145 and miR-224, circ-ITCH activate the inhibitory proteins (RASA1 and PTEN) of Erk and PI3K cascade to suppress the activation of these signaling pathway in BCa, OS, NPC and OC; (C) via sponging miR-17-5p, miR-421, miR-524, miR-660, miR-106a-5p, miR-224-5p, miR-93-5p and miR-199a-5p, circ-ITCH regulates cell cycle-related proteins to inhibit cell proliferation and promote cell apoptosis in PCa, OS, OSCC, HCC, ccRCC, CC, glioma, melanoma and GC; (D) Via sponging miR-184 and miR-199a-5p, circ-ITCH suppresses EMT process in HCC and GC; (E) In addition, circ-ITCH can directly regulate some proteins without specific target miRNAs. In a word, circ-ITCH plays an anti-tumor role by negatively regulating cell proliferation, invasion, migration, and positively regulating cell apoptosis.
Osteoporosis is a systemic bone disease that causes decreased bone density and quality, disturbed bone microarchitecture, and increased bone fragility, all of which increase the risk of fracture (Compston et al., 2019). Zhong et al. (2021) demonstrated that compared to normal tissues circ-ITCH expression was down-regulated in osteoporosis samples, implying that it may play a protective role in bone degenerative diseases. Specifically, circ-ITCH up-regulated the expression of YAP1 by sponging miR-214. YAP1 is a prominent downstream effector of the Hippo pathway, and its up-regulation can stimulate the differentiation of mesenchymal stem cells into osteoblasts, according to previous research (Lorthongpanich et al., 2019). Moreover, YAP1 stimulates osteogenesis though interacting with β-catenin in osteoblasts (Pan et al., 2018). Taken together, the study found that circ-ITCH might enhance osteogenic differentiation in osteoporosis and ameliorate osteoporosis symptoms in mice (Zhong et al., 2021). Similarly, circ-ITCH expression is up-regulated during periodontal ligament stem cell (PDLSC) osteogenic differentiation and may trigger osteogenic differentiation though regulating MAPK pathway (Gu et al., 2017).
Intervertebral disc degeneration (IDD) is a type of degeneration that can cause a variety of minor or self-limiting symptoms. Spinal discomfort is currently thought to be mostly caused by IDD. Degradation of the extracellular matrix (ECM) and apoptosis of the nucleus pulposus (NP) cells are other key markers of IDD development (Wang et al., 2020). Recently, Zhang et al. (2021) discovered that circ-ITCH might sponge miR-17-5p/SOX4 signaling to positively regulate the activation of Wnt/β-catenin pathway in IDD, causing ECM degradation and NP cell apoptosis. However, this finding contradicts prior findings, particularly in tumor research, in that it activates Wnt/β-catenin pathway. After Wnt/β-catenin activation, Zhang et al. were unable to further illustrate the regulatory mechanism. Combined with earlier research (Zimmerman et al., 2013), we speculate that activating Wnt/β-catenin promotes apoptosis by up-regulating the expression level of pro-apoptotic proteins BIM and Bax while down-regulating the expression level of anti-apoptotic proteins Mcl-1 and Bcl-xl.
CircRNAs have been linked to the development of a number of cardiac diseases, including atherosclerosis, myocardial damage, heart failure, and drug-induced cardiotoxicity, according to research (Min et al., 2021). The current emphasis of circ-ITCH research in cardiac diseases is ischaemia-reperfusion (I/R) injury and doxorubicin-induced cardiotoxicity (DOXIC). In I/R damage, a substantial amount of H2O2 can be created, aggravating oxidative stress injury (Wu et al., 2018). H2O2 caused apoptosis in H9c2 rat cardiac cells and reduced viability, ATP levels, and circ-ITCH expression in a recent study (Zhang and Wang, 2020). Furthermore, H2O2 treatment boosted the expression of Wnt3a, Wnt5a, and β-catenin (Zhang and Wang, 2020). Conversely, in H2O2 pretreatment H9c2 cells, overexpression of circ-ITCH reduced apoptosis and Wnt/β-catenin expression, suggesting that its cardioprotective effect is linked to the inactivation of Wnt/β-catenin signaling pathway in I/R injury (Zhang and Wang, 2020). Specifically, circ-ITCH reduced cardiomyocyte apoptosis in I/R injury by sponging miR-17-5p and then inactivating the Wnt/β-catenin signaling pathway (Zhang and Wang, 2020). Doxorubicin is an effective chemotherapeutic agent, but doxorubicin-treated patients are prone to cardiac toxicity and subsequently develop congestive heart failure (Yeh and Bickford, 2009). The two primary pathogenic processes leading to the pathogenesis of doxycycline have been identified in DOXIC as oxidative stress and DNA damage (Zhang et al., 2012). Recently, Han et al. (2020) discovered that overexpressed circ-ITCH reduces doxorubicin-induced oxidative stress and DNA damage in cells and mitochondria. They also discovered that via sponging miR-330-5p in DOXIC, circ-ITCH upregulated sirtuin 6 (SIRT6), Survivin, and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2a (SERCA2a) (Han et al., 2020). SIRT6 has been found to reduce oxidative stress by activating Nrf2 and SOD2 proteins, two important endogenous anti-oxidant defense molecules (Rezazadeh et al., 2019; Tian et al., 2019). In addition, SIRT6 could also ameliorate DNA damage via activating PARP1, a key DNA repair enzyme (Tian et al., 2019). Reportedly, Survivin could inhibit doxorubicin-induced myocardial apoptosis and fibrosis (Lee et al., 2014). Additionally, SERCA2a could catalyze the hydrolysis of ATP and enhance cardiac contractility by binding to calcium translocating from the cytosol to the lumen of the sarcoplasmic reticulum (Reilly et al., 2001). Thus, circ-ITCH can alleviate DOXIC and has good potential as a therapeutic target in DOXIC.
Diabetic patients’ long-term glucose management is suboptimal, which can lead to diabetic microvascular problems such as diabetic neuropathy, diabetic retinopathy (DR) and diabetic nephropathy (DN). One of the most prevalent microvascular consequences of diabetes, diabetic retinopathy (DR), is a chronic, progressive diabetes mellitus-induced leakage and occlusion of retinal micro-vessels, resulting in a series of fundus lesions. DR is a persistent microvascular inflammation and proliferative neovascularization of the retina (Semeraro et al., 2015). And there is an interaction between the two pathological process (Capitão and Soares, 2016). TNF-α has been shown to be an inflammatory factor that plays a major role in high-glucose environments, where it is linked to vascular inflammation, endothelial dysfunction, oxidative stress, and disruption of the blood-retinal barrier, and contributes to the progression of DR synergistically (Capitão and Soares, 2016). The major enzymes responsible for degrading the ECM, matrix metalloproteinase (MMPs), have been linked to inflammatory disorders and diabetes (Kowluru and Mishra, 2017). Among these MMPs, MMP-2 and MMP-9 were both significantly up-regulated in retinal cells under high glucose conditions (Giebel et al., 2005). By suppressing TNF-α, MMP-2 and MMP-9, Zhou et al. (Zhou L. et al., 2021) revealed that overexpression of circ-ITCH might prevent neovascularization and inflammation, hence delaying DR progression. DN refers to the microvascular consequences of diabetes mellitus, with microalbuminuria at its core, and renal impairment in a subgroup of patients. Long-term chronic inflammation has been linked to the advancement of DN in studies (Wada and Makino, 2013). Previous research has shown that overexpression of SIRT6 could stimulate M2 macrophage transformation, inhibit high glucose-induced mitochondrial dysfunction and cell apoptosis by activating AMPK, all of which help to reduce inflammation in DN (Fan et al., 2019; Ji et al., 2019). A recent study in diabetic mice produced with streptozotocin found that circ-ITCH reduced kidney inflammation and fibrosis through modulating the mir-33a-5p/SIRT6 axis (Liu et al., 2021). The role and mechanism of circ-ITCH in uncomplicated diabetes is also worth exploring, according to current research development.
Hirschsprung's disease (HSCR) is caused by a lack of proliferation and migration of intestinal nerve cells (ENCC), which leads to the absence of peristalsis and colon defecation (Heanue and Pachnis, 2007). It then causes the proximal colon to expand and hypertrophy, eventually resulting in the formation of a megacolon. Rearranged during transfection (RET) has recently been identified as a major regulator of ENCC formation, and inactivating mutations in this gene could result in HSCR (Ohgami et al., 2021). Accumulating evidence indicates that circRNAs are dysregulated and play critical roles in the development of HSCR. Xia et al. (2022) revealed that circ-ITCH expression was dramatically reduced in HSCR tissues, and its overexpression greatly promoted the ability of proliferation and migration of 293T, SH-SY5Y cell lines. Mechanistically, circ-ITCH overexpression activated RET by sponging miR-146b-5p, thereby relieving HSCR progression (Xia et al., 2022).
All in all, in non-tumor diseases such as IDD, osteoporosis, I/R injury, DOXIC, DR, DN, HSCR, and others, circ-ITCH plays a more complex regulatory role. It can, for example, regulate distinct target proteins to cause different states of the same signal pathway. These findings imply that the expression level of circ-ITCH and its regulatory mechanism are disease-specific. The role and mechanism of circ-ITCH in non-tumor illnesses and PDLSC osteogenic development are shown in Table 2.
With the increasing popularity of RT-PCR, diagnosing a growing range of diseases has gotten easier. RT-PCR can also be used to determine the degree of circ-ITCH expression. It also shows that it is practical and convenient because it expresses consistently in sample tissues, peripheral blood, and exosomes from patients. As a result, RT-PCR can be employed in practice to detect the level of circ-ITCH expression in patients’ sample tissues, peripheral blood, or exocrine, allowing for early diagnosis. The clinical significance of circ-ITCH in human diseases is shown in Table 3.
Although surgery, chemoradiotherapy, targeted therapy, and the therapeutic outcome of tumor patients have all improved over time, overall survival and quality of life remain a serious problem for patients. Hence, it is critical to diagnose and treat patients as soon as possible. In recent years, scientists have experimented with many approaches to improve the detection and surveillance of early malignant tumors, including radiation, immunology, and biomarkers. Diagnostic biomarkers have received a lot of attention among them. Many circRNAs, notably circ-ITCH, have shown tremendous promise as diagnostic biomarkers in various investigations (Tang et al., 2019).
Many studies have anticipated and proven the clinical application potential of circ-ITCH as a diagnostic biomarker due to its down-regulation in a variety of malignancies (Huang et al., 2019). In GC, circ-ITCH in tissue and serum-derived exosomes were explored separately for their diagnostic value. Among them, the AUC for detecting circ-ITCH in tissues was 0.7055 (sensitivity: 52.71%, specificity: 74.55%); the AUC in serum-derived exosomes was 0.6538 (sensitivity: 42.42%, specificity: 90.91%) (Wang et al., 2021). In multiple myeloma (MM), the AUC was 0.809 (sensitivity: 59.8%, specificity: 80.0%) (Zhou et al., 2020). In PCa, circ-ITCH showed higher diagnostic value (AUC = 0.812 (95% CI: 0.780–0.845)), and its low expression was linked to a higher probability of lymph node metastases (p = 0.047) and an advanced T stage (p = 0.002) (Huang et al., 2019). Furthermore, the expression of circ-ITCH has been linked to tumor size, tumor grade, TNM stage and clinical stage. Specifically, in OC, the expression of circ-ITCH was linked to tumor size (p = 0.0009) and clinical stage (p = 0.0021) (Lin et al., 2020); in TNBC, it was linked to tumor size (p = 0.016), lymphatic metastasis (p = 0.008) and clinical stage (p = 0.002) (Wang S. et al., 2019); in OSCC, it was correlated with clinical stage (p = 0.027) and lymphatic metastasis (p = 0.035) (Hao et al., 2020); in EOC, it was associated with tumor size (p = 0.005) and International Federation of Gynecology and Obstetrics (FIGO) stage (p < 0.001) (Luo et al., 2018a); in GC, it was correlated with tumor grade (p = 0.02), T stage (p = 0.0216) and lymphatic metastasis (p = 0.034) (Ghasemi et al., 2019; Peng and Wang, 2020; Wang et al., 2021); in NPC, it was correlated with lymphatic metastasis (p = 0.0021), clinical stage (p = 0.0028) and bone metastasis (p = 0.0285) (Wang et al., 2022); in non-small cell lung cancer (NSCL), it was linked to tumor size (p < 0.001), lymphatic metastasis (p < 0.001) and clinical stage (p = 0.003) (Li et al., 2019); in MM, it was correlated with international staging system (ISS) stage (p = 0.036) (Zhou et al., 2020). Guo et al. (2017), on the other hand, discovered that the single nucleotide polymorphisms rs10485505 and rs4911154 of circ-ITCH were strongly related with an elevated risk of HCC, suggesting that it might be employed as a biomarker for HCC susceptibility. Similarly, single nucleotide polymorphisms of rs4911154 of circ-ITCH could aggravate the malignant transformation from thyroid nodule (TN) to thyroid cancer (Guo et al., 2021). Furthermore, circ-ITCH's collaboration with established diagnostic indicators like carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19-9) may also boost diagnostic power. All in all, these findings indicate that it has a wide range of diagnostic utility in a variety of tumors, even in cancer susceptibility prediction, since it’s convenient and non-invasive.
With the rapid growth of incidence rate and mortality rate of malignant tumors, its overall prognosis will be the main determinant of global public health and life expectancy. Surgery is the most effective treatment for malignant tumors, but recurrence and metastasis have a significant impact on the prognosis (Lasithiotakis et al., 2014). Recent studies have found that circ-ITCH is closely linked to clinicopathological characteristics and can be employed as a tumor prognostic biomarker, which will aid in tumor treatment. The expression of circ-ITCH is linked to the prognosis of a range of tumors, including HCC, EOC, PCa, BCa, OC, and so on, according to Kaplan-Meier survival analysis (Guo et al., 2017; Luo et al., 2018a; Yang et al., 2018; Wang S. et al., 2019; Wang X. et al., 2019; Huang et al., 2019; Lin et al., 2020). These findings imply that reduced circ-ITCH expression was associated with lower overall survival (OS) and disease-free survival (DFS). Specifically, decrease in circ-ITCH was associated with worse OS (p = 0.018) and DFS (p = 0.017) in MM patients (Zhou et al., 2020); in NSLC, its down-regulation was correlated with worse OS (p = 0.006) and DFS (p = 0.001) (Li et al., 2019); in PCa, its down-regulation was correlated with worse OS and DFS (both p < 0.001) (Huang et al., 2019); in EOC, its down-regulation was correlated with worse OS (p = 0.003) (Luo et al., 2018a). Furthermore, 1604 patients with various malignancies were included in a meta-analysis, which yielded the same results. Patients with reduced circ-ITCH expression had a lower OS (HR = 2.45, 95% CI: 2.07–2.90, p ≤ 0.01, univariate analysis; HR = 2.69, 95% CI: 1.82–3.96, p ≤ 0.01, multivariate analysis) (Sun et al., 2021). Taken together, these results suggest its promising value as a prognostic biomarker.
Many molecules and signal pathways may be appropriate for targeted therapy as our understanding of tumor development improves. Circ-ITCH is a promising therapeutic target since it has an anti-tumor impact that is connected to a range of substances and pathways, as evidenced by recent studies. Up-regulation of circ-ITCH to inhibit proliferation, invasion and migration of HCC cells, for instance, is one of the mechanisms by which lidocaine treats HCC (Zhao et al., 2021). Besides that, it also has great potential in improving chemoresistance and side effects of chemotherapy. Circ-ITCH can decrease MM cell growth and improve MM cell chemosensitivity to bortezomib (BTZ) (Liu et al., 2020). Furthermore, circ-ITCH can also alleviate the symptom of DOXIC, suggesting that it can be used simultaneously with chemotherapeutic drugs to alleviate chemotherapeutic side effects (Han et al., 2020). At present, noncoding RNA (ncRNA) therapy focuses primarily on alternative and inhibitory therapies (Ning et al., 2019). Circ-ITCH's alternative therapy is projected to play a significant role in tumor therapy since it suppresses tumor cell proliferation, increases apoptosis, and slows tumor growth by targeting a range of pathway molecules.
According to recent research, circ-ITCH also has an important role in non-tumor tissue. These findings, together with circ-ITCH's stable expression in patient tissues and peripheral blood, as well as well-defined regulatory mechanisms, point to its potential as a biomarker. In Hepatitis C virus infection, for instance, circ-ITCH expression was positively correlated with liver enzymes AST, ALT (p < 0.001) and child grade. With AUC = 0.661 (sensitivity: 65 percent, specificity: 70 percent), circ-ITCH has diagnostic significance in plasma of Hepatitis C virus infection (Sharkawi et al., 2022). However, no studies have looked at the possibility of circ-ITCH as predictive biomarkers in non-tumor diseases, and this is currently a blank area of research. Circ-ITCH has been reported to play well-defined regulatory roles in bone illnesses, cardiac diseases, diabetic microangiopathy, and Hirschsprung disease, indicating that they could be therapeutically targeted. Specifically, circ-ITCH replacement therapy appears to have promise as a treatment for DOXIC. Moreover, further experiments with circ-ITCH replacement drugs are also worth studying.
The focus of this review is on how circ-ITCH, a circular RNA, regulates gene expression in the post-transcriptional stage by acting as a sponge for miRNAs, blocking them from binding to their target mRNAs. The role of circ-ITCH in cell proliferation, apoptosis, invasion, migration, and EMT regulation, as well as related signaling pathways, is then explored. Circ-ITCH's potential as a diagnostic and predictive biomarker in tumor and non-tumor diseases is then confirmed. Furthermore, circ-ITCH has a lot of potential in disease treatment because of its well-defined regulatory mechanism, notably in terms of enhancing chemosensitivity and reducing chemotherapy adverse effects in malignant tumor. These discoveries not only illuminate the molecular basis of circ-ITCH, but also pave the path for future clinical applications.
Based on the current research, we put forward some promising future research directions. To begin, the up-regulated biomarker is more ideal for clinical detection, but the down-regulated index can still be employed as long as the critical value is evident. As a result, it is crucial to explore the critical value of circ-ITCH in both patients and healthy people. Then, researchers should increase the sample size and AUC detection as much as possible in order to achieve a more accurate association between circ-ITCH and other classic diagnostic markers (CEA, CA19-9) or prognostic indicators (OS, DFS) (Li et al., 2020c). Furthermore, given its critical role in regulating human diseases, more simulations of activators or carrier administration are needed to validate their in vitro and in vivo effects. While circ-ITCH is low expressed in a number of malignancies, it is unclear which tumor has greater specificity and accuracy about circ-ITCH, meriting additional investigation. At the same time, it is also worthwhile to explore the translational applications of circ-ITCH in chemotherapy because of its demonstrated potential in tumor chemotherapy, including chemo-sensitization and alleviation of side effects. Besides, due to the increase of ITCH concentration and regulating epidermal keratinocyte differentiation, the potential of circ-ITCH in decreasing radiotherapy injury also needs to be developed. Finally, it was recently shown that circRNAs are plentiful and stable in exosomes, that they can be produced under a variety of physiological and pathological conditions, and that they can be detected in circulation and urine (He et al., 2021), all of which require further exploration.
TL and TH wrote a draft of the review, MS was involved in literature search and supervision, GH was involved in original idea and critical revision of the manuscript.
This research was supported by the Jilin Science and Technology Department Medical Department project (20160101092JC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bernassola, F., Karin, M., Ciechanover, A., and Melino, G. (2008). The HECT Family of E3 Ubiquitin Ligases: Multiple Players in Cancer Development. Cancer Cell 14 (1), 10–21. doi:10.1016/j.ccr.2008.06.001
Burgess, J., Bolderson, E., Adams, M., Duijf, P., Zhang, S., Gray, S., et al. (2020). SASH1 Is a Prognostic Indicator and Potential Therapeutic Target in Non-small Cell Lung Cancer. Sci. Rep. 10 (1), 18605. doi:10.1038/s41598-020-75625-1
Burotto, M., Chiou, V., Lee, J., and Kohn, E. (2014). The MAPK Pathway across Different Malignancies: a New Perspective. Cancer 120 (22), 3446–3456. doi:10.1002/cncr.28864
Candi, E., Schmidt, R., and Melino, G. (2005). The Cornified Envelope: a Model of Cell Death in the Skin. Nat. Rev. Mol. Cell Biol. 6 (4), 328–340. doi:10.1038/nrm1619
Capitão, M., and Soares, R. (2016). Angiogenesis and Inflammation Crosstalk in Diabetic Retinopathy. J. Cell. Biochem. 117 (11), 2443–2453. doi:10.1002/jcb.25575
Chen, B., Huang, S., Pisanic, II, T. R., Stark, A., Tao, Y., Cheng, B., et al. (2019). Rab8 GTPase Regulates Klotho-Mediated Inhibition of Cell Growth and Progression by Directly Modulating its Surface Expression in Human Non-small Cell Lung Cancer. EBioMedicine 49, 118–132. doi:10.1016/j.ebiom.2019.10.040
Chen, B., Wang, X., Zhao, W., and Wu, J. (2010). Klotho Inhibits Growth and Promotes Apoptosis in Human Lung Cancer Cell Line A549. J. Exp. Clin. Cancer Res. 29 (1), 99. doi:10.1186/1756-9966-29-99
Chen, W., Wu, M., Cui, S., Zheng, Y., Liu, Z., and Luo, L. (2021). CircRNA Circ-ITCH Inhibits the Proliferation and Invasion of Glioma Cells through Targeting the miR-106a-5p/SASH1 Axis. Cell Transpl. 30, 096368972098378. doi:10.1177/0963689720983785
Cheng, S., Qian, K., Wang, Y., Wang, G., Liu, X., Xiao, Y., et al. (2019). PPARγ Inhibition Regulates the Cell Cycle, Proliferation and Motility of Bladder Cancer Cells. J. Cell Mol. Med. 23 (5), 3724–3736. doi:10.1111/jcmm.14280
Chu, P., Xu, L., and Su, H. (2019). RETRACTED ARTICLE: HULC Functions as an Oncogene in Ovarian Carcinoma Cells by Negatively Modulating miR-125a-3p. J. Physiol. Biochem. 75 (2), 163–171. doi:10.1007/s13105-019-00669-5
Compston, J. E., McClung, M. R., and Leslie, W. D. (2019). Osteoporosis. Lancet 393 (10169), 364–376. doi:10.1016/s0140-6736(18)32112-3
Fan, Y., Yang, Q., Yang, Y., Gao, Z., Ma, Y., Zhang, L., et al. (2019). Sirt6 Suppresses High Glucose-Induced Mitochondrial Dysfunction and Apoptosis in Podocytes through AMPK Activation. Int. J. Biol. Sci. 15 (3), 701–713. doi:10.7150/ijbs.29323
Gao, P., Huang, Y., Hou, Y., Li, Q., and Wang, H. (2021). Circular RNA ITCH Is a Tumor Suppressor in Clear Cell Renal Cell Carcinoma Metastasis through miR-106b-5p/PDCD4 Axis. J. Immunol. Res. 2021, 1–10. doi:10.1155/2021/5524344
Ghafouri-Fard, S., Khoshbakht, T., Taheri, M., and Jamali, E. (2021). CircITCH: A Circular RNA with Eminent Roles in the Carcinogenesis. Front. Oncol. 11, 774979. doi:10.3389/fonc.2021.774979
Ghasemi, S., Emadi-Baygi, M., and Nikpour, P. (2019). Down-regulation of Circular RNAITCH and circHIPK3 in Gastric Cancer Tissues. Turk J. Med. Sci. 49 (2), 687–695. doi:10.3906/sag-1806-50
Giebel, S. J., Menicucci, G., McGuire, P. G., and Das, A. (2005). Matrix Metalloproteinases in Early Diabetic Retinopathy and Their Role in Alteration of the Blood-Retinal Barrier. Lab. Invest. 85 (5), 597–607. doi:10.1038/labinvest.3700251
Glöckner, S., Buurman, H., Kleeberger, W., Lehmann, U., and Kreipe, H. (2002). Marked Intratumoral Heterogeneity of C-Myc and cyclinD1 but Not of C-erbB2 Amplification in Breast Cancer. Lab. Invest. 82 (10), 1419–1426. doi:10.1097/01.lab.0000032371.16521.40
Goto, Y., Yajima, I., Kumasaka, M., Ohgami, N., Tanaka, A., Tsuzuki, T., et al. (2016). Transcription Factor LSF (TFCP2) Inhibits Melanoma Growth. Oncotarget 7 (3), 2379–2390. doi:10.18632/oncotarget.6230
Gu, X., Li, M., Jin, Y., Liu, D., and Wei, F. (2017). Identification and Integrated Analysis of Differentially Expressed lncRNAs and circRNAs Reveal the Potential ceRNA Networks during PDLSC Osteogenic Differentiation. BMC Genet. 18 (1), 100. doi:10.1186/s12863-017-0569-4
Guo, W., Zhang, J., Zhang, D., Cao, S., Li, G., Zhang, S., et al. (2017). Polymorphisms and Expression Pattern of Circular RNA Circ-ITCH Contributes to the Carcinogenesis of Hepatocellular Carcinoma. Oncotarget 8 (29), 48169–48177. doi:10.18632/oncotarget.18327
Guo, X., Wang, Z., Deng, X., Lu, Y., Huang, X., Lin, J., et al. (2022). Circular RNA CircITCH (Has-circ-0001141) Suppresses Hepatocellular Carcinoma (HCC) Progression by Sponging miR-184. Cell Cycle, 1–21. doi:10.1080/15384101.2022.2057633
Guo, Y., Zheng, H., Yin, J., and Wang, H. J. S. r. (2021). Rs4911154 of Circ-ITCH Aggravated Tumor Malignancy of Thyroid Nodules via the Circ-ITCH/miR-22-3p/CBL axis. Sci. Rep. 11 (1), 18491. doi:10.1038/s41598-021-97471-5
Hamid, S., Cicek, S., Karamil, S., Ozturk, M., Debelec-Butuner, B., Erbaykent-Tepedelen, B., et al. (2014). HOXB13 Contributes to G1/S and G2/M Checkpoint Controls in Prostate. Mol. Cell. Endocrinol. 383, 38–47. doi:10.1016/j.mce.2013.12.003
Han, D., Wang, Y., Wang, Y., Dai, X., Zhou, T., Chen, J., et al. (2020). The Tumor-Suppressive Human Circular RNA CircITCH Sponges miR-330-5p to Ameliorate Doxorubicin-Induced Cardiotoxicity through Upregulating SIRT6, Survivin, and SERCA2a. Circ. Res. 127 (4), e108–e125. doi:10.1161/circresaha.119.316061
Hao, C., Wangzhou, K., Liang, Z., Liu, C., Wang, L., Gong, L., et al. (2020). Circular RNA ITCH Suppresses Cell Proliferation but Induces Apoptosis in Oral Squamous Cell Carcinoma by Regulating miR-421/PDCD4 Axis. Cmar Vol. 12, 5651–5658. doi:10.2147/cmar.S258887
He, Y., Tao, W., He, T., Wang, B., Tang, X., Zhang, L., et al. (2021). A Urine Extracellular Vesicle circRNA Classifier for Detection of High-Grade Prostate Cancer in Patients with Prostate-specific Antigen 2-10 ng/mL at Initial Biopsy. Mol. Cancer 20 (1), 96. doi:10.1186/s12943-021-01388-6
Heanue, T. A., and Pachnis, V. (2007). Enteric Nervous System Development and Hirschsprung's Disease: Advances in Genetic and Stem Cell Studies. Nat. Rev. Neurosci. 8 (6), 466–479. doi:10.1038/nrn2137
Hermida, M., Dinesh Kumar, J., and Leslie, N. (2017). GSK3 and its Interactions with the PI3K/AKT/mTOR Signalling Network. Adv. Biol. Regul. 65, 5–15. doi:10.1016/j.jbior.2017.06.003
Hu, J., Wang, L., Chen, J., Gao, H., Zhao, W., Huang, Y., et al. (2018). The Circular RNA Circ-ITCH Suppresses Ovarian Carcinoma Progression through Targeting miR-145/RASA1 Signaling. Biochem. biophysical Res. Commun. 505 (1), 222–228. doi:10.1016/j.bbrc.2018.09.060
Huang, E., Chen, X., and Yuan, Y. (2019). Downregulated Circular RNA Itchy E3 Ubiquitin Protein Ligase Correlates with Advanced Pathologic T Stage, High Lymph Node Metastasis Risk and Poor Survivals in Prostate Cancer Patients. Cbm 26 (1), 41–50. doi:10.3233/cbm-182111
Huang, G., Zhu, H., Shi, Y., Wu, W., Cai, H., and Chen, X. (2015). Cir-ITCH Plays an Inhibitory Role in Colorectal Cancer by Regulating the Wnt/β-Catenin Pathway. PloS one 10 (6), e0131225. doi:10.1371/journal.pone.0131225
Huang, J., Wu, D., Shen, J., Wu, P., Ni, C., Chen, J., et al. (2012). Enrichment of Colorectal Cancer Stem Cells through Epithelial-Mesenchymal Transition via CDH1 Knockdown. Mol. Med. Rep. 6 (3), 507–512. doi:10.3892/mmr.2012.938
Iwatsuki, M., Mimori, K., Yokobori, T., Ishi, H., Beppu, T., Nakamori, S., et al. (2010). Epithelial-mesenchymal Transition in Cancer Development and its Clinical Significance. Cancer Sci. 101 (2), 293–299. doi:10.1111/j.1349-7006.2009.01419.x
Ji, L., Chen, Y., Wang, H., Zhang, W., He, L., Wu, J., et al. (2019). Overexpression of Sirt6 Promotes M2 macrophage Transformation, Alleviating Renal Injury in Diabetic Nephropathy. Int. J. Oncol. 55 (1), 103–115. doi:10.3892/ijo.2019.4800
Koch, A., Lang, S., Wild, P., Gantner, S., Mahli, A., Spanier, G., et al. (2015). Glucose Transporter Isoform 1 Expression Enhances Metastasis of Malignant Melanoma Cells. Oncotarget 6 (32), 32748–32760. doi:10.18632/oncotarget.4977
Korać, P., Antica, M., and Matulić, M. (2021). MiR-7 in Cancer Development. Biomedicines 9 (3), 325. doi:10.3390/biomedicines9030325
Kotarba, G., Krzywinska, E., Grabowska, A. I., Taracha, A., and Wilanowski, T. (2018). TFCP2/TFCP2L1/UBP1 Transcription Factors in Cancer. Cancer Lett. 420, 72–79. doi:10.1016/j.canlet.2018.01.078
Kowluru, R. A., and Mishra, M. (2017). Regulation of Matrix Metalloproteinase in the Pathogenesis of Diabetic Retinopathy. Prog. Mol. Biol. Transl. Sci. 148, 67–85. doi:10.1016/bs.pmbts.2017.02.004
Kristensen, L., Andersen, M., Stagsted, L., Ebbesen, K., Hansen, T., and Kjems, J. (2019). The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Kumari, N., Reabroi, S., and North, B. (2021). Unraveling the Molecular Nexus between GPCRs, ERS, and EMT. Mediat. Inflamm. 2021, 1–23. doi:10.1155/2021/6655417
Lasithiotakis, K., Antoniou, S., Antoniou, G., Kaklamanos, I., and Zoras, O. (2014). Gastrectomy for stage IV gastric cancer. a systematic review and meta-analysis. Anticancer Res. 34 (5), 2079–2085.
Lee, P. J., Rudenko, D., Kuliszewski, M. A., Liao, C., Kabir, M. G., Connelly, K. A., et al. (2014). Survivin gene therapy attenuates left ventricular systolic dysfunction in doxorubicin cardiomyopathy by reducing apoptosis and fibrosis. Cardiovasc Res. 101 (3), 423–433. doi:10.1093/cvr/cvu001
Li, F., Zhang, L., Li, W., Deng, J., Zheng, J., An, M., et al. (2015). Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 6 (8), 6001–6013. doi:10.18632/oncotarget.3469
Li, H., Lan, M., Liao, X., Tang, Z., and Yang, C. (2020a). Circular RNA cir-ITCH Promotes Osteosarcoma Migration and Invasion through cir-ITCH/miR-7/EGFR Pathway. Technol. Cancer Res. Treat. 19, 153303381989872. doi:10.1177/1533033819898728
Li, J., Guo, R., Liu, Q., Sun, J., and Wang, H. (2020b). Circular RNA Circ-ITCH Inhibits the Malignant Behaviors of Cervical Cancer by microRNA-93-5p/FOXK2 Axis. Reprod. Sci. 27 (3), 860–868. doi:10.1007/s43032-020-00140-7
Li, J., Song, Y., Wang, J., and Huang, J. (2020c). Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer detection. Am. J. Transl. Res. 12 (11), 7395–7403.
Li, S., Yu, C., Zhang, Y., Liu, J., Jia, Y., Sun, F., et al. (2020d). Circular RNA cir-ITCH Is a Potential Therapeutic Target for the Treatment of Castration-Resistant Prostate Cancer. BioMed Res. Int. 2020, 1–9. doi:10.1155/2020/7586521
Li, Y., Ge, Y., Xu, L., and Jia, R. (2020e). Circular RNA ITCH: A novel tumor suppressor in multiple cancers. Life Sci. 254, 117176. doi:10.1016/j.lfs.2019.117176
Li, Z., Guo, X., and Gao, S. (2019). Circ-ITCH correlates with less advanced tumor features as well as prolonged survival, and it inhibits cells proliferation but promotes apoptosis in non-small cell lung cancer. Transl. Cancer Res. 8 (5), 1672–1679. doi:10.21037/tcr.2019.08.01
Lin, C., Xu, X., Yang, Q., Liang, L., and Qiao, S. (2020). Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 20, 336. doi:10.1186/s12935-020-01420-7
Lin, Q., Jiang, H., and Lin, D. (2021). Circular RNA ITCH downregulates GLUT1 and suppresses glucose uptake in melanoma to inhibit cancer cell proliferation. J. Dermatological Treat. 32 (2), 231–235. doi:10.1080/09546634.2019.1654069
Liu, J., Du, F., Chen, C., Li, D., Chen, Y., Xiao, X., et al. (2020). CircRNA ITCH increases bortezomib sensitivity through regulating the miR-615-3p/PRKCD axis in multiple myeloma. Life Sci. 262, 118506. doi:10.1016/j.lfs.2020.118506
Liu, J., Duan, P., Xu, C., Xu, D., Liu, Y., and Jiang, J. (2021). CircRNA circ-ITCH improves renal inflammation and fibrosis in streptozotocin-induced diabetic mice by regulating the miR-33a-5p/SIRT6 axis. Inflamm. Res. 70 (7), 835–846. doi:10.1007/s00011-021-01485-8
Lorthongpanich, C., Thumanu, K., Tangkiettrakul, K., Jiamvoraphong, N., Laowtammathron, C., Damkham, N., et al. (2019). YAP as a key regulator of adipo-osteogenic differentiation in human MSCs. Stem Cell Res. Ther. 10 (1), 402. doi:10.1186/s13287-019-1494-4
Luo, L., Gao, Y., and Sun, X. (2018a). Circ-ITCH correlates with small tumor size, decreased FIGO stage and prolonged overall survival, and it inhibits cells proliferation while promotes cells apoptosis in epithelial ovarian cancer. Cbm 23 (4), 505–513. doi:10.3233/cbm-181609
Luo, L., Gao, Y., and Sun, X. (2018b). Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR-10a-α. Eur. Rev. Med. Pharmacol. Sci. 22 (23), 8119–8126. doi:10.26355/eurrev_201812_16503
Maass, P. G., Glažar, P., Memczak, S., Dittmar, G., Hollfinger, I., Schreyer, L., et al. (2017). A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 95 (11), 1179–1189. doi:10.1007/s00109-017-1582-9
Melino, G., Gallagher, E., Aqeilan, R. I., Knight, R., Peschiaroli, A., Rossi, M., et al. (2008). Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 15 (7), 1103–1112. doi:10.1038/cdd.2008.60
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 (7441), 333–338. doi:10.1038/nature11928
Min, X., Liu, D. L., and Xiong, X. D. (2021). Circular RNAs as Competing Endogenous RNAs in Cardiovascular and Cerebrovascular Diseases: Molecular Mechanisms and Clinical Implications. Front. Cardiovasc. Med. 8, 682357. doi:10.3389/fcvm.2021.682357
Muralidhar, S., Filia, A., Nsengimana, J., Poźniak, J., O'Shea, S., Diaz, J., et al. (2019). Vitamin D-VDR Signaling Inhibits Wnt/β-Catenin-Mediated Melanoma Progression and Promotes Antitumor Immunity. Cancer Res. 79 (23), 5986–5998. doi:10.1158/0008-5472.Can-18-3927
Nawaz, Z., Patil, V., Paul, Y., Hegde, A., Arivazhagan, A., Santosh, V., et al. (2016). PI3 kinase pathway regulated miRNome in glioblastoma: identification of miR-326 as a tumour suppressor miRNA. Mol. Cancer 15 (1), 74. doi:10.1186/s12943-016-0557-8
Ning, B., Yu, D., and Yu, A. (2019). Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem. Pharmacol. 169, 113638. doi:10.1016/j.bcp.2019.113638
Ohgami, N., Iizuka, A., Hirai, H., Yajima, I., Iida, M., Shimada, A., et al. (2021). Loss-of-function mutation of c-Ret causes cerebellar hypoplasia in mice with Hirschsprung disease and Down's syndrome. J. Biol. Chem. 296, 100389. doi:10.1016/j.jbc.2021.100389
Pan, J. X., Xiong, L., Zhao, K., Zeng, P., Wang, B., Tang, F. L., et al. (2018). YAP promotes osteogenesis and suppresses adipogenic differentiation by regulating β-catenin signaling. Bone Res. 6, 18. doi:10.1038/s41413-018-0018-7
Parravicini, V., Field, A. C., Tomlinson, P. D., Albert Basson, M. A., and Zamoyska, R. (2008). Itch−/−αβ and γδ T cells independently contribute to autoimmunity in Itchy mice. Blood 111 (8), 4273–7282. doi:10.1182/blood-2007-10-115667
Peng, Y., and Wang, H. (2020). Cir-ITCH inhibits gastric cancer migration, invasion and proliferation by regulating the Wnt/β-catenin pathway. Sci. Rep. 10 (1), 17443. doi:10.1038/s41598-020-74452-8
Pichon, X., A. Wilson, L., Stoneley, M., Bastide, A., A King, H., Somers, J., et al. (2012). RNA binding protein/RNA element interactions and the control of translation. Cpps 13 (4), 294–304. doi:10.2174/138920312801619475
Reilly, A. M., Petrou, S., Pancha, R. G., and Williams, D. A. (2001). Restoration of calcium handling properties of adult cardiac myocytes from hypertrophied hearts. Cell Calcium 30 (1), 59–66. doi:10.1054/ceca.2001.0213
Ren, C., Liu, J., Zheng, B., Yan, P., Sun, Y., and Yue, B. (2019). RETRACTED ARTICLE: The circular RNA circ-ITCH acts as a tumour suppressor in osteosarcoma via regulating miR-22. Artif. cells, nanomedicine, Biotechnol. 47 (1), 3359–3367. doi:10.1080/21691401.2019.1649273
Rezazadeh, S., Yang, D., Tombline, G., Simon, M., Regan, S. P., Seluanov, A., et al. (2019). SIRT6 promotes transcription of a subset of NRF2 targets by mono-ADP-ribosylating BAF170. Nucleic Acids Res. 47 (15), 7914–7928. doi:10.1093/nar/gkz528
Sadri Nahand, J., Shojaie, L., Akhlagh, S., Ebrahimi, M., Mirzaei, H., Bannazadeh Baghi, H., et al. (2021). Cell death pathways and viruses: Role of microRNAs. Mol. Ther. - Nucleic Acids 24, 487–511. doi:10.1016/j.omtn.2021.03.011
Semeraro, F., Cancarini, A., dell’Omo, R., Rezzola, S., Romano, M. R., and Costagliola, C. (2015). Diabetic Retinopathy: Vascular and Inflammatory Disease. J. Diabetes Res. 2015, 1–16. doi:10.1155/2015/582060
Sharkawi, F. Z. E., El-Sherbiny, M., Ali, S. A., and Nassif, W. M. H. (2022). The potential value of plasma Circ-ITCH in Hepatocellular carcinoma patients with current Hepatitis C virus infection. Gastroenterol. Hepatol. [Not Available]. doi:10.1016/j.gastrohep.2022.03.006
Shashar, M., Siwak, J., Tapan, U., Lee, S., Meyer, R., Parrack, P., et al. (2016). c-Cbl mediates the degradation of tumorigenic nuclear β-catenin contributing to the heterogeneity in Wnt activity in colorectal tumors. Oncotarget 7 (44), 71136–71150. doi:10.18632/oncotarget.12107
Su, K., Yi, Q., Dai, X., and Liu, O. (2022). Circular RNA ITCH: An Emerging Multifunctional Regulator. Biomolecules 12 (3), 359. doi:10.3390/biom12030359
Sun, X., Huan, C., Sun, D., and Lv, G. (2021). Prognostic and Clinicopathological Significance of Circular RNA circ-ITCH Expression in Cancer Patients: A Meta-analysis. BioMed Res. Int. 2021, 1–13. doi:10.1155/2021/8828299
Sundvall, M., Korhonen, A., Paatero, I., Gaudio, E., Melino, G., Croce, C. M., et al. (2008). Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc. Natl. Acad. Sci. U.S.A. 105, 4162–4167. doi:10.1073/pnas.0708333105
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tang, X., Zhu, J., Liu, Y., Chen, C., Liu, T., and Liu, J. (2019). Current Understanding of Circular RNAs in Gastric Cancer. Cmar Vol. 11, 10509–10521. doi:10.2147/cmar.S223204
Tian, X., Firsanov, D., Zhang, Z., Cheng, Y., Luo, L., Tombline, G., et al. (2019). SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell 177 (3), 622–638. e622. doi:10.1016/j.cell.2019.03.043
Tsuchiya, H., and Oura, S. (2018). Involvement of MAFB and MAFF in Retinoid-Mediated Suppression of Hepatocellular Carcinoma Invasion. Ijms 19 (5), 1450. doi:10.3390/ijms19051450
van der Weyden, L., and Adams, D. (2007). The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochimica Biophysica Acta (BBA) - Rev. Cancer 1776 (1), 58–85. doi:10.1016/j.bbcan.2007.06.003
Verduci, L., Tarcitano, E., Strano, S., Yarden, Y., and Blandino, G. (2021). CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis. 12 (5), 468. doi:10.1038/s41419-021-03743-3
Wada, J., and Makino, H. (2013). Inflammation and the pathogenesis of diabetic nephropathy. Clin. Sci. (Lond) 124 (3), 139–152. doi:10.1042/cs20120198
Wan, L., Zhang, L., Fan, K., Cheng, Z., Sun, Q., and Wang, J. (2016). Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/β-Catenin Pathway. BioMed Res. Int. 2016, 1–11. doi:10.1155/2016/1579490
Wang, L., Sang, J., Zhang, Y., Gao, L., Zhao, D., and Cao, H. (2022). Circular RNA ITCH attenuates the progression of nasopharyngeal carcinoma by inducing PTEN upregulation via miR‐214. J. Gene Med. 24 (1), e3391. doi:10.1002/jgm.3391
Wang, M., Chen, B., Ru, Z., and Cong, L. (2018). CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem. biophysical Res. Commun. 504 (1), 283–288. doi:10.1016/j.bbrc.2018.08.175
Wang, S., Liu, L., Li, X., Wang, Y., Xie, P., Li, Q., et al. (2019a). Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/β-catenin pathway. neo 66 (2), 232–239. doi:10.4149/neo_2018_180710N460
Wang, X., Wang, R., Wu, Z., and Bai, P. (2019b). Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 19, 328. doi:10.1186/s12935-019-0994-8
Wang, Y., Liu, Z., and Shen, J. (2019c). MicroRNA-421-targeted PDCD4 regulates breast cancer cell proliferation. Int. J. Mol. Med. 43 (1), 267–275. doi:10.3892/ijmm.2018.3932
Wang, Y., Wang, H., Zheng, R., Wu, P., Sun, Z., Chen, J., et al. (2021). Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle 20 (5-6), 522–536. doi:10.1080/15384101.2021.1878327
Wang, Z., Han, L., Sun, T., Ma, J., Sun, S., Ma, L., et al. (2020). Extracellular matrix derived from allogenic decellularized bone marrow mesenchymal stem cell sheets for the reconstruction of osteochondral defects in rabbits. Acta Biomater. 118, 54–68. doi:10.1016/j.actbio.2020.10.022
Wei, W., Li, M., Wang, J., Nie, F., and Li, L. (2012). The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting dishevelled protein. Mol. Cell Biol. 32 (19), 3903–3912. doi:10.1128/mcb.00251-12
Wu, M., Deng, X., Zhong, Y., Hu, L., Zhang, X., Liang, Y., et al. (2020). MafF Is Regulated via the circ-ITCH/miR-224-5p Axis and Acts as a Tumor Suppressor in Hepatocellular Carcinoma. Oncol. Res. 28 (3), 299–309. doi:10.3727/096504020x15796890809840
Wu, Z., Wang, H., Fang, S., and Xu, C. (2018). Roles of endoplasmic reticulum stress and autophagy on H2O2-induced oxidative stress injury in HepG2 cells. Mol. Med. Rep. 18 (5), 4163–4174. doi:10.3892/mmr.2018.9443
Xia, R. P., Zhao, F., Ma, T. D., Zou, C. J., Xu, G., and Zhou, C. G. (2022). Circ-ITCH overexpression promoted cell proliferation and migration in Hirschsprung disease through miR-146b-5p/RET axis. Pediatr. Res. doi:10.1038/s41390-021-01860-5
Yan, H., Xiang, H., Sun, B., Feng, F., and Chen, P. (2020). Circular RNA-ITCH Inhibits the Proliferation of Ovarian Carcinoma by Downregulating lncRNA HULC. Reprod. Sci. 27 (1), 375–379. doi:10.1007/s43032-019-00049-w
Yang, B., Zhao, J., Huo, T., Zhang, M., and Wu, X. (2020). Effects of CircRNA-ITCH on proliferation and apoptosis of hepatocellular carcinoma cells through inhibiting Wnt/β-catenin signaling pathway. J. BUON 25 (3), 1368–1374.
Yang, C., Dou, R., Wei, C., Liu, K., Shi, D., Zhang, C., et al. (2021). Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol. Ther. 29 (6), 2088–2107. doi:10.1016/j.ymthe.2021.02.006
Yang, C., Yuan, W., Yang, X., Li, P., Wang, J., Han, J., et al. (2018). Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer 17 (1), 19. doi:10.1186/s12943-018-0771-7
Yeh, E. T., and Bickford, C. L. (2009). Cardiovascular Complications of Cancer Therapy. J. Am. Coll. Cardiol. 53 (24), 2231–2247. doi:10.1016/j.jacc.2009.02.050
Yuan, Y., Chen, X., and Huang, E. (2019). Upregulation of Circular RNA Itchy E3 Ubiquitin Protein Ligase Inhibits Cell Proliferation and Promotes Cell Apoptosis Through Targeting MiR-197 in Prostate Cancer. Technol. Cancer Res. Treat. 18, 153303381988686. doi:10.1177/1533033819886867
Zhang, F., Lin, F., Xu, Z., and Huang, Z. (2021). Circular RNA ITCH promotes extracellular matrix degradation via activating Wnt/β-catenin signaling in intervertebral disc degeneration. Aging 13 (10), 14185–14197. doi:10.18632/aging.203036
Zhang, J., Cai, Y., Sheng, S., Zhao, C., and Jiang, B. (2022). circITCH suppresses cell proliferation and metastasis through miR ‐660/ TFCP2 pathway in melanoma. Cancer Med. 11 (12), 2405–2413. doi:10.1002/cam4.4627
Zhang, N., and Wang, X. (2020). Circular RNA ITCH mediates H 2 O 2 ‐induced myocardial cell apoptosis by targeting miR‐17‐5p via wnt/β‐catenin signalling pathway. Int. J. Exp. Path 102 (1), 22–31. doi:10.1111/iep.12367
Zhang, S., Liu, X., Bawa-Khalfe, T., Lu, L. S., Lyu, Y. L., Liu, L. F., et al. (2012). Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 18 (11), 1639–1642. doi:10.1038/nm.2919
Zhang, Y., Li, Y., Wang, Q., Su, B., Xu, H., Sun, Y., et al. (2020). Role of RASA1 in cancer: A review and update (Review). Oncol. Rep. 44 (6), 2386–2396. doi:10.3892/or.2020.7807
Zhao, L., Ma, N., Liu, G., Mao, N., Chen, F., and Li, J. (2021). Lidocaine Inhibits Hepatocellular Carcinoma Development by Modulating circ_ITCH/miR-421/CPEB3. Dig. Dis. Sci., 66, 4384–4397. doi:10.1007/s10620-020-06787-1
Zhong, D., Xu, G. Z., Wu, J. Z., Liu, H., Tang, J. Y., and Wang, C. G. (2021). Circ-ITCH sponges miR-214 to promote the osteogenic differentiation in osteoporosis via upregulating YAP1. Cell Death Dis. 12 (4), 340. doi:10.1038/s41419-021-03586-y
Zhou, H., Zhang, J., Chen, B., Liu, H., Liu, X., Sun, Z., et al. (2020). Potential of circular RNA itchy E3 ubiquitin protein ligase as a biomarker and treatment target for multiple myeloma. Transl. Cancer Res. TCR 9 (1), 335–345. doi:10.21037/tcr.2019.12.71
Zhou, L., Li, F. F., and Wang, S. M. (2021a). Circ-ITCH restrains the expression of MMP-2, MMP-9 and TNF-α in diabetic retinopathy by inhibiting miR-22. Exp. Mol. Pathology 118, 104594. doi:10.1016/j.yexmp.2020.104594
Zhou, W., Liu, Y., and Wu, X. (2021b). Down‐regulation of circITCH promotes osteosarcoma development and resistance to doxorubicin via the miR‐524/RASSF6 axis. J. Gene Med. 23, e3373. doi:10.1002/jgm.3373
Keywords: circ-ITCH, malignant tumor, non-tumor diseases, mechanism, biomarkers
Citation: Liu T, Huang T, Shang M and Han G (2022) CircRNA ITCH: Insight Into Its Role and Clinical Application Prospect in Tumor and Non-Tumor Diseases. Front. Genet. 13:927541. doi: 10.3389/fgene.2022.927541
Received: 24 April 2022; Accepted: 21 June 2022;
Published: 15 July 2022.
Edited by:
Olanrewaju B. Morenikeji, University of Pittsburgh at Bradford, United StatesReviewed by:
Annie Angers, Université de Montréal, CanadaCopyright © 2022 Liu, Huang, Shang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Han, aGFuZ2FuZ0BqbHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.