
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 23 June 2022
Sec. Computational Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.927222
This article is part of the Research TopicMachine Learning Used in Biomedical Computing and Intelligence Healthcare, Volume IIIView all 9 articles
Multi-modal medical image fusion can reduce information redundancy, increase the understandability of images and provide medical staff with more detailed pathological information. However, most of traditional methods usually treat the channels of multi-modal medical images as three independent grayscale images which ignore the correlation between the color channels and lead to color distortion, attenuation and other bad effects in the reconstructed image. In this paper, we propose a multi-modal medical image fusion algorithm with geometric algebra based sparse representation (GA-SR). Firstly, the multi-modal medical image is represented as a multi-vector, and the GA-SR model is introduced for multi-modal medical image fusion to avoid losing the correlation of channels. Secondly, the orthogonal matching pursuit algorithm based on geometric algebra (GAOMP) is introduced to obtain the sparse coefficient matrix. The K-means clustering singular value decomposition algorithm based on geometric algebra (K-GASVD) is introduced to obtain the geometric algebra dictionary, and update the sparse coefficient matrix and dictionary. Finally, we obtain the fused image by linear combination of the geometric algebra dictionary and the coefficient matrix. The experimental results demonstrate that the proposed algorithm outperforms existing methods in subjective and objective quality evaluation, and shows its effectiveness for multi-modal medical image fusion.
Medical Image fusion technology integrates technologies in many fields such as computer technology, sensor technology, artificial intelligence, and image processing. It comprehensively extracts image information collected by different sensors and concentrates all the information of the image, which can reduce the information redundancy of the image, enhance the readability of the image and provide more specific disease information for diagnosis (Riboni and Murtas, 2019; Li et al., 2021; Wang et al., 2022).
According to the types of fused images, medical image fusion can be divided into unimodal medical image fusion and multi-modal medical image fusion (Tirupal et al., 2021). A unimodal medical image refers to multiple images of a patient’s organ collected by the same device, which are combined into one image by corresponding fusion algorithm. The purpose is to collect image information under different contrasts (Zhang. et al., 2021). Multi-modal medical images refer to images obtained by different imaging methods. Different types of medical images contain different information, and the obtained fused image can summarize various feature information to provide medical staff with more comprehensive pathological information (Zhu et al., 2017). Common medical images include CT images, MR images, and SPECT images (Thieme et al., 2012; Nazir et al., 2021; Engudar et al., 2022).
Multi-modal medical image fusion mainly includes the following methods: morphological methods, knowledge based methods, wavelet based methods, neural network based methods, methods based on fuzzy logic, and so on (James and Dasarathy, 2014). Naeem used discrete wavelet transform (DWT) to fuse images with different details, which changed the uniformity of the details contained in the fused image (Naeem et al., 2016). Guruprasad et al. (2013) proposed an image fusion algorithm based on DWT-DBSS and use the maximum selection rule to obtain detailed fusion coefficients. Bruno presents a novel Wavelet-based method to fuse medical images according to the MRA approach, that aims to put the right “semantic” content in the fused image by applying two different quality indexes: variance and modulus maxima (Alfano et al., 2007). A hierarchical image fusion scheme is presented which preserves the details of the input images of most relevance for visual perception (Marshall and Matsopoulos, 2002).
Sparse representation (Shao et al., 2020) can deal with the natural sparsity of signals by the physiological properties of the human visual system, which is a linear combination of dictionary atoms and sparse coefficients to represent the signal with as few atoms as possible in a given overcomplete dictionary. Bin Yang and Shutao Li (2010) first introduced sparse representation into image fusion, and adopted the sliding window technique to make the fusion process robust to noise and registration. Zong and Qiu (2017) proposed a fusion method based on classified image blocks, which used the directional gradient histogram feature to classify image blocks to establish a sub-dictionary. It can reduce the loss of image details and improve the quality of image fusion.
Traditional sparse representation fusion method usually processes the color channels separately, which easily destroys the correlation between image channels and results in loss of color in the fused image. Geometric algebra (GA) has been considered as one of the most powerful tools in multi-dimensional signal processing and has witnessed great success in a wide range of applications, such as physics, quantum computing, electromagnetism, satellite navigation, neural computing, camera geometry, image processing, robotics, and computer vision, et al. (da Rocha and Vaz, 2006; Wang et al., 2019a; Wang et al., 2021a). Inspired by the paper (Wang et al., 2019b), the geometric algebra-based sparse representation (GA-SR) is introduced for multi-modal medical image fusion in this paper.
The rests of this paper are organized as follows. In Section 2, this paper introduces the basic knowledge of geometric algebra. Section 3 introduces the GA-SR algorithm and the fusion steps of the proposed algorithm. Section 4 provides the experimental analysis including subjective and objective quality evaluations. Finally, Section 5 concludes the papers.
Geometric algebra combines quaternions and Grassmann algebras, which can extend operations to higher-dimensional spaces. The geometric algebra space does not rely on coordinate information for calculation (Batard et al., 2009), and all geometric operators are included in the space. Any multi-modal medical image can be represented by geometric algebraic orthonormal base as a multi-vector for overall processing, which can ensure the correlation between each channel of the image (da Rocha and Vaz, 2006; López-González et al., 2016).
Geometric algebra is generated by quadratic space, and is defined as follows. Let
For example, the orthonormal base of
The orthonormal base in the geometric algebraic space
where
In this section, the GA-SR based multi-modal medical image fusion is provided.
The sparse representation model of GA multi-vector can be defined as
where
For a three-channel multi-modal medical image
For a three-channel multi-modal medical image, its sparse representation model can be defined as shown in the Eq. 9
Where
Therefore, the GA-SR model of the three-channel medical image can be described as follows
The general form of a three-channel medical image sparse coefficient matrix can be obtained by
Any pixel F of a multi-modal medical image can be represented as a multi-vector form in
While
Assuming that each image block of the multi-modal medical image
Let
(1) The sliding window technique is introduced to divide the two source images into several sub-image blocks. The size of the sliding window is generally
(2) The sparse representation coefficients
where D represents the adaptive dictionary of image blocks obtained by dictionary training,
(3) The fused sparse coefficient matrix is obtained by the L1 norm (Yanan et al., 2020) maximum rule. The L1 norm refers to the sum of the absolute values of the elements, and the L1 norm is the optimal convex approximation of the L0 norm, which is more efficient than the L0 norm and is easier to optimize the solution. The L1 coefficient of the corresponding columns of the two sparse coefficient matrices are calculated, and the column with the larger norm is used as the column of the fused sparse coefficient matrix. The fusion rules of the sparse coefficients are as Eq. 17
(4) A dictionary training algorithm is used to obtain the dictionary required for sparse representation. K-SVD is a classic dictionary training algorithm (Fu et al., 2019) in sparse representation. The K-GASVD algorithm in (Wang et al., 2019b) consists of two steps, which are sparse coding (Sandhya et al., 2021) and dictionary update (Thanthrige et al., 2020). The K-GASVD algorithm is used to perform and update dictionary training on the obtained sparse coefficient matrix.
(5) The fusion result of
(6) All image patches are processed in the same way, the image block vector
In order to verify the effectiveness of the GA-SR based multi-modal medical image fusion, the experiments are implemented on four groups of multi-modal medical images selected from Harvard Medical School Database in Matlab with other exiting methods, such as Laplacian Pyramid algorithm (Liu et al., 2019), DWT-DBSS algorithm (Guruprasad et al., 2013), SIDWT-Haar algorithm (Xin et al., 2013) and Morphological Difference Pyramid algorithm (Matsopoulos et al., 1995). The source images are SPECT images obtained with different radionuclide elements, respectively. The spatial resolution of each image is 256 × 256. The source images used in the experiments are shown in Figure 2.
The multi-modal medical images are fused by six different algorithms respectively, and the obtained results are shown in Figures 3–6.
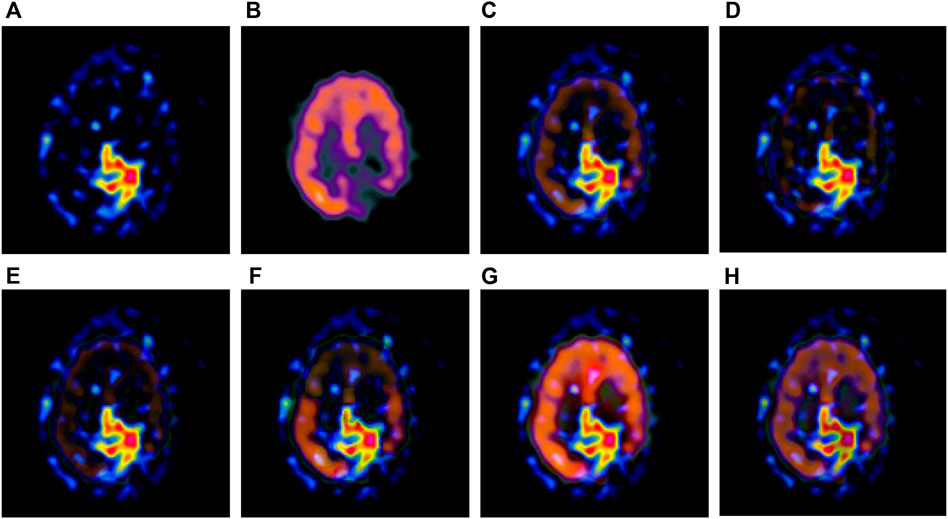
FIGURE 3. Fusion results of the first group. (A) Source image SPECT-TL1 (B) Source image SPECT-TC1 (C) Laplacian Pyramid (D) DWT-DBSS (E) SIDWT-Haar (F) Morphological Difference Pyramid (G) SR (H) GA-SR.
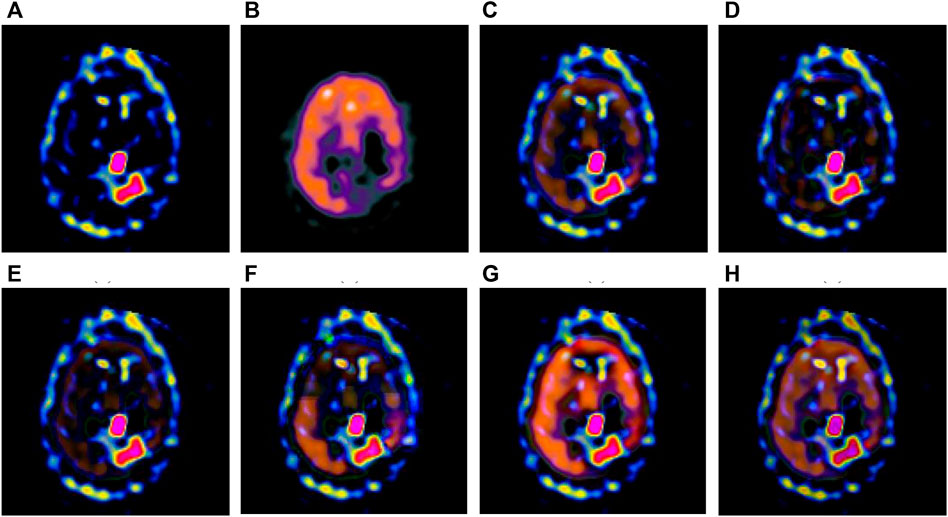
FIGURE 4. Fusion results of the second group. (A) Source image SPECT-TL2 (B) Source image SPECT-TC2 (C) Laplacian Pyramid (D) DWT-DBSS (E) SIDWT-Haar (F) Morphological Difference Pyramid (G) SR (H) GA-SR.
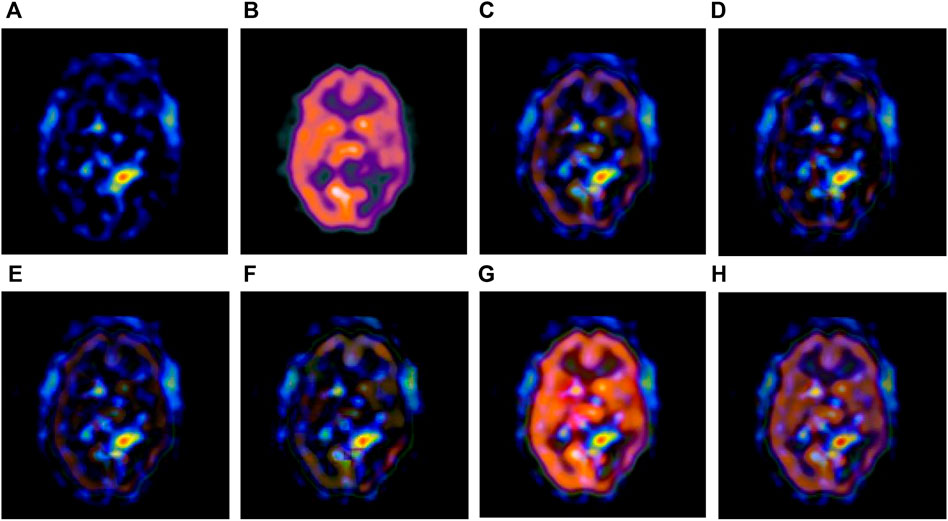
FIGURE 5. Fusion results of the third group. (A) Source image SPECT-TL3 (B) Source image SPECT-TC3 (C) Laplacian Pyramid (D) DWT-DBSS (E) SIDWT-Haar (F) Morphological Difference Pyramid (G) SR (H) GA-SR.
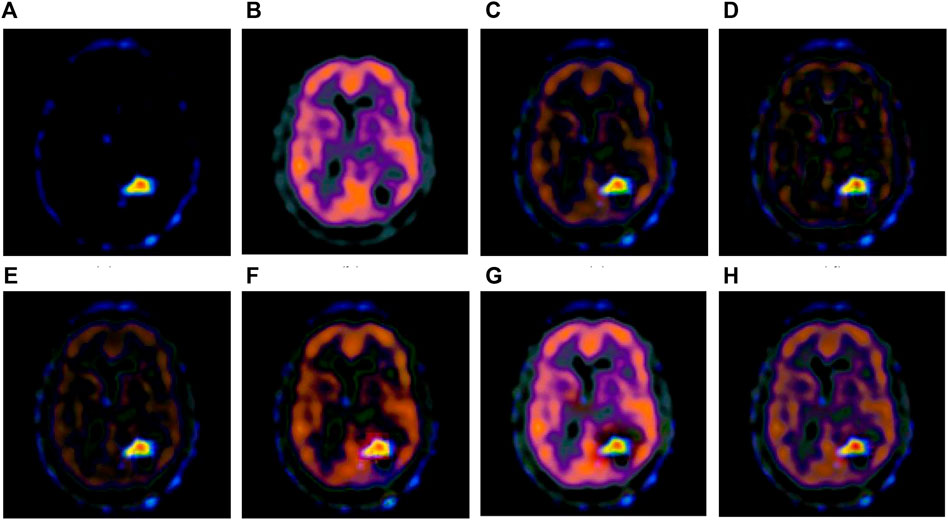
FIGURE 6. Fusion results of the fourth group. (A) Source image SPECT-TL4 (B) Source image SPECT-TC4 (C) Laplacian Pyramid (D) DWT-DBSS (E) SIDWT-Haar (F) Morphological Difference Pyramid (G) SR (H) GA-SR.
Figures 6A,B in each group are the source images used in the experiment, and Figures 6C–H are the fused results obtained by the six different algorithms. Subjectively, it can be seen that the edge of the images obtained by the first four algorithms is relatively complete, but the middle part is darker. The contrast and clarity of the images are low, which indicates that these four algorithms cannot fuse the two source images completely. As a result, the fused image information is incomplete. It can be seen that the fused images obtained by the SR algorithm and GA-SR algorithm are relatively complete, which can comprehensively cover the color and structure information of the two source images, and the fused images obtained are relatively clear. However, there are multiple red spots of different sizes in the images obtained by the SR algorithm, which cause the result to be distorted. The red spots will cover the correct information of the source image, which is not conducive to clinical diagnosis. As can be seen from each group of Figure 6H, the images are relatively clear, and there is no obvious occlusion area. The contrast of the images is relatively high, which indicates that the fused images obtained by the GA-SR algorithm can comprehensively cover the source image. It can provide comprehensive pathological information for medical staff and convenience for clinical medicine.
The evaluation indicators are adopted for objective evaluation of image quality. In this paper, four indicators of CC (Correlation Coefficient) (Li and Dai, 2009), PSNR (Peak Signal to Noise Ratio) (Hore and Ziou, 2010), RMSE (Root Mean Square Error) (Zhao et al., 2020) and Joint-Entropy (Okarma and Fastowicz, 2020) are used for performance analysis with the six fusion algorithms, and four groups of tables are obtained respectively, as shown in Tables 1–4.
For fusion of the four groups, the CC of each group of images obtained by the GA-SR algorithm is higher than that obtained by other algorithms, indicating that the correlation of the images obtained by the GA-SR algorithm with the source image is higher, and the obtained image information is more complete. At the same time, the PSNR and RMSE of the images obtained by the GA-SR algorithm are higher than those obtained by other algorithms, indicating that the fused images obtained by the GA-SR algorithm are closer to the source images and have less distortion and more comprehensive information (Xiao et al., 2021; Gao et al., 2022a; Gao et al., 2022b; Gao et al., 2022c).
Dictionary training is very important for sparse representation, and the quality of the dictionary directly affects the quality of image fusion. The dictionaries training based on the K-SVD and K-GASVD algorithms can be obtained respectively, as shown in Figure 7.
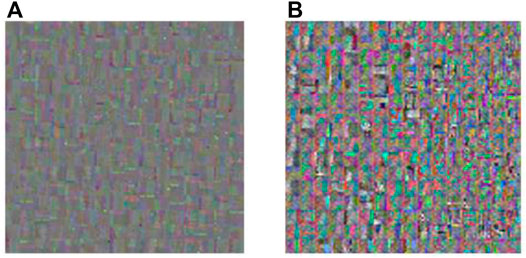
FIGURE 7. Dictionary obtained by K-SVD and K-GASVD. (A) Dictionary obtained by K-SVD algorithm (B) Dictionary obtained by K-GASVD algorithm.
Figure 7A is the dictionary image obtained by the K-SVD algorithm, and Figure 7B is the dictionary image obtained by the K-GASVD algorithm. It is obvious that the color of the dictionary image obtained by the K-SVD algorithm is relatively single, that is because the K-SVD algorithm cannot fully handle the spectral components of the source image, resulting in the generated dictionary image containing a large number of gray image blocks. The dictionary image of K-GASVD contains richer comprehensive information.
In order to verify the effect of the number of dictionary atoms on the quality of the fused image, we change the number of dictionary atoms to obtain different dictionary images, and finally obtain corresponding fused images. The relationship between the PSNR and the atomic number of fused images obtained from dictionaries with different atomic numbers is shown in Figure 8.
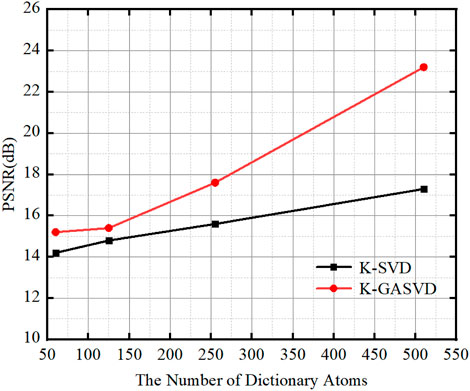
FIGURE 8. The relationship between the PSNR value of the image obtained based on K-SVD and K-GASVD and the number of dictionary atoms.
We can find that the PSNR of the fused images obtained by the K-GASVD model is significantly higher than that of the K-SVD with the increase of the number of dictionary atoms. On the other hand, the number of dictionary atoms required by the K-GASVD model is about 3/10 of the number of atoms required by the K-SVD model if the PSNR is same. Therefore, the number of atoms used in the K-GASVD is significantly reduced in the realization of the same fusion performance, which can present more colorful structures.
For computational complexity, it usually requires longer computational time for multi-modal medical image fusion than other existing real-valued algorithms, because of the non-commutativity of geometric multiplication. Inspired by the work in (Wang et al., 2021b), reduced geometric algebra (RGA) will be introduced to improve our algorithm with lower computational complexity.
In this paper, the multi-modal medical image is represented as a multi-vector, and the GA-SR model is introduced for multi-modal medical image fusion to avoid losing the correlation of channels. And the dictionary learning method based on geometric algebra is provided for more specific disease information for diagnosis. The experimental results validate its rationality and effectiveness. At next steps, we will focus on the analysis and diagnosis of pathological information using GA-SR based multi-modal medical image fusion.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
YL: contributed to the conception of the study; NF: performed the experiment, performed the data analyses and wrote the manuscript; HW: contributed significantly to analysis and manuscript preparation; RW: helped perform the analysis with constructive discussions.
This research was funded by National Natural Science Foundation of China (NSFC) under Grant No. 61771299.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alfano, B., Ciampi, M., and De Pietro, G. (2007). A Wavelet-Based Algorithm for Multimodal Medical Image Fusion. Int. Conf. Samt Semantic Multimedia. DBLP, 117–120. doi:10.1007/978-3-540-77051-0_13
Batard, T., Saint-Jean, C., and Berthier, M. (2009). A Metric Approach to Nd Images Edge Detection with Clifford Algebras. J. Math. Imaging Vis. 33, 296–312. doi:10.1007/s10851-008-0115-0
Bin Yang, B., and Shutao Li, S. (2010). Multifocus Image Fusion and Restoration with Sparse Representation. IEEE Trans. Instrum. Meas. 59, 884–892. doi:10.1109/TIM.2009.2026612
da Rocha, R., and Vaz, J. (2006). Extended Grassmann and Clifford Algebras. Aaca 16, 103–125. doi:10.1007/s00006-006-0006-7
Engudar, G., Rodríguez-Rodríguez, C., Mishra, N. K., Bergamo, M., Amouroux, G., Jensen, K. J., et al. (2022). Metal-ion Coordinated Self-Assembly of Human Insulin Directs Kinetics of Insulin Release as Determined by Preclinical SPECT/CT Imaging. J. Control. Release 343, 347–360. doi:10.1016/J.JCONREL.2022.01.032
Fu, J., Yuan, H., Zhao, R., and Ren, L. (2019). Clustering K-Svd for Sparse Representation of Images. EURASIP J. Adv. Signal Process. 2019, 187–207. doi:10.1186/s13634-019-0650-4
Gao, H., Qiu, B., Duran Barroso, R. J., Hussain, W., Xu, Y., and Wang, X. (2022). TSMAE: A Novel Anomaly Detection Approach for Internet of Things Time Series Data Using Memory-Augmented Autoencoder. IEEE Trans. Netw. Sci. Eng., 1. doi:10.1109/TNSE.2022.3163144
Gao, H., Xiao, J., Yin, Y., Liu, T., and Shi, J. (2022). A Mutually Supervised Graph Attention Network for Few-Shot Segmentation: The Perspective of Fully Utilizing Limited Samples. IEEE Trans. Neural Netw. Learn. Syst., 1–13. doi:10.1109/TNNLS.2022.3155486
Gao, H., Xu, K., Cao, M., Xiao, J., Xu, Q., and Yin, Y. (2022). The Deep Features and Attention Mechanism-Based Method to Dish Healthcare under Social IoT Systems: An Empirical Study with a Hand-Deep Local-Global Net. IEEE Trans. Comput. Soc. Syst. 9 (1), 336–347. doi:10.1109/TCSS.2021.3102.591
Guruprasad, S., Kurian, M. Z., Suma, H. N., and raj, S. (2013). A Medical Multi-Modality Image Fusion of Ct/pet with Pca, Dwt Methods. Ijivp 4, 677–681. doi:10.21917/ijivp.2013.0098
Hore, A., and Ziou, D. (2010). “Image Quality Metrics: PSNR vs. SSIM,” in 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23-26 August 2010 (IEEE Computer Society), 2366–2369. doi:10.1109/ICPR.2010.579
James, A. P., and Dasarathy, B. V. (2014). Medical Image Fusion: a Survey of the State of the Art. Inf. Fusion 19, 4–19. doi:10.1016/j.inffus.2013.12.002
Li, J., and Dai, W. (2009). “Image Quality Assessment Based on Theil Inequality Coefficient and Discrete 2-D Wavelet Transform,” in 2009 IEEE International Conference on Automation and Logistics, Shenyang, China, 05-07 August 2009, 196–199. doi:10.1109/ICIMA.2009.5156594
Li, X., Zhou, F., and Tan, H. (2021). Joint Image Fusion and Denoising via Three-Layer Decomposition and Sparse Representation. Knowledge-Based Syst. 224, 107087. doi:10.1016/j.knosys.2021.107087
Liu, F., Chen, L., Lu, L., Ahmad, A., Jeon, G., and Yang, X. (2019). Medical Image Fusion Method by Using Laplacian Pyramid and Convolutional Sparse Representation. Concurr. Comput. Pract. Exper 32, 97–102. doi:10.1002/cpe.5632
López-González, G., Altamirano-Gómez, G., and Bayro-Corrochano, E. (2016). Geometric Entities Voting Schemes in the Conformal Geometric Algebra Framework. Adv. Appl. Clifford Algebr. 26, 1045–1059. doi:10.1007/s00006-015-0589-y
Marshall, S., and Matsopoulos, G. K. (2002). “Morphological Data Fusion in Medical Imaging,” in IEEE Winter Workshop on Nonlinear Digital Signal Processing, Tampere, Finland, 17-20 January 1993. doi:10.1109/NDSP.1993.767735
Matsopoulos, G. K., Marshall, S., and Brunt, J. (1995). Multiresolution Morphological Fusion of MR and CT Images of the Human Brain. IEEE Vis. Image & Signal Process. Conf. 141, 137–142. doi:10.1049/ic:19950506
Naeem, E. A., Abd Elnaby, M. M., El-Sayed, H. S., Abd El-Samie, F. E., and Faragallah, O. S. (2016). Wavelet Fusion for Encrypting Images with a Few Details. Comput. Electr. Eng. 54, 450–470. doi:10.1016/j.compeleceng.2015.08.018
Nazir, I., Haq, I. U., Khan, M. M., Qureshi, M. B., Ullah, H., and Butt, S. (2021). Efficient Pre-processing and Segmentation for Lung Cancer Detection Using Fused CT Images. Electronics 11, 34. doi:10.3390/ELECTRONICS11010034
Okarma, K., and Fastowicz, J. (2020). Improved Quality Assessment of Colour Surfaces for Additive Manufacturing Based on Image Entropy. Pattern Anal. Applic 23, 1035–1047. doi:10.1007/s10044-020-00865-w
Riboni, D., and Murtas, M. (2019). Sensor-based Activity Recognition: One Picture Is Worth a Thousand Words. Future Gener. Comput. Syst. 101, 709–722. doi:10.1016/j.future.2019.07.020
Sandhya, G., Srinag, A., Pantangi, G. B., and Kanaparthi, J. A. (2021). Sparse Coding for Brain Tumor Segmentation Based on the Non-linear Features. Jbbbe 49, 63–73. doi:10.4028/www.scientific.net/JBBBE.49.63
Shao, L., Wu, J., and Wu, M. (2020). Infrared and Visible Image Fusion Based on Spatial Convolution Sparse Representation. J. Phys. Conf. Ser. 1634, 012113. doi:10.1088/1742-6596/1634/1/012113
Thanthrige, U. S. K. P. M., Barowski, J., Rolfes, I., Erni, D., Kaiser, T., and Sezgin, A. (2020). Characterization of Dielectric Materials by Sparse Signal Processing with Iterative Dictionary Updates. IEEE Sens. Lett. 4, 1–4. doi:10.1109/LSENS.2020.3019924
Thieme, S. F., Graute, V., Nikolaou, K., Maxien, D., Reiser, M. F., Hacker, M., et al. (2012). Dual Energy CT Lung Perfusion Imaging-Correlation with SPECT/CT. Eur. J. radiology 81, 360–365. doi:10.1016/j.ejrad.2010.11.037
Tirupal, T., Mohan, B. C., and Kumar, S. S. (2021). Multimodal Medical Image Fusion Techniques - A Review. Cst 16, 142–163. doi:10.2174/1574362415666200226103116
Wang, R., Fang, N., He, Y., Li, Y., Cao, W., and Wang, H. (2022). Multi-modal Medical Image Fusion Based on Geometric Algebra Discrete Cosine Transform. Adv. Appl. Clifford Algebr. 32, 19. doi:10.1007/S00006-021-01197-6
Wang, R., Shen, M., and Cao, W. (2019). Multivector Sparse Representation for Multispectral Images Using Geometric Algebra. IEEE Access 7, 12755–12767. doi:10.1109/access.2019.2892822
Wang, R., Shen, M., Wang, X., and Cao, W. (2021). RGA-CNNs: Convolutional Neural Networks Based on Reduced Geometric Algebra. Sci. China Inf. Sci. 64, 129101. doi:10.1007/s11432-018-1513-5
Wang, R., Shi, Y., and Cao, W. (2019). GA-SURF: A New Speeded-Up Robust Feature Extraction Algorithm for Multispectral Images Based on Geometric Algebra. Pattern Recognit. Lett. 127, 11–17. doi:10.1016/j.patrec.2018.11.001
Wang, R., Wang, Y., Li, Y., Cao, W., and Yan, Y. (2021). Geometric Algebra-Based Esprit Algorithm for DOA Estimation. Sensors 21, 5933. doi:10.3390/s21175933
Xiao, J., Xu, H., Gao, H., Bian, M., and Li, Y. (2021). A Weakly Supervised Semantic Segmentation Network by Aggregating Seed Cues: The Multi-Object Proposal Generation Perspective. ACM Trans. Multimed. Comput. Commun. Appl. 17, 1–19. doi:10.1145/3419842
Xin, W., You-Li, W., and Fu, L. (2013). “A New Multi-Source Image Sequence Fusion Algorithm Based on Sidwt,” in 2013 seventh international conference on image and graphics, Qingdao, China, 26-28 July 2013, 568–571. doi:10.1109/icig.2013.119
Yanan, G., Xiaoqun, C., Bainian, L., Kecheng, P., Guangjie, W., and Mei, G. (2020). Research on Numerically Solving the Inverse Problem Based on L1 Norm. IOP Conf. Ser. Mater. Sci. Eng. 799, 012044. doi:10.1088/1757-899X/799/1/012044
Zhang., Y., Guo, C., and Zhao, P. (2021). Medical Image Fusion Based on Low-Level Features. Comput. Math. methods Med. 2021–13. doi:10.1155/2021/8798003
Zhao, Y., Zhang, Y., Han, J., and Wang, Y. (2020). “Analysis of Image Quality Assessment Methods for Aerial Images,” in 10th International Conference on Computer Engineering and Networks. Editors Q. Liu, X. Liu, T. Shen, and X. Qiu (Singapore: Springer), 188–195. doi:10.1007/978-981-15-8462-6_19
Zhu, L., Wang, W., Qin, J., Wong, K.-H., Choi, K.-S., and Heng, P.-A. (2017). Fast Feature-Preserving Speckle Reduction for Ultrasound Images via Phase Congruency. Signal Process. 134, 275–284. doi:10.1016/j.sigpro.2016.12.011
Keywords: multi-modal medical image, sparse representation, geometric algebra, image fusion, dictionary learning (DL)
Citation: Li Y, Fang N, Wang H and Wang R (2022) Multi-Modal Medical Image Fusion With Geometric Algebra Based Sparse Representation. Front. Genet. 13:927222. doi: 10.3389/fgene.2022.927222
Received: 24 April 2022; Accepted: 30 May 2022;
Published: 23 June 2022.
Edited by:
Ying Li, Zhejiang University, ChinaReviewed by:
Wei Xiang, La Trobe University, AustraliaCopyright © 2022 Li, Fang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Wang, cndhbmdAc2h1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.