- 1Department of Plastic and Reconstructive Surgery, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of Pathology, Shanghai Ninth People’s Hospital, Affiliated to Shanghai Jiaotong University School of Medicine, Shanghai, China
- 3Department of Dermatology, Shanghai Ninth People’s Hospital, Affiliated to Shanghai Jiaotong University School of Medicine, Shanghai, China
Background: Dermatofibrosarcoma protuberans (DFSP) is a rare cutaneous sarcoma characterized by local invasion and recurrence. RNA sequencing (RNA-seq) allows the qualification of cellular RNA populations and provides information on the transcriptional state. However, few studies have comprehensively analyzed DFSP transcriptional data.
Methods: Fourteen DFSP samples with paired non-neoplastic soft tissue from Chinese patients undergoing Mohs micrographic surgery were used for RNA-seq analysis. Differential expression analysis and enrichment analysis for RNA-seq data were performed to identify fusion genes, biomarkers, and microenvironment characteristics of DFSP.
Results: This study systemically describes the transcriptomic characteristics of DFSP. First, we performed gene fusion analysis and identified a novel FBN1-CSAD fusion event in a DFSP patient with fibrosarcomatous transformation. Then, we identified TLK2 as a biomarker for DFSP based on functional enrichment analysis, and validated its accuracy for diagnosing DFSP by immunohistochemical staining and joint analysis with public data. Finally, microenvironment analysis described the infiltration characteristics of immune and stromal cells in DFSP.
Conclusion: This study demonstrates that RNA-seq can serve as a promising strategy for exploring molecular mechanisms in DFSP. Our results provide new insights into accurate diagnosis and therapeutic targets of DFSP.
Introduction
Dermatofibrosarcoma protuberans (DFSP) is a rare, low-grade cutaneous sarcoma originating from fibroblasts. According to the National Comprehensive Cancer Network (NCCN) guidelines (version 1.2020), the incidence rate of DFSP ranges from 4.2 to 4.5 cases per million people in the United States. Despite the low incidence rate, it is still regarded as the most common skin sarcoma (Ugurel et al., 2019). DFSP most commonly occurs in the trunk, followed by the extremities, head, and neck (Kreicher et al., 2016). It can be divided into dozens of variants based on the composition and microscopic characteristics, such as fibrosarcomatous (FS-DFSP), pigmented, and myxoid (Iwasaki et al., 2019). The rearrangement of chromosomes 17 and 22 leads to the fusion of the PDGFB (beta-type platelet-derived growth factor) gene and the COL1A1 (collagen type 1 alpha 1) gene, which is thought to contribute to tumorigenesis. At the same time, other types of gene fusion, such as COL6A3-PDGFD (Dickson et al., 2018), TNC-PDGFD (Chen et al., 2021), and MAP3K7CL-ERG (Maloney et al., 2019), have been reported in some cases and contribute to the appearance of DFSP.
DFSP is usually characterized by painless, slow-growing nodules, which appear blue-purple or tan in color (Li Y. et al., 2020). However, the specific clinical manifestations of DFSP are lacking. Diagnosis of DFSP mainly depends on hematoxylin–eosin (H&E) and immunohistochemistry stains. Microscopically, tumor cells show a spindle-shaped appearance and are arranged in a storiform pattern. These spindle cells always infiltrate between fat cells and exhibit a ‘honeycomb’ appearance; however, the epidermis is usually not invaded (Kornik et al., 2012). A positive immunohistochemistry for CD34 is considered the only sensitive marker for DFSP and has been used as a diagnostic tool for decades. However, both the microscopic and immunohistochemical features mentioned above are not specific. For example, sclerotic fibroma and solitary fibrous tumors may show similar cellular arrangement features and positive immunohistochemistry for CD34 (Rapini and Golitz, 1989; Westra et al., 1994; Hanft et al., 2000). Thus, it is not easy to make an accurate and timely diagnosis.
TLK2 (tousled-like kinases 2), a member of the tousled-like kinase (TLK) family, encodes a nuclear serine/threonine kinase. In mammals, TLKs affect genome maintenance, cell-cycle progression, and cell division, mainly by regulating histone 3 and histone 4 in S phase (Silljé et al., 1999; Silljé and Nigg, 2001; Han et al., 2005). TLK2 is thought to be associated with the occurrence and progression of multiple diseases. Haploinsufficiency of TLK2 may lead to a series of neurodevelopmental disorders, such as neurodevelopment delay and behavior disorders (Reijnders et al., 2018). At the same time, amplification of TLK2 plays a crucial role in the development of cancers and is correlated with poor prognosis (Stevens et al., 2011; Kim et al., 2016; Mertins et al., 2016; Kuczler et al., 2022). According to the above observations, TLK2 may serve as a biomarker and therapeutic target for multiple diseases.
Next-generation sequencing (NGS) has become a cost-effective technology and is widely used to survey a variety of diseases, allowing the identification of new markers for clinical diagnostics and therapeutics (Kaczkowski et al., 2016; Zhu et al., 2021). RNA sequencing (RNA-seq) allows the detection of all RNA transcripts and provides information on the transcriptional state. Currently, there are millions of RNA-seq samples stored in public databases, such as The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). However, there is an absence of RNA-seq information on DFSP in public databases. Hence, we compiled 27 samples for sequencing, including 14 DFSP samples and 13 normal adjacent tissues. Using R software, we performed gene fusion analysis and identified a novel FBN1-CSAD fusion event. We also screened for the differentially expressed genes (DEGs) and showed that TLK2 served as a specific biomarker for DFSP. Finally, we compared the immune profile of the microenvironment between tumors and normal adjacent tissues, delineating an overall immune landscape of DFSP. Our study uncovered the mechanisms of DFSP at the transcriptome level and provides preliminary data that may allow accurate diagnosis and treatment in the future.
Materials and methods
Clinical samples
A total of 14 formalin-fixed, paraffin-embedded DFSP tissues and paired normal adjacent tissues were collected from patients at Shanghai Ninth People’s Hospital Affiliated Shanghai Jiao Tong University School of Medicine. An expert dermatopathologist confirmed all samples pathologically through hematoxylin–eosin (H&E) staining, immunohistochemistry, and fluorescence in situ hybridization (FISH). The study protocols were granted by the Clinical Research Ethics Committees of Shanghai Ninth People’s Hospital Affiliated Shanghai Jiao Tong University School of Medicine (Approval #: 2017-451-T3347). All the patients signed written informed consent to use their tissue samples.
RNA sequencing and quality control
After standard RNA extraction, quality control was done using an Agilent 2100 Bioanalyzer and a NanoDrop Spectrophotometer. In the 14 paired samples, the normal adjacent tissue of Case 11 was excluded because of its low RNA concentration, and 27 samples were sequenced in total. Clean data were obtained using fastp software (version 0.20.0) and aligned to the reference genome hs37d5 by STAR alignment software (version 2.7.6a). Samples meeting the following criteria were regarded as valid data: 1) more than ∼60% reads uniquely mapped; 2) less than ∼25% reads multiply mapped or unmapped; 3) rRNA rate less than ∼15%. All the expression data were standardized and transformed as fragments per kilobase million (FPKM) and transcripts per kilobase million (TPM) for subsequent analysis.
Gene fusion detection
Based on the results of the STAR alignment, we used Arriba (https://github.com/suhrig/arriba/) to detect fusion genes in the RNA-seq data. We defined a positive fusion event as appearing when >20 supporting reads could be detected in each sample. The tumor sample of Case 3 was omitted from the analysis because of quality issues.
Identifying differentially expressed genes
Principal component analysis (PCA) of the top 5,000 expressed genes was conducted and visualized using the R package ggplot2 to cluster samples with similar gene expression profiles. Differentially expressed genes between tumor and normal adjacent tissue samples were assessed using the R package DESeq2 (version 1.26.0). To make the result more reliable, we only retained genes whose expression could be detected in all samples. We identified the genes which satisfied the following conditions simultaneously as significantly differentially expressed genes: 1) the adjusted p-value (Padj.) was less than 0.05; 2) the basemean was greater than 100.
Functional enrichment analysis
Enrichment analysis allows us to explore gene clusters with similar function or expression trends. We performed weighted gene co-expression network analysis (WGCNA) with the R package WGCNA (version 1.70-3) for all differentially expressed genes. A soft-threshold power of 18 and module size greater than 30 genes were selected to construct a scale-free network. We merged the modules with a similarity greater than 0.75, which were considered to perform similar functions. For genes in selected modules, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were done using the R package clusterProfiler (version 3.14.0). Significant functions and pathways were selected with Padj. < 0.05.
Public data collection and processing
RNA-seq data of the TCGA sarcoma cohort were downloaded from the UCSC Xena database (https://xena.ucsc.edu) and selected by R (version 3.6.3). We selected soft-tissue sarcomas that occurred in the head and neck, trunk, and extremities for subsequent analysis. RUVg function in RUVSeq (version 1.27.0) was used to remove batch effects (parameter k = 1) between TCGA and our RNA-seq data. The receiver operating characteristic (ROC) curves and area under ROC curves (AUC) were plotted and calculated to predict the diagnostic ability of target genes.
Immunohistochemical staining
To immunostain FAM118B and TLK2 protein in DFSP samples, formalin-fixed and paraffin-embedded specimens were cut into 4 µm thick sections and stained with standard protocols. TLK2 polyclonal antibody (1: 100, ab224729, abcam) and FAM118B polyclonal antibody (1:100, PA5-59660, Thermo Fisher Scientific) were used as primary antibodies. Sections were also stained with hematoxylin and eosin for histopathological confirmation of DFSP tissue.
Immune landscape analysis
The abundance of the tumor microenvironment was evaluated by the R package immunedeconv (version 2.0.0) and ssGSEA algorithm. The score of each immune cell type was calculated according to their characteristic gene expression. The immune score, stroma score, and microenvironment score of each sample were evaluated based on xCell algorithms.
Statistical analysis
We used the Wilcoxon rank-sum test for paired testing of the immune landscape analysis between DFSP and normal adjacent tissues. A two-tailed p-value < 0.05 was considered statistically significant. All analyses were performed and visualized using R software (version 3.6.3).
Results
Clinical samples for RNA sequencing
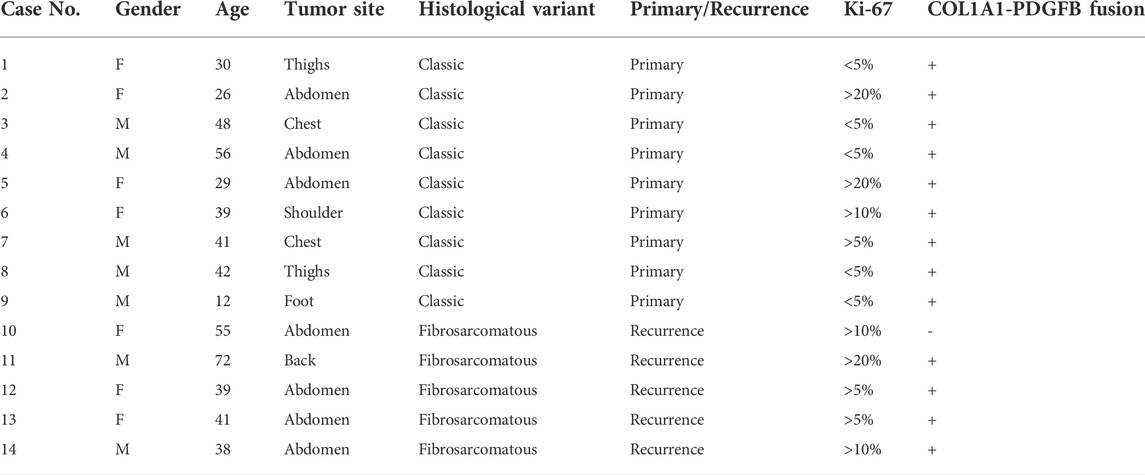
Fourteen formalin-fixed, paraffin-embedded DFSP tissues and paired normal adjacent tissues were retrospectively collected from 14 DFSP patients for RNA-seq analyses. Clinical information on the selected patients is summarized in Table 1. Of the 14 patients, nine were diagnosed with typical DFSP and five with FS-DFSP. Figure 1 shows the histological and immunohistochemical features of typical DFSP and FS-DFSP samples. FISH confirmed that 13 of the 14 patients were positive for the COL1A1-PDGFB fusion. No COL1A1-PDGFB fusion was detected in Case 10. Supplementary Figure S1 shows the histological and immunohistochemical features of the DFSP lesion of Case 10. Normal adjacent tissues were obtained from the negative margins of Mohs micrographic surgery. After quality control, we excluded the normal adjacent tissue of Case 11 owing to its low RNA concentration. Thus, RNA-seq was performed on 27 samples (14 tumors and 13 normal adjacent tissues). The alignment efficiency is presented in Supplementary Table S1. According to the criteria described in the Materials and Methods, 20 samples (13 tumors and 7 normal adjacent tissues) were included in the following analysis.
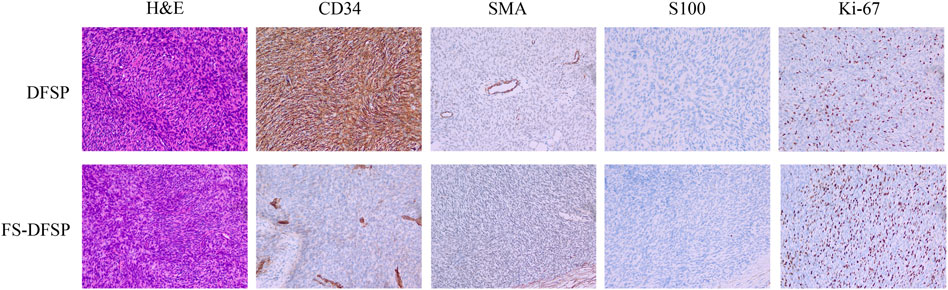
FIGURE 1. Hematoxylin–eosin (H&E) staining and immunohistochemical expression of dermatofibrosarcoma protuberans (DFSP) and dermatofibrosarcoma protuberans of fibrosarcomatous variant (FS-DFSP).
Detection of fusion genes in DFSP lesions
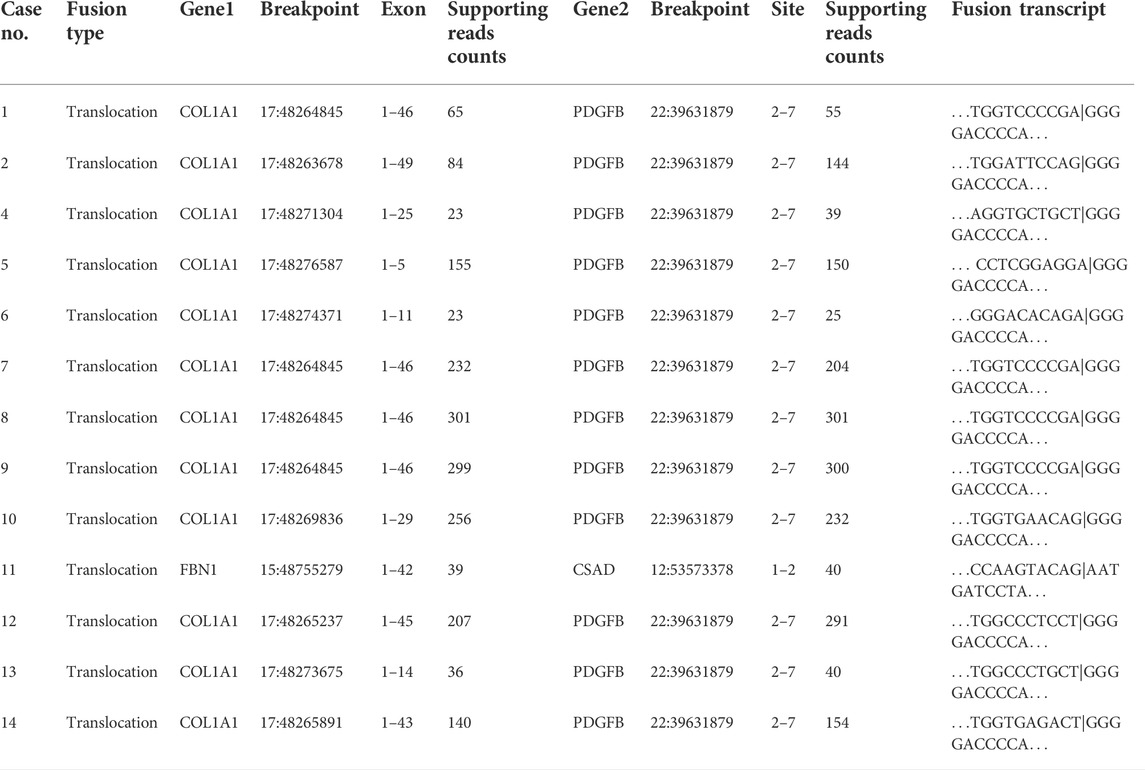
Gene fusions are characteristic of many sarcoma subtypes, including DFSP (Steele et al., 2019). Thus, we focused on fusion gene analysis using Arriba. Consistent with our FISH results, the COL1A1-PDGFB fusion gene was detected in all samples except Case 10 (a DFSP patient with fibrosarcomatous transformation). For patients with the COL1A1-PDGFB fusion gene, the PDGFB breakpoints were all located on chr 22:39631879, while the COL1A1 breakpoints varied from chr 17:48263678 to 17:48276587. We found that case 10 harbored an FBN1-CSAD fusion, which has not been reported previously. The FBN1 breakpoint was located on chr 15:48755279 and the CSAD breakpoint was located on chr 12:5357337. The histological and immunohistochemical features of this lesion are shown in Supplementary Figure S1. Table 2 shows the specific gene fusion profile and breakpoint location for each sample.
Identification of differentially expressed genes
PCA maps of the top 5,000 expressed genes suggested that the gene profiles differed between the DFSP and normal adjacent tissues (Figure 2A). However, we found that the gene profiles were partially overlapping between the cluster of DFSP and FS-DFSP groups (Figure 2B). Compared with genes in normal adjacent tissues, a total of 7,540 DEGs, including 3523 upregulated and 4,017 downregulated genes, were identified in DFSP samples using the screening conditions described in the Materials and methods. The Supplementary Table S2 and Supplementary Table S3 shows the top 50 downregulated genes and up regulated genes respectively according to adjust p-value. The heatmap of the top 500 most differentially expressed genes is shown in Figure 2C.
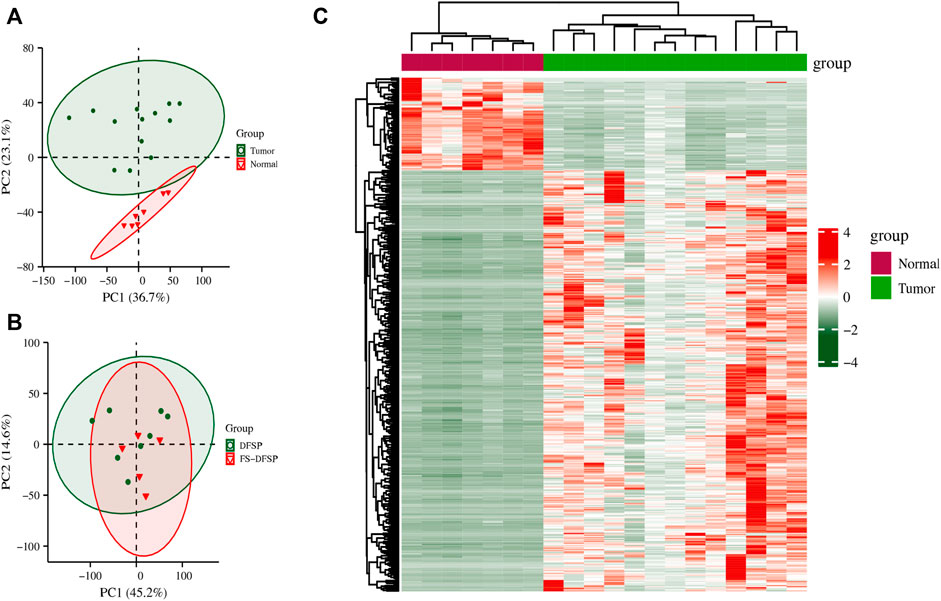
FIGURE 2. Differentially expressed genes. (A) Principal Component Analysis (PCA) of genes between the tumor and normal adjacent tissue groups. (B) PCA of genes between the dermatofibrosarcoma protuberans (DFSP) group and dermatofibrosarcoma protuberans of fibrosarcomatous variant (FS-DFSP) group. (C) The top 500 differentially expressed genes between the tumor and normal adjacent tissue groups. Red, upregulated; green, downregulated.
Construction of WGCNA network and identification of significant modules
We performed WGCNA on all 7,540 DEGs to explore the correlation between gene expression and clinical traits. A soft-thresholding power of 18 made the scale-free fitting index R2 = 0.91, which led to a scale-free network (Figure 3A). Enrichment with a minimum module size of 30 and merging modules with more than 75% similarity yielded eight modules (Figure 3B). Then, each module was associated with the clinical traits (Figure 3C). We found 665 genes in the lightcyan module that correlated to the PDGFB expression with a correlation value of 0.88 (p = 4 × 10–7), which may be associated with DFSP tumorigenesis (Figure 3D). GO/KEGG enrichment analysis for the lightcyan module showed that genes involved in collagen and extracellular organization were enriched (Figure 3E). We also observed a strong positive correlation (r = 0.88, p = 3 × 10–7) between the 617 genes in the black module and Ki-67 expression, which may be associated with DFSP growth and proliferation (Figure 3F). GO/KEGG enrichment analysis for the black module showed that genes involved in cell division were enriched (Figure 3G).
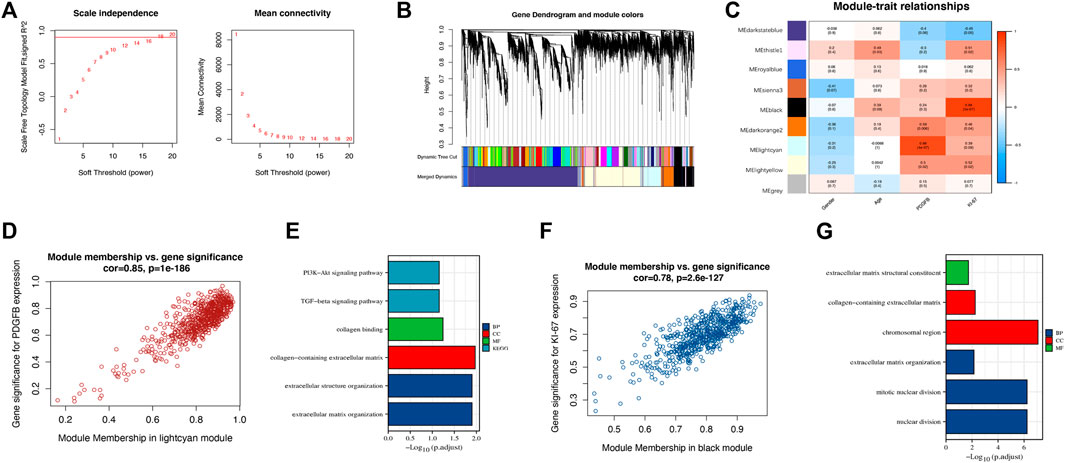
FIGURE 3. Identification of hub genes associated with tumorigenesis. (A) A soft-thresholding power of 18 was selected to construct a scale-free network. (B) Cluster dendrogram of differentially expressed genes. Each branch stands for a single gene, and each color module stands for a gene cluster performing the same function. (C) Correlation heatmap between gene cluster and gender, age, PDGFB, and Ki-67 expression. (D) The scatter plot of gene significance versus module membership in the lightcyan module. (E) Enrichment analysis of the function of genes in the lightcyan module. (F) The scatter plot of gene significance versus module membership in the black module. (G) Enrichment analysis of the function of genes in the black module.
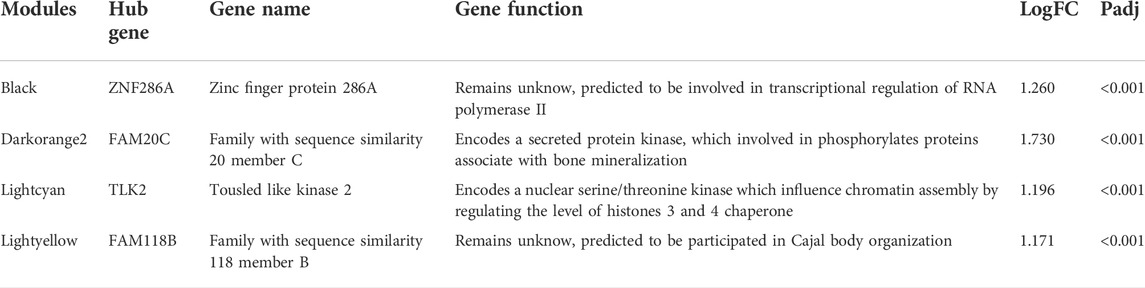
Ultimately, we excluded the darkstateblue, royalblue, thistle1, and sienna3 modules from further analysis. We obtained the hub genes for the remaining modules, including ZNF286A, FAM20C, TLK2, and FAM118B (Table 3).
The diagnostic value of four specifically expressed hub genes in DFSP samples
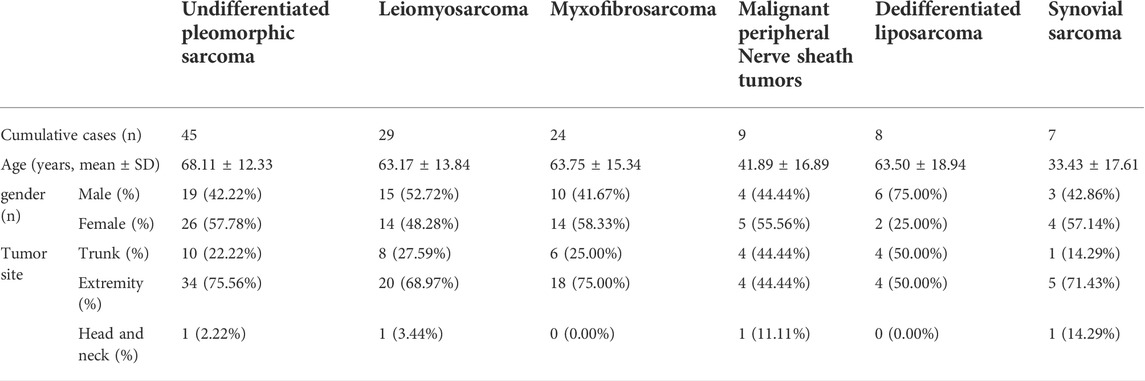
The ROC curve and AUC were used to evaluate the effectiveness of the diagnostic test. A heterogeneous group of patients with superficial tumors from the TCGA database was also enrolled for subsequent analysis to ensure both diagnostic sensitivity and specificity of selected genes. In total, 122 samples were enrolled, including undifferentiated pleomorphic sarcoma (n = 45), leiomyosarcoma (n = 29), myxofibrosarcoma (n = 24), malignant peripheral nerve sheath tumors (n = 9), dedifferentiated liposarcoma (n = 8), and synovial sarcoma (n = 7). Table 4 shows the clinical traits of selected samples in the TCGA database. After removing the batch effect (Figure 4A), the diagnostic values of FAM118B (AUC: 0.837; Figure 4B) and TLK2 (AUC: 0.821; Figure 4C) were greater than 0.8. The diagnostic values of FAM20C (AUC: 0.741; Figure 4D) and ZNF286A (AUC: 0.706; Figure 4E) were weaker. By immunohistochemistry, TLK2 expression was positive while FAM118B was negative in DFSP tissues (Figure 4F). Thus, we hypothesized that TLK2 might be a biomarker for diagnosing DFSP based on our present samples.
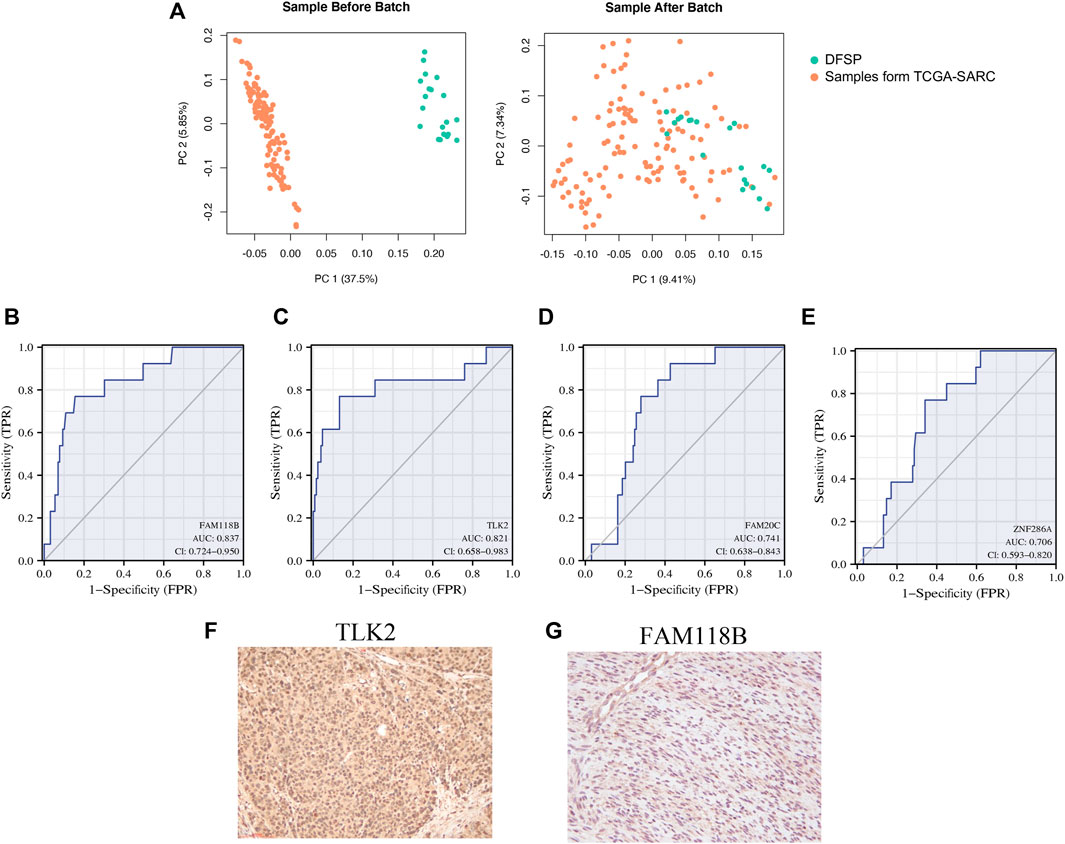
FIGURE 4. ROC curve of four specifically expressed hub genes. (A) Principal Component Analysis (PCA) of genes between public RNA-seq data and DFSP RNA-seq data. Orange, public RNA-seq data; green, our RNA-seq data. (B–E) Receiver operating characteristic (ROC) curves of (B) FAM118B; (C) TLK2; (D) FAM20C; and (E) ZNF286A. (F,G) Immunohistochemical staining of (F) TLK2 and (G) FAM118B.
Immune microenvironment analysis of DFSP
We performed microenvironment analysis to assess the immune and stromal cell abundance in DFSP samples compared with normal tissue (Figure 5). The result of the ssGSEA algorithm showed that the expression of B cells, CD4+ Th2 cells, CD4+ naïve T cells, CD8+ central memory T cells, cancer-associated fibroblasts, and macrophages were significantly higher. CD4+ central memory T cells and eosinophils were expressed significantly lower in tumor cells. However, the immune, stroma, and microenvironment scores showed no statistical differences between the tumor and normal adjacent tissues, indicating that DFSP is an immune-cold tumor.
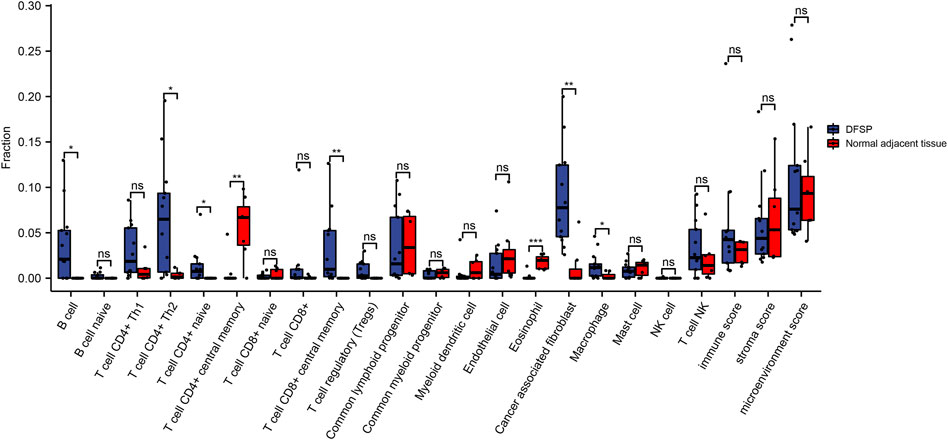
FIGURE 5. Comparison of microenvironment-related cell proportions in DFSP and normal adjacent tissue qualified by xCell algorithm. *p < 0.05, **p < 0.01.
Discussion
DFSP is a rare, aggressive, malignant skin tumor that accounts for about 1% of all sarcoma (Molina et al., 2018). Histologically, the tumor cells exhibit a single spindle pattern with obese or elongated wave-like nuclei, which arranged in a swirling or storiform pattern. Usually, the tumor cells infiltrate diffusely in the dermis and subcutaneous tissue, developing a typical honeycomb appearance, while the epidermis is usually unaffected. Vascular hyperplasia, granulocytic formation, nuclear fenestration and Verocay vesicle can also be found in some cases. Clinically, diagnosis is mainly dependent on morphological characteristics and positive staining for CD34. However, these characteristics significantly overlap with other soft-tissue tumors and lack specificity (Wang et al., 2011). Over decades, several immunohistochemical markers have been implicated for the accurate diagnosis of DFSP in multiple contexts, such as Apo D (Lisovsky et al., 2008), Cthrc1 (Wang et al., 2011), nestin (Sellheyer et al., 2009), the matrix metalloproteinase (MMP) family (Weinrach et al., 2004; Kim et al., 2007; Chen et al., 2012), and WT-1 (Piombino et al., 2021). However, most of these studies have focused on differential diagnosis between dermatofibroma and DFSP, and few markers have been successfully identified.
As sequencing technology develops, NGS has become an effective tool to determine disease mechanisms. Some public data repositories, such as TCGA and GEO, provide sequences and transcript information of millions of samples. In 2003, Linn et al. applied array-based comparative genomic hybridization to nine DFSP samples and identified some highly expressed genes (Linn et al., 2003). In 2020, Köster et al. used NGS to assess the spectrum of DFSP-associated mutations at the chromosome and nucleotide levels (Köster et al., 2020). Unfortunately, we did not find RNA-seq data for DFSP in public databases. Thus, we first carried out RNA-seq on 14 DFSP samples and paired normal adjacent tissues. After quality control, we systematically analyzed the transcriptomic features of DFSP.
The translocation of chromosomes 17 and 22 that generates the COL1A1-PDGFB fusion gene is another characteristic of DFSP. After analyzing the DFSP COL1A1-PDGFB fusion pattern, we found that the PDGFB fusion site was relatively stable (exon 2–7), while the fusion site of the COL1A1 breakpoint varied. At the same time, other rare chromosomal translocations and gene fusions have also been found. The COL6A3-PDGFD fusion gene has been identified in DFSP, mostly associated with DFSP of the breast (Dadone-Montaudié et al., 2018; Dickson et al., 2018). In addition, a novel fusion event between MAP3K7CL and ERG was identified in FS-DFSP (Maloney et al., 2019). Our study included a patient who did not show the COL1A1-PDGFB gene fusion event. We identified a novel FBN1-CSAD fusion event in this patient. Fibrillin 1 (FBN1) is a member of the fibrillin family, which plays a crucial role in extracellular matrix assembly (Chen et al., 2018). FBN1 expression is strongly associated with desmoplasia in ovarian cancer (Millstein et al., 2020). Cysteine sulfinic acid decarboxylase (CSAD) encodes a member of the decarboxylase family and plays a crucial role in cysteine metabolism. However, to the best of our knowledge, no research has shown that CSAD correlates with tumor progression. As the fibrosarcomatous transformation of DFSP is complex, identification of this novel FBN1-CSAD fusion gene is vital in delving into the potential molecular mechanisms of DFSP and its fibrosarcomatous transformation.
Through DEG and WGCNA analyses, we identified four potential biomarkers for DFSP. FAM118B is thought to correlate with the composition and function of Cajal bodies, compartments for the biogenesis of small nuclear ribonucleoproteins (Li et al., 2014). Research found that FAM118B promotes NF2 wild-type meningiomas through the formation of YAP-FAM118B fusion genes (Szulzewsky et al., 2020; Schieffer et al., 2021). As a member of the zinc finger protein family, ZNF286A is predicted to regulate the activity of DNA-binding transcription factors. Although other members of this family, such as ZNF224 (Cesaro et al., 2021) and ZNF217 (Li et al., 2021), play important roles in cancer development, the role of ZNF286A has not been studied in any disease. FAM20C is a secreted protein with atypical kinase activity, which plays important roles in biomineralization, cell migration, and wound healing (Xu et al., 2021). Studies have indicated that FAM20C might be a biomarker for diverse cancers, including triple-negative breast cancer (Tagliabracci et al., 2015), lung adenocarcinoma (Li H. et al., 2020), and stomach adenocarcinoma (Liu et al., 2021). TLK2, a nuclear serine-threonine kinase, binds at the H3 binding site to promote ASF1 (histone chaperone protein anti-silencing factor 1) phosphorylation and cell growth (Simon et al., 2022). TLK2 amplifies and serves as an oncogene in multiple diseases (Kim et al., 2016; Lelieveld et al., 2016; Kuczler et al., 2022). TLK2 depletion activates the cGAS–STRING–TBK1 innate immune axis, and a high TLK2 level contributes to immune evasion of cancers positive for alternative lengthening of telomeres (Segura-Bayona et al., 2020). The mRNA expression levels of these four genes were increased in DFSP samples and were closely related to tumorigenesis through WGCNA and GO/KEGG enrichment analysis. Subsequently, a joint analysis with the other six types of soft-tissue sarcoma samples was performed to verify their diagnostic efficacy. The ROC curves suggested that FAM20C and ZNF286A have a lower specificity for differentiating DFSP from other tumors (AUC <0.75) compared with FAM118B and TLK2 (AUC >0.8). Immunohistochemical staining revealed the presence of TLK2 in DFSP cells while the expression of FAM118B was absent. Therefore, we believe that TLK2 can serve as a biomarker in DFSP.
Furthermore, the microenvironment landscapes showed that the infiltration of non-tumor cells differed between tumor and normal tissues. In general, in most cancers, high infiltration of CD8+ T cells, Th1 cells, and NK cells are indicative of good prognosis, while the infiltration of macrophages, Th17 cells, and Th2 cells are associated with bad prognosis (Bruni et al., 2020). We found increased infiltration of Th2 cells and macrophages (indicating bad prognosis) and no significant difference in CD8+ T cells, Th1 cells, or NK cells (indicating good prognosis), which partially explains the local invasion properties of DFSP. Meanwhile, it is worth noting that cancer-associated fibroblasts were significantly upregulated in DFSP. Cancer-associated fibroblasts are a heterogeneous population of stromal cells that play a vital role in tumor progression and drug resistance (Czekay et al., 2022). They also show intrastromal crosstalk with other types of immune cells, including T cells, dendritic cells, and macrophages (Saw et al., 2022). Targeting cancer-associated fibroblasts directly or indirectly can inhibit tumor malignancy in solid tumors (Saw et al., 2022). Targeted interventions of cancer-associated fibroblasts could serve as a promising measure to inhibit invasion of DFSP.
In conclusion, this study comprehensively describes the transcriptomic characteristics of DFSP and identifies a new FBN1-CSAD fusion gene. At the same time, we have identified TLK2 as a potential biomarker for diagnosis and treatment. Moreover, we have also provided new insight into microenvironment infiltration in DFSP that may identify potential targets for immunotherapy. However, there are several limitations of this study. First, owing to the rarity of this disease, the RNA-seq sample size was limited. Second, validating the diagnostic efficacy of TLK2 in in vivo experiments would be advantageous. In addition, the influence of the tumor microenvironment in DFSP has not been fully clarified and requires further exploration.
Data availability statement
All datasets generated for this study are included in the article/supplementary material.
Ethics statement
The studies involving human participants were reviewed and approved by Clinical Research Ethics Committees of Shanghai Ninth People’s Hospital Affiliated Shanghai Jiao Tong University School of Medicine (Approval #: 2017-451-T3347). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
XZ and DS designed the whole experiments and drafted the manuscript. HZ and YR collected the data and made pathological confirmation. XL and YD performed the data analysis and constructed the figures and tables. JC and JY evaluated and revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by Shanghai Municipal Key Clinical Specialty (NO. shslczdzk00901), Construction Project Clinical Research Project of Multidisciplinary Team, Shanghai Ninth People's Hospital, the Shanghai Jiao Tong University School Medicine (201901), Natural Science Foundation of Shanghai (NO. 21ZR1436800), Interdisciplinary Program of Shanghai Jiao Tong University (No. YG2019QNB13), The National Science Fund for Distinguished Young Scholars (No. 81901962).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.926282/full#supplementary-material
Supplementary Figure S1 | Histological and immunohistochemical features of the lesion in Case 10. The hematoxylin–eosin (H&E) staining shows heterogeneity and irregular arrangement of the spindle tumor cells. The immunohistochemistry shows that the tumor is positive for CD34 and negative for SMA and S100. The positive rate for Ki-67 is approximately 10%.
References
Bruni, D., Angell, H. K., and Galon, J. (2020). The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 20, 662–680. doi:10.1038/s41568-020-0285-7
Cesaro, E., Lupo, A., Rapuano, R., Pastore, A., Grosso, M., and Costanzo, P. (2021). ZNF224 protein: Multifaceted functions based on its molecular partners. Molecules 26, 6296. doi:10.3390/molecules26206296
Chen, M., Yao, B., Yang, Q., Deng, J., Song, Y., Sui, T., et al. (2018). Truncated C-terminus of fibrillin-1 induces Marfanoid-progeroid-lipodystrophy (MPL) syndrome in rabbit. Dis. Model. Mech. 11, dmm031542. doi:10.1242/dmm.031542
Chen, Y.-T., Chen, W.-T., Huang, W.-T., Wu, C.-C., and Chai, C.-Y. (2012). Expression of MMP-2, MMP-9 and MMP-11 in dermatofibroma and dermatofibrosarcoma protuberans. Kaohsiung J. Med. Sci. 28, 545–549. doi:10.1016/j.kjms.2012.04.017
Chen, Y., Shi, Y.-Z., Feng, X.-H., Wang, X.-T., He, X.-L., and Zhao, M. (2021). Novel TNC-PDGFD fusion in fibrosarcomatous dermatofibrosarcoma protuberans: A case report. Diagn. Pathol. 16, 63. doi:10.1186/s13000-021-01123-1
Czekay, R.-P., Cheon, D.-J., Samarakoon, R., Kutz, S. M., and Higgins, P. J. (2022). Cancer-associated fibroblasts: Mechanisms of tumor progression and novel therapeutic targets. Cancers (Basel) 14, 1231. doi:10.3390/cancers14051231
Dadone-Montaudié, B., Alberti, L., Duc, A., Delespaul, L., Lesluyes, T., Pérot, G., et al. (2018). Alternative PDGFD rearrangements in dermatofibrosarcomas protuberans without PDGFB fusions. Mod. Pathol. 31, 1683–1693. doi:10.1038/s41379-018-0089-4
Dickson, B. C., Hornick, J. L., Fletcher, C. D. M., Demicco, E. G., Howarth, D. J., Swanson, D., et al. (2018). Dermatofibrosarcoma protuberans with a novel COL6A3-PDGFD fusion gene and apparent predilection for breast. Genes. Chromosom. Cancer 57, 437–445. doi:10.1002/gcc.22663
Han, Z., Riefler, G. M., Saam, J. R., Mango, S. E., and Schumacher, J. M. (2005). The C. elegans Tousled-like kinase contributes to chromosome segregation as a substrate and regulator of the Aurora B kinase. Curr. Biol. 15, 894–904. doi:10.1016/j.cub.2005.04.019
Hanft, V. N., Shea, C. R., McNutt, N. S., Pullitzer, D., Horenstein, M. G., and Prieto, V. G. (2000). Expression of CD34 in sclerotic (“plywood”) fibromas. Am. J. Dermatopathol. 22, 17–21. doi:10.1097/00000372-200002000-00003
Iwasaki, T., Yamamoto, H., and Oda, Y. (2019). Current update on the molecular biology of cutaneous sarcoma: Dermatofibrosarcoma protuberans. Curr. Treat. Options Oncol. 20, 29. doi:10.1007/s11864-019-0628-3
Kaczkowski, B., Tanaka, Y., Kawaji, H., Sandelin, A., Andersson, R., Itoh, M., et al. (2016). Transcriptome analysis of recurrently deregulated genes across multiple cancers identifies new pan-cancer biomarkers. Cancer Res. 76, 216–226. doi:10.1158/0008-5472.CAN-15-0484
Kim, H. J., Lee, J. Y., Kim, S. H., Seo, Y. J., Lee, J. H., Park, J. K., et al. (2007). Stromelysin-3 expression in the differential diagnosis of dermatofibroma and dermatofibrosarcoma protuberans: Comparison with factor XIIIa and CD34. Br. J. Dermatol. 157, 319–324. doi:10.1111/j.1365-2133.2007.08033.x
Kim, J.-A., Tan, Y., Wang, X., Cao, X., Veeraraghavan, J., Liang, Y., et al. (2016). Comprehensive functional analysis of the tousled-like kinase 2 frequently amplified in aggressive luminal breast cancers. Nat. Commun. 7, 12991. doi:10.1038/ncomms12991
Kornik, R. I., Muchard, L. K., and Teng, J. M. (2012). Dermatofibrosarcoma protuberans in children: An update on the diagnosis and treatment. Pediatr. Dermatol. 29, 707–713. doi:10.1111/j.1525-1470.2012.01767.x
Köster, J., Arbajian, E., Viklund, B., Isaksson, A., Hofvander, J., Haglund, F., et al. (2020). Genomic and transcriptomic features of dermatofibrosarcoma protuberans: Unusual chromosomal origin of the COL1A1-PDGFB fusion gene and synergistic effects of amplified regions in tumor development. Cancer Genet. 241, 34–41. doi:10.1016/j.cancergen.2019.12.001
Kreicher, K. L., Kurlander, D. E., Gittleman, H. R., Barnholtz-Sloan, J. S., and Bordeaux, J. S. (2016). Incidence and survival of primary dermatofibrosarcoma protuberans in the United States. Dermatol. Surg. 42 Suppl 1, S24–S31. doi:10.1097/DSS.0000000000000300
Kuczler, M. D., Zieren, R. C., Dong, L., de Reijke, T. M., Pienta, K. J., and Amend, S. R. (2022). Advancements in the identification of EV derived mRNA biomarkers for liquid biopsy of clear cell renal cell carcinomas. Urology 160, 87–93. doi:10.1016/j.urology.2021.11.002
Lelieveld, S. H., Reijnders, M. R. F., Pfundt, R., Yntema, H. G., Kamsteeg, E.-J., de Vries, P., et al. (2016). Meta-analysis of 2, 104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 19, 1194–1196. doi:10.1038/nn.4352
Li, H., Tong, L., Tao, H., and Liu, Z. (2020a). Genome-wide analysis of the hypoxia-related DNA methylation-driven genes in lung adenocarcinoma progression. Biosci. Rep. 40, BSR20194200. doi:10.1042/BSR20194200
Li, Y., Fong, K.-W., Tang, M., Han, X., Gong, Z., Ma, W., et al. (2014). Fam118B, a newly identified component of Cajal bodies, is required for Cajal body formation, snRNP biogenesis and cell viability. J. Cell. Sci. 127, 2029–2039. doi:10.1242/jcs.143453
Li, Y., Wang, C., Yang, K., Peng, S., Wang, Q., Chen, S., et al. (2020b). Clinical features of dermatofibrosarcoma protuberans and risk factors for local recurrence after Mohs micrographic surgery. J. Am. Acad. Dermatol. 82, 1219–1221. doi:10.1016/j.jaad.2019.09.034
Li, Y., Wu, H., Wang, Q., and Xu, S. (2021). ZNF217: The cerberus who fails to guard the gateway to lethal malignancy. Am. J. Cancer Res. 11, 3378–3405.
Linn, S. C., West, R. B., Pollack, J. R., Zhu, S., Hernandez-Boussard, T., Nielsen, T. O., et al. (2003). Gene expression patterns and gene copy number changes in dermatofibrosarcoma protuberans. Am. J. Pathol. 163, 2383–2395. doi:10.1016/S0002-9440(10)63593-6
Lisovsky, M., Hoang, M. P., Dresser, K. A., Kapur, P., Bhawan, J., and Mahalingam, M. (2008). Apolipoprotein D in CD34-positive and CD34-negative cutaneous neoplasms: A useful marker in differentiating superficial acral fibromyxoma from dermatofibrosarcoma protuberans. Mod. Pathol. 21, 31–38. doi:10.1038/modpathol.3800971
Liu, X., Zhan, Y., Xu, W., Liu, X., Geng, Y., Liu, L., et al. (2021). Prognostic and immunological role of Fam20C in pan-cancer. Biosci. Rep. 41, BSR20201920. doi:10.1042/BSR20201920
Maloney, N., Bridge, J. A., de Abreu, F., Korkolopoulou, P., Sakellariou, S., and Linos, K. (2019). A novel MAP3K7CL-ERG fusion in a molecularly confirmed case of dermatofibrosarcoma protuberans with fibrosarcomatous transformation. J. Cutan. Pathol. 46, 532–537. doi:10.1111/cup.13469
Mertins, P., Mani, D. R., Ruggles, K. V., Gillette, M. A., Clauser, K. R., Wang, P., et al. (2016). Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62. doi:10.1038/nature18003
Millstein, J., Budden, T., Goode, E. L., Anglesio, M. S., Talhouk, A., Intermaggio, M. P., et al. (2020). Prognostic gene expression signature for high-grade serous ovarian cancer. Ann. Oncol. 31, 1240–1250. doi:10.1016/j.annonc.2020.05.019
Molina, A. S., Duprat Neto, J. P., Bertolli, E., da Cunha, I. W., Fregnani, J. H. T. G., Figueiredo, P. H. M., et al. (2018). Relapse in dermatofibrosarcoma protuberans: A histological and molecular analysis. J. Surg. Oncol. 117, 845–850. doi:10.1002/jso.25039
Piombino, E., Broggi, G., Barbareschi, M., Castorina, S., Parenti, R., Bartoloni, G., et al. (2021). Wilms’ tumor 1 (WT1): A novel immunomarker of dermatofibrosarcoma protuberans-an immunohistochemical study on a series of 114 cases of bland-looking mesenchymal spindle cell lesions of the dermis/subcutaneous tissues. Cancers (Basel) 13, E252. doi:10.3390/cancers13020252
Rapini, R. P., and Golitz, L. E. (1989). Sclerotic fibromas of the skin. J. Am. Acad. Dermatol. 20, 266–271. doi:10.1016/s0190-9622(89)70033-5
Reijnders, M. R. F., Miller, K. A., Alvi, M., Goos, J. A. C., Lees, M. M., de Burca, A., et al. (2018). De novo and inherited loss-of-function variants in TLK2: Clinical and genotype-phenotype evaluation of a distinct neurodevelopmental disorder. Am. J. Hum. Genet. 102, 1195–1203. doi:10.1016/j.ajhg.2018.04.014
Saw, P. E., Chen, J., and Song, E. (2022). Targeting CAFs to overcome anticancer therapeutic resistance. Trends Cancer 8, 527–555. doi:10.1016/j.trecan.2022.03.001
Schieffer, K. M., Agarwal, V., LaHaye, S., Miller, K. E., Koboldt, D. C., Lichtenberg, T., et al. (2021). YAP1-FAM118B fusion defines a rare subset of childhood and Young adulthood meningiomas. Am. J. Surg. Pathol. 45, 329–340. doi:10.1097/PAS.0000000000001597
Segura-Bayona, S., Villamor-Payà, M., Attolini, C. S.-O., Koenig, L. M., Sanchiz-Calvo, M., Boulton, S. J., et al. (2020). Tousled-like kinases suppress innate immune signaling triggered by alternative lengthening of telomeres. Cell. Rep. 32, 107983. doi:10.1016/j.celrep.2020.107983
Sellheyer, K., Nelson, P., and Krahl, D. (2009). Dermatofibrosarcoma protuberans: A tumour of nestin-positive cutaneous mesenchymal stem cells? Br. J. Dermatol. 161, 1317–1322. doi:10.1111/j.1365-2133.2009.09363.x
Silljé, H. H., and Nigg, E. A. (2001). Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11, 1068–1073. doi:10.1016/s0960-9822(01)00298-6
Silljé, H. H., Takahashi, K., Tanaka, K., Van Houwe, G., and Nigg, E. A. (1999). Mammalian homologues of the plant Tousled gene code for cell-cycle-regulated kinases with maximal activities linked to ongoing DNA replication. EMBO J. 18, 5691–5702. doi:10.1093/emboj/18.20.5691
Simon, B., Lou, H. J., Huet-Calderwood, C., Shi, G., Boggon, T. J., Turk, B. E., et al. (2022). Tousled-like kinase 2 targets ASF1 histone chaperones through client mimicry. Nat. Commun. 13, 749. doi:10.1038/s41467-022-28427-0
Steele, C. D., Tarabichi, M., Oukrif, D., Webster, A. P., Ye, H., Fittall, M., et al. (2019). Undifferentiated sarcomas develop through distinct evolutionary pathways. Cancer Cell. 35, 441–456. e8. doi:10.1016/j.ccell.2019.02.002
Stevens, K. N., Wang, X., Fredericksen, Z., Pankratz, V. S., Cerhan, J., Vachon, C. M., et al. (2011). Evaluation of associations between common variation in mitotic regulatory pathways and risk of overall and high grade breast cancer. Breast Cancer Res. Treat. 129, 617–622. doi:10.1007/s10549-011-1587-y
Szulzewsky, F., Arora, S., Hoellerbauer, P., King, C., Nathan, E., Chan, M., et al. (2020). Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes. Dev. 34, 1051–1064. doi:10.1101/gad.338681.120
Tagliabracci, V. S., Wiley, S. E., Guo, X., Kinch, L. N., Durrant, E., Wen, J., et al. (2015). A single kinase generates the majority of the secreted phosphoproteome. Cell. 161, 1619–1632. doi:10.1016/j.cell.2015.05.028
Ugurel, S., Kortmann, R.-D., Mohr, P., Mentzel, T., Garbe, C., Breuninger, H., et al. (2019). S1 guidelines for dermatofibrosarcoma protuberans (DFSP) - update 2018. J. Dtsch. Dermatol. Ges. 17, 663–668. doi:10.1111/ddg.13849
Wang, L., Xiang, Y. N., Zhang, Y. H., Tu, Y. T., and Chen, H. X. (2011). Collagen triple helix repeat containing-1 in the differential diagnosis of dermatofibrosarcoma protuberans and dermatofibroma. Br. J. Dermatol. 164, 135–140. doi:10.1111/j.1365-2133.2010.10050.x
Weinrach, D. M., Wang, K. L., Wiley, E. L., and Laskin, W. B. (2004). Immunohistochemical expression of matrix metalloproteinases 1, 2, 9, and 14 in dermatofibrosarcoma protuberans and common fibrous histiocytoma (dermatofibroma). Arch. Pathol. Lab. Med. 128, 1136–1141. doi:10.1043/1543-2165(2004)128<1136:IEOMMA>2.0.CO;2
Westra, W. H., Gerald, W. L., and Rosai, J. (1994). Solitary fibrous tumor. Consistent CD34 immunoreactivity and occurrence in the orbit. Am. J. Surg. Pathol. 18, 992–998. doi:10.1097/00000478-199410000-00003
Xu, R., Tan, H., Zhang, J., Yuan, Z., Xie, Q., and Zhang, L. (2021). Fam20C in human diseases: Emerging biological functions and therapeutic implications. Front. Mol. Biosci. 8, 790172. doi:10.3389/fmolb.2021.790172
Keywords: dermatofibrosarcoma protuberans, RNA sequencing, Tlk2, fusion genes, tumor microenvironment
Citation: Zhang X, Sun D, Zheng H, Rao Y, Deng Y, Liang X, chen J and Yang J (2022) Comprehensive analysis of transcriptome characteristics and identification of TLK2 as a potential biomarker in dermatofibrosarcoma protuberans. Front. Genet. 13:926282. doi: 10.3389/fgene.2022.926282
Received: 22 April 2022; Accepted: 11 August 2022;
Published: 05 September 2022.
Edited by:
Yi Ji, Sichuan University, ChinaReviewed by:
Giuseppe Broggi, University of Catania, ItalySelma Ugurel, University of Duisburg-Essen, Germany
Copyright © 2022 Zhang, Sun, Zheng, Rao, Deng, Liang, chen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Chen, Y2hlbmp1bjEyNTEyNUAxMjYuY29t; Jun yang, eWo1NTU2OUBob3RtYWlsLmNvbQ==
†These authors contributed equally to this work and share first authorship
 Xiao Zhang
Xiao Zhang Di Sun
Di Sun Haiyan Zheng
Haiyan Zheng Yamin Rao
Yamin Rao Yuqi Deng
Yuqi Deng Xiao Liang1
Xiao Liang1 Jun chen
Jun chen