
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 10 August 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.920897
This article is part of the Research TopicMolecular basis of epigenetic regulation in cancer therapiesView all 21 articles
Background: Mitochondria are at the heart of a number of metabolic pathways providing enormous energy for normal cell growth and regulating tumor cell growth as well as survival. Mitochondrial topoisomerase I (TOP1MT) is a type IB topoisomerase found in the mitochondria of vertebrates. However, no pan-cancer analysis of TOP1MT has been reported. This study aims to explore TOP1MT expression in pan-cancer tissues and identify whether it can be a target for mitochondrial anticancer therapy.
Methods and results: The original TOP1MT expression data in 33 different types of cancer patients were downloaded from the TCGA and GTEx databases. TOP1MT was highly expressed in cancer tissues, including BLCA, BRCA, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, PAAD, PCPG, PRAD, READ, SKCM, STAD, THYM, UCEC, and UCS. According to Kaplan-Meier survival curve analysis, high TOP1MT expression in BLCA, HNSC, KIRP, PAAD, UCEC, and LIHC cancer tissues was linked to poor prognosis of cancer patients, i.e., poor OS, disease-specific survival, and PFI. Linkedomics analysis identified a positive correlation of TOP1MT expression with CNA, but a negative correlation with methylation. TOP1MT expression significantly correlated with immune cells and immune checkpoints in the TIMER database. Functional analysis showed a close relationship between TOP1MT expression and ribosomes.
Conclusion: In summary, TOP1MT is a potential biomarker for mitochondrial anticancer therapy and cancer immunotherapy.
The primary role of mitochondria in the human body is the production of Adenosine Triphosphate (ATP) and the synthesis of metabolites necessary for cell bioenergy and biosynthesis (Chandel Navdeep, 2015). Mitochondria generate ATP and metabolites through glycolysis, amino acid decomposition, and fat decomposition, among other processes. These metabolites enter the mitochondria via the Tricarboxylic acid cycle (TCA cycle), producing Nicotinamide adenine dinucleotide (NADH) and reductive molecules, including Flavine adenine dinucleotide, reduced (FADH-2), NADH Subsequently, these energy molecules are phosphorylated via oxidation to produce ATP, whereas the TCA cycle-generated intermediates can be collected and used in other biosynthetic metabolic pathways to produce glucose, amino acids, nucleotides, and other important substances (Spinelli and Haigis, 2018). In this regard, mitochondria regulate several metabolic pathways, supplying enormous energy to cells, which is important for cell growth, survival, and death. Recent evidence indicates that mitochondria promote tumor cell growth and survival in the pathological microenvironment; they also provide tumor microsynthesis and biological energy requirements. This provides a theoretical reference for targeting mitochondria in anticancer therapy. Also, mitochondria are potential targets for the development of novel anticancer drugs (Vyas et al., 2016; Porporato et al., 2017).
Topoisomerase is a ubiquitous enzyme that instantly splits single or double chains in the phosphodiester framework of nucleic acid, regulating the topological structure of nucleic acid (Wang, 2002). Human topoisomerases are classified into two types, i.e., type I and II (Forterre et al., 2007; Forterre and Gadelle, 2009; Singh et al., 2020). Type I topoisomerase instantly cleaves one strand of double-stranded DNA, allowing the other single strand to pass through the gap, thereby disrupting the situation of a DNA superhelix or helix deletion. II-type topoisomerase uniformly cuts both chains and then connects the broken ends. Type I is divided into Type IA (TOP3α and TOP3β) and Type IB (TOP1, TOP1MT), whereas type II is divided into Type IIA (TOP2α and TOP2β) (Hou et al., 2017; Zhang et al., 2017). TOP2α, TOP2β, and TOP3α are located in both nucleus and mitochondria (Low et al., 2003; Zhang et al., 2014; Nicholls et al., 2018). TOP1MT is the only topoisomerase located in the mitochondria (Zhang et al., 2001; Pommier et al., 2016). Unlike other eukaryotic organelles, mitochondria have their circular DNA (mtDNA), encoding 13 oxidative phosphorylation complex proteins. One recent study discovered that TOP1MT is critical in maintaining the integrity of mtDNA and limiting the negative superhelix of mtRNA (Seol et al., 2012; Zhang et al., 2014). TOP1MT maintains tumor cell proliferation and promotes tumor growth in mice models of colon and liver cancers with impaired metabolism (Baechler et al., 2019). TOP1MT is unable to inhibit liver cell proliferation in the liver regeneration model by limiting mtDNA copy number amplification (Khiati et al., 2015). Besides the effects listed above, TOP1MT modulates mitochondrial functional pathways (Zhang and Pommier, 2008; Dalla Rosa et al., 2017; Baechler et al., 2019). Since tumor cells highly depend on mitochondrial biology and TOP1MT could be a target of cancer drugs, inhibiting TOP1MT could be an effective approach for eliminating cancer cells (Scatena et al., 2018).
Herein, we analyzed TOP1MT expression features, prognostic value, correlation with tumor immune infiltrating cells, and biological function using data obtained from cancer genome map (TGCA), TIMER, linkedomics, cBioPortal, and TISIDB databases. TOP1MT regulates tumor development and is a potential biomarker for targeted mitochondrial anticancer therapy.
Gene expression data from normal tissues in the GTEX database (http://gtexportal.org/) were integrated with gene expression data from more than 11,000 tumors of 33 cancer types in the cancer genome atlas (TCGA https://www.cancer.gov/) to analyze and compare the TOP1MT expression pattern in human pan-cancer tissues and normal tissues. Sample size of 33 different tumor types were shown in Supplementary Table S1. Further, we analyzed the correlation of TOP1MT expression in TGCA with different pathological stages of tumor cells. The log2 transformation was used to normalize all expression data. t-test, p < 0.05 was used to determine statistical significance.
The forest graph was used to examine the relationship between TOP1MT expression and the prognosis of 33 different types of cancer patients. The relationship between TOP1MT expression and prognosis of patients with various cancer and pathological stages was further investigated using the Kaplan-Meier curve, which included total survival (OS), disease-specific survival (DSS), and progression-free periods (PFI). The risk ratio (HRS) and 95% confidence interval were calculated using a single factor survival analysis.
The linkedomics database (http://www.linkedomics.org/) was used to investigate the methylation level and copy number variation data levels of TOP1MT in various tumors. Further, the relationship between TOP1MT expression, methylation level, and copy number variation of TOP1MT was analyzed in tumor tissues. Statistically significant differences were estimated by t-test (p<0.05).
Pan-cancer TOP1MT co-expressed genes for gene enrichment analysis were selected to understand the biological function and pathway involved in TOP1MT through TCGA database. The potential pathways were identified by the gene ontology (GO) terminology and the KEGG pathway enrichment analysis. The three categories of GO analysis included biological processes (BP), cellular components (CC), and molecular functions (MF).
The TIMER database (https://cistrome.shinyapps.io/timer/) was used to analyze the relationship between immune infiltrating, TOP1MT expression and through TISIDB (http://cis.hku.hk/TISIDB/) analysis TOP1MT gene expression in the different immune subtypes.
cBioPortal (https://www.cbioportal.org/) is a repository of cancer genome data. TOPIMT copy number alterations (CNA) were investigated in various cancers.
The t-test was used to estimate TOP1MT expression in cancer and normal tissue. Single Factor Cox regression analysis was used to calculate the HR and P. Kaplan-Meier analysis was used to stratify TOPIMT expression. The Spearman correlation analysis was used to evaluate the correlation of TOP1MT expression with the level of methylation and the number of gene amplification. All analyses were performed with R software (version 3.6.3). p <0.05 was used as the significance threshold of statistical analysis.
To explore the role of TOPIMT in cancer, tumor samples from the TGCA were combined with normal samples from GTEx to characterize the TOPIMT mRNA expression. As shown in Figure 1A, TOP1MT expression was consistently higher ion in tumor samples from BLCA, BRCA, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, PAAD, PCPG, PRAD, READ, SKCM, STAD, THYM, UCEC, UCS than in normal tissues (Figure 1B, Supplementary Figure S1A). However, no difference was noted in TOP1MT expression between ACC, CESC, LAML, and TGCT tumor samples and normal tissues (Supplementary Figure S1B). Lower TOP1MT expression was discovered in tumor samples from KICH, THCA, and OV than in normal tissues (Supplementary Figure S1C). The results revealed that TOP1MT is a carcinogenic molecule.
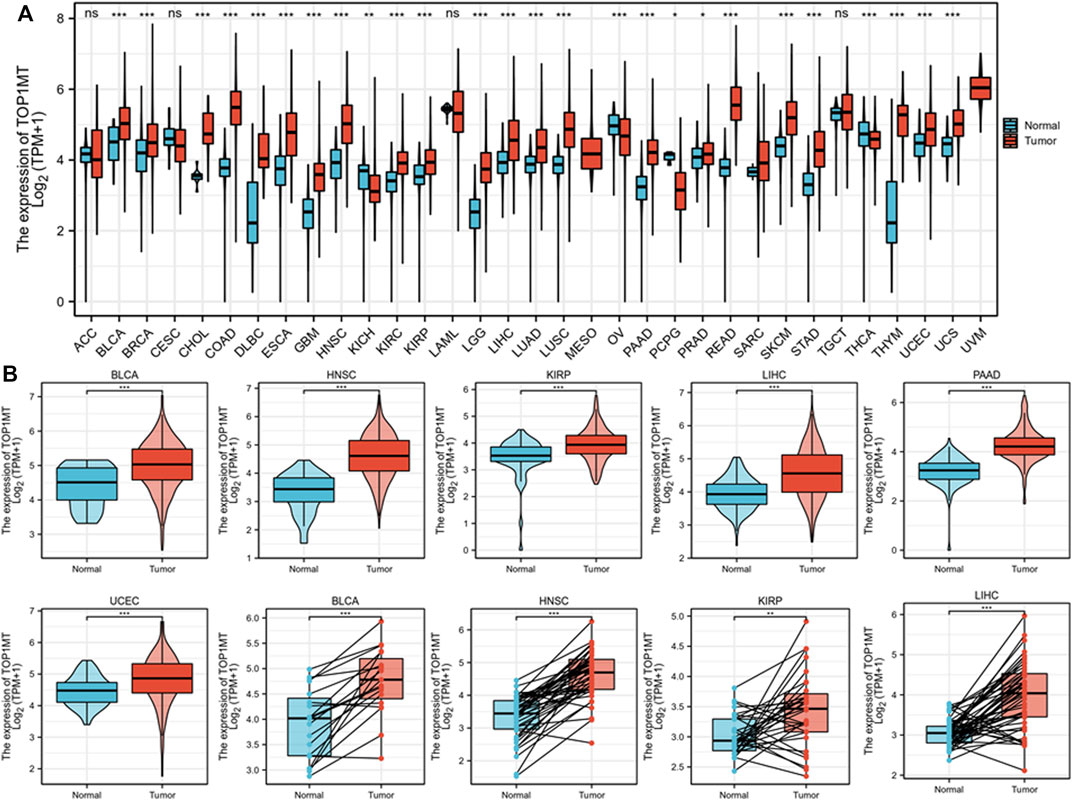
FIGURE 1. Expression pattern of TOP1MT in human generalized carcinoma. (A) Assessing mRNA expression of TOP1MT between tumor and normal tissue using data downloaded from TCGA and GTEx; (B) TOP1MT mRNA expression in patients with BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC from TCGA. Log2 (TPM+1) is used for the logarithmic scale. ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗ p < 0.001.
Further, we investigated the effect of abnormal TOPIMT expression on prognosis. Survival indicators included OS, DSS, and PFI. A Cox regression analysis of 33 cancers revealed that TOP1MT expression significantly correlated with OS in 16, including ACC, BLCA, CESC, DLBC, HNSC, KICH, KIPP, LAML, LIHC, MESO, OV, PAAD, SARC, THYM, UCEC, and UVM (Figure 2A). The Kaplan-Meier survival curve revealed that upregulated TOPIMT expression in BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC significantly correlated with poor overall survival (Figure 2B). Moreover, up-regulation of TOPIMT expression was significantly associated with low OS in ACC, CESC, DLBC, KICH, LAML, MESO, OV, SARC, THYM, and UVM cancer tissues; whereas high TOP1MT expression was significantly associated with high prognosis in DLBC and THYM cancer tissues (Supplementary Figure S2A). This work further investigated the relationship between TOPIMT expression and DSS in cancer patients. TOPIMT expression affected DSS in 13 cancers, including ACC, BLCA, COAD, HNSC, KICH, KIPP, LIHC, MESO, OV, SARC, UCUE, UCS, and UVM (Figure 3A). Kaplan-Meier analysis revealed that upregulated TOPIMT expression was associated with poor DSS in patients with BLCA, HNSC, KIPP, UCEC, and LIHC, but without any significant prognostic value for PAAD (Figure 3B). Also, ACC, COAD, KICH, MESO, OV, SARC, and UVM had poor prognostic values (Supplementary Figure S2B). Cox regression analysis of PFI revealed that upregulated TOPIMT expression was a risk factor for ACC, BLCA, COAD, HNSC, KICH, KIRC, KIPP, LGG, LIHC, OV, PAAD, PRAD, STAD, UCEC, and UVM (Figure 4A). Kaplan-Meier analysis demonstrated a relationship between upregulated TOPIMT expression and poor prognosis in six types of cancer, including BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC (Figure 4B). Furthermore, upregulated TOPIMT expression contributed to adverse prognosis in ACC, COAD, KICH, KIRC, LGG, OV, PAAD, STAD, and UVM (Supplementary Figure S2C). Eventually, Kaplan-Meier Plotter database analysis revealed that high expression in BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC tumor tissues confirmed poor prognosis (Supplementary Figure S3). We identified the six most significant cancers by combining the expression features of TOP1MT mRNA in pan-cancerous tissues and prognostic analysis of OS, DSS, and PFI. These cancers include BLCA, HNSC, KIRP, PAAD, UCEC, and LIHC. High TOP1MT expression in these six types of tumor tissues was significantly linked to poor prognosis.
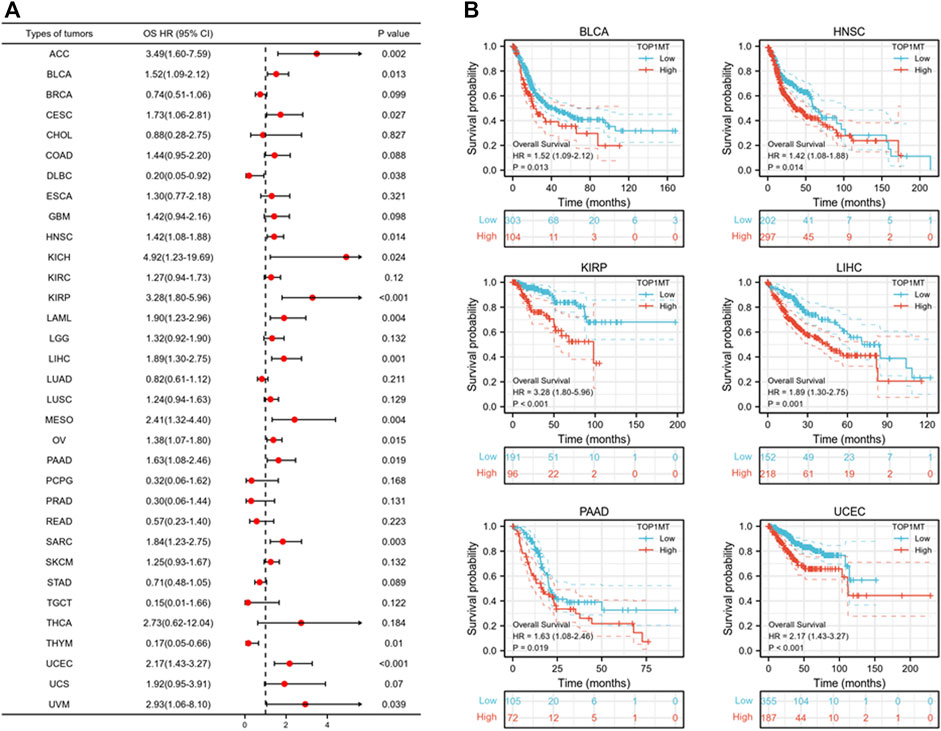
FIGURE 2. Relationship between TOP1MT expression and OS in cancer patients. (A) Forest map of TOP1MT risk ratios in 33 tumors; (B) Kaplan-Meier survival curves of OS in BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC patients. ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗ p < 0.001.
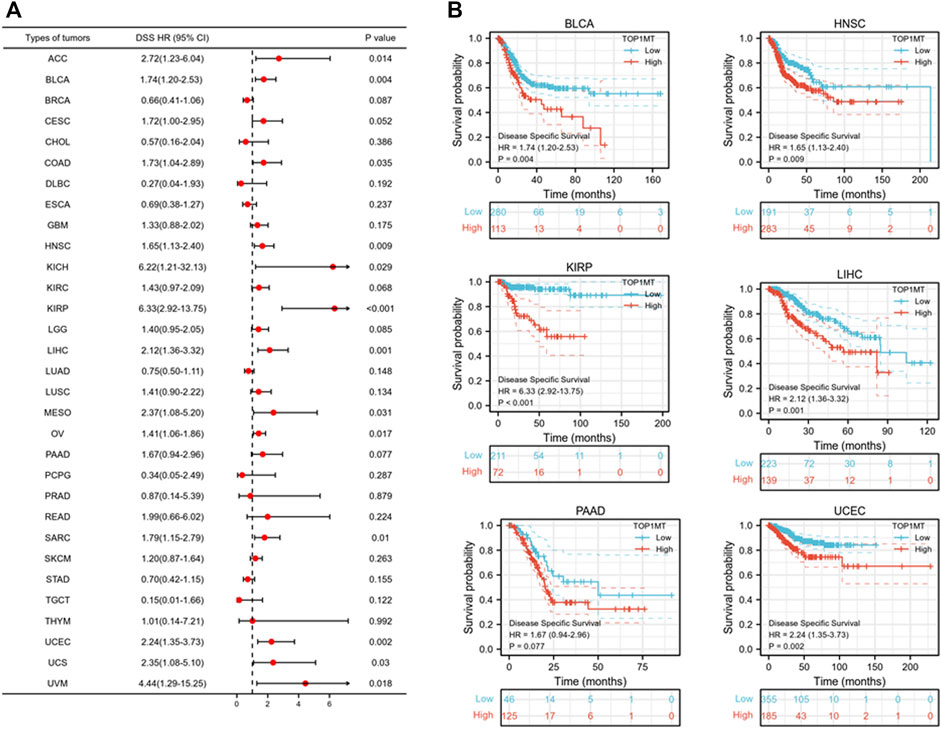
FIGURE 3. Relationship between TOP1MT expression and DSS in cancer patients. (A) Forest map of TOP1MT risk ratios in 33 tumors; (B) Kaplan-Meier survival curve of DSS in PATIENTS with BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC. ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗ p < 0.001.
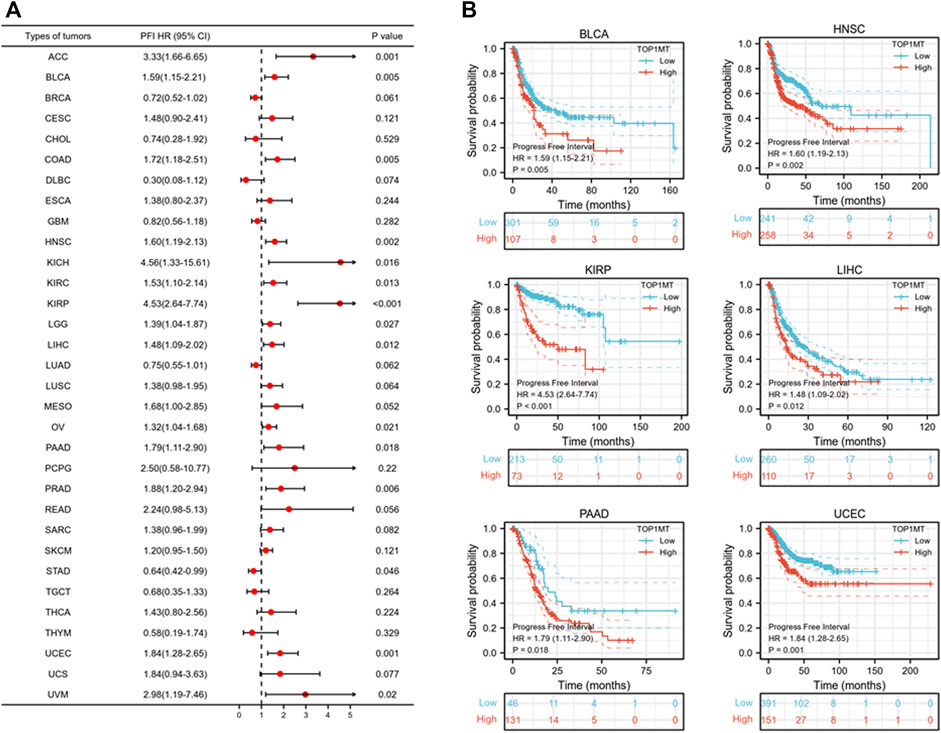
FIGURE 4. Relationship between TOP1MT expression and PFI in cancer patients. (A) Forest map of TOP1MT risk ratios in 33 tumors; (B) Kaplan-Meier survival curves of PFI in PATIENTS with BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC. ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗ p < 0.001.
Secondly, TOPIMT expression was examined in cancer patients with various pathological stages and grades to investigate the expressionist relationship with clinic pathological features. The TCGA database was used to evaluate TOP1MT expression in patients with stage I, II, III, and IV cancers, as well as grades G1, G2, G3, and G4. TOP1MT expression was significantly up-regulated in poor pathological features of BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC (Figure 5A). The Kaplan Meier survival curve analysis revealed that poor pathological stages and grades exhibited a low overall survival (Figure 5B). Finally, the expression features of tumor size, lymph node status, and distant metastasis were investigated between TOP1MT and BLCA, HNSC, KIRP, PAAD, and LIHC using the TCGA database. The higher TOP1MT expression in BLCA, HNSC, KIRP, PAAD, and LIHC tumor tissues, the larger tumor diameter, the wider range of lymph node involvement, and the distant organ metastasis (Supplementary Figure S4A). Further, the relationship between TOP1MT expression and TNM stage prognosis was investigated using the Kaplan-Meier survival curve. The specific manifestations include: High expression of T1&T2 and N0 in BLCA patients indicates a poor prognosis. High expression in T3&T4, lymph node-positive, and M0 patients in HNSA suggested a poor prognosis. High expression in T1&T2 and T3&T4 in KIPP suggested a poor prognosis. High expression in T1&T2, T3&T4, N0, and M0 patients in LIHC suggests a poor prognosis. Patients with PAAD expressing high levels of T1&T2 and N1 had a poor prognosis (Supplementary Figure S4B).
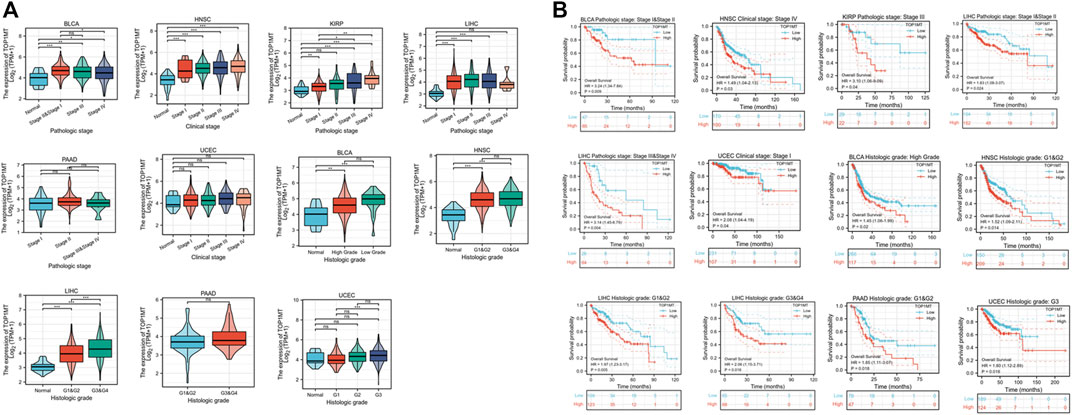
FIGURE 5. Correlation between TOP1MT expression, primary pathological stages, and prognosis. (A) Relationship between TOP1MT mRNA expression and different pathological stages in different cancer patients from TCGA; (B) Kaplan-Meier survival curves of TOP1MT mRNA expression in different pathological stages. Log2 (TPM+1) is used for the logarithmic scale. ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗ p < 0.001.
Linkedomics database was used to analyze the methylation level and copy number changes of TOP1MT in BLCA, HNSC, KIRP, PAAD, UCEC, and LIHC tumor tissues to understand the mechanism of the TOP1MT gene during tumorigenesis. Consequently, tumor tissues including BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC significantly correlated with TOPIMT methylation and gene copy number levels (Figure 6A). TOP1MT expression positively correlated with CNA in BLCA, HNSC, KIRP, PAAD, UCEC, and LIHC tumors, whereas negatively correlated with methylation in BLCA, HNSC, KIRP, PAAD, UCEC, and LIHC tumors (Figure 6B). The relationship between TOP1MT genetic changes and mRNA expression on the cBioPortal website was explored using the GISTIC algorithm. As illustrated in Figure 6C, increased TOP1MT copies (gain and amplification) in tumor tissues significantly increased TOP1MT mRNA expression unlike copy neutral (diploid) and copy loss (shallow deletion). These results show that TOP1MT mRNA expression is affected by changes in its DNA copy number. Thus, abnormal TOP1MTmRNA expression in BLCA, HNSC, KIRP, PAAD, UCEC, and LIHC tumor tissues could be attributed to a high copy number and low methylation level.
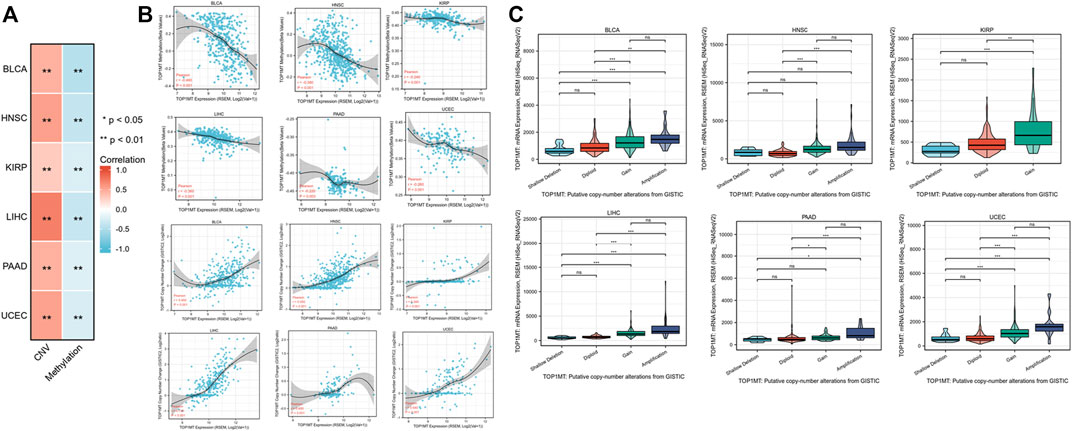
FIGURE 6. CNA and DNA methylation of TOP1MT in human cancer. (A,B) Linkedomics analysis correlation analysis between TOP1MT expression and TOP1MT methylation level and copy number variation data level; (C) cBioPortal analysis of TOP1MT CAN data in different cancer studies. ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗ p < 0.001.
We established the prognostic value of TOPIMT and investigated its relationship with tumor-infiltrating immune cells using the TIMER database. TOPIMT expression significantly correlated with the abundance of infiltrated immune cells (Figure 7A). The clustering heat map of the relationship between TOP1MT and immune cells demonstrated a positive correlation of TOP1MT with B cells and CD4+T lymphocytes in KIPP and LIHC cancers and with purity in BLCA and HNCC. TOP1MT negatively correlated with B cells and CD8+T lymphocytes in HNSC and UCUE, neutrophil cells and dendritic cells in BLCA, HNSC, and UCUE, CD4+T lymphocytes in BLCA and HNSC, and macrophages in BLCA and PAAD. Supplementary Figure S5 shows a specific correlation between each tumor and immune cells. Immune surveillance is widely recognized as important for cancer prognosis, and tumors can evade immune responses utilizing immune checkpoint genes. The correlation of TOP1MT with the expression of immune checkpoint genes was examined to investigate the relationship between TOP1MT expression and the degree of immune invasion in BLCA, HNSC, KIRP, and PAAD, UCEC, and LIHC. TOP1MT expression negatively correlated with common immune checkpoints in BLCA, HNSC, and UCUE (Figure 7B). Also, TOP1MT expression positively correlated with LAG3 in KIRP and negatively correlated with LAG3 and PDCD1LG2 in PAAD. TOP1MT positively correlated with CTLA4 and PDCD1 and negatively correlated with CD274 and PDCD1LG2 in LIHC. These findings suggest that high TOP1MT expression modulates immune invasion of tumor tissues, especially BLCA, HNSC, and UCUE tumor tissues.
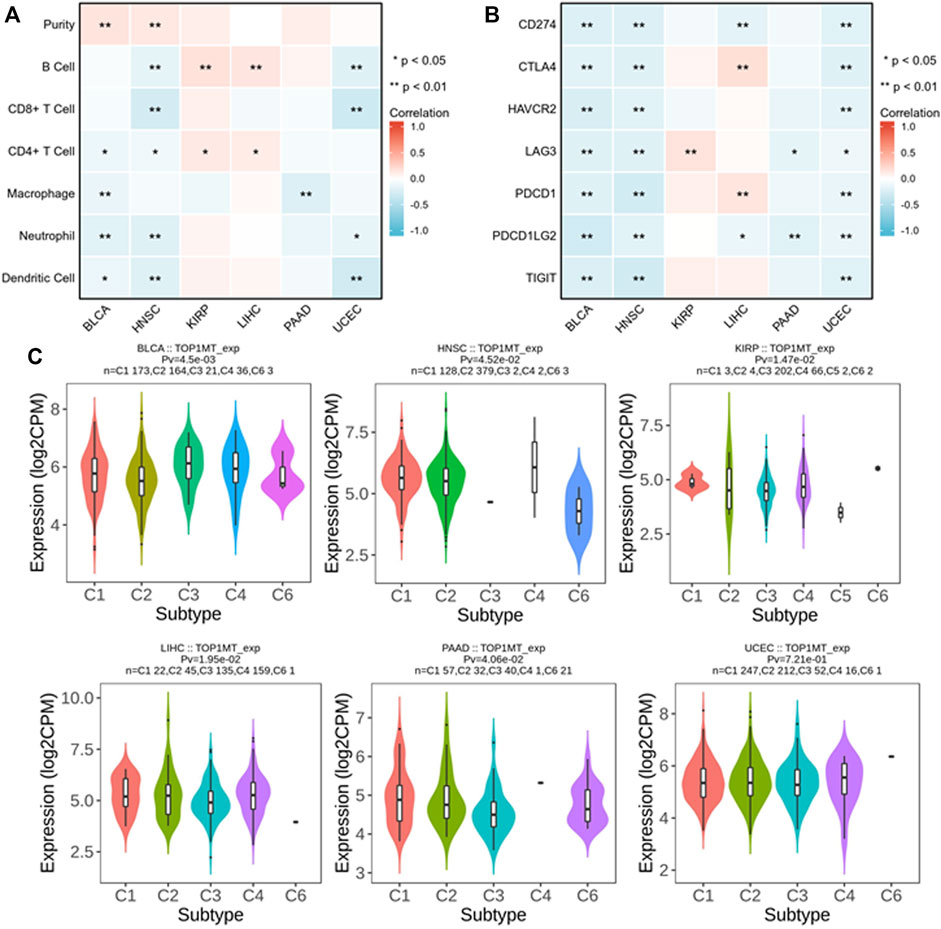
FIGURE 7. Correlation analysis between TOP1MT expression and tumor immune invasion. (A) TOP1MT expression significantly correlates with infiltration level of various immune cells in the TIMER database; (B) TOP1MT expression significantly correlates with various immune checkpoints in the TIMER database; (C) TOP1MT expression in different molecular subtypes of cancer by TISIDB. ∗ p < 0.05, ∗∗ p < 0.01 and ∗∗ p < 0.001.
The expression of the TOP1MT gene in different immune subtypes was analyzed using the TISIDB database. Significant differences in TOPIMT expression characteristics were noted in the C1 (wound healing), C2 (IFN-γ dominant), C3 (inflammation), C4 (lymphocytopenia), C5 (immune silencing), and C6 (TGF-β dominant) subtypes in the six cancer tissues (Figure 7C). TOP1MT was highly expressed in C1 of BLCA, PAAD, UCEC, C2 of HNSC, C3 of KIRP, and C4 of LIHC. Differential TOP1MT expression in different immune subtypes predicts that it regulates cancer prognosis (Thorsson et al., 2019).
KEGG and GO pathway analyses were performed on multiple cancer types to reveal the potential function of TOPIMT. First, we mapped genes expressed in pan-cancer tissues using the TCGA database and identified 122 co-expressed genes in six cancer tissues (Figure 8A). Co-expressed genes were used to examine KEGG and GO pathways. As shown in Figure 8B, TOP1MT modulates the ribosomal pathway of genetic information. As per the GO analysis, TOP1MT affected ribosomal associated BP, CC, and MF in most cancers, including BP (nuclear transcriptional mRNA catabolism process, membrane co-transfer protein, membrane-targeted SRP-dependent co-translation protein, and other biological processes); CC (cytoplasmic part, ribosomal subunit, cytoplasmic ribosome, and other cellular components); MF (rRNA linkage, structural composition of ribosomes, regulatory activity of ubiquitin-protein transferase and other molecular functions).
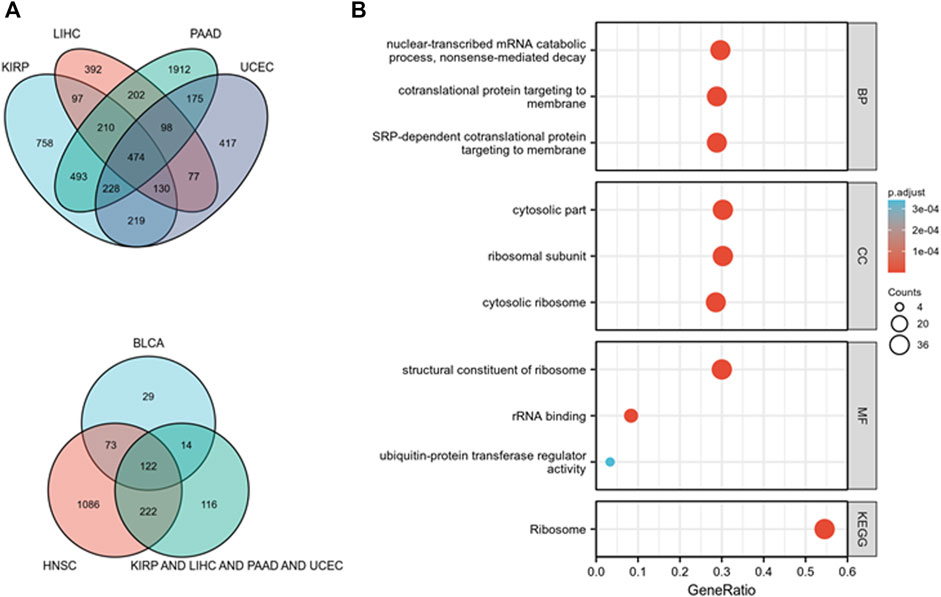
FIGURE 8. TOP1MT functional analysis. (A) Co-expressed genes in BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC tumors; (B) KEGG and GO pathway analysis of co-expressed genes in BLCA, HNSC, KIPP, PAAD, UCEC, and LIHC tumor tissues.
Mitochondrial topoisomerase I (TOP1MT) is a type IB topoisomerase that exists in vertebrates and specifically targets mitochondria (Zhang et al., 2001; Pommier et al., 2016; Baechler et al., 2019). TOP1MT relaxes the mitochondrial DNA (mtDNA) superhelix by introducing instantaneous cutting complexes, revolving broken DNA strands around intact strands (Zhang et al., 2001; Dalla Rosa et al., 2014). In addition to regulating mtDNA, TOP1MT directly affects mitochondrial translation through protein interactions with small mitochondrial glycosomal subunits. TOP1MT inhibitors may be an alternative approach for targeting mitochondrial DNA due to their role in mitochondrial protein synthesis and their up-regulation in several tumors (Khiati et al., 2014). Nonetheless, we did not conduct a pan-cancer analysis on the function of TOP1MT in different cancers.
This work examined TOP1MT expression in pan-cancer datasets. In an analysis of 33 cancer datasets obtained from the TCGA, TOP1MT expression was significantly higher in BLCA, BRCA, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, PAAD, PCPG, PRAD, READ, SKCM, TAD, THYM, UCEC, UC than in normal tissues. No significant difference was noted between TOP1MT and normal tissue in tumor samples from ACC, CESC, LAML, and TGCT. Low TOP1MT expression was discovered in tumor samples from KICH, THCA, and OV than that in normal tissues. This means that TOP1MT could play different roles in different cancer types. Gene mutation is crucial in human cancer development. TOP1MT expression positively correlates with CNV. Moreover, TOP1MT upregulation was linked to the poor OS, DSS, and PFI in BLCA, HNSC, KIPP, PAAD, UCEC, as well as LIHC. DSS exhibited no prognostic value in PAAD cancer patients with upregulated TOPIMT expression. Additional analysis revealed that high TOP1MT expression was linked to poor overall survival in patients with poor clinic pathologic typing. These findings reveal the TOP1MT role in promoting tumor progression and that high TOP1MT expression could impair survival in cancer patients.
The tumor microenvironment (TME) encompasses tumor cells, immune cells, and stromal cells; it is affected by various factors including cytokines, reactive oxygen species, extracellular Matrix, metabolites, and inflammation (Chen et al., 2015; Balta et al., 2021). TME is fundamental in tumor inhibition or progression (Roma-Rodrigues et al., 2019). Humans have innate and adaptive immunity that combat various diseases, including cancer (Schreiber et al., 2011; Vesely et al., 2011; Seager et al., 2017). Tumor cells have evolved immune-evasion mechanisms among them loss of antigenicity, immunogenicity, and generation of immunosuppressive TME (Khong and Restifo, 2002; Blank et al., 2005; Thomas and Massague, 2005; Drake et al., 2006). Consequently, immunotherapy continues to face significant hurdles in cancer treatment (Murciano-Goroff et al., 2020; Zhang and Zhang, 2020). A total of seven common immune cells and immune checkpoints were selected to analyze the relationship between TOP1MT expression immune cells, and immune checkpoints. We found that TOP1MT expression is linked to immune invasion and checkpoint markers in BlCA, HNSC, KIPP, PAAD, UCEC, and LIHC cancers. TOP1MT is associated with tumor purity, B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in various cancers. Previous studies have shown that tumors with higher levels of Programmed cell death-Ligand 1(PD-L1) are more malignant and less survivable; besides, suppressing the PD-1/PD-L1 pathway promotes tumor cell survival (Nakanishi et al., 2007; Okudaira et al., 2009). Pai and colleagues proposed an anti-CTLA4 blocking technique that simultaneously reduces tumor invasion, maintains the anti-tumor effect, and minimizes toxicity (Pai et al., 2019). As a consequence, the expression of the checkpoint genes programmed cell death 1(PD-1), PD-L1, and cytotoxic T-lymphocyte-associated protein 4 (CTL-4) is a predictive biomarker of immunosuppressive response. Notably, TOP1MT and checkpoint gene expression were significantly correlated. TOP1MT expression negatively correlated with most immunosuppressive sites. These results indicate a potential relationship between TOP1MT and immunologic invasion in cancer patients. Collectively, our research sheds light on the application of TOP1MT as a potential prognostic biomarker for several cancers in the context of immuno-oncology, as well as a theoretical reference for new targets.
DNA methylation is an epigenetic mechanism necessary for gene transcription (Feinberg and Vogelstein, 1983; Hsieh and Gage, 2004). Of note, 5-methylcytosine is synthesized by attaching methyl groups to cytosine residues in cytosine-guanine (CG) (Patil et al., 2014; Mahmoud and Ali, 2019). Since most of the CpG sites in the human genome are methylated, low methylation of CpG sites in these regions induces genomic instability and loss of genomic imprinting. This ultimately results in tumor cell development (Kulis et al., 2013). Also, hypermethylation in the same promoter region silences or inactivates the tumor suppressor gene in cancer cells (Bakshi et al., 2018). Studies on TOP1MT methylation in cancer are limited. Our findings suggest a negative correlation between TOP1MT expression and DNA methylation, indicating TOP1MT expression in BlCA, HNSC, KIPP, PAAD, UCEC, and LIHC with low methylation. Moreover, the findings of this work act as a reference for further research on TOP1MT methylation-related roles, and expound on the TOP1MT mechanism in tumorigenesis and development.
TOP1MT ribosome functions are linked to the biological function of TOP1MT in patients with liver cancer (Baechler et al., 2019). Evidence shows that the biogenesis of mitochondrial ribosomes depends on the coordinated synthesis of 80 mitochondrial ribosomal proteins encoded in nuclear DNA, which must be translated into mitochondria via the cytoplasmic ribosomes (Bogenhagen et al., 2014). Therefore, the formation of mitochondrial glycosomes is critical for subsequent mitochondrial function. Biological function analysis revealed that the function of TOP1MT is primarily related to ribosome-related activities. Notably, TOP1MT significantly modulates mitochondrial function.
Although we used data from multiple databases to analyze the significance of TOP1MT, this work has compelling limitations. First, we searched the database for phenotypic features of TOP1MT expression in various cancers, yet no additional evidence was discovered in the cell, animal, or clinical samples. Secondly, we verified that TOP1MT expression is associated with immunologic invasion. Nevertheless, we did not find any in-depth study on TOP1MT and immune infiltration in relevant articles, which lacked certain basis -. Thus, the specific approach by which TOP1MT participates in immune infiltration is required. Furthermore, while we found that the biological functions of TOP1MT of 122 co-expressed genes in six cancer tissues were related to ribosomal functions, limitations were noted. Therefore, additional studies on other promising biological roles of TOP1MT are necessary.
In conclusion, this is the first comprehensive examination of TOP1MT in pan-cancer. The results demonstrate a heterologous TOP1MT expression in various cancers; besides, upregulated TOP1MT expression is associated with poor prognosis. Moreover, TOP1MT expression significantly correlates with immune invasion. Thus, TOP1MT is a potential biomarker for mitochondrial anticancer therapy and cancer immunotherapy.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
LF, ZL, YX, and GH designed and performed the experiments, analyzed and interpreted the results and created Figures. GH and LF wrote the manuscript. GH supervised the project. All authors revised the content and approved the final manuscript.
This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ22H160009, Medicine and Health Technology Plan Project of Zhejiang Province (2022KY371), Jiaxing Key Laboratory of Oncology radiotherapy (2021-zlzdsys), 2019 Jiaxing Key Discipline of Medicine-Oncology (Supporting Subject) (2019-zc-11).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.920897/full#supplementary-material
Baechler, S. A., Factor, V. M., Dalla Rosa, I., RAvji, A., Becker, D., Khiati, S., et al. (2019). The mitochondrial type IB topoisomerase drives mitochondrial translation and carcinogenesis. Nat. Commun. 10 (1), 83. doi:10.1038/s41467-018-07922-3
Bakshi, A., Bretz, C. L., Cain, T. L., and Kim, J. (2018). Intergenic and intronic DNA hypomethylated regions as putative regulators of imprinted domains. Epigenomics 10, 445–461. doi:10.2217/epi-2017-0125
Balta, E., Wabnitz, G. H., and Samstag, Y. (2021). Hijacked immune cells in the tumor microenvironment: Molecular mechanisms of immunosuppression and cues to improve T cell-based immunotherapy of solid tumors. Int. J. Mol. Sci. 22 (11), 5736. doi:10.3390/ijms22115736
Blank, C., Gajewski, T. F., and Mackensen, A. (2005). Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: Implications for tumor immunotherapy. Cancer Immunol. Immunother. 54, 307–314. doi:10.1007/s00262-004-0593-x
Bogenhagen, D. F., Martin, D. W., and Koller, A. (2014). Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell. Metab. 19, 618–629. doi:10.1016/j.cmet.2014.03.013
Chandel Navdeep, S. (2015). Evolution of mitochondria as signaling organelles. Cell. Metab. 22 (2), 204–206. doi:10.1016/j.cmet.2015.05.013
Chen, F., Zhuang, X., Lin, L., Yu, P., Wang, Y., Shi, Y., et al. (2015). New horizons in tumor microenvironment biology: Challenges and opportunities. BMC Med. 13, 45. doi:10.1186/s12916-015-0278-7
Dalla Rosa, I., Huang, S. Y., Agama, K., Khiati, S., Zhang, H., and Pommier, Y. (2014). Mapping topoisomerase sites in mitochondrial DNA with a poisonous mitochondrial topoisomerase I (Top1mt). J. Biol. Chem. 289 (26), 18595–18602. doi:10.1074/jbc.M114.555367
Dalla Rosa, I., Zhang, H., Khiati, S., Wu, X., and Pommier, Y. (2017). Transcription profiling suggests that mitochondrial topoisomerase IB acts as a topological barrier and regulator of mitochondrial DNA transcription. J. Biol. Chem. 292 (49), 20162–20172. doi:10.1074/jbc.M117.815241
Drake, C. G., Jaffee, E., and Pardoll, D. M. (2006). Mechanisms of immune evasion by tumors. Adv. Immunol. 90, 51–81. doi:10.1016/S0065-2776(06)90002-9
Feinberg, A. P., and Vogelstein, B. (1983). Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92. doi:10.1038/301089a0
Forterre, P., and Gadelle, D. (2009). Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 37 (3), 679–692. doi:10.1093/nar/gkp032
Forterre, P., Gribaldo, S., Gadelle, D., and Serre, M. C. (2007). Origin and evolution of DNA topoisomerases. Biochimie 89 (4), 427–446. doi:10.1016/j.biochi.2006.12.009
Hou, G. X., Liu, P., Yang, J., and Wen, S. (2017). Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLoS One 12 (3), e0174515. doi:10.1371/journal.pone.0174515
Hsieh, J., and Gage, F. H. (2004). Epigenetic control of neural stem cell fate. Curr. Opin. Genet. Dev. 14, 461–469. doi:10.1016/j.gde.2004.07.006
Khiati, S., Baechler, S. A., Factor, V. M., Zhang, H., Huang, S. y. N., Dalla Rosa, I., et al. (2015). Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc. Natl. Acad. Sci. U. S. A. 112 (36), 11282–11287. doi:10.1073/pnas.1511016112
Khiati, S., Seol, Y., Agama, K., Dalla Rosa, I., Agrawal, S., Fesen, K., et al. (2014). Poisoning of mitochondrial topoisomerase I by lamellarin D. Mol. Pharmacol. 86 (2), 193–199. doi:10.1124/mol.114.092833
Khong, H. T., and Restifo, N. P. (2002). Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat. Immunol. 3, 999–1005. doi:10.1038/ni1102-999
Kulis, M., Queiros, A. C., Beekman, R., and Martin-Subero, J. I. (2013). Intragenic DNA methylation in transcriptional regulation, normal differentiation, and cancer. Biochim. Biophys. Acta 1829, 1161–1174. doi:10.1016/j.bbagrm.2013.08.001
Low, R. L., Orton, S., and Friedman, D. B. (2003). A truncated form of DNA topoisomerase IIbeta associates with the mtDNA genome in mammalian mitochondria. Eur. J. Biochem. 270 (20), 4173–4186. doi:10.1046/j.1432-1033.2003.03814.x
Mahmoud, A. M., and Ali, M. M. (2019). Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients 11 (3), 608. doi:10.3390/nu11030608
Murciano-Goroff, Y. R., Warner, A. B., and Wolchok, J. D. (2020). The future of cancer immunotherapy: Microenvironment-targeting combinations. Cell. Res. 30 (6), 507–519. doi:10.1038/s41422-020-0337-2
Nakanishi, J., Wada, Y., Matsumoto, K., Azuma, M., Kikuchi, K., Ueda, S., et al. (2007). Overexpression of B7-H1 (PD-L1) significantly associates with tumor grade and postoperative prognosis in human urothelial cancers. Cancer Immunol. Immunother. 56, 1173–1182. doi:10.1007/s00262-006-0266-z
Nicholls, T. J., Nadalutti, C. A., Motori, E., Sommerville, E. W., Gorman, G. S., Basu, S., et al. (2018). Topoisomerase 3α is required for decatenation and segregation of human mtDNA. Mol. Cell. 69 (1), 9–23. doi:10.1016/j.molcel.2017.11.033
Okudaira, K., Hokari, R., Tsuzuki, Y., Okada, Y., Komoto, S., Watanabe, C., et al. (2009). Blockade of B7-H1 or B7-DC induces an anti-tumor effect in a mouse pancreatic cancer model. Int. J. Oncol. 35, 741–749. doi:10.3892/ijo_00000387
Pai, C. S., Simons, D. M., Lu, X., Evans, M., Wei, J., Wang, Y. H., et al. (2019). Tumor-conditional anti-CTLA4 uncouples antitumor efficacy from immunotherapy-related toxicity. J. Clin. Invest 129 (1), 349–363. doi:10.1172/JCI123391
Patil, V., Ward, R. L., and Hesson, L. B. (2014). The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 9, 823–828. doi:10.4161/epi.28741
Pommier, Y., Sun, Y., Huang, S. N., and Nitiss, J. L. (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell. Biol. 17, 703–721. doi:10.1038/nrm.2016.111
Porporato, P. E., Filigheddu, N., Pedro, J. M. B-S., Kroemer, G., and Galluzzi, L. (2017). Mitochondrial metabolism and cancer. Cell. Res. 28, 265–280. doi:10.1038/cr.2017.155
Roma-Rodrigues, C., Mendes, R., Baptista, P. V., and Fernandes, A. R. (2019). Targeting tumor microenvironment for cancer therapy. Int. J. Mol. Sci. 20 (4), 840. doi:10.3390/ijms20040840
Scatena, C., Roncella, M., Di Paolo, A., Aretini, P., Menicagli, M., Fanelli, G., et al. (2018). Doxycycline, an inhibitor of mitochondrial biogenesis, effectively reduces cancer stem cells (CSCs) in early breast cancer patients: A clinical pilot study. Front. Oncol. 8, 452. doi:10.3389/fonc.2018.00452
Schreiber, R. D., Old, L. J., and Smyth, M. J. (2011). Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570. doi:10.1126/science.1203486
Seager, R. J., Hajal, C., Spill, F., Kamm, R. D., and Zaman, M. H. (2017). Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg. Sci. Phys. Oncol. 3, 034002. doi:10.1088/2057-1739/aa7e86
Seol, Y., Zhang, H., Pommier, Y., and Neuman, K. C. (2012). A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc. Natl. Acad. Sci. U. S. A. 109 (40), 16125–16130. doi:10.1073/pnas.1206480109
Singh, S., Pandey, V. P., Yadav, K., Yadav, A., and Dwivedi, U. N. (2020). Natural products as anti-cancerous therapeutic molecules targeted towards topoisomerases. Curr. Protein Pept. Sci. 21 (11), 1103–1142. doi:10.2174/1389203721666200918152511
Spinelli, J. B., and Haigis, M. C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell. Biol. 20 (7), 745–754. doi:10.1038/s41556-018-0124-1
Thomas, D. A., and Massague, J. (2005). TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 8, 369–380. doi:10.1016/j.ccr.2005.10.012
Thorsson, V., Gibbs, D. L., Brown, S. D., Wolf, D., Bortone, D. S., Ou Yang, T. H., et al. (2019). The immune landscape of cancer. Immunity 51 (2), 411–412. doi:10.1016/j.immuni.2019.08.004
Vesely, M. D., Kershaw, M. H., Schreiber, R. D., and Smyth, M. J. (2011). Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29, 235–271. doi:10.1146/annurev-immunol-031210-101324
Vyas, S., Zaganjor, E., and Haigis, M. C. (2016). Mitochondria and cancer. Cell. 166 (3), 555–566. doi:10.1016/j.cell.2016.07.002
Wang, J. C. (2002). Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell. Biol. 3 (6), 430–440. doi:10.1038/nrm831
Zhang, H., Barceló, J. M., Lee, B., KohlhaGen, G., Zimonjic, D. B., Popescu, N. C., et al. (2001). Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. U. S. A. 98 (19), 10608–10613. doi:10.1073/pnas.191321998
Zhang, H., and Pommier, Y. (2008). Mitochondrial topoisomerase I sites in the regulatory D-loop region of mitochondrial DNA. Biochemistry 47 (43), 11196–11203. doi:10.1021/bi800774b
Zhang, H., Seol, Y., Agama, K., Neuman, K. C., and Pommier, Y. (2017). Distribution bias and biochemical characterization of TOP1MT single nucleotide variants. Sci. Rep. 7 (1), 8614. doi:10.1038/s41598-017-09258-2
Zhang, H., Zhang, Y-W., Yasukawa, T., Dalla Rosa, I., Khiati, S., and Pommier, Y. (2014). Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIα and IIβ in vertebrate mitochondria. Nucleic Acids Res. 42 (11), 7259–7267. doi:10.1093/nar/gku384
Keywords: TOP1MT, pan-cancer, prognosis, biomarker, immunity
Citation: Fei L, Lu Z, Xu Y and Hou G (2022) A comprehensive pan-cancer analysis of the expression characteristics, prognostic value, and immune characteristics of TOP1MT. Front. Genet. 13:920897. doi: 10.3389/fgene.2022.920897
Received: 15 April 2022; Accepted: 12 July 2022;
Published: 10 August 2022.
Edited by:
Ángeles Carlos Reyes, Instituto Nacional de Enfermedades Respiratorias-México (INER), MexicoReviewed by:
Claudia Calabrese, University of Cambridge, United KingdomCopyright © 2022 Fei, Lu, Xu and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yufen Xu, MTM1ODgwNjA3QDE2My5jb20=; Guoxin Hou, Z3hob3U5MUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.