
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 13 June 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.916867
Class III peroxidases (PODs) are plant-specific enzymes that play significant roles in plant physiological processes and stress responses. However, a comprehensive analysis of the POD gene family in tobacco has not yet been conducted. In this study, 210 non-redundant POD gene members (NtPODs) were identified in tobacco (Nicotiana tabacum) and distributed unevenly throughout 24 tobacco chromosomes. Phylogenetic analysis clustered these genes into six subgroups (I-VI). Gene structure and motif analyses showed the structural and functional diversity among the subgroups. Segmental duplication and purifying selection were the main factors affecting NtPOD gene evolution. Our analyses also suggested that NtPODs might be regulated by miRNAs and cis-acting regulatory elements of transcription factors that are involved in various biological processes. In addition, the expression patterns in different tissues and under various stress treatments were investigated. The results showed that the majority of NtPODs had tissue-specific expression patterns and may be involved in many biotic and abiotic responses. qRT-PCR analyses of different tissues and stress treatments were performed to verify transcriptome patterns. Expression of a green fluorescent protein-NtPOD fusion confirmed the plasma membrane localization of NtPOD121 and NtPOD4. Furthermore, 3D structures provided evidences of membrane-bound peroxidase. These findings provide useful information to better understand the evolution of the NtPOD gene family and lay the foundation for further studies on POD gene function in tobacco.
Peroxidases (EC 1.11.1.X) are a large family of enzymes that are widely distributed in living organisms and catalyze the oxidation of various substrates with hydrogen peroxide (H2O2) as an electron acceptor (Welinder, 1992; Kidwai et al., 2020). Peroxidase performs three distinct cycles: the peroxide cycle, the oxidation cycle, and the hydroxyl cycle, leading to the elimination and generation of reactive oxygen species (ROS) (H2O2, O2−, and OH−) (Kidwai et al., 2020). Therefore, peroxidase can be considered as a bifunctional enzyme maintaining intracellular ROS levels by oxidizing various substrates with H2O2 or generating ROS (Passardi et al., 2004). According to their structure, they can be divided into heme or non-heme containing proteins. The heme-containing peroxidases can be further subdivided into animal and non-animal groups (Mathé et al., 2010). Previous study contended that non-animal peroxidases contain three large families: class I, II, and III (Cosio and Dunand, 2009). Class III peroxidases (guaiacol peroxidases, EC 1.11.1.7) are heme oxidoreductase enzymes that exist in various plants as a multigenic family (Mathé et al., 2010). In previous studies, different abbreviations, such as POX, Px, PER, POD, and Prx, have been used for class III peroxidases (e.g.) (Almagro et al., 2009).
Class III peroxidases are plant-specific glycoproteins and have been extensively studied in higher plants. PODs have been shown to play a role in a broad range of physiological and developmental processes, including cell wall metabolism, lignification, auxin metabolism, stress tolerance, and defense response (Pandey et al., 2017). To date, class III peroxidases have been investigated and characterized in a variety of plant species, including Arabidopsis (Tognolli et al., 2002), cassava (Wu et al., 2019), Chinese pear (Cao et al., 2016), maize (Wang Y et al., 2015), rice (Passardi et al., 2004), orange (Li et al., 2020), grapevine (Xiao et al., 2020), and soybean (Aleem et al., 2022). Some genetic evidences suggested that class III peroxidase is an abiotic stress-responsive enzyme in different plant species. For example, the overexpression of AtPrx64 could enhance the tolerance of tobacco to aluminum stress (Wu et al., 2017), and the overexpression of TaPRX-2A in wheat exhibited an enhanced tolerance to salt stress (Su et al., 2020). Similarly, the overexpression of CrPRX, a Catharanthus roseus peroxidase, improved tolerance of tobacco to salt and drought stress (Jaggi et al., 2011). Peroxidases also play a role in resistance to pathogens. It has been reported that TpoxN1 expression was induced in tobacco within 20 min of wounding, and the response could be sustained for longer than 2 weeks, indicating that TpoxN1 may involve in wounding-healing (Sasaki et al., 2002). The overexpression of CaPO2 in Arabidopsis enhanced the accumulation of H2O2, the expression of many pathogen-responsive genes, and resistance to Xanthomonas campestris pv. vesicatoria (Choi and Hwang, 2012). Hence, plant PODs may play an essential role in the response to biotic and abiotic stresses.
Tobacco (Nicotiana tabacum) is an important commercial crop in the world and is often used as a model organism to conduct plant genetic researches. During growth, tobacco is often affected by adverse stresses, including pathogenic bacteria, drought, cold, salinization, and heavy metals (Dana et al., 2006; Wei et al., 2008). However, the POD gene family in tobacco remains unexplored. In this study, we conducted a comprehensive analysis of the structure and function of class III peroxidases in tobacco, including phylogenetic relationships, duplication events, functional domain characterization, and cis-acting elements. The 3D structures of eight PODs were explored to elucidate their mechanisms. Additionally, the expression patterns in eight typical tissues and in response to different abiotic and biotic stresses were investigated. Our results provide comprehensive insights into the biological functions of class III peroxidases in tobacco plants.
To identify POD genes in tobacco, the protein sequences of 73 Arabidopsis (Tognolli et al., 2002) and 138 rice PODs (Passardi et al., 2004) were searched in the Sol Genomics Network (Fernandez-Pozo et al., 2015) and our unpublished tobacco genome using the TBLASTN (v2.12.0) (McGinnis and Madden, 2004) program with default parameters. HMMER (v3.3.2) (Finn et al., 2011) was employed to identify the candidate tobacco POD genes using the Hidden Markov Model (HMM) based on the POD conserved domain (PF00141). All candidate sequences were confirmed using PFAM (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/). The physicochemical properties of the POD proteins, including molecular weight (MW) and isoelectric points (pI), were predicted using the online ExPASy tool (http://web.expasy.org/protparam) (Gasteiger et al., 2003).
A phylogenetic tree of the NtPOD genes was constructed using the neighbor-joining (NJ) method with a bootstrap value of 1,000 in MEGA X (Kumar et al., 2018). To reveal the exon-intron organization of the POD genes in tobacco, TBtools (v1.09867) (Chen et al., 2020) were used to determine the gene structures of each NtPOD gene. The motifs were analyzed using the MEME program (http://meme-suite.org/tools/meme). The maximum number of motifs was set to 10, the optimum width of motifs was set to 6–200 amino acid residues, and the remaining settings were kept at default values (Bailey et al., 2015). Subsequently, the identified motifs were annotated using InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) (Quevillon et al., 2005).
All NtPOD genes were mapped to their respective chromosomes. Chromosome maps of all identified NtPODs were drawn using TBtools (v1.09867) software (Chen et al., 2020). For gene duplication analysis, the amino acid sequences of NtPODs were aligned using BLAST (v2.12.0, e-value = 1e−5) (McGinnis and Madden, 2004). Gene pairs were considered as duplicated only if their similarity was ≥90%. Duplicated gene pairs located adjacent to the same chromosome were defined as tandemly duplicated, whereas those positioned on separate chromosomes were defined as segmental duplicates. The non-synonymous (Ka) and synonymous substitution (Ks) rates of duplicated NtPOD genes were calculated using ParaAT2.0 program (Zhang et al., 2012). Furthermore, the Ka/Ks ratio was calculated to evaluate selection pressure.
The 1,500 bp upstream sequence from the transcription start site was used to analyze the cis-elements. Putative stress or hormone-responsive cis-elements in the promoter region of the NtPOD genes were obtained using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Mature tobacco miRNAs were downloaded from the miRBase database (Kozomara et al., 2019). The regulatory relationship between miRNAs and NtPODs were searched using PsRNATarget (http://plantgrn.noble.org/psRNATarget/) (Dai and Zhao, 2011) with the default settings. The regulatory relationship between NtPODs and transcription factors was retrieved from the Plant Transcription Factor Database PlantTFDB (http://planttfdb.cbi.pku.edu.cn) (Jin et al., 2016) (specie setting with Nicotiana tabacum) by searching for promoter sequences. Cytoscape (Smoot et al., 2011) was used to visualize the interaction networks.
Transcriptome data from eight distinct tissues (leaves, veins, blades, stems, roots, callus, axillary buds, and seeds) of tobacco were used to investigate the expression patterns of the NtPOD genes. The sampling stages for each tissue are listed in Supplementary Table S1. Raw sequence data of different tissues were obtained from PLncDB (http://plncdb.tobaccodb.org/) (Jin et al., 2021) and our unpublished data. Raw RNA-seq datasets, including cold (Jin et al., 2017), drought (Yang H et al., 2017), cadmium (Cd) (He et al., 2016), topping (Chen et al., 2019), NaCl, abscisic acid (ABA) (Wu et al., 2021), cucumber mosaic virus (CMV) (Liu et al., 2019), and Phytophthora nicotianae (Yang J. K et al., 2017) were obtained from the SRA database (Leinonen et al., 2010). All clean reads were mapped to the tobacco reference genome using Hisat2 (2.2.1) (Kim et al., 2019). The FPKM values were computed using the StringTie software (2.1.7) (Pertea et al., 2015).
The cultivated tobacco variety K326 was used to analyze the expression of POD genes in various tissues and stress treatments. The seedlings were cultivated in plastic pots with a 16 h light photoperiod at 28°C during the day and at 23°C at night. Plant root, stem, leaf, vein, axillary bud, terminal bud, flower, and seed samples were collected as described in our previous study (Wang Z et al., 2015). For salinity stress, the tobacco seeds were first germinated and cultivated in plastic pots under normal conditions for 14 days, and then exposed to salt (150 mM NaCl) for 7 days (Cheng et al., 2009). After 1 month of germination, the tobacco seedlings were treated for 6 h with plant hormones, including ABA (10 µM), IAA (10 µM) (Braybrook, 2017), salicylic acid (SA) (10 µM) (Kawano and Muto, 2000), and jasmonic acid (JA) (10 µM). Untreated plantlets were used as the control (CK). The treated and control plantlets were collected after treatment, and then all the samples were immediately frozen in liquid nitrogen and stored at −80°C.
A SuperPure Plantpoly RNA Kit (Gene Answer, Beijing, China) was used to extract total RNA from the plant samples. DNA contamination was eliminated by digestion with RNase-free DNase I (Gene Answer). Reverse transcriptase M-MLV (Takara Biomedical Technology, Beijing, China) was used to synthesize first-strand cDNA using 1 μg of total RNA as a template. The cDNA was diluted to 50 ng/µl. RT-PCR was performed using a SYBR Green kit (Imagene, Beijing, China) in a 20-μl reaction solution. The PCR program was as follows: 95°C for 30 s, 40 cycles of 95°C for 10 s, and 60°C for 30 s. The expression levels of target genes were standardized to the expression level of the NtL25 (Volkov et al., 2003) gene using the 2−∆∆Ct method. Three independent biological replicates were used for each gene. The gene specific primers used for qRT-PCR are listed in Supplementary Table S2
Signal peptide sequence and potential cleavage site of NtPODs were conducted by SignalP 5.0 program (Almagro Armenteros et al., 2019). The transmembrane domains of POD proteins were analyzed by the online tool TMHMM server v2.0 program (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001), and their subcellular localization were predicted by WoLF PSORT (https://www.genscript.com/wolf-psort.html) (Horton et al., 2007). To verify subcellular localization of NtPODs, we generated C-terminal green fluorescent protein (GFP) fusions for two POD proteins and visualized their subcellular location by confocal microscopy after transient expression of the fusions in Nicotiana benthamiana. The three-dimensional (3D) structures of NtPOD proteins were predicted using AlphaFold2 (Jumper et al., 2021), and displayed using PyMOL software (http://pymol.org/) (DeLano, 2002). The quality of the predicted 3D structures was measured using ERRAT test scores on the SAVES server (https://saves.mbi.ucla.edu/). Molecular docking was performed using AutoDock Vina (Trott and Olson, 2010).
Based on 73 PODs from Arabidopsis and 138 PODs from rice, we used BLAST and HMMER to search for PODs against the tobacco genome. A total of 210 non-redundant NtPODs were identified as class III peroxidases, denoted as NtPOD1 to NtPOD210 (Supplementary Table S3). The length of NtPOD proteins ranged from 250 (NtPOD116) to 500 (NtPOD26) amino acid residues, with an average of 326 amino acids (Supplementary Table S4). The relative molecular weight varied from 27.45 (NtPOD11) to 54.50 kDa (NtPOD26), with isoelectric points ranging from 4.57 (NtPOD192) to 10.03 (NtPOD138) (Supplementary Table S4, Supplementary Figure S1).
Phylogenetic analysis revealed that NtPOD genes could be divided into six subgroups (Figure 1A). Large subgroups I and II consisted of 67 and 66 NtPOD members, respectively, whereas small subgroups III, IV, and VI contained 14, 17, and 15 NtPOD members, respectively. Subgroup V comprised of 31 NtPOD members. The ten most conserved motifs for NtPODs were explored using the MEME program and annotated using InterProScan (Supplementary Table S5). Seven motifs (1, 2, 3, 4, 7, 8, and 10) were annotated as peroxidase domains, which were present in most of the NtPODs (92.4%, 94.3%, 99.0%, 98.1%, 98.6%, 93.3%, and 62.86%) (Figure 1B), suggesting that these motifs have been preserved for a long time. In total, 189 NtPODs in subgroups I-V contained at least seven motifs (1, 3, 4, 6, 7, 8, and 9), except for NtPOD16 (in subgroup I), NtPOD190 (in subgroup II) and NtPOD179, NtPOD97, NtPOD72, NtPOD158 (in subgroup III), (Figure 1B). Interestingly, subgroup VI was distinct from the other groups, and most members contained only motifs 3, 4, and 7 (Figure 1B). Furthermore, some unknown motifs (5, 6, and 9) were found in some subgroups.
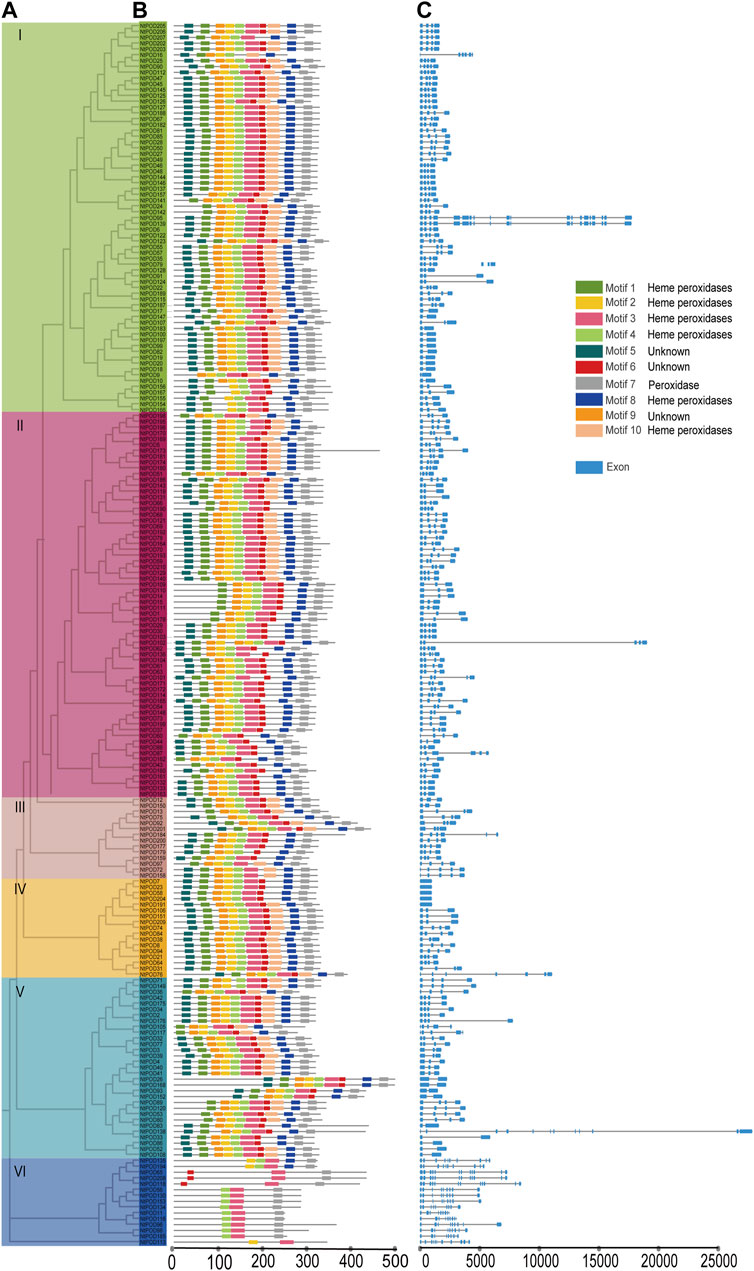
FIGURE 1. Phylogenetic tree, conserved motif, and gene structure of NtPOD genes. (A) Phylogenetic relationship among the NtPOD genes based on the amino acid sequence alignment. (B) Conserved motifs in amino acid sequence of different sub-groups (I-VI) of NtPODs. The 10 different colored boxes on the right represent diverse conserved motifs. The detailed sequences of motifs are listed in Supplementary Table S5 (C) Exon-intron analysis of six subgroups of NtPODs.
To obtain further insights into POD evolution, we examined the exon-intron structures of the NtPOD genes. The results demonstrated structural variation among these NtPOD genes, ranging from 1 to 12 exons, whereas most NtPODs contained four exons and three introns (Figure 1C). Among the NtPOD genes, 50.0% (105/210) consisted of four exons, and 58.2% of subgroup I (39/67) had four exons (Figure 1C). Subgroup VI was exon-rich with 9–12 exons, whereas subgroup IV contained fewer exons (between one and four) (Figure 1C). Generally, NtPOD genes in the same subgroup exhibited similar exon-intron features, providing further evidence of their phylogenetic relationships.
To explore the distribution of NtPODs, a physical map was constructed using TBtools. In total, 155 NtPODs were mapped onto tobacco chromosomes, while the others were mapped onto scaffolds (Supplementary Figure S2). Notably, there was no POD gene distribution on chromosomes 21 or 24. The NtPOD genes were unevenly distributed throughout the 22 chromosomes (Supplementary Figure S2). Gene duplication is an important mechanism for the evolution of novel gene functions. Segmental and tandem duplications are considered as the two major mechanisms of gene family expansion in plants. To further investigate the expansion of POD genes in tobacco, we aligned the nucleotide sequences of NtPOD genes to identify duplication events (Figure 2). Finally, 103 duplication events consisting of 109 paralogs were identified, including 25 tandem duplications and 78 segmental duplications. Similar to previous findings (Cai et al., 2021; Meng et al., 2021), segmental duplication was also 2-3-fold higher than tandem duplication in the NtPOD family, which was indicative of its contribution to the evolution and expansion of the NtPOD family.
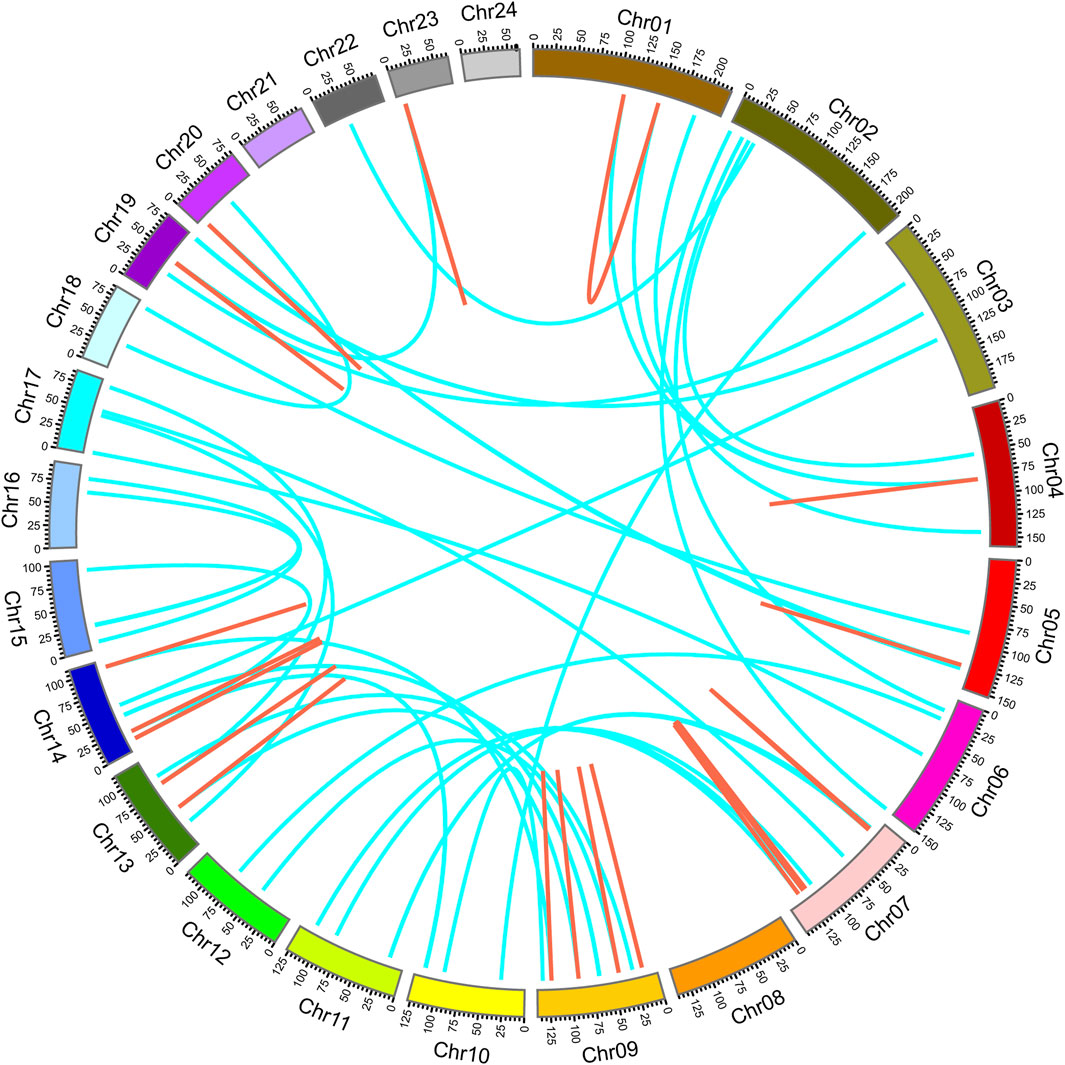
FIGURE 2. Distribution of tandem and segmental duplicated POD genes in the tobacco genome generated by Circos. Tandem and segmental duplicated gene pairs are linked by the red in same chromosome and cyan lines between chromosomes, respectively.
Next, using the non-synonymous (Ka) and synonymous (Ks) ratios, we attempted to understand the evolutionary selection for the duplicated NtPOD genes (Supplementary Table S6). In general, a ratio of Ka/Ks greater than 1 indicates positive selection, while a ratio of Ka/Ks less than 1 indicates purifying selection and Ka/Ks equal to 1 indicates neutral selection. Among the 103 duplicated events, we found that most Ka/Ks ratios were less than 0.47 (Supplementary Table S6 and Supplementary Figure S3), indicating that these genes underwent purifying selection during evolution. Positive selection was observed in only two duplication events (NtPOD112/NtPOD90 and NtPOD90/NtPOD25) (Supplementary Table S6).
The upstream promoter regions of genes possess many cis-acting elements that can regulate gene expression. To better understand the potential regulatory mechanisms of NtPOD genes, we identified the presence of cis-elements in the promoter regions of NtPOD genes. The identified cis-acting elements were further classified into five distinct groups, based on their putative functions (Figure 3 and Supplementary Table S7). The most abundant elements were light-responsive elements, including Box 4, TCT-motif, and GT1-motif. Regulatory elements related to abiotic and biotic stresses were also found to be abundant, comprising 10 cis-elements, including anoxic, cold, anaerobic, drought, disease, and wound responsive elements. A total of 155 NtPOD genes contained cis-acting elements involved in the anaerobic induction (ARE) (Figure 3 and Supplementary Table S7). Hormone-responsive elements included 19 members, most of which were associated with ABA, and methyl jasmonate (MeJA), followed by gibberellic acid (GA), auxin, and SA (Figure 3 and Supplementary Table S7). Notably, the promoter regions of 163 NtPOD genes contained cis-acting elements related to ABA (ABRE) and 121 genes were involved in MeJA response elements (TGACG-motif). These results indicate that the NtPOD family may be involved in the complex hormone regulatory network. Furthermore, the elements regulating plant development had 6 elements: seed, root, endosperm, palisade mesophyll cells, and meristem-specific expression elements. Moreover, a number of site-binding related elements were also identified in the promoter regions of NtPODs (Figure 3 and Supplementary Table S7). Hence, diverse cis-elements among NtPOD genes may reflect their potential functional variation.
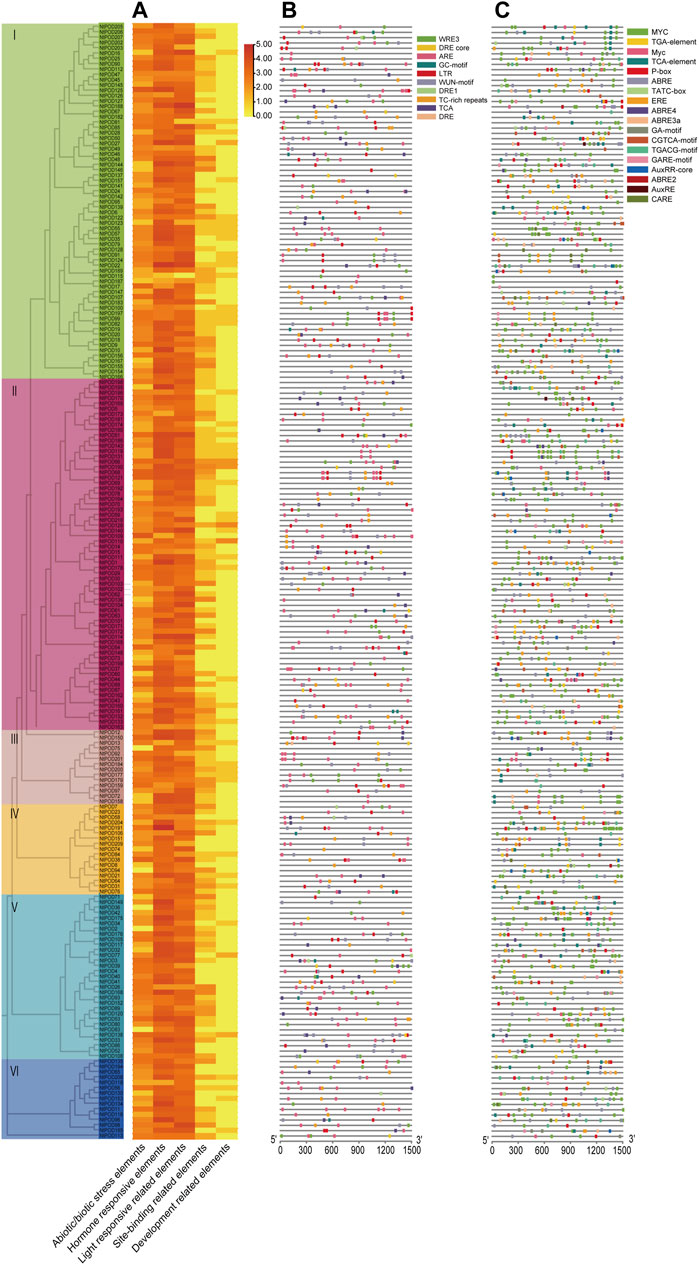
FIGURE 3. Prediction of cis-acting elements in the promoter sequences of NtPOD genes. (A) The number (log2 (count)) of cis-acting elements detected in the promoter region of each NtPOD gene. (B) Distribution of abiotic/biotic stress-related elements in NtPODs. (C) Distribution of hormone-responsive elements in NtPODs.
Due to abundant cis-elements enriched in the promoter regions of NtPOD genes, we speculated that the corresponding transcription factors (TFs) may directly regulate NtPOD genes in tobacco. Therefore, we explored the regulatory relationship between transcription factors and NtPODs using PlantTFDB (Jin et al., 2016). In total, 349 TF members from 39 families may play important roles in the regulation of NtPODs (Figure 4). Among these, MIKC_MADS, Dof, MYB, AP2 and C2H2 transcription factors were the most abundant. We also investigated potential miRNA-binding sites for NtPODs using PsRNATarget (Dai and Zhao, 2011). Finally, 49 miRNA families, consisting of 129 miRNAs, may have regulatory relationships with NtPODs (Figure 4 and Supplementary Table S8). Most miRNAs had several NtPOD targets, including nta-miR6156 which could target 26 NtPOD genes (Figure 4 and Supplementary Table S8). In contrast, some NtPODs could be targeted by several miRNAs. For example, NtPOD102 can be targeted by several miRNAs, including miR166 and miR168 (Figure 4 and Supplementary Table S8). The relationship between NtPODs and TF/miRNAs requires further study.
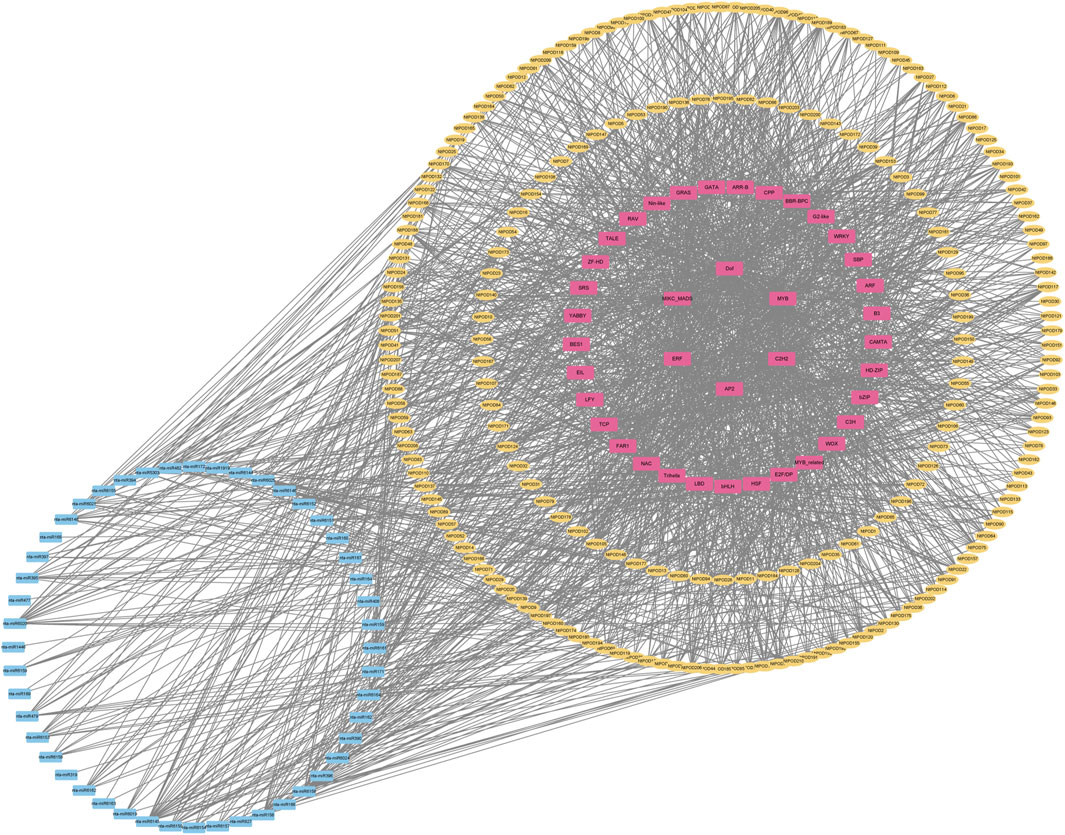
FIGURE 4. Regulation network of NtPODs in tobacco. The yellow, pink, and blue boxes represent NtPOD genes, TFs, and miRNAs, respectively. The black line represents the interaction.
To further study the potential function of each NtPOD gene, the expression patterns in eight tissues (root, stem, leaf, blade, vein, axillary bud, callus, and seed) were explored. Except for 21 NtPODs that not or weakly expressed in the eight tissues, the remaining 189 genes were expressed in at least one tissue (Supplementary Figure S4 and Supplementary Table S9). As shown in Figure 5A, a number of NtPODs exhibited distinct tissue-specific expression patterns. It is worth noting that the number of NtPOD genes expressed in the roots was the highest, suggesting that most NtPOD genes play important roles in the root (Supplementary Figure S4). Interestingly, several NtPOD genes, such as NtPOD12 and NtPOD90, were specifically expressed in the seed (Figure 5A). Furthermore, NtPOD199 may play an important role in callus redifferentiation due to its high expression in callus. Nine NtPOD genes (NtPOD4, 11, 77, 96, 126, 134, 140, 177, and 184) with tissue-specific expression were randomly selected for further qRT-PCR analysis in different tissues, and the results were similar to those of the RNA-seq analyses (Figure 5B). NtPOD126 and NtPOD4 showed higher relative transcription levels in the roots and veins, respectively. Consistent with phylogenetic and motif analyses, the specific and varied expression profiles of NtPOD genes in different tissues also suggested their diverse roles. Moreover, the expression of NtPODs also varied in three developmental stages (seedling, mature, and 2 days after the topping) (Figure 5A and Supplementary Figure S4). For the genes expressed in leaves, six (NtPOD27, 78, 79, 98, 115 and 206) showed relatively stronger expression during seedling stages, suggesting that these genes may play specific roles during the early stages of leaf development. However, the other six (NtPOD2, 52, 75, 101, 108, and 157) and five (NtPOD69, 72, 158, 192, and 201) genes were relatively higher in roots and leaves during the 2 days after the topping stage, respectively.
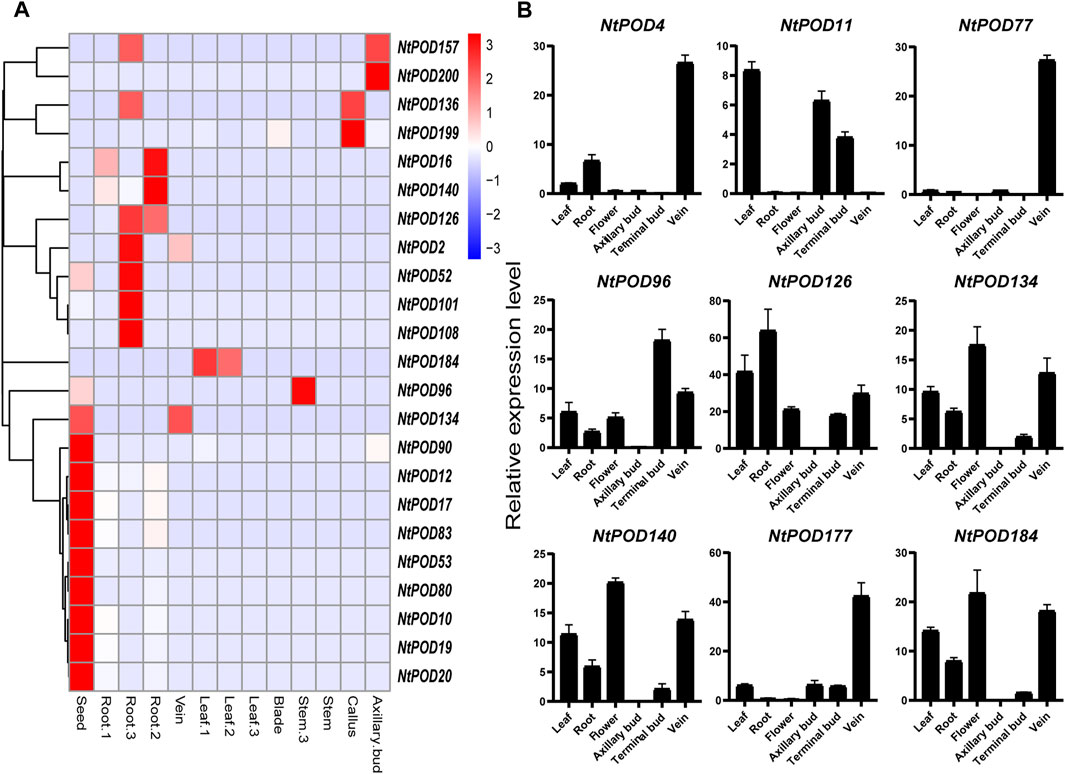
FIGURE 5. Expression profiles of tissue-specific NtPOD genes in different tissues. (A) Heatmap of the expression in different tissues at three development stages [seedling (1), mature (2), and 2 days after topping (3)], scaled by rows. (B) qRT-PCR analysis of nine selected NtPODs in axillary bud, flower, leaf, root, terminal bud, and vein.
To further explore the response of NtPODs during various stress responses, the expression patterns of all NtPODs were investigated using publicly available transcriptome data (Supplementary Table S10). As shown in Figure 6A, three genes (NtPOD68, 121, and 192) were significantly up-regulated after salt treatment. Some NtPOD genes were extremely sensitive to the cold stress and exhibited significant down-regulation, such as NtPOD86, 92, 104, 136, 154, 155, and 166. In addition, ABA treatment significantly reduced the expression of 11 NtPODs. Among the various biotic stresses, 59 NtPODs showed a significant response upon inoculation with P. nicotianae, whereas only a few genes changed slightly under CMV treatment. Interestingly, most NtPODs were specifically involved in individual stress treatment rather than a universal response. To compare the transcription of NtPOD genes between different hormone treatments including ABA, IAA, SA, and JA, we selected eight significantly up-regulated (NtPOD5, 8, 68, 121, 123, 176, 183 and 202) and two significantly down-regulated gene (NtPOD13 and 209) under salt stress to further explore the effects of various plant hormone treatments by qRT-PCR (Figure 6B). Five genes (NtPOD5, 8, 68, 121, 176 and 202) were found to be extremely sensitive to salinity. Their expression levels increased more than 7-fold compared to the control. Consistent with the RNA-seq analysis, the expression level of NtPOD13 and NtPOD209 decreased after salt stress. Under ABA treatment, NtPOD176 were up-regulated, whereas NtPOD8 and NtPOD209 were down-regulated. However, the expression levels of NtPOD13, 176 and NtPOD183 increased slightly upon SA and IAA treatment, whereas JA treatment induced the expression of NtPOD202 and NtPOD209. In summary, these results demonstrate that NtPOD genes are involved in various abiotic and biotic stress treatments.
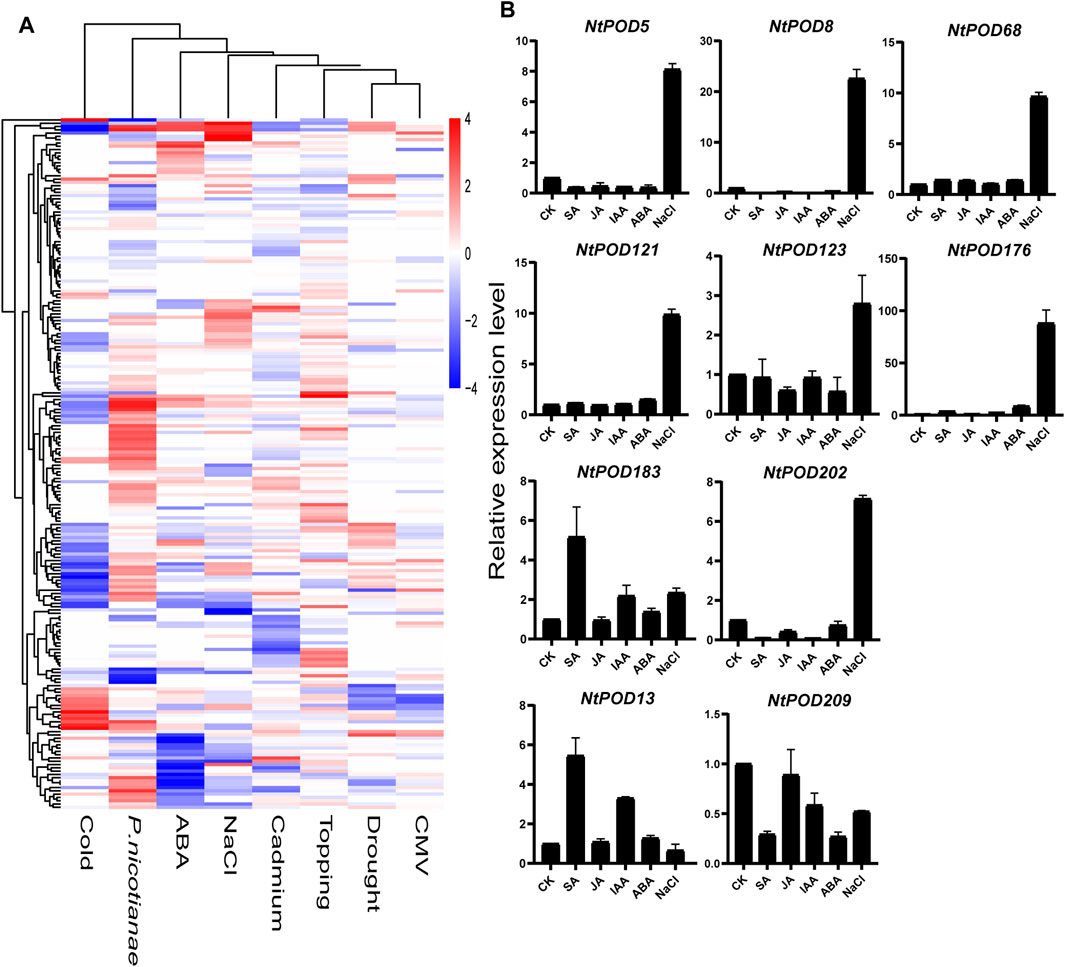
FIGURE 6. Expression patterns of NtPOD genes under various stress treatments. (A) Expression changes of NtPOD genes under cold, ABA, NaCl, cadmium, topping, P. nicotiana, drought, and CMV stresses. The expression change is indicated by the ratio of FPKM value of the treatment to that of the control (CK). (B) Results of qRT-PCR analysis of ten NtPOD genes in response to NaCl and hormones (ABA, IAA, SA, JA) treatment.
The analysis based on the NtPOD amino acid sequences indicated 162 (77.1%) NtPODs contain an N-terminal signal peptide with a putative cleavage site (Supplementary Table S4). After the identification of peroxidase sequences with transmembrane domains, NtPODs were classified as 119 secreted and 91 membrane-bound peroxidases (Supplementary Figure S5A). Furthermore, 91.2% (83) of the membrane-bound peroxidases contained N-terminal signal peptides (Supplementary Table S4). In addition, the majority of NtPODs were predicted a chloroplast location (n = 92) and extracellular regions (n = 63) (Supplementary Figure S5B). Subcellular localization prediction revealed that NtPOD4 contained a putative signal peptide with a predicted plasma membrane (PM) location, whereas NtPOD121 contained a putative signal peptide with a predicted chloroplast location (Supplementary Table S4). However, result of subcellular localization assay using transient expression of NtPOD4-GFP and NtPOD121-GFP in Nicotiana benthamiana leaves, suggested both the plasma membrane localization for these two peroxidases (Figure 7A).
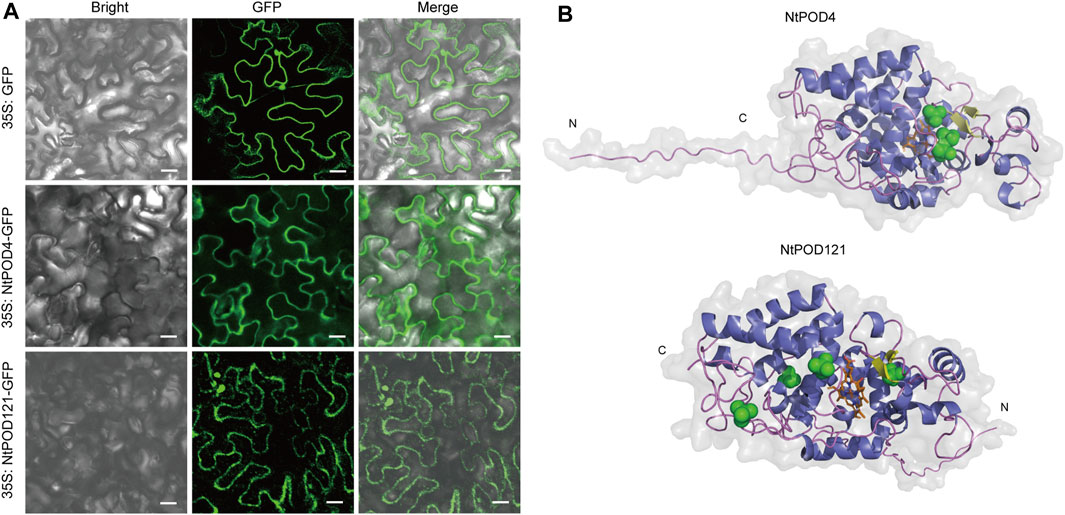
FIGURE 7. Subcellular localization and 3D structure of NtPOD4 and NtPOD121. (A) Subcellular localization of NtPOD4 and NtPOD121. Scale bars: 20 µM. (B) Putative 3D structures of two class III peroxidases: POD4 (predicted location in plasma membrane) and NtPOD121 (predicted location in chloroplast). Colors indicate secondary structures: α-helixes (blue), β-sheets (yellow), loops (purple), disulfides (green), and heme group (orange).
In order to gather additional information about the NtPOD proteins, the structural models of eight peroxidases were built using Alphafold2 (Supplementary Figure S6). The ERRAT test scores by the SAVES server for the eight NtPOD 3D models above 88, which indicated that the predicted 3D models were reliable. Our results showed that NtPOD4 and NtPOD121 were monomers that contained two β-sheets with 17 (NtPOD4) and 20 (NtPOD121) α-helices. Similarly, both NtPOD4 and NtPDO121 had characteristic loops at their N-termini (Figure 7B). As the classical peroxidase structure, NtPOD121 and NtPOD4 contain eight Cys residues, which are necessary for the formation of four conserved disulfide bridges.
Environmental pressure poses a considerable challenge to crop production. Class III peroxidases are widely distributed among terrestrial plant species and play an essential role in plant resistance to different stresses, such as salt, drought, and metal toxicity (Almagro et al., 2009). Our study is the first comprehensive and systematic report to characterize the NtPOD gene family in N. tabacum. PODs encoded by large multigene family have been found in many plants; however, the number of PODs varies greatly among different plants (Kidwai et al., 2020). In total, 210 class III peroxidase genes were identified in the tobacco genome. This number is larger than that of most reported species, such as Arabidopsis (73) (Tognolli et al., 2002), rice (138) (Passardi et al., 2004), maize (119) (Wang Y et al., 2015), soybean (124) (Aleem et al., 2022), polar (93) (Ren et al., 2014), Chinese pear (94) (Cao et al., 2016), carrot (102) (Meng et al., 2021), and other diploid genome plants, but fewer than that in hexaploid wheat (374) (Su et al., 2020). This indicates a considerable expansion of the POD gene family in tobacco and wheat compared to other plant species. In plants, gene family expansion is typically the result of polyploidy and gene duplication. Tobacco (N. tabacum) is an allotetraploid (2n = 48) that originates from chromosome doubling after an interspecific hybridization event between N. tomentosiformis (2n = 24) and N. sylvestris (2n = 24), which occurred nearly 200,000 years ago (Lim et al., 2004). Tobacco POD genes may undergo duplication along with the whole genome duplication event. Gene duplication can help organisms adapt to various environmental conditions (Kondrashov, 2012). Gene family expansion primarily occurs via three modes: segmental duplication of multiple genes, tandem duplication of individual genes, and whole-genome duplication (Panchy et al., 2016). We identified 103 duplication events in NtPOD gene family, involving 109 paralogs. Segmental duplication is thought to be the main driver of NtPOD evolution, as 71.6% (78) of the duplications were segmental events (Supplementary Table S6). Ka and Ks analyses revealed that the evolution of POD genes was driven mainly by purifying selection and that only a few POD sequences may have experienced positive selection during the evolutionary period. Similar results were found in the study on the evolution of PODs in maize (Wang Y et al., 2015), sweet orange (Li et al., 2020), while in contrast with results in soybean (Aleem et al., 2022), Arabidopsis (Tognolli et al., 2002), and rice (Passardi et al., 2004). Based on phylogenetic relationships, the NtPOD family was categorized into six subgroups. Notably, subgroup VI was characterized by multiple exons and different motifs from the other five subgroups, which was consistent with previous studies in cassava (Wu et al., 2019), Chinese pear (Cao et al., 2016), and watermelon (Yang et al., 2022). Therefore, we inferred that POD subgroup VI may have similar origins with other plants and have some specific functions in plants.
Peroxidases can be classified into secreted and membrane-bound peroxidases. Some are secreted into the extracellular space and cell walls under the guidance of N-terminal signal peptides, while some membrane-bound peroxidases are targeted to the ER, vacuole, plasma membrane, or thylakoid (Ren et al., 2014; Lüthje and Martinez-Cortes, 2018). Correspondingly, our results showed that NtPODs were located in the extracellular space, PM, and intracellular spaces, including the thylakoid, cytoplasm, ER, and nucleus. In this study, 43% of the NtPODs were considered as membrane-binding proteins because their N-terminus contained transmembrane domains, which is consistent with previous reports (Lüthje and Martinez-Cortes, 2018). The presence of a cleavable signal peptide at the N-terminus, which varies in cleavage sites from 14 to 34 residues, indicated the secretory nature of 162 NtPODs. Interestingly, members of subgroup VI were predicted to be localized in the cytoplasm, endoplasmic reticulum, and chloroplast, and all lacked N-terminal signal peptides. However, the subcellular localization result showed that NtPOD4 (predicted location in plasma membrane) and NtPOD121 (predicted location in chloroplast) was both localized to the plasma membrane, inconsistent with the predictions. Due to limitation of prediction tools/methods, inconsistent results for subcellular location between prediction and experiment validation are widespread (Nielsen, 2015). Hence, more experimental work might need to explore whether there were some specific domains or structures for specific NtPOD.
The structure of class III peroxidases is well-conserved (Welinder et al., 2002; Mathé et al., 2010). These proteins contain N-terminal signal peptides, binding sites for heme and calcium, and eight conserved Cys residues required for four conserved disulfide bridges (Welinder et al., 2002). The 3D structure of a protein is important for understanding its detailed functional mechanism. Deriving the protein 3D structure could facilitate a mechanistic understanding of POD function. Hence, we attempted to predict the 3D structures of NtPODs using AlphaFold2 (Supplementary Figure S6). The results revealed the diversity of their N-terminal structures, which may explain their different subcellular localizations. Although Class III peroxidases share structural features, which indicated the likelihood of similar mechanisms, the high degree of similarity in protein structure was relatively inconsistent with the functional diversity of these enzymes. These data support the idea that, due to the low specificity of class III peroxidase substrates, the response to specific environment may be the key to determine the role of individual peroxidase isomer in plants.
In plants, a complex gene regulatory network consists of transcription factors, regulatory RNAs, and enzymes that regulate plant growth and development (Ibraheem et al., 2010; Chen et al., 2018). Transcription is initiated by the interaction of TFs, which usually combine with cis-acting regulatory elements in genes in response to environmental changes (Priest et al., 2009). An analysis of the cis-elements in NtPOD gene promoters resulted in the detection of six major types of cis-elements associated with biotic/abiotic stress, hormone response, light response, and developmental processes. Based on these cis-elements, we found 349 TF members from 39 families might play important roles in the regulation of NtPODs (Figure 4). Many TF families implicated in stress responses have been identified, including WRKY (Niu et al., 2012), MYB (Chen et al., 2015), NAC (Nakashima et al., 2007), and bZIP (Hsieh et al., 2010). Moreover, identifying the potential target sites of miRNAs provides valuable insights into the biological functions of miRNAs and target genes related to plant growth and development, stress response and adaptation, and signaling mechanisms. Finally, 49 miRNA families, consisting of 129 miRNAs, may have regulatory relationships with NtPODs (Figure 4 and Supplementary Table S8). Among this regulatory network, NtPOD93 could be targeted by miR172, which was one of well-known miRNAs involved in drought response (Han et al., 2013). Meanwhile, one well-known drought response TF, Dof (Li et al., 2021), might also regulate NtPOD93 (Figure 4). Another example was miR395-NtPOD202-AP2 module, and many evidences suggested that both miR395 (Kim et al., 2010; Çakır et al., 2021) and AP2 (Zhang et al., 2009) might play important roles in salt treatment. Hence, the POD interaction network constructed in our study provides insights into the regulation of peroxides in plants in response to various stresses.
Peroxidases are involved in cell wall-related reactions, metabolic pathways, and stress-related processes (Veljović Jovanović et al., 2018). These enzymes play key roles in the scavenging of ROS (Das and Roychoudhury, 2014). Although it is challenging to study individual POD genes owing to their functional redundancy and low substrate specificity (Cosio and Dunand, 2009), accumulating evidences suggest that the overexpression of POD genes results in increased plant tolerance to stresses. To understand the potential functions of the NtPOD genes in stress resistance, public RNA-seq data were used to investigate their expression patterns. The complexity and diversity of NtPOD expression patterns under various biotic and abiotic stresses were observed. In tobacco, the expression levels of only few POD genes increased after drought, cold, salt, topping, and cadmium treatments, while the expression levels of many members of the POD family increased after P. nicotianae infection. Meanwhile, individual POD genes are usually sensitive to one specific external stress, and few genes are widely responsive to various biotic and abiotic stresses. For example, NtPOD166 was responsive only to cold treatment. NtPOD93 was localized in vacuoles and induced only by drought stress. Moreover, the regulatory network constructed in our study implied that NtPOD93 may be regulated by miR172 during drought response. NtPOD131 was located in the chloroplast and associated with Cd stress. Salinization stress is one of the environmental factors that limit tobacco yield and quality, and some plant hormones, such as ABA, IAA, SA, and JA, are considered as salt stress response hormones (Yu et al., 2020). The qRT-PCR results were consistent with the RNA-seq gene expression patterns. Salt treatment resulted in increased transcript levels of NtPOD5, 8, 68, 176, 121, and 202, while the expression level of NtPOD209 and NtPOD13 was decreased. NtPOD68 and NtPOD121 significantly responded to hormone signals, such as ABA, IAA, SA, and JA, suggesting that these hormones exhibit complex signaling regulation to control plant responses to salt stress. NtPOD5, 8, 68, 176, 121, and 202 could be regarded as candidates that participate in the salt stress response. However, more work need to be performed to investigate the detailed mechanism for these candidates. In summary, the expression patterns of NtPODs under various abiotic and biotic stresses are complex and diverse, and may be related to the functional diversity of POD gene family members. These results provide useful insights into the potential capabilities of NtPODs under various abiotic and biotic stresses.
In the present study, we identified, characterized, and analyzed the members of the class III peroxidase family in tobacco by investigating phylogeny, protein properties, 3D models, and expression patterns. Many NtPOD genes were found to be expressed in a tissue-specific manner, with showing involvement in specific biotic and abiotic stresses. The function of these NtPOD genes is of great significance for the improvement of resistance to stresses in tobacco plants, and will need to be elucidated further in future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material
JJ and PC conceived and designed the experiments. LC and HS performed bioinformatics data analysis. LC and LM did the qRT-PCR experiments. LM performed the subcellular localization experiments. LC, PC, and JJ wrote the manuscript and all authors read and approved the final version.
This work was supported by the Zhengzhou Tobacco Research Institute [CNTC: 110202001020(JY-03), 110201901024(SJ-03)]; the Joint Laboratory of HNTI and ZTRI for Tobacco Gene Research and Utilization.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage (www.editage.com) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.916867/full#supplementary-material
Aleem, M., Riaz, A., Raza, Q., Aleem, M., Aslam, M., Kong, K., et al. (2022). Genome-wide Characterization and Functional Analysis of Class III Peroxidase Gene Family in Soybean Reveal Regulatory Roles of GsPOD40 in Drought Tolerance. Genomics 114 (1), 45–60. doi:10.1016/j.ygeno.2021.11.016
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 37 (4), 420–423. doi:10.1038/s41587-019-0036-z
Almagro, L., Gómez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barceló, A., and Pedreño, M. A. (2009). Class III Peroxidases in Plant Defence Reactions. J. Exp. Bot. 60 (2), 377–390. doi:10.1093/jxb/ern277
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43 (W1), W39–W49. doi:10.1093/nar/gkv416
Braybrook, S. A. (2017). Analyzing Cell Wall Elasticity after Hormone Treatment: an Example Using Tobacco BY-2 Cells and Auxin. Plant Horm. 1497, 125–133. doi:10.1007/978-1-4939-6469-7_12
Cai, K., Liu, H., Chen, S., Liu, Y., Zhao, X., and Chen, S. (2021). Genome-wide Identification and Analysis of Class III Peroxidases in Betula Pendula. BMC Genomics 22 (1), 1–19. doi:10.1186/s12864-021-07622-1
Çakır, Ö., Arıkan, B., Karpuz, B., and Turgut-Kara, N. (2021). Expression Analysis of miRNAs and Their Targets Related to Salt Stress in Solanum lycopersicum H-2274. Biotechnol. Biotechnol. Equip. 35 (1), 283–290. doi:10.1080/13102818.2020.1870871
Cao, Y., Han, Y., Meng, D., Li, D., Jin, Q., Lin, Y., et al. (2016). Structural, Evolutionary, and Functional Analysis of the Class III Peroxidase Gene Family in Chinese Pear (Pyrus Bretschneideri). Front. Plant Sci. 7, 1874–1885. doi:10.3389/fpls.2016.01874
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, D., Yan, W., Fu, L. Y., and Kaufmann, K. (2018). Architecture of Gene Regulatory Networks Controlling Flower Development in Arabidopsis thaliana. Nat. Commun. 9 (1), 4534. doi:10.1038/s41467-018-06772-3
Chen, T., Li, W., Hu, X., Guo, J., Liu, A., and Zhang, B. (2015). A Cotton MYB Transcription Factor, GbMYB5, Is Positively Involved in Plant Adaptive Response to Drought Stress. Plant Cell Physiology 56 (5), 917–929. doi:10.1093/pcp/pcv019
Chen, X., Sun, S., Liu, F., Shen, E., Liu, L., Ye, C., et al. (2019). A Transcriptomic Profile of Topping Responsive Non-coding RNAs in Tobacco Roots (Nicotiana Tabacum). BMC Genomics 20 (1), 856. doi:10.1186/s12864-019-6236-6
Cheng, Y., Qi, Y., Zhu, Q., Chen, X., Wang, N., Zhao, X., et al. (2009). New Changes in the Plasma-Membrane-Associated Proteome of Rice Roots under Salt Stress. Proteomics 9 (11), 3100–3114. doi:10.1002/pmic.200800340
Choi, H. W., and Hwang, B. K. (2012). The Pepper Extracellular Peroxidase CaPO2 Is Required for Salt, Drought and Oxidative Stress Tolerance as Well as Resistance to Fungal Pathogens. Planta 235 (6), 1369–1382. doi:10.1007/s00425-011-1580-z
Cosio, C., and Dunand, C. (2009). Specific Functions of Individual Class III Peroxidase Genes. J. Exp. Bot. 60 (2), 391–408. doi:10.1093/jxb/ern318
Dai, X., and Zhao, P. X. (2011). PsRNATarget: a Plant Small RNA Target Analysis Server. Nucleic Acids Res. 39 (Suppl. l_2), W155–W159. doi:10.1093/nar/gkr319
Dana, M. d. l. M., Pintor-Toro, J. A., and Cubero, B. (2006). Transgenic Tobacco Plants Overexpressing Chitinases of Fungal Origin Show Enhanced Resistance to Biotic and Abiotic Stress Agents. Plant Physiol. 142 (2), 722–730. doi:10.1104/pp.106.086140
Das, K., and Roychoudhury, A. (2014). Reactive Oxygen Species (ROS) and Response of Antioxidants as ROS-Scavengers during Environmental Stress in Plants. Front. Environ. Sci. 2, 53–65. doi:10.3389/fenvs.2014.00053
DeLano, W. L. (2002). Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 40 (1), 82–92.
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The Sol Genomics Network (SGN)-from Genotype to Phenotype to Breeding. Nucleic Acids Res. 43 (D1), D1036–D1041. doi:10.1093/nar/gku1195
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 39 (Suppl. l_2), W29–W37. doi:10.1093/nar/gkr367
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the Proteomics Server for In-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 31 (13), 3784–3788. doi:10.1093/nar/gkg563
Han, Y., Zhang, X., Wang, Y., and Ming, F. (2013). The Suppression of WRKY44 by GIGANTEA-miR172 Pathway Is Involved in Drought Response of Arabidopsis thaliana. PLoS One 8 (11), e73541. doi:10.1371/journal.pone.0073541
He, X., Zheng, W., Cao, F., and Wu, F. (2016). Identification and Comparative Analysis of the microRNA Transcriptome in Roots of Two Contrasting Tobacco Genotypes in Response to Cadmium Stress. Sci. Rep. 6 (1), 32805–32814. doi:10.1038/srep32805
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 35 (Suppl. l_2), W585–W587. doi:10.1093/nar/gkm259
Hsieh, T.-H., Li, C.-W., Su, R.-C., Cheng, C.-P., Sanjaya, Y.-C., Tsai, Y.-C., et al. (2010). A Tomato bZIP Transcription Factor, SlAREB, Is Involved in Water Deficit and Salt Stress Response. Planta 231 (6), 1459–1473. doi:10.1007/s00425-010-1147-4
Ibraheem, O., Botha, C. E., and Bradley, G. (2010). In Silico analysis of Cis-Acting Regulatory Elements in 5′ Regulatory Regions of Sucrose Transporter Gene Families in Rice (Oryza Sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 34 (5-6), 268–283. doi:10.1016/j.compbiolchem.2010.09.003
Jaggi, M., Kumar, S., and Sinha, A. K. (2011). Overexpression of an Apoplastic Peroxidase Gene CrPrx in Transgenic Hairy Root Lines of Catharanthus Roseus. Appl. Microbiol. Biotechnol. 90 (3), 1005–1016. doi:10.1007/s00253-011-3131-8
Jin, J., Zhang, H., Zhang, J., Liu, P., Chen, X., Li, Z., et al. (2017). Integrated Transcriptomics and Metabolomics Analysis to Characterize Cold Stress Responses in Nicotiana Tabacum. BMC Genomics 18 (1), 496. doi:10.1186/s12864-017-3871-7
Jin, J., Lu, P., Xu, Y., Li, Z., Yu, S., Liu, J., et al. (2021). PLncDB V2.0: a Comprehensive Encyclopedia of Plant Long Noncoding RNAs. Nucleic Acids Res. 49 (D1), D1489–D1495. doi:10.1093/nar/gkaa910
Jin, J., Tian, F., Yang, D.-C., Meng, Y.-Q., Kong, L., Luo, J., et al. (2016). PlantTFDB 4.0: toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 45 (D1), D1040–D1045. doi:10.1093/nar/gkw982
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kawano, T., and Muto, S. (2000). Mechanism of Peroxidase Actions for Salicylic Acid‐induced Generation of Active Oxygen Species and an Increase in Cytosolic Calcium in Tobacco Cell Suspension Culture. J. Exp. Bot. 51 (345), 685–693. doi:10.1093/jxb/51.345.685
Kidwai, M., Ahmad, I. Z., and Chakrabarty, D. (2020). Class III Peroxidase: An Indispensable Enzyme for Biotic/abiotic Stress Tolerance and a Potent Candidate for Crop Improvement. Plant Cell Rep. 39 (11), 1381–1393. doi:10.1007/s00299-020-02588-y
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 37 (8), 907–915. doi:10.1038/s41587-019-0201-4
Kim, J. Y., Lee, H. J., Jung, H. J., Maruyama, K., Suzuki, N., and Kang, H. (2010). Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta 232 (6), 1447–1454. doi:10.1007/s00425-010-1267-x
Kondrashov, F. A. (2012). Gene Duplication as a Mechanism of Genomic Adaptation to a Changing Environment. Proc. R. Soc. B 279 (1749), 5048–5057. doi:10.1098/rspb.2012.1108
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). MiRBase: from microRNA Sequences to Function. Nucleic Acids Res. 47 (D1), D155–D162. doi:10.1093/nar/gky1141
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 305 (3), 567–580. doi:10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Leinonen, R., Sugawara, H., Shumway, M., and International Nucleotide Sequence Database, C. (2010). The Sequence Read Archive. Nucleic Acids Res. 39 (1), D19–D21.
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Li, G., Xu, W., Jing, P., Hou, X., and Fan, X. (2021). Overexpression of VyDOF8, a Chinese Wild Grapevine Transcription Factor Gene, Enhances Drought Tolerance in Transgenic Tobacco. Environ. Exp. Bot. 190, 104592. doi:10.1016/j.envexpbot.2021.104592
Li, Q., Dou, W., Qi, J., Qin, X., Chen, S., and He, Y. (2020). Genomewide Analysis of the CIII Peroxidase Family in Sweet Orange (Citrus Sinensis) and Expression Profiles Induced by Xanthomonas Citri Subsp. Citri and Hormones. J. Genet. 99 (1), 1–13. doi:10.1007/s12041-019-1163-5
Lim, K. Y., Matyasek, R., Kovarik, A., and Leitch, A. R. (2004). Genome Evolution in Allotetraploid Nicotiana. Biol. J. Linn. Soc. 82 (4), 599–606. doi:10.1111/j.1095-8312.2004.00344.x
Liu, D., Cheng, Y., Gong, M., Zhao, Q., Jiang, C., Cheng, L., et al. (2019). Comparative Transcriptome Analysis Reveals Differential Gene Expression in Resistant and Susceptible Tobacco Cultivars in Response to Infection by Cucumber Mosaic Virus. Crop J. 7 (3), 307–321. doi:10.1016/j.cj.2018.11.008
Lüthje, S., and Martinez-Cortes, T. (2018). Membrane-bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions. Int. J. Mol. Sci. 19 (10), 2876–2897. doi:10.3390/ijms19102876
Mathé, C., Barre, A., Jourda, C., and Dunand, C. (2010). Evolution and Expression of Class III Peroxidases. Arch. Biochem. Biophys. 500 (1), 58–65. doi:10.1016/j.abb.2010.04.007
McGinnis, S., and Madden, T. L. (2004). BLAST: at the Core of a Powerful and Diverse Set of Sequence Analysis Tools. Nucleic Acids Res. 32 (Suppl. l_2), W20–W25. doi:10.1093/nar/gkh435
Meng, G., Fan, W., and Rasmussen, S. K. (2021). Characterisation of the Class III Peroxidase Gene Family in Carrot Taproots and its Role in Anthocyanin and Lignin Accumulation. Plant Physiology Biochem. 167, 245–256. doi:10.1016/j.plaphy.2021.08.004
Nakashima, K., Tran, L.-S. P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional Analysis of a NAC-type Transcription Factor OsNAC6 Involved in Abiotic and Biotic Stress-Responsive Gene Expression in Rice. Plant J. 51 (4), 617–630. doi:10.1111/j.1365-313x.2007.03168.x
Nielsen, H. (2015). “Predicting Subcellular Localization of Proteins by Bioinformatic Algorithms,” in Protein and Sugar Export and Assembly in Gram-Positive Bacteria. Cham: Springer, 129–158. doi:10.1007/82_2015_5006
Niu, C.-F., Wei, W., Zhou, Q.-Y., Tian, A.-G., Hao, Y.-J., Zhang, W.-K., et al. (2012). Wheat WRKY Genes TaWRKY2 and TaWRKY19 Regulate Abiotic Stress Tolerance in Transgenic Arabidopsis Plants. Plant, Cell & Environ. 35 (6), 1156–1170. doi:10.1111/j.1365-3040.2012.02480.x
Panchy, N., Lehti-Shiu, M., and Shiu, S.-H. (2016). Evolution of Gene Duplication in Plants. Plant Physiol. 171 (4), 2294–2316. doi:10.1104/pp.16.00523
Pandey, V. P., Awasthi, M., Singh, S., Tiwari, S., and Dwivedi, U. N. (2017). A Comprehensive Review on Function and Application of Plant Peroxidases. Biochem. Anal. Biochem. 6 (1), 308–324. doi:10.4172/2161-1009.1000308
Passardi, F., Longet, D., Penel, C., and Dunand, C. (2004). The Class III Peroxidase Multigenic Family in Rice and its Evolution in Land Plants. Phytochemistry 65 (13), 1879–1893. doi:10.1016/j.phytochem.2004.06.023
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 33 (3), 290–295. doi:10.1038/nbt.3122
Priest, H. D., Filichkin, S. A., and Mockler, T. C. (2009). Cis-regulatory Elements in Plant Cell Signaling. Curr. Opin. Plant Biol. 12 (5), 643–649. doi:10.1016/j.pbi.2009.07.016
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: Protein Domains Identifier. Nucleic Acids Res. 33 (Suppl. l_2), W116–W120. doi:10.1093/nar/gki442
Ren, L.-L., Liu, Y.-J., Liu, H.-J., Qian, T.-T., Qi, L.-W., Wang, X.-R., et al. (2014). Subcellular Relocalization and Positive Selection Play Key Roles in the Retention of Duplicate Genes ofPopulusClass III Peroxidase Family. Plant Cell 26 (6), 2404–2419. doi:10.1105/tpc.114.124750
Sasaki, K., Hiraga, S., Ito, H., Seo, S., Matsui, H., and Ohashi, Y. (2002). A Wound-Inducible Tobacco Peroxidase Gene Expresses Preferentially in the Vascular System. Plant Cell Physiology 43 (1), 108–117. doi:10.1093/pcp/pcf013
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L., and Ideker, T. (2011). Cytoscape 2.8: New Features for Data Integration and Network Visualization. Bioinformatics 27 (3), 431–432. doi:10.1093/bioinformatics/btq675
Su, P., Yan, J., Li, W., Wang, L., Zhao, J., Ma, X., et al. (2020). A Member of Wheat Class III Peroxidase Gene Family, TaPRX-2A, Enhanced the Tolerance of Salt Stress. BMC Plant Biol. 20 (1), 392. doi:10.1186/s12870-020-02602-1
Tognolli, M., Penel, C., Greppin, H., and Simon, P. (2002). Analysis and Expression of the Class III Peroxidase Large Gene Family in Arabidopsis thaliana. Gene 288 (1-2), 129–138. doi:10.1016/s0378-1119(02)00465-1
Trott, O., and Olson, A. J. (2010). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Veljović Jovanović, S., Kukavica, B., Vidović, M., Morina, F., and Menckhoff, L. (2018). “Class III Peroxidases: Functions, Localization and Redox Regulation of Isoenzymes,” in Antioxidants and Antioxidant Anzymes in Higher Plants (Cham: Springer), 269–300.
Volkov, R. A., Panchuk, I. I., and Schöffl, F. (2003). Heat-stress-dependency and Developmental Modulation of Gene Expression: the Potential of House-Keeping Genes as Internal Standards in mRNA Expression Profiling Using Real-Time RT-PCR. J. Exp. Bot. 54 (391), 2343–2349. doi:10.1093/jxb/erg244
Wang, Y., Wang, Q., Zhao, Y., Han, G., and Zhu, S. (2015). Systematic Analysis of Maize Class III Peroxidase Gene Family Reveals a Conserved Subfamily Involved in Abiotic Stress Response. Gene 566 (1), 95–108. doi:10.1016/j.gene.2015.04.041
Wang, Z., Wei, P., Wu, M., Xu, Y., Li, F., Luo, Z., et al. (2015). Analysis of the Sucrose Synthase Gene Family in Tobacco: Structure, Phylogeny, and Expression Patterns. Planta 242 (1), 153–166. doi:10.1007/s00425-015-2297-1
Wei, W., Zhang, Y., Han, L., Guan, Z., and Chai, T. (2008). A Novel WRKY Transcriptional Factor from Thlaspi Caerulescens Negatively Regulates the Osmotic Stress Tolerance of Transgenic Tobacco. Plant Cell Rep. 27 (4), 795–803. doi:10.1007/s00299-007-0499-0
Welinder, K. G., Justesen, A. F., Kjaersgård, I. V. H., Jensen, R. B., Rasmussen, S. K., Jespersen, H. M., et al. (2002). Structural Diversity and Transcription of Class III Peroxidases fromArabidopsis Thaliana. Eur. J. Biochem. 269 (24), 6063–6081. doi:10.1046/j.1432-1033.2002.03311.x
Welinder, K. G. (1992). Superfamily of Plant, Fungal and Bacterial Peroxidases. Curr. Opin. Struct. Biol. 2 (3), 388–393. doi:10.1016/0959-440x(92)90230-5
Wu, C., Ding, X., Ding, Z., Tie, W., Yan, Y., Wang, Y., et al. (2019). The Class III Peroxidase (Pod) Gene Family in Cassava: Identification, Phylogeny, Duplication, and Expression. Int. J. Mol. Sci. 20 (11), 2730–2743. doi:10.3390/ijms20112730
Wu, H., Li, H., Zhang, W., Tang, H., and Yang, L. (2021). Transcriptional Regulation and Functional Analysis of Nicotiana Tabacum under Salt and ABA Stress. Biochem. Biophysical Res. Commun. 570, 110–116. doi:10.1016/j.bbrc.2021.07.011
Wu, Y., Yang, Z., How, J., Xu, H., Chen, L., and Li, K. (2017). Overexpression of a Peroxidase Gene (AtPrx64) of Arabidopsis thaliana in Tobacco Improves Plant's Tolerance to Aluminum Stress. Plant Mol. Biol. 95 (1), 157–168. doi:10.1007/s11103-017-0644-2
Xiao, H., Wang, C., Khan, N., Chen, M., Fu, W., Guan, L., et al. (2020). Genome-wide Identification of the Class III POD Gene Family and Their Expression Profiling in Grapevine (Vitis vinifera L). BMC Genomics 21 (1), 444. doi:10.1186/s12864-020-06828-z
Yang, H., Zhao, L., Zhao, S., Wang, J., and Shi, H. (2017). Biochemical and Transcriptomic Analyses of Drought Stress Responses of LY1306 Tobacco Strain. Sci. Rep. 7 (1), 17442. doi:10.1038/s41598-017-17045-2
Yang, J. K., Tong, Z. J., Fang, D. H., Chen, X. J., Zhang, K. Q., and Xiao, B. G. (2017). Transcriptomic Profile of Tobacco in Response to Phytophthora Nicotianae Infection. Sci. Rep. 7 (1), 401–407. doi:10.1038/s41598-017-00481-5
Yang, T., Zhang, P., Pan, J., Amanullah, S., Luan, F., Han, W., et al. (2022). Genome-wide Analysis of the Peroxidase Gene Family and Verification of Lignin Synthesis-Related Genes in Watermelon. Int. J. Mol. Sci. 23 (2), 642–666. doi:10.3390/ijms23020642
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 25 (11), 1117–1130. doi:10.1016/j.tplants.2020.06.008
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the Soybean GmERF3 Gene, an AP2/ERF Type Transcription Factor for Increased Tolerances to Salt, Drought, and Diseases in Transgenic Tobacco. J. Exp. Bot. 60 (13), 3781–3796. doi:10.1093/jxb/erp214
Keywords: plant peroxidases, tobacco, expression pattern, 3D model, stress
Citation: Cheng L, Ma L, Meng L, Shang H, Cao P and Jin J (2022) Genome-Wide Identification and Analysis of the Class III Peroxidase Gene Family in Tobacco (Nicotiana tabacum). Front. Genet. 13:916867. doi: 10.3389/fgene.2022.916867
Received: 10 April 2022; Accepted: 30 May 2022;
Published: 13 June 2022.
Edited by:
Fatemeh Maghuly, University of Natural Resources and Life Sciences, AustriaReviewed by:
Changbo Dai, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2022 Cheng, Ma, Meng, Shang, Cao and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingjing Jin, amluamluZ2ppbmcxMjE4QDEyNi5jb20=; Peijian Cao, cGVpamlhbmNhb0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.