- 1Department of Genetics and Plant Breeding, College of Agriculture, Professor Jayashankar Telangana State Agricultural University (PJTSAU), Hyderabad, India
- 2Administrative Office, PJTSAU, Hyderabad, India
- 3ICAR-Indian Institute of Rice Research (IIRR), Hyderabad, India
- 4CGIAR Excellence in Breeding (EiB), CIMMYT-ICRISAT, Hyderabad, India
- 5International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, India
- 6Agricultural Research Station, PJTSAU, Peddapalli, India
- 7Institute of Biotechnology, PJTSAU, Hyderabad, India
Rice (Oryza sativa L.) is an important source of nutrition for the world’s burgeoning population that often faces yield loss due to infestation by the brown planthopper (BPH, Nilaparvata lugens (Stål)). The development of rice cultivars with BPH resistance is one of the crucial precedences in rice breeding programs. Recent progress in high-throughput SNP-based genotyping technology has made it possible to develop markers linked to the BPH more quickly than ever before. With this view, a genome-wide association study was undertaken for deriving marker-trait associations with BPH damage scores and SNPs from genotyping-by-sequencing data of 391 multi-parent advanced generation inter-cross (MAGIC) lines. A total of 23 significant SNPs involved in stress resistance pathways were selected from a general linear model along with 31 SNPs reported from a FarmCPU model in previous studies. Of these 54 SNPs, 20 were selected in such a way to cover 13 stress-related genes. Kompetitive allele-specific PCR (KASP) assays were designed for the 20 selected SNPs and were subsequently used in validating the genotypes that were identified, six SNPs, viz, snpOS00912, snpOS00915, snpOS00922, snpOS00923, snpOS00927, and snpOS00929 as efficient in distinguishing the genotypes into BPH-resistant and susceptible clusters. Bph17 and Bph32 genes that are highly effective against the biotype 4 of the BPH have been validated by gene specific SNPs with favorable alleles in M201, M272, M344, RathuHeenati, and RathuHeenati accession. These identified genotypes could be useful as donors for transferring BPH resistance into popular varieties with marker-assisted selection using these diagnostic SNPs. The resistant lines and the significant SNPs unearthed from our study can be useful in developing BPH-resistant varieties after validating them in biparental populations with the potential usefulness of SNPs as causal markers.
Introduction
Insect pests are important causal factors for major yield losses in rice crops and are detrimental to food security worldwide. Among the various insect pests, the brown planthopper (BPH; Nilaparvata lugens Stål) is extremely notorious and destructive that has advanced from non-significant to a prominent state and can cause up to 80% yield losses in rice (Rashid et al., 2017; Balachiranjeevi et al., 2019) with curtailment in the photosynthetic rate, nitrogen content, chlorophyll pigment, leaf area, and dry matter accumulation in most susceptible rice cultivars (Cagampang et al., 1974). Apart from serving as a carrier for ragged stunt and grassy stunt diseases (Jena and Kim, 2010; Sarao et al., 2016), the BPH causes hopperburn symptoms (Backus et al., 2005) which leads to major yield losses in farmer fields. Four different biotypes of the BPH that vary in virulence against different genotypes have been reported in rice ecologies (Khush et al., 1985; Brar et al., 2009). Of these, biotype 4 is exclusive to the Indian subcontinent (Liu et al., 2015; Ren et al., 2016; Prahalada et al., 2017).
Conventional measures to reduce BPH damage with the application of chemical insecticides are not only costly but also environmentally hazardous, polluting the ecosystem and disrupting the natural balance of BPH predators such as mirid bugs viz., Cyrtorhinus lividipennis (Reuter) and Tytthus parviceps (Reuter) that hold the pest population (JhansiLakshmi et al., 2010a and b). For building a sustainable pest management strategy, the correct combination of breeding for resistance and control measures that must be established to diminish the BPH’s ecological fitness while maintaining the pest below economic threshold levels was suggested by Bosque-Perez and Buddenhagen (1992). Of the different strategies, the use of resistant varieties as a measure of host plant resistance (HPR) has been considered among the most economic and potent methods to protect against virulent BPH populations to achieve long-term and broad-spectrum resistance (Khush, 2001). As the vast majority of BPH-resistant genes do not confer broad-spectrum resistance to different biotypes of the BPH, the production of new rice cultivars with a broad range of resistance to BPH populations derived from diverse genetic sources is the need of the hour. With the constant efforts of rice scientists since the 1960s, around 42 BPH resistance genes and more than 70 quantitative trait loci (QTLs) have been identified and assigned to different rice chromosomes (Akanksha et al., 2019; Balachiranjeevi et al., 2019; Muduli et al., 2021; Tan et al., 2021) but only few genes, mostly Bph17, Bph3/32, Bph31, and Bph33(t) showed broad-spectrum resistance to Indian biotype 4 (Liu et al., 2015; Ren et al., 2016; Prahalada et al., 2017; Naik et al., 2018).
Over the past few decades, crop breeding has been altered to a great extent and taken into a positive direction with cutting-edge contemporary genomic technologies (Bohar et al., 2020; Ahmar et al., 2021). Genomics-assisted breeding led to the development of several crop varieties in rice (Fukuoka et al., 2010), groundnut (Varshney et al., 2013), chickpea (Varshney et al., 2013), pigeon pea (Varshney et al., 2013; Bohra and Kumar Varshney, 2020), wheat (De et al., 2009), and maize (Xu et al., 2009; Leng et al., 2017) with the introduction of molecular approaches in varietal development. Recent breakthroughs in next-generation sequencing (NGS) technology and high-throughput SNP marker-based genotyping technologies have become the unprecedented tools for the accelerated identification of markers linked to desirable traits in the field of plant breeding (Peng et al., 2020). Since their discovery, SNPs have become the most promising tool for the incorporation of desired genes because of their wide dispersal within genomes and suitability for high-throughput automated genotyping (Tamura et al., 2014; Yang et al., 2019). With several re-sequencing projects, QTL mapping, gene mapping, and GWAS in rice provide abundant information about an enormous number of genetic loci and SNPs that accord to BPH resistance. However, trait-linked SNPs need to be perfectly validated to use them as diagnostic markers in future marker-assisted breeding (MAB) programs. The identified SNPs through different approaches can be converted to KASP assays and validated for the linked traits (Gouda et al., 2021).
Kompetitive allele-specific PCR (KASP) assay is one of the most widely used uniplex genotyping platforms which is simple, fast, and economical. The method permits the detection of SNPs with high precision and has potential usefulness in MAS related to many breeding programs (He et al., 2014; Yang et al., 2019). KASP-based genotyping services are offered through several private and public service providers. For instance, the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) has led the High-Throughput Genotyping Project (HTPG) for SNP genotyping using KASP in collegiality with International Rice Research Institute (IRRI), International Maize and Wheat Improvement Center (CIMMYT), and the CGIAR EiB (Excellence in Breeding) platform with the financial support of the Bill & Melinda Gates Foundation. The HTPG facilitated shared industrial-scale low-cost, high-throughput SNP genotyping for CGIAR, NARS, and small–medium private sector organizations, mainly for forward breeding applications through collaboration, knowledge allocation, and new technologies adoption in 18 crops including rice (Bohar et al., 2020). Presently, the HTPG has transitioned into an EIB platform, and the shared services are offered as EiB low-density genotyping service (EiB-LDSG) (CGIAR, 2021). KASP-based genotyping was well used through the HTPG/EiB-LDSG with the successful development of markers for target traits along with the use of the existing KASP markers for introgression of target traits in a number of crops including groundnut (Parmar et al., 2021), sorghum (Mwamahonje et al., 2021), potato (Kante et al., 2021; Sood et al., 2022), cassava (Thuy et al., 2021), and rice (Arbelaez et al., 2019). Among the marker panels of rice with the HTPG, few SNPs related to BPH resistance for genes Bph17, Bph32, and Bph9 are available (Excellence in breeding, 2021) and can be genotyped for further use as diagnostic markers.
Authors have performed a genome-wide SNP discovery using GBS data of MAGIC indica population and identified SNPs associated with BPH resistance. To validate the identified SNPs through the KASP assay using HTPG services, the present study was undertaken with an aim to confirm the presence of Bph17, Bph32, and Bph9 genes in the germplasm and to validate the newly identified SNPs to use them as diagnostic markers in institutional MAB programs.
Materials and Methods
Plant Materials Used for Genome-Wide SNP Discovery
The panel comprised 391 MAGIC indica lines (Supplementary Table S1) that were produced at IRRI, Philippines, by inter-crossing eight elite indica founder parents through 8-way F1 inter-crossing (Bandillo et al., 2013; Satturu et al., 2020). The BPH damage score and GBS data of these lines were used for genome-wide association studies. Serial numbers for the 391 lines were given from M1 to M395 and four lines were not included as the GBS data were missing.
Plant Materials Used for Validation
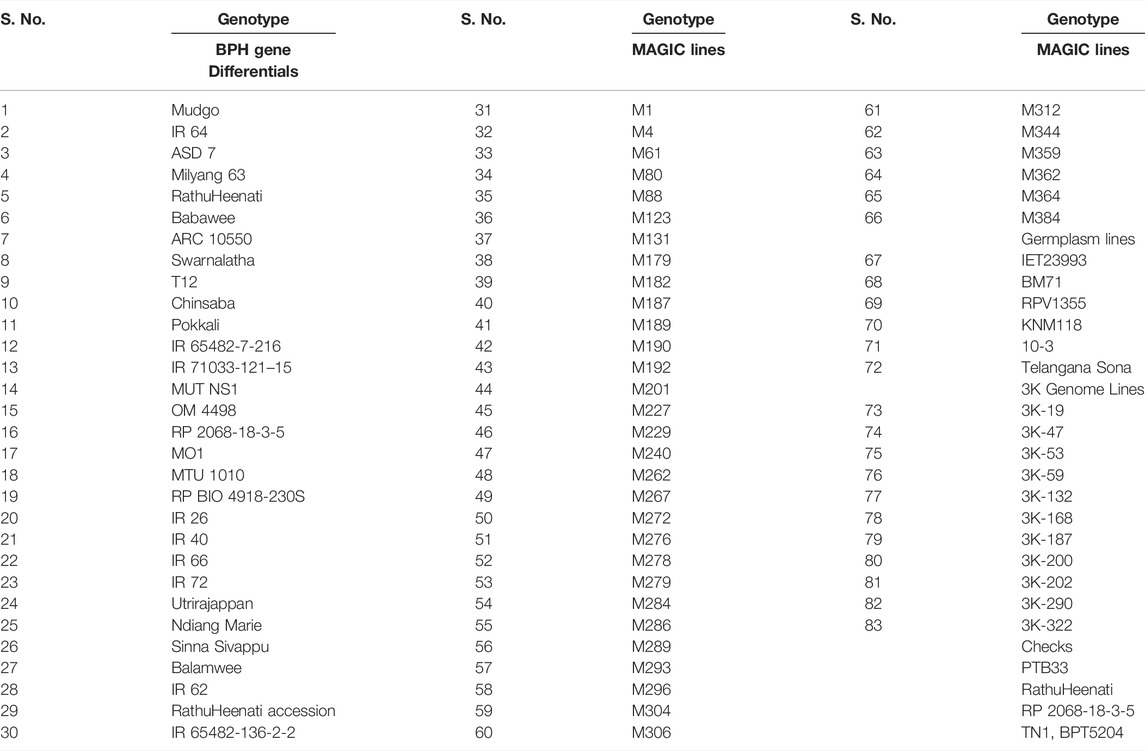
The validation panel comprised 83 rice genotypes comprising 30 BPH gene differentials, 36 MAGIC lines, 11 3K genome lines, six germplasm lines (including popular varieties and landraces), three resistant (PTB33, RathuHeenati, RP 2068-18-3-5), and two susceptible checks (TN1 and BPT5204) (Table 1). The materials were acquired from the Institute of Biotechnology, PJTSAU, Hyderabad, and ICAR-Indian Institute of Rice Research (IIRR), Hyderabad.
Brown Planthopper Insects and Mass Rearing
BPH adults were collected from the rice fields of ICAR-IIRR, Rajendranagar, Hyderabad, and the pure colonies were reared and managed at a temperature of 25 ± 5°C with a relative humidity of 70 ± 5% on 60 days old potted plants of the susceptible variety (TN1) under glasshouse conditions. Cages of 70 × 62 × 75 cm dimensions were mounted on wooden benches with glass-paneled doors on one side and a wire mesh on all the other sides for mass rearing. This was followed by the collection of adult gravid female hoppers and their release on pre-cleaned potted plants of TN1. The process was repeated after 3–4 days of egg-laying and the hatched nymphs were used for screening when they had attained the appropriate age.
Evaluation of Rice Genotypes for BPH Response
The standard seedbox screening technique (SSST) (Heinrichs et al., 1985) was used to assess the extent of BPH resistance across the 83 rice genotypes at the seedling stage. Seeds of the 83 genotypes were pre-soaked and sown in rows of 60 × 45 × 10 cm in seed boxes accommodating 20– 25 seedlings per row in augmented block design conducted during Kharif 2018 and Rabi 2018–19 (Figure 1) at the ICAR-Indian Institute of Rice Research, Entomology glasshouse. Four screening trays (blocks) comprising 20 test entries each and three entries in the fifth tray were evaluated with the checks being replicated in all trays at random. Twelve days after sowing, first-instar nymphs were delivered on the seedlings at 6–8 nymphs/seedling. The tray was turned 180° when TN1 plants on one side showed symptoms to have even reactions on both sides. The Standard Evaluation System (SES) for rice (Standard Evaluation System for Rice, 2013) was used to rate the damage of the test lines (Table 2) when 90% of the TN1 plants were killed. The plants with scores 0–1.0 were considered as highly resistant, 1.1–3.0 as resistant, 3.1– 5.0 as moderately resistant, 5.1– 7.0 as moderately susceptible, 7.1– 8.9 as susceptible, and 9.0 as highly susceptible.
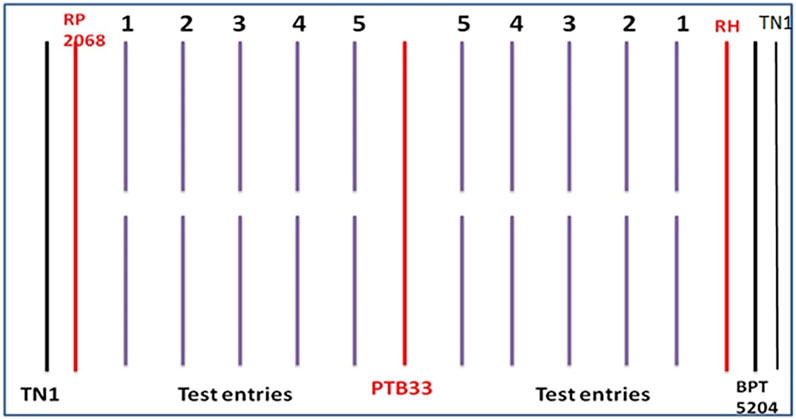
FIGURE 1. Layout of entries along with resistant and susceptible checks evaluated for BPH resistance using SSST method (RH = RathuHeenati).

TABLE 2. Classification of resistance based on the damage reaction (Standard Evaluation System for Rice, 2013).
Genotyping-by-Sequencing (GBS)
The genotyping-by-sequencing (GBS) method was followed for genotyping 391 MAGIC lines through Illumina HiSeq (Elshire et al., 2011). The raw genotyping data of 391 MAGIC lines (Raghavan et al., 2017) were retrieved (SNPs and Phenotypes Data, 2021) and processed through the TASSEL GBS pipeline to obtain polymorphic SNPs. These SNPs were curated by the GBS pipeline using TASSEL 3.0.169 software (Glaubitz et al., 2014) based on <30% missing, locus homozygosity, and minor allele frequency (MAF) of 0.05.
Genome-Wide Association Studies (GWAS) and Candidate Genes
During 2016 and 2017, 391 MAGIC indica lines were screened for resistance against the BPH using the SSST at the ICAR-Indian Institute of Rice Research and IBT, PJTSAU, Hyderabad, by Satturu et al. (2020). The GWAS analysis was conducted using the FarmCPU model that identified 31 annotated significant SNPs associated with 13 stress-related genes (Satturu et al., 2020).
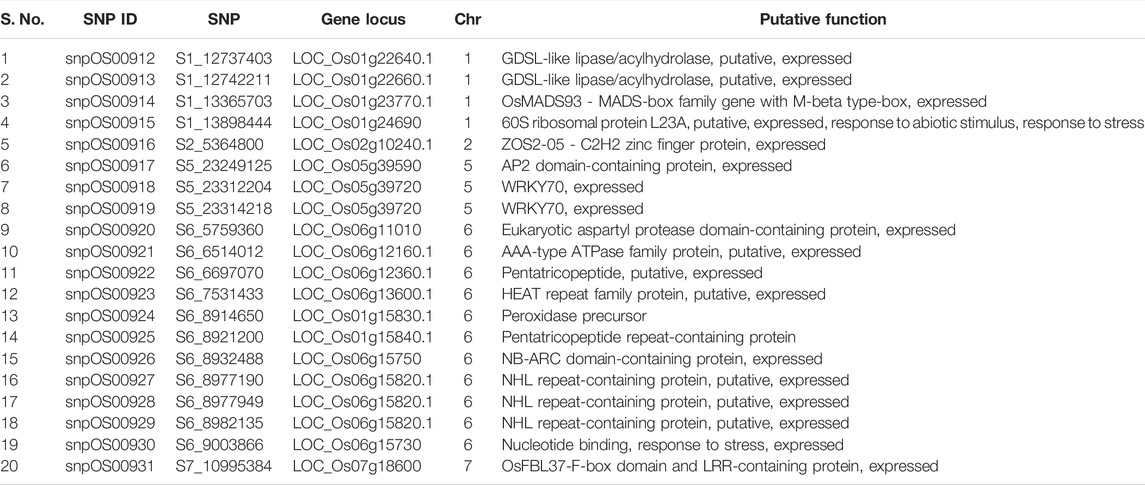
For our study, the average BPH score values of all the 391 MAGIC indica lines previously evaluated by Satturu et al. (2020) were considered for calculating the best linear unbiased predictions (BLUPs). The cured polymorphic SNPs obtained from GBS and across-year phenotypic data were engaged for conducting the GWAS through R software-based GAPIT (Genetic Association and Prediction Integrated Tools) (Lipka et al., 2012) analysis package for identifying marker-trait associations. The MAGIC lines were treated as unrelated individuals in the GWAS since the MAGIC population has an insignificant population structure (Bossa-Castro et al., 2018; Satturu et al., 2020). To account for the relatedness among accessions of the panel, the kinship matrix was established using the Centered IBS method. Three single-locus models such as the general linear model (GLM) (Price et al., 2006), mixed linear model (MLM) (Zhang et al., 2005), and Settlement of MLM Under Progressively Exclusive Relationship (SUPER) (Wang et al., 2014) and three multi-locus models viz., multi-locus mixed-model (MLMM) (Segura et al., 2012), Fixed and Random Model Circulating Probability Unification (FarmCPU) (Zhang et al., 2010), and Bayesian-information and Linkage-disequilibrium Iteratively Nested Keyway (BLINK) (Huang et al., 2019) were engaged to derive the marker-trait associations. According to the general distribution of all p-values of the SNPs for BPH resistance, a suggestive significance threshold of p-values < 0.001 was considered. The results from all the models were studied to find out the consistency and repeatability of the associations. Based on the deviation of the observed statistic values from the expected statistic values in Q-Q plots, the significant SNPs identified through the GLM model were selected for further annotation studies. To spot the candidate genes underlying the haplotypes of interest, rice genome browsers including the Rice Genome Annotation Project-Michigan State University Rice Genome Annotation Project database (Osa1) Release 7 were searched and 23 SNPs involved in stress resistance pathways were collected using SNPEff 4.3T software (SnpEff and SnpSift, 2021). Of these 23 SNPs, 20 SNPs along with the previously identified 31 SNPs from the FarmCPU model (Satturu et al., 2020) were selected in such a way to cover all the 13 stress-related genes for designing KASP assays in our validation studies.
DNA Extraction for Uniplex SNP Validation
For validation studies, leaf samples of the 83 rice genotypes along with checks raised at the College farm, PJTSAU, Rajendranagar, Hyderabad, were collected after 21 days of planting. Two leaf discs of 4 mm diameter were collected from each genotype using the paper punching machine and placed into a 12 x 8-well strip tube of 96-well microtiter plate provided by Intertek-Agritech laboratory, Hyderabad (Intertek AgriTech, 2021), by taking enough care to avoid DNA contamination and for robust genotypic data quality. The leaf discs were then oven-dried at 45°C for 12 h to remove moisture and stored at room temperature. These leaf discs were subsequently used for genomic DNA isolation, quantification, and genotyping. DNA isolation was carried out using LGC oKtopure™ automated high-throughput sbeadex™ (surface-coated superparamagnetic beads) DNA extraction and purification system (Biosearch technologies, 2021b) and the steps were followed according to the manufacturer’s instructions at AgriTech-Intertek Pvt. Ltd., Hyderabad, India. The leaf samples were homogenized by steel bead grinding in 96-deep-well plates and an extraction buffer in the sbeadex™ kit (Biosearch technologies, 2021b) was added. Purification of the extracted DNA was performed using sbeadex™ coated super paramagnetic particles with a surface chemistry that catches nucleic acids from a sample. The purified DNA was eluted and used for quantification and genotyping experiments. The genotyping data were generated using the KASP assay.
Genotyping Using KASP Assay
KASP genotyping was carried out using the HTPG services provided at Intertek, Hyderabad, through the EiB platform funded by the CGIAR and Bill & Melinda Gates Foundation (Bohar et al., 2020). To develop KASP genotyping assays for SNPs related to BPH resistance, flanking sequences of the selected 20 SNPs were retrieved using SAMtools1.1 software (Li et al., 2009) by aligning the genomic sequences with chromosome positions to the reference genome. A minimum of 100 bases of sequences for each of the identified SNPs having 50 bases on either flanking side was extracted. These sequences were further used to design KASP primers/assays (LGC, Biosearch Technologies) and were used for validating the genotypes used in the study by using the HTPG platform (Supplementary Table S2). Along with these 20 SNPs, SNPs specific to Bph17, Bph32, and Bph9 (Table 3) available with HTPG services (Excellence in breeding, 2021) were used for validating the rice panel as these are stable resistance genes providing valuable defense against a BPH attack.
SNPviewer Software
The cluster plots generated from the SNP genotype datasets were graphically viewed in SNPviewer v.4.1.2 (Biosearch technologies, 2021a). The HTPG genotyping result files present in a CSV (comma-separated value) format were given as input for grouping the allele calls to differentiate the resistant and susceptible genotypes based on the presence of favorable SNP alleles. Accordingly, homozygotes and heterozygotes for the SNP loci were distinguished based on the difference in fluorescence using the SNPviewer software.
Results
Analysis of Variance for BPH Resistance
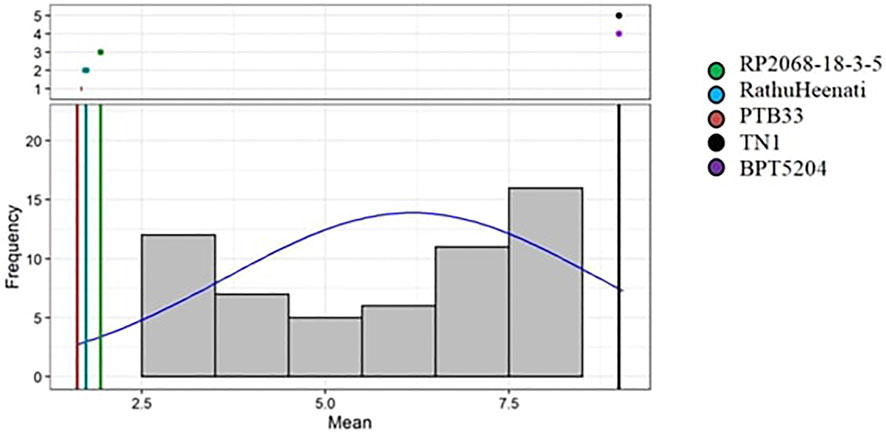
The phenotypic reaction of 83 rice genotypes and checks evaluated using SSST exhibited varied levels of resistance response (Supplementary Table S3). The analysis of variance for damage scores revealed significant differences among the genotypes for various sources of variation as shown in Table 4. The block effect (unadjusted), treatment effects (adjusted and unadjusted), and effects due to checks, varieties, and checks versus varieties were all significant, whereas the adjusted block effects were non-significant. On comparing the damage scores of both the seasons, 13 of the 83 entries were resistant, 15 were moderately resistant, 12 genotypes were moderately susceptible, 34 were susceptible, and the remaining nine genotypes were found to be highly susceptible. The 13 resistant genotypes (Supplementary Figure S1, Table 5) included nine MAGIC lines (M4, M88, M179, M182, M192, M229, M240, M312, and M344), three gene differentials (RP 2068-18-3-5, RP Bio4918-230S, and RathuHeenati), and a landrace (10-3) with a low damage score varying between 1.3 and 3.0. Considerable skewness was observed for the genotypes more toward susceptibility with most of the genotypes showing damage scores between 7.1 and 8.9 as depicted by frequency distribution-based adjusted mean values (Figure 2).
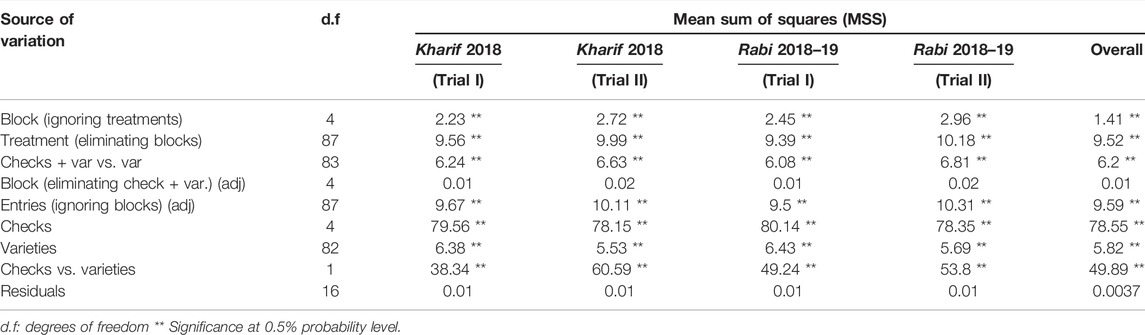
TABLE 4. Analysis of variance for brown planthopper scoring of 83 rice genotypes and checks in augmented RCBD.
Identification of Candidate SNPs for BPH Resistance Using GWAS
Raw genotyping data of the 391 MAGIC panel were used to perform the GWAS analysis which detected an aggregate of 27,000 polymorphic SNPs after curation of the data. The GWAS analysis using the BLUPs and cured polymorphic SNPs obtained from GBS data identified the common SNPs related to the resistance to BPH from three single-locus (GLM, MLM, and SUPER) and three multi-locus models (MLMM, BLINK, and FarmCPU). Manhattan plots (Supplementary Figures S2,S3) were used to display the p-value (0.001) distributions of SNPs with significant relationships across the chromosomes and the deviation of the observed statistical values from the expected statistical values was visualized in Q–Q plots (Figure 3). A total of 23 annotated stress-related SNPs linked to BPH resistance were identified from the GLM model on chromosomes 1, 2, and 6 from the model (Supplementary Table S4). Of the 23 annotated significant SNPs, 11 SNPs were found to cluster on chromosome 1 and were associated with six genes, viz., LOC_Os01g22640.1, LOC_Os01g23610.1, LOC_Os01g23680.1, LOC_Os01g23770.1, LOC_Os01g24050.1, and LOC_Os01g24950.1. These SNPs spanned from 12.7 to 14.0 Mb. One SNP (S2_5364800) was detected at a physical distance of 5.3 Mb on chromosome 2, while 11 significant SNPs identified on chromosome 6 were found to be associated with six genes viz., LOC_Os06g07420.1, LOC_Os06g07620.1, LOC_Os06g15730.1, LOC_Os06g15740.1, LOC_Os06g15810.1, and LOC_Os06g15850.1. These SNPs were located between 3.5–3.6 Mb and 8.9–9.0 Mb.
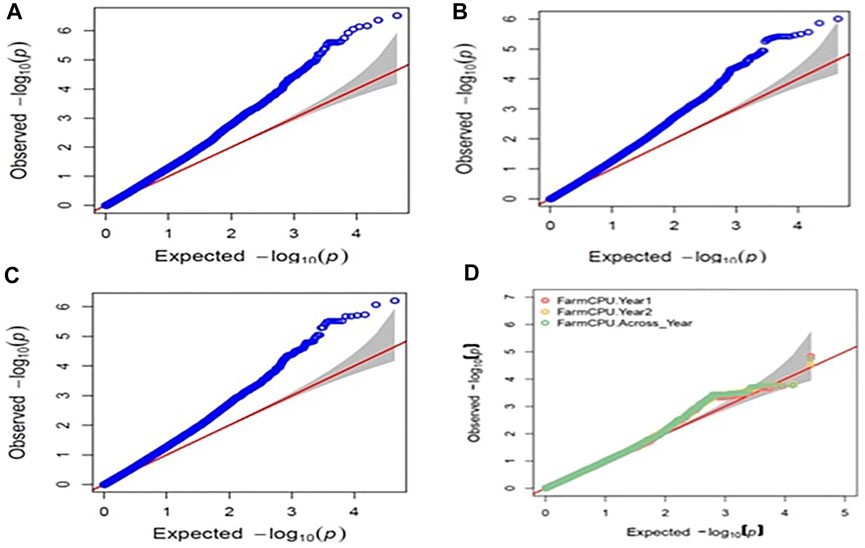
FIGURE 3. Q-Q plots obtained for BPH resistance in GWAS for (A) 2016 GLM model (B) 2017 GLM model (C) pooled years using GLM model (D) pooled years using FarmCPU model.
While considering the results obtained from the GLM model (from the present study) and FarmCPU model from earlier studies (Satturu et al., 2020), it was observed that significant SNPs were identified at the same positions on chromosome 1 (1.3 Mb) and chromosome 6 (8.9 Mb). Of the total 54 significant SNPs identified from both the models, 20 SNPs (Table 6) were selected for genotyping in such a way to cover all the annotated genes representing functional defense-related mechanisms.
Annotated Candidate Genes Related to Resistance Against BPH
Among the four annotated genes on chromosome 1 (Table 6), LOC_Os01g24690 associated with SNP snpOS00915 was found to encode 60S ribosomal protein L23A, while LOC_Os01g22640.1 (snpOS00912) and LOC_Os01g22660.1 (snpOS00913) were associated with GDSL-like lipase/acylhydrolase protein and LOC_Os01g23770.1 (snpOS00914) with the OsMADS93-MADS-box family. LOC_Os02g10240.1 (snpOS00916) on chromosome 2 was found to encode the ZOS2-05-C2H2 zinc finger protein, while on chromosome 5, the significant SNPs, snpOS00917, snpOS00918, and snpOS00919 were found in the genic regions of LOC_Os05g39720 and LOC_Os05g39590 associated with the AP2 domain-containing protein and transcription factor-WRKY70. Furthermore, on chromosome 6, genes LOC_Os06g12360.1 and LOC_Os01g15840.1 co-located with snpOS00922 and snpOS00925, respectively, were found to be related to pentatricopeptide repeat proteins; SNPs snpOS00927, snpOS00928, and snpOS00929 (LOC_Os06g15820.1) with NHL repeat-containing protein, snpOS00926 was close to LOC_Os06g15750 with the NB-ARC functional ATPase protein, LOC_Os06g13600.1 (snpOS00923) was related to the HEAT repeat protein, while LOC_Os06g11010 (snpOS00920) and LOC_Os01g15830.1(snpOS00924) were linked to the eukaryotic aspartyl protease domain-containing protein and peroxidase precursors. On chromosome 7, LOC_Os07g18600 (snpOS00931) was found to be associated with the OsFBL37-F-box domain and LRR-containing protein.
KASP assays for these 20 SNPs were designed using 100 base pair sequences and were used in the validation studies along with five SNP markers specific to Bph17, Bph32, and Bph9 available with the HTPG platform.
Validation of Designed SNPs Associated With BPH Resistance
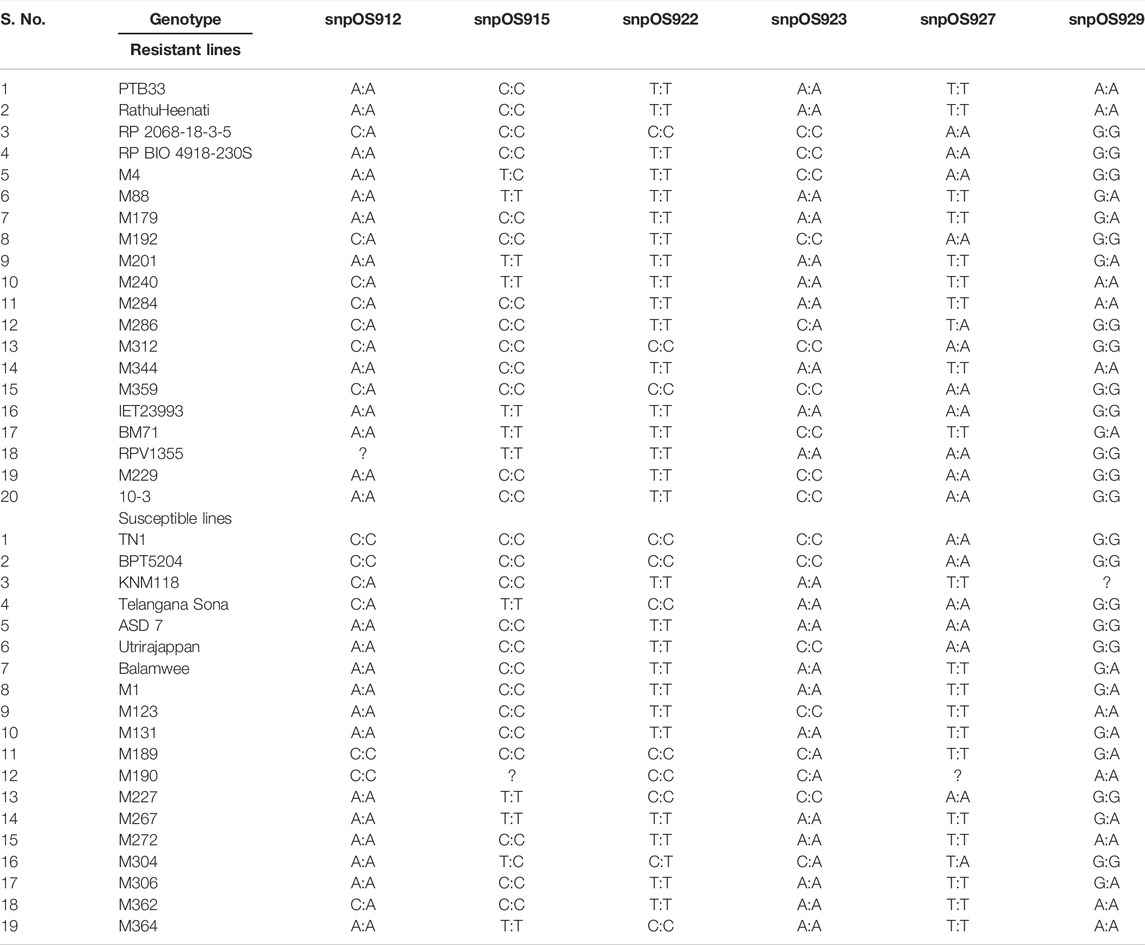
Among the 20 designed SNPs for BPH resistance, six SNPs, viz, snpOS00912, snpOS00915, snpOS00922, snpOS00923, snpOS00927, and snpOS00929 were able to distinguish the resistant and susceptible checks in the presence and absence of favorable alleles, respectively (Table 7), while four SNPs (snpOS00913, snpOS00914, snpOS00916, and snpOS00925) were completely monomorphic (Supplementary Table S5). Among all the SNPs, snpOS00922 on chromosome 6 had been the most efficient marker with high power of distinguishing the genotypes into resistant and susceptible clusters. The resistant lines included M4, M88, M179, M192, M201, M229, M240, M284, M286, M344, IET23993, BM71, PTB33, 10-3, RathuHeenati, RathuHeenati accession, and RPV1355 with favorable allele T:T. snpOS00923, snpOS00912, and snpOS00929 pertaining to chromosome 6 differentiated resistant and susceptible genotypes in the presence and absence of favorable allele A:A, respectively (Figure 4). Correspondingly, snpOS00927and snpOS00915 had T:T and C:C favorable alleles, respectively, which also exhibited a moderate mode of demarcating the genotypes based on resistance reaction.
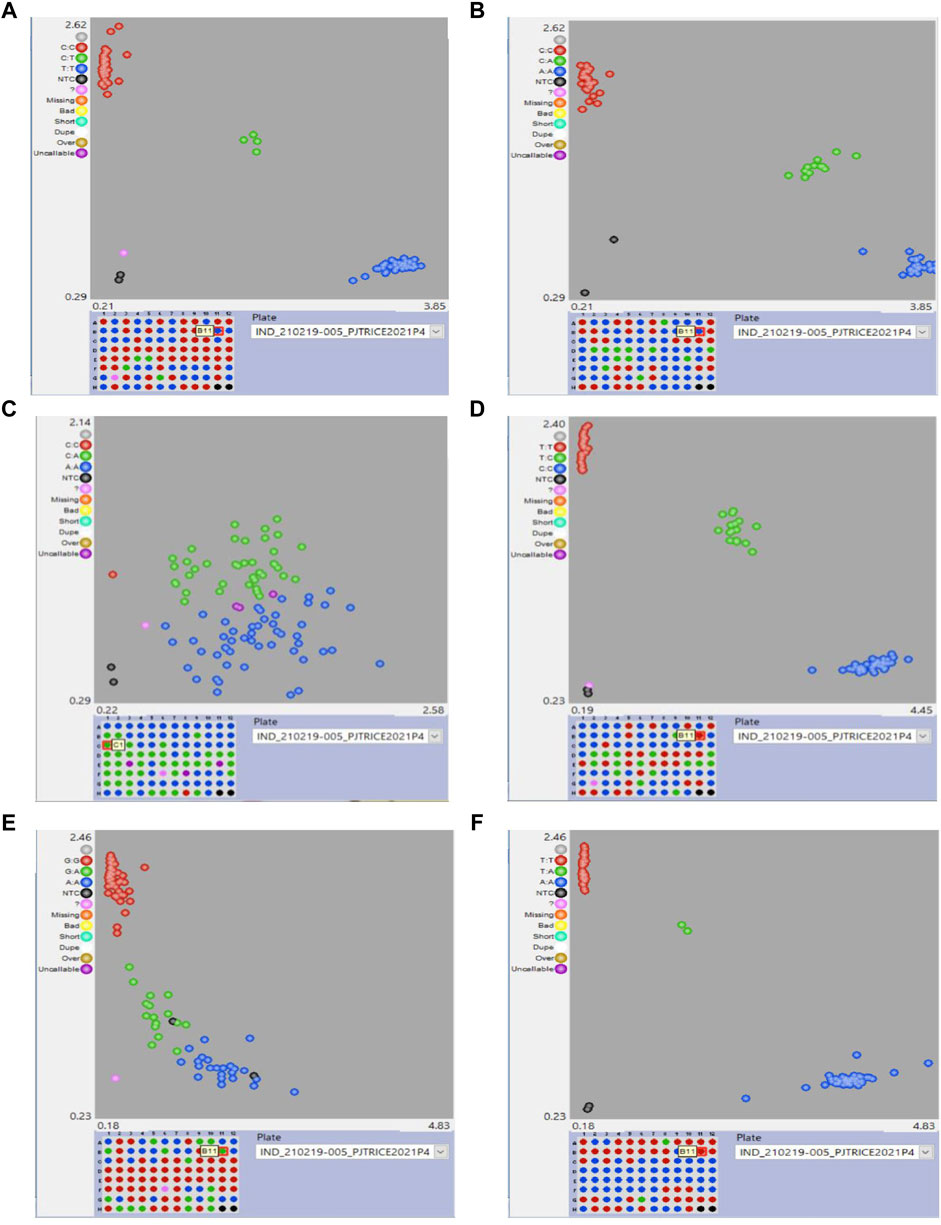
FIGURE 4. Clustering of genotypes for the SNPs (A) snpOS00922, (B) snpOS00923, (C) snpOS00912 (D) snpOS00915 (E) snpOS00929 and (F) snpOS00927.
Validation of the SNPs Specific to Bph17, Bph32, and Bph9 in a Pre-Defined Panel of Genotypes
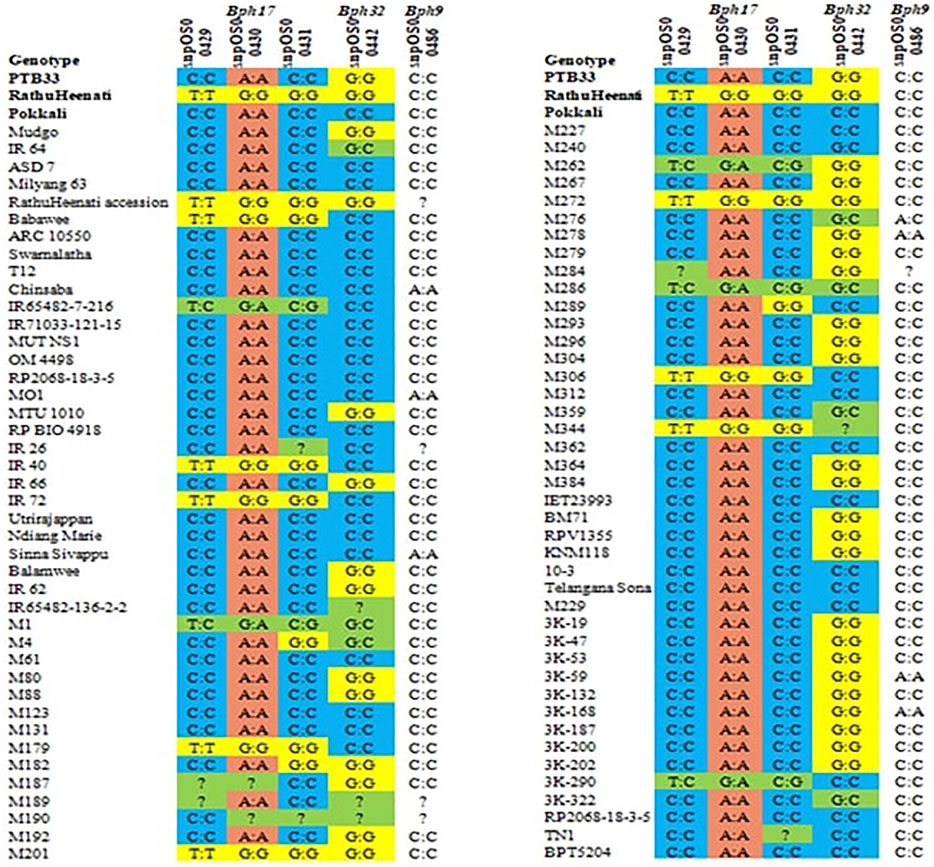
A total of five SNP markers reported to be specific to Bph17, Bph32, and Bph9 genes were used for validating the panel of genotypes for BPH resistance (Figure 5). Of the 83 genotypes and checks evaluated for the Bph17 gene, 10 lines were having the favorable alleles for snpOS00429 (T:T) and snpOS00430 (G:G), while the favorable allele (G:G) for snpOS00431 was present in nine genotypes (Supplementary Table S6). Altogether, five MAGIC lines (M201, M179, M306, M272, and M344) and four gene differentials (RathuHeenati, RathuHeenati accession, Babawee, and IR72) were positive for all the three Bph17 gene-specific SNPs. Lines M1, M262, M286, and 3K-290 and gene differential IR65482-7-216 were heterozygous for these loci. Regarding the Bph32 gene, 38 accessions including 17 MAGIC lines, seven gene differentials, and nine 3K lines along with PTB33, KNM118, RDR1200, RPV1355, and BM71 were detected to contain the positive allele (G:G) in homozygous condition (Supplementary Table S7). The original donor PTB33 was found with the positive allele for the Bph32 gene while RathuHeenati and RathuHeenati accession confirmed the presence of Bph17 and Bph32 genes.
Three MAGIC lines viz., M201, M272, and M344 had the favorable alleles for both Bph17 and Bph32 genes (Table 8). MAGIC lines M1 and M286 were noted to be heterozygous for both the gene loci (Bph17 and Bph32). Although the SNP for Bph9 (snpOS00486) was detected in lines M278, 3K-18, 3K-41, 3K-59, 3K-60, 3K-80, 3K-168, 3K-182, 3K-184, 3K-292, 3K-321, SinnaSivappu, MO1, and Chinsaba (Supplementary Table S8) with favorable allele A:A, its absence in the original donor, Pokkali, determines its ineffectiveness in working as a diagnostic marker as observed in the present study.
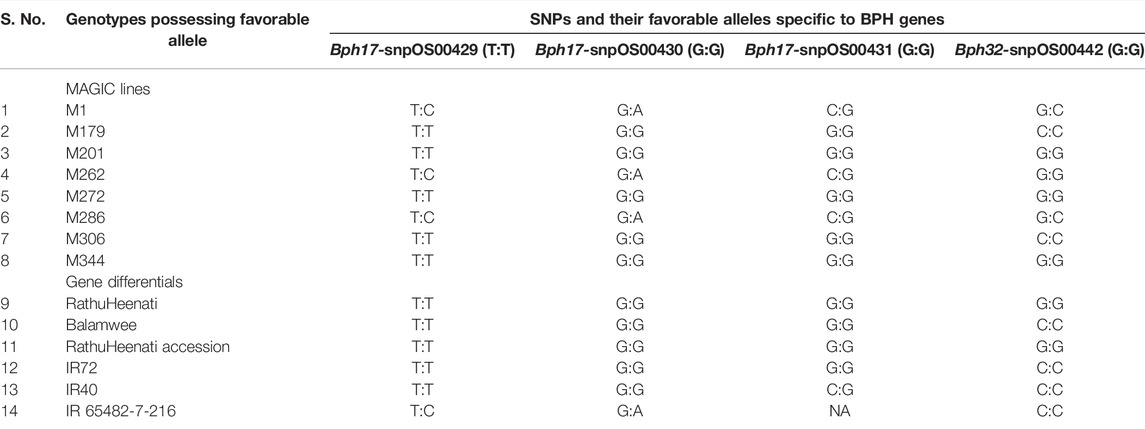
TABLE 8. List of functional SNPs and favorable alleles specific to Bph17 and Bph32 genes validated in the panel of genotypes.
Discussion
The brown planthopper is one of the utmost devastating rice pests, producing significant crop losses. Identification of resistant donors and effective screening approaches for evaluating breeding lines are essential for transferring BPH resistance genes into high-yielding cultivars. In addition, a high amount of genetic diversity lowers the risk of widespread insect epidemics (Newton et al., 2009). The most often used technique at the IRRI and by the National Agricultural Research and Extension Systems (NARES) has been the mass screening of the BPH involving the evaluation of rice genotypes at the seedling stage. Using SSST, the IRRI evaluated many germplasm lines for resistance to three BPH biotypes as mentioned by Jackson (1997). SSST is a rapid and most widely used mass screening method for evaluating seedling susceptibility to the BPH (Fujita et al., 2013; Horgan et al., 2015). In the present study, ANOVA disclosed a significant mean sum of squares for BPH damage scores for different sources of variation. The block effect (unadjusted) and the treatment effects (adjusted and unadjusted) were significant for indicating the presence of a considerable amount of genetic variability in the material studied. Similarly, the effects due to checks, varieties, and checks versus varieties were significant, suggesting the test entries to be significantly different from the checks. The adjusted block effects were non-significant for BPH damage scores indicating homogeneity of evaluation blocks.
The phenotypic responses of the 83 genotypes with checks indicated varying levels of resistance reaction to BPH screening. A comparison of the damage scores of both seasons revealed 13 genotypes as resistant to the BPH with a low damage score extending from 1.3 to 3.0. The resistant lines that comprised gene differentials, RathuHeenati, RP 2068-18-3-5, and RP Bio4918-230S were previously reported to be resistant (Sunil et al., 2018; Akanksha et al., 2019) along with nine MAGIC lines (Durga Sai, 2017). Susceptible checks TN1 and BPT5204 exhibited absolute susceptibility (9.0 score), while the resistant checks PTB33, RathuHeenati, and RP 2068-18-3-5 revealed damage scores of ≤2.0. Several researchers reported PTB33 as resistant to BPH (Harini et al., 2013; Ramulamma et al., 2015; Reddy et al., 2016; Thamarai and Soundararajan, 2017; Udayasree et al., 2018), along with RathuHeenati and RP 2068-18-3-5 that were reported to exhibit durable resistance by Sarao and Bentur (2016); Sunil et al. (2018) and Akanksha et al. (2019). Similar studies were also taken up by Gangaram et al. (2019) who evaluated 74 rice genotypes of Sikkim and Tripura against the BPH in glasshouse conditions along with resistant (PTB33) and susceptible (TN1) checks. The polarity in the resistance response of the rice accessions might be due to the variation in the toxin or antibiotic produced by the rice plant (Qiu et al., 2011; Singh et al., 2019), notably alkaloids or other organic compounds that have repellent effects against the BPH (Sodiq, 2009; Qiu et al., 2011), thus exhibiting varying levels of reaction to the BPH infestation. Furthermore, a thorough investigation of their HPR mechanisms is indispensable to uncover knowledge about the types of resistance, such as antibiosis, antixenosis, and tolerance in each of the genotypes. The production of resistant cultivars that can withstand and prevail over BPH damage in the field for longer periods of time with fewer pesticide applications will result from the research-based application of these HPR mechanisms.
The GWAS is an efficient and reliable method for the excavation of genetic loci and candidate genes that are accountable for natural variability in a polygenic trait (Ishimaru et al., 2013). The density of molecular markers, population size, trait of interest, phenotype-based evaluation, and statistical techniques are the factors that influence the power of GWAS to find related loci for a target trait (Zhang et al., 2015). In the present investigation, based on the consistency and power of associations, 23 corresponding significant SNPs found by the GLM statistical approach were considered for further annotations. The 11 SNPs associated with six candidate genes on chromosome 1 were just 6 Mb away from the reported Bph38(t) that is located between 20.7 and 21.2 Mb (Balachiranjeevi et al., 2019), while a single SNP on chromosome 2 was detected at a physical distance of 5.3 Mb away from the reported Bph13(t) gene located at 31 Mb (Liu et al., 2001), Qbph2 at 22 Mb (Sun et al., 2007), and qBph2 at 17 Mb (Huang et al., 2012). Similarly, 11 significant SNPs identified on chromosome 6 were in close proximity to Bph22(t) located at 3.4 Mb (Harini et al., 2010), Bph3 and bph4 (1.4–1.6 Mb; Jairin et al., 2007, 2010), Bph20(t) (9.3 Mb; Rahman et al., 2009), bph29 (5.3Mb; Wang et al., 2015), bph25(t) (1.7 Mb; Myint et al., 2012), and Bph32 (1.2–1.5 Mb; Ren et al., 2016). Co-localization of the SNPs that were identified to be significant in close proximity to the reported resistance genes/QTLs such as Bph22(t) and Bph20(t) provides a clue about the relatedness of the parents of these MAGIC lines with the original BPH gene differentials carrying these genes/QTLs.
Primarily, of the 42 BPH genes reported, 14 genes have been cloned and characterized to expedite the development of broad-spectrum and durable insect-resistant rice varieties (Du et al., 2020). These genes code for proteins that detect insect effectors and activate defense-related pathways. Similarly, several genes identified in the present investigation have functions pertinent to BPH resistance. The 60S ribosomal protein L23A encoded by LOC_Os01g24690 (snpOS00915) was found to have multiple functions and differential regulation during stress conditions as suggested by Fromont-Racine et al. (2003). GDSL-like lipase/acylhydrolase associated with LOC_Os01g22640.1 (snpOS00912) and LOC_Os01g22660.1 (snpOS00913) acts as an elicitor of systemic resistance through ethylene signaling, thus playing a critical part for both local and systemic resistance in plant immunity systems (Kwon et al., 2010). These family genes were found to be significantly increased in pest-infested leaves and were correlated with different kinds of biotic stress responses (Truong et al., 2017). Similarly, LOC_Os01g23770.1 (snpOS00914) encoding the OsMADS93-MADS-box family gene protein was related to abiotic stress tolerance (Castelán-Munoz et al., 2019), while LOC_Os02g10240.1 (snpOS00916) on chromosome 2 was found to encode the ZOS2-05-C2H2 zinc finger protein that functions as a transcriptional activator in regulating abiotic stress signaling pathways as mentioned by Han et al. (2020). LOC_Os05g39590 and LOC_Os05g39720 (snpOS00917, snpOS00918, and snpOS00919) on chromosome 5 associated with AP2 domain-containing protein and WRKY70 transcription factor were found to be engaged in a stress-tolerant system with the control over ABA-dependent/independent stress-responsive pathways (Eulgem and Somssich, 2007; Khong et al., 2008) that could play a major role in BPH resistance. A similar class of transcription factors (WRKY46 and WRKY72) responsible for BPH resistance were reported by Hu et al. (2017) for the Bph14 gene, which, by over-expressing RLCK281 and callose synthase genes coupled with trypsin secretion, caused the inhibition of phloem sucking, thereby providing resistance to the BPH.
In the same way, LOC_Os06g12360.1 (snpOS00922) and LOC_Os01g15840.1 (snpOS00925) were associated with pentatricopeptide repeat proteins that have implications as mitochondrion-localized proteins in the defense against biotic pathogens (Laluk et al., 2011). LOC_Os06g15820.1 (snpOS00927, snpOS00928, and snpOS00929) associated with the NHL repeat-containing protein was found to encode a plasma membrane protein whose over-expression was correlated with increased resistance to biotic stress as reported by Varet et al. (2003), and LOC_Os06g15750 (snpOS00926) with the NB-ARC functional ATPase protein was found possessing regulatory activity of the R protein that triggers the induction of plant defenses to restrict pathogen proliferation achieving resistance as stated by Ooijen et al. (2008). LOC_Os06g13600.1 gene (snpOS00923) related to the HEAT repeat protein has been identified to play a critical part in the immunity of plant systems by facilitating protein–protein interactions (Monaghan and Li, 2010), while LOC_Os06g11010 (snpOS00920) and LOC_Os01g15830.1(snpOS00924) were associated with the eukaryotic aspartyl protease domain-containing protein and peroxidase precursors that might be useful in providing resistance to the BPH. LOC_Os07g18600 (snpOS00931) on chromosome 7 was linked to the OsFBL37-F-box domain and LRR-containing protein that aids in recognizing pathogen-associated molecular patterns or effectors and turning on the host-resistance pathways. A similar protein conferring resistance to the BPH was reported for Bph26, Bph18, and Bph9 genes that acted as a sensor for signal transduction in response to BPH attack and inhibiting the pest from sucking the phloem sap (Tamura et al., 2014; Ji et al., 2016). Annotation works such as these may benefit researchers by intuiting the genic level of rice-BPH interactions along with the production of new insect-resistant rice cultivars, resulting in better long-term control of the BPH.
To validate the suitability of the set of SNPs, these were converted into a KASP assay, which is a uniplex SNP genotyping platform. Previous research has been reported on the generation of KASP markers in a variety of crop species, including rice, wheat, maize, bajra, sorghum, tomato, potato, cotton, peanut, and rubus species (Rasheed et al., 2016; Zhao et al., 2017; Ryu et al., 2018; Steele et al., 2018; Burow et al., 2019; Devran and Kahveci, 2019; Chen et al., 2021; Kante et al., 2021). A total of six of our designed SNPs were able to distinguish the resistant and susceptible lines, which correlated with our previous phenotyping results (Lakshmi et al., 2021). The SNPs were associated with defense-related genes with putative functions viz., GDSL-like lipase/acylhydrolase, 60S ribosomal protein, pentatricopeptide protein, HEAT repeat family protein, and NHL repeat-containing protein. Altogether, these designed SNPs having functional defense mechanisms and moderate efficiency in classifying the BPH-resistant and susceptible genotypes could be used as diagnostic markers in MAB studies after validating them in biparental populations developed for BPH resistance.
The BPH biotype 4 is widely spread across Asian countries for which Bph17 and Bph32 genes were found to be effective (Liu et al., 2015; Ren et al., 2016). Bph17 was found to possess three tandem duplicated genes encoding for plasma membrane-localized lectin receptor kinase. Analysis of the sequence revealed the polymorphism in a single nucleotide of three genes (OsLecRK1, OsLecRK2, and OsLecRK3) to be accountable for controlling resistance in RathuHeenati (Liu et al., 2015). Similarly, Bph32 encodes an unknown protein holding a signal peptide and SCOP d1gkna2 domain (Ren et al., 2016). SNPs associated with these genes contained positive alleles in the original donors PTB33 (Bph32) and RathuHeenati (Bph17 and Bph32), confirming the presence of these loci as reported in previous studies by Liu et al. (2015), Ren et al. (2016), Jena et al. (2017), and Kusumawati et al. (2018). As the combination of multiple BPH-specific genes is an effective strategy to develop cultivars with broad-spectrum resistance, the five digenic lines viz, M201, M272, M344, RathuHeenati, and RathuHeenati accession with Bph17 and Bph32 genes could also be used for introducing BPH resistance into popular varieties through the MAB approach. Also, these lines are highly useful for studying the effects of each gene either singly or in combination with yield and other agronomic traits.
Conclusion
The present study reported the identification of BPH-resistant donors by phenotypic screening followed by validation with BPH-related SNPs that could contribute to the development of new BPH-resistant cultivars. Screening of germplasm lines for BPH resistance identified 13 genotypes as the best with a low damage score comparable to the resistant checks. Molecular and morphological validation of the germplasm lines and checks with the reported SNPs pinned down MAGIC lines, M201, M272, and M344 for having favorable alleles for both Bph17 and Bph32, indicating their use as donors for introducing BPH resistance. Since the gene-specific markers for Bph17 and Bph32 which proved to be effective against Asian biotype 4 were validated in the original donors PTB33 and RathuHeenati, the purity of the lines maintained was confirmed. Accompanying these reported SNPs, validation with our designed SNPs has successfully identified six robust SNP markers that can distinguish the genotypes based on resistance reaction to the BPH and could be used as diagnostic markers for their routine use in rice improvement programs targeting BPH resistance.
Remarks
With respect to the genotypes used in the study, RathuHeenati is a Sri Lankan indica rice cultivar, while RathuHeenati accession is a derived line of the original RathuHeenati cultivar.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
VI conducted the research and drafted the manuscript. SV, MS, CG, and SD conceptualized the research and designed the experiment. SV, VJ, and SS contributed to the experimental material and screening facilities. RB helped in providing HTPG services and checking the manuscript. SR analyzed the data and aided in interpretation. RL and KK helped in collecting information on candidate genes and designing SNPs. All the authors contributed to the manuscript and approved the submitted version.
Funding
The authors acknowledge the genotyping support from the “Shared Industrial-Scale Low-Density SNP Genotyping for CGIAR and Partner Breeding Programs Serving SSA and SA” (High-Throughput Genotyping-HTPG) BMGF-funded project (OPP1130244).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to PJTSAU and ICAR-IIRR, Hyderabad, for providing experimental fields and glasshouse facilities. The authors acknowledge the Excellence in Breeding (EiB) platform through HTPG services for supporting the KASP genotyping at the Intertek-Agritech Laboratory, Hyderabad.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.914131/full#supplementary-material
References
Ahmar, S., Mahmood, T., Fiaz, S., Mora-Poblete, F., Shafique, M. S., Chattha, M. S., et al. (2021). Advantage of Nanotechnology-Based Genome Editing System and its Application in Crop Improvement. Front. Plant Sci. 12, 849. doi:10.3389/fpls.2021.663849
Akanksha, S., JhansiLakshmi, V., Singh, A. K., Deepthi, Y., Chirutkar, P. M., Balakrishnan, D., et al. (2019). Genetics of Novel Brown Planthopper Nilaparvata Lugens (Stål) Resistance Genes in Derived Introgression Lines from the Interspecific Cross O. Sativa Var. Swarna × O. Nivara. J. Genet. 98 (5), 1–10. doi:10.1007/s12041-019-1158-2
Arbelaez, J. D., Tandayu, E., Reveche, M. Y., Jarana, A., van Rogen, P., Sandager, L., et al. (2019). Methodology: Ssb-MASS: a Single Seed-Based Sampling Strategy for Marker-Assisted Selection in Rice. Plant Methods 15 (1), 78–11. doi:10.1186/s13007-019-0464-2
Backus, E. A., Serrano, M. S., and Ranger, C. M. (2005). MECHANISMS of HOPPERBURN: An Overview of Insect Taxonomy, Behavior, and Physiology. Annu. Rev. Entomol. 50 (1), 125–151. doi:10.1146/annurev.ento.49.061802.123310
Balachiranjeevi, C. H., Prahalada, G. D., Mahender, A., Jamaloddin, M., Sevilla, M. A. L., Marfori-Nazarea, C. M., et al. (2019). Identification of a Novel Locus, Bph38(t), Conferring Resistance to Brown Planthopper (Nilaparvata Lugens Stal.) Using Early Backcross Population in Rice (Oryza Sativa L.). Euphytica 215 (11), 185–214. doi:10.1007/s10681-019-2506-2
Bandillo, N., Raghavan, C., Muyco, P. A., Sevilla, M. A., Lobina, I. T., Dilla-Ermita., C. J., et al. (2013). Multi-parent Advanced Generation Inter-cross (MAGIC) Populations in Rice: Progress and Potential for Genetics Research and Breeding. Rice (N Y) 6 (1), 11–15. doi:10.1186/1939-8433-6-11
Biosearch technologies (2021b). Genomic Analysis by LGC. Available at: https://www.biosearchtech.com/.
Biosearch technologies (2021a). SNP Viewer. Available at: https://www.biosearchtech.com/support/tools/genotyping-software/snpviewer.
Bohar, R., Chitkineni, A., and Varshney, R. K. (2020). Genetic Molecular Markers to Accelerate Genetic Gains in Crops. Biotechniques 69 (3), 158–160. doi:10.2144/btn-2020-0066
Bohra, A., Chand Jha, U., Godwin, I. D., and Kumar Varshney, R. (2020). Chand Jha, U., Godwin, IGenomic Interventions for Sustainable Agriculture. Plant Biotechnol. J. 18 (12), 2388–2405. doi:10.1111/pbi.13472
Bosque-Pérez, N. A., and Buddenhagen, I. W. (1992). “The Development of Host-Plant Resistance to Insect Pests: Outlook for the Tropics,” in Proceedings of 8th International Symposium: Insect–Plant Relationships. Editors S. B. J. Menken, J. H. Visser, and P. Harrewijn (Dordrecht: Kluwer), 235–249. doi:10.1007/978-94-011-1654-1_74
Bossa-Castro, A. M., Tekete, C., Raghavan, C., Delorean, E. E., Dereeper, A., Dagno, K., et al. (2018). Allelic Variation for Broad-Spectrum Resistance and Susceptibility to Bacterial Pathogens Identified in a Rice MAGIC Population. Plant Biotechnol. J. 16 (9), 1559–1568. doi:10.1111/pbi.12895
Brar, D. S., Virk, P. S., Jena, K. K., and Khush, G. S. (2009). Plant Hoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia.” in Breeding for Resistance to Planthoppers in Rice Los Banos: International Rice Research Institute, 401
Burow, G., Chopra, R., Hughes, H., Xin, Z., and Burke, J. (2019). “Marker Assisted Selection in Sorghum Using KASP Assay for the Detection of Single Nucleotide Polymorphism/insertion Deletion,” in Sorghum (New York, NY: Humana Press), 75–84. doi:10.1007/978-1-4939-9039-9_6
Cagampang, G. B., Pathak, M. D., and Juliano, B. O. (1974). Metabolic Changes in the Rice Plant during Infestation by the Brown Planthopper, Nilaparvata Lugens STÅL (Hemiptera : Delphacidae). Appl. Entomol. Zool. 9, 174–184. doi:10.1303/aez.9.174
Castelán-Muno, N., Joel, H., Cajero-Sánchez., W, Maite, A., Carlos, T., García-Ponce, B., et al. (2019). MADS-box Genes Are Key Components of Genetic Regulatory Networks Involved in Abiotic Stress and Plastic Developmental Responses in Plants. Front. Plant Sci. 10, 853, 1–18. doi:10.3389/fpls.2019.00853
CGIAR (2021). Low-density Genotyping Services. Available at: https://excellenceinbreeding.org/toolbox/services/low-density-genotyping-services-introduction.
Chen, Z., Tang, D., Ni, J., Li, P., Wang, L., Zhou, J., et al. (2021). Development of Genic KASP SNP Markers from RNA-Seq Data for Map-Based Cloning and Marker-Assisted Selection in Maize. BMC Plant Biol. 21 (1), 1–11. doi:10.1186/s12870-021-02932-8
De, L. M., Martinant, J. P., Duborjal, H., Laffaire, J. B., and Beugnot, R. (2009). “High Throughput SNP Discovery in Wheat Using Methylation-Sensitive Digestion and Next-Generation Sequencing,” in 19th International Triticeae Mapping Initiative Meeting (France: Clermont-Ferrand).
Devran, Z., and Kahveci, E. (2019). Development and Validation of a User-Friendly KASP Marker for the Sw-5 Locus in Tomato. Australas. Plant Pathol. 48 (5), 503–507. doi:10.1007/s13313-019-00651-1
Du, B., Chen, R., Guo, J., and He, G. (2020). Current Understanding of the Genomic, Genetic, and Molecular Control of Insect Resistance in Rice. Mol. Breed. 40 (2), 1–25. doi:10.1007/s11032-020-1103-3
Durga Sai, J. (2017). Marker Trait Association Studies for Blast and BPH Tolerance in MAGIC Indica Plus Population Rice (Oryza Sativa L.). M.Sc. Thesis. Hyderabad, India: Professor Jayashankar Telangana State Agricultural University.
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A Robust, Simple Genotyping-By-Sequencing (GBS) Approach for High Diversity Species. PloS one 6 (5), e19379. doi:10.1371/journal.pone.0019379
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY Transcription Factors in Defense Signaling. Curr. Opin. Plant Biol., 10, 366–371. doi:10.1016/j.pbi.2007.04.020
Excellence in breeding (2021). Consultative Group on International Agricultural Research (CGIAR). Available at: https://excellenceinbreeding.org/sites/default/files/u5/List%20Trait-based%20SNPs%20GSL-HTPG.pdf.
Fromont-Racine, M., Senger, B., Saveanu, C., and Fasiolo, F. (2003). Ribosome Assembly in Eukaryotes. Gene 313, 17–42. doi:10.1016/s0378-1119(03)00629-2
Fujita, D., Kohli, A., and Horgan, F. G. (2013). Rice Resistance to Planthoppers and Leafhoppers. Crit. Rev. Plant Sci. 32 (3), 162–191. doi:10.1080/07352689.2012.735986
Fukuoka, S., Ebana, K., Yamamoto, T., and Yano, M. (2010). Integration of Genomics into Rice Breeding. Rice 3 (2), 131–137. doi:10.1007/s12284-010-9044-9
Gangaram, B. N., Gowda, B., Shaw, S. S., Behera, S. K., Pandi, G. G. P., Pati, P., et al. (2019). Evaluation of Rice Genotypes of Sikkim and Tripura for Resistance to Brown Planthopper, Nilaparvata Lugens (Stal). Int. J. Curr. Microbiol. Appl. Sci. 8 (8), 2185–2200. doi:10.20546/ijcmas.2019.808.254
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., et al. (2014). TASSEL-GBS: a High Capacity Genotyping by Sequencing Analysis Pipeline. PloS one 9 (2), e90346. doi:10.1371/journal.pone.0090346
Gouda, A. C., Warburton, M. L., Djedatin, G. L., Kpeki, S. B., Wambugu, P. W., Gnikoua, K., et al. (2021). Development and Validation of Diagnostic SNP Markers for Quality Control Genotyping in a Collection of Four Rice (Oryza) Species. Sci. Rep. 11 (1), 18617–18711. doi:10.1038/s41598-021-97689-3
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 11, 115. doi:10.3389/fpls.2020.00115
Harini, A. S. A. I., Lakshmi, S. S., Kumar, S. S. A. I., Sivaramakrishnan, S., and Foundation, B. (2010). Validation and Fine-Mapping of Genetic Locus Associated with Resistance to Brown Plant Hopper [Nilaparvata Lugens (Stal.)] in Rice (Oryza Sativa L.). Asian J. Bio Sci. 5 (1), 32
Harini, A. S., Kumar, S. S., Balaravi, P., Sharma, R., Dass, M. A., and Shenoy, V. (2013). Evaluation of Rice Genotypes for Brown Planthopper (BPH) Resistance Using Molecular Markers and Phenotypic Methods. Afr. J. Biotechnol. 12 (19), 2515–2525. doi:10.5897/AJB2013.11980
He, C., Holme, J., and Anthony, J. (2014). “SNP Genotyping: the KASP Assay,” in Crop Breeding (New York, NY: Humana Press), 75–86. doi:10.1007/978-1-4939-0446-4_7
Heinrichs, E. A., Medrano, E. G., and Rapusas, H. R. (1985). Genetic Evaluation for Insect Resistance in Rice. Int. Rice Res. Inst. IRRI, Los Baños, 356.
Horgan, F. G., Ramal, A. F., Bentur, J. S., Kumar, R., Bhanu, K. V., Sarao, P. S., et al. (2015). Virulence of Brown Planthopper (Nilaparvata Lugens) Populations from South and South East Asia against Resistant Rice Varieties. Crop Prot. 78, 222–231. doi:10.1016/j.cropro.2015.09.014
Hu, L., Wu, Y., Wu, D., Rao, W., Guo, J., Ma, Y., et al. (2017). The Coiled-Coil and Nucleotide Binding Domains of BROWN PLANTHOPPER RESISTANCE14 Function in Signaling and Resistance against Planthopper in Rice. Plant Cell. 29 (12), 3157–3185. doi:10.1105/tpc.17.00263
Huang, D.-R., Chen, J., Lai, F.-X., Liu, G.-J., and Zhuang, J.-Y. (2012). Analysis of Quantitative Trait Loci for Resistance to Brown Planthopper in Dongxiang Wild Rice (Oryza Rufipogon Griff.). Acta Agron. Sin. 38 (2), 210–214. doi:10.1016/s1875-2780(11)60101-6
Huang, M., Liu, X., Zhou, Y., Summers, R. M., and Zhang, Z. (2019). BLINK: a Package for the Next Level of Genome-wide Association Studies with Both Individuals and Markers in the Millions. GigaScience 8 (2), 154. doi:10.1093/gigascience/giy154
Intertek AgriTech (2021). Intertek India Pvt. Ltd., Hyderabad. Available at: http://www.intertek.com/agriculture/agritech/.
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of Function of the IAA-Glucose Hydrolase Gene TGW6 Enhances Rice Grain Weight and Increases Yield. Nat. Genet. 45 (6), 707–711. doi:10.1038/ng.2612
Jackson, M. T. (1997). Conservation of Rice Genetic Resources: the Role of the International Rice Genebank at IRRI. Plant Mol. Biol. 35, 61–67. doi:10.1007/978-94-011-5794-0_6
Jairin, J., Phengrat, K., Teangdeerith, S., Vanavichit, A., and Toojinda, T. (2007). Mapping of a Broad-Spectrum Brown Planthopper Resistance Gene, Bph3, on Rice Chromosome 6. Mol. Breed. 19 (1), 35
Jairin, J., Sansen, K., Wongboon, W., and Kothcharerk, J. (2010). Detection of a Brown Planthopper Resistance Gene Bph4 at the Same Chromosomal Position of Bph3 Using Two Different Genetic Backgrounds of Rice. Breed. Sci. 60, 71–75. doi:10.1270/jsbbs.60.71
Jena, K. K., Hechanova, S. L., Verdeprado, H., Prahalada, G. D., and Kim, S.-R. (2017). Development of 25 Near-Isogenic Lines (NILs) with Ten BPH Resistance Genes in Rice (Oryza Sativa L.): Production, Resistance Spectrum, and Molecular Analysis. Theor. Appl. Genet. 130 (11), 2345–2360. doi:10.1007/s00122-017-2963-8
Jena, K. K., and Kim, S. M. (2010). Current Status of Brown Planthopper (BPH) Resistance and Genetics. Rice 3 (2-3), 161–171. doi:10.1007/s12284-010-9050-y
JhansiLakshmi, V., Krishnaiah, N. V., Katti, G. R., Pasalu, I. C., and Vasantha Bhanu, K. (2010b). Development of Insecticide Resistance in Rice Brown Planthopper and White Backed Planthopper in Godavari Delta of Andhra Pradesh. Indian J. Plant Prot. 38 (1), 35
JhansiLakshmi, V., Krishnaiah, N. V., and Katti, G. R. (2010a). Potential Toxicity of Selected Insecticides to Rice Leafhoppers and Planthoppers and Their Important Natural Enemies. J. Biol. Control 24 (3), 244–252. doi:10.18311/jbc/2010/3648
Ji, H., Kim, S. R., Kim, Y. H., Suh, J. P., Park, H. M., Sreenivasulu, N., et al. (2016). Map-based Cloning and Characterization of the Bph18 Gene from Wild Rice Conferring Resistance to Brown Planthopper (BPH) Insect Pest. Sci. Rep. 6 (1), 34376–34414. doi:10.1038/srep34376
Kante, M., Lindqvist-Kreuze, H., Portal, L., David, M., and Gastelo, M. (2021). Kompetitive Allele Specific PCR (KASP) Markers for Potato: An Effective Tool for Increased Genetic Gains. Agronomy 11 (11), 2315. doi:10.3390/agronomy11112315
Khong, G. N., Richaud, F., Coudert, Y., Pati, P. K., Santi, C., Périn, C., et al. (2008). Modulating Rice Stress Tolerance by Transcription Factors. Biotechnol. Genet. Eng. Rev. 25 (1), 381–404. doi:10.5661/bger-25-381
Khush, G. S. (2001). Green Revolution: the Way Forward. Nat. Rev. Genet. 2, 815–822. doi:10.1038/35093585
Khush, G. S., Karim, A. N. M. R., and Angeles, E. R. (1985). Genetics of Resistance of Rice Cultivar ARC10550 to Bangladesh Brown Planthopper Biotype. J. Genet. 64 (2), 121–125. doi:10.1007/bf02931140
Kusumawati, L., Chumwong, P., Jamboonsri, W., Wanchana, S., Siangliw, J. L., Siangliw, M., et al. (2018). Candidate Genes and Molecular Markers Associated with Brown Planthopper (Nilaparvata Lugens Stal) Resistance in Rice Cultivar RathuHeenati. Mol. Breed. 38 (7), 1–16. doi:10.1007/s11032-018-0847-5
Kwon, Y. S., Ryu, C.-M., Lee, S., Park, H. B., Han, K. S., Lee, J. H., et al. (2010). Proteome Analysis of Arabidopsis Seedlings Exposed to Bacterial Volatiles. Planta 232 (6), 1355–1370. doi:10.1007/s00425-010-1259-x
Lakshmi, V. G. I., Sreedhar, M., Lakshmi, V. J., Gireesh, C., Rathod, S., and Vanisri, S. (2021). Molecular Diversity Assessment of Rice Genotypes for Brown Planthopper Resistance Using Microsatellite Markers. Electron. J. Plant Breed. 12 (2), 499–506. doi:10.37992/2021.1202.070
Laluk, K., AbuQamar, S., and Mengiste, T. (2011). The Arabidopsis Mitochondria-Localized Pentatricopeptide Repeat Protein PGN Functions in Defense against Necrotrophic Fungi and Abiotic Stress Tolerance. Plant Physiol. 156 (4), 2053–2068. doi:10.1104/pp.111.177501
Leng, P., Ji, Q., Asp, T., Frei, U. K., Ingvardsen, C. R., Xing, Y., et al. (2017). Auxin Binding Protein 1 Reinforces Resistance to Sugarcane Mosaic Virus in Maize. Mol. Plant 10 (10), 1357–1360. doi:10.1016/j.molp.2017.07.013
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/map Format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi:10.1093/bioinformatics/btp352
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: Genome Association and Prediction Integrated Tool. Bioinformatics 28, 2397–2399. doi:10.1093/bioinformatics/bts444
Liu, G., Yan, H., Fu, Q., Qian, Q., Zhang, Z., Zhai, W., et al. (2001). Mapping of a New Gene for Brown Planthopper Resistance in Cultivated Rice Introgressed fromOryza Eichingeri. Chin. Sci. Bull. 46 (17), 1459–1462. doi:10.1007/bf03187031
Liu, Y., Wu, H., Chen, H., Liu, Y., He, J., Kang, H., et al. (2015). A Gene Cluster Encoding Lectin Receptor Kinases Confers Broad-Spectrum and Durable Insect Resistance in Rice. Nat. Biotechnol. 33 (3), 301–305. doi:10.1038/nbt.3069
Monaghan, J., and Li, X. (2010). The HEAT Repeat Protein ILITYHIA Is Required for Plant Immunity. Plant Cell. physiology 51 (5), 742–753. doi:10.1093/pcp/pcq038
Muduli, L., Pradhan, S. K., Mishra, A., Bastia, D. N., Samal, K. C., Agrawal, P. K., et al. (2021). Understanding Brown Planthopper Resistance in Rice: Genetics, Biochemical and Molecular Breeding Approaches. Rice Sci. 28 (6), 532–546. doi:10.1016/j.rsci.2021.05.013
Mwamahonje, A., Eleblu, J. S. Y., Ofori, K., Feyissa, T., Deshpande, S., Garcia-Oliveira, A. L., et al. (2021). Introgression of QTLs for Drought Tolerance into Farmers' Preferred Sorghum Varieties. Agriculture 11 (9), 883. doi:10.3390/agriculture11090883
Myint, K. K. M., Fujita, D., Matsumura, M., Sonoda, T., Yoshimura, A., and Yasui, H. (2012). Mapping and Pyramiding of Two Major Genes for Resistance to the Brown Planthopper (Nilaparvata Lugens [Stål]) in the Rice Cultivar ADR52. Theor. Appl. Genet. 124 (3), 495–504. doi:10.1007/s00122-011-1723-4
Naik, S. B., Divya, D., Sahu, N., Sundaram, R. M., Sarao, P. S., Singh, K., et al. (2018). A New Gene Bph33 (T) Conferring Resistance to Brown Planthopper (BPH), Nilaparvata Lugens (Stål) in Rice Line RP2068-18-3-5. Euphytica 214 (3), 1–12. doi:10.1007/s10681-018-2131-5
Newton, A. C., Begg, G. S., and Swanston, J. S. (2009). Deployment of Diversity for Enhanced Crop Function. Ann. Appl. Biol. 154, 309–322. doi:10.1111/j.1744-7348.2008.00303.x
Parmar, S., Deshmukh, D. B., Kumar, R., Manohar, S. S., Joshi, P., Sharma, V., et al. (2021). Single Seed-Based High-Throughput Genotyping and Rapid Generation Advancement for Accelerated Groundnut Genetics and Breeding Research. Agronomy 11 (6), 1226. doi:10.3390/agronomy11061226
Peng, H., Wang, K., Chen, Z., Cao, Y., Gao, Q., Li, Y., et al. (2020). MBKbase for Rice: an Integrated Omics Knowledgebase for Molecular Breeding in Rice. Nucleic Acids Res. 48, D1085. doi:10.1093/nar/gkz921
Prahalada, G. D., Shivakumar, N., Lohithaswa, H. C., Sidde Gowda, D. K., Ramkumar, G., Kim, S.-R., et al. (2017). Identification and Fine Mapping of a New Gene, BPH31 Conferring Resistance to Brown Planthopper Biotype 4 of India to Improve Rice, Oryza Sativa L. Rice 10 (1), 41. doi:10.1186/s12284-017-0178-x
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., and Reich, D. (2006). Principal Components Analysis Corrects for Stratification in Genome-wide Association Studies. Nat. Genet. 38, 904–909. doi:10.1038/ng1847
Qiu, Y., Guo, J., Jing, S., Tang, M., Zhu, L., and He, G. (2011). Identification of Antibiosis and Tolerance in Rice Varieties Carrying Brown Planthopper Resistance Genes. Entomologia Exp. Appl. 141 (3), 224–231. doi:10.1111/j.1570-7458.2011.01192.x
Raghavan, C., Mauleon, R., Lacorte, V., Jubay, M., Zaw, H., Bonifacio, J., et al. (2017). Approaches in Characterizing Genetic Structure and Mapping in a Rice Multiparental Population. G3 Genes, Genomes, Genet. 7 (6), 1721–1730. doi:10.1534/g3.117.042101
Rahman, M. L., Jiang, W., Chu, S. H., Qiao, Y., Ham, T.-H., Woo, M.-O., et al. (2009). High-resolution Mapping of Two Rice Brown Planthopper Resistance Genes, Bph20(t) and Bph21(t), Originating from Oryza Minuta. Theor. Appl. Genet. 119 (7), 1237–1246. doi:10.1007/s00122-009-1125-z
Ramulamma, A., Sridevi, D., JhansiLakshmi, V., Nagendra Reddy, B., and Bhat., B (2015). Evaluation of Different Rice Germplasm Accessions for Resistance to Brown Planthopper, Nilaparvata Lugens (Stal). Progressive J. Res. 10 (3), 223
Rasheed, A., Wen, W., Gao, F., Zhai, S., Jin, H., Liu, J., et al. (2016). Development and Validation of KASP Assays for Genes Underpinning Key Economic Traits in Bread Wheat. Theor. Appl. Genet. 129 (10), 1843–1860. doi:10.1007/s00122-016-2743-x
Rashid, M. M., Jahan, M., Islam, K. S., and Latif, M. (2017). Effects of Nitrogen, Phosphorous and Potassium on Host-Choice Behavior of Brown Planthopper, Nilaparvata Lugens (Stål) on Rice Cultivar. J. Insect Behav. 30 (1), 1–15. doi:10.1007/s10905-016-9594-9
Reddy, N. B., JhansiLakshmi, V., Uma Maheswari, T., Ramulamma, A., and Katti, G. R. (2016). Identification of Resistant Sources to Brown Planthopper, Nilaparvata Lugens (Stal) (Delphacidae: Hemiptera). Progressive Res. – Int. J. 11 (1), 5
Ren, J., Gao, F., Wu, X., Lu, X., Zeng, L., Lv, J., et al. (2016). Bph32, a Novel Gene Encoding an Unknown SCR Domain-Containing Protein, Confers Resistance against the Brown Planthopper in Rice. Sci. Rep. 6 (1), 37645–37714. doi:10.1038/srep37645
Ryu, J., Kim, W. J., Im, J., Kim, S. H., Lee, K.-S., Jo, H.-J., et al. (2018). Genotyping-by-sequencing Based Single Nucleotide Polymorphisms Enabled Kompetitive Allele Specific PCR Marker Development in Mutant Rubus Genotypes. Electron. J. Biotechnol. 35, 57–62. doi:10.1016/j.ejbt.2018.08.001
Sarao, P. S., and Bentur, J. S. (2016). Antixenosis and Tolerance of Rice Genotypes against Brown Planthopper. Rice Sci. 23 (2), 96–103. doi:10.1016/j.rsci.2016.02.004
Sarao, P. S., Sahi, G. K., Neelam, K., Mangat, G. S., Patra, B. C., and Singh, K. (2016). Donors for Resistance to Brown Planthopper Nilaparvata Lugens (Stål) from Wild Rice Species. Rice Sci. 23 (4), 219–224. doi:10.1016/j.rsci.2016.06.005
Satturu, V., Vattikuti, J. L., J, D. S., Kumar, A., Singh, R. K., M, S. P., et al. (2020). Multiple Genome Wide Association Mapping Models Identify Quantitative Trait Nucleotides for Brown Planthopper (Nilaparvata Lugens) Resistance in MAGIC Indica Population of Rice. Vaccines 8 (4), 608. doi:10.3390/vaccines8040608
Segura, V., Vilhjálmsson, B. J., Platt, A., Korte, A., Seren, Ü., Long, Q., et al. (2012). An Efficient Multi-Locus Mixed-Model Approach for Genome-wide Association Studies in Structured Populations. Nat. Genet. 44 (7), 825–830. doi:10.1038/ng.2314
Singh, A., Jaswal, A., and Sarkar, S. (2019). Incidence of Brown Plant Hopper in the Rice Field with the Use of Different Doses of Fertilizers. Agric. Sci. Dig. 39 (2), 108–113. doi:10.18805/ag.d-4775
SnpEff and SnpSift (2021). Genomic Variant Annotations and Functional Effect Prediction Toolbox. Available at: http://snpeff.sourceforge.net/.
SNPs and Phenotypes Data (2021). Data Repository for the 3K RGP and OryzaSNP Project, Hosted by IRRI. Available at: https://snp-seek.irri.org/download.zul.
Sodiq, M. (2009). Resistance of Plant to Pests. [Thesis]. Surabaya, Jawa Timur [Indonesia]: Universitas Pembangunan Nasional.
Sood, S., Bhardwaj, V., Chourasia, K. N., Kaur, R. P., Kumar, V., Kumar, R., et al. (2022). KASP Markers Validation for Late Blight, PCN and PVY Resistance in a Large Germplasm Collection of Tetraploid Potato (Solanum tuberosum L.). Sci. Hortic. 295, 110859. doi:10.1016/j.scienta.2021.110859
Standard Evaluation System (SES) for Rice (2013). Manila, Philippines: International Rice Research Institute, Vol. 5, 38–39.
Steele, K. A., Quinton-Tulloch, M. J., Amgai, R. B., Dhakal, R., Khatiwada, S. P., Vyas, D., et al. (2018). Accelerating Public Sector Rice Breeding with High-Density KASP Markers Derived from Whole Genome Sequencing of Indica Rice. Mol. Breed. 38 (4), 38–13. doi:10.1007/s11032-018-0777-2
Sun, L., Liu, Y., Jiang, L., Su, C., Wang, C., Zhai, H., et al. (2007). Identification of Quantitative Trait Loci Associated with Resistance to Brown Planthopper in the Indica Rice Cultivar Col.5 Thailand. Hereditas 144 (2), 48–52. doi:10.1111/j.2006.0018-0661.01932.x
Sunil, V., Lakshmi, V. J., Chiranjeevi, K., Sampathkumar, M., Bentur, J. S., and Katti, G. R. (2018). Rice Genotypes Resistant to Brown Planthopper, Nilaparvata Lugens (Stål) Population of West Godavari, Andhra Pradesh. Ann. Plan. Prote. Scien. 26 (2), 249–255. doi:10.5958/0974-0163.2018.00057.5
Tamura, Y., Hattori, M., Yoshioka, H., Yoshioka, M., Takahashi, A., Wu, J., et al. (2014). Map-based Cloning and Characterization of a Brown Planthopper Resistance Gene Bph26 from Oryza Sativa L. Ssp. Indica Cultivar ADR52. Sci. Rep. 4, 5872. doi:10.1038/srep05872
Tan, H. Q., Sreekanth, P., Gouda, J., Kumar, P., and Kumar, S. C. (2021). Identification of Two QTLs, BPH41 and BPH42, and Their Respective Gene Candidates for Brown Planthopper Resistance in Rice by a Combinatorial Approach of QTL Mapping and RNA-Seq. (pre-print). doi:10.21203/rs.3.rs-979035/v1
Thamarai, M., and Soundararajan, R. P. (2017). Reaction of Rice Genotypes against Specific Population of Brown Planthopper, Nilaparvata Lugens (Stal.). Ann. Plant Prot. Sci. 25 (1), 74
Thuy, C. T. L., Lopez-Lavalle, L. A. B., Vu, N. A., Hy, N. H., Nhan, P. T., Ceballos, H., et al. (2021). Identifying New Resistance to Cassava Mosaic Disease and Validating Markers for the CMD2 Locus. Agriculture 11 (9), 829. doi:10.3390/agriculture11090829
Truong, H. A., Jeong, C. Y., Lee, W. J., Lee, B. C., Chung, N., Kang, C.-S., et al. (2017). Evaluation of a Rapid Method for Screening Heat Stress Tolerance Using Three Korean Wheat (Triticum aestivum L.) Cultivars. J. Agric. Food Chem. 65 (28), 5589–5597. doi:10.1021/acs.jafc.7b01752
Udayasree, M., Rajanikanth, P., Varma, N. R. G., and Sreedhar, M. (2018). Screening of Selected Rice Genotypes for Their Resistance against Brown Planthopper, Nilaparvata Lugens (Stal). Int. J. Curr. Microbiol. App. Sci. 7 (11), 3138–3143. doi:10.20546/ijcmas.2018.711.360
van Ooijen, G., Mayr, G., Kasiem, M. M. A., Albrecht, M., Cornelissen, B. J. C., and Takken, F. L. W. (2008). Structure-function Analysis of the NB-ARC Domain of Plant Disease Resistance Proteins. J. Exp. Bot. 59 (6), 1383–1397. doi:10.1093/jxb/ern045
Varet, A., Hause, B., Hause, G., Scheel, D., and Lee, J. (2003). The Arabidopsis NHL3 Gene Encodes a Plasma Membrane Protein and its Overexpression Correlates with Increased Resistance to Pseudomonas syringae Pv. Tomato DC3000. Plant physiol. 132 (4), 2023–2033. doi:10.1104/pp.103.020438
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G., et al. (2013). Draft Genome Sequence of Chickpea (Cicer Arietinum) Provides a Resource for Trait Improvement. Nat. Biotechnol. 31 (3), 240–246. doi:10.1038/nbt.2491
Wang, Q., Tian, F., Pan, Y., Buckler, E. S., and Zhang, Z. (2014). A Super Powerful Method for Genome Wide Association Study. PLoS ONE. 107684. do.doi:10.1371/journal.pone.0107684
Wang, Y., Cao, L., Zhang, Y., Cao, C., Liu, F., Huang, F., et al. (2015). Map-based Cloning and Characterization ofBPH29, a B3 Domain-Containing Recessive Gene Conferring Brown Planthopper Resistance in Rice. Exbotj 66 (19), 6035–6045. doi:10.1093/jxb/erv318
Xu, Y., Skinner, D. J., Wu, H., Palacios-Rojas, N., Araus, J. L., Yan, J., et al. (2009). Advances in Maize Genomics and Their Value for Enhancing Genetic Gains from Breeding. Int. J. Plant Genomics 2009, 957602. doi:10.1155/2009/957602
Yang, G., Chen, S., Chen, L., Sun, K., Huang, C., Zhou, D., et al. (2019). Development of a Core SNP Arrays Based on the KASP Method for Molecular Breeding of Rice. Rice 12, 21–18. doi:10.1186/s12284-019-0272-3
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of Allotetraploid Cotton (Gossypium Hirsutum L. Acc. TM-1) Provides a Resource for Fiber Improvement. Nat. Biotechnol. 33, 531–537. doi:10.1038/nbt.3207
Zhang, Y.-M., Mao, Y., Xie, C., Smith, H., Luo, L., and Xu, S. (2005). Mapping Quantitative Trait Loci Using Naturally Occurring Genetic Variance Among Commercial Inbred Lines of Maize (Zea mays L.). Genetics 169, 2267–2275. doi:10.1534/genetics.104.033217
Zhang, Z., Ersoz, E., Lai, C.-Q., Todhunter, R. J., Tiwari, H. K., Gore, M. A., et al. (2010). Mixed Linear Model Approach Adapted for Genome-wide Association Studies. Nat. Genet. 42, 355–360. doi:10.1038/ng.546
Keywords: rice, brown planthopper, SNP, GWAS, GBS, KASP
Citation: Ishwarya Lakshmi VG, Sreedhar M, JhansiLakshmi V, Gireesh C, Rathod S, Bohar R, Deshpande S, Laavanya R, Kiranmayee KNSU, Siddi S and Vanisri S (2022) Development and Validation of Diagnostic KASP Markers for Brown Planthopper Resistance in Rice. Front. Genet. 13:914131. doi: 10.3389/fgene.2022.914131
Received: 06 April 2022; Accepted: 06 June 2022;
Published: 08 July 2022.
Edited by:
Vikas Kumar Singh, International Rice Research Institute (IRRI), PhilippinesReviewed by:
Sachin Kumar, Chaudhary Charan Singh University, IndiaManivannan Narayana, Tamil Nadu Agricultural University, India
Copyright © 2022 Ishwarya Lakshmi, Sreedhar, JhansiLakshmi, Gireesh, Rathod, Bohar, Deshpande, Laavanya, Kiranmayee, Siddi and Vanisri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. Vanisri, dmFuaXNyZWVkaGFyMTk5NEBnbWFpbC5jb20=
 V. G. Ishwarya Lakshmi
V. G. Ishwarya Lakshmi M. Sreedhar
M. Sreedhar V. JhansiLakshmi3
V. JhansiLakshmi3 C. Gireesh
C. Gireesh Santosha Rathod
Santosha Rathod Rajaguru Bohar
Rajaguru Bohar Santosh Deshpande
Santosh Deshpande R. Laavanya
R. Laavanya K. N. S. Usha Kiranmayee
K. N. S. Usha Kiranmayee S. Vanisri
S. Vanisri