
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 04 July 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.912251
This article is part of the Research Topic Genetics and Epigenetics: Plausible Role in Development of Climate Resilient Crops View all 17 articles
 Amna Abdul Rahim1,2
Amna Abdul Rahim1,2 Muhammad Uzair2
Muhammad Uzair2 Nazia Rehman2
Nazia Rehman2 Obaid Ur Rehman2
Obaid Ur Rehman2 Nageen Zahra2
Nageen Zahra2 Muhammad Ramzan Khan1,2*
Muhammad Ramzan Khan1,2*Receptor-like protein kinase 1 (RPK1) genes play crucial roles in plant growth and development processes, root architecture, and abiotic stress regulation. A comprehensive study of the RPK1 gene family has not been reported in bread wheat (Triticum aestivum). Here, we reported the genome-wide identification, characterization, and expression patterns of the RPK1 gene family in wheat. Results confirmed 15 TaRPK1 genes, classified mainly into three sub-clades based on a phylogenetic tree. The TaRPK1 genes were mapped on chromosomes 1–3 in the respective A, B, and D genomes. Gene structure, motif conservation, collinearity prediction, and synteny analysis were carried out systematically. A Gene ontology study revealed that TaRPK1 genes play a vital role during molecular and biological processes. We also identified 18 putative miRNAs targeting TaRPK1 genes, suggesting their roles in growth, development, and stress responses. Cis-Regulatory elements interpreted the presence of light-related elements, hormone responsiveness, and abiotic stress-related motifs in the promoter regions. The SWISS_MODEL predicted the successful models of TaRPK1 proteins with at least 30% identity to the template, a widely accepted threshold for successful modeling. In silico expression analysis in different tissues and stages suggested that TaRPK1 genes exhibited the highest expression in root tissues. Moreover, qRT-PCR further validated the higher expression of TaRPK1 genes in roots of drought-tolerant varieties compared to the drought-susceptible variety. Collectively, the present study renders valuable information on the functioning of TaRPK1 genes in wheat that will be useful in further functional validation of these genes in future studies.
Globally, wheat is a staple food and a source of nutrition. In the last 2 decades, the production of wheat increased by up to 1% annually (Manès et al., 2012), but this increase is not enough to meet the demand of the population, which will increased from 7.8 billion to 9.7 billion in 2050 (Roser and Ortiz-Ospina, 2013). The climatic changes including abiotic and biotic stresses are the main causes which extremely effect the quality and yield of the crops. To face these challenges, it is vital to explore the crop genotypes that can stand up to all of these hurdles. Plants are immobile in nature; they don’t move here and there in search of food, but their roots do. The root is the major organ that has a crucial role in the adaptation of the plant to its unfavorable environment. Root systems captivate the water and nutrients essential for the growth and maintenance of plant (Alahmad et al., 2019; Grzesiak et al., 2019). Hence, improved root system overcomes the challenges of the harsh environment and might enhance crop production (Djanaguiraman et al., 2019; Danakumara et al., 2021; Rasool et al., 2021).
Drought is one of the major abiotic stresses caused due to scarce rainfall that affects productivity. An increase in drought in the coming 30 years will have adverse effects on crop yield with 6–12 bushels/acre (Zargar et al., 2017). Creation of drought tolerance is a very complicated because many genes such as TaER1, 2, and 3, TaZFP34, TaWRKY1, 10, 33, 44, and 93, TaDR O 1, and TaRAP2.1 directly or indirectly involved. In animals, receptor protein kinases (RPKs) are the genes which play a significant role in the stimulation of hormones and other growth factors (Fantl et al., 1993). In plants, similar to animals’ RPKs, there is a receptor-like protein kinase (RLK) gene family. The RLK family is a huge family of genes found in many plants. The typical RLK structure comprises an extracellular domain at the N-terminal, a membrane helix, and an intracellular conserved kinase domain (KD) at the C-terminal. The extracellular domains of the RLK family are highly diverged, which results in the differentiation of RLKs into 17 distinct subfamilies, including the receptor-like kinases (Mishra et al., 2021). The LRRKs (leucine-rich repeat kinase) represents biggest subfamily of RLK with 531 TaLRRK genes in wheat (Sharma et al., 2016), comprising of ECD (extracellular domain) to receive signals, TM (transmembrane) region to bound it to cell membrane and cytoplasmic kinase domain for phosphorylation of substrate (Gou et al., 2010; Dievart et al., 2020). The LRRKs has numerous roles in plants as it is involved in initiating innate defense at front-line against microbial pathogens (Nejat and Mantri, 2017), morphogenesis, organogenesis, hormone signaling, abiotic, and biotic stress regulation in plants (Diévart and Clark, 2003; Li and Tax, 2013; Dufayard et al., 2017). Later on the role of LRR-RLKs in pathogen sensing and activation of downstream defense response has been reviewed deeply (Nejat and Mantri, 2017). Due to the indispensable roles of LRR-RLKs in plants, they have been classified into two main classes (Diévart and Clark, 2003). First, the LRR-RLK is crucial for morphogenesis, organogenesis, hormone signaling, signifying development, and growth regulation. Secondly, numerous LRR-RLK members respond to biotic and abiotic stresses like Fusarium wilt, drought, salt, and cold, and hence are associated with defense (Afzal et al., 2008; Cao et al., 2020). Some of the LRR-RLKs have dual roles that might be because of the cross-talk among development and defense cascades or due to the binding of several ligands to a receptor (Afzal et al., 2008).
The RPK1 gene is a calcium independent Serine-Threonine (Ser-Thr) kinase that belongs to the subfamily of leucine-rich receptor kinases (LRR kinases) and family of Receptor-Like Kinases (RLK) (Zou et al., 2014). RPK1 is one of the short subfamilies with few genes that regulates abiotic stresses and root system architecture. The RPK1 comprises of extracellular six LRR motifs, a transmembrane domain, extracellular ligand-binding domain, and single cytoplasmic kinase conserved domain in rice (Hong et al., 1997; Cheng et al., 2009; Motte et al., 2014). Studies in rice have shown that RPK1 is involved in root system architecture (RSA) via regulating negatively polar auxin transport (PAT) and accumulation of auxin in roots (Zou et al., 2014). In other studies of rice, it was also reported that auxin defective mutants showed stunted growth and shorter roots (Uzair et al., 2021). Down-regulation of RPK1 endorsed the growth and enhanced the height of the plant and number of tillers, whereas up-regulation resulted in immature lateral roots, adventitious roots, and a decreased apical meristem of roots (Zou et al., 2014). In Arabidopsis, the inhibition of AtRPK1 displayed greater salt tolerance than normal plants, while overexpressed plants exhibited lesser salt tolerance degrees (Shi et al., 2014). The levels of AtRPK1 were enhanced ominously under less water, abscisic acid (ABA), high salt and lower temperature (Hong et al., 1997). In Arabidopsis thaliana, inhibition of RPK1 delayed ABA-induced senescence significantly (Lee et al., 2011). AtRPK1 is also prerequisite for cotyledon primordial initiation of cotyledons during embryogenesis in Arabidopsis thaliana (Nodine et al., 2007; Nodine and Tax, 2008). AtRPK1 positively regulates CaM1 gene expression, which in turn regulates ROS (reactive oxygen species) production, leaf senescence, and ABA response (Dai et al., 2018).
Triticum aestivum L., commonly known as “bread wheat”, is a cereal and staple food grain all over the globe. Being a most consumed cereal crop, it was grown on a large scale of 240 million hectares in 2016 (Milner et al., 2018). However, due to water scarcity, nutrient deficiency, and abiotic stresses, wheat yield is curtailed (Mondal et al., 2015; Abbas et al., 2022). Wheat is a drought sensitive crop. Therefore, in order to meet the global demand, that is 50% of the grain in 20 years approximately, the varieties of wheat with effective utilization of minerals and water are requisite (Odegard and Van der Voet, 2014). Since roots are the main structures for the minerals and water uptake and decipher stress stimuli from soil (Fang et al., 2017). Hence, identification of stress-tolerant genes within the root system could be propitious.
Since the genome of T. aestivum has been sequenced, it is feasible to carry out a genome-wide analysis of different genes. In this study, 15 TaRPK1 genes were analyzed for their structure, chromosomal location within the genome, phylogenetic relationships, conserved motifs, synteny, and cis-regulatory elements. Additionally, the patterns of expression of all 15 TaRPK1 members were also studied in silico. RT-PCR expression analysis of TaRPK1 members was also performed in Pakistan-13, Galaxy (drought tolerant), and Shafaq (drought susceptible) wheat varieties under normal and drought conditions. The current study enlightens the role of TaRPK1 genes in plant developmental processes under drought conditions and provides a solid foundation for the functional characterization of the wheat RPK1 gene family.
The sequence IDs of Arabidopsis and rice RPK1 genes were acquired from the available literature (Shi et al., 2014; Zou et al., 2014). These sequences were retrieved from Ensembl plants and NCBI, which were then used as queries for the Basic-Local Alignment Search tool (BlastP and BlastN) against IWGSC (INSDC Assembly GCA_900519105.1 July 2018 database version 106.4), NCBI (https://www.ncbi.nlm.nih.gov/) and Ensembl plants (plantshttps://plants.ensembl.org/index.html) for T. aestivum. For all of the candidate RPK1 genes, the kinase domain presence was substantiated with Pfam (http://pfam.sanger.ac.uk), and by SMART (http://smart.embl-heidelberg.de/) (Letunic and Bork, 2018) databases. The sequences in which the kinase domain was absent were removed (Supplementary Table S1). In silico based putative protein information of RPK1 genes (physio-chemical) was analyzed through the Protparam (https://web.expasy.org/protparam/) tool. The subcellular localization of RPK1 proteins was predicted via Plant-mSubP and pLoc-mPlant (http://bioinfo.usu.edu/Plant-mSubP/; http://www.jci-bioinfo.cn/pLoc-mPlant/) (Cheng et al., 2017; Sahu et al., 2020).
The chromosomal locations of all candidate RPK1 genes in T. aestivum were acquired from Ensembl (http://plants.ensembl.org/Triticum_aestivum/Info/Index). The gene map of TaRPK1 genes was drawn with the help of MapChart and confirmed through TBtools.
To retrieve the RPK1 protein sequences, the amino acid sequences of 15 TaRPK1 members were used as queries to blast (BLASTP) against the Triticum turgidium, Triticum dicoccoides, Titicum urartu, Triticum speltoides, Aegilops tauschii, Hordeum vulgare, Arabidopsis thaliana, and different species of Oryza (rufipigon, japonica, indica, and glaberrima). The sequences with > 60% identities were retrieved from Ensembl (http://plants.ensembl.org). The phylogenetic trees were made by means of MEGA-X software with NJ (neighbor-joining method) (Kumar et al., 1994). The parameter Poisson model and pairwise deletion were used with replicates of 1,000 bootstraps for assessment of node significance.
The number of exons and introns was predicted by the gene structure display server (GSDS, http://gsds.cbi.pku.edu.cn/) and the genomic sequences and coding sequences were aligned using ClustalW. Conserved motifs in RPK1 proteins of T. aestivum were analyzed using MEME, a multiple-EM for motif elicitation program (http://meme-suite.org/tools/meme) (Bailey et al., 2009). The execution of MEME search was done with default parameters apart from motif maximum number, which was set to 10, and optimum motif width of ≥6 and ≤200 was selected.
The analysis of TaRPK1 gene ontology was performed by TaRPK1 protein sequences via the online gProfiler tool (https://biit.cs.ut.ee/gprofiler/gost) with default parameters (Raudvere et al., 2019).
The miRNA prediction was performed as mentioned formerly (Yan et al., 2019). The TaRPK1 sequences were submitted for potential miRNA prediction through a search against the available wheat miRNA reference by means of the psRNATarget Server (https://www.zhaolab.org/psRNATarget/), using default settings (Dai and Zhao, 2011). The visualization of the interaction network of the predicted miRNA with their corresponding TaRPK1 target genes was done by Cytoscape software (https://cytoscape.org/) with default settings (Shannon et al., 2003).
The sequence size of 2 kb in the upstream region were dug out from all TaRPK1 genes of T. aestivum that acted as promoters for the regulatory cis acting elements prediction through the PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database (Lescot et al., 2002).
The GFF3 files and proteomes of Triticum aestivum and its ancestors, including Aegilopes tauschii, Triticum spelta, Triticum turgidum, and Triticum dicoccoides, were used from the Ensembl Plants database for collinearity prediction via the MCScanX algorithm (Wang et al., 2012). Synteny scrutiny of RPK1 family members was performed via Tbtools (Chen et al., 2018).
The TaRPK1 protein structures were modeled via amino acid sequence using the SWISS-MODEL database (https://www.swissmodel.expasy.org) (Biasini et al., 2014), and for visualization of 3D structure Pymol software (https://pymol.org/2/) was applied. The verification and validation of the predicted 3D structures of TaRPK1 proteins were assessed using the Ramachandran Plot—Zlab, (https://zlab.umassmed.edu/bu/rama/) (Anderson et al., 2005).
In silico expression analysis was performed using the wheat-expression browser (www.wheat-expression.com) at different wheat stages (Kaur et al., 2017). The data were unruffled in the course of developing seedling, vegetative, and reproductive stages from different organs of wheat such as roots, leaf sheath, leaf blade, shoot, spike, and grain. The heatmap was then created from the composed data, based on the expression values of genes (in TPM) by means of Tbtool.
For interaction network studies, String (https://string-db.org/) was used by selecting Triticum aestivum as a platform species. For visualization of the molecular library, Cytoscape was used. Correlation coefficients on the basis of verities, treatments, and tissues were calculated in R 3.4.0. These coefficients indicate the degree of association among the terms and provide linkages among the TaRPK1 members.
Previously, Pakistan-13, Galaxy, and Shafaq were studied under drought stress and categorized as drought tolerant and susceptible varieties, respectively (Shabbir et al., 2015; Ulfat et al., 2017; Ahmad et al., 2021; Wasaya et al., 2021; Iqra et al., 2022). So, seeds of these varieties were obtained and sown under controlled glass-house conditions at the National Institute for Genomics and Advanced Biotechnology (NIGAB), National Agriculture Research Center (NARC), Islamabad, Pakistan. After 2 weeks of sowing (seedling stage), the roots and leaves tissues were collected. At growth stage 8 (tillering stage), roots, stems, and leaf tissues were collected. At the grain filling stage (14 days after flowering), sampling for roots, stems, leaf, and grains was done (Hyles et al., 2020). For expression profile analysis under drought stress, seeds of selected varieties were first surface sterilized with sodium hypochlorite followed by three washings, then soaked in distilled water in a growth chamber (16 h light/8 h dark cycle at 22°C). After 2 weeks, young seedlings were treated with 20% polyethylene glycol (PEG) 6,000 (v/v). The root and leaf tissues of seedlings were harvested after 12 h of exposure to stress conditions. All the samples were collected in three replicates, and samples were frozen immediately in liquid nitrogen, and placed in −80°C storage for RNA extraction.
Approximately 100 mg of tissues were taken for total RNA extraction using an RNA mini kit (Cat # 12183018A, Invitrogen, Thermo Fischer Scientific) followed by the manufacturer’s instructions. Through agarose gel electrophoresis, the quality and concentration of RNA were determined, followed by optical density measurement through a spectrophotometer. With the help of the RT Prime-Script Reagent Kit, the cDNA was made from 1 ug of RNA. Specific primers were designed for TaRPK1 genes manually, followed by confirmation via NCBI Primer Blast software (http://www.ncbi.nlm.gov/tools/primer-blast), provided in Supplemental Table S2. The qRT-PCR was accomplished with SYBR Green I (Roche) Master Mix. Wheat β-Actin was used as a control reference gene. Three independent biological replicates were analyzed for each sample. The values were means and standard deviations (SD) were calculated from biological replicates. The relative expression levels of each gene were studied by means of 2−∆∆Ct (Schmittgen and Livak, 2008).
A set of 15 candidate RPK1 genes were retrieved from Triticum aestivum based on BlastP and BlastN. A domain search by the SMART tool with the corresponding RPK1 candidate amino acid sequences confirmed the S_TKc Domain (SM00220). Thus, a total of 15 TaRPK1 with complete structures were analyzed in T. aestivum (Table 1). Subsequent sequence identification of 15 TaRPK1 showed the protein length of 609–1,123 amino acids and a molecular mass ranged from 67–120 kDa. The iso-electric points (PI) of these proteins were 6–9. The Instability Index (II) ranged from 28.37–52.17, the Aliphatic Index (AI) was 87.93–106.38, and the grand-average of hydropathicity (GRAVY) −0.032–0.138. The instability index of group I was less than 40, representing stable proteins, whereas proteins of groups II and III showed instability index values of more than 40, indicating unstable proteins. The AI signified that all of the TaRPK1 proteins are thermally stable. The GRAVY indicated TaRPK1 proteins to be hydrophilic proteins except for TaRPK10, TaRPK11, and TaRPK12, which showed a value less than zero, representing them as hydrophobic proteins. The sub-cellular localizations of the TaRPK1 were anticipated, which showed that all the TaRPK1 were localized to the cell membrane (Table 2).
A detailed protein alignment of structural predictions showed that all TaRPK1 proteins are composed of the leucine-rich repeat N terminal (LRRNT_2) domain, leucine-rich repeat (LRR) domains, transmembrane domain I, and a serine–threonine kinase (S_TKc) domain. However, the LRR domains were missing in TaRPK1, TaRPK2, and TaRPK3 sequences (Supplemental Figure S1).
The physical location of RPK genes in T. aestivum, to the corresponding chromosomes, is shown in Figure 1. A total of 15 RPK genes were mapped on 9 out of 21 chromosomes in wheat. The genes were mainly mapped on chromosomes 1, 2, and 3 on the respective A, B, and D genomes. No RPK genes were found on the rest of the chromosomes.
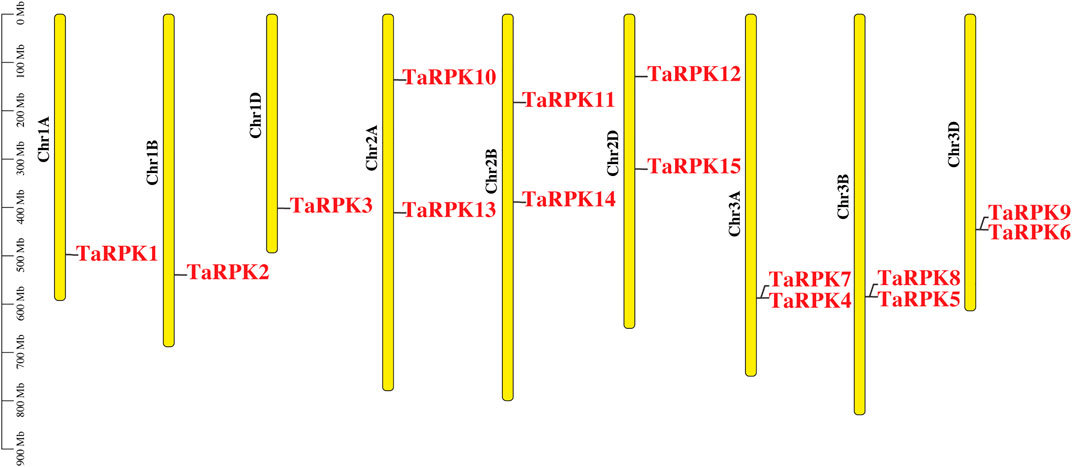
FIGURE 1. Chromosomal location of T. aestivum RPK genes on chromosomes in A, B, and D genomes. Respective chromosome numbers are written as Chr IA to Chr 3D on the top of each chromosome. Gene position can be estimated using the scale (in megabase; Mb) on the left of the figure.
Of the 15 identified TaRPK1 genes in this study in Triticum aestivum, two RPK genes from Arabidopsis thaliana, 16 RPK genes from rice, seven RPK genes from Triticum dicoccoides, three RPK genes from Triticum urata, seven RPK genes from Triticum turgidium, four RPK genes from Aegilops tauschii, 11 RPK genes from Triticum speltoides, and four RPK genes from Hordeum vulgare were used to construct a neighbor-joining based tree with MEGA X software in order to study the evolutionary relationships (Figure 2). The phylogenetic tree generated on the basis of similarities with protein sequences distributed RPK members into four main groups, with TaRPK1 members in three groups. Overall group I possessed nine TaRPK1 members (TaRPK1-9), that were closely associated with RPK members of rice. Group II (TaRPK10-12) and Group III (TaRPK13-15) exhibited three TaRPK1 members each, that exhibited close association with Triticum turgidium, Triticum speltoides, Aegilops tauschii and Triticum dicoccoides.
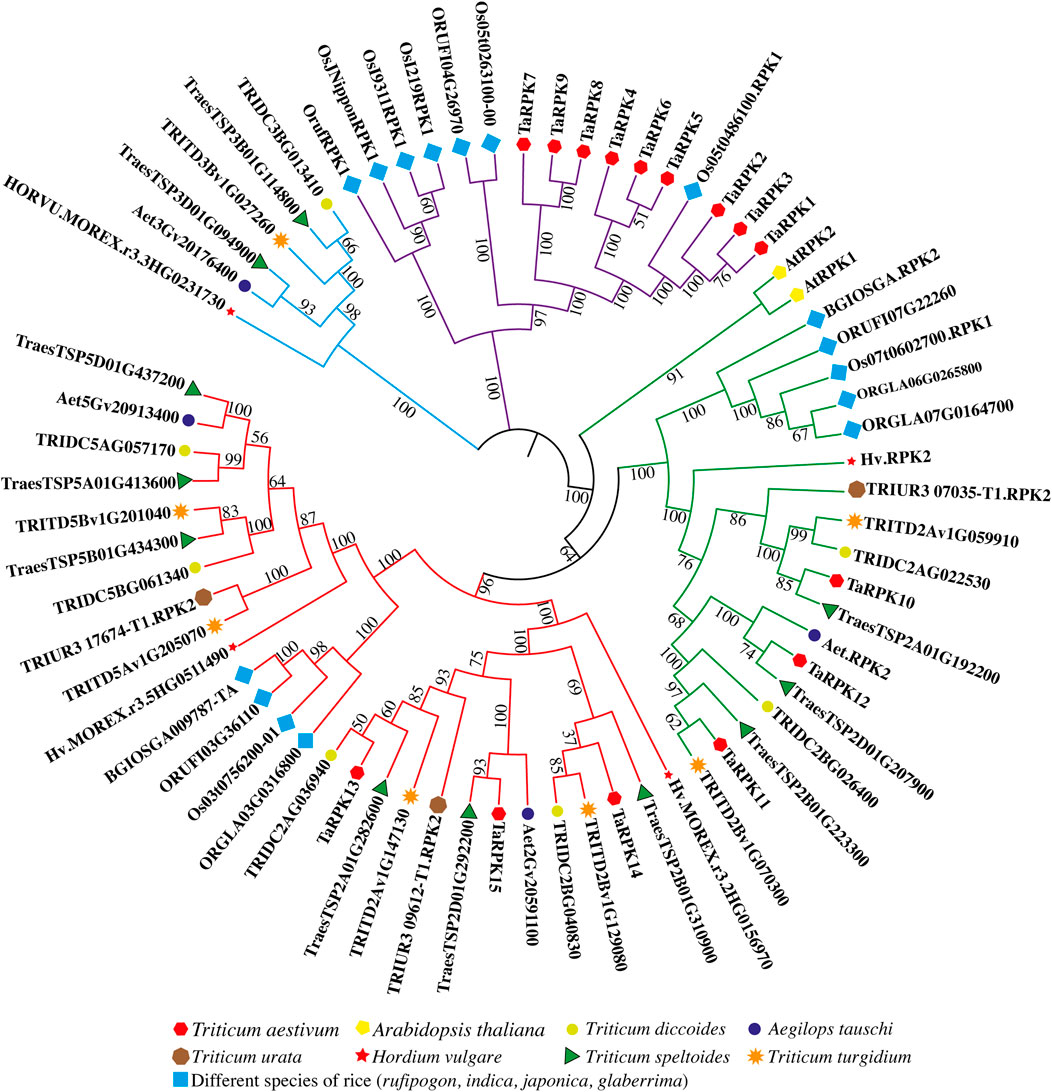
FIGURE 2. Comparative phylogenetic tree of RPK genes between Triticum aestivum, Triticum dicoccoides, Triticum turgidum, Triticum speltoides, Aegilops tauschii, Triticum urata, Hordeum vulgare, Arabidopsis thaliana, and different species of Oryza (rufipigon, indica, japonica, and glaberrima). 1,000 replicates were used for the bootstrap test, and the replication percentage is presented next to the branches.
The intron–exon number and arrangements of the RPK1 members were envisaged through comparing the coding sequence with the genomic DNA sequence. All of the TaRPK1 genes in group I consisted of 16–18 introns, except for the groups II and III that contained 0 and 1 intron (Figure 3A). Furthermore, the conserved motifs within TaRPK1 proteins were predicted by online MEME software. Ten conserved motifs (1-10) were analyzed (Figure 3B and Supplementary Figure S2). The motifs 1, 3, 4, 7, 8, and 10 were present in all of the RPK1 sequences. However, group II did not display motifs 3 and 10, and the motif three was also missing in group III sequences.

FIGURE 3. (A) Exon–intron structures of T. aestivum RPK1 genes. Yellow boxes denote exons, straight black lines represent introns, and blue boxes denote upstream/downstream. (B) Schematic representation of identified motifs in T. aestivum RPK1 proteins using the MEME motif search tool. Different colors indicate different motifs.
GO annotation analysis was conducted for the functional analysis of RPK1 genes. In-silico functional prediction was performed, and the results displayed two types of processes involved, that is, molecular processes (MPs) and biological processes (BPs) (Figure 4 and Supplementary Table S3). Biological processes indicate that RPK members are involved actively in various metabolic processes. The molecular processes suggested the RPK1 member’s catalytic activity. Such outcomes clearly denote RPK1 genes’ significant role in growth and development via modulation of molecular and biological processes.
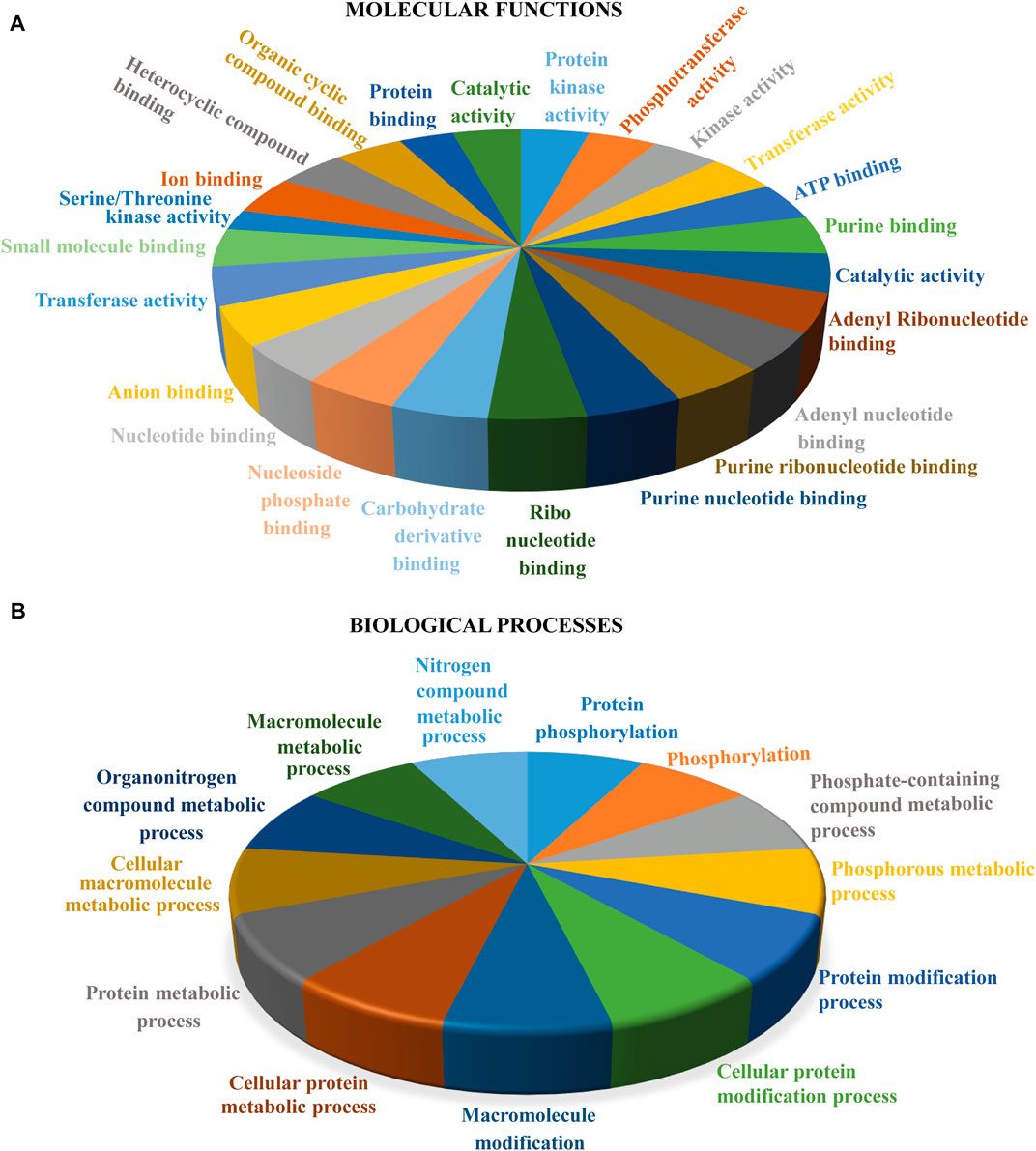
FIGURE 4. Gene Ontology prediction of RPK1 genes. The data represent (A) molecular functions and (B) biological processes.
We also identified putative 18 miRNAs targeting TaRPK1 genes for the generation of interaction networks by Cytoscape software in order to better understand the underlying miRNA mechanism involved in the modulation of TaRPK1 genes (Figure 5 and Supplementary Table S4). In the connection distribution and regulation network, TaRPK1, TaRPK2, and TaRPK3 were found targeted by single miRNAs, which are tae-miR9782, tae-miR9776, and tae-miR1122c-3p, respectively. TaRPK10 and TaRPK11 are the most targeted RPK1 wheat genes by tae-miR1134, tae-miR9774, tae-miR9661-5p, tae-miR9664-3p and tae-miR9777 targeting TaRPK10, and tae-miR9774, tae-miR9777, tae-miR9664-3p, tae-miR395a and tae-miR9661-5p targeting TaRPK11 genes. However, no miRNA was found targeting TaRPK13, TaRPK14, and TaRPK15 genes.
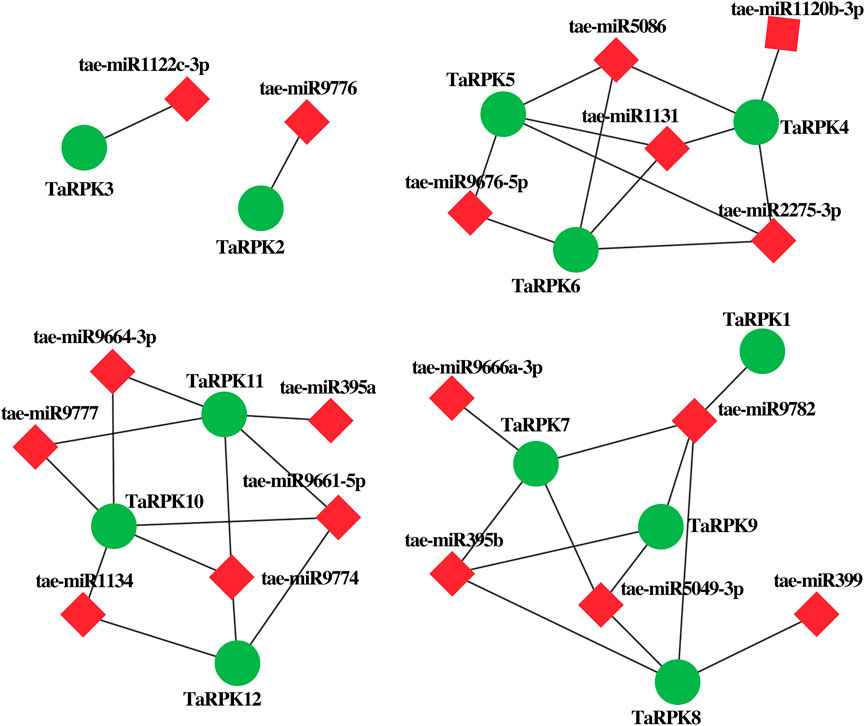
FIGURE 5. MicroRNA and their targeted TaRPK1 genes. Regulatory network relationship between miRNA and their targeted TaRPK1 genes.
The promoter regions contain cis-modulatory elements which are critical for the binding of transcription factors for transcription initiation, which has an essential function in the expression of genes. The promoter regions of RPK1 members were used for the cis-regulatory element prediction (Figure 6A). The results indicated that the cis-regulatory elements can be distributed into several categories, such as hormone related elements, light-related elements, developmental responsive elements, abiotic stress responsive elements, promoter-related motifs, and other motifs. Amid them, the elements chiefly present were associated with photoreaction, hormone responsiveness, and abiotic stress-related motifs. The photoreaction responsive cis-regulatory elements included ACE, AE-Box, ATCT, G-Box, GATA, GT1, SP1, AT1, Box 4, Box II, I-Box, TCT, GA, L-Box, TCCC, and ATC motif. The most abundant light-responsive elements were found in TaRPK11 and TaRPK13, which had 17 and 12 members, respectively. Hormone responsive elements were also copiously present in the RPK1 promoter, mostly comprising abscisic acid response elements. The three extensively distributed cis elements were related to abiotic stress response, among which drought responsive elements were profuse. Other elements correlated to abiotic stress were also identified.
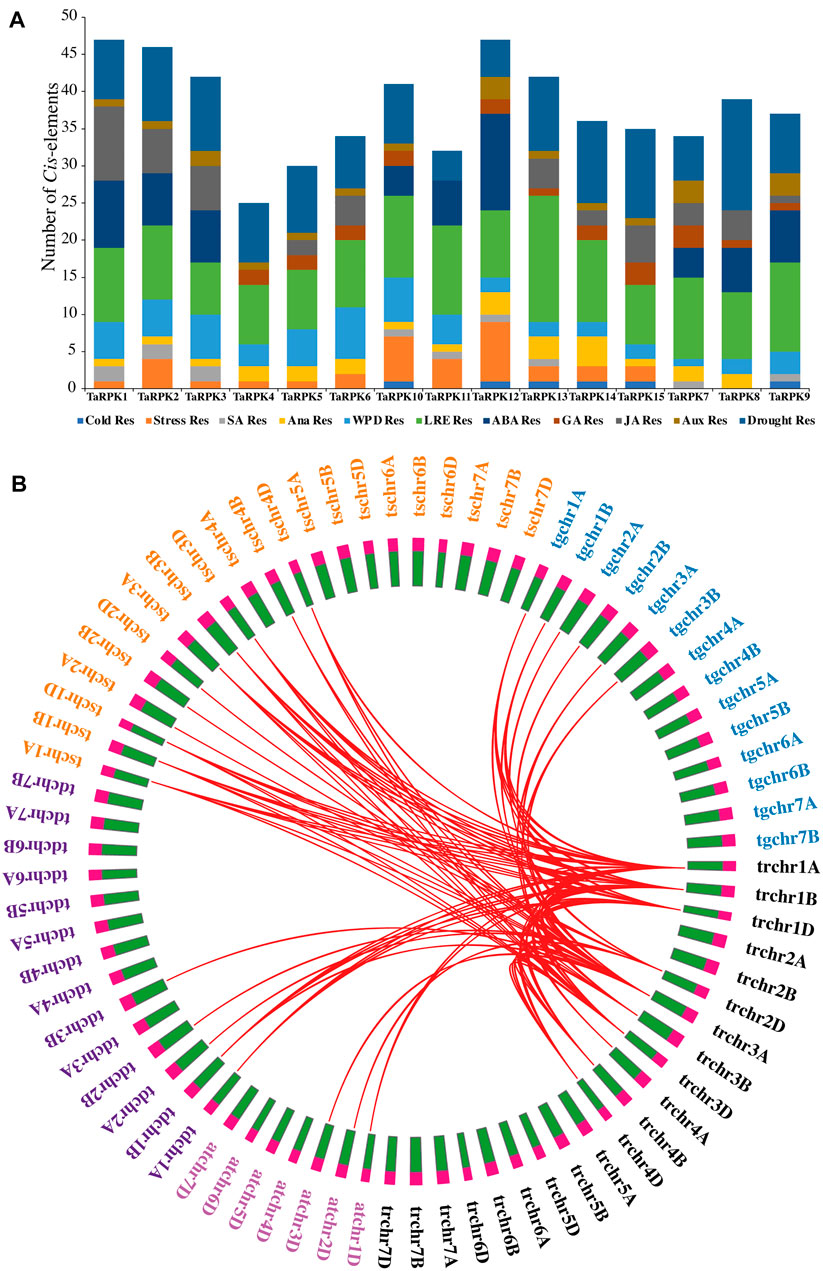
FIGURE 6. Cis-Elements and evolutionary conservation. (A) Regulatory Cis-element prediction of 2-Kb sequence upstream of RPK1 genes in T. aestivum. (B) Syntenic relationship between Triticum aestivum (tr, black), Triticum dicoccoides (td, purple), Triticum turgidum (tg, blue), Aegilops tauschii (at, pink), and Triticum speltoides (ts, orange).
In order to understand the evolutionary relationship and origin of Triticum aestivum (tr) with Triticum turgidum (tg), Aegilops tauschii (at), Triticum speltoides (ts) and Tritium dicoccoides (td), a comparative synteny scrutiny of RPK protein sequences was performed. The proteins were closely related among five species and exhibited significant similarity in analysis of evolutionary correlation. It was observed that the TaRPK1 genes of T. aestivum have similar origins of evolution to other Triticum species (Figure 6B and Supplementary Table S5).
Three-dimensional (3D) structures of TaRPK1 proteins were predicted by using SWISS_MODEL online computational software. 3D structures of target proteins were anticipated based on homology modeling. The SWISS MODEL predicted 15 successful models of TaRPK1 proteins with at least 30% identity to the template (4mn8.1. A, 5hyx.1. A, 5xkj.1. C, 6mOu.1. A, 4mna.1. A, 4oh4.1. A, 6cth.1. A, 7brc.1. A, and 5tos.1. A) that was a widely recognized threshold for effective modeling (Xiang, 2006). However, TaRPK2 and TaRPK3 showed sequence identity of 27.84% and 29.47%, respectively, with the template, which was less than 30%. The highest sequence identity of 45% with the template was observed by TaRPK4, TaRPK5 and TaRPK6 (Figure 7). The verification and validation of the predicted 3D structure of TaRPK1 were assessed via Ramachandran Plots (Anderson et al., 2005) that validated the backbone diahedral angles of the targeted protein. The Ramachandran plot assessment showed that 92–98% of the regions of TaRPK1 protein showed highly favorable regions, which indicates the stability and good quality of the predicted protein structure (Supplemental Table S6).

FIGURE 7. 3D structure of TaRPK1 proteins along with Ramachandran plots in T. aestivum. In all 3D protein structures, the spirals are helices, broad strips with arrow heads are beta-pleated sheets, and thin loops are coils. In Ramachandran plots, dark black, gray, and light gray represent highly preferred conformations with Delta ≥ −2. White with a black grid denotes preferred conformations with −2 > Delta ≥ −4. White with gray grid symbolizes questionable conformations with Delta < −4. The green crosses signify highly preferred observations, brown triangles specify preferred observations, and red circles represent unfavorable observations.
The data of RNA-seq for all of the 15 RPK sequences were obtained from online database. A heatmap was generated showing expression levels of RPK members at different stages, namely seedling stage, vegetative stage, and reproductive stage (Figure 8 and Supplementary Table S7) and in various organs (root, leaf, shoot, spike, and grain) of wheat. The highest expression of TaRPK1 members was observed in root tissues compared to other tissues. TaRPK1, TaRPK2, and TaRPK3 exhibited the highest expression patterns in roots at seedling, vegetative, and reproductive stages. Higher to moderate expression was observed in grain at the developing reproductive stage by TaRPK13 and TaRPK14, respectively. Spikes, leaves, and shoots showed moderate to low expression in all of the TaRPK1 members in wheat.
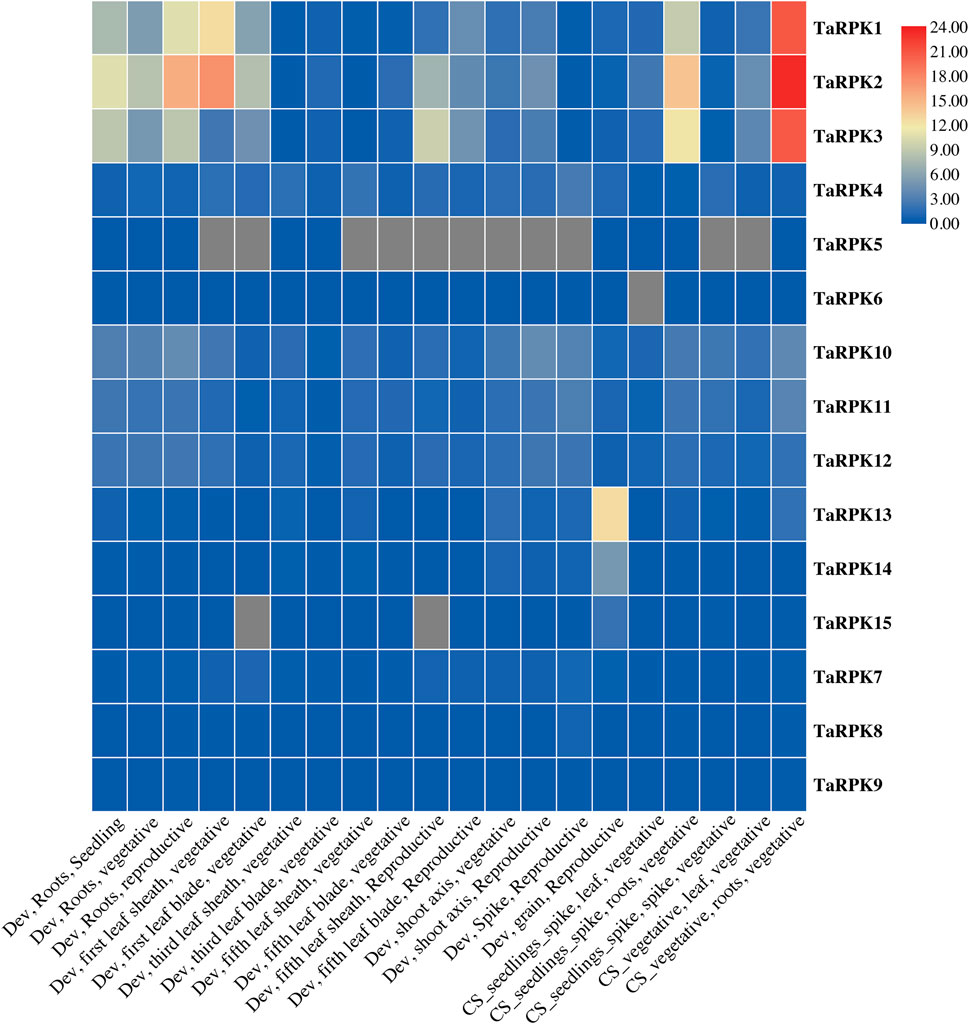
FIGURE 8. In silico analysis of RPK members in different tissues (root, shoot, leaf, grain, and spike) was generated using TB tool. Dev = developmental stage, CS = seedlings spike; Chinese Spring seedlings (leaves, roots) and spikes at anthesis and CS vegetative; Chinese Spring leaves and roots from seven leaf stages. The heatmap was constructed from transcripts per 10 million values with the scale bar displaying expression of the genes. The blue and red colors denote lower and higher expressions of the transcripts, respectively.
The TaRPK1 gene expression was determined in drought-tolerant (Pakistan 13 and Galaxy) and drought-susceptible (Shafaq) varieties under normal growth conditions in order to get a baseline expression profile. The expression pattern in all of the three varieties was examined in various developmental stages, including seedling stage, tillering stage, and heading stage and in different tissues such as root, stem, leaf, and grain (Figure 9). The TaRPK1, TaRPK2, and TaRPK3 showed significant expression in the roots at the heading and seedling stages of the Pakistan-13 and Galaxy varieties. The TaRPK13 exhibited higher expression in grain tissues of all varieties compared to other TaRPK1. The TaRPK1 genes displayed higher expression in roots whereas they showed less expression in leaves and stems compared to the grain and root expression in developmental stages. Our results indicated that TaRPK1 genes had similar expression patterns in both Pakistan 13 and Galaxy varieties, unlike the Shafaq variety. The higher expression of TaRPK1 genes was observed in the heading > seedling > tillering stages in Pakistan 13, Galaxy, and Shafaq varieties. Overall, TaRPK1 exhibited significant expression in root tissues compared to leaf, shoot, and grain tissues.
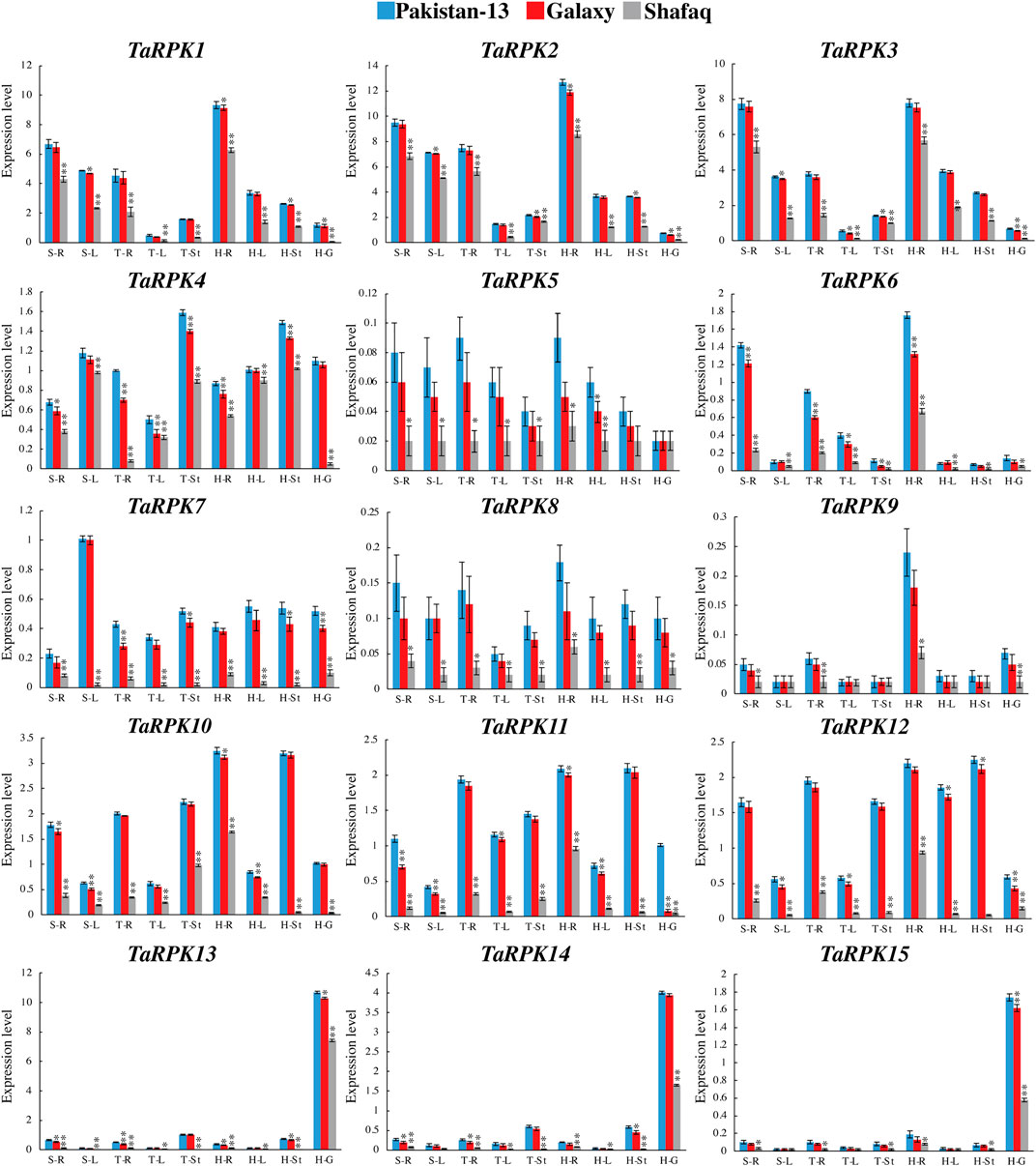
FIGURE 9. Real time PCR-expression analysis of TaRPK1 genes in wheat varieties; Pakistan 13 (blue), Galaxy (red), and Shafaq (gray). The wheat plant was germinated and grown in soil under normal conditions. Expressions of TaRPK1 genes were determined in root (R), stem (St), leave (L), and grain (G) at seedling (S), tillering (T), and heading (H) stages. Standard deviation (SD) of three biological replicates is represented by the error bars. Significance was assessed by using a t-test (*p < 0.05, **p < 0.01, and ns = non-significant).
Roots are a good source to study the drought mechanism. To further confirm this, qRT-PCR showed the expression of TaRPK1 members in the leaves and roots of two-week-old seedlings with drought stress through PEG simulation. PEG-6000 treatment induced an upregulated expression in roots and leaf tissues in comparison to the susceptible genotype. Higher expression was observed in root seedlings in comparison to the leaf seedlings, except for TaRPK4 and TaRPK7, where higher expression was detected in the leaf tissues compared to the root tissues under drought stress (Figure 10). The TaRPK1 genes displayed higher expression in Pakistan 13 > Galaxy > Shafaq varieties. Furthermore, we also performed co-expression (Supplementary Figure S3) and interaction network (Supplementary Figure S4 and Supplementary Table S8) analyses and the results revealed that all the RPK1 members showed highly significant associations. These results indicate TaRPK1 gene involvement in drought stress regulation.
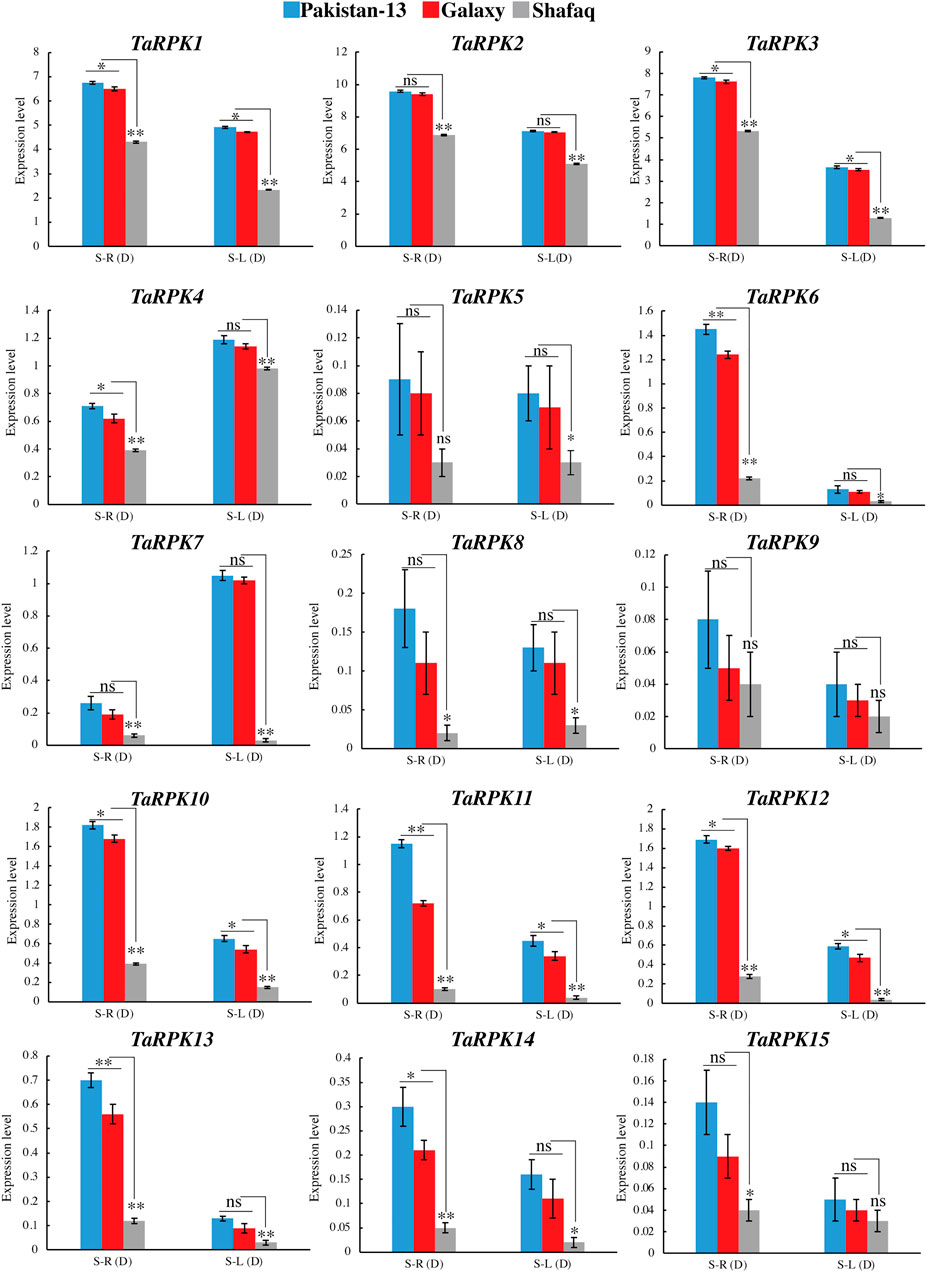
FIGURE 10. Expression profiling of TaRPK1 genes under 20% PEG stress in Pakistan 13, Galaxy, and Shafaq varieties. Expressions of TaRPK1 genes were determined in root (S–R) and leave (S–L) at the seedling stage (S). Error bars denote standard errors of three biological replicates. Significance was assessed by using a t-test (*p < 0.05, **p < 0.01, and ns = non-significant).
RPK1 is a serine/threonine protein kinase and belongs to the subfamily LRRKs, which is the largest subfamily of RLK. The LRRKs play a crucial role in a large number of biological activities, from development and growth to stress management in plants (Dufayard et al., 2017). RPK genes play significant roles in root system architecture (RSA), plant height, number of tillers, salt tolerance, cotyledon primordial initiation of cotyledons during embryogenesis, ABA-induced senescence, and ROS production (Shi et al., 2014; Zou et al., 2014; Dai et al., 2018). The functional characterization of RPK1 members in wheat has not been reported in previous studies. The reason for it could be a complex allohexaploid (2n = 6x = 42) genome and other factors. Since RPK1 genes are accountable for essential roles in plants, therefore a comprehensive study was performed to determine the chromosomal location, phylogenetic analysis, gene structure and expression of these genes in T. aestivum.
The standard process for the sequence identification of a new gene family is by a BLAST search of sequences of known proteins in model plants. A similar BLAST method was used to characterize two RPK genes from Arabidopsis thaliana, 16 RPK genes from different species of rice, seven RPK genes from Triticum dicoccoides, three RPK genes from Triticum urata, seven RPK genes from Triticum turgidum, four RPK genes from Aegilops tauschii, 11 RPK genes from Triticum speltoides, four RPK genes from Hordeum vulgare, and 15 TaRPK1 genes in Triticum aestivum. The number of TaRPKs identified in T. aestivum is similar to that of RPKs in Triticum speltoides (11). The identified RPK genes were confirmed for the conserved domains by the SMART database. A higher number of TaRPK1 genes might be because of the large allohexaploid nature of the bread wheat genome.
The allohexaploid T. aestivum genome was originated due to the 3A, B, and D diploid sub genomes hybridization (Marcussen et al., 2014). Three homoeologous genes at a minimum should be for each T. aestivum gene, that is, one from each sub genome, also named as homoeologous genes for their homologous chromosomal localization (Sharma et al., 2016). The genome wide analysis displayed that TaRPK1 genes along with the homoeologous genes were located mainly on chromosomes 1, 2, and 3 on A, B, and D sub-genomes, which showed that there might be no deletion of TaRPK1 genes in the course of the acclimatization and evolution process of T. aestivum. The TaRPK1 genes were found to be with maximum number on chromosome 2 and 3 (Figure 1) which was very similar to other studied crops. Crops such as Triticum dicoccoides, Aegilops tauschii, Hordeum vulgare, Triticum speltoides, and Triticum turgidum also showed the distribution of RPKs on chromosome 2 and 3, in addition to chromosome 5. However, the RPKs were distributed on chromosomes 3, 4, and 7 in rice, and in Arabidopsis thaliana they were on chromosomes 1 and 3.
The phylogenetic relationship was studied using complete TaRPK1 protein sequences, as it indicated evolutionary inference. The known homoeologous sequences were clustered closely (Figure 2), which indicated further evolutionary relationships and homology of sequences among them. The putative paralogous sequences were grouped together by those that specified similar origins. Similarity in organization and architecture of domains and motifs in clades designates functional association between these proteins. The gene structure analysis revealed intron numbers in TaRPK1 genes that ranged from 0 to 1 and 16-18 (Figure 3A). The difference in the number of exons in TaRPK1 was analogous to the one observed in other crops. Triticum dicoccoides, Aegilops tauschii, Hordeum vulgare, and Triticum speltoides exhibited one to two coding exons, and Triticum turgidum had one to three coding exons. Oryza species also exhibited one to three exons except for Oryza rufipogon; ORUFI04G26970 had 102 exons and Os05t0486100-01 RPK1 exhibited 18 exons. This points toward evolutionary conservation and hence expression of genes between these species.
Prediction of protein domain configuration revealed the similarity to the previously studied RPK proteins (Cheng et al., 2009), with conserved C-terminal Ser/Thr kinase, a transmembrane domain suggesting membrane-bound features of TaRPK1 proteins and LRR domains. The LRR domains were absent in TaRPK1, TaRPK2, and TaRPK3 proteins. However, all TaRPK1 members showed an additional LRRNT_2 (leucine-rich repeat N-terminal) domain in the N-terminal region of the amino acid (Supplemental Figure S1). In addition to sequence alignment, motif analysis also displayed the conservation of the motif at the initial N-terminal region and kinase domain with the motif that remained conserved in all of the 15 TaRPK1 protein sequences (Figure 3B). For the functional analysis of TaRPK1 genes, the gene ontology enrichment analysis was performed (Figure 4). In silico prediction showed that TaRPK1 members were involved in several processes of development through regulation of molecular functions (MFs) and biological processes (BPs), and exhibited response to environmental stresses. Several prior studies also described that through monitoring expression of genes, microRNAs respond to stress stimuli (Yan et al., 2019; Rasool et al., 2021; Rehman et al., 2022). The microRNAs are 21–24 nucleotides long endogenous non-coding RNAs that regulates development, growth, and adaptive response against abiotic stresses via monitoring target genes at posttranscriptional level or translation level of protein synthesis (Bai et al., 2017). In this study, we recognized microRNAs and their target genes in order to explore specific transcripts involved in development and growth processes and in response to different stress environments. We identified that miRNAs are majorly involved in cleavage mechanisms rather than translation inhibition (Figure 5).
The cis-regulatory elements identified in TaRPK1 were mostly related to light responsiveness (Figure 6A). Other distributed cis-regulatory elements were related to stress factors, such as drought, cold stress, anaerobic response, wounding pathogens, and defensive elements. Functional relation of other cis elements was linked to plant hormones comprising auxins, abscisic, gibberellin and salicylic acid. Thus, the occurrence of various groups of cis-regulatory elements functioning in diverse physiological processes is suggestive of the dynamic RPK1 gene regulation in T. aestivum. Synteny analysis with other ancestral Triticum species revealed that the RPK1 gene family converges to a single ancestor (Figure 6B). This relationship validates that RPKs with analogous evolutionary status might have similar functions in plant growth and development. Homology models for 15 TaRPK1 proteins were made and evaluated with homologous templates. The TaRPK1 proteins exhibited 28%–45% identity to the template, which is a widely accepted threshold for successful modeling. The Ramachandran plots verification and validation displayed that a very higher percentage of all 15 TaRPK1 protein regions showed highly favorable regions that denote good quality protein structure prediction (Figure 7). Previous studies have shown similar 3D structure of TATrx proteins in wheat through homology modeling along with Ramachandran plot. The proteins were compared to 2iwt.1. A, 2vlt.1. A, 1fb0.1. A, 3d22.1. A, 2vm1.3. A, and 1faa.1. A templates, and the Ramachandran plot showed more than 95% of the thioredoxin amino acids lying in the most favored area (Bhurta et al., 2022). Another study in wheat has shown similar three-dimensional structure prediction of twenty-one TaEIL proteins via SWISS-MODEL along with Ramachandran plot analysis. The prediction model on the basis of templates heuristically enhanced percentage identification, alignment range, and confidence score of test sequences. The Ramachandran plot analysis confirmed 80% of residuals in the allowed area, signifying the quality of the model (Yi-Qin et al., 2020).
The gene expression in a specific tissue can be used as an information source for function identification in that tissue. Studies have revealed that OsRPK1 overexpression altered the total architecture of roots in transgenic seedlings along with height, tillering numbers, and apical meristem of roots (Zou et al., 2014). The larger root system might result in a substantial upsurge in water and nutrient uptake. The relative expression level in different tissues of OsRPK1 was studied, which indicated higher to lower expression in the pattern of root tips > leaf blades > roots > leaf sheath > stem (Zou et al., 2014). Alike expression pattern was also detected in TaRPK1 genes. The heatmap generated showed significant expression of RPK1 genes in the root tissIes in comparison to the other tissues studied (Figure 9). As gene expression profiles are always related to their function, we further investigated their expression profiles in various tissues and varieties under normal and drought stress responses. Results of real-time quantitative PCR indicated that TaRPK1 showed higher expression levels in root tissues at seedling and heading stages under normal conditions. TaRPK1 exhibited tissue specific expression and showed higher expression in drought stress treatment in root tissues (Figure 10). This high expression in particular organs like roots indicates their particular roles in the root development and function of that tissue. The higher expression of TaRPK1 genes was observed in Pakistan 13 > Galaxy > Shafaq varieties, which indicates the vital role of TaRPK1 in plant growth and development. The sequence similarity and conserved domains of these protein kinases from Arabidopsis, rice, and wheat combined with the evidence from in-silico expression analysis and RT-PCR suggest that TaRPK1 might share analogous functions in root development and hence yield. Future functional validation of these genes will be required.
We completely investigated the properties, developmental, location on chromosomes, cis-components, synteny, and expression profiles of TaRPK1 members. An aggregate of 15 TaRPK1s were distinguished in the T. aestivum genome. This work can fill in as an initial phase in the complete useful portrayal of RPK1 genes by reversible genetic methodologies. This study provides helpful assets to future investigations on the design and function of RPK1 genes and for distinguishing and describing these genes in different species. Consequently, the outcomes might offer important data to examine the role of TaRPK1 genes being developed and stress reactions through present-day practical genomics tools (next-generation sequencing) and genome editing, henceforth clearing the way toward genetic improvement of wheat.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
MK and AR perceived the idea and planned the experiments. AR performed the research and wrote the manuscript. NZ and OR helped in data analysis. NR and MU helped in manuscript revision. MK supervised the research and acquired resources.
This research was conducted with funds from the Agricultural Linkages Program of Pakistan (ALP project number CS-399).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge the research facilities provided by the National Institute for Genomics and Advanced Biotechnology (NIGAB), Pakistan. We also thank the Ensembl plants, NCBI, and various other tools for the availability of sequences and data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.912251/full#supplementary-material
Abbas, H., Naeem, M. K., Rubab, M., Widemann, E., Uzair, M., Zahra, N., et al. (2022). Role of Wheat Phosphorus Starvation Tolerance 1 Genes in Phosphorus Acquisition and Root Architecture. Genes. 13, 487. doi:10.3390/genes13030487
Afzal, A. J., Wood, A. J., and Lightfoot, D. A. (2008). Plant Receptor-like Serine Threonine Kinases: Roles in Signaling and Plant Defense. Mpmi 21, 507–517. doi:10.1094/mpmi-21-5-0507
Ahmad, Z., Ishaq, M., Tahir, I., Khan, A., and Jehan, K. (2021). Heat Tolerance Stability of Bread Wheat Genotypes under Early and Late Planting Environments through Stress Selection Indices. Curr. Res. Agric. Sci. 8, 37–46. doi:10.18488/journal.68.2021.81.37.46
Alahmad, S., El Hassouni, K., Bassi, F. M., Dinglasan, E., Youssef, C., Quarry, G., et al. (2019). A Major Root Architecture QTL Responding to Water Limitation in Durum Wheat. Front. Plant Sci. 10, 436. doi:10.3389/fpls.2019.00436
Anderson, R. J., Weng, Z., Campbell, R. K., and Jiang, X. (2005). Main-chain Conformational Tendencies of Amino Acids. Proteins 60, 679–689. doi:10.1002/prot.20530
Bai, J.-F., Wang, Y.-K., Wang, P., Duan, W.-J., Yuan, S.-H., Sun, H., et al. (2017). Uncovering Male Fertility Transition Responsive miRNA in a Wheat Photo-Thermosensitive Genic Male Sterile Line by Deep Sequencing and Degradome Analysis. Front. Plant Sci. 8, 1370. doi:10.3389/fpls.2017.01370
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Bhurta, R., Hurali, D. T., Tyagi, S., Sathee, L., Adavi, B., Singh, D., et al. (2022). Genome-Wide Identification and Expression Analysis of the Thioredoxin (Trx) Gene Family Reveals its Role in Leaf Rust Resistance in Wheat (Triticum aestivum L.). Front. Genet., 563. doi:10.3389/fgene.2022.836030
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic acids Res. 42, W252–W258. doi:10.1093/nar/gku340
Cao, Y., Liu, M., Long, H., Zhao, Q., Jiang, L., and Zhang, L. (2020). Hidden in Plain Sight: Systematic Investigation of Leucine-Rich Repeat Containing Genes Unveil the Their Regulatory Network in Response to Fusarium Wilt in Tung Tree. Int. J. Biol. Macromol. 163, 1759–1767. doi:10.1016/j.ijbiomac.2020.09.106
Chen, C., Chen, H., He, Y., and Xia, R. (2018). TBtools, a Toolkit for Biologists Integrating Various Biological Data Handling Tools with a User-Friendly Interface. BioRxiv 289660.
Cheng, X., Xiao, X., and Chou, K.-C. (2017). pLoc-mPlant: Predict Subcellular Localization of Multi-Location Plant Proteins by Incorporating the Optimal GO Information into General PseAAC. Mol. Biosyst. 13, 1722–1727. doi:10.1039/c7mb00267j
Cheng, Y., Qi, Y., Zhu, Q., Chen, X., Wang, N., Zhao, X., et al. (2009). New Changes in the Plasma-Membrane-Associated Proteome of Rice Roots under Salt Stress. Proteomics 9, 3100–3114. doi:10.1002/pmic.200800340
Dai, C., Lee, Y., Lee, I. C., Nam, H. G., and Kwak, J. M. (2018). Calmodulin 1 Regulates Senescence and ABA Response in Arabidopsis. Front. Plant Sci. 9, 803. doi:10.3389/fpls.2018.00803
Dai, X., and Zhao, P. X. (2011). psRNATarget: a Plant Small RNA Target Analysis Server. Nucleic acids Res. 39, W155–W159. doi:10.1093/nar/gkr319
Danakumara, T., Kumari, J., Singh, A. K., Sinha, S. K., Pradhan, A. K., Sharma, S., et al. (2021). Genetic Dissection of Seedling Root System Architectural Traits in a Diverse Panel of Hexaploid Wheat through Multi-Locus Genome-wide Association Mapping for Improving Drought Tolerance. Ijms 22, 7188. doi:10.3390/ijms22137188
Diévart, A., and Clark, S. E. (2003). Using Mutant Alleles to Determine the Structure and Function of Leucine-Rich Repeat Receptor-like Kinases. Curr. Opin. Plant Biol. 6 (5), 507–516. doi:10.1016/s1369-5266(03)00089-x
Dievart, A., Gottin, C., Périn, C., Ranwez, V., and Chantret, N. (2020). Origin and Diversity of Plant Receptor-like Kinases. Annu. Rev. Plant Biol. 71, 131–156. doi:10.1146/annurev-arplant-073019-025927
Djanaguiraman, M., Prasad, P. V. V., Kumari, J., Sehgal, S. K., Friebe, B., Djalovic, I., et al. (2019). Alien Chromosome Segment from Aegilops Speltoides and Dasypyrum Villosum Increases Drought Tolerance in Wheat via Profuse and Deep Root System. BMC Plant Biol. 19, 242–315. doi:10.1186/s12870-019-1833-8
Dufayard, J. F., Bettembourg, M., Fischer, I., Droc, G., Guiderdoni, E., Périn, C., et al. (2017). New Insights on Leucine-Rich Repeats Receptor-like Kinase Orthologous Relationships in Angiosperms. Front. Plant Sci. 8, 381. doi:10.3389/fpls.2017.00381
Fang, Y., Du, Y., Wang, J., Wu, A., Qiao, S., Xu, B., et al. (2017). Moderate Drought Stress Affected Root Growth and Grain Yield in Old, Modern and Newly Released Cultivars of Winter Wheat. Front. Plant Sci. 8, 672. doi:10.3389/fpls.2017.00672
Fantl, W. J., Johnson, D. E., and Williams, L. T. (1993). Signalling by Receptor Tyrosine Kinases. Annu. Rev. Biochem. 62, 453–481. doi:10.1146/annurev.bi.62.070193.002321
Gou, X., He, K., Yang, H., Yuan, T., Lin, H., Clouse, S. D., et al. (2010). Genome-wide Cloning and Sequence Analysis of Leucine-Rich Repeat Receptor-like Protein Kinase Genes in Arabidopsis thaliana. BMC genomics 11, 19–15. doi:10.1186/1471-2164-11-19
Grzesiak, M. T., Hordyńska, N., Maksymowicz, A., Grzesiak, S., and Szechyńska-Hebda, M. (2019). Variation Among Spring Wheat (Triticum aestivum L.) Genotypes in Response to the Drought Stress. II-Root System Structure. Plants 8, 584. doi:10.3390/plants8120584
Hong, S. W., Jon, J. H., Kwak, J. M., and Nam, H. G. (1997). Identification of a Receptor-like Protein Kinase Gene Rapidly Induced by Abscisic Acid, Dehydration, High Salt, and Cold Treatments in Arabidopsis thaliana. Plant physiol. 113, 1203–1212. doi:10.1104/pp.113.4.1203
Hyles, J., Bloomfield, M. T., Hunt, J. R., Trethowan, R. M., and Trevaskis, B. (2020). Phenology and Related Traits for Wheat Adaptation. Heredity 125, 417–430. doi:10.1038/s41437-020-0320-1
Iqra, T., Ullah, K. N., Samrin, G., Sheraz, A., Yusra, A., Amina, H., et al. (2022). Late Planting Induced Heat Stress Tolerance in Wheat. Pak. J. Bot. 54.
Kaur, S., Dhugga, K. S., Beech, R., and Singh, J. (2017). Genome-wide Analysis of the Cellulose Synthase-like (Csl) Gene Family in Bread Wheat (Triticum aestivum L.). BMC Plant Biol. 17, 193–217. doi:10.1186/s12870-017-1142-z
Kumar, S., Tamura, K., and Nei, M. (1994). MEGA: Molecular Evolutionary Genetics Analysis Software for Microcomputers. Bioinformatics 10, 189–191. doi:10.1093/bioinformatics/10.2.189
Lee, I. C., Hong, S. W., Whang, S. S., Lim, P. O., Nam, H. G., and Koo, J. C. (2011). Age-dependent Action of an ABA-Inducible Receptor Kinase, RPK1, as a Positive Regulator of Senescence in Arabidopsis Leaves. Plant Cell. physiology 52, 651–662. doi:10.1093/pcp/pcr026
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2018). 20 Years of the SMART Protein Domain Annotation Resource. Nucleic acids Res. 46, D493–D496. doi:10.1093/nar/gkx922
Li, J., and Tax, F. E. (2013). Receptor‐like Kinases: Key Regulators of Plant Development and Defense. Wiley Online Library.
Manès, Y., Gomez, H., Puhl, L., Reynolds, M., Braun, H., and Trethowan, R. (2012). Genetic Yield Gains of the CIMMYT International Semi‐arid Wheat Yield Trials from 1994 to 2010. Crop Sci. 52, 1543–1552.
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., Jakobsen, K. S., et al. International Wheat Genome Sequencing Consortium (2014). Ancient Hybridizations Among the Ancestral Genomes of Bread Wheat. science 345, 1250092. doi:10.1126/science.1250092
Milner, M. J., Howells, R. M., Craze, M., Bowden, S., Graham, N., and Wallington, E. J. (2018). A PSTOL-like Gene, TaPSTOL, Controls a Number of Agronomically Important Traits in Wheat. BMC Plant Biol. 18, 115–214. doi:10.1186/s12870-018-1331-4
Mishra, D., Suri, G. S., Kaur, G., and Tiwari, M. (2021). Comprehensive Analysis of Structural, Functional, and Evolutionary Dynamics of Leucine Rich Repeats-RLKs in Thinopyrum Elongatum. Int. J. Biol. Macromol. 183, 513–527. doi:10.1016/j.ijbiomac.2021.04.137
Mondal, S., Singh, R. P., Huerta-Espino, J., Kehel, Z., and Autrique, E. (2015). Characterization of Heat- and Drought-Stress Tolerance in High-Yielding Spring Wheat. Crop Sci. 55, 1552–1562. doi:10.2135/cropsci2014.10.0709
Motte, H., Vercauteren, A., Depuydt, S., Landschoot, S., Geelen, D., Werbrouck, S., et al. (2014). Combining Linkage and Association Mapping Identifies RECEPTOR-LIKE PROTEIN KINASE1 as an Essential Arabidopsis Shoot Regeneration Gene. Proc. Natl. Acad. Sci. U.S.A. 111, 8305–8310. doi:10.1073/pnas.1404978111
Nejat, N., and Mantri, N. (2017). Plant Immune System: Crosstalk between Responses to Biotic and Abiotic Stresses the Missing Link in Understanding Plant Defence. Curr. Issues Mol. Biol. 23, 1–16. doi:10.21775/cimb.023.001
Nodine, M. D., and Tax, F. E. (2008). Two Receptor-like Kinases Required Together for the Establishment of Arabidopsis Cotyledon Primordia. Dev. Biol. 314, 161–170. doi:10.1016/j.ydbio.2007.11.021
Nodine, M. D., Yadegari, R., and Tax, F. E. (2007). RPK1 and TOAD2 Are Two Receptor-like Kinases Redundantly Required for Arabidopsis Embryonic Pattern Formation. Dev. Cell. 12, 943–956. doi:10.1016/j.devcel.2007.04.003
Odegard, I. Y. R., and Van Der Voet, E. (2014). The Future of Food - Scenarios and the Effect on Natural Resource Use in Agriculture in 2050. Ecol. Econ. 97, 51–59. doi:10.1016/j.ecolecon.2013.10.005
Rasool, F., Uzair, M., Naeem, M. K., Rehman, N., Afroz, A., Shah, H., et al. (2021). Phenylalanine Ammonia-Lyase (PAL) Genes Family in Wheat (Triticum aestivum L.): Genome-wide Characterization and Expression Profiling. Agronomy 11, 2511. doi:10.3390/agronomy11122511
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). g:Profiler: a Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic acids Res. 47, W191–W198. doi:10.1093/nar/gkz369
Rehman, O. U., Uzair, M., Chao, H., Fiaz, S., Khan, M. R., and Chen, M. (2022). Role of the Type‐B Authentic Response Regulator Gene Family in Fragrant Rice under Alkaline‐salt Stress. Physiol. Plant. 174, e13696. doi:10.1111/ppl.13696
Sahu, S. S., Loaiza, C. D., and Kaundal, R. (2020). Plant-mSubP: a Computational Framework for the Prediction of Single- and Multi-Target Protein Subcellular Localization Using Integrated Machine-Learning Approaches. AoB Plants 12, plz068. doi:10.1093/aobpla/plz068
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Shabbir, R. N., Ashraf, M. Y., Waraich, E., Ahmad, R., and Shahbaz, M. (2015). Combined Effects of Drought Stress and NPK Foliar Spray on Growth, Physiological Processes and Nutrient Uptake in Wheat. Pak. J. Bot. 47, 1207–1216.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shi, C.-C., Feng, C.-C., Yang, M.-M., Li, J.-L., Li, X.-X., Zhao, B.-C., et al. (2014). Overexpression of the Receptor-like Protein Kinase Genes AtRPK1 and OsRPK1 Reduces the Salt Tolerance of Arabidopsis thaliana. Plant Sci. 217-218, 63–70. doi:10.1016/j.plantsci.2013.12.002
Shumayla, S., Sharma, S., Kumar, R., Mendu, V., Singh, K., and Upadhyay, S. K. (2016). Genomic Dissection and Expression Profiling Revealed Functional Divergence in Triticum aestivum Leucine Rich Repeat Receptor like Kinases (TaLRRKs). Front. Plant Sci. 7, 1374. doi:10.3389/fpls.2016.01374
Ulfat, A., Majid, S. A., and Hameed, A. (2017). Hormonal Seed Priming Improves Wheat (Triticum aestivum L.) Field Performance under Drought and Non-stress Conditions. Pak. J. Bot. 49, 1239–1253.
Uzair, M., Long, H., Zafar, S. A., Patil, S. B., Chun, Y., Li, L., et al. (2021). Narrow Leaf21, Encoding Ribosomal Protein RPS3A, Controls Leaf Development in Rice. Plant Physiol. 186, 497–518. doi:10.1093/plphys/kiab075
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic acids Res. 40, e49. doi:10.1093/nar/gkr1293
Wasaya, A., Manzoor, S., Yasir, T. A., Sarwar, N., Mubeen, K., Ismail, I. A., et al. (2021). Evaluation of Fourteen Bread Wheat (Triticum aestivum L.) Genotypes by Observing Gas Exchange Parameters, Relative Water and Chlorophyll Content, and Yield Attributes under Drought Stress. Sustainability 13, 4799. doi:10.3390/su13094799
Xiang, Z. (2006). Advances in Homology Protein Structure Modeling. Cpps 7, 217–227. doi:10.2174/138920306777452312
Yan, F., Li, H., and Zhao, P. (2019). Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.). Genes. 10, 46. doi:10.3390/genes10010046
Yi‐Qin, H., Wen‐Di, H., Lei, Y., Yi‐Ting, L., Chen, L., Yong‐Xing, Z., et al. (2020). Genome‐wide Analysis of Ethylene‐insensitive3 (EIN3/EIL) in Triticum aestivum. Crop Sci. 60, 2019–2037.
Zargar, S. M., Gupta, N., Nazir, M., Mahajan, R., Malik, F. A., Sofi, N. R., et al. (2017). Impact of Drought on Photosynthesis: Molecular Perspective. Plant gene. 11, 154–159. doi:10.1016/j.plgene.2017.04.003
Zou, Y., Liu, X., Wang, Q., Chen, Y., Liu, C., Qiu, Y., et al. (2014). OsRPK1, a Novel Leucine-Rich Repeat Receptor-like Kinase, Negatively Regulates Polar Auxin Transport and Root Development in Rice. Biochimica Biophysica Acta (BBA) - General Subj. 1840, 1676–1685. doi:10.1016/j.bbagen.2014.01.003
Keywords: receptor-like protein kinase 1 (RPK1), abiotic stress, genome-wide studies, phylogenetic relationship, expression patterns, Triticum aestivum
Citation: Rahim AA, Uzair M, Rehman N, Rehman OU, Zahra N and Khan MR (2022) Genome-Wide Identification and Characterization of Receptor-Like Protein Kinase 1 (RPK1) Gene Family in Triticum aestivum Under Drought Stress. Front. Genet. 13:912251. doi: 10.3389/fgene.2022.912251
Received: 04 April 2022; Accepted: 24 May 2022;
Published: 04 July 2022.
Edited by:
Vijay Gahlaut, Institute of Himalayan Bioresource Technology (CSIR), IndiaReviewed by:
Shumayla, Panjab University, IndiaCopyright © 2022 Rahim, Uzair, Rehman, Rehman, Zahra and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Ramzan Khan, bXJraGFuQHBhcmMuZ292LnBr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.