- 1Centre for Research on Environmental Ecology and Fish Nutrition of Ministry of Agriculture and Rural Affairs, Shanghai Ocean University, Shanghai, China
- 2Key Laboratory of Marine Biotechnology of Jiangsu Province, Jiangsu Ocean University, Lianyungang, China
- 3Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
- 4National Demonstration Centre for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai, China
Ovarian development is a key physiological process that holds great significance in the reproduction of the Chinese mitten crab (Eriocheir sinensis), which is an economically important crab species for aquaculture. However, there is limited knowledge for the regulatory mechanisms of ovarian development. To study the molecular mechanisms of its ovarian development, transcriptome analysis was performed in the ovary and hepatopancreas of E. sinensis during ovarian stages I (oogonium proliferation), II (endogenous vitellogenesis), and III (exogenous vitellogenesis). The results showed that 5,520 and 226 genes were differentially expressed in the ovary and hepatopancreas, respectively. For KEGG enrichment analysis, the differentially expressed genes in the ovary were significantly clustered in phototransduction-fly, phagosome, and ECM-receptor interaction. Significantly enriched pathways in the hepatopancreas included fatty acid biosynthesis, fatty acid metabolism, and riboflavin metabolism. Further analysis showed that 25 genes and several pathways were mainly involved in oogenesis, including the ubiquitin-proteasome pathway, cyclic AMP-protein kinase A signaling pathway, and mitogen-activated protein kinase signaling pathway. Twenty-five candidate genes involved in vitellogenesis and endocrine regulation were identified, such as vitellogenin, vitellogenin receptor, estrogen sulfotransferase, ecdysone receptor, prostaglandin reductase 1, hematopoietic prostaglandin D synthase and juvenile hormone acid O-methyltransferase. Fifty-six genes related to nutritional metabolism were identified, such as fatty acid synthase, long-chain-fatty-acid-CoA ligase 4, 1-acyl-sn-glycerol-3-phosphate acyltransferase 4, fatty acid-binding protein, and glycerol-3-phosphate acyltransferase 1. These results highlight the genes involved in ovarian development and nutrition deposition, which enhance our understanding of the regulatory pathways and physiological processes of crustacean ovarian development.
Introduction
Ovarian development is a complex physiological process in crustacean reproduction (Meusy and Payen, 1988). Female crustaceans usually begin vitellogenesis and ovarian development after the puberty molt (Wang et al., 2014; Wu et al., 2017). Ovarian development can be divided into oogonium proliferation stage, endogenous vitellogenic stage, exogenous vitellogenic stage, and oocyte maturation stage (Wu et al., 2017). Previous studies have shown that the ovary and hepatopancreas are the primary vitellogenesis site in the endogenous and exogenous vitellogenic stages, respectively (Li et al., 2006). During crustacean ovarian development, large amounts of nutrients in the hepatopancreas would be transported to developing oocytes, especially vitellin (Vn) and lipids (Cheng et al., 1998). Previous studies have revealed that vitellogenin (Vg) synthesized in the hepatopancreas would be transported to oocytes by vitellogenin receptor (VgR) synthesized in the ovary (Bai et al., 2016; Guo et al., 2019). However, the physiological process of nutrient metabolism and deposition, including digestion, absorption, transportation, storage, anabolism, and catabolism of nutrients, are highly complicated during the ovarian maturation cycle of crustaceans (Vogt, 1994). To date, gene expression patterns related to nutritional metabolism during ovarian maturation remain unknown.
Various hormonal factors have been employed to positively control ovarian development in crustaceans (Subramoniam, 2000; Nagaraju, 2011). These hormonal factors originating from different endocrine organs are involved in the ovarian development of crustaceans, either individually or in synergy with one another. These organs mainly include the brain ganglion, eyestalk, Y-organ (YO), and mandibular organ (MO), which jointly regulate ovarian development of crustaceans by secreting hormones such as peptides, biogenic amines, steroid hormones, prostaglandins, and methyl farnesoate (MF) (Nagaraju, 2011). The main targets of these hormones are the ovaries and hepatopancreas. Endocrine hormones have complex regulatory pathways involved in ovarian development in crustaceans. For example, ecdysteroids can promote vitellogenesis of crustacean (Gong et al., 2015), whereas MF regulates the vitellin synthesis pathway (Homola and Chang, 1997). However, the regulatory mechanism underlying this phenomenon remains unclear.
The Chinese mitten crab Eriocheir sinensis is an economically important aquaculture species favored by consumers due to their delicacy (China Fisheries Yearbook, 2021). The status of ovarian development directly affects the quality and subsequent reproduction of adult E. sinensis and cannot be controlled artificially (Wu et al., 2017). Therefore, it is crucial to understand the mechanism of ovarian development in E. sinensis. The transcriptome can be used to mine gene expression patterns, obtain abundant genetic resources, and initially understand physiological processes. Advances in transcriptome technology have greatly improved our ability to analyze non-model species (Mardis, 2008). In the present study, changes in genes in the ovary and hepatopancreas were identified during the ovarian development of E. sinensis by transcriptomic analysis. The results of this study not only reveal the genes involved in ovarian development, nutrition transport and accumulation, but will also enhance the understanding of molecular regulation pathways and physiological processes of ovarian development in crustaceans.
Materials and Methods
Sample Collection
Between July 2015 and November 2015, healthy and intact female crabs (body weights of 70–135 g) were obtained monthly from the experimental ponds of the Chongming Research Base of Shanghai Ocean University. The crabs were maintained in aquarium tanks and acclimated for at least 7 days before sacrifice, as described previously (Wu et al., 2014). Based on the study of Wu et al. (2017), the ovarian development of E. sinensis was classified into five stages. Specifically, the stage I ovary was small and the dominant gametocyte were oogonia and previtellogenic oocytes. The dominant type of gametocytes in stage II ovary was endogenous vitellogenic oocytes. The dominant type of gametocytes in stage III ovary was exogenous vitellogenic oocyte. The dominant type of gametocytes in stage IV ovary was nearly mature oocytes. The stage V ovary was filled with mature oocytes (Table 1). Transcriptome analysis was performed on individuals at different ovarian stages, namely, the oogonium proliferation stage (Stage I), endogenous vitellogenic stage (Stage II), and exogenous vitellogenic stage (Stage III). Three individual crabs at each stage were sampled and anesthetized on ice, and then the ovaries and hepatopancreas were removed from the crabs for transcriptome analysis. Three biological replicates were performed per tissue at each stage for a total of nine crabs.

TABLE 1. The main features of the different development stages during the ovarian maturation of E. sinensis.
RNA Extraction and Transcriptomic Sequencing
Total RNA was isolated from each tissue using TRIzol reagent (Invitrogen, USA). RNA library construction and sequencing were performed as previously described (Xu et al., 2020). RNA purity, concentration, and integrity were determined using a NanoPhotometer spectrophotometer (IMPLEN, CA, USA), Qubit 2.0 Fluorometer (Life Technologies, CA, USA), and Agilent 2,100 Bioanalyzer (Agilent Technologies, CA, USA), respectively. Next, the NEBNext Ultra RNA Library Prep Kit for Illumina platform was used to generate sequencing libraries according to the manufacturer’s instructions. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads, and then mRNA was randomly fragmented in fragmentation buffer. The mRNA fragments were used as templates to synthesize double-stranded cDNA, which was further purified with the AMPure XP system (Beckman Coulter, Beverly, USA). Next, the ends of the double-stranded cDNA fragments were repaired using poly (A) and adapter ligation. The cDNA fragment size was selected using the AMPure XP system (Beckman Coulter, Beverly, USA) to obtain fragments preferentially 250–300 bp in length. Sequencing libraries were created after PCR amplification and sequenced using an Illumina HiSeq2500 platform.
Transcriptome Assembly
The original image data were transferred to raw sequence data and saved as FASTQ files. Clean reads were obtained by removing those with adapters, ploy-N reads, and low-quality reads from the raw sequence data. All subsequent analyzes were based on high-quality clean data. Clean reads were aligned to the reference genome assembly using HISAT v2.0.4 program. HTSeq v0.6.1 program was used to count the read numbers mapped to each gene. Finally, gene expression levels were normalized as FPKM (expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced). Raw data were uploaded to the National Center for Biotechnology Information (NCBI) (accession number is PRJNA660118).
Differential Expression Analysis and Functional Annotation
Differential expression analysis was performed using the DESeq package (1.10.1) (Anders and Huber, 2010). The resulting p-values were adjusted using Benjamini and Hochberg corrections to control the false discovery rate. Genes with an adjusted p-value (Padj) < 0.05 were assigned as differentially expressed.
In this study, GO enrichment analysis of differentially expressed genes (DEGs) was performed using GOseq (Young et al., 2010). GO terms with corrected p-value < 0.05 were considered significantly enriched by DEGs. The KEGG database was used to further interpret the functions of the genes. The statistical enrichment of DEGs in KEGG pathways was performed using KOBAS software (Mao et al., 2005).
Validation of DEGs by Quantitative Real-Time PCR
Eight genes involved in ovarian development and triacylglycerol (TAG) metabolism were randomly selected for validation using quantitative real-time PCR (qRT-PCR). A reverse first strand cDNA synthesis kit (RR036A, Takara Bio, Japan) was used to synthesize first-strand cDNA. Primer pairs were designed using Primer 6.0, and all the primer sequences are listed in Supplementary Table S1. β-actin was used as an internal control to normalize target gene expression. qRT-PCR was performed in a FAST-7500 system (ABI-7500, Thermo Fisher, Singapore) using a TB-green ® Premix Ex TaqTM II kit (RR420A, Takara Bio, Japan). The reaction system and PCR protocols were performed as previously described (Pan et al., 2018). Each ovarian stage had three replicate crabs for each gene, and the relative expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The qRT-PCR results were compared to the FPKM value of each stage in RNA-seq for RNA-Seq validation.
Statistical Analysis
Statistical analyzes were performed using SPSS Statistics V22.0 (IBM Corporation, NY, USA). The data are presented as the mean for each ovarian stage, and homogeneity of data variance was evaluated by Levene’s test. The Student’s t-test was used to determine significant differences between the two ovarian stages. When homogeneity of variances was not achieved, the data were subjected to Welch’s t-test with Bonferroni correction. Statistical significance was set at p < 0.05.
Result
Sequencing and De Novo Assembly
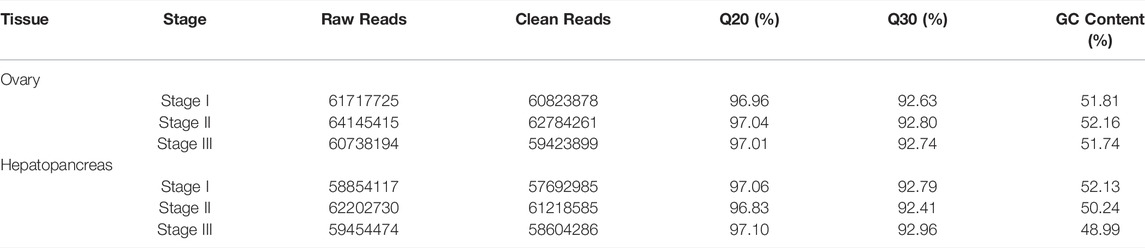
After adaptor sequences and low-quality reads were removed, a total of 360 million clean reads were obtained from the three stages of the E. sinensis transcriptome, including 60,823,878 (Stage I), 62,784,261 (Stage II), and 59,423,899 (Stage III) reads from the ovary libraries; and 57,692,985 (Stage I), 61,218,585 (Stage II), and 58,604,286 (Stage III) reads from the hepatopancreas libraries (Table 2).
Identification of DEGs
In this study, the analysis of ovary transcriptome data revealed that 5,520 DEGs were annotated successfully across the three stages, of which 209 DEGs were annotated in all stages (Figure 1A). By comparing the databases of stages I and II, 1,872 DEGs were identified, of which 972 and 900 DEGs were upregulated and downregulated, respectively. The comparison results between stages I and III showed that 4,930 DEGs were identified, of which 2,419 and 2,511 DEGs were upregulated and downregulated, respectively. Moreover, the comparison results between stages II and III identified 532 upregulated and 450 downregulated genes (Figure 2A).
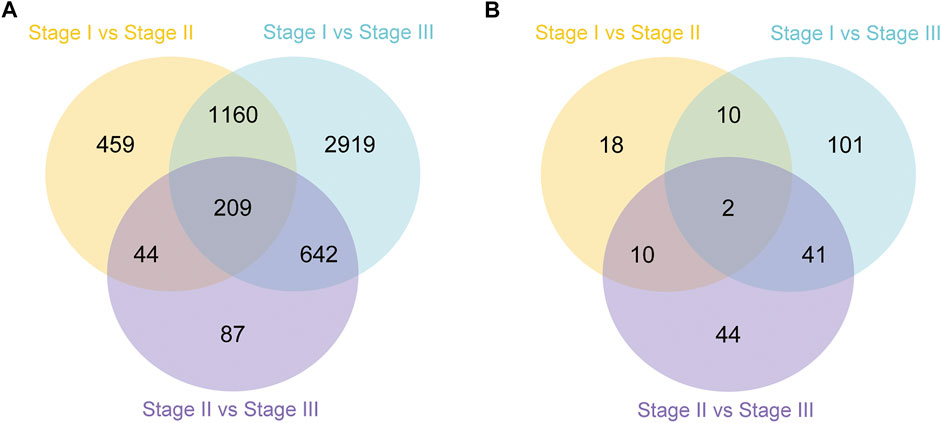
FIGURE 1. Summary of differentially expressed genes (DEGs) in transcriptome of ovary (A) and hepatopancreas (B) from different ovarian development stage of E. sinensis.
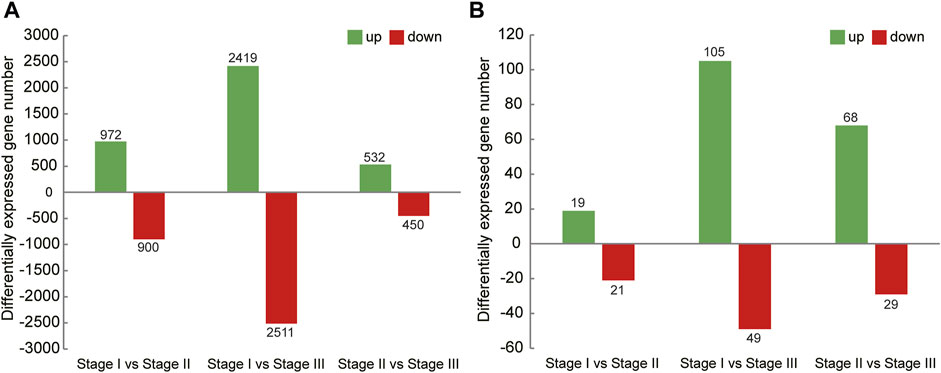
FIGURE 2. Summary of upregulated and downregulated genes in transcriptome of ovary (A) and hepatopancreas (B) from different ovarian development stage of E. sinensis.
A total of 226 DEGs were annotated successfully in the hepatopancreas transcriptome data, of which only two DEGs were annotated in all stages (Figure 1B). We found that the DEGs between stages I and II were relatively few, with only 40 genes identified, of which 19 were upregulated and 21 were downregulated. A total of 154 DEGs were identified between stages I and III, of which 105 and 49 genes were upregulated and downregulated, respectively. Moreover, 97 DEGs were detected between stages II and III, of which 68 and 29 genes were upregulated and downregulated, respectively (Figure 2B).
Figures Functional Classification of DEGs
GO assignments were performed to analyse the functions of these DEGs. These transcripts were assigned to one or more GO terms. In the ovary, GO annotation of the DEGs between the stages I and II showed that both DEGs were categorized mainly into translation and peptide biosynthetic process for biological processes, ribosome for cellular components, and structural constituent of ribosome for molecular functions (Supplementary Figure S1). When stage I and stage III were compared, DEGs were categorized mainly into intracellular signal transduction for biological processes and nucleoside binding for molecular functions (Supplementary Figure S2). Compared to stages II and III, organic acid transport was the most abundant term in biological processes. Cell cortex and sodium:dicarboxylate symporter activity were the top terms in the cellular components and molecular functions, respectively (Supplementary Figure S3). In the hepatopancreas, compared to stages I and II, DEGs were categorized mainly into alcohol metabolic process for biological processes, proteinaceous extracellular matrix for cellular components, and alcohol O-acetyltransferase activity for molecular functions (Supplementary Figure S4). When stage I and stage III were compared, the DEGs were categorized mainly into lipid transport for biological processes, and lipid transporter activity for molecular functions (Supplementary Figure S5). The GO result of stages II and III was similar with that between stages I and III (Supplementary Figure S6).
To further analyze the possible pathways involved in ovarian development and nutritional metabolism, KEGG analysis was performed based on the transcriptome results. The top 16 most enriched metabolic pathways in the ovary and hepatopancreas are shown in Table 3. In the ovary, the top 3 most enriched pathways included phototransduction-fly, phagosome and ECM-receptor interaction. Furthermore, there are many biosynthesis and metabolism enriched pathway of amino acids were obtained in ovary, such as cysteine and methionine metabolism, beta-Alanine metabolism, and phenylalanine metabolism. In the hepatopancreas, DEGs between the stages I and II were significantly clustered in fatty acid biosynthesis and fatty acid metabolism. On comparing stages I and III, riboflavin metabolism, tyrosine metabolism, and metabolic pathways were the most enriched pathways, which were similar results between stages II and III.
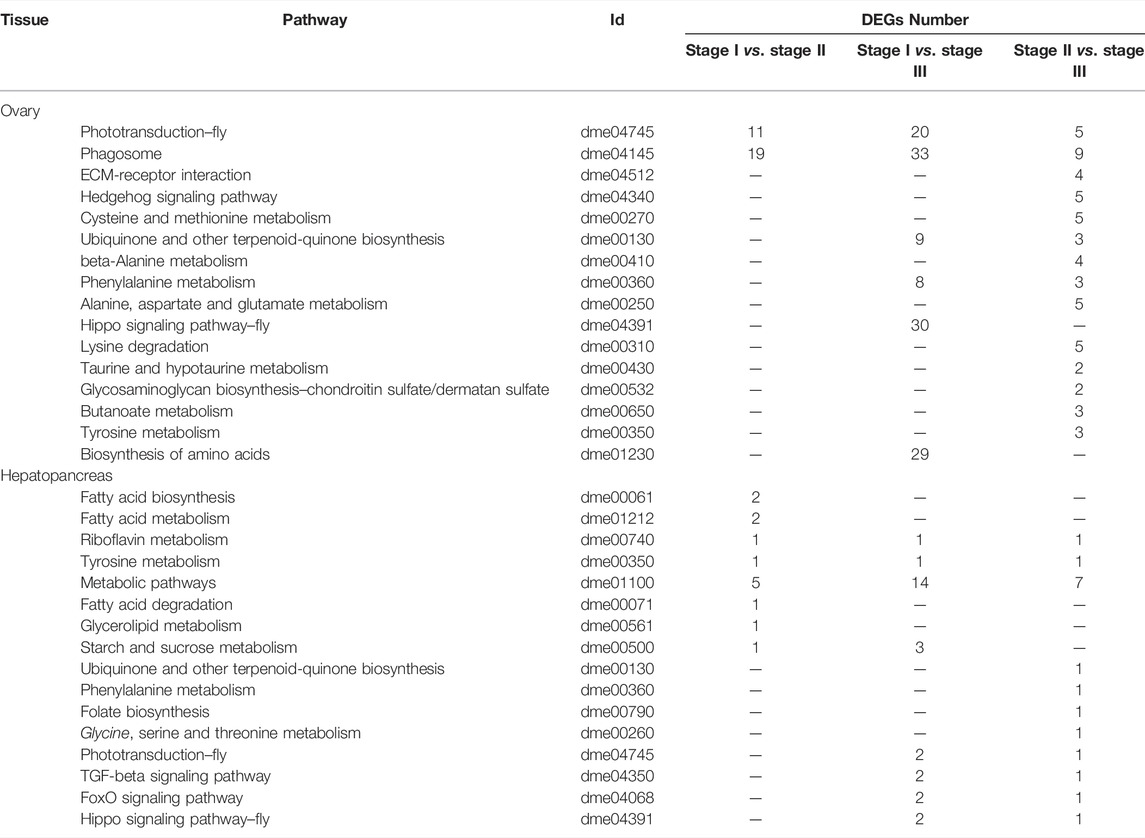
TABLE 3. Most enriched pathways obtained with KEGG, using the differentially expressed genes from ovary and hepatopancreas of female E. sinensis.
Key Genes Involved in Ovarian Development and Nutrition Metabolism
The genes involved in various processes of ovarian development in the ovary and hepatopancreas are shown in Table 4. Twenty-five key genes were mainly involved in oogenesis, including the ubiquitin-proteasome pathway (UPP), cyclic AMP-protein kinase A (cAMP-PKA) signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway. There were four candidate genes for vitellogenesis in the ovary, including Vg, VgR, vitelline membrane outer layer protein 1 homolog (VM O 1) and bone morphogenetic protein 3, of which Vg and VM O 1 were also present in the hepatopancreas. In the ovary, Vg expression was downregulated in stages I to II but upregulated in stages II to III. The expression level of VgR was the highest in stage II. However, the expression level of Vg was upregulated in stages I to III in the hepatopancreas. Nineteen key genes were mainly involved in the process of ovarian development, modulated by a variety of hormones in the ovary and hepatopancreas. Specifically, four genes encode steroid hormones, including estrogen sulfotransferase (EST), ecdysone receptor (EcR), ecdysteroid-regulated 16 kDa protein and estradiol 17-beta-dehydrogenase 8. Three genes belong to the MF pathway, including 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR), juvenile hormone acid O-methyltransferase (JHAMT), and mevalonate kinase (MevK). The expression levels of EST and EcR were the highest in ovarian stage III. However, its expression level was the highest in the stage II in the hepatopancreas. The expression levels of estradiol 17-beta-dehydrogenase 8 and ecdysteroid-regulated 16 kDa protein were upregulated from stage I to stage III in the ovary and hepatopancreas. The expression level of prostaglandin reductase 1 was the highest in stage III in the ovary and hepatopancreas. The expression level of hematopoietic prostaglandin D synthase (H-PGDS) was the highest in the stage III ovary. The expression level of prostaglandin G/H synthase 2 was the highest in stage II in the hepatopancreas.
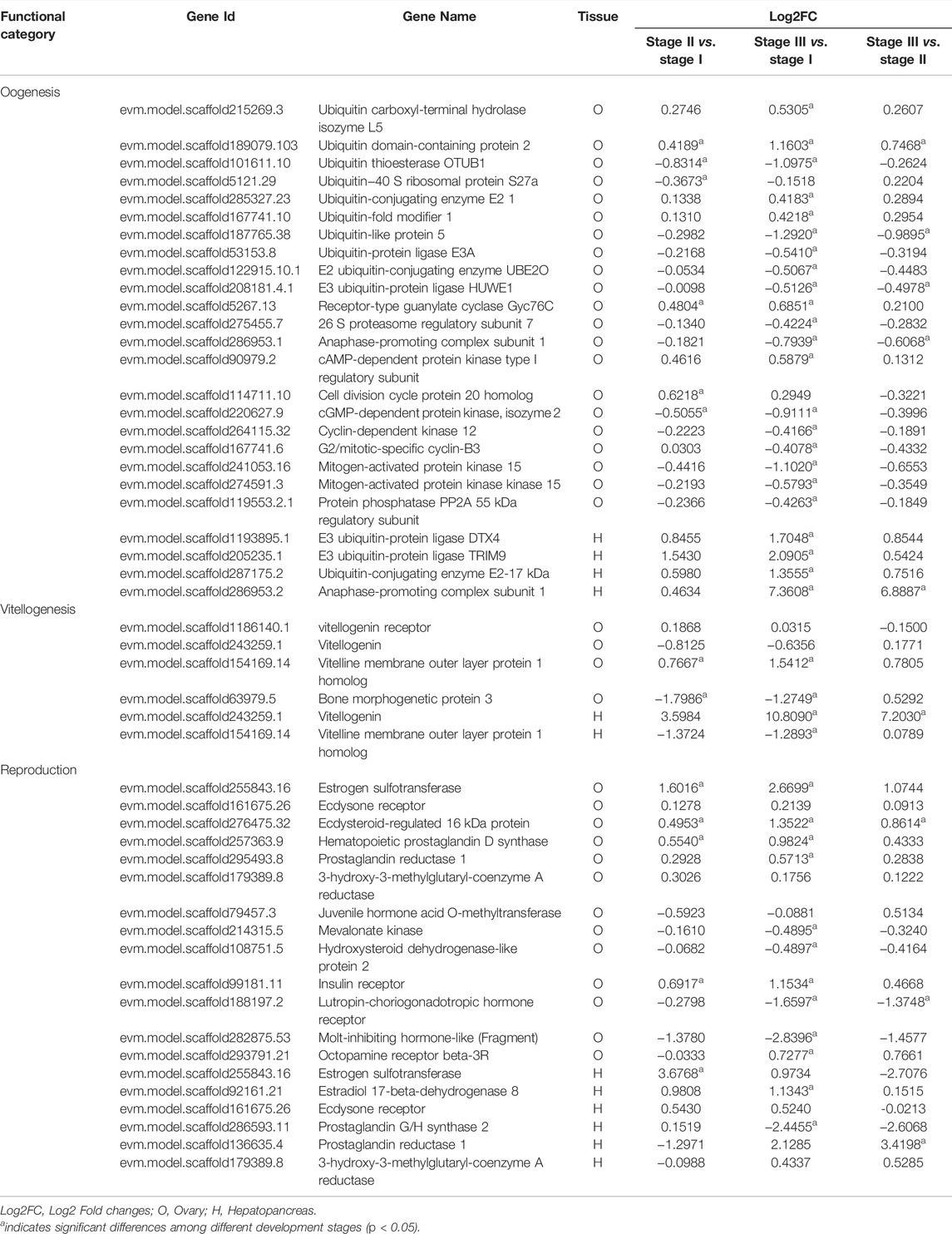
TABLE 4. Summary of differential and non-differentially expressed genes related to ovarian development in transcriptome of female E. sinensis.
The genes related to nutrition metabolism in the ovary and hepatopancreas are listed in Table 5. There were fifty-six genes related to lipid metabolism in the ovaries and hepatopancreas. Twenty-eight genes involved in lipid catabolism and anabolism were identified in the ovary and hepatopancreas. Among them, 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 (AGPAT4), AGPAT2, AGPAT3, glycerol-3-phosphate acyltransferase 1 (GPAT1), GPAT3, GPAT4 and diacylglycerol O-acyltransferase 1 (DGAT1) are associated with TAG anabolism. Eight genes involved in lipid oxidation and decomposition were identified in the ovary and hepatopancreas, including acyl-CoA synthetase family member 2 (ACSF2), acyl-CoA-binding protein, fatty acid-binding protein (FABP) and FABP homolog 9. Twenty genes involved in lipid digestion and transportation were identified in the ovary and hepatopancreas. Specifically, in the ovary, eight genes were related to lipid digestion, including phospholipase, monoacylglycerol lipase (MGL), hormone-sensitive lipase (HSL) and pancreatic triacylglycerol lipase. Three genes were related to lipid transportation were identified: apolipoprotein D, very low-density lipoprotein receptor and low-density lipoprotein receptor 1. In the hepatopancreas, seven genes were related to lipid digestion, such as pancreatic lipase-related protein 2, phospholipase, MGL and HSL. Moreover, the expression levels of microsomal triglyceride transfer protein (MTP) large subunit and low-density lipoprotein receptor-related protein 1 (LRP1), which were related to lipid transportation, were upregulated from stage I to stage III in the hepatopancreas.
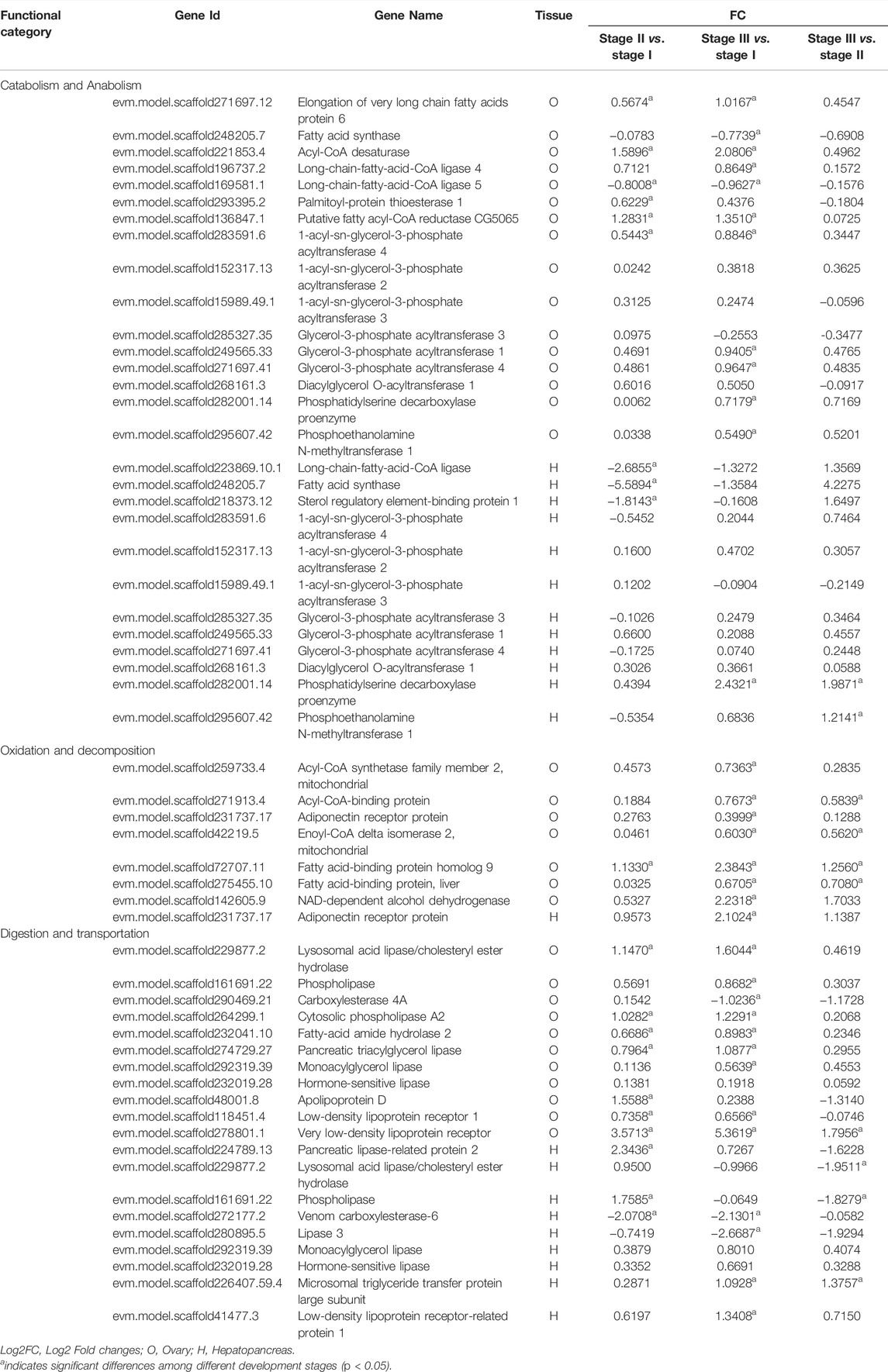
TABLE 5. Summary of differential and non-differentially expressed genes related to nutrition metabolism in transcriptome of female E. sinensis.
Validation of Gene Expression by Quantitative Real-Time PCR
Eight selected genes involved in ovarian development (Figure 3A) and TAG metabolism (Figure 3B) were determined using qRT-PCR to validate the RNA-seq results. The relative expression patterns of these seven genes were consistent with the RNA-seq results. Notably, the qRT-PCR results showed that the expression level of VgR was highest in stage III. However, the RNA-seq results showed that the expression level of VgR was highest in stage II.
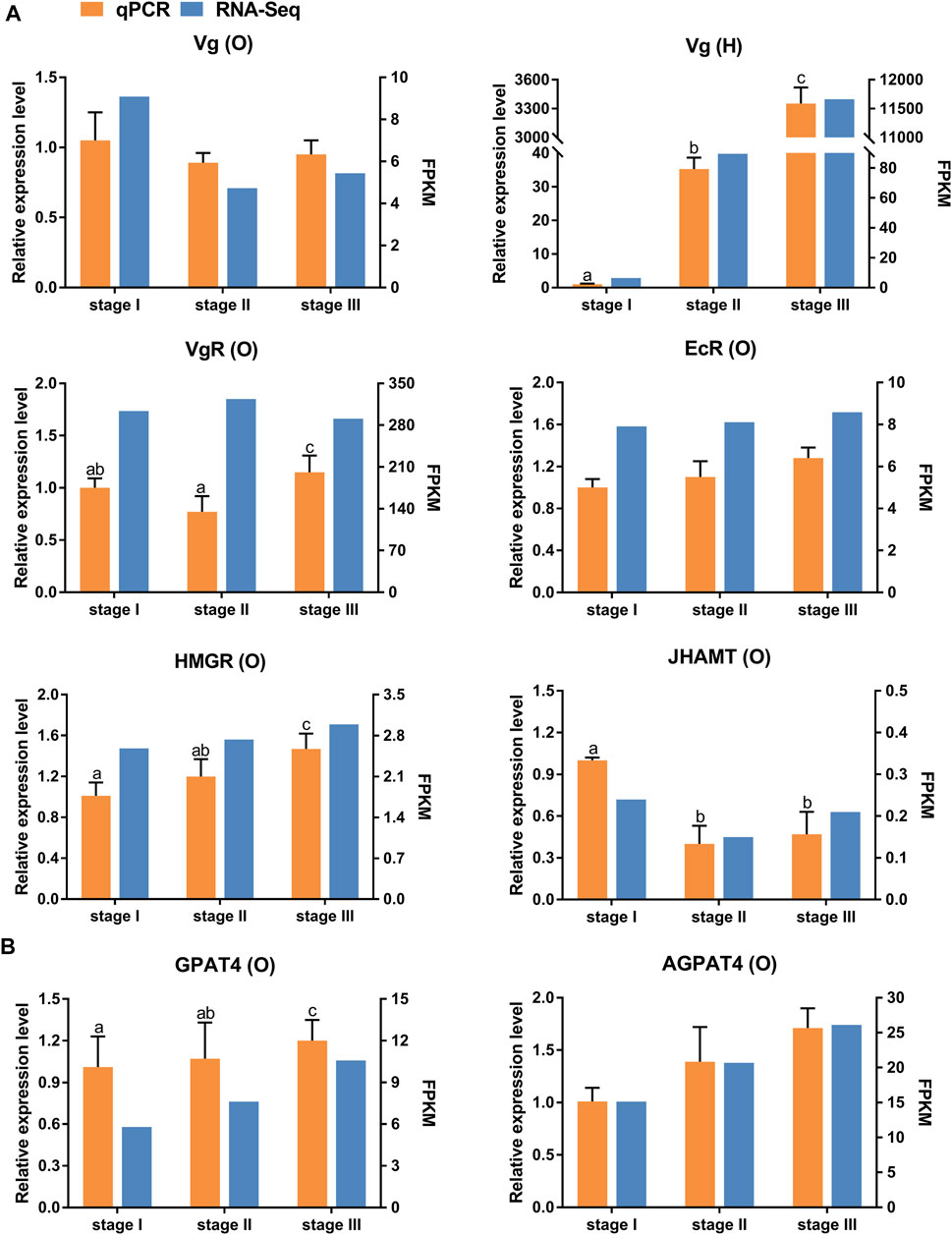
FIGURE 3. Quantitative expression of eight genes were determined by qPCR in ovary and hepatopancreas from different ovarian development stage of E. sinensis. (A) genes involved in ovary development. (B) genes involved in triacylglycerol metabolism. Vg: vitellogenin; VgR: vitellogenin receptor; EcR: ecdysone receptor; HMGR: 3-hydroxy-3-methylglutaryl-coenzyme A reductase; JHAMT: juvenile hormone acid O-methyltransferase; AGPAT4: 1-acyl-sn-glycerol-3-phosphate acyltransferase 4; GPAT4: glycerol-3-phosphate acyltransferase 4. O: ovary; H: hepatopancreas. Different letters on the top of each bar indicate significant differences between the different ovarian stages (p < 0.05).
Discussion
Ovarian development in crustaceans is regulated by several organs and hormones. During the ovarian development of crustacean, protein is involved in many synthetic reactions, such as synthesis of enzymes, hormones and yolk (Wouters et al., 2001). Protein is the first important nutrient for animal growth and reproduction, amino acids are its basic components. Ovarian GO annotation between stages I and II showed the translation and peptide biosynthetic process was main biological processes. The KEGG analysis of the ovary also revealed that many pathways for amino acid synthesis and metabolism were obtained in the ovary, which indicates that the reproduction and growth process in the ovary of this crab need the involvement of proteins and amino acids. In addition, the result of GO annotation showed that lipid transport was the most abundant GO term in the biological processes in the hepatopancreas. And KEGG analysis of the hepatopancreas also showed that DEGs were clustered in the metabolic pathways of various nutrients. This is consistent with role of the hepatopancreas as an absorption and storage organ. As the primary organ of reproductive and nutritional metabolism, the ovary and hepatopancreas play crucial roles during ovarian development (Cheng et al., 1997; Jiang et al., 2009; Nagaraju, 2011; Wang et al., 2018). Furthermore, the conversion from endogenous to exogenous vitellogenesis in crustaceans might be related to hormone regulation (Subramoniam, 2000). Thus, we identified changes in gene expression levels in the ovary and hepatopancreas using transcriptomic analysis during the ovarian development of E. sinensis. Both GO and KEGG analyses will facilitate the annotation of these unigenes and investigations of gene function in future studies. These data will provide information to further investigate the molecular regulatory mechanisms involved in oogenesis, vitellogenesis, and nutritional metabolism and transportation in crustaceans.
Key Genes Related to Ovarian Development
Ovarian formation and maturation are complex processes that require the cooperation of a series of functional pathways (Yang et al., 2015). Oogenesis and vitellogenesis are two finely regulated processes during ovarian maturation in crustaceans (Tsukimura B, 2001). Both mitosis and meiosis are essential for oogenesis. UPP is involved in oocyte meiotic maturation as a key intracellular protein degradation system in the cell cycle (Bebington et al., 2001). In crustaceans, Ubiquitin C-terminal hydrolases L3 (UCHL3), UCHL5, ubiquitin ribosomal protein S27 (UbS27), ubiquitin ribosomal protein L40, and UPP system-related factors participate in the regulation of oogenesis (Han et al., 2010; Wang et al., 2012; Han et al., 2018). In this study, we detected many genes in the pathway that changed in the ovary and hepatopancreas during ovarian development, such as UbS27a, ubiquitin-conjugating enzyme, ubiquitin carboxyl-terminal hydrolase isozyme L5, and ubiquitin-protein ligase. Thus, UPP may be involved in oocyte meiosis in E. sinensis. Moreover, previous studies have revealed that the cAMP-PKA signaling pathway could regulate the meiosis process of oocytes by activating downstream genes, such as cyclin-dependent kinase 1 (CDK1), cyclin B, anaphase-promoting complex/cyclosome (APC/C) and maturation promoting factor (MPF) (Feng and Thompson, 2018; Firmani et al., 2018). APC/C combines with the cell division cycle protein 20 homolog 1 (Cdh1) and cell division cycle protein 20, which play a vital role in oocyte first meiotic prophase resumption (Marangos et al., 2007). We detected the expression levels of Cdh1 and anaphase-promoting complex subunit 1 were highest in stage II, which indicated that they were related to the resumption of the first meiotic prophase. The 26 S proteasome hydrolyses cyclin B to maintain the inactive state of MPF, which blocks the oocyte in the diplotene stage of the first meiosis prophase (King et al., 1996; Zachariae and Nasmyth, 1999). In this study, the expression level of 26 S proteasome regulatory subunit 7 was highest in stage 1, which proved that it was related to oocyte first meiotic arrest. In addition, previous studies have indicated that the MAPK signaling pathway participates in the regulation of oocyte meiosis (Gao et al., 2014). In this study, the expression levels of MAPK 15, mitogen-activated protein kinase kinase 15, and protein phosphatase PP2A (55 kDa regulatory subunit) belonging to the MAPK signaling pathway were highest in stage I of the ovary, which demonstrated that the MAPK signaling pathway is required in E. sinensis to promote oocyte meiosis.
Vitellogenesis is the process of yolk formation via the accumulation Vn and nutrients in oocytes (Chen et al., 2018). Abundant expression of Vg, a Vn precursor, is an indicator of the onset of gonadal maturation in female crustaceans (Tsutsui et al., 2004; Arcos et al., 2011). In crustaceans, the major Vg synthesis site is the ovary in endogenous vitellogenic stage, and the major site of Vg synthesis is the hepatopancreas in exogenous vitellogenic stage (Li et al., 2006). Our study showed that the expression level of ovarian Vg was lower than that in the hepatopancreas from stage II to stage III, which indicated that the primary site of Vg synthesis changed from the ovary to the hepatopancreas at stage III. In this study, the expression level of Vg dramatically increased from stage I to stage III in the hepatopancreas, further supporting the above hypothesis. Similar results have also been reported for other crabs (Zmora et al., 2007; Jia et al., 2013). Moreover, Vg synthesized in the hepatopancreas is immediately transported into developing oocytes by VgR synthesized in the ovary to achieve quick yolk accumulation in oocytes (Bai et al., 2016; Guo et al., 2019). In the present study, the expression level of VgR was highest in the stage II transcriptome data. However, the qRT-PCR results showed that the expression level of VgR was highest in stage III. Studies by other scholars have suggested that the expression level of VgR in stage III was higher than that in stage II during different ovarian developmental stages in E. sinensis (Pan et al., 2018; Sun et al., 2020). Therefore, we speculated that this is because the sample of stage III used for transcriptome sequencing was in early-stage III, resulting in not reaching the highest value of VgR-mRNA.
The development of crustacean ovaries is controlled by various hormones (Nagaraju, 2011). Estrogen is an important steroid hormone in most crustacean and promotes vitellogenesis and ovarian development (Rodrigue et al., 2002). Previous studies have shown that EST is responsible for the sulfonation and inactivation of estrogens, and plays a crucial role in estrogen homeostasis (Gao et al., 2012). In our study, the EST expression level was positively correlated with ovarian stages, indicating that E. sinensis may have a self-regulation mechanism to maintain estrogen homeostasis. High levels of ecdysteroids can induce high levels of EcR expression during the ovarian maturation of crustaceans, thereby initiating the ecdysone signaling pathway to be involved in reproduction (Okumura et al., 1992; Subramoniam, 2000). EcR may be involved in vitellogenesis by regulating Vg synthesis (Girish et al., 2015; Gong et al., 2015; Girish et al., 2018). Su et al. (2020) reported that four EsEcR isoforms were detected in the ovaries and hepatopancreas in E. sinensis. We obtained EsEcR1 via NCBI BLAST analysis in our study. The EsEcR1 expression level was concurrently upregulated in the ovary during ovarian development, suggesting that EsEcR1 may participate in vitellogenesis. MF is a sesquiterpenoid hormone that mediates Vg synthesis in crustacean and is produced by the crustacean MO (Rodriguez et al., 2002; Nagaraju, 2007; Miyakawa et al., 2014). The biosynthesis of crustacean MF is similar to that of insect JH. HMGR is a rate-limiting enzyme in the early steps of JH and MF biosynthesis and is responsible for mevalonic acid biosynthesis (Bellés et al., 2005; Li et al., 2010). The HMGR expression was specific to MO (Li et al., 2003; Qiu et al., 2014). In crustaceans, methyl esterification in the final steps of MF biosynthesis is thought to be catalyzed by farnesoic acid O-methyltransferase, and this step is catalyzed by JHAMT in insects (Gunawardene et al., 2002; Holford et al., 2004; Hui et al., 2008; Li et al., 2013). JHAMT plays an essential role in the methyl esterification of fatty acids to MF in crustaceans and is highly specific to MO (Xie et al., 2016). In this study, HMGR and JHAMT showed low expression levels in the ovary and hepatopancreas. We speculated that the two genes, both highly specific to MO, were difficult to detect in the ovary and hepatopancreas. Furthermore, prostaglandins (PGs) play essential roles in reproductive functions, such as the release of hatching factors and ovarian maturation in crustaceans (Sagi et al., 1995; Reddy et al., 2004). Among them, prostaglandin D2 was synthesized by prostaglandin D synthase (PGDS) (Narumiya et al., 1999). In this study, the expression level of H-PGDS was upregulated in the ovary, indicating that it is related to ovarian maturation.
Key Genes Related to Nutrition Metabolism
Lipids play an important role in the biochemical metabolism and reproduction of decapod crustaceans (Cheng et al., 1997). As a neutral lipid, TAG is the major energy lipid that supports energy needs for reproduction (Yao et al., 2008; Pasquevich et al., 2011). TAG can be synthesized in the ovaries or be transported to the developing ovaries from the hepatopancreas (Alava et al., 2007). Glycerol-3-phosphate acyltransferase (GPAT) is a key enzyme that catalyzes the first step in TAG synthesis (Kennedy, 1961). Four subtypes of GPAT with different functions have been identified in mammals: GPAT1 and GPAT2 participate in TAG synthesis, and GPAT3 is involved in adipocyte differentiation, while GPAT4 plays a pivotal role in mammary lipid droplet (Chen et al., 2008; Gimeno and Cao, 2008; Cattaneo et al., 2012). Zhu et al. (2019) found that two EsGPATs subtypes (EsGPAT1 and EsGPAT2), which are more similar to mammalian GPAT3 and GPAT4, exist in E. sinensis. In our study, high expression levels of GPAT1 were found in ovary and hepatopancreas, suggesting that it could participate in TAG synthesis in ovary and hepatopancreas. Although, the expression levels of GPAT3 and GPAT4 increased to varying degrees during the ovarian development of E. sinensis, its specific function in lipid metabolism requires further investigation. 1-acyl-sn-glycerol-3-phosphate acyltransferase (AGPAT) enzymes catalyze the second acylation step TAG synthesis (Kennedy, 1961). In mammals, several subtypes of AGPAT have exhibited different substrate preferences and physiological functions (Takeuchi and Reue, 2009). In this study, the expression levels of AGPAT2 and AGPAT4 were highest in stage III in the ovary and hepatopancreas, whereas the highest expression of AGPAT3 was found in stage II in the ovary and hepatopancreas. It has been speculated that AGPATs may play different roles during oocyte maturation in E. sinensis. Diacylglycerol acyltransferase (DGAT) enzymes mediate the last committed step in TAG synthesis by converting diacylglycerol (DAG) to TAG (Harris et al., 2011). In mammals, two DGAT enzymes (DGAT1 and DGAT2) belong to two separate protein families (Coleman and Lee, 2004). In this study, the expression level of ovarian DGAT1 decreased from stage II to III; however, it increased from stage II to stage III in the hepatopancreas. We speculate that TAGs were mainly synthesized in the hepatopancreas during exogenous vitellogenesis, resulting in decreased ovarian TAG synthesis.
For fatty acid synthesis, fatty acid synthase (FAS) is the key enzyme to endogenous fatty acid synthesis (Chirala and Wakil, 2004). Elongation of very long chain fatty acids (Elovl) is responsible for chain elongation to produce long-chain fatty acids (LCFAs) (Wu et al., 2010). Similar to the study of Sun et al. (2014), the transcription levels of hepatopancreatic FAS increased from stage II to III during the ovarian development of E. sinensis. These results confirmed that FAS is involved in fatty acid synthesis for hepatopancreas-mediated lipid transport during the ovarian development of E. sinensis. In our study, Elovl6 was upregulated in the ovary, demonstrating that Elovl6 might synthesise LCFAs in the ovary to meet ovarian nutritional requirements. Furthermore, fatty acid-CoA ligase converts free fatty acids into fatty acyl-CoA esters (Cao et al., 2000). The long-chain-fatty-acid-CoA ligase 4 (FACL4) preferentially controls the level of free arachidonic acid (ARA) (Cao et al., 2000). ARA is a precursor of eicosanoids, which plays vital roles in reproduction (Harrison, 1990; Masoudi Asil et al., 2017). In this study, the expression level of ovarian FACL4 continuously increased from stage I to III, suggesting that it participates in the conversion of ARA in E. sinensis. Four genes related to phosphatide metabolism were identified in this study. These results highlight the importance of lipid metabolism during ovarian development in E. sinensis.
For lipid oxidation and decomposition, ACSF2 was identified in the ovary of E. sinensis. Acyl-CoA synthetase (ACS) would specifically activate fatty acids, essential for β-oxidation (Xu et al., 2012; Gondim et al., 2018). ACSF2 belongs to the ACS family, activating fatty acids by forming a thioester bond with CoA (Yu et al., 2016). In this study, the expression level of ACSF2 was upregulated from stage I to stage III in the ovary, suggesting that it was related to fatty acids β-oxidation, thereby providing energy during the ovarian development of E. sinensis. Furthermore, FABPs transport fatty acids from the cells to various organelles or nuclei for β-oxidation or lipid synthesis (Kaikaus et al., 1990; Jordal et al., 2006). Gong et al. (2010) reported that ovarian Es-FABP expression increases with ovarian development in E. sinensis to meet the nutritional requirements of ovarian lipids. Coincidentally, the expression level of FABPs was also increased in ovarian stage I to III in this study, which further indicated that FABPs participate in fatty acid transport during ovarian development of E. sinensis.
As mentioned in this study, absorbed nutrients are transported to the ovary from the hepatopancreas during the ovarian development of crustacean (Jiang et al., 2009). In vertebrates, the apolipoprotein (APO) family transports plasma TAGs and cholesterol (Shelness and Ledford, 2005; Wu et al., 2013). MTP transfers intracellular lipid molecules to the APO family, forming lipoprotein particles such as very low-density lipoproteins (Hussain et al., 2003). Lipoprotein receptors can mediate the endocytosis of lipoproteins (Van Hoof et al., 2005). The transfer of APO provides yolk protein precursors and lipid sources to oocytes in the mud crab Scylla paramamosain (Zeng et al., 2021). In the present study, the expression level of apolipoprotein D increased from stage I to II in the ovary, suggesting that it might be involved in lipid transportation to facilitate oocyte development. The expression levels of the MTP large subunit and LRP1 consecutively increased from stage I to III in the hepatopancreas. It was speculated that MTP and LRP1 are probably involved in lipid transportation during nutrients production and/or transferred to the oocytes in E. sinensis. Lipids, mainly TAG, are taken up and packed into lipid droplets stored in oocytes during ovarian maturation in decapod species (Lee and Walker, 1995). HSL and MGL are intracellular lipases that play a principal role in the hydrolysis of TAG in crustaceans (Miled et al., 2000; Rivera-Perez and Garcia-Carreno, 2011). Specifically, HSL is involved in the hydrolysis of TAG and DAG (Lafontan and Langin, 2009), while MGL hydrolyses monoacylglycerol (MAG) to free fatty acids (Karlsson et al., 1997). In this study, the expression levels of HSL and MGL were increased from stage I to III in the ovary and hepatopancreas, which indicated that they play an important role in mobilizing energy reserves to promote ovarian development of E. sinensis.
Conclusion
In this study, we performed a comparative transcriptome analysis of the ovaries and hepatopancreas during three vital ovarian stages of E. sinensis. The results identified 56 and 43 genes involved in ovarian development and nutritional metabolism, respectively, which are mainly related to the ubiquitin-proteasome pathway, cAMP-PKA signaling pathway, MAPK signaling pathway, triacylglycerol metabolism, and lipid oxidation and transportation. This study will provide new insights into the mechanisms of oogenesis, vitellogenesis, nutritional metabolism and transportation in crustaceans. In the future, the role of these genes and signaling pathways need to be elucidated during the process of crustacean ovarian development and nutritional metabolism.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XW contributed to the conception and design of the study. QF performed the experiments and wrote the drafting of the manuscript. QF, ML, and XW analyzed the data and interpreted the results. ML and XW reviewed and edited the manuscript. YC and XW contributed to the project administration. All authors approved the manuscript.
Funding
This work was supported by two general projects (grant numbers 31572630 and 41876190) from the National Natural Science Foundation of China and a Key R&D Program (grant number 2018YFD0900100) from the Ministry of Science and Technology of China. Infrastructure costs were partially supported by the Shanghai Talent Development Fund for Young Scientists (grant number 2018100) from Shanghai Municipal Human Resources and Social Security Bureau and Special Fund (grant number CARS-48) of the Chinese Agriculture Research System from the Ministry of Agriculture and Rural Affairs of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Dr. Xiaowen Long, Pengsheng Dong and Zehua Liu for their help with crab culture and sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.910682/full#supplementary-material
References
Alava, V. R., Quinitio, E. T., De Pedro, J. B., Priolo, F. M. P., Orozco, Z. G. A., and Wille, M. (2007). Lipids and Fatty Acids in Wild and Pond-Reared Mud Crab Scylla serrata (Forsskål) during Ovarian Maturation and Spawning. Aquac. Res. 38, 1468–1477. doi:10.1111/j.1365-2109.2007.01793.x
Anders, S., and Huber, W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11, R106. doi:10.1186/gb-2010-11-10-r106
Arcos, F. G., Ibarra, A. M., and Racotta, L. S. (2011). Vitellogenin in Hemolymph Predicts Gonad Maturity in Adult Female Litopenaeus (Penaeus) Vannamei Shrimp. Aquaculture 316, 93–98. doi:10.1016/j.aquaculture.2011.02.045
Bai, H., Qiao, H., Li, F., Fu, H., Jiang, S., Zhang, W., et al. (2016). Molecular and Functional Characterization of the Vitellogenin Receptor in Oriental River Prawn, Macrobrachium Nipponense. Comp. Biochem. Physiology Part A Mol. Integr. Physiology 194, 45–55. doi:10.1016/j.cbpa.2015.12.008
Bebington, C., Doherty, F. J., and Fleming, S. D. (2001). The Possible Biological and Reproductive Functions of Ubiquitin. Hum. Reprod. Update 7, 102–111. doi:10.1093/humupd/7.1.102
Bellés, X., Martín, D., and Piulachs, M.-D. (2005). The Mevalonate Pathway and the Synthesis of Juvenile Hormone in Insects. Annu. Rev. Entomol. 50, 181–199. doi:10.1146/annurev.ento.50.071803.130356
Cao, Y., Murphy, K. J., McIntyre, T. M., Zimmerman, G. A., and Prescott, S. M. (2000). Expression of Fatty Acid-CoA Ligase 4 during Development and in Brain. FEBS Lett. 467, 263–267. doi:10.1016/S0014-5793(00)01159-5
Cattaneo, E. R., Pellon-Maison, M., Rabassa, M. E., Lacunza, E., Coleman, R. A., and Gonzalez-Baro, M. R. (2012). Glycerol-3-Phosphate Acyltransferase-2 Is Expressed in Spermatic Germ Cells and Incorporates Arachidonic Acid into Triacylglycerols. PLoS One 7, e42986. doi:10.1371/journal.pone.0042986
Chen, T., Ren, C., Jiang, X., Zhang, L., Li, H., Huang, W., et al. (2018). Mechanisms for Type-II Vitellogenesis-Inhibiting Hormone Suppression of Vitellogenin Transcription in Shrimp Hepatopancreas: Crosstalk of GC/cGMP Pathway with Different MAPK-dependent Cascades. PLoS One 13, e0194459. doi:10.1371/journal.pone.0194459
Chen, Y. Q., Kuo, M.-S., Li, S., Bui, H. H., Peake, D. A., Sanders, P. E., et al. (2008). AGPAT6 Is a Novel Microsomal Glycerol-3-Phosphate Acyltransferase. J. Biol. Chem. 283, 10048–10057. doi:10.1074/jbc.M708151200
Cheng, Y. X., Du, N. S., and Lai, W. (1998). Lipid Composition in Hepatopancreas of Chinese Mitten Crab Eriocheir Sinensis at Different Stages (In Chinese). Acta Zool. Sin. 44, 420–429. doi:10.1016/0730-725X(91)90483-3
Cheng, Y. X., Lai, W., and Du, N. S. (1997). Lipid Accumulation and Hepatopancreatic Lipid Changes during Ovarian Development in Decapod Crustaceans (In Chinese). Chin. J. Zool. 32, 58–61. doi:10.13859/j.cjz.1997.02.022
Chirala, S. S., and Wakil, S. J. (2004). Structure and Function of Animal Fatty Acid Synthase. Lipids 39, 1045–1053. doi:10.1007/s11745-004-1329-9
Coleman, R., and Lee, D. P. (2004). Enzymes of Triacylglycerol Synthesis and Their Regulation. Prog. Lipid Res. 43, 134–176. doi:10.1016/S0163-7827(03)00051-1
Feng, H., and Thompson, E. M. (2018). Specialization of CDK1 and Cyclin B Paralog Functions in a Coenocystic Mode of Oogenic Meiosis. Cell cycle 17, 1425–1444. doi:10.1080/15384101.2018.1486167
Firmani, L. D., Uliasz, T. F., and Mehlmann, L. M. (2018). The Switch from cAMP-independent to cAMP-dependent Arrest of Meiotic Prophase Is Associated with Coordinated GPR3 and CDK1 Expression in Mouse Oocytes. Dev. Biol. 434, 196–205. doi:10.1016/j.ydbio.2017.12.014
Fishery Statistical Yearbook, Chinese. (2021). Fisheries Agency of China Agriculture Ministry. Beijing: China Agriculture Press, 24.
Gao, J., He, J., Shi, X., Stefanovic-Racic, M., Xu, M., O’Doherty, R. M., et al. (2012). Sex-Specific Effect of Estrogen Sulfotransferase on Mouse Models of Type 2 Diabetes. Diabetes 61, 1543–1551. doi:10.2337/db11-1152
Gao, J., Wang, X., Zou, Z., Jia, X., Wang, Y., and Zhang, Z. (2014). Transcriptome Analysis of the Differences in Gene Expression between Testis and Ovary in Green Mud Crab (Scylla Paramamosain). BMC Genomics 15, 585. doi:10.1186/1471-2164-15-585
Gimeno, R. E., and Cao, J. (2008). Thematic Review Series: Glycerolipids. Mammalian Glycerol-3-Phosphate Acyltransferases: New Genes for an Old Activity. J. Lipid Res. 49, 2079–2088. doi:10.1194/jlr.R800013-JLR200
Girish, B. P., Swetha, C., and Reddy, P. S. (2015). Expression of RXR, EcR, E75 and VtG mRNA Levels in the Hepatopancreas and Ovary of the Freshwater Edible Crab, Oziothelphusa Senex Senex (Fabricius, 1798) during Different Vitellogenic Stages. Sci. Nat. 102, 20. doi:10.1007/s00114-015-1272-9
Girish, B. P., Swetha, C., Srilatha, M., Hemalatha, M., and Sreenivasula Reddy, P. (2018). Evidence for Retinoic Acid Involvement in the Regulation of Vitellogenesis in the Fresh Water Edible Crab, Oziotelphusa Senex Senex. Comp. Biochem. Physiology Part A Mol. Integr. Physiology 222, 1–6. doi:10.1016/j.cbpa.2018.04.003
Gondim, K. C., Atella, G. C., Pontes, E. G., and Majerowicz, D. (2018). Lipid Metabolism in Insect Disease Vectors. Insect Biochem. Mol. Biol. 101, 108–123. doi:10.1016/j.ibmb.2018.08.005
Gong, J., Ye, H., Xie, Y., Yang, Y., Huang, H., Li, S., et al. (2015). Ecdysone Receptor in the Mud Crab Scylla Paramamosain: a Possible Role in Promoting Ovarian Development. J. Endocrinol. 224, 273–287. doi:10.1530/JOE-14-0526
Gong, Y.-N., Li, W.-W., Sun, J.-L., Ren, F., He, L., Jiang, H., et al. (2010). Molecular Cloning and Tissue Expression of the Fatty Acid-Binding Protein (Es-FABP) Gene in Female Chinese Mitten Crab (Eriocheir Sinensis). BMC Mol. Biol. 11, 71. doi:10.1186/1471-2199-11-71
Gunawardene, Y. I. N. S., Tobe, S. S., Bendena, W. G., Chow, B. K. C., Yagi, K. J., and Chan, S.-M. (2002). Function and Cellular Localization of Farnesoic Acid O -methyltransferase (FAMeT) in the Shrimp, Metapenaeus ensis. Eur. J. Biochem. 269, 3587–3595. doi:10.1046/j.1432-1033.2002.03048.x
Guo, H., Chen, L.-l., Li, G.-l., Deng, S.-p., and Zhu, C.-h. (2019). Accumulation and Depuration of Nonylphenol and its Effect on the Expressions of Vitellogenin and Vitellogenin Receptor in Freshwater Prawn Macrobrachium Rosenbergii. Bull. Environ. Contam. Toxicol. 103, 729–733. doi:10.1007/s00128-019-02714-x
Han, K., Dai, Y., Zhang, Z., Zou, Z., and Wang, Y. (2018). Molecular Characterization and Expression Profiles of Sp-Uchl3 and Sp-Uchl5 during Gonad Development of Scylla Paramamosain. Molecules 23, 213. doi:10.3390/molecules23010213
Han, K. H., Zhang, Z. P., Wang, Y. L., and Zou, Z. H. (2010). Cyclin-CDK-CKI and UPP Participate in the Regulation of Reproduction and the Progression of Gonad Development in Crustacean (In Chinese). Biotechnol. Bull. 1(7), 48–54. doi:10.13560/j.cnki.biotech.bull.1985.2010.07.042
Harris, C. A., Haas, J. T., Streeper, R. S., Stone, S. J., Kumari, M., Yang, K., et al. (2011). DGAT Enzymes Are Required for Triacylglycerol Synthesis and Lipid Droplets in Adipocytes. J. Lipid Res. 52, 657–667. doi:10.1194/jlr.M013003
Harrison, K. E. (1990). The Role of Nutrition in Maturation, Reproduction and Embryonic Development of Decapod Crustaceans: A Review. J. Shellfish Res. 9, 1–28.
Holford, K. C., Edwards, K. A., Bendena, W. G., Tobe, S. S., Wang, Z., and Borst, D. W. (2004). Purification and Characterization of a Mandibular Organ Protein from the American Lobster, Homarus Americanus: a Putative Farnesoic Acid O-Methyltransferase. Insect Biochem. Mol. Biol. 34, 785–798. doi:10.1016/j.ibmb.2004.04.003
Homola, E., and Chang, E. S. (1997). Methyl Farnesoate: Crustacean Juvenile Hormone in Search of Functions. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 117, 347–356. doi:10.1016/S0305-0491(96)00337-9
Hui, J. H. L., Tobe, S. S., and Chan, S.-M. (2008). Characterization of the Putative Farnesoic Acid O-Methyltransferase (LvFAMeT) cDNA from White Shrimp, Litopenaeus Vannamei: Evidence for its Role in Molting. Peptides 29, 252–260. doi:10.1016/j.peptides.2007.08.033
Hussain, M. M., Shi, J., and Dreizen, P. (2003). Microsomal Triglyceride Transfer Protein and its Role in apoB-Lipoprotein Assembly. J. Lipid Res. 44, 22–32. doi:10.1194/jlr.R200014-JLR200
Jia, X., Chen, Y., Zou, Z., Lin, P., Wang, Y., and Zhang, Z. (2013). Characterization and Expression Profile of Vitellogenin Gene from Scylla Paramamosain. Gene 520, 119–130. doi:10.1016/j.gene.2013.02.035
Jiang, H., Yin, Y., Zhang, X., Hu, S., and Wang, Q. (2009). Chasing Relationships between Nutrition and Reproduction: A Comparative Transcriptome Analysis of Hepatopancreas and Testis from Eriocheir Sinensis. Comp. Biochem. Physiology Part D Genomics Proteomics 4, 227–234. doi:10.1016/j.cbd.2009.05.001
Jordal, A.-E. O., Hordvik, I., Pelsers, M., Bernlohr, D. A., and Torstensen, B. E. (2006). FABP3 and FABP10 in Atlantic Salmon (Salmo salar L.)- General Effects of Dietary Fatty Acid Composition and Life Cycle Variations. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 145, 147–158. doi:10.1016/j.cbpb.2006.05.007
Kaikaus, R. M., Bass, N. M., and Ockner, R. K. (1990). Functions of Fatty Acid Binding Proteins. Experientia 46, 617–630. doi:10.1007/BF01939701
Karlsson, M., Contreras, J. A., Hellman, U., Tornqvist, H., and Holm, C. (1997). cDNA Cloning, Tissue Distribution, and Identification of the Catalytic Triad of Monoglyceride Lipase. J. Biol. Chem. 272, 27218–27223. doi:10.1074/jbc.272.43.27218
Kennedy, E. P. (1961). Biosynthesis of Complex Lipids. Fed. Proc. 20, 934–940. doi:10.1079/pns19610014
King, R. W., Deshaies, R. J., Peters, J.-M., and Kirschner, M. W. (1996). How Proteolysis Drives the Cell Cycle. Science 274, 1652–1659. doi:10.1126/science.274.5293.1652
Lafontan, M., and Langin, D. (2009). Lipolysis and Lipid Mobilization in Human Adipose Tissue. Prog. Lipid Res. 48, 275–297. doi:10.1016/j.plipres.2009.05.001
Lee, R. F., and Walker, A. (1995). Lipovitellin and Lipid Droplet Accumulation in Oocytes during Ovarian Maturation in the Blue Crab,Callinectes sapidus. J. Exp. Zool. 271, 401–412. doi:10.1002/jez.1402710510
Li, K., Chen, L., Zhou, Z., Li, E., Zhao, X., and Guo, H. (2006). The Site of Vitellogenin Synthesis in Chinese Mitten-Handed Crab Eriocheir Sinensis. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 143, 453–458. doi:10.1016/j.cbpb.2005.12.019
Li, S., Friesen, J. A., Holford, K. C., and Borst, D. W. (2010). Methyl Farnesoate Synthesis in the Lobster Mandibular Organ: the Roles of HMG-CoA Reductase and Farnesoic Acid O-Methyltransferase. Comp. Biochem. Physiology Part A Mol. Integr. Physiology 155, 49–55. doi:10.1016/j.cbpa.2009.09.016
Li, S., Wagner, C. A., Friesen, J. A., and Borst, D. W. (2003). 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase in the Lobster Mandibular Organ: Regulation by the Eyestalk. General Comp. Endocrinol. 134, 147–155. doi:10.1016/S0016-6480(03)00246-6
Li, W., Huang, Z. Y., Liu, F., Li, Z., Yan, L., Zhang, S., et al. (2013). Molecular Cloning and Characterization of Juvenile Hormone Acid Methyltransferase in the Honey Bee, Apis mellifera, and its Differential Expression during Caste Differentiation. PLoS One 8, e68544. doi:10.1371/journal.pone.0068544
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 21, 3787–3793. doi:10.1093/bioinformatics/bti430
Marangos, P., Verschuren, E. W., Chen, R., Jackson, P. K., and Carroll, J. (2007). Prophase I Arrest and Progression to Metaphase I in Mouse Oocytes Are Controlled by Emi1-dependent Regulation of APCCdh1. J. Cell Biol. 176, 65–75. doi:10.1083/jcb.200607070
Mardis, E. R. (2008). The Impact of Next-Generation Sequencing Technology on Genetics. Trends Genet. 24, 133–141. doi:10.1016/j.tig.2007.12.007
Masoudi Asil, S., Abedian Kenari, A., Rahimi Miyanji, G., and Van Der Kraak, G. (2017). The Influence of Dietary Arachidonic Acid on Growth, Reproductive Performance, and Fatty Acid Composition of Ovary, Egg and Larvae in an Anabantid Model Fish, Blue Gourami ( Trichopodus Trichopterus ; Pallas, 1770). Aquaculture 476, 8–18. doi:10.1016/j.aquaculture.2017.03.048
Meusy, J. J., and Payen, G. G. (1988). Female Reproduction in Malacostracan Crustacea. Zool. Sci. 5, 217–265. doi:10.1111/j.1096-3642.1988.tb01730.x
Miled, N., Canaan, S., Dupuis, L., Roussel, A., Rivière, M., Carriere, F., et al. (2000). Digestive Lipases: from Three-Dimensional Structure to Physiology. Biochimie 82, 973–986. doi:10.1016/S0300-9084(00)01179-2
Miyakawa, H., Toyota, K., Sumiya, E., and Iguchi, T. (2014). Comparison of JH Signaling in Insects and Crustaceans. Curr. Opin. Insect Sci. 1, 81–87. doi:10.1016/j.cois.2014.04.006
Nagaraju, G. P. C. (2007). Is Methyl Farnesoate a Crustacean Hormone? Aquaculture 272, 39–54. doi:10.1016/j.aquaculture.2007.05.014
Nagaraju, G. P. C. (2011). Reproductive Regulators in Decapod Crustaceans: an Overview. J. Exp. Biol. 214, 3–16. doi:10.1242/jeb.047183
Narumiya, S., Sugimoto, Y., and Ushikubi, F. (1999). Prostanoid Receptors: Structures, Properties, and Functions. Physiol. Rev. 79, 1193–1226. doi:10.1152/physrev.1999.79.4.1193
Okumura, T., Han, C. H., Suzuki, Y., Aida, K., and Hanyu, I. (1992). Changes in Hemolymph Vitellogenin and Ecdysteroid Levels during the Reproductive and Non-reproductive Molt Cycles in the Freshwater Prawn Macrobrachium Nipponense. Zool. Sci. 9, 37–45.
Pan, J., Liu, M., Chen, T., Cheng, Y., and Wu, X. (2018). Immunolocalization and Changes of 17beta-Estradiol during Ovarian Development of Chinese Mitten Crab Eriocheir Sinensis. Cell Tissue Res. 373, 509–520. doi:10.1007/s00441-018-2834-x
Pasquevich, M. Y., Dreon, M. S., Lavarías, S., and Heras, H. (2011). Triacylglycerol Catabolism in the Prawn Macrobrachium Borellii (Crustacea: Palaemoniade). Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 160, 201–207. doi:10.1016/j.cbpb.2011.08.006
Qiu, X. E., Zhu, D. F., Cui, X. Y., Tang, J., and Xie, X. (2014). Cloning and Expression Analysis of HMGR during Molt Cycle in Portunus Trituberculatus (In Chinese). Oceanol. Limnol. Sin. 45, 1192–1201.
Rivera-Pérez, C., and García-Carreño, F. (2011). Effect of Fasting on Digestive Gland Lipase Transcripts Expression in Penaeus Vannamei. Mar. Genomics 4, 273–278. doi:10.1016/j.margen.2011.07.002
Rodríguez, E. M., Medesani, D. A., Greco, L. S. L., and Fingerman, M. (2002). Effects of Some Steroids and Other Compounds on Ovarian Growth of the Red Swamp crayfish,Procambarus clarkii, during Early Vitellogenesis. J. Exp. Zool. 292, 82–87. doi:10.1002/jez.1144
Sagi, A., Silkovsy, J., Fleisher-Berkovich, S., Danon, A., and Chayoth, R. (1995). Prostaglandin E2 in Previtellogeic Ovaries of the Prawn Macrobrachium Rosenbergii: Synthesis and Effect on the Level of cAMP. General Comp. Endocrinol. 100, 308–313. doi:10.1006/gcen.1995.1161
Shelness, G. S., and Ledford, A. S. (2005). Evolution and Mechanism of Apolipoprotein B-Containing Lipoprotein Assembly. Curr. Opin. Lipidol. 16, 325–332. doi:10.1097/01.mol.0000169353.12772.eb
Sreenivasula Reddy, P., Ramachandra Reddy, P., and Purna Chandra Nagaraju, G. (2004). The Synthesis and Effects of Prostaglandins on the Ovary of the Crab Oziotelphusa Senex Senex. General Comp. Endocrinol. 135, 35–41. doi:10.1016/j.ygcen.2003.08.002
Su, Y., Guo, Q., Gong, J., Cheng, Y., and Wu, X. (2020). Functional Expression Patterns of Four Ecdysteroid Receptor Isoforms Indicate Their Different Functions during Vitellogenesis of Chinese Mitten Crab, Eriocheir Sinensis. Comp. Biochem. Physiology Part A Mol. Integr. Physiology 248, 110754. doi:10.1016/j.cbpa.2020.110754
Subramoniam, T. (2000). Crustacean Ecdysteriods in Reproduction and Embryogenesis. Comp. Biochem. Physiology Part C Pharmacol. Toxicol. Endocrinol. 125, 135–156. doi:10.1016/S0742-8413(99)00098-5
Sun, W., Li, L., Li, H., Zhou, K., Li, W., and Wang, Q. (2020). Vitellogenin Receptor Expression in Ovaries Controls Innate Immunity in the Chinese Mitten Crab (Eriocheir Sinensis) by Regulating Vitellogenin Accumulation in the Hemolymph. Fish Shellfish Immunol. 107, 480–489. doi:10.1016/j.fsi.2020.09.007
Sun, Y., Zhang, Y., Liu, Y., Xue, S., Geng, X., Hao, T., et al. (2014). Changes in the Organics Metabolism in the Hepatopancreas Induced by Eyestalk Ablation of the Chinese Mitten Crab Eriocheir Sinensis Determined via Transcriptome and DGE Analysis. PLoS One 9, e95827. doi:10.1371/journal.pone.0095827
Takeuchi, K., and Reue, K. (2009). Biochemistry, Physiology, and Genetics of GPAT, AGPAT, and Lipin Enzymes in Triglyceride Synthesis. Am. J. Physiology-Endocrinology Metabolism 296, E1195–E1209. doi:10.1152/ajpendo.90958.2008
Tsukimura, B. (2001). Crustacean Vitellogenesis: Its Role in Oocyte Development. Am. Zool. 41, 465–476. doi:10.1668/0003-1569(2001)041
Tsutsui, N., Saido-Sakanaka, H., Yang, W.-J., Jayasankar, V., Jasmani, S., Okuno, A., et al. (2004). Molecular Characterization of a cDNA Encoding Vitellogenin in the Coonstriped shrimp,Pandalus Hypsinotus and Site of Vitellogenin mRNA Expression. J. Exp. Zool. 301A, 802–814. doi:10.1002/jez.a.53
Van Hoof, D., Rodenburg, K. W., and Van der Horst, D. J. (2005). Receptor-mediated Endocytosis and Intracellular Trafficking of Lipoproteins and Transferrin in Insect Cells. Insect Biochem. Mol. Biol. 35, 117–128. doi:10.1016/j.ibmb.2004.09.009
Vogt, G. (1994). Life-cycle and Functional Cytology of the Hepatopancreatic Cells of Astacus Astacus (Crustacea, Decapoda). Zoomorphology 114, 83–101. doi:10.1007/BF00396642
Wang, Q., Chen, L., Wang, Y., Li, W., He, L., and Jiang, H. (2012). Expression Characteristics of Two Ubiquitin/Ribosomal Fusion Protein Genes in the Developing Testis, Accessory Gonad and Ovary of Chinese Mitten Crab, Eriocheir Sinensis. Mol. Biol. Rep. 39, 6683–6692. doi:10.1007/s11033-012-1474-6
Wang, W., Wu, X., Liu, Z., Zheng, H., and Cheng, Y. (2014). Insights into Hepatopancreatic Functions for Nutrition Metabolism and Ovarian Development in the Crab Portunus Trituberculatus: Gene Discovery in the Comparative Transcriptome of Different Hepatopancreas Stages. PLoS One 9, e84921. doi:10.1371/journal.pone.0084921
Wang, Z., Sun, L., Guan, W., Zhou, C., Tang, B., Cheng, Y., et al. (2018). De Novo Transcriptome Sequencing and Analysis of Male and Female Swimming Crab (Portunus Trituberculatus) Reproductive Systems during Mating Embrace (Stage II). BMC Genet. 19, 3. doi:10.1186/s12863-017-0592-5
Wouters, R., Lavens, P., Nieto, J., and Sorgeloos, P. (2001). Penaeid Shrimp Broodstock Nutrition: An Updated Review on Research and Development. Aquaculture 202, 1–21. doi:10.1016/S0044-8486(01)00570-1
Wu, L. T., Hui, J. H. L., and Chu, K. H. (2013). Origin and Evolution of Yolk Proteins: Expansion and Functional Diversification of Large Lipid Transfer Protein Superfamily1. Biol. Reprod. 88, 102. doi:10.1095/biolreprod.112.104752
Wu, X., Chang, G., Cheng, Y., Zeng, C., Southgate, P. C., and Lu, J. (2010). Effects of Dietary Phospholipid and Highly Unsaturated Fatty Acid on the Gonadal Development, Tissue Proximate Composition, Lipid Class and Fatty Acid Composition of Precocious Chinese Mitten crab,Eriocheir Sinensis. Aquacult. Nutr. 16, 25–36. doi:10.1111/j.1365-2095.2008.00637.x
Wu, X., Chen, H., Liu, Z., and Cheng, Y. (2014). Immunorecognition and Distribution of Progesterone Receptors in the Chinese Mitten CrabEriocheir sinensisDuring Ovarian Development. J. Shellfish Res. 33, 35–43. doi:10.2983/035.033.0105
Wu, X., Liu, M., Pan, J., Chen, H., Zeng, C., and Cheng, Y. (2017). The Ovarian Development Pattern of Pond-Reared Chinese Mitten Crab, Eriocheir Sinensis H. Milne-Edwards, 1853. Crustac 90, 449–470. doi:10.1163/15685403-00003662
Xie, X., Tao, T., Liu, M., Zhou, Y., Liu, Z., and Zhu, D. (2016). The Potential Role of Juvenile Hormone Acid Methyltransferase in Methyl Farnesoate (MF) Biosynthesis in the Swimming Crab, Portunus Trituberculatus. Animal Reproduction Sci. 168, 40–49. doi:10.1016/j.anireprosci.2016.02.024
Xu, X., Gopalacharyulu, P., Seppänen-Laakso, T., Ruskeepää, A.-L., Aye, C. C., Carson, B. P., et al. (2012). Insulin Signaling Regulates Fatty Acid Catabolism at the Level of CoA Activation. PLoS Genet. 8, e1002478. doi:10.1371/journal.pgen.1002478
Xu, Z., Liu, A., Li, S., Wang, G., and Ye, H. (2020). Hepatopancreas Immune Response during Molt Cycle in the Mud Crab, Scylla Paramamosain. Sci. Rep. 10, 13102. doi:10.1038/s41598-020-70139-2
Yang, Y., Wang, J., Han, T., Liu, T., Wang, C., Xiao, J., et al. (2015). Ovarian Transcriptome Analysis of Portunus Trituberculatus Provides Insights into Genes Expressed during Phase III and IV Development. PLoS One 10, e0138862. doi:10.1371/journal.pone.0138862
Yao, G. G., Wu, X. G., Cheng, Y. X., Yang, X. Z., and Wang, C. L. (2008). The Changes of Histology and Main Biochemical Composition in the Hepatopancreas at the Different Physiological Stages of Portunus Trituberculatus in East China Sea (In Chinese). Acta Oceanol. Sin. 30, 122–131.
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 11, R14. doi:10.1186/gb-2010-11-2-r14
Yu, S., Wei, W., Xia, M., Jiang, Z., He, D., Li, Z., et al. (2016). Molecular Characterization, Alternative Splicing and Expression Analysis ofACSF2and its Correlation with Egg-Laying Performance in Geese. Anim. Genet. 47, 451–462. doi:10.1111/age.12435
Zachariae, W., and Nasmyth, K. (1999). Whose End Is Destruction: Cell Division and the Anaphase-Promoting Complex. Genes & Dev. 13, 2039–2058. doi:10.1101/gad.13.16.2039
Zeng, X., Wan, H., Zhong, J., Feng, Y., Zhang, Z., and Wang, Y. (2021). Large Lipid Transfer Proteins in Hepatopancreas of the Mud Crab Scylla Paramamosain. Comp. Biochem. Physiology Part D Genomics Proteomics 38, 100801. doi:10.1016/j.cbd.2021.100801
Zhu, W., Guo, Q., Cheng, Y., and Wu, X. (2019). Identification and Functional Expression of Two Subtypes of Glycerol-3-Phosphate Acyltransferase Differently Regulating Triacylglyceride Synthesis during Ovary Development in Chinese Mitten crab,Eriocheir Sinensis. J. Exp. Zool. 331, 494–505. doi:10.1002/jez.2316
Keywords: Eriocheir sinensis, ovarian development, hormones, nutrient deposition, comparative transcriptome
Citation: Feng Q, Liu M, Cheng Y and Wu X (2022) Comparative Transcriptome Analysis Reveals the Process of Ovarian Development and Nutrition Metabolism in Chinese Mitten Crab, Eriocheir Sinensis. Front. Genet. 13:910682. doi: 10.3389/fgene.2022.910682
Received: 01 April 2022; Accepted: 09 May 2022;
Published: 24 May 2022.
Edited by:
Sonia Andrade, University of São Paulo, BrazilReviewed by:
Yong Zhang, Sun Yat-sen University, ChinaJitao Li, Chinese Academy of Fishery Sciences, China
Copyright © 2022 Feng, Liu, Cheng and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xugan Wu, eGd3dXl4QDE2My5jb20=; Meimei Liu, bW1saXVAam91LmVkdS5jbg==
 Qiangmei Feng1
Qiangmei Feng1 Yongxu Cheng
Yongxu Cheng Xugan Wu
Xugan Wu