A New Prognostic Risk Score: Based on the Analysis of Autophagy-Related Genes and Renal Cell Carcinoma
- 1Department of Urology, The Second Affiliated Hospital, School of Medicine, Xi’an Jiaotong University, Xi’an, China
- 2School of Medicine, Xi’an Jiaotong University, Xi’an, China
A Corrigendum on
A New Prognostic Risk Score: Based on the Analysis of Autophagy-Related Genes and Renal Cell Carcinoma
by He, M, Li, M, Guan, Y, Wan, Z, Tian, J, Xu, F, Zhou, H, Gao, M, Bi, H and Chong, T (2022). Front. Genet. 12:820154. doi: 10.3389/fgene.2021.820154
In the original article, there was a mistake in the legend for Figure 1A. The threshold of DEGs was wrongly depicted in the legend. The correct legend is presented as follows:
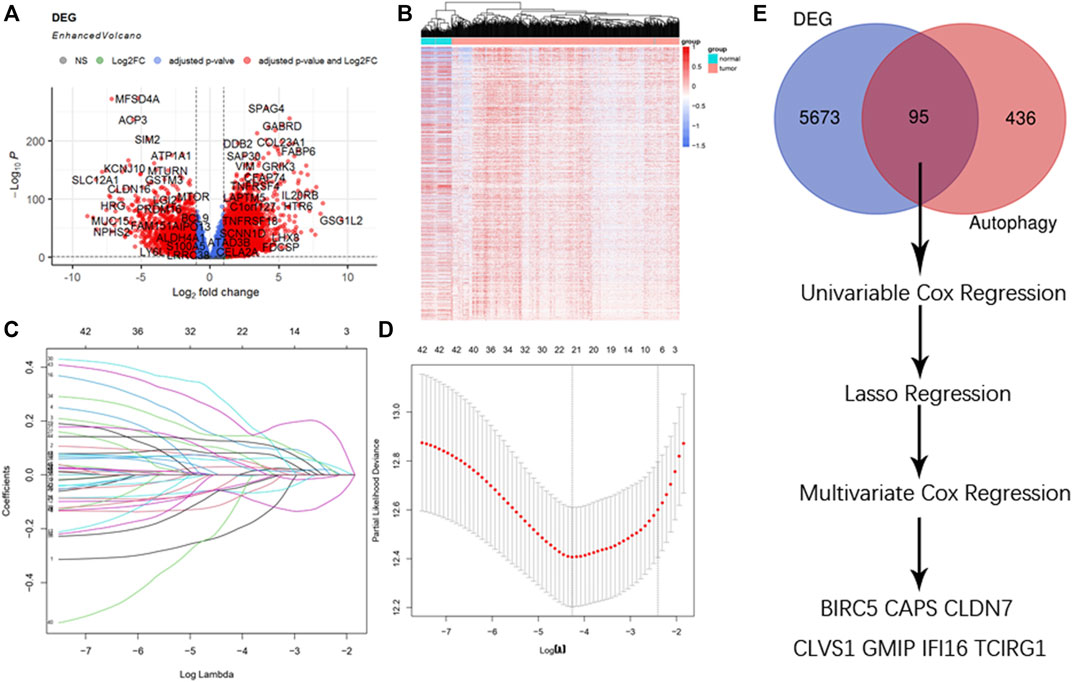
FIGURE 1. Selection of DEGs. (A) Enhanced volcano plot of DEGs when comparing ccRCC with normal tissue. Red nodes represented genes with |log 2 FC| ≥ 1 and adjusted p < 0.05, blue nodes represented genes with adjusted p < 0.05 only, and grey nodes represented genes that were neither eligible in conditions of adjusted p value nor |log 2 FC|. (B) Heatmap of DEGs in ccRCC. (C) Lasso coefficients profiles of 95 genes significant in univariate cox regression. (D) Lasso regression obtained 21 prognostic genes using minimum lambda value. (E) Selecting procedure of DEGs, venn gram showed 95 genes in the intersection of 5,768 DEGs and 531 ATGs. These genes then underwent univariate cox regression, lasso cox regression, and multivariate cox regression, and finally 7 DEGs were selected to construct the risk score formula.
“(A) Enhanced volcano plot of DEGs when comparing ccRCC with normal tissue. Red nodes represented genes with |log 2 FC| ≥ 1 and adjusted p < 0.05, blue nodes represented genes with adjusted p < 0.05 only, and gray nodes represented genes that were neither eligible in conditions of adjusted p-value nor |log 2 FC|.”
In the original article, there was a mistake in Figure 1 as published. The threshold of DEGs was wrongly set when drawing enhanced volcano plots the corrected Figure 1 is included here.
In the original article, the method of correlation analysis was wrongly typed as “pearson” in “Correlation analysis of risk score and other clinical signatures was performed by the method of “pearson”.” A correction has been made to Materials and Methods, Construction of Risk Score, Paragraph 2:
The sentence “Correlation analysis of risk score and other clinical signatures was performed by the method of “pearson”.” should be corrected as “Correlation analysis of risk score and other clinical signatures was performed by the method of “Spearman”.”
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: risk score, prognosis, bioinformatics analysis, renal cell carcinoma, autophagy
Citation: He M, Li M, Guan Y, Wan Z, Tian J, Xu F, Zhou H, Gao M, Bi H and Chong T (2022) Corrigendum: A New Prognostic Risk Score: Based on the Analysis of Autophagy-Related Genes and Renal Cell Carcinoma. Front. Genet. 13:904512. doi: 10.3389/fgene.2022.904512
Received: 25 March 2022; Accepted: 23 May 2022;
Published: 04 July 2022.
Edited and reviewed by:
Farhad Maleki, McGill University, CanadaCopyright © 2022 He, Li, Guan, Wan, Tian, Xu, Zhou, Gao, Bi and Chong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tie Chong, chongtie@126.com
 Minxin He
Minxin He Mingrui Li1,2
Mingrui Li1,2 Yibing Guan
Yibing Guan Juanhua Tian
Juanhua Tian Fangshi Xu
Fangshi Xu Tie Chong
Tie Chong