- 1Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Wuhan, China
- 2College of Fisheries, Huazhong Agricultural University, Wuhan, China
The Asian swamp eel (Monopterus albus) is an excellent model species for studying sex change and chromosome evolution. M. albus is also widely reared in East Asia and South-East Asia because of its great nutritional value. The low fecundity of this species (about 300 eggs per fish) greatly hinders fries production and breeding programs. Interestingly, about 3–5% of the eels could remain as females for 3 years and lay more than 3,000 eggs per fish, which are referred to as non-sex-reversal (NSR) females. Here, we presented a new chromosome-level genome assembly of such NSR females using Illumina, HiFi, and Hi-C sequencing technologies. The new assembly (Mal.V2_NSR) is 838.39 Mb in length, and the N50 of the contigs is 49.8 Mb. Compared with the previous assembly obtained using the continuous long-read sequencing technology (Mal.V1_CLR), we found a remarkable increase of continuity in the new assembly Mal.V2_NSR with a 20-times longer contig N50. Chromosomes 2 and 12 were assembled into a single contig, respectively. Meanwhile, two highly contiguous haplotype assemblies were also obtained, with contig N50 being 14.54 and 12.13 Mb, respectively. BUSCO and Merqury analyses indicate completeness and high accuracy of these three assemblies. A comparative genomic analysis revealed substantial structural variations (SVs) between Mal.V2_NSR and Mal.V1_CLR and two phased haplotype assemblies, as well as whole chromosome fusion events when compared with the zig-zag eel. Additionally, our newly obtained assembly provides a genomic view of sex-related genes and a complete landscape of the MHC genes. Therefore, these high-quality genome assemblies would provide great help for future breeding works of the swamp eel, and it is a valuable new reference for genetic and genomic studies of this species.
Introduction
Monopterus albus, commonly known as the Asian swamp eel or rice field eel, is widely distributed throughout Asia, from northern India and Burma to China, Asiatic Russia, Japan, and the Indo-Malayan Archipelago. M. albus is an economical fish in China (Yang et al., 2018), Vietnam (Khanh and Ngan, 2010), Thailand (Amornsakun et al., 2018), Philippines (Silva et al., 2020), and Indonesia (Herawati et al., 2018) due to its flavor and nutritional value. One local strain of M. albus mainly farmed in Jianghan Plain has been subjected to consecutive selective breeding programs recently due to its preferred body color and superiority in growth rate and fecundity (Yang et al., 2009), and the high-quality genome assembly of the selecting breed had been sequenced recently (Tian et al., 2021).
As a famous hermaphroditic protogynous freshwater fish, every individual of M. albus develops first as a functional female and then transforms into a functional male by sex reversal after their first spawning during its life cycle (Liu, 1944; Liem, 1963). The sex-changing phenomenon has attracted significant research interest, and the underlying sex-change mechanisms became a hot topic in past decades, hence making M. albus an excellent model for studying sex change (Cheng et al., 2003). Previously, the sex change in M. albus was found to be accompanied by changes in steroid levels (Yeung and Chan, 1987) and activity of enzymes involved in steroid pathways (Tang et al., 1975). Recently, some sex-related genes and microRNAs have been characterized, and their expressions seem to be varied among three genders during the sex-change progress, such as Dmrt (Sheng et al., 2014), AMH (Hu et al., 2015), Foxl2 (Hu et al., 2014), dax1 (Hu et al., 2015), p450 (Zhang et al., 2008), G2 (Qu et al., 2015), Cyp19a1a (Liu et al., 2009; Zhang et al., 2013), gonadal soma-derived factor (gsdf) (Zhu et al., 2016), Sox9 (Zhou et al., 2003), Dmrt1 (Huang et al., 2005), Jnk1 (Xiao et al., 2010), and mal-miR-430a, mal-miR-430b, and mal-miR-430c (Gao et al., 2014; Zhang et al., 2021). Moreover, a batch number of genes with different expression profiles between females and males was characterized through high-throughput RNA-seq (Chi et al., 2017). Lastly, epigenetic modification has been suggested playing important roles in sex reversal, such as the histone deacetylation and methylation of cyp19a1a (Zhang et al., 2013) and histone acetylation, methylation, and ubiquitination of dmrt1 (Lai et al., 2021). No heteromorphic chromosome has been found in M. albus (Ma and Shi, 1987). Altogether, the underlying mechanism of sex changes in M. albus remains unknown yet (Hilles et al., 2018). The classification of these triggering sex-reversal mechanisms would help understand the diversity and evolution of such an intriguing mechanism and would provide significant help to future breeding programs of this aquaculture species. Furthermore, M. albus has the smallest haploid number (2n = 24) of chromosomes among most freshwater fishes (2n = 24–446), and it is also an ideal material for chromosomal evolution studies (Supiwong et al., 2019; Cheng et al., 2020).
Generally, female individuals of M. albus obtain well-developed ova during 1–1.5 years of age (∼20 cm long), and intersex individuals have a body length of 30–40 cm (Susatyo et al., 2020; Cheng et al., 2021a). The fecundity of M. albus is about 200–300 eggs per fish at about 1 year of age (Susatyo et al., 2020; Yue et al., 2021). Such low fecundity greatly hinders large-scale aquaculture production and breeding works of swamp eel. Generally, the majority of the females of swamp eel complete sex reversal stage in about 8–30 weeks when they are 2 and 3 years old. However, few female individuals remain as functional female when they are over 55 cm and 10 years old (Chan and Phillips, 1967; Yang et al., 2008; Yang and Xiong, 2010). During our breeding work, we also found that few (3–5%) individuals remain as females and could consecutively spawn three more years after their first spawning, and they were referred to as non-sex-reversal (NSR) females. Such NSR females could reach more than 50 cm in total length and lay more than 3,000 eggs per fish (our unpublished observations). They are excellent materials for studying the mechanisms of sex reversal and breeding works. Although a collapsed haplotype assembly genome of M. albus (Mal.V1_CLR) has been obtained using the continuous long-read sequencing technology (Tian et al., 2021), improvement in the de novo assembly of genomes is still to be desired.
Recent advances in the long-read sequencing technology have allowed a single molecule to be sequenced multiple times to produce long high-fidelity reads (HiFi) with a base level accuracy of 99.9% and hence drastically improve the base quality of the assembly (Wenger et al., 2019) and enable to phase the genome into haplotypes (Porubsky et al., 2021). Mastacembelus armatus, a closely related species that belongs to the order Synbranchiformes, also experiences female-to-male sex changes and is proved to possess neo-sex chromosomes, although cytogenetically all chromosome pairs are homomorphic (Xue et al., 2021). Hence, we presented a de novo chromosome-scale assembly of NSR females using HiFi reads with the Hifiasm assembly tool. The final assembly Mal.V2_NSR is 838.39 Mb in length with the contig and scaffold N50s of 49.8 and 68.68 Mb, respectively, showing a significant improvement in continuity compared with the assembly Mal.V1_CLR. Meanwhile, two highly contiguous haplotype assemblies were also obtained, with the contig N50 being 14.54 and 12.13 Mb, respectively. The new assembly Mal.V2_NSR is annotated with 22,704 protein-coding genes and contains 97.3% of the core set of conserved actinopterygian orthologs, and there are 331 more predicted genes in it than in the previous assembly Mal.V1_CLR. BUSCO and Merqury analyses indicate completeness and high accuracy of all three assemblies. The comparative genomic analysis revealed substantial structural variations (SVs) between Mal.V2_NSR and Mal.V1_CLR and two phased haplotype assemblies, as well as whole chromosome fusion events when compared with the zig-zag eel. The newly almost complete chromosome-level assemblies would provide resources for the future genomics-assisted breeding program and would be a valuable new reference for genetic and genomic studies of this species.
Materials and Methods
Animals and Tissue Sampling
The experimental procedure was approved by the Animal Experimental Ethical Inspection of Laboratory Animal Centre, Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences (ID Number: 2020-THF-01).
Twelve non-sex-reversal female individuals were obtained from a breeding farm in Xiantao City, Hubei province. Their total length is 61.53 (±5.09) cm, and their weight is 289.28 (±93.98) grams (Supplementary Table S1). The fish were immediately dissected after anesthesia with MS-222. The tissue was flash-frozen in liquid nitrogen and stored at −80°C until DNA isolation and sequencing were performed. The muscle tissue of one fish was collected for whole-genome sequencing and Hi-C library construction, and different tissues (the brain, muscle, skin, liver, spleen, heart, stomach, intestine, kidney, and ovary) of another fish were collected and used for transcriptome sequencing.
Library Preparation and Sequencing
Total genomic DNA was extracted from the muscle using the standard phenol/chloroform method, and the integrity of the genomic DNA molecules was checked by agarose gel electrophoresis. Both the PacBio Sequel II platform and the Illumina NovaSeq platform were applied for genomic sequencing to generate long and short genomic reads, respectively.
For the Illumina NovaSeq platform (San Diego, CA, USA), a pair-end library was constructed with an insert size of 350 base pairs (bp), according to the protocol provided by the manufacturer. Large-insert (15 kb) single-molecule real-time circular consensus sequencing (HiFi CCS) library preparation was conducted, following the Pacific Biosciences recommended protocols as described in our previous work (Tian et al., 2021). The constructed library was sequenced on one SMRT cell on the PacBio RS II machine (Pacific Biosciences of California, Inc.). To obtain a chromosome-level genome, 1 g of muscle tissue from the same individual was used for Hi-C library construction. The Hi-C library was prepared from cross-linked chromatins using a standard Hi-C protocol; then, the library was sequenced using the Illumina NovaSeq 6000 instrument (Illumina), as in our previous study (Tian et al., 2021).
We also performed RNA sequencing to generate transcriptome data on the Illumina NovaSeq platform for gene model prediction. RNA was extracted from nine tissues mentioned previously using the TRIzol reagent (Invitrogen). RNA purity was checked using the kaiaoK5500VR spectrophotometer (Kaiao, Beijing, China), and RNA integrity and concentration were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total amount of 2 μg RNA per sample was mixed, and sequencing libraries were generated according to the manufacturer’s recommendations and sequenced using the Illumina NovaSeq 6000 platform.
Genome Assembly and Phasing
The genome size, heterozygosity, and repeat content were first estimated through a k-mer analysis with Jellyfish v2.2.10 (Marcais and Kingsford, 2011). HiFi reads were assembled by Hifiasm, version 0.15 (r327) (https://github.com/chhylp123/hifiasm/, last accessed on 16 July 2021) (Cheng et al., 2021b). gfatools (https://github.com/lh3/gfatools) was used to convert sequence graphs from the GFA to FASTA format. Duplicated haplotigs and artifacts were identified and removed using the purge_haplotigs (https://bitbucket.org/mroachawri/purge_haplotigs, last accessed on 10 October 2021) (Roach et al., 2018). To order and orient contigs into pseudomolecules, Hi-C reads were mapped to the draft assembly with Juicer v1.5.6 (Durand et al., 2016b); then, the candidate chromosome-length assembly was obtained using the 3D-DNA (v201008) (Dudchenko et al., 2017) pipeline to correct misjoins, order, orient, and anchor contigs from the draft assembly. Last, the assembly was manually checked and refined with Juicebox v. 1.11.08 (Durand et al., 2016a) for quality control and interactive correction. This newly obtained genome assembly was named Mal.V2_NSR. Moreover, two sets of haplotype-resolved, phased contig (haplotig) assemblies were generated using hifiasm with a combination of HiFi reads and paired-end Hi-C reads, and they were scaffolded into a chromosome-scale assembly as mentioned previously. These two phased haplotype assemblies were referred to as Hap1 and Hap 2, respectively.
Assessment of Assembly Quality
First, the HiFi reads and Illumina paired-end reads were mapped to the assembly by Minimap2 and BWA, respectively, to evaluate the completeness (Li, 2018). The mapping rates and the genome coverages were calculated by Samtools (version 0.1.19) (Li et al., 2009). Picard, GATK (version 3.1-1) was used to call SNPs (McKenna et al., 2010). Second, the completeness of the primary assembly was assessed by Benchmarking Universal Single-Copy Orthologs (BUSCO) (v4.0.6) (Simao et al., 2015) using the Actinopterygii_odb9 database as the reference data set. Third, the consensus quality values (QVs) of the obtained three assemblies were also computed by Merqury version 1.3 (https://github.com/marbl/merqury, last accessed on 21 October 2021) (Rhie et al., 2020). Last, to assess the quality of the three assemblies, CHROMEISTER (Perez-Wohlfeil et al., 2019) was used to perform a pairwise comparison between Mal.V2_NSR and Mal.V1_CLR, and Hap1 and Hap2.
Repeat Characterization and Genome Annotation
Repetitive elements were identified by homolog searching using RepeatMasker and RepeatProteinMask (Chen, 2004) with Repbase (version 23.08) libraries (Jurka et al., 2005; Bao et al., 2015) and identified de novo assemblies by using RepeatModeler, Tandem Repeats Finder (TRF) (Benson, 1999), and LTR-FINDER (Xu and Wang, 2007).
Based on the repetitive-element masked genome, genes were predicted using ab initio approach, and homolog searching. For ab initio gene prediction, AUGUSTUS v3.3.2 (Stanke et al., 2006), GeneScan (Aggarwal and Ramaswamy, 2002), and GlimmerHMM were used for the prediction of genes in the repeat-masked genome. For homology-based predictions, protein sequences of six closely related teleost species, including Acanthochromis polyacanthus (GCF_002109545.1), Amphiprion ocellaris (GCF_002776465.1), Anabas testudineus (GCF_900324465.1), Astatotilapia calliptera (GCF_900246225.1), Mastacembelus armatus (GCF_900324485.1), and our previously assembled genome of M. albus (Mal.V1_CLR) (GWHBEHV00000000) were downloaded from the National Center for Biotechnological Information (NCBI) database and National Genomics Data Center (https://ngdc.cncb.ac.cn/) and were mapped onto the newly assembled M. albus genome using BLASTN. The alignment hits were joined by Solar software (Yu et al., 2006). Subsequently, GeneWise v2.2.0 (Birney et al., 2004) with default options was used for homologous annotation. In addition, RNA-seq reads were directly mapped to the assembled genome to identify putative exon regions using the TopHat package (v2.1.1) (Trapnell et al., 2009) and Cufflinks (v2.2.1) (Ghosh and Chan, 2016). Finally, all the gene models were merged, and redundancy was removed by MAKER (Cantarel et al., 2008) and integrating the CEGMA (Parra et al., 2007) results based on the HiCESAP process.
All predicted genes were functionally annotated, as previously described (Tian et al., 2021). Various databases, including NCBI non-redundant protein (nr), SwissProt (Boeckmann et al., 2003), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Ogata et al., 1999), TrEMBL (Boeckmann et al., 2003), InterPro (Zdobnov and Apweiler, 2001), PFAM (Mistry et al., 2021), eukaryotic orthologous groups of proteins (KOG) (Tatusov et al., 2001), TFs, and Gene Ontology (GO) (Ashburner et al., 2000), were used to functionally annotate the predicted protein-coding genes.
For non-coding RNAs, microRNA (miRNA) and small nuclear RNA (snRNA) were predicted using INFERNAL v1.1.2 (Nawrocki and Eddy, 2013) and the Rfam database (release 13.0) (Griffiths-Jones et al., 2003; Kalvari et al., 2018). Transfer RNA (tRNA) and ribosomal RNA (rRNA) were identified using tRNAscan-SE v1.3.1 (Lowe and Eddy, 1997) and RNAmmer (v1.2, http://www.cbs.dtu.dk/services/RNAmmer/), respectively.
Comparative Genomic Analysis
Synteny and Rearrangement Identifier (SyRI) (https://github.com/schneebergerlab/syri) v1.5 (Goel et al., 2019) was used to identify structural variations (SVs) between the newly obtained assembly Mal.V2_NSR and the previous assembly Mal.V1_CLR and the two phased assemblies Hap 1 and Hap 2. Minimap2 v2.23-r1111 was used to perform the alignment with the following parameter setting “-ax asm5 –eqx,” and then, SVs were detected. The output of SyRI was plotted with the script plotsr. The SVs from SyRI were reclassified as presence-absence variations (PAVs) as follows: the CPL, DEL, DUP/INVDP (loss) variants, and the sequences in NOTAL were converted as Absence SVs, and the CPG, INS, DUP/INVDP (gain) variants, and the query sequences in NOTAL were converted as Presence SVs. Genes affected by these PAVs were identified with the BEDTools (v2.30.0) (Quinlan and Hall, 2010) by comparing their absolute positions to the genome annotations. We performed a Gene Ontology enrichment analysis to find enriched biological functions by using TBtools (Chen et al., 2020).
The pairwise genome comparison between Mal.V2_NSR and the genome of M. armatus (fMasArm1.2, GCF_900324485.2) (Rhie et al., 2021) was performed by MCScanX (Python version) (Tang et al., 2008; Wang et al., 2012). Syntenic regions were identified with the LAST results, and dot plots for genome pairwise synteny visualization were generated using the command “python -m jcvi.graphics.dotplot.” The synteny pattern of the two compared genomes was generated using the command “python -m jcvi.compara.synteny depth -histogram.”
Sex-Related Genes and Major Histocompatibility Complex Genes
Previous studies have revealed multiple sex-related genes that might be involved in sex change, gonadal differentiation in M. albus, and other teleosts (Zhang et al., 2008; Wu et al., 2012; Hu et al., 2014; Hu et al., 2015; Gao et al., 2016; Cai et al., 2017; Yi et al., 2019; Ding et al., 2020; Chen et al., 2022; Feng et al., 2022). To investigate a genomic survey of these genes in the assembled genome, these genes were directly searched in the annotated genome using gene name or identified with tBlastn (v2.9.0+) (Camacho et al., 2009) in the annotated genome. Major histocompatibility complex (MHC) genes play important roles in the immune response of vertebrates and exhibit high degrees of genetic diversity, and their genome organization varies greatly among fish (Hu et al., 2022). To obtain an overview of the genome organization of MHC genes in swamp eels, MHC genes were searched by their gene names and characterized by domain analysis and then classified as class I and II by searching against a non-redundant (nr) protein database with BlastP (last accessed on 10 March 2022).
Results
Genome Assembly and Assessment
A total of 89.84-gigabyte (Gb) Illumina clean reads were obtained by using Illumina paired-end libraries (Supplementary Table S2), and the genome size was estimated to be 830 Mb based on 17-nt k-mer length with a mean k-mer coverage depth of 71-fold (Supplementary Figure S1). A total of 1,764,134 HiFi reads consisting of 29,369,894,529 bp were obtained (Supplementary Table S2) on the PacBio Sequel II platform, with the average lengths of 16,648.34 bp and N50 16,790 bp. Using 35.4× high-fidelity (HiFi) reads, an 843-Mb primary assembly was obtained with Hifiasm, including 384 contigs. The contig N50 of the primary assembly was 45.38 Mb. Then, a total of 116.59 Gb clean data obtained from the Hi-C library (Supplementary Table S2) were used to scaffold the contigs to a chromosome-scale assembly. According to the aforementioned mapping strategy, the proportion of valid interaction pairs was 87.6%, indicating the high quality of the Hi-C library. As a result, 384 contigs were successfully anchored and oriented into 12 chromosomes (Figure 1B). More than 98.27% of contigs longer than 100 kb were anchored on chromosomes, exhibiting an excellent anchoring rate for the chromosome assembly. The final assembly was 838.39 Mb (Figure 1C), with a contig N50 of 49.8 Mb and scaffold N50 of 68.68 Mb, and included 38.66 Mb additional sequences (Table 1). This new assembly was named Mal.V2_NSR, and the 12 chromosomes were numbered based on similarity to the Mal.V1_CLR assembly (Accession No. GWHBEHV00000000). The Mal.V2_NSR assembly was highly concordant with Mal.V1_CLR, according to the synteny plot (Supplementary Figure S2), and it is more contiguous than the Mal.V1_CLR (Figure 1D), with a 20- fold increase in contig N50 values (Table 1). Noticeably, chromosomes 2 and 12 were assembled into a single contig (Figure 1D; Table 1). Meanwhile, with the help of the hifiasm assembler, two sets of haplotigs (phased haplotype-resolved contigs) were obtained and scaffolded into pseudochromosomes with Hi-C data. The obtained two phased haplotype assemblies (referred to as Hap1 and Hap2) are ultra-continuous, with contig N50s of 14.53 Mb for Hap1 and 12.13 Mb for Hap2, respectively (Table 1). The quantitative statistics for these three assemblies are listed in Table 1.
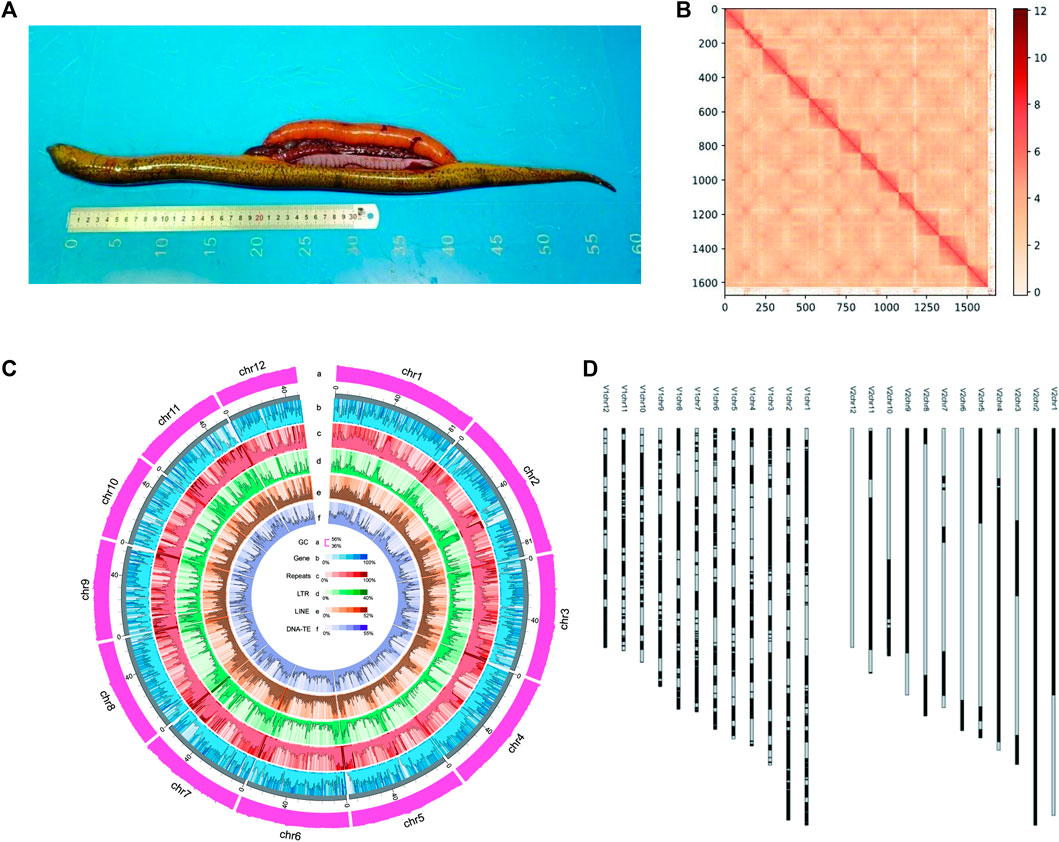
FIGURE 1. Genome assembly of non-sex-reversal (NSR) female of M. albus. (A) Picture of an NSR swamp eel. (B) Heatmap of the M. albus Hi-C assembly. Red blocks refer to the intensity of interaction among sequences. (C) Genome features of the primary assembly (Mal.V2_NSR). From a to f: a, GC content; b, Gene density; c, Repeats; d, LTR; e, LINE, and f, DNA transposable elements (TEs) and RNA transposable elements (RNA-TEs). (D) Visualization of improvements in assembly contiguity. Ideogram of Mal.V1_CLR (left) and Mal.V2_NSR (right) assemblies.
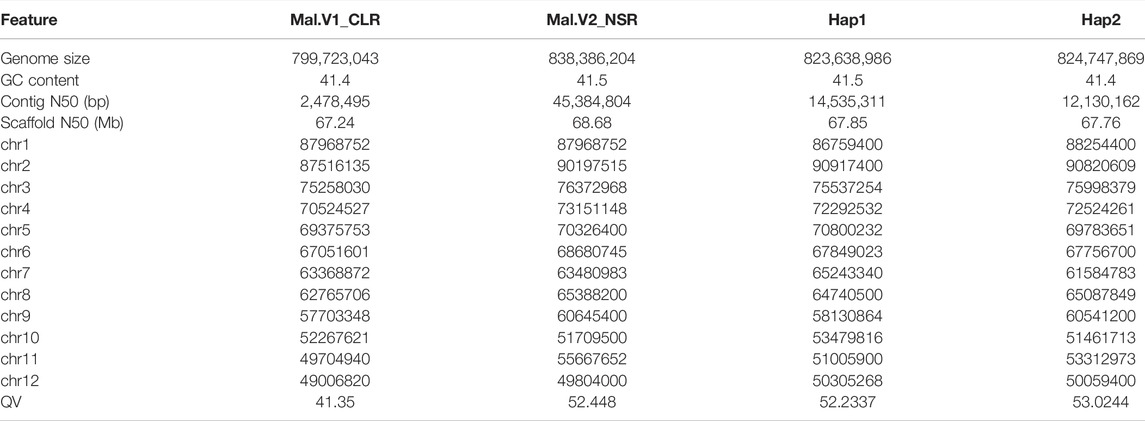
TABLE 1. Comparisons of the genomic features for the Mal.V2_NSR, Hap1, Hap2, and Mal.V1_CLR assemblies.
The mapping rate of CCS reads was 99.98% (covering 99.97% of the total length of the genome, among which, 99.78% with a coverage depth ≥4×, 99.18% with a coverage depth ≥10×, and 92.02% with a coverage depth ≥20×) (Supplementary Table S3). The mapping rate of the whole Illumina short reads by BWA was 99.8% (Supplementary Table S3), which means that almost all sequencing data were represented (covering 99.96% of the total genome length, among which, 99.91% with a coverage depth ≥4×, 99.82% with a coverage depth ≥10×, and 99.65% with a coverage depth ≥20×, showing high coverage). Both coverage rates of PacBio sequencing and Illumina sequencing were consistent and relatively high. The homozygous rate of SNPs and indels were both as low as 0%, suggesting the accuracy of the genome assembly was very high; the heterozygous rate of SNPs and indels were as low as 0.144% and 0.027%, respectively, indicating that genome heterozygosity was low. The BUSCO analysis showed that the primary assembly covered 97.3% of complete BUSCOs (C) and 94.4% of universal single-copy genes, and the Hap 1 and Hap 2 assemblies covered 97.1% and 97.3 of complete BUSCOs, respectively (Supplementary Table S4, and Supplementary Figure S3). The whole-genome comparison between Mal.V2_NSR and Mal.V1_CLR and two phased haplotype assemblies (Hap 1 and Hap2) indicated the distance scores are very low, and very significant signals are found on the diagonals (Supplementary Figure S2), indicating the good quality of our assemblies. Additionally, the consensus quality values (QVs), which represent the Phred-scaled probability of the base error in the assembly, were 52.48 for Mal.V2_NSR, 52.23 for Hap1, and 53.02 for Hap2, indicating a relatively high assembly accuracy for the newly obtained V2-NSR and two phased assemblies (Table 1).
Genome Annotation
A complete summary of the repeat element composition identified in the Mal.V2_NSR assembly is available in Supplementary Table S5. DNA transposons (21.01%), long interspersed nuclear elements (LINEs, 18.07%), and long terminal repeats (LTRs, 10.32%) are the top three categories of repetitive elements in this assembly (Supplementary Figure S4). Altogether, a total of 47.99% of the genome comprised repeats, which is slightly higher than our previously published Mal.V1_CLR assembly. After the analysis, 273 miRNAs, 42,869 tRNAs, 3,933 rRNAs, and 964 snRNAs were annotated in the Mal.V2_NSR assembly (Supplementary Table S6).
Finally, a total of 22,704 protein-coding genes were identified (Supplementary Table S7), of which 19,516, 21,813, 22,066, 20,356, and 15,457 protein-coding genes were annotated in the SwissProt, KEGG, TrEMBL, InterPro, and GO databases, respectively (Supplementary Table S8). BUSCO was also used to test the completeness of the genome annotation with the actinopterygii_odb9 database, which showed that 93.9% complete and 1.0% fragmented conserved single-copy orthologs were predicted for the Mal.V2_NSR assembly.
Comparative Genomic Analysis
Comparing the new assembly (Mal.V2_NSR) with the reference Mal.V1_CLR assembly using the whole-genome comparison tool SyRI, we found 703–707 Mb of collinear regions in each of these two assemblies (Figure 2A). In addition, the SyRI analysis identified 268 inversions (∼15 Mb), 540 translocations (∼5 Mb), and 137 duplications in Mal.V1_CLR (1.5 Mb) and 890 duplications in Mal.V2_NSR (4.38 Mb), and we also identified that 78.73 Mb of Mal.V1_CLR and 88.50 Mb of Mal.V2_NSR DNA sequences were not aligned (Supplementary Table S9). The large PAVs (>1,000 bp) contain 2,474 genes in Mal.V1_CLR and 2,616 genes in Mal.V2_NSRv. Gene ontology enrichment analysis for these PAV genes identified 19 GO terms including six biological processes, nine molecular functions and four cellular components that were significantly enriched (Supplementary Figure S5). Meanwhile, comparing the two phased haplotype assemblies allowed us to gain insights into the haplotype diversity within NSR female individuals, including 224 inversions (∼3.8 Mb), 276 translocations (∼1.75 Mb), and 25 duplications (∼0.22 Mb) in Hap1, and inversions (∼3.8 Mb), 276 translocations (∼1.75 Mb), and 272 duplications (∼1.74 Mb) in Hap2, making up about 8% of the assembled sequence (66.04 and 69.64 Mb) in each of the haplotype assemblies (Figure 2B, Supplementary Table S10).
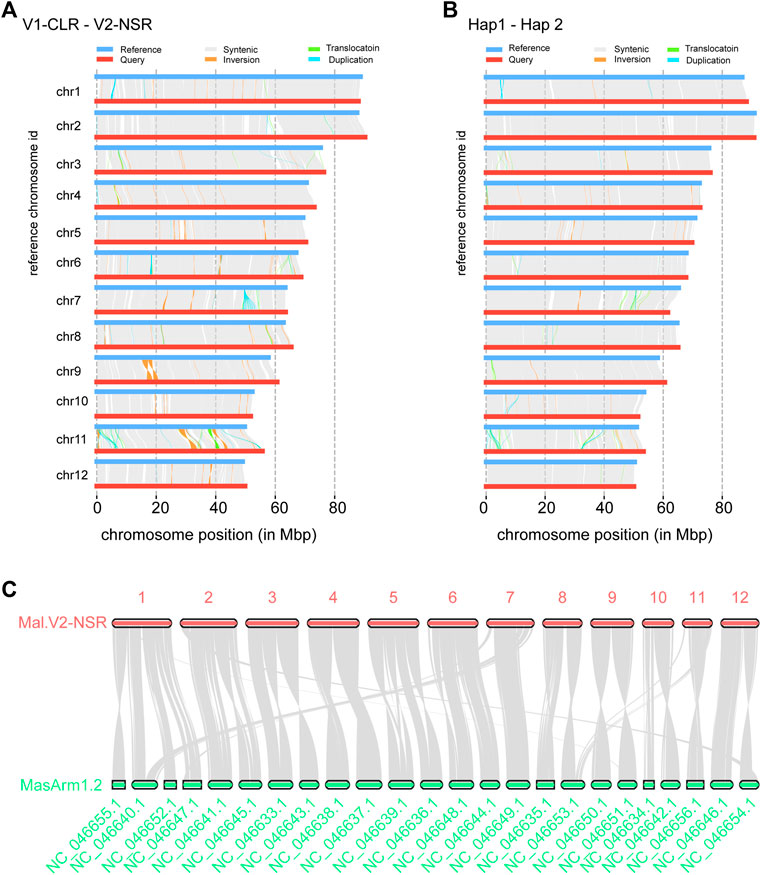
FIGURE 2. Genome comparative analysis. Synteny and rearrangement (SyRI) plot of chromosomes of the Mal.V2_NSR assembly (Query) against Mal.V1_CLR assembly (Ref) (A), and Hap1 (Query) against Hap2 (Ref) (B). (C) Chromosome-conserved synteny between Mal.V2_NSR and M. armatus (MasArm 1.2). Syntenic blocks are linked by shaded bands, and non-syntenic (non-matching) regions are shown as white gaps between Ref and Query.
Moreover, the synteny analysis for the Mal.V2_NSR assembly and M. armatus v1.2 (GCF_900324485.2) using homolog gene pairs was performed (Figure 2C). As a result, a total of 176 collinear blocks and 16,553 gene pairs were identified between swamp eel and M. armatus. Noticeably, nine chromosomes (chromosomes 2, 3, 4, 5, 6, 8, 9, 10, and 12) of M. albus harbored one-to-two synteny to the chromosomes of M. armatus, and chromosome 1 of M. albus showed conserved synteny with four chromosomes of M. armatus.
Sex-Related and Major Histocompatibility Complex Genes
A total of 59 sex-related genes were found and confirmed by using Blast against the nr database, and they were mapped onto 12 chromosomes (Supplementary Table S11) (Figure 3). Noticeably, a conserved dmrt1–dmrt3–dmrt2 (2a) gene cluster was found on chromosome 2. In addition, two copies of four sex-related genes, Tsh (Feng et al., 2022), Vasa (Ye et al., 2007), Smad2 (He et al., 2020), and cyp17 (Yu et al., 2003)were characterized with only one coding gene previously, and they were mapped to different chromosome positions. Twenty-three MHC genes were identified and classified as class I and class II, respectively (Supplementary Table S12), and they were mapped onto chromosomes 3, 6, 9, and 10 (Figure 3).

FIGURE 3. Distributions of sex-related genes and MHC genes on the newly obtained assembly (Mal.V2_NSR). The conserved dmrt1-dmrt3-dmrt2 gene cluster is shown in blue, MHC genes are shown in red, and sex genes with two copies are shown in green.
Discussion
As a widely distributed species throughout Asia, M. albus is widely cultured in China and other countries of Asia as aquaculture fish because of its economic value. M. albus undergoes sequential sex change from female to male after its first spawning at about 1.5 years old. The average fecundity of one-year-old females is only about 300 eggs per fish, and such low fecundity greatly hindered the production of fries and the breeding program, leading to the shortage of functional females in production. Few 10-years-old female individuals of swamp eel found previously (Yang et al., 2008) suggest some individuals of swamp eel could maintain its gender during whole life history. About 3–5% females could remain their gender 3 years or more and lay more than 3,000 eggs per fish in production. These females are referred to as non-sex reversal (NSR) females. According to the relationships between total length with age and von Bertalanffy growth curve (Yang and Xiong, 2010), these NSR female might be over 6 years old. Hence, such NSR female individuals of swamp eel would be valuable resources for studying mechanisms of sex change.
In the present study, we have combined the benefit of highly accurate reads with an improved assembly tool Hifiasm (Cheng et al., 2021b) and Hi-C sequencing technologies, a de novo assembly (Mal.V2_NSR), and two haplotype-resolved assemblies (Hap1 and Hap2) of one natural NSR female of M. albus were obtained. The higher contig N50 values indicate that all these three assemblies were significantly more continuous than the Mal.V1_CLR assembly. All these analyses indicate that these assemblies were significantly more continuous than the Mal.V1_CLR assembly. The slightly higher BUSCO percentages and QV values indicate the completeness and accuracy of the three assemblies, and the conserved similarity detected using CHROMEISTER further confirmed the quality of these three assemblies.
Genomic structural variation are important contributor to genetic diversity, and have been found facilitating adaptation, shaping complex trait, and even involved in speciation (Feulner and De-Kayne, 2017; Chakraborty et al., 2019; Hamala et al., 2021). Excepting massive syntenic blocks detected by Syri, we found 268 inversions (∼15 Mb), 540 translocations (∼5 Mb), and 137 duplications in Mal.V1_CLR (1.5 Mb) and 890 duplications in Mal.V2_NSR (4.38 Mb). We also identified 78.73 Mb of Mal.V1_CLR and 88.50 Mb of Mal.V2_NSR DNA sequences that were not aligned (Supplementary Table S9). Meanwhile, comparing the two haplotype assemblies allowed us to gain insights into the haplotype diversity within NSR female individuals, including 224 inversions, 276 translocations, and 25 duplications in Hap1 and 272 duplications in Hap2, making up about 8% of the assembled sequence (66.04 and 69.64 Mb) in each of the haplotype assemblies. Structural variation refers to genomic alterations with a wide size range, including inversions, translocations, and duplications (or deletions). A total of 19 GO terms were enriched for the PAV genes specifically for the Mal.V1_CLR and Mal.V2_NSR assembly. Interestingly, threonine-type peptidase activity and threonine-type endopeptidase activity, which are enriched molecular functions for bovine seminal plasma proteins (Gomes et al., 2020), are also enriched in the Mal.V2_NSR assembly, suggesting the genes involved in semen development might be affected by these PAVs in NSR females. Besides, the uncovered heterozygosity between two haplotype assemblies would also be valuable resources to uncovering potential sex-determination regions and/or allele-specific expression that might lead to large phenotypic variations (Zhang et al., 2020).
M. albus has 12 acrocentric chromosome pairs (Ji et al., 2003), the lowest chromosome number among teleost fish. Whole-chromosome fusions have been found in this lineage after being separated from medaka (Cheng et al., 2020). Through a comparison with the annotated genome of M. armatus, a total of 176 collinear blocks and 16,553 gene pairs were identified between swamp eel and M. armatus, and multiple 2:1 chromosome level syntenic alignments between M. armatus and M. albus were found (Figure 2B). Considering the chromosome number of other species in Synbranchids, for example, M. cuchia (2n = 42) (Rishi and Haobam, 1984), Synbranchus marmoratus (2n = 42–46) (Carvalho et al., 2012; Utsunomia et al., 2014), Ophisternon aenigmaticum (2n = 46) (Nirchio et al., 2011), and O. bengalense (2n = 46) (Carvalho et al., 2012) and a reduction in the chromosome number from 2n = 24 to 2n = 18 found in one Asian swamp population native to central Thailand (Supiwong et al., 2019), the chromosome-wide fusion might have occurred very recently and frequently after the divergence of M. albus from other species of Synbranchids during evolution and led to the great reduction in the chromosome number of this species.
Fish sex can be very plastic, with the combination of genetic and environmental influences, and the sex determination mechanism shows a high degree of plasticity and complexity correspondingly (Ribas et al., 2017). As a protogynous hermaphroditic fish, no heteromorphic sex chromosome has been found in M. albus yet. Batch sex-related genes have been characterized in M. albus previously and/or other teleosts and seem to be equally distributed on 12 chromosomes. The dmrt1-dmrt3-dmrt2 gene cluster conserved among teleosts (Dong et al., 2020) was found and mapped on chromosome 2, which confirmed a previous postulation based on the Southern blot analysis (Sheng et al., 2014). M. armatus, a closely related species of M. albus belonging to Synbranchiformes, which is also a sequentially hermaphroditic fish, and a young sex chromosome pair and an almost identical sex-linked region (SLR) between X and Y chromosomes were identified recently (Xue et al., 2021). Moreover, protogynous hermaphrodites have also been widely found in Synbranchidae, such as Synbranchus marmoratus, S. bengalensis, and Typhlosynbranchus boueti (Liem, 1968; Barros et al., 2012; Barros et al., 2017; Mazzoni et al., 2018). A closer inspection of the underlying sex-reversal mechanisms in Synbranchiformes would help understand the diversity and evolution of sex-determining modes.
The major histocompatibility complex (MHC) genes play key roles in antigen recognition and presentation in adaptive immunity, which are believed to have evolved in response to pathogens. The genomic organization of MHC genes varies greatly in teleosts (Hu et al., 2022). Because of the high degree of gene duplication and diversification of MHC genes, only two MHC genes have been characterized (Li et al., 2014; Pan et al., 2018). The high-quality chromosome-level genome assembly provide a new opportunity to address this problem. Here, a total of 23 MHC genes were characterized in the high continuous assembly, and they were mainly mapped on chromosomes 3, 6, 9, and 10 separately. This distribution pattern is similar to the pattern of other teleosts, such as D. rerio and rare minnow (Gobiocypris rarus) (Dijkstra et al., 2013; Hu et al., 2022). These results would help carry out future studies on the functional diversity of MHC genes.
Conclusion
In this study, we assembled a chromosome-scale de novo genome assembly and two haplotype-resolved genome assemblies of one natural non–sex reversal female of M.albus based on Illumina, HiFi, and Hi-C sequencing data. Both the sequence continuity (contig N50) and genome quality (base accuracy and completeness) were higher than those of the previously released swamp eel genome, implicating the advantages of HiFi reads on the de novo genome assembly. A comparative genome analysis revealed amounts of SVs between the NSR female and previous assembly, two haplotype assemblies, and the recent massive chromosome fusion in M. albus. This chromosome-scale unphased and two phased haplotype genome assemblies will help the characterization of phenotypic- and haplotype-specific mutations and their functions. Furthermore, we provided a genomic survey on the 59 genes potentially associated with the sex change and 23 MHC genes. Altogether, these data provide valuable resources for further biological studies and molecular breeding of this species to obtain the genetic improvement of this economically important teleost fish.
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Wang et al., 2017) in the National Genomics Data Center (National Genomics Data Center and Partners, 2020), Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under the accession number CRA006424 that are publicly accessible at https://ngdc.cncb.ac.cn/gsa/. Datasets generated during this study have been deposited in the Genome Warehouse in National Genomics Data Center (https://ngdc.cncb.ac.cn/gwh/), under GWHBHOS00000000, GWHBHOR00000000, and GWHBHOQ00000000.
Ethics Statement
The animal study was reviewed and approved by the Animal Experimental Ethical Inspection of Laboratory Animal Centre, Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Author Contributions
ZL conceived the project. H-FT and QH collected, identified, and photographed the specimens. H-FT and QH analyzed the genome data. H-YL performed gene analysis. H-FT and H-YL wrote the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by the Central Public-Interest Scientific Institution Basal Research Fund, the Chinese Academy of Fishery Sciences (CAFS) (Nos 2020XT0801 and 2020TD33), and the Central Public-Interest Scientific Institution Basal Research Fund (No. YFI202204).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.903185/full#supplementary-material
References
Aggarwal, G., and Ramaswamy, R. (2002). Ab Initio gene Identification: Prokaryote Genome Annotation with GeneScan and GLIMMER. J. Biosci. 27 (1), 7–14. doi:10.1007/Bf02703679
Amornsakun, T., Promkaew, P., and bin Hassan, A. (2018). Female Biology and Early Life Aspects of the Swamp Eel (Monopterus albus). Songklanakarin J. Sci. Tech. 40 (6), 1420–1427.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 25 (1), 25–29. doi:10.1038/75556
Bao, W., Kojima, K. K., and Kohany, O. (2015). Repbase Update, a Database of Repetitive Elements in Eukaryotic Genomes. Mobile DNA 6, 11. doi:10.1186/s13100-015-0041-9
Barros, N. H. C., de Souza, A. A., and Chellappa, S. (2012). Histological Aspects of Gonad Development in the Diandric Protogynous Sequential Hermaphrodite Fish Synbranchus Marmoratus (Osteichthyes: Synbranchidae). Anim. Biol. J. 3 (4), 159.
Barros, N. H. C., de Souza, A. A., Peebles, E. B., and Chellappa, S. (2017). Dynamics of Sex Reversal in the Marbled Swamp Eel (Synbranchus Marmoratus Bloch, 1795), a Diandric Hermaphrodite from Marechal Dutra Reservoir, Northeastern Brazil. J. Appl. Ichthyol. 33 (3), 443–449. doi:10.1111/jai.13273
Benson, G. (1999). Tandem Repeats Finder: a Program to Analyze DNA Sequences. Nucleic Acids Res. 27 (2), 573–580. doi:10.1093/nar/27.2.573
Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and Genomewise. Genome Res. 14 (5), 988–995. doi:10.1101/gr.1865504
Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., et al. (2003). The SWISS-PROT Protein Knowledgebase and its Supplement TrEMBL in 2003. Nucleic Acids Res. 31 (1), 365–370. doi:10.1093/nar/gkg095
Cai, J., Yang, W., Chen, D., Zhang, Y., He, Z., Zhang, W., et al. (2017). Transcriptomic Analysis of the Differentiating Ovary of the Protogynous Ricefield Eel Monopterus albus. Bmc Genomics 18, 573. doi:10.1186/S12864-017-3953-6
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and Applications. Bmc Bioinformatics 10, 421. doi:10.1186/1471-2105-10-421
Cantarel, B. L., Korf, I., Robb, S. M. C., Parra, G., Ross, E., Moore, B., et al. (2008). MAKER: an Easy-To-Use Annotation Pipeline Designed for Emerging Model Organism Genomes. Genome Res. 18 (1), 188–196. doi:10.1101/gr.6743907
Carvalho, N. D. M., Gross, M. C., Schneider, C. H., Terencio, M. L., Zuanon, J., and Feldberg, E. (2012). Cytogenetics of Synbranchiformes: a Comparative Analysis of Two Synbranchus Bloch, 1795 Species from the Amazon. Genetica 140 (4-6), 149–158. doi:10.1007/s10709-012-9666-5
Chakraborty, M., Emerson, J. J., Macdonald, S. J., and Long, A. D. (2019). Structural Variants Exhibit Widespread Allelic Heterogeneity and Shape Variation in Complex Traits. Nat. Commun. 10. doi:10.1038/S41467-019-12884-1
Chan, S. T. H., and Phillips, J. G. (1967). The Structure of the Gonad during Natural Sex Reversal in Monopterus albus (Pisces: Teleostei). J. Zool. 151 (1), 129–141. doi:10.1111/j.1469-7998.1967.tb02868.x
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, J., Zhu, Z., and Hu, W. (2022). Progress in Research on Fish Sex Determining Genes. Water Biol. Security 1 (1), 100008. doi:10.1016/j.watbs.2022.100008
Chen, N. (2004). Using RepeatMasker to Identify Repetitive Elements in Genomic Sequences. Curr. Protoc. Bioinformatics Chapter 4 (1), Unit.10.11. doi:10.1002/0471250953.bi0410s05
Cheng, H., Guo, Y., Yu, Q., and Zhou, R. (2003). The rice Field Eel as a Model System for Vertebrate Sexual Development. Cytogenet. Genome Res. 101 (3-4), 274–277. doi:10.1159/000074348
Cheng, Y., Shang, D., Luo, M., Huang, C., Lai, F., Wang, X., et al. (2020). Whole Genome-wide Chromosome Fusion and New Gene Birth in the Monopterus albus Genome. Cell Biosci 10, 67. doi:10.1186/s13578-020-00432-0
Cheng, H., He, Y., and Zhou, R. (2021a). Swamp Eel (Monopterus albus). Trends Genetics: TIG S0168-9525, 00268–00267. doi:10.1016/j.tig.2021.09.007
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H., and Li, H. (2021b). Haplotype-resolved De Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat. Methods 18 (2), 170–175. doi:10.1038/s41592-020-01056-5
Chi, W., Gao, Y., Hu, Q., Guo, W., and Li, D. (2017). Genome-wide Analysis of Brain and Gonad Transcripts Reveals Changes of Key Sex Reversal-Related Genes Expression and Signaling Pathways in Three Stages of Monopterus albus. Plos One 12 (3), e0173974. doi:10.1371/journal.pone.0173974
Dijkstra, J. M., Grimholt, U., Leong, J., Koop, B. F., and Hashimoto, K. (2013). Comprehensive Analysis of MHC Class II Genes in Teleost Fish Genomes Reveals Dispensability of the Peptide-Loading DM System in a Large Part of Vertebrates. BMC Evol. Biol. 13, 260. doi:10.1186/1471-2148-13-260
Ding, W., Cao, L., Cao, Z., and Bing, X. (2020). Transcriptome Analysis of Blood for the Discovery of Sex-Related Genes in Ricefield Eel Monopterus albus. Fish. Physiol. Biochem. 46 (4), 1507–1518. doi:10.1007/s10695-020-00809-5
Dong, J., Li, J., Hu, J., Sun, C., Tian, Y., Li, W., et al. (2020). Comparative Genomics Studies on the Dmrt Gene Family in Fish. Front. Genet. 11, 563947. doi:10.3389/Fgene.2020.563947
Dudchenko, O., Batra, S. S., Omer, A. D., Nyquist, S. K., Hoeger, M., Durand, N. C., et al. (2017). De Novo assembly of the Aedes aegypti Genome Using Hi-C Yields Chromosome-Length Scaffolds. Science 356 (6333), 92–95. doi:10.1126/science.aal3327
Durand, N. C., Robinson, J. T., Shamim, M. S., Machol, I., Mesirov, J. P., Lander, E. S., et al. (2016a). Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cel Syst. 3 (1), 99–101. doi:10.1016/j.cels.2015.07.012
Durand, N. C., Shamim, M. S., Machol, I., Rao, S. S. P., Huntley, M. H., Lander, E. S., et al. (2016b). Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cel Syst. 3 (1), 95–98. doi:10.1016/j.cels.2016.07.002
Feng, K., Zhu, K., Wu, Z., Su, S., and Yao, W. (2022). Identification and Expression Analysis of Thyroid-Stimulating Hormone β Subunit, and Effects of T3 on Gonadal Differentiation-Related Gene Expression in rice Field Eel, Monopterus albus. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 258, 110681. doi:10.1016/j.cbpb.2021.110681
Feulner, P. G. D., and De-Kayne, R. (2017). Genome Evolution, Structural Rearrangements and Speciation. J. Evol. Biol. 30 (8), 1488–1490. doi:10.1111/jeb.13101
Gao, Y., Guo, W., Hu, Q., Zou, M., Tang, R., Chi, W., et al. (2014). Characterization and Differential Expression Patterns of Conserved microRNAs and mRNAs in Three Genders of the Rice Field Eel (Monopterus albus). Sex. Dev. 8 (6), 387–398. doi:10.1159/000369181
Gao, Y., Jia, D., Hu, Q., and Li, D. (2016). Foxl3, a Target of miR-9, Stimulates Spermatogenesis in Spermatogonia during Natural Sex Change in Monopterus albus. Endocrinology 157 (11), 4388–4399. doi:10.1210/en.2016-1256
Ghosh, S., and Chan, C.-K. K. (2016). “Analysis of RNA-Seq Data Using TopHat and Cufflinks,” in Plant Bioinformatics: Methods and Protocols. Editor D. Edwards (New York, NY: Springer New York), 339–361. doi:10.1007/978-1-4939-3167-5_18
Goel, M., Sun, H., Jiao, W.-B., and Schneeberger, K. (2019). SyRI: Finding Genomic Rearrangements and Local Sequence Differences from Whole-Genome Assemblies. Genome Biol. 20 (1), 277. doi:10.1186/S13059-019-1911-0
Gomes, F. P., Park, R., Viana, A. G., Fernandez-Costa, C., Topper, E., Kaya, A., et al. (2020). Protein Signatures of Seminal Plasma from Bulls with Contrasting Frozen-Thawed Sperm Viability. Sci. Rep. 10 (1). doi:10.1038/S41598-020-71015-9
Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A., and Eddy, S. R. (2003). Rfam: an RNA Family Database. Nucleic Acids Res. 31 (1), 439–441. doi:10.1093/nar/gkg006
Hamala, T., Wafula, E. K., Guiltinan, M. J., Ralph, P. E., dePamphilis, C. W., and Tiffin, P. (2021). Genomic Structural Variants Constrain and Facilitate Adaptation in Natural Populations of Theobroma cacao, the Chocolate Tree. Proc. Nat. Acad. Sci. USA 118 (35). doi:10.1073/pnas.2102914118
He, Z., Deng, F., Xiong, S., Cai, Y., He, Z., Wang, X., et al. (2020). Expression and Regulation of Smad2 by Gonadotropins in the Protogynous Hermaphroditic Ricefield Eel (Monopterus albus). Fish. Physiol. Biochem. 46 (3), 1155–1165. doi:10.1007/s10695-020-00778-9
Herawati, V. E., Nugroho, R. A., PinandoyoHutabarat, J., Hutabarat, J., Prayitno, B., and Karnaradjasa, O. (2018). The Growth Performance and Nutrient Quality of Asian Swamp Eel Monopterus albus in Central Java Indonesia in a Freshwater Aquaculture System with Different Feeds. J. Aquat. Food Product. Tech. 27 (6), 658–666. doi:10.1080/10498850.2018.1483990
Hilles, A. R., Mahmood, S., and Hashim, R. (2018). The Secret behind the Natural Sex Reversal in Ricefield Eel (Monopterus albus) Remains Unknown. Adv. Complement Alt. Med. 2 (2), ACAM.000533.2018. doi:10.31031/ACAM.2018.02.000533
Hu, Q., Guo, W., Gao, Y., Tang, R., and Li, D. (2014). Molecular Cloning and Analysis of Gonadal Expression of Foxl2 in the rice-field Eel Monopterus albus. Sci. Rep. 4, 6884. doi:10.1038/Srep06884
Hu, Q., Guo, W., Gao, Y., Tang, R., and Li, D. (2015). Molecular Cloning and Characterization of Amh and Dax1 Genes and Their Expression during Sex Inversion in rice-field Eel Monopterus albus. Sci. Rep. 5 (1), 16667. doi:10.1038/srep16667
Hu, X. D., Li, H. R., Lin, Y. S., Wang, Z. K., Feng, H. H., Zhou, M., et al. (2022). Genomic Deciphering of Sex Determination and Unique Immune System of a Potential Model Species Rare Minnow (Gobiocypris rarus). Sci. Adv. 8 (5), eabl7253. doi:10.1126/sciadv.abl7253
Huang, X., Guo, Y., Shui, Y., Gao, S., Yu, H., Cheng, H., et al. (2005). Multiple Alternative Splicing and Differential Expression of Dmrt1 during Gonad Transformation of the Rice Field Eel1. Biol. Reprod. 73 (5), 1017–1024. doi:10.1095/biolreprod.105.041871
Ji, F. Y., Yu, Q. X., Li, K., and Ren, X. H. (2003). Ag-staining Pattern, FISH and ISH with rDNA Probes in the rice Field Eel (Monopterus albus Zuiew) Chromosomes. Hereditas 138 (3), 207–212. doi:10.1034/j.1601-5223.2003.01643.x
Jurka, J., Kapitonov, V. V., Pavlicek, A., Klonowski, P., Kohany, O., and Walichiewicz, J. (2005). Repbase Update, a Database of Eukaryotic Repetitive Elements. Cytogenet. Genome Res. 110 (1-4), 462–467. doi:10.1159/000084979
Kalvari, I., Argasinska, J., Quinones-Olvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., et al. (2018). Rfam 13.0: Shifting to a Genome-Centric Resource for Non-coding RNA Families. Nucleic Acids Res. 46 (D1), D335–D342. doi:10.1093/nar/gkx1038
Khanh, N., and Ngan, H. (2010). Current Practices of rice Field Eel Monopterus albus (Zuiew, 1973) Culture in Vietnam. Aquac. Asia Mag. 15 (3), 26–29.
Lai, F., Cheng, Y., Zou, J., Wang, H., Zhu, W., Wang, X., et al. (2021). Identification of Histone Modifications Reveals a Role of H2b Monoubiquitination in Transcriptional Regulation of Dmrt1 in Monopterus albus. Int. J. Biol. Sci. 17 (8), 2009–2020. doi:10.7150/ijbs.59347
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi:10.1093/bioinformatics/btp352
Li, H. (2018). Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 34 (18), 3094–3100. doi:10.1093/bioinformatics/bty191
Li, W., Sun, W.-x., Meng, L., and Hong, D.-w. (2014). Molecular Cloning, Genomic Structure, Polymorphism and Expression Analysis of Major Histocompatibility Complex Class IIA Gene of Swamp Eel Monopterus albus. Biologia 69 (2), 236–246. doi:10.2478/s11756-013-0307-y
Liem, K. F. (1968). Geographical and Taxonomic Variation in the Pattern of Natural Sex Reversal in the Teleost Fish Order Synbranchiformes. J. Zoolog. 156 (2), 225–238. doi:10.1111/j.1469-7998.1968.tb05930.x
Liem, K. F. (1963). Sex Reversal as a Natural Process in the Synbranchiform Fish Monopterus albus. Copeia 1963 (2), 303–312. doi:10.2307/1441348
Liu, C. K. (1944). Rudimentary Hermaphroditism in the Symbranchoid Eel, Monopterus Javanensis. Sinensia 15, 1–8.
Liu, J.-F., Guiguen, Y., and Liu, S.-J. (2009). Aromatase (P450arom) and 11β-Hydroxylase (P45011β) Genes Are Differentially Expressed during the Sex Change Process of the Protogynous rice Field Eel, monopterus Albus. Fish. Physiol. Biochem. 35 (3), 511–518. doi:10.1007/s10695-008-9255-9
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 25 (5), 955–964. doi:10.1093/nar/25.5.955
Ma, K., and Shi, L.-m. (1987). Meiosis and Synaptonemal Complex Karyotype of the Eel (Monopterus albus Zuiew). Zoolog. Res. 8 (2), 159–163.
Marçais, G., and Kingsford, C. (2011). A Fast, Lock-free Approach for Efficient Parallel Counting of Occurrences of K-Mers. Bioinformatics 27 (6), 764–770. doi:10.1093/bioinformatics/btr011
Mazzoni, T., Lo Nostro, F., Antoneli, F., and Quagio-Grassiotto, I. (2018). Action of the Metalloproteinases in Gonadal Remodeling during Sex Reversal in the Sequential Hermaphroditism of the Teleostei Fish Synbranchus Marmoratus (Synbranchiformes: Synbranchidae). Cells 7 (5), 34. doi:10.3390/Cells7050034
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 20 (9), 1297–1303. doi:10.1101/gr.107524.110
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 49 (D1), D412–D419. doi:10.1093/nar/gkaa913
National Genomics Data Center, and Partners (2020). Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 48 (D1), D24–D33. doi:10.1093/nar/gkaa1022
Nawrocki, E. P., and Eddy, S. R. (2013). Infernal 1.1: 100-fold Faster RNA Homology Searches. Bioinformatics 29 (22), 2933–2935. doi:10.1093/bioinformatics/btt509
Nirchio, M., Mariguela, T. C., Ferreira, I. A., Foresti, F., and Oliveira, C. (2011). Karyotype and Nucleolus Organizer Regions of Ophisternon Aenigmaticum (Teleostei: Synbranchiformes: Synbranchidae) FROM VENEZUELA. Interciencia 36 (3), 229–233.
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27 (1), 29–34. doi:10.1093/nar/27.1.29
Pan, T., Jiang, H., Duan, G., Hu, Y., Ling, J., and Zhou, H. (2018). Molecular Cloning and Expression of Major Histocompatibility Complex Class Iα Gene of Swamp Eel (Monopterus albus). The Israeli J. Aquac. – Bamidgeh 1468, 1–9. doi:10.46989/001c.20949
Parra, G., Bradnam, K., and Korf, I. (2007). CEGMA: a Pipeline to Accurately Annotate Core Genes in Eukaryotic Genomes. Bioinformatics 23 (9), 1061–1067. doi:10.1093/bioinformatics/btm071
Pérez-Wohlfeil, E., Diaz-del-Pino, S., and Trelles, O. (2019). Ultra-fast Genome Comparison for Large-Scale Genomic Experiments. Sci. Rep. 9, 10274. doi:10.1038/S41598-019-46773-W
Porubsky, D., Ebert, P., Ebert, P., Audano, P. A., Vollger, M. R., Harvey, W. T., et al. (2021). Fully Phased Human Genome Assembly without Parental Data Using Single-Cell Strand Sequencing and Long Reads. Nat. Biotechnol. 39 (3), 302–308. doi:10.1038/s41587-020-0719-5
Qu, X. C., Jiang, J. Y., Cheng, C., Feng, L., and Liu, Q. G. (2015). Cloning and Transcriptional Expression of a Novel Gene during Sex Inversion of the rice Field Eel (Monopterus albus). Springerplus 4 (1), 1–10. doi:10.1186/S40064-015-1544-Z
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 26 (6), 841–842. doi:10.1093/bioinformatics/btq033
Rhie, A., Walenz, B. P., Koren, S., and Phillippy, A. M. (2020). Merqury: Reference-free Quality, Completeness, and Phasing Assessment for Genome Assemblies. Genome Biol. 21 (1), 245. doi:10.1186/s13059-020-02134-9
Rhie, A., McCarthy, S. A., Fedrigo, O., Damas, J., Formenti, G., Koren, S., et al. (2021). Towards Complete and Error-free Genome Assemblies of All Vertebrate Species. Nature 592 (7856), 737–746. doi:10.1038/s41586-021-03451-0
Ribas, L., Liew, W. C., Díaz, N., Sreenivasan, R., Orbán, L., and Piferrer, F. (2017). Heat-induced Masculinization in Domesticated Zebrafish Is Family-specific and Yields a Set of Different Gonadal Transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 114 (6), E941–E950. doi:10.1073/pnas.1609411114
Rishi, K., and Haobam, M. (1984). Somatic Chromosomes in a marine Fish, Megalops cyprinoides (Broussonet) (Megalopidae: Elopiformes). Chromosome Inf. Serv. 36, 22–24.
Roach, M. J., Schmidt, S. A., and Borneman, A. R. (2018). Purge Haplotigs: Allelic Contig Reassignment for Third-Gen Diploid Genome Assemblies. BMC Bioinformatics 19 (1), 460. doi:10.1186/s12859-018-2485-7
Sheng, Y., Chen, B., Zhang, L., Luo, M., Cheng, H., and Zhou, R. (2014). Identification of Dmrt Genes and Their Up-Regulation during Gonad Transformation in the Swamp Eel (Monopterus albus). Mol. Biol. Rep. 41 (3), 1237–1245. doi:10.1007/s11033-013-2968-6
Silva, A. A., Suay, E. S., Silva, W. S., and Albarico, F. P. J. B. (2020). Notes on Freshwater Clam and Land Snail as Alternative Food for rice Paddy Eel, Monopterus albus. Int. J. Fish. Aquat. Stud. 8 (3), 223–226.
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 31 (19), 3210–3212. doi:10.1093/bioinformatics/btv351
Stanke, M., Keller, O., Gunduz, I., Hayes, A., Waack, S., and Morgenstern, B. (2006). AUGUSTUS: Ab Initio Prediction of Alternative Transcripts. Nucleic Acids Res. 34 (Web Server issue), W435–W439. doi:10.1093/nar/gkl200
Supiwong, W., Pinthong, K., Seetapan, K., Saenjundaeng, P., Bertollo, L. A. C., de Oliveira, E. A., et al. (2019). Karyotype Diversity and Evolutionary Trends in the Asian Swamp Eel Monopterus albus (Synbranchiformes, Synbranchidae): a Case of Chromosomal Speciation? BMC Evol. Biol. 19 (1), 73. doi:10.1186/s12862-019-1393-4
Susatyo, P., Umami, R., and Sukmaningrum, S. (2020). Reproductive Characters of the Ricefield Eel (Monopterus albus Zuieuw) in Babakan Village, Karang Lewas District, Banyumas, Central Java. IOP Conf. Ser. Earth Environ. Sci. 593, 012015. doi:10.1088/1755-1315/593/1/012015
Tang, F., Chan, S. T. H., and Lofts, B. (1975). A Study on the 3β‐ and 17β‐hydroxysteroid Dehydrogenase Activities in the Gonads of Monopterus albus (Pisces: Teleostei) at Various Sexual Phases during Natural Sex Reversal. J. Zoolog. 175 (4), 571–580. doi:10.1111/j.1469-7998.1975.tb01418.x
Tang, H., Wang, X., Bowers, J. E., Ming, R., Alam, M., and Paterson, A. H. (2008). Unraveling Ancient Hexaploidy through Multiply-Aligned Angiosperm Gene Maps. Genome Res. 18 (12), 1944–1954. doi:10.1101/gr.080978.108
Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., et al. (2001). The COG Database: New Developments in Phylogenetic Classification of Proteins from Complete Genomes. Nucleic Acids Res. 29 (1), 22–28. doi:10.1093/Nar/29.1.22
Tian, H.-F., Hu, Q.-M., and Li, Z. (2021). A High-Quality De Novo Genome Assembly of One Swamp Eel (Monopterus albus) Strain with PacBio and Hi-C Sequencing Data. G3-Genes Genomes Genet. 11 (1), 1–9. doi:10.1093/g3journal/jkaa032
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: Discovering Splice Junctions with RNA-Seq. Bioinformatics 25 (9), 1105–1111. doi:10.1093/bioinformatics/btp120
Utsunomia, R., Pansonato-Alves, J. C., Scacchetti, P. C., Oliveira, C., and Foresti, F. (2014). Scattered Organization of the Histone Multigene Family and Transposable Elements in Synbranchus. Genet. Mol. Biol. 37 (1), 30–36. doi:10.1590/S1415-47572014000100007
Wang, Y., Song, F., Zhu, J., Zhang, S., Yang, Y., Chen, T., et al. (2017). GSA: Genome Sequence Archive *. Genomics, Proteomics & Bioinformatics 15 (1), 14–18. doi:10.1016/j.gpb.2017.01.001
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wenger, A. M., Peluso, P., Rowell, W. J., Chang, P.-C., Hall, R. J., Concepcion, G. T., et al. (2019). Accurate Circular Consensus Long-Read Sequencing Improves Variant Detection and Assembly of a Human Genome. Nat. Biotechnol. 37 (10), 1155–1162. doi:10.1038/s41587-019-0217-9
Wu, Y., He, Z., Zhang, L., Jiang, H., and Zhang, W. (2012). Ontogeny of Immunoreactive Lh and Fsh Cells in Relation to Early Ovarian Differentiation and Development in Protogynous Hermaphroditic Ricefield Eel Monopterus albus1. Biol. Reprod. 86 (3), 95646. doi:10.1095/biolreprod.111.095646
Xiao, Y.-M., Chen, L., Liu, J., Liu, W.-B., Chen, H.-G., Zou, L.-J., et al. (2009). Contrast Expression Patterns of JNK1 during Sex Reversal of the rice-field Eel. J. Exp. Zool. 9999B (3), a–n. doi:10.1002/jez.b.21332
Xu, Z., and Wang, H. (2007). LTR_FINDER: an Efficient Tool for the Prediction of Full-Length LTR Retrotransposons. Nucleic Acids Res. 35, W265–W268. doi:10.1093/nar/gkm286
Xue, L., Gao, Y., Wu, M., Tian, T., Fan, H., Huang, Y., et al. (2021). Telomere-to-telomere Assembly of a Fish Y Chromosome Reveals the Origin of a Young Sex Chromosome Pair. Genome Biol. 22 (1), 203. doi:10.1186/s13059-021-02430-y
Yang, D., Chen, F., and Ruan, G. (2018). “Aquaculture of the Paddy Eel, Monopterus albus,” in Aquaculture in China: Success Stories and Modern Trends (Wiley Online Library), 283–296. doi:10.1002/9781119120759.ch3_9
Yang, D., Chen, F., Ruan, G., and Su, Y.-b. (2008). Relationship between Sex Reversal, Body Weight and Age of Monopterus albus. J. Yangtze Univ.(Nat. Sci. Edit) 5 (4), 45–47.
Yang, D., Chen, F., Ruan, G. L., and Li, T. M. (2009). Comparative Study on Fecundity of Different Strains of Monopterus albus. J. Hydroecology 2 (4), 133–135.
Yang, M. S., and Xiong, B. X. (2010). Age and Growth of Monopterus albus Zuiew, 1793 (Synbranchidae). J. Appl. Ichthyol. 26 (3), 488–490. doi:10.1111/j.1439-0426.2009.01358.x
Ye, D., Lv, D., Song, P., Peng, M., Chen, Y., Guo, M., et al. (2007). Cloning and Characterization of a rice Field Eel Vasa-like Gene cDNA and its Expression in Gonads during Natural Sex Transformation. Biochem. Genet. 45 (3-4), 211–224. doi:10.1007/s10528-006-9066-6
Yeung, W. S. B., and Chan, S. T. H. (1987). The Plasma Sex Steroid Profiles in the Freshwater, Sex-Reversing Teleost Fish, Monopterus albus (Zuiew). Gen. Comp. Endocrinol. 65 (2), 233–242. doi:10.1016/0016-6480(87)90171-7
Yi, T. L., Pei, M. T., and Yang, D. Q. (2019). Expression Patterns of Kiss2 and Gpr54‐2 in Monopterus albus Suggest These Genes May Play a Role in Sex Reversal in Fish. Febs Open Bio 9 (10), 1835–1844. doi:10.1002/2211-5463.12727
Yu, H., Cheng, H., Guo, Y., Xia, L., and Zhou, R. (2003). Alternative Splicing and Differential Expression of P450c17 (CYP17) in Gonads during Sex Transformation in the rice Field Eel. Biochem. Biophysical Res. Commun. 307 (1), 165–171. doi:10.1016/S0006-291x(03)01137-9
Yu, X.-J., Zheng, H.-K., Wang, J., Wang, W., and Su, B. (2006). Detecting Lineage-specific Adaptive Evolution of Brain-Expressed Genes in Human Using Rhesus Macaque as Outgroup. Genomics 88 (6), 745–751. doi:10.1016/j.ygeno.2006.05.008
Yue, H., Fu, P., Ruan, R., Ye, H., Li, Z., and Li, C. (2021). Influence of Broodstock Diets on Growth, Fecundity and Spawning Performance of Swamp Eel Monopterus albus. Aquac. Res. 52 (5), 1935–1944. doi:10.1111/are.15042
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan - an Integration Platform for the Signature-Recognition Methods in InterPro. Bioinformatics 17 (9), 847–848. doi:10.1093/bioinformatics/17.9.847
Zhang, X. T., Wu, R. X., Wang, Y. B., Yu, J. X., and Tang, H. B. (2020). Unzipping Haplotypes in Diploid and Polyploid Genomes. Comput. Struct. Biotechnol. J. 18, 66–72. doi:10.1016/j.csbj.2019.11.011
Zhang, Y., Zhang, W., Yang, H., Zhou, W., Hu, C., and Zhang, L. (2008). Two Cytochrome P450 Aromatase Genes in the Hermaphrodite Ricefield Eel Monopterus albus: mRNA Expression during Ovarian Development and Sex Change. J. Endocrinol. 199 (2), 317–331. doi:10.1677/Joe-08-0303
Zhang, Y., Zhang, S., Liu, Z., Zhang, L., and Zhang, W. (2013). Epigenetic Modifications during Sex Change Repress Gonadotropin Stimulation of Cyp19a1a in a Teleost Ricefield Eel (Monopterus albus). Endocrinology 154 (8), 2881–2890. doi:10.1210/en.2012-2220
Zhang, L., Yang, Q., Xu, W., Wu, Z., and Li, D. (2021). Integrated Analysis of miR-430 on Steroidogenesis-Related Gene Expression of Larval Rice Field Eel Monopterus albus. Int. J. Mol. Sci. 22 (13), 6994. doi:10.3390/Ijms22136994
Zhou, R., Liu, L., Guo, Y., Yu, H., Cheng, H., Huang, X., et al. (2003). Similar Gene Structure of twoSox9a Genes and Their Expression Patterns during Gonadal Differentiation in a Teleost Fish, rice Field Eel (Monopterus albus). Mol. Reprod. Dev. 66 (3), 211–217. doi:10.1002/mrd.10271
Keywords: Monopterus albus, non-sex-reversal female, phased haplotype, genome assembly, HIFI
Citation: Tian H-F, Hu Q, Lu H-Y and Li Z (2022) Chromosome-Scale, Haplotype-Resolved Genome Assembly of Non-Sex-Reversal Females of Swamp Eel Using High-Fidelity Long Reads and Hi-C Data. Front. Genet. 13:903185. doi: 10.3389/fgene.2022.903185
Received: 24 March 2022; Accepted: 04 April 2022;
Published: 18 May 2022.
Edited by:
Xiangshan Ji, Shandong Agricultural University, ChinaReviewed by:
Guangfu Hu, Huazhong Agricultural University, ChinaDapeng Li, Huazhong Agricultural University, China
Copyright © 2022 Tian, Hu, Lu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong Li, bGl6aG9uZ0B5ZmkuYWMuY24=
 Hai-Feng Tian
Hai-Feng Tian Qiaomu Hu
Qiaomu Hu Hong-Yi Lu
Hong-Yi Lu Zhong Li
Zhong Li