
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet. , 20 May 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.900324
This article is part of the Research Topic Harnessing Cytokinin Biology in Crop Biofortification and Enhanced Food Security View all 16 articles
 Rohit Kumar Mahto1
Rohit Kumar Mahto1 Ambika2
Ambika2 Charul Singh3
Charul Singh3 B S. Chandana1
B S. Chandana1 Rajesh Kumar Singh1
Rajesh Kumar Singh1 Shruti Verma4
Shruti Verma4 Vijay Gahlaut5
Vijay Gahlaut5 Murli Manohar6
Murli Manohar6 Neelam Yadav7
Neelam Yadav7 Rajendra Kumar1*
Rajendra Kumar1*Globally more than two billion people suffer from micronutrient malnutrition (also known as “hidden hunger”). Further, the pregnant women and children in developing nations are mainly affected by micronutrient deficiencies. One of the most important factors is food insecurity which can be mitigated by improving the nutritional values through biofortification using selective breeding and genetic enhancement techniques. Chickpea is the second most important legume with numerous economic and nutraceutical properties. Therefore, chickpea production needs to be increased from the current level. However, various kind of biotic and abiotic stresses hamper global chickpea production. The emerging popular targets for biofortification in agronomic crops include targeting cytokinin dehydrogenase (CKX). The CKXs play essential roles in both physiological and developmental processes and directly impact several agronomic parameters i.e., growth, development, and yield. Manipulation of CKX genes using genome editing tools in several crop plants reveal that CKXs are involved in regulation yield, shoot and root growth, and minerals nutrition. Therefore, CKXs have become popular targets for yield improvement, their overexpression and mutants can be directly correlated with the increased yield and tolerance to various stresses. Here, we provide detailed information on the different roles of CKX genes in chickpea. In the end, we discuss the utilization of genome editing tool clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) to engineer CKX genes that can facilitate trait improvement. Overall, recent advancements in CKX and their role in plant growth, stresses and nutrient accumulation are highlighted, which could be used for chickpea improvement.
Micronutrient malnutrition, often known as “hidden hunger” affects more than half of the world’s population, with pregnant women and children in developing nations bearing the brunt of the burden. According to the World Health Organisation (WHO), more than 2 billion people are suffering from hidden hunger and one of the most important responsible factors for malnutrition is food insecurity which affects almost a billion people worldwide (FAO, 2013). Food security is defined as when all people, at all times, have physical, social and economic access to sufficient, safe and nutritious food that meets their dietary needs and food preferences for active and healthy life (FAO, 2016). This is particularly an issue in developing countries where families cannot afford or get access to sufficient and nutritious food. Each year twenty million infants are born with low body weight. As per the malnutrition report around 150.8 million stunted, 50.5 million wasted, and 38.3 million overweight children under five years of age are found (WHO, 2018; Kumar and Pandey, 2020; Ahmed et al., 2022). The reason for malnutrition is due to the insufficient or poor-quality supply or uptake of nutrition. The consumption of legumes in daily diets, especially in developing countries (Afro-Asian countries), could majorly eradicate protein malnutrition. Legumes could act as a base for the development of many functional foods to promote health benefits in humans (Maphosa and Jideani, 2017; Kumar and Pandey, 2020; Ahmed et al., 2022).
Chickpea (Cicer arietinum L.) is the world’s second largest, cool season food legume. It is in high demand owing to its high nutritional value. Chickpea is considered invaluable because it provides food for human consumption and feed to livestock. Owing to these astounding properties of chickpea, its production needs to be enhanced to feed and ensure nutritional health and well-being of world’s population. It is also essential to involve the screening techniques, which is useful to promote the breeding program for increasing the growth of chickpea (Talip et al., 2018). However, various factors hamper the yield of this crop. Climate variability has altered plant physiology in a variety of ways. Multiple stressors on plants are increased as a result of environmental extremes and climate unpredictability (Thornton et al., 2014). Heat stress reduces grain yield and productivity, cold stress causes sterility, and drought stress has a deleterious impact on plant morpho-physiology (Barlow et al., 2015). The generation of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), superoxide (.O2), and hydroxyl radicals (.OH−) are accelerated in plant tissues due to harsh circumstances. In addition to other stress hormones like abscisic acid (ABA), jasmonate, and salicylic acid, cytokinins (CKs) also play a significant role in increasing plant stress tolerance and regulate the action of plants defensive mechanisms. Thus, understanding the roles of cytokinin in plant responses to abiotic stressors is very critical imperative.
The cytokinins have been largely involved in the regulation of plant yield, particularly by influencing the grain related traits i.e., number, size and growth of the root (Yamburenko et al., 2017). Moreover, they are found to be involved in stress regulations (Cortleven et al., 2019) and plant mineral concentration status (Gao et al., 2019). It has also been reported that cytokines negatively affect micronutrient uptake regulations (Gao et al., 2019). Cytokinin dehydrogenase (CKXs) is a key enzyme that regulates the cytokinin hormone level in plants (Sakakibara, 2006). It is a small gene family, for instance in Arabidopsis only seven CKX genes were reported (Werner et al., 2003). Several studies in Arabidopsis showed that CKX genes regulate plant growth and developmental processes including shoot and root development, reproductive meristem activity (Werner et al., 2006; Bartrina et al., 2011), and mineral [phosphorus (P), calcium (Ca), sulfur (S) and microelements like copper (Cu), manganese (Mn), iron (Fe), and zinc (Zn)] accumulations (Werner et al., 2010; Bartrina et al., 2011; Chang et al., 2015). However, related information and functional studies about CKX genes in chickpea and their utilization to improve agronomic traits are lacking. Various techniques are being employed to develop a sustainable agriculture system to decrease food insecurity. One such method is Genome editing (GE) for crop improvement that has the potential to create a climate-resilient agriculture system on a global scale (Liu et al., 2013). GE technologies have had a significant impact on plant breeding techniques including new strategies for making rapid and precise changes in crop genomes to protect plants from various challenges and enhance crop outputs (Taranto et al., 2018). In genome editing methods, site-specific endonucleases such as zinc-finger nucleases (ZFNs), transcription activator like effector nucleases (TALENs), and CRISPR-Cas9 are used (Zhu et al., 2017). Unlike the ZENs and TALENs genome editing tools, the CRISPR/Cas9 system is proving to be the most effective GE technology since it is cost-efficient, rapid, accurate, and allows for various site-specific genome editing (Abdelrahman et al., 2018a). In comparison to other genome editing methods like TALENs/ZFNs, CRISPR-based techniques have been intensively investigated in plant genomes. It also offers a lot of potential for assisting crop breeders in developing high-yielding, stress-resistant varieties (Abdelrahman et al., 2018b). Most importantly, CRISPR/Cas9 is coming up as a straight forward, environmentally benign strategy for making genome-edited non-transgenic plants to combat environmental extremes and maintain food security (Haque et al., 2018).
Apart from genome editing tools like CRISPR-Cas9 which are being increasingly used nowadays to develop stress tolerant varieties in different crops, there are some alternative strategies as well. Selection from landraces, hybridization to develop novel variety followed by pedigree selection, mutation breeding, and exploitation of hybrid vigour are examples of traditional breeding procedures that have resulted in considerable improvements in stress tolerance and nutritional quality. Applications of PGPRs are being used to alleviate abiotic stresses and increase productivity in economically important crops including rice, soybean, lettuce, tomato, maize and wheat. Nano-biotechnology has also proven to be a promising tool for sustainable agriculture, seed treatment and germination, plant growth and development, disease diagnostics, and detection of harmful agrochemicals (Nuruzzaman et al., 2016).
In this comprehensive review article, we emphasize the importance of legumes with reference to chickpea, malnutrition and food security, biotic and abiotic stresses, and the role of cytokinin. We also discuss how recently developed genome editing technologies such as CRISPR/Cas9 are being utilized to engineer CKX genes to improve agricultural traits and biofortification in chickpea.
Legumes are plants of the Leguminosae/Fabaceae family that bear seeds in pods (Staniak et al., 2014; Kouris-Blazos and Belski, 2016) and as a distinguishing feature fix atmospheric N2 in symbiosis with suitable rhizobia. Agriculturally significant legumes fix 40–60 million metric tonnes N2, along with an additional 3–5 million tonnes by wild legumes, annually (Smil, 1999). The principal edible legumes are bean, broad bean, chickpea, cowpea, pea, pigeon peaand lentil (National Academy of Science, 1994). However, peas, broad beans, lentils, soybeans, lupins, sprouts, mung bean, green beans, and peanuts are common legumes utilized for human consumption and are known as grain or grain food legumes (Yorgancilar and Bilgicli, 2014). Food legumes are classified into two types: oilseeds and pulses. The oilseeds are high-oil-content legumes such as soybeans and peanuts, whereas the pulses are all dry seeds of cultivated legumes eaten as traditional food. These seeds are recognized globally as a low-cost meat substitute and regarded as the second most important dietary source after cereals (Kouris-Blazos and Belski, 2016). Legumes are high inprotein, essential amino acids, complex carbohydrates, dietary fibre, unsaturated fats, vitamins, and critical minerals, all of which are important in the human diet (Bouchenak and Lamri-Senhadji, 2013; Clark et al., 2014; Rebello et al., 2014). Due to the abundance of useful bioactive chemicals, legumes also have been assigned economic, cultural, physiological, and therapeutic functions in addition to their nutritional excellence.
Legumes are an excellent source of high-quality protein, including 20–45% protein and particularly high in the important amino acid lysine (Philips, 1993). Peas and beans contain 17–20% protein, whilst lupins and soybeans contain 38–45% protein (Mlyneková et al., 2014; Kouris-Blazos and Belski, 2016). Legumes contain twice the protein level of cereals and are richer in protein than most of the other plant diets (Leonard, 2012; FAO, 2016; Kouris-Blazos and Belski, 2016). Leguminous proteins, with the exception of soybean protein, are poor in the important sulphur-containing amino acids namely methionine, cysteine, and cysteine, as well as tryptophan, and are thus regarded as an inadequate source of protein (Kouris-Blazos and Belski, 2016). The primary components of leguminous protein are albumins and globulins, which are further subdivided into vialin and legumin. Vialin is the primary protein group in most of the legumes and is defined by a low quantity of sulphur-containing amino acids, which reflects those legumes have small amounts of sulphur-containing amino acids (FAO, 2016). In terms of protein, legumes and cereals complement each other because cereals are high in sulphur-containing amino acids (poor in legumes) and low in lysine (high in legumes) (Staniak et al., 2014). As a result, when beans are combined with grains, the protein quality improves dramatically (FAO, 2016).
Legumes contain up to 60% carbohydrates by dry weight and are source of complex energy-giving carbohydrates (Leonard, 2012). Leguminous starch digests more slowly than cereal and tuber starch. As a result, beans have a low glycemic index (GI) rating for blood glucose control (Philips, 1993; Khalid and Elharadallou, 2013) making them ideal for diabetic patients and those at greater risk of acquiring diabetes. In general, legumes are beneficial for people who want to live a healthy, disease-free lifestyle (Bouchenak and Lamri-Senhadji, 2013). Legumes are also a rich source of dietary fiber (5–37%), with large levels of both soluble and insoluble fibers (Philips, 1993; Leonard, 2012; Kouris-Blazos and Belski, 2016). Diets high in dietary fiber have been linked to plenty of health advantages. Constipation, obesity, diabetes, heart issues, piles, and various malignancies are among the various diseases and ailments that can be prevented and treated (Maphosa and Jideani, 2016; Tamang et al., 2016).
Except for peanuts (45%) and soybeans (47%), legumes have no cholesterol and are generally low in fat, with 5% calories from fat (Messina, 2016). Legumes have a high concentration of mono- and polyunsaturated fatty acids (PUFA) and almost no saturated fatty acids (Kouris-Blazos and Belski, 2016). Kidney beans and chickpeas have the highest levels of PUFA (71.1%) and MUFA (34%), respectively (Kouris-Blazos and Belski, 2016). Because the human body cannot synthesize these PUFAs, they must be consumed through the diet (FAO, 2016). Legumes are high in B-complex vitamins like foliate, thiamin, and riboflavin, but low in fat-soluble vitamins and vitamin C. (Kouris-Blazos and Belski, 2016). Folate is an important nutrient shown to minimize the likelihood of neural tube abnormalities such as spina bifida in newborns (FAO, 2016; Messina, 2016). Zinc, iron, calcium, selenium, phosphorus, copper, potassium, magnesium, and chromium are also found in legumes (Brigide et al., 2014; Kouris-Blazos and Belski, 2016). These micronutrients serve critical physiological functions in bone health (calcium), enzyme activity and iron metabolism (copper), carbohydrate and lipid metabolism (chromium, zinc), hemoglobin production (iron), antioxidative action, protein synthesis, and plasma membrane stability (zinc) (Mogobe et al., 2015). Legumes are often low in sodium, which is ideal given current developments advocating sodium reduction (Leonard, 2012).
Chickpea has been identified as the second most important legume and it has numerous economic facilities. According to Chen et al. (2020), India has been identified as the largest chickpea producer with nearly 65% of total global chickpea production. It is being cultivated on about 12 million hectares and its annual production rate is 9 million tonnes. Chickpea contains high protein content, and is also rich in dietary fibres, calcium, zinc, phosphorus and magnesium. Due to a heavy breeding programme, the production rate of chickpea is gradually increasing in the last thirty years. Determination of the effectiveness and importance of yield components have been identified as the main target. According to Coyne et al. (2020), it has been reported that there is a positive relationship between plant height, seed mass, number of pods per plant and number of branches. The grain yield of chickpea is dependent on different quantitative characteristics, such as environmental location and genetic factors.
Chickpea is a true self-pollinated diploid (2 n = 2 x = 16) with an estimated genome size of 738 Mb having 28,269 genes (Varshney et al., 2013). It is the second most economically important pulse crop with the production of 14.24 million metric tons (FAOSTAT, 2019). Chickpea is valuable because it provides both human food and livestock feed and there is a growing demand for chickpea due to its nutritional value. It is an effective source of protein, carbohydrates, minerals and vitamins, dietary fiber, folate, β-carotene, anti-oxidants, micronutrients (phosphorus, calcium, magnesium, iron and zinc) and health-promoting fatty acids (Sharma et al., 2013a; 2013b). It is rich in carbohydrates and proteins, which altogether account for 80% of the total mass of dry seed (Chibbar et al., 2012). Chickpea’s protein content trends vary considerably by17–22% as a % of the total dry seed mass before dehulling and 25.3–28.9% after dehulling (Hulse, 1989; Misra et al., 2016) and is about 2-3 folds more than cereals. The composition of amino acid in chickpea is well balanced having minimal amino acid containing sulphur i.e., methionine, cysteine and a considerable amount of lysine making it an excellent combination with cereals, which are good source of sulfur-containing amino acids. It is rich in carotenoids responsible for the yellow color of the cotyledon. The prominent and widely distributed carotenoid in chickpea is β-carotene and is more efficiently transformed to vitamin A than any other carotenoids. Chickpea has a higher amount of β-carotene on a dry seed weight basis than “golden rice” endosperm or red wheat. It has a higher dietary fiber content (∼18–22 g), particularly in comparison to wheat (∼12.7 g) and a higher fat content, especially in comparison to other pulses or cereals and two polyunsaturated fatty acids (PUFAs) namely, linoleic and oleic acids that constitute approximately ∼50–60% of chickpea fat, therefore works as cholesterol reducer food.
External factors that negatively affect plant growth, development, or productivity are referred to as stress in plants (Verma et al., 2013). Plants face abiotic and biotic the two main types of stresses and as sessile organisms are continually confronted with a variety of biotic and abiotic stressors. They require continual alterations at the molecular level in order to adapt to changing conditions. Epigenetic regulators provide efficient and effective controls to promote plant survival by increasing their tolerance to stresses (Richards, 2006; Hirayama and Shinozaki, 2010). Different chemical alterations at the molecular level that regulate gene expression are involved in epigenetic control. Today, epigenetics refers primarily to alterations that are related to chemical modifications not to changes in DNA sequence and can be passed on through generations (Feng et al., 2010; Fujimoto et al., 2012). Plants use three types of epigenetic regulatory systems to resist severe conditions caused due to stress conditions: DNA methylation, histone modification, and RNA interference (RNAi). Plants respond to stresses in a variety of ways, including changes in gene expression, cellular metabolism, growth rates, crop yields, and so on. Severe stresses cause crop plants to die by inhibiting flowering, seed development, and inducing senescence (Verma et al., 2013). Abiotic stress is the adverse effect on biological organisms in a particular environment caused by non-living elements. Toxic abiotic stressors, such as hyper drought and salinity, low or high temperatures, depleted or surplus water, high salt levels, heavy metals, and UV radiation are examples of abiotic stress which pose a threat to plant development and growth, resulting in a significant agricultural production penalty throughout the globe. These stresses also adversely affect plant nutrition, for instance when water availability is limited due to drought, total nutrient intake is lowered, and mineral nutrient concentrations in agricultural plants are frequently reduced. Water shortages have the most significant impact on nutrient transport to the root, root growth and extension. The interference of nutrient uptake and unloading mechanisms as well as lower transpiration flow results in reduced nutritional absorption (Marschner 1995; Baligar et al., 2001). Due to lower tissue nutrient concentrations, root development is inhibited when the soil temperature is too low. Nutrient insufficiency may lead to stunted or dying of plant tissue as well as yellowing of leaves. A reduction in crop output or a decline in plant quality growth might be the outcome of nutrients shortage. Traditional and contemporary methods of plant breeding that aim to improve stress tolerance would benefit from a better knowledge of how plants respond to abiotic stresses.
Apart from the abiotic stress, there are many other biotic stresses, such as insect pests, plant-parasitic nematodes and chickpea diseases. Identification of these diseases is accountable for reducing the risk of chickpea cultivation. According to Kumar and Naik (2020), the major disease of chickpeas, such as ascochyta blight (Ascochyta rabie), phytophthora root rot (Phytophthora medicaginis) and Botrytis cinerea is accountable for decreasing the growth of chickpea. There are some major insect pests, such as Helicoverpa punctigera and Helicoverpa armigera, which are accountable for reducing the nutritive value of chickpea (Abdelrahman et al., 2018a). Moreover, the plant-parasitic nematodes, such as root-lesion nematodes and Crystormin nematodes are accountable for decreasing the 14% growth of chickpea.
The pod borer (Helicoverpa armigera) is the most dangerous, followed by the pod fly. Nematodes, which have been efficiently managed by bio-agents, are another key pest impacting chickpea. Wilt and root-knot nematodes are crucial in terms of distribution and chickpea yield damage. Chickpeas are typically grown as rainfed crops since they require less irrigation than competitive crops such as cereal. Post-harvest losses are responsible for 9.5%of total chickpea production. Among post-harvest processes, storage accounts for the greatest amount of loss (7.5%). Processing, threshing, and transportation all result in 1%, 0.50%, and 0.50% losses, respectively. Chickpeas are likewise the most vulnerable to insect damage (5%) among storage losses, compared to wheat (2.5%), rice (2%), and maize (3.5%) (Deshpande and Singh, 2001). Unfortunately, improvements in legumes yield have lagged as those of cereals.
Worldwide, plant productivity is hampered by a lack of water and high salinity. Plants have evolved sophisticated and sensitive defense systems that allow them to signal immediately, respond to, and adapt to a variety of challenges, including drought and excessive salt (Yamaguchi-Shinozaki and Shinozaki, 2006; Tran L. S. P. et al., 2007; Tran L.-S. P. et al., 2007). Plant’s defensive responses to abiotic and biotic stress factors are regulated by various phytohormones.
Cytokinins appear to be implicated in stress reactions, according to growing evidence (Tran L.-S. P. et al., 2007; Argueso et al., 2009) and regulate various aspects of root growth, architecture, and function and plays a crucial regulatory role in a variety of developmental and physiological plant processes (Werner and Schmulling, 2009; Hwang et al., 2012). Cytokinins have been identified as a key signal that passes from roots to shoots (Letham, 1994). According to recent research, the abscisic acid (ABA) and cytokinin ratios in xylem sap are critical for stress signaling (Alvarez et al., 2008; Schachtman and Goodger, 2008). Drought, for example, reduces the generation and distribution of cytokinins from roots. The major enzymes involved in cytokinin metabolism in plants such as Arabidopsis thaliana are adenosine phosphate-isopentenyltransferases (IPTs) and cytokinin oxidases/dehydrogenases (CKX) (Hirose et al., 2008; Werner and Schmulling, 2009). Cytokinin oxidases/dehydrogenases accelerate irreversible cytokinin breakdown by selectively cleaving unsaturated isoprenoid side chains, culminating in the synthesis of adenine/adenosine and the associated side-chain aldehyde (Sakakibara, 2006; Werner et al., 2006).
Plant cytokinin levels have been altered in genetic experiments assuming usually a negative participant in stress response (Nishiyama et al., 2011). For example, in transgenic tobacco plants, overexpression of the cytokinin degrading enzyme cytokininoxidase/dehydrogenase improved drought and heat stress tolerance (Macková et al., 2013). However, recent research suggests that cytokinins (CK) are N6-substituted adenine derivatives that were first identified as a major regulator in plant developmental processes such as organ formation, apical dominance, leaf senescence (El-Showk et al., 2013) and may play a significant role in drought stress adaption as a positive regulator (Hai et al., 2020). For example, in transgenic cotton (Kuppu et al., 2013), creeping bentgrass (Xu et al., 2016), eggplant (Xiao et al., 2017), and tropical maize (Leta et al., 2016), ectopic expression of the isopentenyltransferase gene (IPT), which encodes a rate-limiting enzyme in cytokinin biosynthesis, increases endogenous cytokinin levels. According to a new rice cytokinin-responsive transcriptome analysis, a substantial number of genes are implicated in both biotic and abiotic stressors (Raines et al., 2016). Temperature, drought, osmotic stress, salinity, nutritional stress, plant diseases, and herbivores are among the environmental conditions where cytokinin is said to be essential for responses (Raines et al., 2016; Cortleven et al., 2019).
Accordingly, multiple functional studies were undertaken to determine the CKX-mutant derived tolerance mechanisms. For example, partial root-zone drying resulted in lower cytokinin concentrations in leaves, buds, and shoot tips. This increased apical dominance and aided in overcoming drought stress, particularly when combined with ABA modulation of stomatal apertures. In plants exposed to drought stress, tolerance responses may be induced by manipulating endogenous cytokinin levels, either by deletion of the biosynthesis genes isopentyltransferase or by overexpression of cytokinin oxidase (CKK)-encoding degradation genes. Meanwhile, heat stress is known to lower cytokinin levels, and thus exogenous cytokinin application has generally been shown to improve plant heat stress responses, combating the negative effects of heat stress on photosynthesis and chloroplast growth. Additionally, N6-(D2-isopentenyl) adenine (iP) and trans-zeatin (tZ), the biologically active free-base forms of cytokinins, were found to play a key role in tolerance mechanisms, thought to be via yielding higher relative abundances and affinities for cytokinin receptors. Thus, CKX enzymes play a key role in controlling cytokinin concentrations, which influences plant growth and development. Plants evolve through structural and metabolic adaptations to cope with stress, such as increased root area and leaf curling when subjected to dryness, and increased production of antioxidant chemicals like carotenoids, proline, and ascorbic acid. Plants with bigger root systems have a higher chance of competing for nutrients and surviving in low-nutrient environments (Passioura, 1981; Brown et al., 1989; Saxena et al., 1993; Morita and Nemoto, 1995; Kondo et al., 1999; Steele et al., 2006; Henry, 2013). Root biomass and the availability of soil resources like water and minerals have a big impact on seed output and quality. W31:CaCKX6 expressions in chickpea roots have shown to boost root biomass, shoot biomass and yield (Khandal et al., 2020). The broader root network, obtaining more nutrients from the soil and enhancing the plant’s lifetime, are ascribed to the increased vegetative and reproductive growth of shoots in chickpea lines with W31:CaCKX6.
Chickpea lines expressing W31:CaCKX6 had higher relative water content (RWC) in their leaves, indicating that they were more drought tolerant. Better leaf RWC mixed with lower ABA levels may have contributed to higher carbon assimilation under long-term drought conditions. Chickpea plant introgressed with W31:CaCKX6 in chickpea root produced the seeds having better concentrations of zinc, iron, copper, phosphorus, magnesium, and potassium (Khandal et al., 2020). Thus, increasing the root network through local biofortification of cytokinin and lowering the ABA content employing genome editing in combination with classical breeding may be an effective approach for maintaining the balance for enhanced yield, grain quality and stress tolerance.
A search of the annotated Medicago truncatula genome assembly turned up nine CKX-encoding genes (Young et al., 2011). A comparable search of publicly available annotated chickpea genome and transcriptome sequences (Garg et al., 2011; Varshney et al., 2013) revealed the presence of 10 non-redundant genes that encode proteins with sequence similarities to seven Arabidopsis CKX proteins (Schmulling et al., 2003). They were annotated as C. arietinum cytokinin oxidases/dehydrogenases (CaCKX) and as a result, the chickpea genome has ten CaCKX genes. Climate change and population growth have put pressure on the agriculture industry to enhance productivity, resulting in the development of new, improved technologies aimed at improving crop’s ability to remain productive in conditions such as high temperatures and low moisture availability.
The current review highlights the effectiveness of the spatio-temporal regulation of cytokinin, which is significant for nodule development. Investigating the root manipulation for cytokinin is essential for managing the growth of chickpea. Promoter-driven CaWRKY31 in chickpeas, CaCKX6 expression resulted in a larger root system, increased CKX activity in the root, and increased seed yield. With the help of W31:CaCKX6 construct and the chickpea cultivar Pusa 362, T4 transgenic chickpea plants were created (IC296139). Root nodulation and nitrogen fixation were not affected while increasing the CKX activity. When grown in soil rite pots in a controlled growth environment, chickpea transgenic plants showed up to 1.8-fold increase in root length and lateral root numbers in the 10 days post germination (dpg) stage. In soil-grown 30 dpg plants, CKX activity was measured, and only the root showed a 2.1–3.7 fold increase over the untransformed plant, whereas CKX activity in the shoot tissue remained unchanged. The total length of the roots rose by 1.5–1.85 times. The average amount of biomass in the shoots increased by up to 20%. In transgenic chickpea lines, the root-to-shoot biomass ratio was raised by up to 1.7 times. According to two-year growth statistics, average seed number per plant increased by 20%–25%, with no significant variance in 100 seed weight. Statistical significance was often poor due to variance in seed counts between individual plants of a line. CaCKX6 expression in the roots also increased mineral content in seeds like the concentrations of Zn (27 %–62%), Cu (26 %–61%), Fe (22 %–48%) etc. were all greater in transgenic lines’ seeds (Khandal et al., 2020).
Due to the presence of effective nutrients, the global economic demand and importance of chickpea are gradually increasing. It has been detected that chickpea is one of the good sources of multivitamins, such as niacin, riboflavin, thiamin, vitamin A (β carotene) and folate for the fulfilment of nutritional requirements. There are three primary components, such as inadequate supply, food accessibility and inappropriate food, which are accountable for food insecurity. Maintenance of sustainability in the cultivation of chickpea to fulfil nutritional requirements and increase economic values has enhanced food security. However, it has been identified that the production of chickpea is dependent on several challenging situations, different abiotic stresses, such as high and low temperature and drought.
Agronomic practices and plant breeding is accountable for providing sufficient nutrients to people. However, one approach towards achieving greater food security is through improving the nutrient value of the food that is consumed. This may be achieved through biofortification approaches via breeding for enhanced concentrations of bioavailable nutrients within staple food crops. Several efficacy studies have demonstrated that the biofortification of staple crops can effectively alleviate micronutrient malnutrition or “hidden hunger” among vulnerable populations across the world. Biofortification is one of the innovative techniques, which is used to increase the level of nutrients such as minerals, vitamins and minerals for the enhancement of product’s demand. The nutritional value of legumes including other crops can be increased with the help of various methods such as traditional breeding, molecular technologies, transgenic approaches or genome editing approaches, thereby preventing malnutrition. The former is a well-established, albeit is a labor-intensive and long-term operation.
The efforts have mainly focused on cereal grain staple species, whereas the application of this approach to grain legume/pulse crops has been largely overlooked. The process of biofortification in agronomic crops includes targeting the cytokinin gene family. The cytokinins are one of the phyto hormones that play essential roles in both physiological and developmental processes and directly impact several agronomic parameters, including growth, development and yield, including root extension and branching during post-embryonic advancement. The root-specific degradation of cytokinin was used to engineer maize genetically (Zea mays L.) plants to have a larger root system. Root-specific expression of a cytokinin oxidase (CKK)/dehydrogenase (CKX) gene of Arabidopsis caused the formation of up to 46% more root dry weight while shoot growth of the same transgenic lines was similar to the control plants. Meanwhile, the concentrations of K, P, Mo and Zn were significantly increased in the leaves of the transgenic plants. Subsequently, fine-tuning of cytokinin metabolism by root-specific expression of a cytokinin degradation enzyme was undertaken to improve both Zn nutrient level and yield traits.
Chickpea has been identified as one of the effective nutritious crops, for decreasing the negative impact of nutritional deficiencies. According to Yadav et al. (2019), there are nearly 40–60% of low digestible carbohydrates, 4–8% of essential fats, 15–22% of proteins and a sufficient range of vitamins and minerals (Wang et al., 2020). The presence of these nutrients is essential for increasing the nutritional value of chickpeas. Henceforth, the fatty acid composition is accountable for increasing the value of the seed. It has been identified that fat is essential for governing the texture, flavour, shelf life, nutritional composition and aroma. Therefore, the involvement of the biofortification of essential fatty acids is significant for the fulfilment of the nutritive value of crops including chickpea (Abdelrahman et al., 2018b). Some potential area for biofortification in chickpea is given in Figure 1.
Malnutrition has been considered as the cause of global calamity in Asia and Africa. According to Ninan et al. (2019), the current biofortification efforts focus on the enrichment of significant micronutrients and decreasing the anti-nutrient factors. Implementation of the Agronomic approaches, such as fertilizer application is essential for the enrichment of different minerals, such as Zn, Se and Fe. The combined application of Zn, Fe and urea is accountable for increasing the Zn and Fe concentration in the chickpea. It has been detected that the implementation of the transgenic approaches is one of the most efficient for iron biofortification in chickpea. According to Lastochkina, (2019), over expression of the nicotinamide synthesis, such as ferritin (GmFER) and 2 (CaNAS2) is essential for increasing the Fe concentration rate in chickpea (Toğay et al., 2019). The biofortification process focused on the macro nutritional traits. Linoleic acid (LA; ω-6) has been identified as the essential fatty acid to facilitate human health. Whereas (α-linolenic acid) ALA is the other most essential fatty acid for managing human health benefits (Talip et al., 2018). It has been identified that there are about 3.8–10.2% of general facts in chickpeas. Enhancement of the nutritional values in chickpea is essential for managing the growth of chickpeas, simultaneously its quality and economic value.
Conversely, developments in molecular technologies based biofortification and the availability of improved species-specific genomic resources have led to the evolution of gene editing methods with targeted precision and validated outcomes within a relatively short time frame. Emerging popular genomic targets for the focus of biofortification efforts in food crop species are members of the cytokinin gene family expression pathway, phytohormones essential for many varied physiological and developmental processes.
Genomic editing or genetic engineering is an important aspect of today’s world, in which the DNA is inserted, modified or deleted. First genome editing technologies were developed in the 1900s (Kiran and Chimmad, 2018). These technologies act like scissors and cut the DNA at specific sites. The most efficient tool for genome editing is the CRISPR/cas9 system. This system is mainly used in the production of genetically modified organisms (GMOs) and genomic engineering. CRISPR/cas9 has extended the scope of agricultural research allowing for new potentials to generate novel plant types with undesirable features removed or significant characters added such as acrylamide-free potatoes (Halterman et al., 2015); non-browning apples, mushrooms and potatoes; low phytic acid maize (Liang et al., 2014); blast disease resistant rice (Wang et al., 2016) and powdery mildew resistant wheat (Wang et al., 2014). CRISPR technology is continually improving, allowing for more genetic manipulations such as creating knockouts, precise changes, multiplex genome engineering, and target gene activation and repression. With more precision and simplicity, CRISPR targets endogenous genes that are unable to target specifically using RNAi technology the mechanism of which can beseen in Figure 2. CRISPR/Cas9 uses a 100 nucleotide (nt) guide RNA (gRNA) sequence to target specific genomic loci. Using Watson and Crick base pairing through 17–20 nt at the gRNA 5′-end, sgRNA binds to the protospacer adjacent motif (PAM) on targeted DNA and guides Cas9 for selective cleavage. Cas9 accelerates DNA repair by causing DSBs (Double Stranded Breaks) in the target DNA. To induce genomic changes, gene knockouts, and gene insertions, the repair mechanism uses error-prone non-homologous end joining (NHEJ) or homologous recombination (HR). NHEJ makes random insertions or deletions in the coding area, resulting in frame shift mutations and gene knockouts. Thus, loss-of-function, gain-of-function, and gene expression analysis are possible with CRISPR technology enabling it as one of the effective plant breeding tool, which focuses on different gene action by acquiring knowledge on gene family members (GFMs) and is desperately needed.
According to Kumar et al. (2017),CRISPR/Cas9is known as one of the effective gene-editing tools, which is accountable for manipulating cytokinin dehydrogenase (Kumar et al., 2017; Jha et al., 2018). The GFMs expression is essential for cytokine biosynthesis and destruction for managing the gene factors. According to Ninan et al. (2019), cytokines have been identified as the enhancement of sink activities in chickpea leaves. The primary steps in cytokine biosynthesis can be controlled by isopentenyltransferase (IPT). Henceforth, the cytokinin dehydrogenase or oxidase is accountable to control the process of cytokinin degradation. Involvement of the DNA sequencing technology is essential for gathering delta knowledge in the gene concept. Cytokinin is identified as one of the effective plant hormones, which is accountable for regulating plant development. The PsCKX7 (Pisum sativum cytokinin dehydrogenase) gene when down-regulated, cytokinin levels increased in roots, shoots and leaves also involves delaying of senescence. It is noteworthy that PsCKX5 and PsCKX7 express in the sink and mature leaves respectively (Ninan et al., 2019). In rice, Zhang et al. (2021) performed CRISPR/Cas9 editing to target serval CKX genes. They found that OsCKX11 (Oryza sativa cytokinin dehydrogenase) have simultaneously regulates cytokinin-mediated leaf senescence and grain number (Figure 3).
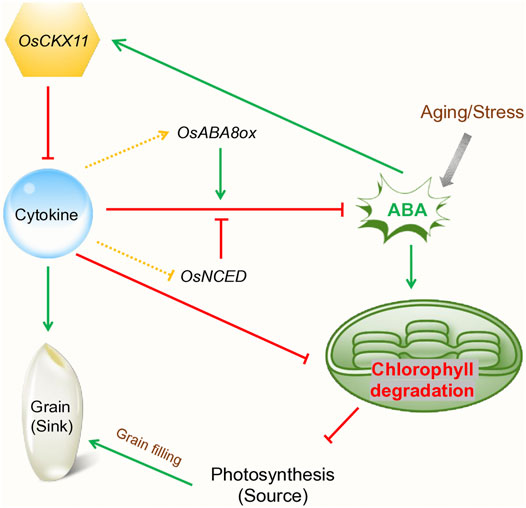
FIGURE 3. Schematic presentation showing the role of OsCKX11 in cytokinin-mediated leaf senescence and grain number in rice (Modified from Zhang et al., 2021)
The cytokine dehydrogenase (CKXs) is known as an essential protein for an irreversible breakdown of cytokinin’s. It is significant for the molecular evolution for the determination of the homologous protein. Uses of these gene-editing tools are essential for detecting the presence of CKX in prokaryotic and eukaryotic. Apart from this, it has been identified that CKX plays a significant role in the improvement of plant life. Controlling the plant development process is beneficial for managing the abiotic and biotic stress for influencing the nutritive value of chickpea (Abdelrahman et al., 2018a). Cytokinin dehydrogenase is an essential plant hormone for promoting cell division. Promotion of primary cell growth and differentiation is essential for increasing the growth of this hormone. The involvement of the gene-editing tools helped in gene formation for improving the growth of its products. Apart from cytokinin, ethylene exhibition is equally important for managing the growth of plants. Involvement of the photosynthetic machinery process is essential for stimulating the growth of chickpea (Jyothi et al., 2018). Cytokinin is accountable for increasing the grain size and grain numbers for yielding the components of this plant.
In recent years, the Clustered Regularly Interspaced Short Palindromic Repeats Cas9 (CRISPR/Cas9) genome editing method has revolutionized targeted gene editing in plants (Woo et al., 2015; Baek et al., 2016; Malnoy et al., 2016; Liang et al., 2017; Kim et al., 2018; Lin et al., 2018; Murovec et al., 2018; Osakabe et al., 2018; Johansen et al., 2019; Petersen et al., 2019). CRISPR/Cas9 genome editing has a wide range of applications in agricultural improvement, including the development of designer genetically modified non-GM crops. The application of this strategy to plant breeding for the production of new crop varieties with greater tolerance to environmental challenges is a major focus of agricultural scientists (Khatodia et al., 2016; Noman et al., 2016). CRISPR/Cas9 gene-editing tools have been utilized for gene activation, repression, knockout, knockdown, repression, and for altering epigenetic modifications in several plants crops such as Arabidopsis (Feng et al., 2014), apple (Osakabe et al., 2018), citrus, carrot (Klimek-Chodacka et al., 2018), grape (Nakajima et al., 2017), tomato (Wang et al., 2019), rice (Zhang et al., 2014), sorghum (Liu et al., 2019), maize and soybean (Chilcoat et al., 2017), and wheat (Zhang et al., 2016). CRISPR/Cas9 genome editing was utilized to discover abiotic stress response in Arabidopsis plants; the findings revealed that OST2 (proton pump), a mutant allele produced through editing, changed stomatal closure under environmental stress (Osakabe et al., 2018). Another recent maize work employed the CRISPR/Cas9 method to create unique allelic variants that could be exploited to engineer drought-tolerant crops. This system genetically modified ARGOS8, whose over expression can result in lower ethylene sensitivity. Field investigations demonstrated that ARGOS8 variants had higher grain yield under drought stress; further, no yield loss was documented under well-watered conditions (Shi et al., 2017).
This present review highlights the role of CKX genes in chickpea growth and development traits, biotic and abiotic stress regulation, and biofortification. Chickpea is the most economically important product all over the world. There are various types of stress like heat, cold, drought and so on those are faced by the crop plant. Due to global warming, a temperature rise is a frequent event in today’s world that causes the drought condition. Heat stress causes severe damage to the leaves and also ruptures the membrane. All these factors adversely affect the agronomic traits of chickpea. CKs play many crucial roles in plants when they experience any kind of stress. Phytohormones in the cytokinin family control root length and branching in the post-embryonic stages. Cytokinin oxidases or dehydrogenases (CKXs) are enzymes that degrade cytokinin in order to study its biological functions and engineer root development. A chickpea root-specific promoter of CaWRKY31 may be used to explore how cytokinin depletion affects root development and drought tolerance in Arabidopsis thaliana and chickpea with definite and indeterminate growth patterns, respectively. In Arabidopsis and chickpea, root specific expressions of CaCKX6 increased lateral root number and plant biomass without affecting shoot vegetative and reproductive development. Root CKX activity was elevated in transgenic chickpea lines. The root-to-shoot biomass ratio was greater in soil-grown advanced chickpea transgenic lines, and the plants had improved long-term drought resistance. Nutrient fixation in the roots and leaves of these chickpea varieties was unaffected. In certain transgenic lines, the seed output was up to 25% greater with enhanced concentrations of zinc, iron, potassium, and copper without corresponding decrease in protein content. Apart from this other phytohormones also play an important role in alleviating stress condition in chickpea. ABA plays an important role to reduce oxidative damage in chickpeas. It interacts with the various types of antioxidants to reduce stress and reduces the ROS production in the plant body which harms the plants. It has also introduced some heat shock protein to provide tolerance against the heat. Salicylic acid also plays an important role against abiotic and biotic stress as well as against pathogens and herbivores. However, in chickpea, functional characterization studies of CKX genes have just started. Gene editing tools such as TALENs or CRISPR/Cas9 can play crucial role in this context. Still, less functional studies exist in the case of stress regulation and biofortification. This is a potential area for research to unravel the CK signaling networks and their cross talk elucidating its biochemical pathways which will draw a detailed picture and pave the road towards developing tolerant crops, and in the long-term, more sustainable agriculture. Similarly, many more such genes are hidden in the plant genome, which are required to be explored and investigated to harness and develop cultivars with a higher yield, better abiotic stress resistance and biofortification.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
RK conceptualized and supervised the manuscript writing. RM, Ambika, CS, BC, RS and SV collected the related literature and contributed to the original writing. RK, VG, MM and NY extended their assistance in inference, review, and editing of the manuscript. All authors went through the final manuscript draft and approved.
The study was supported by ICAR-Indian Agricultural Research Institute, New Delhi.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The 1st and lead author gratefully acknowledges the DST-Science and Engineering Research Board for providing the financial support to carry out the manuscript preparation. All authors acknowledge the ICAR-Indian Agricultural Research Institute, New Delhi; UAS-GKVK, Bangalore, India; GGS Indraprastha University, Delhi, India; CSIR-IHBT, Palampur, India; CFT, University of Allahabad, India for providing necessary support for the study along with Prof Rebecca Ford, Griffith University, Queensland, Australia and Prof Syed S H Rizvi, Cornell University for reviewing and meticulous scientific editing of the 1st draft of the manuscript.
Abdelrahman, M., Jogaiah, S., Burritt, D. J., and Tran, L. P. (2018b). Legume Genetic Resources and Transcriptome Dynamics under Abiotic Stress Conditions. Plant Cell. Environ. 41 (9), 1972–1983. doi:10.1111/pce.13123
Abdelrahman, M., Al-Sadi, A. M., Pour-Aboughadareh, A., Burritt, D. J., and Tran, L.-S. P. (2018a). Genome Editing Using CRISPR/Cas9-targeted Mutagenesis: An Opportunity for Yield Improvements of Crop Plants Grown under Environmental Stresses. Plant Physiology Biochem. 131, 31–36. doi:10.1016/j.plaphy.2018.03.012
Ahmed, M. H., Vasas, D., Hassan, A., and Molnár, J. (2022). The Impact of Functional Food in Prevention of Malnutrition. Pharma Nutr. 19, 100288. doi:10.1016/j.phanu.2022.100288
Alvarez, S., Marsh, E. L., Schroeder, S. G., and Schachtman, D. P. (2008). Metabolomic and Proteomic Changes in the Xylem Sap of Maize under Drought. Plant Cell. Environ. 31, 325–340. doi:10.1111/j.1365-3040.2007.01770.x
Argueso, C. T., Ferreira, F. J., and Kieber, J. J. (2009). Environmental Perception Avenues: the Interaction of Cytokinin and Environmental Response Pathways. Plant Cell. Environ. 32, 1147–1160. doi:10.1111/j.1365-3040.2009.01940.x
Baek, K., Kim, D. H., Jeong, J., Sim, S. J., Melis, A., Kim, J.-S., et al. (2016). DNA-free Two-Gene Knockout in Chlamydomonas Reinhardtii via CRISPR-Cas9 Ribonucleoproteins. Sci. Rep. 6, 30620. doi:10.1038/srep30620
Baligar, V. C., Fageria, N. K., and He, Z. L. (2001). Nutrient Use Efficiency in Plants. Commun. Soil Sci. Plant Anal.32 (7-8), 921–950. doi:10.1081/css-100104098
Barlow, K. M., Christy, B. P., O’Leary, G. J., Riffkin, P. A., and Nuttall, J. G. (2015). Simulating the Impact of Extreme Heat and Frost Events on Wheat Crop Production: A Review. Field crops Res. 171, 109–119. doi:10.1016/j.fcr.2014.11.010
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield inArabidopsis Thaliana. Plant Cell. 23, 69–80. doi:10.1105/tpc.110.079079
Bouchenak, M., and Lamri-Senhadji, M. (2013). Nutritional Quality of Legumes, and Their Role in Cardiometabolic Risk Prevention: A Review. J. Med. Food 16 (3), 185–198. doi:10.1089/jmf.2011.0238
Brigide, P., Canniatt-Brazaca, S. G., and Silva, M. O. (2014). Nutritional Characteristics of Biofortified Common Beans. Food Sci. Technol. (Campinas) 34 (3), 493–500. doi:10.1590/1678-457x.6245
Brown, S. C., Gregory, P. J., Cooper, P. J. M., and Keatinge, J. D. H. (1989). Root and Shoot Growth and Water Use of Chickpea (Cicer Arietinum) Grown in Dryland Conditions: Effects of Sowing Date and Genotype. J. Agric. Sci. 113 (1), 41–49. doi:10.1017/s0021859600084598
Chang, L., Ramireddy, E., and Schmülling, T. (2015). Cytokinin as a Positional Cue Regulating Lateral Root Spacing inArabidopsis. Exbotj 66, 4759–4768. doi:10.1093/jxb/erv252
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2020). Cytokinin Dehydrogenase: a Genetic Target for Yield Improvement in Wheat. Plant Biotechnol. J. 18 (3), 614–630. doi:10.1111/pbi.13305
Chilcoat, D., Liu, Z.-B., and Sander, J. (2017). “Use of CRISPR/Cas9 for Crop Improvement in Maize and Soybean,” in Progress in Molecular Biology and Translational Science. Editors D. P. Weeks, and B. Yang (Cambridge, MA, USA: Academic Press), 149, 27–46. doi:10.1016/bs.pmbts.2017.04.005
S. Clark, S. Jung, and B. &Lamsal (Editors) (2014). Food Processing: Principles and Applications (John Wiley & Sons).
Cortleven, A., Leuendorf, J. E., Frank, M., Pezzetta, D., Bolt, S., and Schmülling, T. (2019). Cytokinin Action in Response to Abiotic and Biotic Stresses in Plants. Plant Cell. Environ. 42, 998–1018. doi:10.1111/pce.13494
Coyne, C. J., Kumar, S., von Wettberg, E. J., Marques, E., Berger, J. D., Redden, R. J., and &Smýkal, P. (2020). Potential and Limits of Exploitation of Crop Wild Relatives for Pea, Lentil, and Chickpea Improvement. Legume Sci. 2 (2), e36. doi:10.1002/leg3.36
Deshpande, S. D., and Singh, G. (2001). “Long Term Storage Structures in Pulses,” in National symposium on pulses for sustainable agriculture and nutritional security, 17-19 April (New Delhi: Indian Institute of Pulses Research).
El-Showk, S., Ruonala, R., and Helariutta, Y. (2013). Crossing Paths: Cytokinin Signalling and Crosstalk. Development 140, 1373–1383. doi:10.1242/dev.086371
FAO (2016). Legumes Can Help Fight Climate Change, Hunger and Obesity in Latin America and the Caribbean. Rome: Food and Agriculture Organisation of the United Nations.
FAOSTAT (2019) http://faostat.fao.org/site (Accessed December 27, 2021).
Feng, S., Jacobsen, S. E., and Reik, W. (2010). Epigenetic Reprogramming in Plant and Animal Development. Science 330 (6004), 622–627. doi:10.1126/science.1190614
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D.-L., et al. (2014). Multigeneration Analysis Reveals the Inheritance, Specificity, and Patterns of CRISPR/Cas-induced Gene Modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 (12), 4632–4637. doi:10.1073/pnas.1400822111
Fujimoto, R., Sasaki, T., Ishikawa, R., Osabe, K., Kawanabe, T., and Dennis, E. S. (2012). Molecular Mechanisms of Epigenetic Variation in Plants. Ijms 13 (8), 9900–9922. doi:10.3390/ijms13089900
Gao, S., Xiao, Y., Xu, F., Gao, X., Cao, S., Zhang, F., et al. (2019). Cytokinin‐dependent Regulatory Module Underlies the Maintenance of Zinc Nutrition in Rice. New Phytol. 224 (1), 202–215. doi:10.1111/nph.15962
Garg, R., Patel, R. K., Jhanwar, S., Priya, P., Bhattacharjee, A., Yadav, G., et al. (2011). Gene Discovery and Tissue-specific Transcriptome Analysis in Chickpea with Massively Parallel Pyrosequencing and Web Resource Development. Plant Physiol. 156 (4), 1661–1678. doi:10.1104/pp.111.178616
Hai, N. N., Chuong, N. N., Tu, N. H. C., Kisiala, A., Hoang, X. L. T., and Thao, N. P. (2020). Role and Regulation of Cytokinins in Plant Response to Drought Stress. Plants 9, 422. doi:10.3390/plants9040422
Halterman, D., Guenthner, J., Collinge, S., Butler, N., and Douches, D. (2015). Biotech Potatoes in the 21st Century: 20 Years since the First Biotech Potato. Am. J. Potato Res. 93, 1–20. doi:10.1007/s12230-015-9485-1
Haque, E., Taniguchi, H., Hassan, M. M., Bhowmik, P., Karim, M. R., Śmiech, M., et al. (2018). Application of CRISPR/Cas9 Genome Editing Technology for the Improvement of Crops Cultivated in Tropical Climates: Recent Progress, Prospects, and Challenges. Front. Plant Sci. 9, 617. doi:10.3389/fpls.2018.00617
Henry, A. (2013). IRRI's Drought Stress Research in Rice with Emphasis on Roots: Accomplishments over the Last 50 Years. Plant Root 7, 92–106. doi:10.3117/plantroot.7.92
Hirayama, T., and Shinozaki, K. (2010). Research on Plant Abiotic Stress Responses in the Post-genome Era: Past, Present and Future. Plant J. 61 (6), 1041–1052. doi:10.1111/j.1365-313x.2010.04124.x
Hirose, N., Takei, K., Kuroha, T., Kamada-Nobusada, T., Hayashi, H., and Sakakibara, H. (2008). Regulation of Cytokinin Biosynthesis, Compartmentalization and Translocation. J. Exp. Bot. 59, 75–83. doi:10.1093/jxb/erm157
Hulse, J. H. (1989). Nature, Composition and Utilization of Grain Legumes, 27. Uses of tropical grain legumes.11
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin Signaling Networks. Annu. Rev. Plant Biol. 63, 353–380. doi:10.1146/annurev-arplant-042811-105503
Jha, U. C., Jha, R., Singh, N. P., Shil, S., and Kole, P. C. (2018). Heat Tolerance Indices and Their Role in Selection of Heat Stress Tolerant Chickpea Cicerarietinum Genotypes. Ind. J. Agricul Sci. 88 (2), 260–267.
Johansen, I. E., Liu, Y., Jørgensen, B., Bennett, E. P., Andreasson, E., Nielsen, K. L., et al. (2019). High Efficacy Full Allelic CRISPR/Cas9 Gene Editing in Tetraploid Potato. Sci. Rep. 9, 17715. doi:10.1038/s41598-019-54126-w
Jukanti, A. K., Gaur, P. M., Gowda, C. L., and Chibbar, R. N. (2012). Nutritional Quality and Health Benefits of Chickpea (Cicer Arietinum L.): a Review. Br. J. Nutr. 108 Suppl 1 (1), S11–S26. doi:10.1017/S0007114512000797
Jyothi, V., Saifulla, M., and Teli, K. (2018). Biochemical Analysis of Chickpea Genotypes against Dry Root Rots (Macrophominaphaseolina (Tassi) Goid). J. Pharmacog. Phytochem. 1, 1895–1901.
Khalid, I. I., and Elharadallou, S. B. (2013). Functional Properties of Cowpea (Vigna UngiculataL.Walp), and Lupin (Lupinus Termis) Flour and Protein Isolates. J. Nutr. Food Sci. 3, 234. doi:10.4172/2155-9600.1000234
Khandal, H., Gupta, S. K., Dwivedi, V., Mandal, D., Sharma, N. K., Vishwakarma, N. K., Pal, L., Choudhary, M., Francis, A., Malakar, P., Singh, N. P., Sharma, K., Sinharoy, S., Singh, N. P., Sharma, R., and Chattopadhyay, D. (2020). Root‐specific Expression of Chickpea Cytokinin Oxidase/dehydrogenase 6 Leads to Enhanced Root Growth, Drought Tolerance and Yield without Compromising Nodulation. Plant Biotechnol. J. 18 (11), 2225–2240. doi:10.1111/pbi.13378
Khatodia, S., Bhatotia, K., Bhatotia, K., Passricha, N., Khurana, S. M. P., and Tuteja, N. (2016). The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 7, 506. doi:10.3389/fpls.2016.00506
Kim, D., Alptekin, B., and Budak, H. (2018). CRISPR/Cas9 Genome Editing in Wheat. Funct. Integr. Genomics 18, 31–41. doi:10.1007/s10142-017-0572-x
Kiran, B., and Chimmad, V. (2018). Studies on Morpho-Phenological Traits and Heat Unit Accumulation in Chickpea Genotypes under Different Temperature Regimes. J. Pharmacogn. Phytochem. 7, 2956–2961.
Klimek-Chodacka, M., Oleszkiewicz, T., Lowder, L. G., Qi, Y., and Baranski, R. (2018). Efficient CRISPR/Cas9-based Genome Editing in Carrot Cells. Plant Cell. Rep. 37, 575–586. doi:10.1007/s00299-018-2252-2
Kondo, M., Murty, M. V. R., Aragones, D. V., Okada, K., Winn, T., and Kwak, K. S. (2000). Characteristics of Root Growth and Water Uptake from Soil in Upland Rice and Maize Under Water Stress. Soil Sci. Plant Nutr. 46 (3), 721–732. doi:10.1080/00380768.2000.10409137
Kouris-Blazos, A., and Belski, R. (2016). Health Benefits of Legumes and Pulses with a Focus on Australian Sweet Lupins. Asia Pac J. Clin. Nutr. 25 (1), 1–17. doi:10.6133/apjcn.2016.25.1.23
Kumar, P., and Naik, M. (2020). Biotic Symbiosis and Plant Growth Regulators as a Strategy against Cadmium and Lead Stress in Chickpea. Plant Arch. 20 (2), 2495–2500. Available at: http://www.plantarchives.org/SPL%20ISSUE%2020-2/413__2495-2500.
Kumar, R., Yadav, R., Soi, S., Yadav, S. S., Yadav, A., Mishra, J. P., et al. (2017). Morpho-molecular Characterization of Landraces/wild Genotypes of Cicer for Biotic/Abiotic Stresses. Legume Res. 40 (6), 974-984. doi:10.18805/lr.v0iof.9100
Kumar, S., and Pandey, G. (2020). Biofortification of Pulses and Legumes to Enhance Nutrition. Heliyon 6 (3), e03682. doi:10.1016/j.heliyon.2020.e03682
Kuppu, S., Mishra, N., Hu, R., Sun, L., Zhu, X., Shen, G., et al. (2013). Water-deficit Inducible Expression of a Cytokinin Biosynthetic Gene IPT Improves Drought Tolerance in Cotton. PLoS One 8, e64190. doi:10.1371/journal.pone.0064190
Lastochkina, O. (2019). “Bacillus Subtilis-Mediated Abiotic Stress Tolerance in Plants,” in Bacilli and Agrobiotechnology: Phytostimulation and Biocontrol. Editors G. Haesaert, M. Mahfuz Rahman, M. D. Tofazzal Islam, M. Henry Boehme, and P. Pandey (Cham: Springer), 97–133. doi:10.1007/978-3-030-15175-1_6
Leonard, E. (2012). Cultivating Good Health. Grains And Legumes Nutrition Council. Adelaide: Cadillac Printing, 3–18.
Leta, T. B., Miccah, S. S., Steven, M. R., Wondyifraw, T., Charless, M., Clet, W. M., et al. (2016). Drought Tolerant Tropical Maize (Zea mays L.) Developed through Genetic Transformation with Isopentenyltransferase Gene. Afr. J. Biotechnol. 15, 2447–2464. doi:10.5897/AJB2016.15228
Letham, D. S. (1994). “Cytokinins as Phytohormones-Sites of Biosynthesis, Translocation and Function of Translocated Cytokinin,” in Cytokinins: Chemistry, Activity and Function. Editors D. W. S. Mok, and M. C. Mok (Boca Raton, FL: CRC Press), 57–80.
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free Genome Editing of Bread Wheat Using CRISPR/Cas9 Ribonucleoprotein Complexes. Nat. Commun. 8 (1), 14261–14265. doi:10.1038/ncomms14261
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J. Genet. Genomics 41, 63–68. doi:10.1016/j.jgg.2013.12.001
Lin, C.-S., Hsu, C.-T., Yang, L.-H., Lee, L.-Y., Fu, J.-Y., Cheng, Q.-W., et al. (2018). Application of Protoplast Technology to CRISPR/Cas9 Mutagenesis: from Single-Cell Mutation Detection to Mutant Plant Regeneration. Plant Biotechnol. J. 16 (7), 1295–1310. doi:10.1111/pbi.12870
Liu, G., Li, J., and Godwin, I. D. (2019). in Genome Editing by CRISPR/Cas9 in Sorghum through Biolistic Bombardment in Sorghum: Methods and Protocols. Editors Z. Y. Zhao, and J. Dahlberg (New York, NY, USA: Springer), 169–183. doi:10.1007/978-1-4939-9039-9_12
Liu, W., Yuan, J. S., and Stewart Jr, C. N. (2013). Advanced Genetic Tools for Plant Biotechnology. Nat. Rev. Genet. 14, 781–793. doi:10.1038/nrg3583
Macková, H., Hronková, M., Dobrá, J., Turečková, V., Novák, O., Lubovská, Z., et al. (2013). Enhanced Drought and Heat Stress Tolerance of Tobacco Plants with Ectopically Enhanced Cytokinin Oxidase/dehydrogenase Gene Expression. J. Exp. Bot. 64, 2805–2815. doi:10.1093/jxb/ert131
Malnoy, M., Viola, R., Jung, M.-H., Koo, O.-J., Kim, S., Kim, J.-S., et al. (2016). DNA-free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 7, 1904. doi:10.3389/fpls.2016.01904
Maphosa, Y., and Jideani, V. A. (2016). Physicochemical Characteristics of Bambara Groundnut Dietary Fibres Extracted Using Wet Milling. S. Afr. J. Sci. 112 (1/2), 1–8. doi:10.17159/sajs.2016/20150126
Maphosa, Y., and Jideani, V. A. (2017). “The Role of Legumes in Human Nutrition,”. Editor M. C. Hueda, 1, 13. doi:10.5772/intechopen.69127Funct. food-improve health through adequate food
Messina, M. J. (2016). Legumes and Soybeans: Overview of Their Nutritional Profiles and Health Effects. Asia Pac. J. Clin.Nutr. 25 (1), 1–17.
Misra, J. P., Yadav, A., Kumar, A., Renu, Y., Shami, V., and Kumar, R. (2016). Bio-chemical Characterization of Chickpea Genotypes with Special Reference to Protein. Res. J. Chem. Environ. 20 (8), 38–43.
Mlyneková, Z., Chrenková, M., and Formelová, Z. (2014). Cereals and Legumes in Nutrition of People with Celiac Disease. Int. J. Celiac Dis. 2 (3), 105–109. doi:10.12691/ijcd-2-3-3
Mogobe, M., Mosepele, K., and Masamba, W. R. L. (2015). Essential Mineral Content of Common Fish Species in Chanoga, Okavango Delta, Botswana. Afr. J. Food Sci. 9 (9), 480–486. doi:10.5897/ajfs2015.1307
Morita, S., and Nemoto, K. (1995). “Morphology and Anatomy of Rice Roots with Special Reference to Coordination in Organo- and Histogenesis,” in Structure and Function of Roots (Dordrecht: Springer), 75–86. doi:10.1007/978-94-017-3101-0_9
Murovec, J., Guček, K., Bohanec, B., Avbelj, M., and Jerala, R. (2018). DNA-free Genome Editing of Brassica oleracea and B. Rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 9, 1594. doi:10.3389/fpls.2018.01594
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9-mediated Targeted Mutagenesis in Grape. PLoS ONE 12, e0177966. doi:10.1371/journal.pone.0177966
Ninan, A., Grant, J., Song, J., and Jameson, P. (2019). Expression of Genes Related to Sugar and Amino Acid Transport and Cytokinin Metabolism during Leaf Development and Senescence in Pisum Sativum L. Plants 8 (3), 76. doi:10.3390/plants8030076
Nishiyama, R., Watanabe, Y., Fujita, Y., Le, D. T., Kojima, M., Werner, T., et al. (2011). Analysis of Cytokinin Mutants and Regulation of Cytokinin Metabolic Genes Reveals Important Regulatory Roles of Cytokinins in Drought, Salt and Abscisic Acid Responses, and Abscisic Acid Biosynthesis. Plant Cell. 23, 2169–2183. doi:10.1105/tpc.111.087395
Noman, A., Aqeel, M., and He, S. (2016). CRISPR-Cas9: Tool for Qualitative and Quantitative Plant Genome Editing. Front. Plant Sci. 7, 1740. doi:10.3389/fpls.2016.01740
Nuruzzaman, M., Rahman, M. M., Liu, Y., and Naidu, R. (2016). Nanoencapsulation, Nano-Guard for Pesticides: a New Window for Safe Application. J. Agric. Food Chem. 64 (7), 1447–1483. doi:10.1021/acs.jafc.5b05214
Osakabe, Y., Liang, Z., Ren, C., Nishitani, C., Osakabe, K., Wada, M., et al. (2018). CRISPR-Cas9-mediated Genome Editing in Apple and Grapevine. Nat. Protoc. 13 (12), 2844–2863. doi:10.1038/s41596-018-0067-9
Passioura, J. B. (1981). College, Laguna (Philippines).The Role of Root System Characteristics in Drought Resistance of Crop PlantsSpecial International Symposium on Principles and Methods of Crop Improvement for Drought Resistance: With Emphasis on Rice4-8 May 1981
Petersen, B. L., Möller, S. R., Mravec, J., Jørgensen, B., Christensen, M., Liu, Y., et al. (2019). Improved CRISPR/Cas9 Gene Editing by Fluorescence Activated Cell Sorting of Green Fluorescence Protein Tagged Protoplasts. BMC Biotechnol. 19, 36. doi:10.1186/s12896-019-0530-x
Philips, R. D. (1993). Starchy Legumes in Human Nutrition and Culture. Plant Foods Hum. Nutr. 44 (3), 195–211. doi:10.1007/BF01088314
Raines, T., Blakley, I. C., Tsai, Y.-C., Worthen, J. M., Franco-Zorrilla, J. M., Solano, R., et al. (2016). Characterization of the Cytokinin-Responsive Transcriptome in Rice. BMC Plant Biol. 16, 260. doi:10.1186/s12870-016-0932-z
Rebello, C. J., Greenway, F. L., and Finley, J. W. (2014). A Review of the Nutritional Value of Legumes and Their Effects on Obesity and its Related Co-morbidities. Obes. Rev. 15 (5), 392–407. doi:10.1111/obr.12144
Richards, E. J. (2006). Inherited Epigenetic Variation - Revisiting Soft Inheritance. Nat. Rev. Genet. 7 (5), 395–401. doi:10.1038/nrg1834
Sakakibara, H. (2006). Cytokinins: Activity, Biosynthesis, and Translocation. Annu. Rev. Plant Biol. 57, 431–449. doi:10.1146/annurev.arplant.57.032905.105231
Saxena, N. P., Krishnamurthy, L., and Johansen, C. (1993). Registration of a Drought‐Resistant Chickpea Germplasm. Crop Sci. 33 (6), 1424. doi:10.2135/cropsci1993.0011183x003300060088x
Schachtman, D. P., and Goodger, J. Q. D. (2008). Chemical Root to Shoot Signaling under Drought. Trends Plant Sci. 13, 281–287. doi:10.1016/j.tplants.2008.04.003
Schmülling, T., Werner, T., Riefler, M., Krupková, E., Bartrina y Manns, I., and Manns, I. (2003). Structure and Function of Cytokinin Oxidase/dehydrogenase Genes of Maize, Rice, Arabidopsis and Other Species. J. Plant. Res. 116, 241–252. doi:10.1007/s10265-003-0096-4
Sharma, S., Yadav, N., Singh, A., and Kumar, R. (2013a). Antioxidant Activity Nutraceutical Profile and Health Relevant Functionality of Nine Newly Developed Chickpea Cultivars (Cicer Arietinum L.). Int. J. Nat. Prod. Res. 3 (2), 44–53.
Sharma, S., Yadav, N., Singh, A., and Kumar, R. (2013b). Nutritional and Antinutritional Profile of Newly Developed Chickpea (Cicer Arietinum L.) Varieties. Int. Food Res. J. 20 (2), 805–810.
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 15, 207–216. doi:10.1111/pbi.12603
Smil, V. (1999). Nitrogen in Crop Production: An Account of Global Flows. Glob. Biogeochem. Cycles 13, 647–662. doi:10.1029/1999gb900015
Staniak, M., Księżak, J., and Bojarszczuk, J. (2014). “Mixtures of Legumes with Cereals as a Source of Feed for Animals,” in Organic Agriculture towards Sustainability. Editor V. Pilipavicius (Croatia: Tech), 123–145. doi:10.5772/58358
Steele, K. A., Price, A. H., Shashidhar, H. E., and Witcombe, J. R. (2006). , 112, 208–221. doi:10.1007/s00122-005-0110-4Marker-assisted Selection to Introgress Rice QTLs Controlling Root Traits into an Indian Upland Rice VarietyTheor. Appl. Genet.2
Talip, M., Adak, A., Kahraman, A., Berger, J., Sari, D., Sari, H., et al. (2018). Agro-morphological Traits of Cicer Reticulatum Ladizinsky in Comparison to C. Echinospermum P.H. Davis in Terms of Potential to Improve Cultivated Chickpea (C. Arietinum L.). Genet. Resour. Crop Evol. 65 (3), 951–962. doi:10.1007/s10722-017-0587-0
Tamang, J. P., Shin, D.-H., Jung, S.-J., and Chae, S.-W. (2016). Functional Properties of Microorganisms in Fermented Foods. Front. Microbiol. 7, 578. doi:10.3389/fmicb.2016.00578
Taranto, F., Nicolia, A., Pavan, S., De Vita, P., and D’Agostino, N. (2018). Biotechnological and Digital Revolution for Climate-Smart Plant Breeding. Agronomy 8, 277. doi:10.3390/agronomy8120277
Thornton, P. K., Ericksen, P. J., Herrero, M., and Challinor, A. J. (2014). Climate Variability and Vulnerability to Climate Change: a Review. Glob. Change Biol. 20 (11), 3313–3328. doi:10.1111/gcb.12581
Toğay, Y., Toğay, N., Çiğ, F., and &Akkoç, G. (2019). Determination of Some Quality Criteria and Nutrient Contents of Local Black Chickpea Genotypes Growth in Different Locations. doi:10.5897/IJGMB2017.0158
Tran, L.-S. P., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., et al. (2007b). Functional Analysis of AHK1/ATHK1 and Cytokinin Receptor Histidine Kinases in Response to Abscisic Acid, Drought, and Salt Stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628. doi:10.1073/pnas.0706547105
Tran, L. S. P., Nakashima, K., Shinozaki, K., and Yamaguchi‐Shinozaki, K. (2007a). Plant Gene Networks in Osmotic Stress Response: from Genes to Regulatory Networks. Meth. Enzymol. 428, 109–128. doi:10.1016/s0076-6879(07)28006-1
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G., et al. (2013). Draft Genome Sequence of Chickpea (Cicer Arietinum) Provides a Resource for Trait Improvement. Nat. Biotechnol. 31 (3), 240–246. doi:10.1038/nbt.2491
Verma, S., Nizam, S., and Verma, P. K. (2013). “Biotic and Abiotic Stress Signaling in Plants,” in Stress Signaling in Plants: Genomics and Proteomics Perspective. Editors A. Ahmad, M. Z. Abdin, and M. Sarwat (New York, NY: Springer), Vol. 1, 25–49. doi:10.1007/978-1-4614-6372-6_2
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced Rice Blast Resistance by CRISPR/Cas9-targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLOS ONE 11, e0154027. doi:10.1371/journal.pone.0154027
Wang, J., Song, L., Gong, X., Xu, J., and Li, M. (2020). Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Ijms 21 (4), 1446. doi:10.3390/ijms21041446
Wang, T., Zhang, H., and Zhu, H. (2019). CRISPR Technology Is Revolutionizing the Improvement of Tomato and Other Fruit Crops. Hortic. Res. 6, 77. doi:10.1038/s41438-019-0159-x
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat. Biotechnol. 32, 947–951. doi:10.1038/nbt.2969
Werner, T., Köllmer, I., Bartrina, I., Holst, K., and Schmülling, T. (2006). New Insights into the Biology of Cytokinin Degradation. Plant Biol. 8, 371–381. doi:10.1055/s-2006-923928
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell. 15 (11), 2532–2550. doi:10.1105/tpc.014928
Werner, T., Nehnevajova, E., Köllmer, I., Novák, O., Strnad, M., Krämer, U., et al. (2010). Root-Specific Reduction of Cytokinin Causes Enhanced Root Growth, Drought Tolerance, and Leaf Mineral Enrichment in Arabidopsis and Tobacco. Plant Cell. 22, 3905–3920. doi:10.1105/tpc.109.072694
Werner, T., and Schmülling, T. (2009). Cytokinin Action in Plant Development. Curr. Opin. Plant Biol. 12, 527–538. doi:10.1016/j.pbi.2009.07.002
WHO (World Health Organisation) (2018). Global Nutrition Report. Available at: https://globalnutritionreport.org/reports/global-nutrition-report-2018/executive-summary/(accessed on February 4, 2022).
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free Genome Editing in Plants with Preassembled CRISPR-Cas9 Ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi:10.1038/nbt.3389
Xiao, X. O., Zeng, Y. M., Cao, B. H., Lei, J. J., Chen, Q. H., Meng, C. M., et al. (2017). PSAG12-IPT Overexpression in Eggplant Delays Leaf Senescence and Induces Abiotic Stress Tolerance. J. Hortic. Sci. Biotechnol. 92, 1–9. doi:10.1080/14620316.2017.1287529
Xu, Y., Burgess, P., Zhang, X., and Huang, B. (2016). Enhancing Cytokinin Synthesis by Overexpressingiptalleviated Drought Inhibition of Root Growth through Activating ROS-Scavenging Systems inAgrostis Stolonifera. Exbotj 67, 1979–1992. doi:10.1093/jxb/erw019
Yadav, R., Jain, V., Hegde, V. V. S., Yadav, N., and Kumar, R. (2019). Bio-Physico-Chemical Response of Drought Tolerant Chickpeas to Nickel. Lr 43 (3), 345–352. doi:10.18805/LR-4179
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 57, 781–803. doi:10.1146/annurev.arplant.57.032905.105444
Yamburenko, M. V., Kieber, J. J., and Schaller, G. E. (2017). Dynamic Patterns of Expression for Genes Regulating Cytokinin Metabolism and Signaling during Rice Inflorescence Development. PLoS ONE 12, e0176060. doi:10.1371/journal.pone.0176060
Yorgancilar, M., and Bilgiçli, N. (2014). Chemical and Nutritional Changes in Bitter and Sweet Lupin Seeds (Lupinus Albus L.) during Bulgur Production. J. Food Sci. Technol. 51 (17), 1384–1389. doi:10.1007/s13197-012-0640-0
Young, N. D., Debellé, F., Oldroyd, G. E. D., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago Genome Provides Insight into the Evolution of Rhizobial Symbioses. Nature 480 (7378), 520–524. doi:10.1038/nature10625
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 System Produces Specific and Homozygous Targeted Gene Editing in Rice in One Generation. Plant Biotechnol. J. 12 (6), 797–807. doi:10.1111/pbi.12200
Zhang, W., Peng, K., Cui, F., Wang, D., Zhao, J., Zhang, Y., et al. (2021). Cytokinin Oxidase/dehydrogenase OsCKX11 Coordinates Source and Sink Relationship in Rice by Simultaneous Regulation of Leaf Senescence and Grain Number. Plant Biotechnol. J. 19 (2), 335–350. doi:10.1111/pbi.13467
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and Transgene-free Genome Editing in Wheat through Transient Expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi:10.1038/ncomms12617
Keywords: biofortification, chickpea, cytokinindehydrogenase (CKX), genome-editing, stress
Citation: Mahto RK, Ambika , Singh C, Chandana BS, Singh RK, Verma S, Gahlaut V, Manohar M, Yadav N and Kumar R (2022) Chickpea Biofortification for Cytokinin Dehydrogenase via Genome Editing to Enhance Abiotic-Biotic Stress Tolerance and Food Security. Front. Genet. 13:900324. doi: 10.3389/fgene.2022.900324
Received: 20 March 2022; Accepted: 22 April 2022;
Published: 20 May 2022.
Edited by:
Santosh Kumar Gupta, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Surendra Pratap Singh, Chhatrapati Shahu Ji Maharaj University, IndiaCopyright © 2022 Mahto, Ambika, Singh, Chandana, Singh, Verma, Gahlaut, Manohar, Yadav and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajendra Kumar, cmFqZW5kcmFrNjRAeWFob28uY28uaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.