- 1Department of Obstetrics and Gynecology, Key Laboratory for Major Obstetric Diseases of Guangdong Province, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 2Key Laboratory of Reproduction and Genetics of Guangdong Higher Education Institutes, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 3Guangzhou Key Laboratory for Clinical Rapid Diagnosis and Early Warning of Infectious Diseases, KingMed School of Laboratory Medicine, Guangzhou Medical University, Guangzhou, China
Advances in induced pluripotent stem cell (iPSC) techniques have opened up new perspectives in research on developmental biology. Compared with other sources of human cellular models, iPSCs present a great advantage in hosting the unique genotype background of donors without ethical concerns. A wide spectrum of cellular and organoid models can be generated from iPSCs under appropriate in vitro conditions. The pluripotency of iPSCs is orchestrated by external signalling and regulated at the epigenetic, transcriptional and posttranscriptional levels. Recent decades have witnessed the progress of studying tissue-specific expressions and functions of microRNAs (miRNAs) using iPSC-derived models. MiRNAs are a class of short non-coding RNAs with regulatory functions in various biological processes during development, including cell migration, proliferation and apoptosis. MiRNAs are key modulators of gene expression and promising candidates for biomarker in development; hence, research on the regulation of human development by miRNAs is expanding. In this review, we summarize the current progress in the application of iPSC-derived models to studies of the regulatory roles of miRNAs in developmental processes.
Introduction
MiRNAs are short RNA molecules with 20–24 nucleotides that regulate the posttranscriptional silencing of target genes (Krol et al., 2010; Fabian and Sonenberg, 2012; Luo and Zhu, 2014; Lu and Rothenberg, 2018). MiRNAs exhibit a complex regulatory network resulting from a particular miRNA targeting multiple mRNAs and multiple miRNAs targeting the same mRNA, and affecting the expression levels of many protein-coding genes involved in functional pathways (Liu et al., 2014; Barwari et al., 2016; Luo et al., 2016; Rupaimoole and Slack, 2017). Over the past few decades, the role of miRNAs has been evaluated in a variety of biological processes (Ambros, 2004; Luo et al., 2015b; Lopez et al., 2017; Song et al., 2019). To date, numerous studies have delineated the regulatory role of miRNAs in development. For instance, miRNAs are regulating cell differentiation, proliferation, apoptosis and migration during B cell development by regulating a spectrum of signalling pathways, including BCR, MAPK/ERK, PI3K/AKT and NFκB pathways (Katsaraki et al., 2021). Thus, miRNAs have been characterized as valuable modulators of human development.
The investigation of miRNAs in development requires in vitro models derived from human pluripotent stem cells to simulate the tissue developmental procedures. Nevertheless, there are a number of shortages of human embryonic stem cell (hESC) techniques, such as ethical issues and complicated manipulation, thus preventing its wide application in clinical and basic research (Barker and de Beaufort, 2013; Luo et al., 2014). In 2006, studies were conducted to reprogram somatic cells into pluripotent stem cells with a cocktail of transcriptional factors, such as the combination of OCT4, KLF4, SOX2 and c-Myc (Takahashi and Yamanaka, 2006). This method avoids moral controversies and has led to the application of cellular programming techniques in human developmental research (Lo Sardo et al., 2017). Thus, the emergence of human induced pluripotent stem cells (hiPSC) has solved these problems (Luo et al., 2015a).
Remarkable progress has been created within the area of hiPSC over the past decade (Luo et al., 2018; Luo et al., 2021a). At present, hiPSCs can specifically differentiate into cardiomyocytes, endothelial cells, insulin-producing cells, germ cells, neuronal cells, osteoblasts, retinal pigment epithelium and so on (Figure 1). These cells could be ultilized for research of human development and diseases (Luo et al., 2021c). Hence, this review aims to systematically summarize the regulatory roles of miRNAs in development identified by iPSC-derived models.
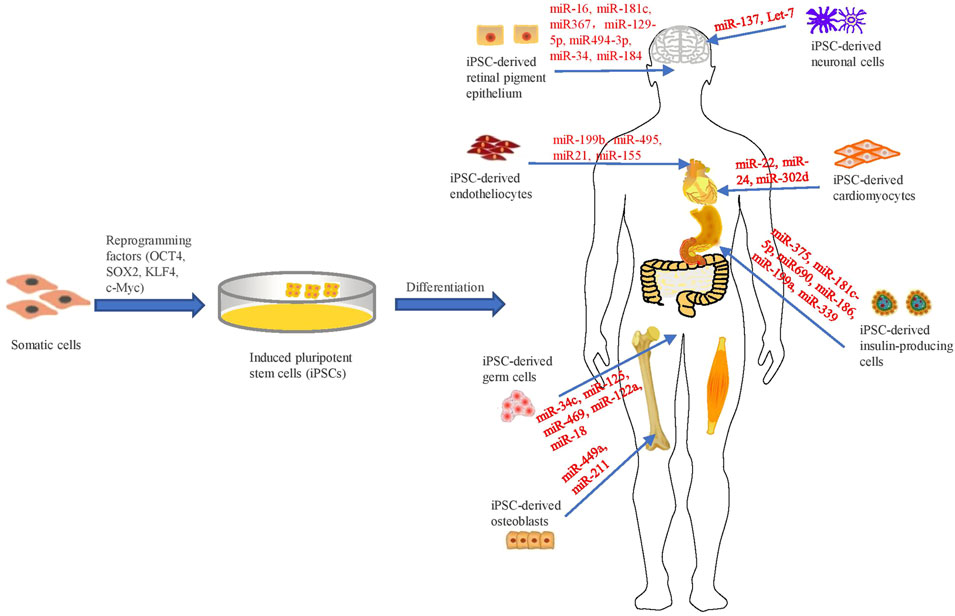
FIGURE 1. Schematic representation of investigating the regulatory roles of miRNAs in iPSC-derived cellular models.
Cardiomyocytes
Cardiovascular diseases (CVDs), such as myocardial infarction (MI) and cardiomyopathy, are recognized as the leading lethal causes around the world and are often associated with degeneration of cardiomyocytes (CMs). CMs are fully differentiated cells with minimal proliferative potential. Given the restricted effectiveness of drug therapy in treating myocardial injuries, the development of novel therapeutic approaches for curing these disorders is of urgency. HiPSC-derived CMs (iPS-CMs) introduce a new prospect for CVD treatment. However, the molecular mechanisms regulating the development of these cells is a pivotal problem that should be solved prior to clinical usage.
For instance, a study has compared the mRNA and miRNA expression profiles of iPS-CMs and biopsies from fetal, adult and hypertensive hearts to find out the core miRNA network, which revealed miRNAs associated with human heart development (Babiarz et al., 2012). Further studies profiled the miRNAs in human iPS-CMs and revealed 96 miRNAs that could promote CM proliferation (Diez-Cuñado et al., 2018). The CM proliferation-associated miRNAs in human were quite different from those of rodent (Eulalio et al., 2012). Most human CM proliferation-associated miRNAs function by targeting the Hippo pathway, an evolutionarily conserved pathway regulating organ size (Yu and Guan, 2013). Another study also confirmed that the mRNAs encoding most components of the Hippo pathway were recruited into the RNA-induced silencing complex (RISC) in iPS-CMs (Diez-Cuñado et al., 2018). In addition, some studies have demonstrated that miR-302d promoted CM proliferation by inhibiting LATS2 of the Hippo pathway (Xu F. et al., 2019).
Recently, miR-24 has been demonstrated as an important regulator for human heart development by using iPS-CM models (Guo et al., 2015). This is an execllent example of the complex regulatory roles of miRNAs in human development. On one hand, miR-24 has been demonstrated to suppress CM apoptosis. It is shown that delivery of miR-24 into CMs significantly alleviates cardiomyopathies, suggesting that modulating miRNA levels might be a novel therapeutic means for cardiac diseases (Qian et al., 2011; Guo et al., 2015). One study showed that miR-24 promoted functional implantation of cardiovascular progenitor cells (CPCs), in which miR-24 was utilized as a component of the antiapoptotic cocktail to enhance the survival of CPCs implantated into the MI heart tissues (Hu et al., 2011). Other studies have also identified other prosurvival roles of miR-24 in cardiac fibrosis and found that overexpression of miR-24 through lentivirus-mediated transduction reduces fibrosis and improves cardiac function in MI hearts, confirming the beneficial role of miR-24 (Guo et al., 2015). On the other hand, miR-24 has been demonstarted to exert proapoptotic effects. MiR-24 is characterized as a proapoptotic miRNA in cardiac endothelial cells, and blocking its function by injection of miR-24 antagonists can prevent apoptosis, enhance vascular distribution, and improve cardiac function after MI (Fiedler et al., 2011). However, these experiments in earlier studies were performed by viral transduction or polymeric transfection of miR-24 mimics or inhibitors, in which the protective effect observed might be partly caused by their off-target effects in non-CM cells (Guo et al., 2015).
In addition, some studies have also found that pri-miR-22/miR-22-3p is the top-ranked expressed primary miRNA transcript in heart tissues and iPS-CMs, and contributes to myocardial ischemia/reperfusion injury (Du et al., 2016; Sun et al., 2019). Some studies have confirmed the pro-apoptotic effect of miR-22 in iPS-CM (Pan and Zhu, 2018; Sun et al., 2019). Hypoxia-mediated apoptosis was augmented by miR-22 overexpression but resuced by miR-22 knockdown in iPS-CMs (Gidlöf et al., 2020). Meanwhile, this study also demonstrated that the long non-coding RNA Neat1 in the paraspeckles is the essential factor for pri-miR-22 processing in CMs. Knockdown of Neat1 could lead to significant accumulation of pri-miR-22 and consumption of mature ones in iPS-CMs (Gidlöf et al., 2020).
Vascular Endothelial Cells
MiR-199b is a highly conserved miRNA across species and capable of guiding the hiPSCs to differentiate into vascular endothelial cells (ECs) by regulating key molecular pathways, such as Notch signaling, in response to angiogenic signals. In particular, miR-199b regulates EC fate by targeting the Notch ligand JAG1, which leads to expression and secretion of VEGF via STAT3-mediated trascription (Chen et al., 2015). Nevertheless, the molecular mechanism underlying the upstream regulation remains unclear. Moreover, VEGF-induced miR-155 promotes angiogenesis by directly silencing E2F2, a E2F family transcriptional factor involved in cell proliferation, apoptosis and death (Dimova and Dyson, 2005), during EC differentiation from hiPSCs (Yang et al., 2016).
MiR-495 is a member of the DLK1-Dio3 miRNA cluster and exerts antiangiogenic effects. It is abundant in the non-EC portion while downregulated in the EC portion. It induces endothelial or angiogenic gene expression by downregulating VEZF1, a major transcriptional factor regulating EC genes, such as IGF1 and CD31, during EC differentiation and angiogenesis (Zou et al., 2010). In contrast, increasing VEZF1 expression via miR-495 blockage promotes angiogenesis post implantation of hiPSCs via enhancement of EC production. Studies have shown that the derived ECs significantly augmented the formation of new blood vessels in infarcted hearts, prevented functional deterioration and restricted the expansion of infarcted areas post transplantation in MI mice (Liang et al., 2017).
Additionally, miR-21 overexpression could enhance the Akt/TGF-β2 signal by downregulating PTEN on chromosome 10, thereby increasing the amount of ECs derived from hiPSCs (Zeng et al., 2018). Overexpression of miR-21 increased the mRNA and protein levels of TGF-β2, which is an essential cytokine for cell survival proliferation, migration and differentiation (Vargel et al., 2016). Neutralizing TGF-β2 by antibodies prohibits the expression of miR-21-induced EC markers, such as VE-CAD and CD31 (Di Bernardini et al., 2014).
Insulin-Producing Cells
The generation of insulin-producing cells (IPCs) from hiPSCs is a promising approach to investigate the molecular mechanisms of pancreatic development and a potential source of treatment for type I diabetes (Zeng et al., 2018). MiRNAs are major posttranscriptional regulators of gene expression and thus might involve in the control of β cell development in the pancreas.
For examples, miR-375 is essential for pancreatic endocrine function as its blockage results in glucose imbalance, α cell increment and β cell reduction (Poy et al., 2009). MiR-375 and miR-186 overexpression in hiPSCs leads to differentiation into insulin-secreting β-like cells that expressing pancreatic endocrine markers, such as PDX1, GLUT2, NGN3, PAX4 and PAX6. Despite secreting less insulin than natural β cells, these hiPSC-derived β-like cells could rescue blood glucose levels after transplantation into diabetic mice (Shaer et al., 2014). In addition, miR-375 affects insulin secretion by regulating the expression of muscular dystrophy protein in MIN6 cells (Poy et al., 2004; Krek et al., 2005).
The development of organisms is a result of the reprogramming of gene regulatory networks (Hornstein and Shomron, 2006). Some studies have described miR-375 as a key regulator of pancreatic development in humans (Poy et al., 2004; Lynn et al., 2007; Bravo-Egana et al., 2008; Avnit-Sagi et al., 2012). Mice lacking miR-375 showed α/β cell imbalance and reduced β cell propagation in spite of insulin insufficiency (Avnit-Sagi et al., 2012). Studies have shown that miR-7, miR-9, miR-375 and miR-376 are dramatically upregulated throughout the islet development (Wei et al., 2013). Some studies have found that miR-186, miR-199a and miR-339 are also upregulated during the formation of IPCs in vitro. The target genes of these three microRNAs include LIN28, PRDM1, CALB1, GCNB2, RBM47, PLEKHH1, RBPMS2 and PAK6 mRNA. (Joglekar et al., 2009; Chen et al., 2011; Kredo-Russo et al., 2012).
Studies have shown that miR-181c-5p accumulates gradually during the derivation of IPCs from hiPSCs. Increased phosphorylation of Smad2/3 is observed in iPSC-derived cells, and treatment with a Smad2/3 inhibitor after overexpression of miR-181c-5p had the opposite effect on IPC formation (Li et al., 2020). Similarily, other studies have also shown that miR-181c-5p is abundant in the late differentiation steps of hESC-derived IPCs, fetal pancreas, and adult islets (Liao et al., 2013; Fogel et al., 2015). Furthermore, miR-181c-5p was differentially expressed between the pancreas and the liver despite the common developmental origin of both tissues, with upregulation in the former and downregulation in latter (Porciuncula et al., 2013). Therefore, it is speculated that miR-181c-5p might play a pancreatic-specific role.
On the other hand, miR-690 overexpression dramatically delayed iPSC-derived IPC maturation and reduced insulin secretion in vitro and in vivo. Bioinformatic analysis suggested that its putative targets, such as CTNNB1, STAT3 and SOX9, were essential factors for of pancreatic endocrine development. Elevated miR-690 expression levels disrupt IPC differentiation by directly binding to Sox9. Subsequent experimental studies suggest that miR-690 could negatively modulate the Wnt signalling pathway during the pancreatic developmental process (Xu Y. et al., 2019).
In conclusion, these findings may help us better understand the process of pancreatic differentiation of hiPSCs in vitro and the underlying mechanisms involving miRNAs. As miRNAs could modulate certain transcriptional factors throughout the pancreatic developmental process, they could serve as novel therapeutic targets for diabetes treatment.
Neuronal Cells
HiPSC-derived neurons and neural progenitor cells (NPCs) are important models for investigating neurogenesis and synaptogenesis as well as their disruption in disorder statuses. Moreover, they are promising therapeutic vectors for brain disorders in the future (Zhu et al., 2013; Zhu et al., 2014). HiPSC-derived cellular and organoid models serve as an important bridge between model organism research and human postmortem brain research by providing living human cells, consisting of hiPSCs and their derived NPCs and neurons, with the composite genetic background present in patients. Hence, there is a quickly growing body of research projects using patient-specific iPSC-derived neurons to investigate neurogenesis.
For instance, iPSCs with mutations in the LRRK2 and α synuclein gene families were used to generate dopamine (DA) neurons, which exhibited higher sensitivity to oxidative stress and susceptibility to apoptosis (Byers et al., 2011; Reinhardt et al., 2013). Such phenotypes were also observed in iPSC-derived DA neurons from idiopathic Parkinson’s disease (PD) patients (Sánchez-Danés et al., 2012); meanwhile, apoptotic markers were also detected in the postmortem brain of PD patients (Hartmann et al., 2000; Mogi et al., 2000). Apoptosis-related miRNAs are also associated with neuronal differentiation (Aranha et al., 2011). For examples, miR-14, let-7a and miR-34a are elevated during neural stem cell differentiation (Heman-Ackah et al., 2013).
A large number of investigations have demonstrated that miRNAs play important roles in neural development (Hsu et al., 2012; Jimenez-Mateos et al., 2012; Liu et al., 2012). In addition, abundant molecular evidences support the essential roles of miRNAs in development of schizophrenia and other neural diseases (Green et al., 2013). For instance, miRNA-seq analysis was performed to distinguish differentially expressed miRNAs in iPSC-derived neurons from schizophrenia patients with 22q11.2 deletions compared to those from healthy donors (Yang et al., 2010). They discovered that miRNA expression levels in the deleted region decreased to approximately half the normal levels, and the levels were also altered in several other miRNAs out of the deleted region. The functional annotations of the putative targets of these dysregulated miRNAs were enrich in neurological diseases, neuronal development, axon formation and other important pathways relevant with the nervous system (Zhao et al., 2015).
Finally, posttranscriptional modifications could be identified by transcriptome analysis. RNA editing is a posttranscriptional event. Adenosine to inosine (A-to-I) transition is the dominate RNA editing process and happens most frequently in RNA molecules relevant with neurotransmission (Sanjana et al., 2012), especially in a lot of brain-specific miRNAs (Nishikura, 2010). Intriguingly, comparing postmortem cerebellum of autism patients with the control ons discovered that RNA editing was more abundant in the autism samples (Eran et al., 2013). Total transcriptome analysis can also detect fusion genes (Zhang Y. et al., 2014), which is valuable for building coexpression networks that can help researchers discover gene networks and pathways that are disrupted in neuropsychiatric disorders.
MiRNAs are believed to exert key regulatory effects in a wide spectrum of neural developmental processes, such as neurogenesis, neuronal maturation, axon regeneration, synaptic development and brain plasticity (Giraldez et al., 2005; Weston et al., 2006). Let-7 family miRNAs are the highest expressed miRNAs in the mammalian brains (Lagos-Quintana et al., 2002). They were firstly identified in Caenorhabditis elegans (Roush and Slack, 2008) and highly conserved across species. They are the key regulators for organism development, such as cell proliferation, cell specification and terminal differentiation (Nishikura, 2010). In the developing brain, Let-7 miRNAs participate in control of various developmental processes, such as neuronal differentiation (Schwamborn et al., 2009), neuronal subtype specification (Weick et al., 2013), neuronal regeneration (Li et al., 2015) and synaptic formation (Edbauer et al., 2010). Albeit in silico models suggest that the Let-7 miRNAs are involved in modulating postsynaptic gene expression (Paschou et al., 2012), their direct functions in mature human neurons remain unclear.
One of the Let-7 members, Let-7c is located on chromosome 21; thus, it exists in an extra copy of trisomy 21 (T21) and is associated with the symptoms of mild to moderate mental retardatoin featured in this neurodevelopmental syndrome (Antonarakis, 2017). It has been reported that miRNAs encoded by chromosome 21 may be important for a comprehensive understanding of the pathophysiology of T21-related neural diseases (Izzo et al., 2017). Taken together, these investigations indicate that the Let-7 family plays an important role in modulating human neurodevelopment and provide clues to illustrating the complicated molecular aetiology of neurodevelopmental syndromes (McGowan et al., 2018).
Germ Cells
In humans, genetic information is passed on to their offspring via germ cells (Luo et al., 2021b; Zhou et al., 2022). At present, ESCs, iPSCs and spermatogonial stem cells are the major cell sources used for generation of male germ cells expressing functional genes (Saito et al., 2015). However, their clinical utility is still challenged by several safety issues (Zhang D. et al., 2014). MiRNAs have recently emerged as important factors in translation regulation and the epigenetic control of stem cell self-renewal and pluripotent capacities (Gangaraju and Lin, 2009). Key roles of miRNA pathways in germline stem cell maintenance have been reported in vertebrate iPSCs (Gangaraju and Lin, 2009). In addition, miRNAs are very important in spermatogenesis and might play key roles in sperm mitosis, meiosis and postmeiotic stages (Wang and Xu, 2015).
The role of miRNAs in germ cell development has been functionally proven (Fernández-Pérez et al., 2018). For examples, RNA binding protein Lin28 blocks Let-7 and desuppresses Blimp1 translation in the initial stage of germ cell development (West et al., 2009). In addition, miR-125 posttranscriptionally suppress Oct4 during sperm meiosis in males (Medrano et al., 2013).
Further experimental evidence should be pursued to identify specific microRNAs that are regulating the three stages of human spermatogenesis, pachytene spermatocytes, spermatogonial cells and round spermatozoa cells (Liu Y. et al., 2015). For examples, miR-34c increased in pachytene spermatocytes and round sperm cells and prohibited survival by targeting the transcription factor ATF1 (Romero et al., 2011). In addition, miR-469 inhibited protamine and transition protein 2 (TP2) mRNA in pachytene spermatocytes and round sperm cells (Dai et al., 2011). Moreover, during spermatogenesis, miR-122a and miR-18 downregulate TP2 and heat shock factor 2, respectively (Chen et al., 2017).
Retinal Pigment Epithelium
The retinal pigment epithelium (RPE) is a special layer arranged at the rear of retina. Injury or RPE dysfunction can severely affect the health of photoreceptors and visual function, which is a result of potential RPE pathological blinding disease. Examples include age-related macular degeneration (AMD), Stargardt disease and retinitis pigmentosa (Greene et al., 2014). So far, there is no efficient therapy to rescue the vision; thus, iPSC-derived RPE (iPS-RPE) cells might be a source of cells to regenerate the disrupted RPE. However, before iPS-RPE cells can be used clinically, as much information as possible about the factors that modulate RPE development is of urgency to increase the production and quality of the cells for therapeutic use (Greene et al., 2014).
A study has identified 155 potential miRNA markers in iPS-RPE cells (Wang et al., 2014). Upregulated miRNAs, such as miR-181c and miR-129-5p, might drive cell specification (Naguibneva et al., 2006; Ryan et al., 2006), while downregulated miRNAs, including miR-367, miR-18b and miR-20b, are associated with mitotic division (Budde et al., 2010; Murakami et al., 2013). Putative targets of these miRNAs are relevant with cell survival, cell cycle and development.
It is of interest to evaluate the possible role of iPS-RPE miRNAs in tumorigenesis. On one hand, some iPS-RPE-upregulated miRNAs are tumour suppressors. For instance, miR-34 is a typicall tumour suppressor that prohibits tumor growth, metastasis, invasion and epithelial-mesenchymal transformation (EMT) via downregulating TP53 (Zhang et al., 2007; Nana-Sinkam and Croce, 2013). MiR-34 is generally silenced in multiple cancer types. MiR-34 expression was amplified in iPS-RPE cells by 5-fold, indicating an extremely low proliferative capacity in these terminally differentiated cells (Hermeking, 2012). Similarly, miR-16 is a tumour suppressive miRNA targeting multiple oncogenes, including EGFR, JUN and BCL2. In contrast, many iPS-RPE-downregulated miRNAs are oncogenic miRNAs (Wang et al., 2014).
MiRNAs in extracellular vesicles (EVs) derived from RPE cells might exert effects in the malignant inflammatory cycle. A specific enrichment of miR-494-3p was identified in EVs secreted from iPS-RPE cells after interaction with MPs, which might be a potential therapeutic target for the treatment of AMD (Mukai et al., 2021). AMD is the first and the third top causes of blindness in developed countries and around the world, respectively (Kuo et al., 2012). MiR-184 on chromosome 15q25.1 is a highly conserved miRNA across species (Nomura et al., 2008). MiR-137 is gradually upregulated during the differentiation of hiPSCs into RPE cells and it will downregulate PKBβ (also known as Akt2), the major downstream effector of rapamycin (mTOR) signalling pathway (Jiang et al., 2016). Hence, dysregulation of miR-137 is an important molecular event during the progression of AMD (Jiang et al., 2016).
Osteoblasts
HiPSCs could provide a rich cell source for regenerative medicine and to create patient-specific cellular and organoid models to investigate both intracellular and extracellular agents in bone repair and osteoarthritis (Diekman et al., 2012). Several histone deacetylase (HDAC) inhibitors have been shown to promote osteoblast maturation and specific gene expression by upregulating Runx2 gene expression in bone marrow stem cells (Hu et al., 2013). HDAC1 changes the expression of many genes associate with cell growth, survival, subtype specification and genome integrity (Buurman et al., 2012). One miRNA, miR-449a, specifically interferes with HDAC1 expression (Jeon et al., 2012; Okamoto et al., 2012). Exogenous miR-449a silencing endogenous HDAC1 expression keeps histone acetylation, induces Runx2 expression, which is a regulator of osteoblast genes (Nishimura et al., 2012), and accelerates osteoblast derivation from iPSCs (Liu T. et al., 2015).
In addition, an independent study demonstrated that a group of six miRNAs, miR-10a/b, miR-19b, miR-9, miR-124a, and miR-181a, are key regulators of the iPSC differentiation into osteoblasts (Okamoto et al., 2012). Moreover, another study has shown that miR-211 promoted iPSC differentiation into osteoblast-like cells via upregulating the expression of autophagy-related genes like ATG14 (Ozeki et al., 2017).
Discussion
MiRNAs are functioning within the RNA-protein complexes known as RNA-induced silencing complexes (RISC), which regulates gene expression posttranscriptionally in higher eukaryotes (Ameres and Zamore, 2013). Their roles in human development are rapidly being discovered (Table 1). MiRNAs are undoubtedly involved in many stages of normal cell development through their ability to block or promote development. They can be regulated by epigenetics, which may lead to other regulatory effects. In addition, they could serve as valuable markers for patient diagnosis and prognosis, as well as promising therapeutic targets. Although the multifaceted role of miRNAs in some diseases has been extensively studied over the past few years, important information is still missing, and no single molecule has been proven to be an effective regulator of the many pathogenic pathways of disease (Katsaraki et al., 2021).
IPSC-derived models are promising tools for deepening the understanding of early developmental processes (Dvash and Benvenisty, 2004). The major advantage of iPSC-derived models over primary cells is their capacity of repeatly generating cells with specific genetic background of the donors. With this property along with their pluripotency, hiPSCs can serve as a powerful tool for human cell replacement therapies and as an in vitro platform for personalized drug screening and discovery (Pouton and Haynes, 2007; Stadtfeld and Hochedlinger, 2010).
The reprogramming of somatic cells derived from patients and healthy donors into iPSCs is an important step to establish human-relevant models for illustrating the molecular and cellular mechanisms underlying the disease pathology. Notably, iPSCs can also be used to develop and test new therapies in vitro. Here, we discuss the regulatory role of miRNAs in iPSC-derived models for human development. In the future, miRNA-related studies need to be further improved to utilize hiPSCs as powerful tools in research of developmental biology. To address this issue, new methods, such as employing ectopic miRNAs as epigenetic modulators, should also be developed to optimize existing cell reprogramming and differentiation protocols (Ferreira et al., 2018).
There is a need to more thoroughly explore the role of miRNAs in human developoment. Given their relevance, we expect miRNAs to be exploited as diagnostic markers and as therapeutic targets for developmental diseases soon.
Author Contributions
YoC conceived the study. HC and ZM prepared the figure and table. HC, MZ, JZ, YpC, YZ, ZX, GZ, SC and YoC wrote and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Guangzhou City Science, Technology and Innovation Commission (202002030077).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ambros, V. (2004). The Functions of Animal microRNAs. Nature 431 (7006), 350–355. doi:10.1038/nature02871
Ameres, S. L., and Zamore, P. D. (2013). Diversifying microRNA Sequence and Function. Nat. Rev. Mol. Cell Biol. 14 (8), 475–488. doi:10.1038/nrm3611
Antonarakis, S. E. (2017). Down Syndrome and the Complexity of Genome Dosage Imbalance. Nat. Rev. Genet. 18 (3), 147–163. doi:10.1038/nrg.2016.154
Aranha, M. M., Santos, D. M., Solá, S., Steer, C. J., and Rodrigues, C. M. P. (2011). miR-34a Regulates Mouse Neural Stem Cell Differentiation. PLoS One 6 (8), e21396. doi:10.1371/journal.pone.0021396
Avnit-Sagi, T., Vana, T., and Walker, M. D. (2012). Transcriptional Mechanisms Controlling miR-375 Gene Expression in the Pancreas. Exp. Diabetes Res. 2012, 1–5. doi:10.1155/2012/891216
Babiarz, J. E., Ravon, M., Sridhar, S., Ravindran, P., Swanson, B., Bitter, H., et al. (2012). Determination of the Human Cardiomyocyte mRNA and miRNA Differentiation Network by Fine-Scale Profiling. Stem Cells Dev. 21 (11), 1956–1965. doi:10.1089/scd.2011.0357
Barker, R. A., and de Beaufort, I. (2013). Scientific and Ethical Issues Related to Stem Cell Research and Interventions in Neurodegenerative Disorders of the Brain. Prog. Neurobiol. 110, 63–73. doi:10.1016/j.pneurobio.2013.04.003
Barwari, T., Joshi, A., and Mayr, M. (2016). MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 68 (23), 2577–2584. doi:10.1016/j.jacc.2016.09.945
Bravo-Egana, V., Rosero, S., Molano, R. D., Pileggi, A., Ricordi, C., Domínguez-Bendala, J., et al. (2008). Quantitative Differential Expression Analysis Reveals miR-7 as Major Islet microRNA. Biochem. Biophysical Res. Commun. 366 (4), 922–926. doi:10.1016/j.bbrc.2007.12.052
Budde, H., Schmitt, S., Fitzner, D., Opitz, L., Salinas-Riester, G., and Simons, M. (2010). Control of Oligodendroglial Cell Number by the miR-17-92 Cluster. Development 137 (13), 2127–2132. doi:10.1242/dev.050633
Buurman, R., Gürlevik, E., Schäffer, V., Eilers, M., Sandbothe, M., Kreipe, H., et al. (2012). Histone Deacetylases Activate Hepatocyte Growth Factor Signaling by Repressing microRNA-449 in Hepatocellular Carcinoma Cells. Gastroenterology 143 (3), 811–820. e815. doi:10.1053/j.gastro.2012.05.033
Byers, B., Cord, B., Nguyen, H. N., Schüle, B., Fenno, L., Lee, P. C., et al. (2011). SNCA Triplication Parkinson's Patient's iPSC-Derived DA Neurons Accumulate α-Synuclein and Are Susceptible to Oxidative Stress. PLoS One 6 (11), e26159. doi:10.1371/journal.pone.0026159
Chen, B. Z., Yu, S. L., Singh, S., Kao, L. P., Tsai, Z. Y., Yang, P. C., et al. (2011). Identification of microRNAs Expressed Highly in Pancreatic Islet-like Cell Clusters Differentiated from Human Embryonic Stem Cells. Cell. Biol. Int. 35 (1), 29–37. doi:10.1042/CBI20090081
Chen, T., Margariti, A., Kelaini, S., Cochrane, A., Guha, S. T., Hu, Y., et al. (2015). MicroRNA-199b Modulates Vascular Cell Fate during iPS Cell Differentiation by Targeting the Notch Ligand Jagged1 and Enhancing VEGF Signaling. Stem Cells 33 (5), 1405–1418. doi:10.1002/stem.1930
Chen, X., Li, X., Guo, J., Zhang, P., and Zeng, W. (2017). The Roles of microRNAs in Regulation of Mammalian Spermatogenesis. J. Anim. Sci. Biotechnol. 8, 35. doi:10.1186/s40104-017-0166-4
Dai, L., Tsai-Morris, C.-H., Sato, H., Villar, J., Kang, J.-H., Zhang, J., et al. (2011). Testis-specific miRNA-469 Up-Regulated in Gonadotropin-Regulated Testicular RNA Helicase (GRTH/DDX25)-null Mice Silences Transition Protein 2 and Protamine 2 Messages at Sites within Coding Region. J. Biol. Chem. 286 (52), 44306–44318. doi:10.1074/jbc.M111.282756
Di Bernardini, E., Campagnolo, P., Margariti, A., Zampetaki, A., Karamariti, E., Hu, Y., et al. (2014). Endothelial Lineage Differentiation from Induced Pluripotent Stem Cells Is Regulated by MicroRNA-21 and Transforming Growth Factor β2 (TGF-Β2) Pathways. J. Biol. Chem. 289 (6), 3383–3393. doi:10.1074/jbc.M113.495531
Diekman, B. O., Christoforou, N., Willard, V. P., Sun, H., Sanchez-Adams, J., Leong, K. W., et al. (2012). Cartilage Tissue Engineering Using Differentiated and Purified Induced Pluripotent Stem Cells. Proc. Natl. Acad. Sci. U.S.A. 109 (47), 19172–19177. doi:10.1073/pnas.1210422109
Diez-Cuñado, M., Wei, K., Bushway, P. J., Maurya, M. R., Perera, R., Subramaniam, S., et al. (2018). miRNAs that Induce Human Cardiomyocyte Proliferation Converge on the Hippo Pathway. Cell Rep. 23 (7), 2168–2174. doi:10.1016/j.celrep.2018.04.049
Dimova, D. K., and Dyson, N. J. (2005). The E2F Transcriptional Network: Old Acquaintances with New Faces. Oncogene 24 (17), 2810–2826. doi:10.1038/sj.onc.1208612
Du, J.-K., Cong, B.-H., Yu, Q., Wang, H., Wang, L., Wang, C.-N., et al. (2016). Upregulation of microRNA-22 Contributes to Myocardial Ischemia-Reperfusion Injury by Interfering with the Mitochondrial Function. Free Radic. Biol. Med. 96, 406–417. doi:10.1016/j.freeradbiomed.2016.05.006
Dvash, T., and Benvenisty, N. (2004). Human Embryonic Stem Cells as a Model for Early Human Development. Best Pract. Res. Clin. Obstetrics Gynaecol. 18 (6), 929–940. doi:10.1016/j.bpobgyn.2004.06.005
Edbauer, D., Neilson, J. R., Foster, K. A., Wang, C.-F., Seeburg, D. P., Batterton, M. N., et al. (2010). Regulation of Synaptic Structure and Function by FMRP-Associated microRNAs miR-125b and miR-132. Neuron 65 (3), 373–384. doi:10.1016/j.neuron.2010.01.005
Eran, A., Li, J. B., Vatalaro, K., McCarthy, J., Rahimov, F., Collins, C., et al. (2013). Comparative RNA Editing in Autistic and Neurotypical Cerebella. Mol. Psychiatry 18 (9), 1041–1048. doi:10.1038/mp.2012.118
Eulalio, A., Mano, M., Ferro, M. D., Zentilin, L., Sinagra, G., Zacchigna, S., et al. (2012). Functional Screening Identifies miRNAs Inducing Cardiac Regeneration. Nature 492 (7429), 376–381. doi:10.1038/nature11739
Fabian, M. R., and Sonenberg, N. (2012). The Mechanics of miRNA-Mediated Gene Silencing: a Look under the Hood of miRISC. Nat. Struct. Mol. Biol. 19 (6), 586–593. doi:10.1038/nsmb.2296
Fernández-Pérez, D., Brieño-Enríquez, M. A., Isoler-Alcaraz, J., Larriba, E., and Del Mazo, J. (2018). MicroRNA Dynamics at the Onset of Primordial Germ and Somatic Cell Sex Differentiation during Mouse Embryonic Gonad Development. Rna 24 (3), 287–303. doi:10.1261/rna.062869.117
Ferreira, A. F., Calin, G. A., Picanço-Castro, V., Kashima, S., Covas, D. T., and de Castro, F. A. (2018). Hematopoietic Stem Cells from Induced Pluripotent Stem Cells - Considering the Role of microRNA as a Cell Differentiation Regulator. J. Cell Sci. 131 (4), jcs203018. doi:10.1242/jcs.203018
Fiedler, J., Jazbutyte, V., Kirchmaier, B. C., Gupta, S. K., Lorenzen, J., Hartmann, D., et al. (2011). MicroRNA-24 Regulates Vascularity after Myocardial Infarction. Circulation 124 (6), 720–730. doi:10.1161/circulationaha.111.039008
Fogel, G. B., Kai, Z. S., Zargar, S., Hinton, A., Jones, G. A., Wong, A. S., et al. (2015). MicroRNA Dynamics during Human Embryonic Stem Cell Differentiation to Pancreatic Endoderm. Gene 574 (2), 359–370. doi:10.1016/j.gene.2015.08.027
Gangaraju, V. K., and Lin, H. (2009). MicroRNAs: Key Regulators of Stem Cells. Nat. Rev. Mol. Cell Biol. 10 (2), 116–125. doi:10.1038/nrm2621
Gidlöf, O., Bader, K., Celik, S., Grossi, M., Nakagawa, S., Hirose, T., et al. (2020). Inhibition of the Long Non-coding RNA NEAT1 Protects Cardiomyocytes from Hypoxia In Vitro via Decreased Pri-miRNA Processing. Cell Death Dis. 11 (8), 677. doi:10.1038/s41419-020-02854-7
Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., et al. (2005). MicroRNAs Regulate Brain Morphogenesis in Zebrafish. Science 308 (5723), 833–838. doi:10.1126/science.1109020
Green, M. J., Cairns, M. J., Wu, J., Dragovic, M., Jablensky, A., Tooney, P. A., et al. (2013). Genome-wide Supported Variant MIR137 and Severe Negative Symptoms Predict Membership of an Impaired Cognitive Subtype of Schizophrenia. Mol. Psychiatry 18 (7), 774–780. doi:10.1038/mp.2012.84
Greene, W. A., Muñiz, A., Plamper, M. L., Kaini, R. R., and Wang, H.-C. (2014). MicroRNA Expression Profiles of Human iPS Cells, Retinal Pigment Epithelium Derived from iPS, and Fetal Retinal Pigment Epithelium. JoVE 88, e51589. doi:10.3791/51589
Guo, C., Deng, Y., Liu, J., and Qian, L. (2015). Cardiomyocyte‐specific Role of miR‐24 in Promoting Cell Survival. J. Cell. Mol. Med. 19 (1), 103–112. doi:10.1111/jcmm.12393
Hartmann, A., Hunot, S., Michel, P. P., Muriel, M.-P., Vyas, S., Faucheux, B. A., et al. (2000). Caspase-3: A Vulnerability Factor and Final Effector in Apoptotic Death of Dopaminergic Neurons in Parkinson's Disease. Proc. Natl. Acad. Sci. U.S.A. 97 (6), 2875–2880. doi:10.1073/pnas.040556597
Heman-Ackah, S. M., Hallegger, M., Rao, M. S., and Wood, M. J. A. (2013). RISC in PD: the Impact of microRNAs in Parkinson's Disease Cellular and Molecular Pathogenesis. Front. Mol. Neurosci. 6, 40. doi:10.3389/fnmol.2013.00040
Hermeking, H. (2012). MicroRNAs in the P53 Network: Micromanagement of Tumour Suppression. Nat. Rev. Cancer 12 (9), 613–626. doi:10.1038/nrc3318
Hornstein, E., and Shomron, N. (2006). Canalization of Development by microRNAs. Nat. Genet. 38 (6), S20–S24. doi:10.1038/ng1803
Hsu, R., Schofield, C. M., Dela Cruz, C. G., Jones-Davis, D. M., Blelloch, R., and Ullian, E. M. (2012). Loss of microRNAs in Pyramidal Neurons Leads to Specific Changes in Inhibitory Synaptic Transmission in the Prefrontal Cortex. Mol. Cell. Neurosci. 50 (3-4), 283–292. doi:10.1016/j.mcn.2012.06.002
Hu, S., Huang, M., Nguyen, P. K., Gong, Y., Li, Z., Jia, F., et al. (2011). Novel microRNA Prosurvival Cocktail for Improving Engraftment and Function of Cardiac Progenitor Cell Transplantation. Circulation 124 (11 Suppl. l), S27–S34. doi:10.1161/circulationaha.111.017954
Hu, X., Zhang, X., Dai, L., Zhu, J., Jia, Z., Wang, W., et al. (2013). Histone Deacetylase Inhibitor Trichostatin A Promotes the Osteogenic Differentiation of Rat Adipose-Derived Stem Cells by Altering the Epigenetic Modifications on Runx2 Promoter in a BMP Signaling-dependent Manner. Stem Cells Dev. 22 (2), 248–255. doi:10.1089/scd.2012.0105
Izzo, A., Manco, R., Cristofaro, T. d., Bonfiglio, F., Cicatiello, R., Mollo, N., et al. (2017). Overexpression of Chromosome 21 miRNAs May Affect Mitochondrial Function in the Hearts of Down Syndrome Fetuses. Int. J. Genomics 2017, 1–10. doi:10.1155/2017/8737649
Jeon, H. S., Lee, S. Y., Lee, E. J., Yun, S. C., Cha, E. J., Choi, E., et al. (2012). Combining microRNA-449a/b with a HDAC Inhibitor Has a Synergistic Effect on Growth Arrest in Lung Cancer. Lung Cancer 76 (2), 171–176. doi:10.1016/j.lungcan.2011.10.012
Jiang, C., Qin, B., Liu, G., Sun, X., Shi, H., Ding, S., et al. (2016). MicroRNA-184 Promotes Differentiation of the Retinal Pigment Epithelium by Targeting the AKT2/mTOR Signaling Pathway. Oncotarget 7 (32), 52340–52353. doi:10.18632/oncotarget.10566
Jimenez-Mateos, E. M., Engel, T., Merino-Serrais, P., McKiernan, R. C., Tanaka, K., Mouri, G., et al. (2012). Silencing microRNA-134 Produces Neuroprotective and Prolonged Seizure-Suppressive Effects. Nat. Med. 18 (7), 1087–1094. doi:10.1038/nm.2834
Joglekar, M. V., Joglekar, V. M., and Hardikar, A. A. (2009). Expression of Islet-specific microRNAs during Human Pancreatic Development. Gene Expr. Patterns 9 (2), 109–113. doi:10.1016/j.gep.2008.10.001
Katsaraki, K., Karousi, P., Artemaki, P. I., Scorilas, A., Pappa, V., Kontos, C. K., et al. (2021). MicroRNAs: Tiny Regulators of Gene Expression with Pivotal Roles in Normal B-Cell Development and B-Cell Chronic Lymphocytic Leukemia. Cancers 13 (4), 593. doi:10.3390/cancers13040593
Kredo-Russo, S., Mandelbaum, A. D., Ness, A., Alon, I., Lennox, K. A., Behlke, M. A., et al. (2012). Pancreas-enriched miRNA Refines Endocrine Cell Differentiation. Development 139 (16), 3021–3031. doi:10.1242/dev.080127
Krek, A., Grün, D., Poy, M. N., Wolf, R., Rosenberg, L., Epstein, E. J., et al. (2005). Combinatorial microRNA Target Predictions. Nat. Genet. 37 (5), 495–500. doi:10.1038/ng1536
Krol, J., Loedige, I., and Filipowicz, W. (2010). The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat. Rev. Genet. 11 (9), 597–610. doi:10.1038/nrg2843
Kuo, C.-H., Deng, J. H., Deng, Q., and Ying, S.-Y. (2012). A Novel Role of miR-302/367 in Reprogramming. Biochem. Biophysical Res. Commun. 417 (1), 11–16. doi:10.1016/j.bbrc.2011.11.058
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of Tissue-specific microRNAs from Mouse. Curr. Biol. 12 (9), 735–739. doi:10.1016/s0960-9822(02)00809-6
Li, N., Jiang, D., He, Q., He, F., Li, Y., Deng, C., et al. (2020). microRNA-181c-5p Promotes the Formation of Insulin-Producing Cells from Human Induced Pluripotent Stem Cells by Targeting Smad7 and TGIF2. Cell Death Dis. 11 (6), 462. doi:10.1038/s41419-020-2668-9
Li, S., Wang, X., Gu, Y., Chen, C., Wang, Y., Liu, J., et al. (2015). Let-7 microRNAs Regenerate Peripheral Nerve Regeneration by Targeting Nerve Growth Factor. Mol. Ther. 23 (3), 423–433. doi:10.1038/mt.2014.220
Liang, J., Huang, W., Cai, W., Wang, L., Guo, L., Paul, C., et al. (2017). Inhibition of microRNA-495 Enhances Therapeutic Angiogenesis of Human Induced Pluripotent Stem Cells. Stem Cells 35 (2), 337–350. doi:10.1002/stem.2477
Liao, X., Xue, H., Wang, Y.-C., Nazor, K. L., Guo, S., Trivedi, N., et al. (2013). Matched miRNA and mRNA Signatures from a hESC-Based In Vitro Model of Pancreatic Differentiation Reveal Novel Regulatory Interactions. J. Cell Sci. 126 (Pt 17), 3848–3861. doi:10.1242/jcs.123570
Liu, B., Li, J., and Cairns, M. J. (2014). Identifying miRNAs, Targets and Functions. Briefings Bioinforma. 15 (1), 1–19. doi:10.1093/bib/bbs075
Liu, J., Githinji, J., McLaughlin, B., Wilczek, K., and Nolta, J. (2012). Role of miRNAs in Neuronal Differentiation from Human Embryonic Stem Cell-Derived Neural Stem Cells. Stem Cell Rev Rep 8 (4), 1129–1137. doi:10.1007/s12015-012-9411-6
Liu, T., Hou, L., Zhao, Y., and Huang, Y. (2015a). Epigenetic Silencing of HDAC1 by miR-449a Upregulates Runx2 and Promotes Osteoblast Differentiation. Int. J. Mol. Med. 35 (1), 238–246. doi:10.3892/ijmm.2014.2004
Liu, Y., Niu, M., Yao, C., Hai, Y., Yuan, Q., Liu, Y., et al. (2015b). Fractionation of Human Spermatogenic Cells Using STA-PUT Gravity Sedimentation and Their miRNA Profiling. Sci. Rep. 5, 8084. doi:10.1038/srep08084
Lo Sardo, V., Ferguson, W., Erikson, G. A., Topol, E. J., Baldwin, K. K., and Torkamani, A. (2017). Influence of Donor Age on Induced Pluripotent Stem Cells. Nat. Biotechnol. 35 (1), 69–74. doi:10.1038/nbt.3749
Lopez, J. P., Fiori, L. M., Cruceanu, C., Lin, R., Labonte, B., Cates, H. M., et al. (2017). MicroRNAs 146a/b-5 and 425-3p and 24-3p Are Markers of Antidepressant Response and Regulate MAPK/Wnt-system Genes. Nat. Commun. 8, 15497. doi:10.1038/ncomms15497
Lu, T. X., and Rothenberg, M. E. (2018). MicroRNA. J. Allergy Clin. Immunol. 141 (4), 1202–1207. doi:10.1016/j.jaci.2017.08.034
Luo, Y., Chen, Y., Zhang, M., Ma, X., Zhu, D., and Chen, Y. (2021a). Generation of an Induced Pluripotent Stem Cell Line GZHMCi008-A Derived from a Patient with SRY-Positive 46,XX Testicular Disorder of Sex Development. Stem Cell Res. 57, 102583. doi:10.1016/j.scr.2021.102583
Luo, Y., Li, J., Zhu, D., Fan, Y., Li, S., and Sun, X. (2014). High-resolution Chromosomal Microarray Analysis of Early-Stage Human Embryonic Stem Cells Reveals an Association between X Chromosome Instability and Skewed X Inactivation. Cell Biosci. 4 (1), 74. doi:10.1186/2045-3701-4-74
Luo, Y., Wu, S., Yuan, J., Zhou, H., Zhong, Y., Zhang, M., et al. (2021b). Evaluation of Prognostic Factors for Clinical Pregnancy Rate Following Artificial Insemination by Husband in the Chinese Population. Front. Med. 8, 638560. doi:10.3389/fmed.2021.638560
Luo, Y., Xu, X., An, X., Sun, X., Wang, S., and Zhu, D. (2016). Targeted Inhibition of the miR-199a/214 Cluster by CRISPR Interference Augments the Tumor Tropism of Human Induced Pluripotent Stem Cell-Derived Neural Stem Cells under Hypoxic Condition. Stem Cells Int. 2016, 1–8. doi:10.1155/2016/3598542
Luo, Y., Zhang, M., Chen, Y., Chen, Y., and Zhu, D. (2021c). Application of Human Induced Pluripotent Stem Cell-Derived Cellular and Organoid Models for COVID-19 Research. Front. Cell Dev. Biol. 9, 720099. doi:10.3389/fcell.2021.720099
Luo, Y., and Zhu, D. (2014). Combinatorial Control of Transgene Expression by Hypoxia-Responsive Promoter and Microrna Regulation for Neural Stem Cell-Based Cancer Therapy. BioMed Res. Int. 2014, 1–9. doi:10.1155/2014/751397
Luo, Y., Zhu, D., Du, R., Gong, Y., Xie, C., Xu, X., et al. (2015a). Uniparental Disomy of the Entire X Chromosome in Turner Syndrome Patient-specific Induced Pluripotent Stem Cells. Cell Discov. 1, 15022. doi:10.1038/celldisc.2015.22
Luo, Y., Zhu, D., Lam, D. H., Huang, J., Tang, Y., Luo, X., et al. (2015b). A Double-Switch Cell Fusion-Inducible Transgene Expression System for Neural Stem Cell-Based Antiglioma Gene Therapy. Stem Cells Int. 2015, 1–8. doi:10.1155/2015/649080
Luo, Y., Zhu, D., Xu, X., Ge, L., Sun, X., Chen, G., et al. (2018). Generation of an Induced Pluripotent Stem Cell Line from an Adult Male with 45,X/46,XY Mosaicism. Stem Cell Res. 27, 42–45. doi:10.1016/j.scr.2018.01.003
Lynn, F. C., Skewes-Cox, P., Kosaka, Y., McManus, M. T., Harfe, B. D., and German, M. S. (2007). MicroRNA Expression Is Required for Pancreatic Islet Cell Genesis in the Mouse. Diabetes 56 (12), 2938–2945. doi:10.2337/db07-0175
McGowan, H., Mirabella, V. R., Hamod, A., Karakhanyan, A., Mlynaryk, N., Moore, J. C., et al. (2018). hsa-let-7c miRNA Regulates Synaptic and Neuronal Function in Human Neurons. Front. Synaptic Neurosci. 10, 19. doi:10.3389/fnsyn.2018.00019
Mogi, M., Togari, A., Kondo, T., Mizuno, Y., Komure, O., Kuno, S., et al. (2000). Caspase Activities and Tumor Necrosis Factor Receptor R1 (P55) Level Are Elevated in the Substantia Nigra from Parkinsonian Brain. J. Neural Transm. 107 (3), 335–341. doi:10.1007/s007020050028
Mukai, A., Otsuki, Y., Ito, E., Fujita, T., Ueno, M., Maeda, T., et al. (2021). Mitochondrial miRNA494-3p in Extracellular Vesicles Participates in Cellular Interplay of iPS-Derived Human Retinal Pigment Epithelium with Macrophages. Exp. Eye Res. 208, 108621. doi:10.1016/j.exer.2021.108621
Murakami, Y., Tamori, A., Itami, S., Tanahashi, T., Toyoda, H., Tanaka, M., et al. (2013). The Expression Level of miR-18b in Hepatocellular Carcinoma Is Associated with the Grade of Malignancy and Prognosis. BMC Cancer 13, 99. doi:10.1186/1471-2407-13-99
Naguibneva, I., Ameyar-Zazoua, M., Polesskaya, A., Ait-Si-Ali, S., Groisman, R., Souidi, M., et al. (2006). The microRNA miR-181 Targets the Homeobox Protein Hox-A11 during Mammalian Myoblast Differentiation. Nat. Cell Biol. 8 (3), 278–284. doi:10.1038/ncb1373
Nana-Sinkam, S. P., and Croce, C. M. (2013). Clinical Applications for microRNAs in Cancer. Clin. Pharmacol. Ther. 93 (1), 98–104. doi:10.1038/clpt.2012.192
Nishikura, K. (2010). Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 79, 321–349. doi:10.1146/annurev-biochem-060208-105251
Nishimura, R., Wakabayashi, M., Hata, K., Matsubara, T., Honma, S., Wakisaka, S., et al. (2012). Osterix Regulates Calcification and Degradation of Chondrogenic Matrices through Matrix Metalloproteinase 13 (MMP13) Expression in Association with Transcription Factor Runx2 during Endochondral Ossification. J. Biol. Chem. 287 (40), 33179–33190. doi:10.1074/jbc.M111.337063
Nomura, T., Kimura, M., Horii, T., Morita, S., Soejima, H., Kudo, S., et al. (2008). MeCP2-dependent Repression of an Imprinted miR-184 Released by Depolarization. Hum. Mol. Genet. 17 (8), 1192–1199. doi:10.1093/hmg/ddn011
Okamoto, H., Matsumi, Y., Hoshikawa, Y., Takubo, K., Ryoke, K., and Shiota, G. (2012). Involvement of microRNAs in Regulation of Osteoblastic Differentiation in Mouse Induced Pluripotent Stem Cells. PLoS One 7 (8), e43800. doi:10.1371/journal.pone.0043800
Ozeki, N., Hase, N., Hiyama, T., Yamaguchi, H., Kawai-Asano, R., Nakata, K., et al. (2017). RETRACTED: MicroRNA-211 and Autophagy-Related Gene 14 Signaling Regulate Osteoblast-like Cell Differentiation of Human Induced Pluripotent Stem Cells. Exp. Cell Res. 352 (1), 63–74. doi:10.1016/j.yexcr.2017.01.018
Pan, H., and Zhu, L. (2018). RETRACTED: Angelica Sinensis Polysaccharide Protects Rat Cardiomyocytes H9c2 from Hypoxia-Induced Injury by Down-Regulation of microRNA-22. Biomed. Pharmacother. 106, 225–231. doi:10.1016/j.biopha.2018.06.120
Paschou, M., Paraskevopoulou, M. D., Vlachos, I. S., Koukouraki, P., Hatzigeorgiou, A. G., and Doxakis, E. (2012). miRNA Regulons Associated with Synaptic Function. PLoS One 7 (10), e46189. doi:10.1371/journal.pone.0046189
Pera, R., Simón, C., and Medrano, J. (2013). Germ Cell Differentiation from Pluripotent Cells. Semin. Reprod. Med. 31 (1), 014–023. doi:10.1055/s-0032-1331793
Porciuncula, A., Zapata, N., Guruceaga, E., Agirre, X., Barajas, M., and Prosper, F. (2013). MicroRNA Signatures of iPSCs and Endoderm-Derived Tissues. Gene Expr. Patterns 13 (1-2), 12–20. doi:10.1016/j.gep.2012.08.002
Pouton, C. W., and Haynes, J. M. (2007). Embryonic Stem Cells as a Source of Models for Drug Discovery. Nat. Rev. Drug Discov. 6 (8), 605–616. doi:10.1038/nrd2194
Poy, M. N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., MacDonald, P. E., et al. (2004). A Pancreatic Islet-specific microRNA Regulates Insulin Secretion. Nature 432 (7014), 226–230. doi:10.1038/nature03076
Poy, M. N., Hausser, J., Trajkovski, M., Braun, M., Collins, S., Rorsman, P., et al. (2009). miR-375 Maintains Normal Pancreatic α- and β-cell Mass. Proc. Natl. Acad. Sci. U.S.A. 106 (14), 5813–5818. doi:10.1073/pnas.0810550106
Qian, L., Van Laake, L. W., Huang, Y., Liu, S., Wendland, M. F., and Srivastava, D. (2011). miR-24 Inhibits Apoptosis and Represses Bim in Mouse Cardiomyocytes. J. Exp. Med. 208 (3), 549–560. doi:10.1084/jem.20101547
Reinhardt, P., Schmid, B., Burbulla, L. F., Schöndorf, D. C., Wagner, L., Glatza, M., et al. (2013). Genetic Correction of a LRRK2 Mutation in Human iPSCs Links Parkinsonian Neurodegeneration to ERK-dependent Changes in Gene Expression. Cell Stem Cell 12 (3), 354–367. doi:10.1016/j.stem.2013.01.008
Romero, Y., Meikar, O., Papaioannou, M. D., Conne, B., Grey, C., Weier, M., et al. (2011). Dicer1 Depletion in Male Germ Cells Leads to Infertility Due to Cumulative Meiotic and Spermiogenic Defects. PLoS One 6 (10), e25241. doi:10.1371/journal.pone.0025241
Roush, S., and Slack, F. J. (2008). The Let-7 Family of microRNAs. Trends Cell Biol. 18 (10), 505–516. doi:10.1016/j.tcb.2008.07.007
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA Therapeutics: towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov. 16 (3), 203–222. doi:10.1038/nrd.2016.246
Ryan, D. G., Oliveira-Fernandes, M., and Lavker, R. M. (2006). MicroRNAs of the Mammalian Eye Display Distinct and Overlapping Tissue Specificity. Mol. Vis. 12, 1175–1184.
Saito, S., Lin, Y.-C., Murayama, Y., Nakamura, Y., Eckner, R., Niemann, H., et al. (2015). Retracted Article: In Vitro Derivation of Mammalian Germ Cells from Stem Cells and Their Potential Therapeutic Application. Cell. Mol. Life Sci. 72 (23), 4545–4560. doi:10.1007/s00018-015-2020-1
Sánchez‐Danés, A., Richaud‐Patin, Y., Carballo‐Carbajal, I., Jiménez‐Delgado, S., Caig, C., Mora, S., et al. (2012). Disease‐specific Phenotypes in Dopamine Neurons from Human iPS‐based Models of Genetic and Sporadic Parkinson's Disease. EMBO Mol. Med. 4 (5), 380–395. doi:10.1002/emmm.201200215
Sanjana, N. E., Levanon, E. Y., Hueske, E. A., Ambrose, J. M., and Li, J. B. (2012). Activity-dependent A-To-I RNA Editing in Rat Cortical Neurons. Genetics 192 (1), 281–287. doi:10.1534/genetics.112.141200
Schwamborn, J. C., Berezikov, E., and Knoblich, J. A. (2009). The TRIM-NHL Protein TRIM32 Activates microRNAs and Prevents Self-Renewal in Mouse Neural Progenitors. Cell 136 (5), 913–925. doi:10.1016/j.cell.2008.12.024
Shaer, A., Azarpira, N., and Karimi, M. H. (2014). Differentiation of Human Induced Pluripotent Stem Cells into Insulin-like Cell Clusters with miR-186 and miR-375 by Using Chemical Transfection. Appl. Biochem. Biotechnol. 174 (1), 242–258. doi:10.1007/s12010-014-1045-5
Song, Y., Zhang, C., Zhang, J., Jiao, Z., Dong, N., Wang, G., et al. (2019). Localized Injection of miRNA-21-Enriched Extracellular Vesicles Effectively Restores Cardiac Function after Myocardial Infarction. Theranostics 9 (8), 2346–2360. doi:10.7150/thno.29945
Stadtfeld, M., and Hochedlinger, K. (2010). Induced Pluripotency: History, Mechanisms, and Applications. Genes Dev. 24 (20), 2239–2263. doi:10.1101/gad.1963910
Sun, H., Shi, K., Xie, D., Zhang, H., and Yu, B. (2019). Long Noncoding RNA C2dat1 Protects H9c2 Cells against Hypoxia Injury by Downregulating miR‐22. J. Cell. Physiology 234 (11), 20623–20633. doi:10.1002/jcp.28667
Takahashi, K., and Yamanaka, S. (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126 (4), 663–676. doi:10.1016/j.cell.2006.07.024
Vargel, Ö., Zhang, Y., Kosim, K., Ganter, K., Foehr, S., Mardenborough, Y., et al. (2016). Activation of the TGFβ Pathway Impairs Endothelial to Haematopoietic Transition. Sci. Rep. 6, 21518. doi:10.1038/srep21518
Wang, H.-C., Greene, W. A., Kaini, R. R., Shen-Gunther, J., Chen, H.-I. H., Car, H., et al. (2014). Profiling the microRNA Expression in Human iPS and iPS-Derived Retinal Pigment Epithelium. Cancer Inf. 13s5 (Suppl. 5), CIN.S14074–35. doi:10.4137/cin.S14074
Wang, L., and Xu, C. (2015). Role of microRNAs in Mammalian Spermatogenesis and Testicular Germ Cell Tumors. Reproduction 149 (3), R127–R137. doi:10.1530/rep-14-0239
Wei, R., Yang, J., Liu, G.-q., Gao, M.-j., Hou, W.-f., Zhang, L., et al. (2013). Dynamic Expression of microRNAs during the Differentiation of Human Embryonic Stem Cells into Insulin-Producing Cells. Gene 518 (2), 246–255. doi:10.1016/j.gene.2013.01.038
Weick, J. P., Held, D. L., Bonadurer, G. F., Doers, M. E., Liu, Y., Maguire, C., et al. (2013). Deficits in Human Trisomy 21 iPSCs and Neurons. Proc. Natl. Acad. Sci. U.S.A. 110 (24), 9962–9967. doi:10.1073/pnas.1216575110
West, J. A., Viswanathan, S. R., Yabuuchi, A., Cunniff, K., Takeuchi, A., Park, I.-H., et al. (2009). A Role for Lin28 in Primordial Germ-Cell Development and Germ-Cell Malignancy. Nature 460 (7257), 909–913. doi:10.1038/nature08210
Weston, M. D., Pierce, M. L., Rocha-Sanchez, S., Beisel, K. W., and Soukup, G. A. (2006). MicroRNA Gene Expression in the Mouse Inner Ear. Brain Res. 1111 (1), 95–104. doi:10.1016/j.brainres.2006.07.006
Xu, F., Yang, J., Shang, J., Lan, F., Li, M., Shi, L., et al. (2019a). MicroRNA-302d Promotes the Proliferation of Human Pluripotent Stem Cell-Derived Cardiomyocytes by Inhibiting LATS2 in the Hippo Pathway. Clin. Sci. (Lond) 133 (13), 1387–1399. doi:10.1042/cs20190099
Xu, Y., Huang, Y., Guo, Y., Xiong, Y., Zhu, S., Xu, L., et al. (2019b). microRNA-690 Regulates Induced Pluripotent Stem Cells (iPSCs) Differentiation into Insulin-Producing Cells by Targeting Sox9. Stem Cell Res. Ther. 10 (1), 59. doi:10.1186/s13287-019-1154-8
Yang, D., Wang, J., Xiao, M., Zhou, T., and Shi, X. (2016). Role of Mir-155 in Controlling HIF-1α Level and Promoting Endothelial Cell Maturation. Sci. Rep. 6, 35316. doi:10.1038/srep35316
Yang, J., Cai, J., Zhang, Y., Wang, X., Li, W., Xu, J., et al. (2010). Induced Pluripotent Stem Cells Can Be Used to Model the Genomic Imprinting Disorder Prader-Willi Syndrome. J. Biol. Chem. 285 (51), 40303–40311. doi:10.1074/jbc.M110.183392
Yu, F.-X., and Guan, K.-L. (2013). The Hippo Pathway: Regulators and Regulations. Genes Dev. 27 (4), 355–371. doi:10.1101/gad.210773.112
Zeng, Z.-L., Lin, X.-l., Tan, L.-L., Liu, Y.-M., Qu, K., and Wang, Z. (2018). MicroRNAs: Important Regulators of Induced Pluripotent Stem Cell Generation and Differentiation. Stem Cell Rev Rep 14 (1), 71–81. doi:10.1007/s12015-017-9785-6
Zhang, B., Pan, X., Cobb, G. P., and Anderson, T. A. (2007). microRNAs as Oncogenes and Tumor Suppressors. Dev. Biol. 302 (1), 1–12. doi:10.1016/j.ydbio.2006.08.028
Zhang, D., Liu, X., Peng, J., He, D., Lin, T., Zhu, J., et al. (2014a). Potential Spermatogenesis Recovery with Bone Marrow Mesenchymal Stem Cells in an Azoospermic Rat Model. Ijms 15 (8), 13151–13165. doi:10.3390/ijms150813151
Zhang, Y., Chen, K., Sloan, S. A., Bennett, M. L., Scholze, A. R., O'Keeffe, S., et al. (2014b). An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci. 34 (36), 11929–11947. doi:10.1523/jneurosci.1860-14.2014
Zhao, D., Lin, M., Chen, J., Pedrosa, E., Hrabovsky, A., Fourcade, H. M., et al. (2015). MicroRNA Profiling of Neurons Generated Using Induced Pluripotent Stem Cells Derived from Patients with Schizophrenia and Schizoaffective Disorder, and 22q11.2 Del. PLoS One 10 (7), e0132387. doi:10.1371/journal.pone.0132387
Zhu, D., Chen, C., Purwanti, Y. I., Du, S., Lam, D. H., Wu, C., et al. (2014). Induced Pluripotent Stem Cell-Derived Neural Stem Cells Transduced with Baculovirus Encoding CD40 Ligand for Immunogene Therapy in Mouse Models of Breast Cancer. Hum. Gene Ther. 25 (8), 747–758. doi:10.1089/hum.2013.160
Zhu, D., Lam, D. H., Purwanti, Y. I., Goh, S. L., Wu, C., Zeng, J., et al. (2013). Systemic Delivery of Fusogenic Membrane Glycoprotein-Expressing Neural Stem Cells to Selectively Kill Tumor Cells. Mol. Ther. 21 (8), 1621–1630. doi:10.1038/mt.2013.123
Zhu, D., Luo, Y., Zhou, H., Wu, S., Tang, X., Zhou, G., et al. (2022). Chlamydia trachomatis Infection in the Genital Tract Is Associated with Inflammation and Hypospermia in the Infertile Male of China. Asian J. Androl. 24 (1), 56–61. doi:10.4103/aja.aja_54_21
Keywords: microRNA, induced pluripotent stem cell, cellular model, develoment, gene regulaiton
Citation: Chen H, Zhang M, Zhang J, Chen Y, Zuo Y, Xie Z, Zhou G, Chen S and Chen Y (2022) Application of Induced Pluripotent Stem Cell-Derived Models for Investigating microRNA Regulation in Developmental Processes. Front. Genet. 13:899831. doi: 10.3389/fgene.2022.899831
Received: 19 March 2022; Accepted: 06 May 2022;
Published: 26 May 2022.
Edited by:
Jian-Hong Fang, Sun Yat-sen University, ChinaReviewed by:
Qihua Fu, Shanghai Children’s Medical Center, ChinaWenmin Sun, Sun Yat-sen University, China
Copyright © 2022 Chen, Zhang, Zhang, Chen, Zuo, Xie, Zhou, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoyong Chen, eWNoZW5AZ3pobXUuZWR1LmNu
†These authors have contributed equally to this work
 Hongyu Chen1,2†
Hongyu Chen1,2† Mimi Zhang
Mimi Zhang Yapei Chen
Yapei Chen Yabo Zuo
Yabo Zuo Guanqing Zhou
Guanqing Zhou Yaoyong Chen
Yaoyong Chen