- 1Department of Hematology, Pingxiang People’s Hospital, Pingxiang, China
- 2Department of Cardiology, Pingxiang People’s Hospital, Pingxiang, China
Background: Fourteen meta-analyses reported the individual effects of the GSTM1 and GSTT1 polymorphisms on leukemia risk. However, over 40 studies were not included in previously published meta-analyses. Moreover, one key aspect was that previous meta-analyses did not conduct the false-positive test on the aforementioned issues. Furthermore, previous meta-analyses did not observe the combined effects of GSTM1 present/null and GSTT1 present/null polymorphism with leukemia risk. Therefore, we conducted the current study to further analyze these associations.
Objectives: This study aimed to investigate the association between the individual and combined effects of the GSTM1 present/null and GSTT1 present/null polymorphisms and the risk of leukemia.
Methods: A meta-analysis was performed applying Meta-analyses of Observational Studies in Epidemiology (MOOSE) guidelines. Moreover, false-positive report probability (FPRP) and Bayesian false discovery probability (BFDP) were applied to investigate the false-positive results.
Results: The individual GSTM1 and GSTT1 null genotypes and combined effects of the two genes were associated with a significantly increased leukemia risk in overall and several subgroup analyses, such as Asians, Caucasians, and so on. Then, further analysis was conducted using FPRP and BFDP. Significant associations were considered as “positive” results on the GSTM1 null genotype with leukemia risk in overall populations (FPRP < 0.001 and BFDP = 0.006), Asians (FPRP < 0.001 and BFDP < 0.001), and East Asian population (FPRP < 0.001 and BFDP = 0.002). For the GSTT1 null genotype, significant associations were regarded “positive” results in overall populations, acute myeloid leukemia (AML), Asians, and East Asian population. For the combined effects of the GSTM1 and GSTT1 polymorphisms, significant associations were also considered “positive” results in the overall analysis of Asians, Indians, and East Asian population.
Conclusion: This study strongly indicates that the individual GSTM1 and GSTT1 null genotypes and combined effects of the two genes are associated with increased leukemia risk in Asians, especially in the East Asian population; the GSTT1 null genotype is associated with increased AML risk; the combined effects of the two genes are associated with increased leukemia risk in Indians.
Introduction
Leukemia, commonly diagnosed in childhood, is a complex and heterogeneous disease caused by irreversible genetic lesions in initially normal hematopoietic cells (Bloomfield et al., 2001). Chronic myeloid leukemia (CML) is a clonal, myeloproliferative disease characterized by the accumulation of myeloid precursors in the bone marrow, blood, and body tissues. It is a relatively rare disease worldwide, accounting for approximately 14% of all types of leukemia (Quintás-Cardama and Cortes, 2006). The highest incidence rate is found in males of all age groups, and the fact remains to be explained (Henderson et al., 1990; Goyette et al., 1994; Pui, 2000; Pui et al., 2000; Pui et al., 2002; Pui et al., 2006; Hirschhorn et al., 2002).
The glutathione S-transferases (GSTs) are a family of multifunctional enzymes, which play an important role in the detoxification of toxic, potentially carcinogenic compounds and a series of basic physiological processes of the human body (Benjamini and Hochberg, 1995; Hayes et al., 2005; Udomsinprasert et al., 2005). The GST family is divided into seven categories of genes in humans according to their primary structure (Curran et al., 2000).
The GSTM1 and GSTT1 polymorphisms have been identified, resulting in possibly impaired activity for the elimination of carcinogenic compounds and increased risk of cancer (Hayes and Strange, 2000). The GSTM1 and GSTT1 genes are located on chromosome 1 (1p13.3) and chromosome 22 (22q11.2), respectively (Hayes and Strange, 2000). Polymorphisms in both GSTM1 and GSTT1 result in gene deletions (null genotype), resulting in loss of expression and enzyme activity loss (Seidegård et al., 1988; Hayes and Strange, 2000). The lack of enzymatic activity may lead to the occurrence of cancer.
Fourteen meta-analyses (Ye and Song, 2005; Das et al., 2009; Zintzaras, 2009; Vijayakrishnan and Houlston, 2010; Tang et al., 2013; He et al., 2014a; He et al., 2014b; Ma et al., 2014; Moulik et al., 2014; Tang et al., 2014; Xu and Cao, 2014; Li et al., 2018; Zhao et al., 2018; Wang et al., 2019) reported the individual effects of the GSTM1 and GSTT1 polymorphisms with leukemia risk. However, over forty studies were not included in previously published meta-analyses. Moreover, one key question was that previous meta-analyses did not conduct the false-positive test on the above issues. Furthermore, previous meta-analyses did not perform the combined effects of GSTM1 present/null and GSTT1 present/null polymorphisms with leukemia risk. Therefore, we conducted the current study to further analyze these associations.
Materials and Methods
Identification and Eligibility of Relevant Studies
A comprehensive literature search was conducted applying the PubMed, EMBASE, ISI, CNKI, and WanFang databases for relevant articles published (the last search update was 26 February 2022). The search strategy (it was designed to be sensitive and broad) was as follows (glutathione S-transferase T1 OR GSTT1 OR glutathione S-transferase M1 OR GSTM1) AND (polymorphism OR genotype OR allele OR variant OR mutation) AND (leukemia OR leukaemia). In addition, studies were also identified by a search of the reference lists of reviews and retrieved studies. Moreover, all eligible studies were retrieved, and their bibliographies were checked for other relevant publications.
Inclusion Criteria
Inclusion criteria were as listed below: 1) Case–control or cohort studies; 2) publications on the individual or combined effects of GSTM1 present/null and GSTT1 present/null polymorphisms with leukemia risk; and 3) complete genotype data between leukemia cases and controls. Exclusion criteria were as listed below: 1) Duplicate genotype data; 2) no case–control studies; 3) Meta-analyses, reviews, or letters; and 4) other SNPs.
Data Extraction and Quality Score Assessment
Data were extracted independently by two investigators according to the inclusion criteria. Supplementary Table S1 lists the information on data extraction. Ethnicity was categorized as “Caucasian,” “Asian,” “Indian,” “African,” and mixed populations. “Indian” mainly came from India and Pakistan. The ethnicity was considered as “mixed population” when one study did not state which ethnic groups were included or if it was impossible to separate participants based on phenotype.
The scale of quality assessment criteria was designed based on one previous meta-analysis (Thakkinstian et al., 2011) (Supplementary Table S2). Studies scoring > 9 were considered high quality.
Statistical Analysis
Crude odds ratios (ORs) with 95% confidence intervals (CIs) were applied to evaluate the associations between the individual and combined effects of GSTM1 and GSTT1 polymorphisms on leukemia risk. Between-study heterogeneity was assessed by applying the Q statistic and I2 value. A random-effect model (DerSimonian–Laird model) (DerSimonian and Laird, 1986) was applied if p < 0.10 and/or I2 > 50%; otherwise, a fixed-effect model (Mantel–Haenszel method) was used (Mantel and Haenszel, 1959). Subgroups were conducted by ethnicity, geographic region, and type of leukemia. In addition, a meta-regression analysis was performed to explore the source of heterogeneity. Sensitivity analysis was performed by removing a single study each time and excluding low-quality studies. Publication bias was calculated using Begg’s funnel plot (Begg and Mazumdar, 1994) and Egger’s regression asymmetry test (Egger et al., 1997). If publication bias existed, a nonparametric “trim and fill” method (Dual and Tweedie, 2000) was applied to add missing studies. Moreover, we used the following criteria to investigate the false significant results: false-positive report probability (FPRP) < 0.2 and Bayesian false discovery probability (BFDP) < 0.8 because FPRP and BFDP values can clarify the probability of no true association between genetic association and disease risk. All statistical analyses were calculated using STATA version 12.0 (STATA Corporation, College Station, TX, United States).
Results
Study Characteristics
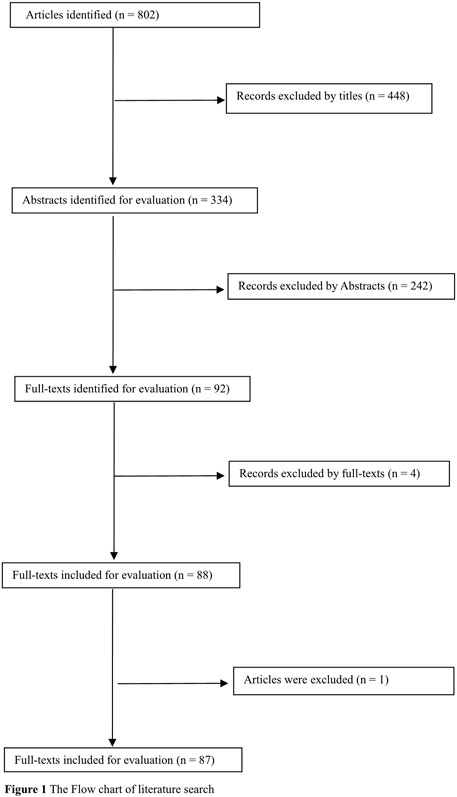
Overall, 802 articles were identified. Of these, 694 were excluded by carefully reading titles, abstracts, and full text. In addition, one study (Jiang et al., 2008) was excluded because another publication (Jiang and Tan, 2010) included their cases and controls. Therefore, 87 publications (Zintzaras, 2009; Aydin-Sayitoglu et al., 2006; Al-Achkar et al., 2014; Arruda et al., 2001; Allan et al., 2001; Abdalhabib et al., 2021; Alves et al., 2002; Al-Eitan et al., 2016; Bajpai et al., 2007; Balta et al., 2003; Bhat et al., 2012; Bhatla et al., 2008; Barnette et al., 2004; Bolufer et al., 2007; Baba et al., 2021; Bănescu et al., 2016; Bănescu et al., 2014; Chen et al., 1997; Chen et al., 2008; Crump et al., 2000; Chauhan et al., 2011; Chauhan et al., 2012; Canalle et al., 2004; Clavel et al., 2005; Chan et al., 2011; D'Alò et al., 2004; Davies et al., 2000; Davies et al., 2002; Dunna et al., 2013; Eyada et al., 2007; Feng et al., 2004; Farasani, 2019; Gra et al., 2008; Guven et al., 2015; Haase et al., 2002; Hishida et al., 2005; Haranatha Reddy and Jamil, 2006; Idris et al., 2020; Jiang and Tan, 2010; Joseph et al., 2004; Krajinovic et al., 1999; Lemos et al., 1999; Sasai et al., 1999; Naoe et al., 2000; Rollinson et al., 2000; Saadat and Saadat, 2000; Woo et al., 2000; Loffler et al., 2001; Wang et al., 2002; Yuille et al., 2002; Zhang et al., 2003; Seedhouse et al., 2004; Wang et al., 2004; Wu et al., 2004; Zou et al., 2004; Liu et al., 2005; Lourenco et al., 2005; Mondal et al., 2005; Pakakasama et al., 2005; Yang et al., 2005; Pigullo et al., 2007; Majumdar et al., 2008; Müller et al., 2008; Rimando et al., 2008; Souza et al., 2008; Suneetha et al., 2008; Taspinar et al., 2008; Ovsepian et al., 2010; Mandegary et al., 2011; Ouerhani et al., 2011; Suneetha et al., 2011; Kim et al., 2012; Li et al., 2012; Lordelo et al., 2012; Özten et al., 2012; Zhou et al., 2013; Zi et al., 2014; Kassogue et al., 2015; Lopes et al., 2015; Nasr et al., 2015; Weich et al., 2015; Kreile et al., 2016; Weich et al., 2016; Liu et al., 2017; Zehra et al., 2018; Muddathir et al., 2019; Rostami et al., 2019) were selected for the current study (Figure 1). Of these, there were 104 studies from 86 publications (14,100 leukemia cases and 23,793 controls, Table 1, Supplementary Table S1) for the GSTM1 null genotype, 94 studies from 79 publications (12,928 leukemia cases and 22,036 controls, Table 2, Supplementary Table S1) for the GSTT1 null genotype, and 33 studies from 30 publications (4,613 leukemia cases and 6,826 controls, Supplementary Table S1) for the combined effects of the GSTM1 present/null and GSTT1 present/null polymorphisms. In addition, there were 74 high-quality studies for the GSTM1 null genotype, 71 high-quality studies for the GSTT1 null genotype, and 25 high-quality studies for the combined effects of the GSTM1 and GSTT1 polymorphisms, as shown in Supplementary Table S1.
Quantitative Synthesis
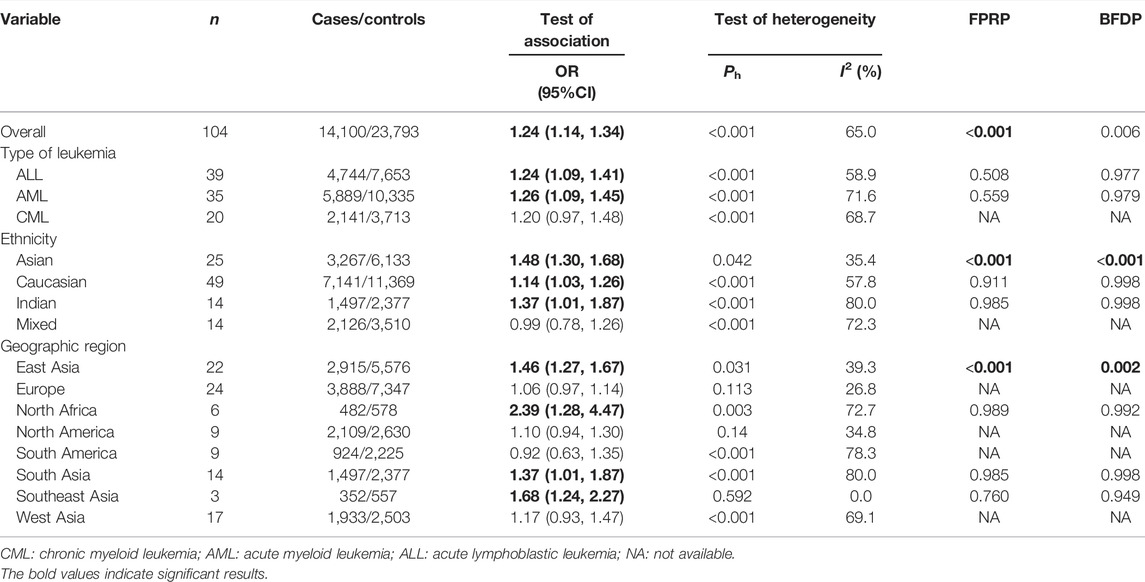
Overall, the GSTM1 null genotype was associated with a significantly increased leukemia risk (OR = 1.24, 95% CI: 1.14–1.34, Table 1) when all the eligible studies were merged. Then, subgroup analysis was conducted by type of leukemia, and significantly increased acute lymphoblastic leukemia (ALL) (OR = 1.24, 95% CI: 1.09–1.41) and acute myeloid leukemia (AML) (OR = 1.26, 95% CI: 1.09–1.45) risk were also observed for the GSTM1 null genotype. In addition, the GSTM1 null genotype was associated with a significantly increased leukemia risk in Asians (OR = 1.48, 95% CI: 1.30–1.68), Caucasians (OR = 1.14, 95% CI: 1.03–1.26), and Indians (OR = 1.37, 95% CI: 1.01–1.87). Moreover, significantly increased leukemia risk was found for the GSTM1 null genotype among countries of East Asia (OR = 1.46, 95% CI: 1.27–1.67), North Africa (OR = 2.39, 95% CI: 1.28–4.47), South Asia (OR = 1.37, 95% CI: 1.01–1.87), and Southeast Asia (OR = 1.68, 95% CI: 1.24–2.27), as shown in Table 1. Moreover, we found that the GSTM1 null genotype frequencies were different in the different populations (Africans: 29.7%, Asians: 53.7%, Caucasians: 49.5%, and Indians: 35.8%) for the control groups.
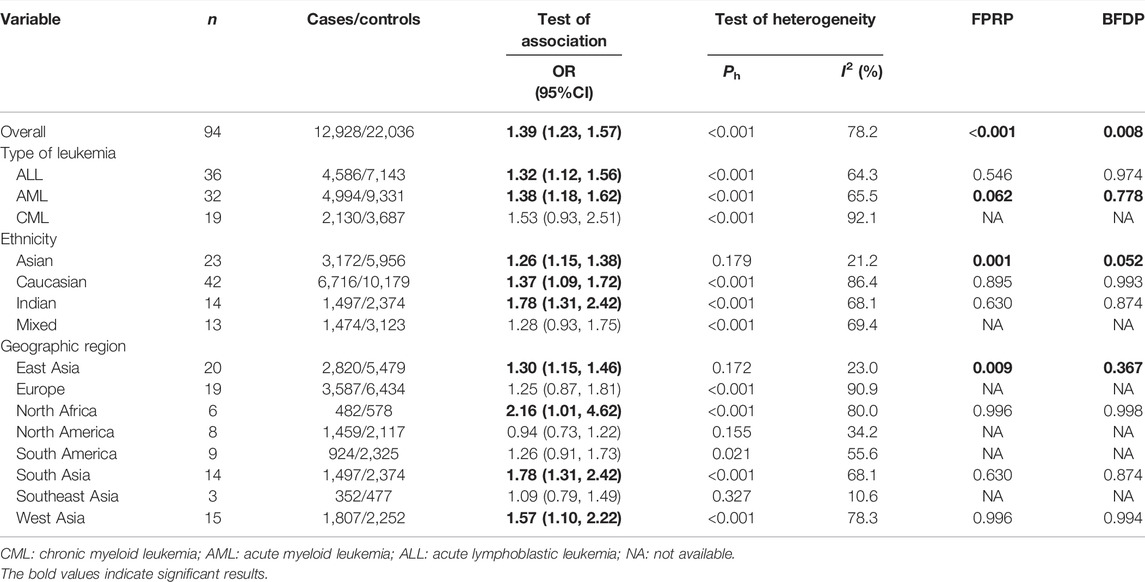
Overall, the GSTT1 null genotype was associated with a significantly increased leukemia risk (OR = 1.39, 95% CI: 1.23–1.57, Table 2). Then, subgroup analysis was conducted by type of leukemia, and significantly increased ALL (OR = 1.32, 95% CI: 1.12–1.56) and AML (OR = 1.38, 95% CI: 1.18–1.62) risk were observed for the GSTT1 null genotype. In addition, the GSTT1 null genotype was associated with significantly increased leukemia risk in Asians (OR = 1.26, 95% CI: 1.15–1.38), Caucasians (OR = 1.37, 95% CI: 1.09–1.72), and Indians (OR = 1.78, 95% CI: 1.31–2.42). Moreover, significantly increased leukemia risk was found for the GSTT1 null genotype among countries of East Asia (OR = 1.30, 95% CI: 1.15–1.46), North Africa (OR = 2.16, 95% CI: 1.01–4.62), South Asia (OR = 1.78, 95% CI: 1.31–2.42), and West Asia (OR = 1.57, 95% CI: 1.10–2.22), as shown in Table 2. Moreover, we also observed that GSTT1 null genotype frequencies were also different in the different races (Africans: 25.9%, Asians: 44.5%, Caucasians: 19.5%, and Indians: 15.6%) for the control groups.
Overall, the combined effects of the GSTM1 present/null and GSTT1 present/null genotypes were associated with a significantly increased leukemia risk (OR = 1.49, 95% CI: 1.27–1.74, Figure 2). Then, subgroup analysis was conducted by type of leukemia, and significantly increased chronic myeloid leukemia (CLL) (OR = 1.76, 95% CI: 1.26–2.46) and AML (OR = 1.50, 95% CI: 1.11–2.02) risk was observed (Figure 2). In addition, the combined effects were associated with significantly increased leukemia risk in Asians (OR = 1.66, 95% CI: 1.35–2.03), Caucasians (OR = 1.47, 95% CI: 1.14–1.89), and Indians (OR = 1.83, 95% CI: 1.42–2.35), as shown in Figure 3. Moreover, significantly increased leukemia risk was found for the combined effects among countries of East Asia (OR = 1.66, 95% CI: 1.35–2.03), South Asia (OR = 1.83, 95% CI: 1.42–2.35), and West Asia (OR = 1.64, 95% CI: 1.13–2.38), as shown in Figure 4. Moreover, we also found that the risk genotypes frequencies of the combined effects of the GSTM1 and GSTT1 polymorphisms were also different in the different races (Asians: 46.8%, Caucasians: 54.0%, and Indians: 39.3%) for the control groups.
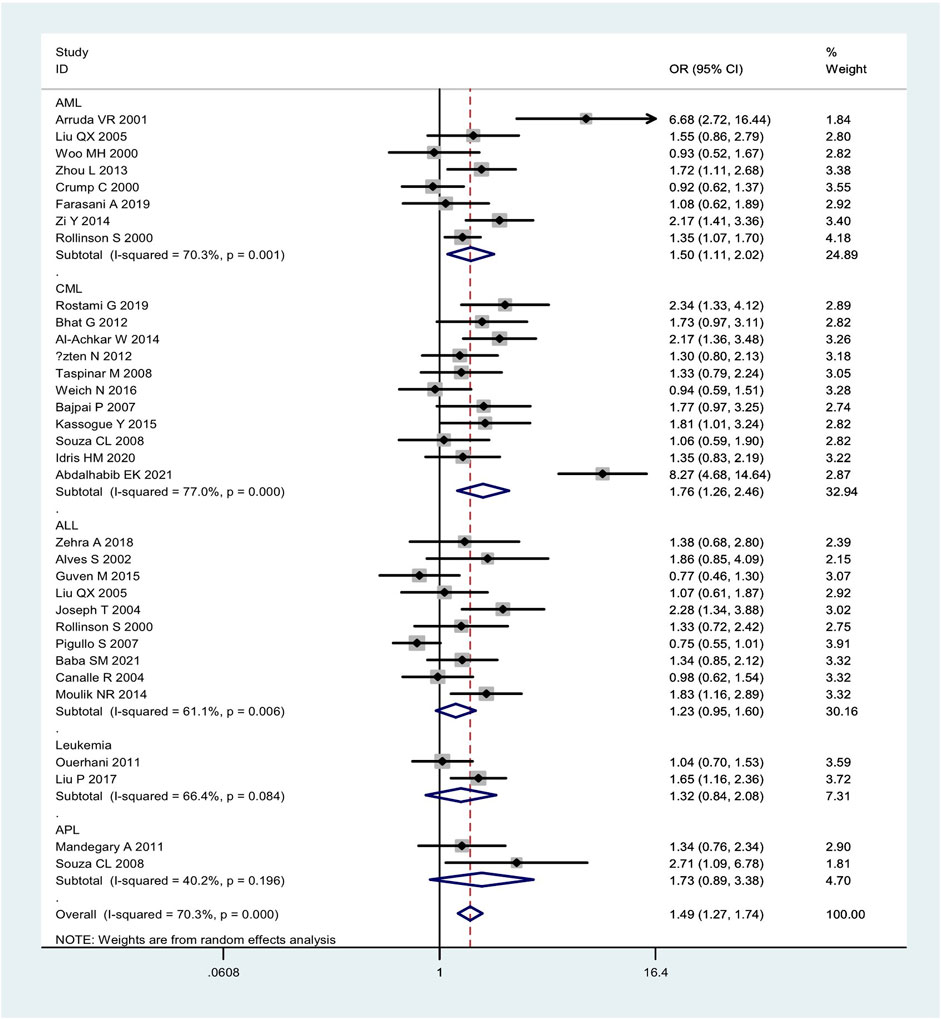
FIGURE 2. Forest plot of the combined effects of the GSTMI and GSTTI null genotypes with risk of leukemia in overall analysis and subgroup analysis by type of leukemia.
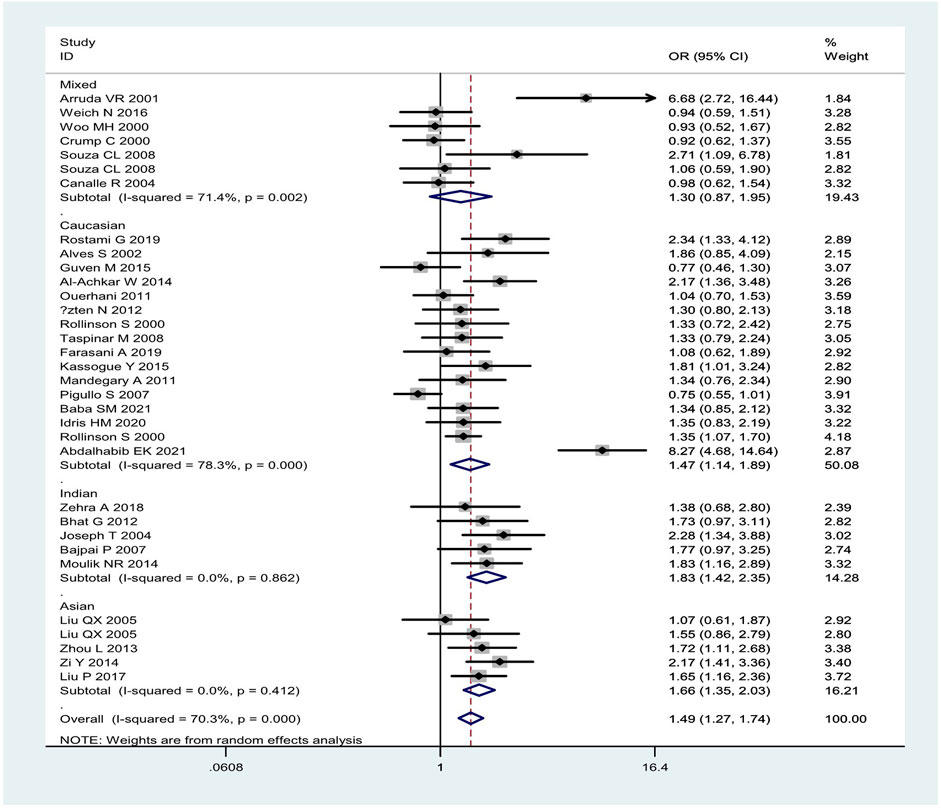
FIGURE 3. Forest plot of the combined effects of the GSTM1 and GSTT1 null genotypes with risk of leukemia in overall analysis and subgroup analysis by ethnicity.
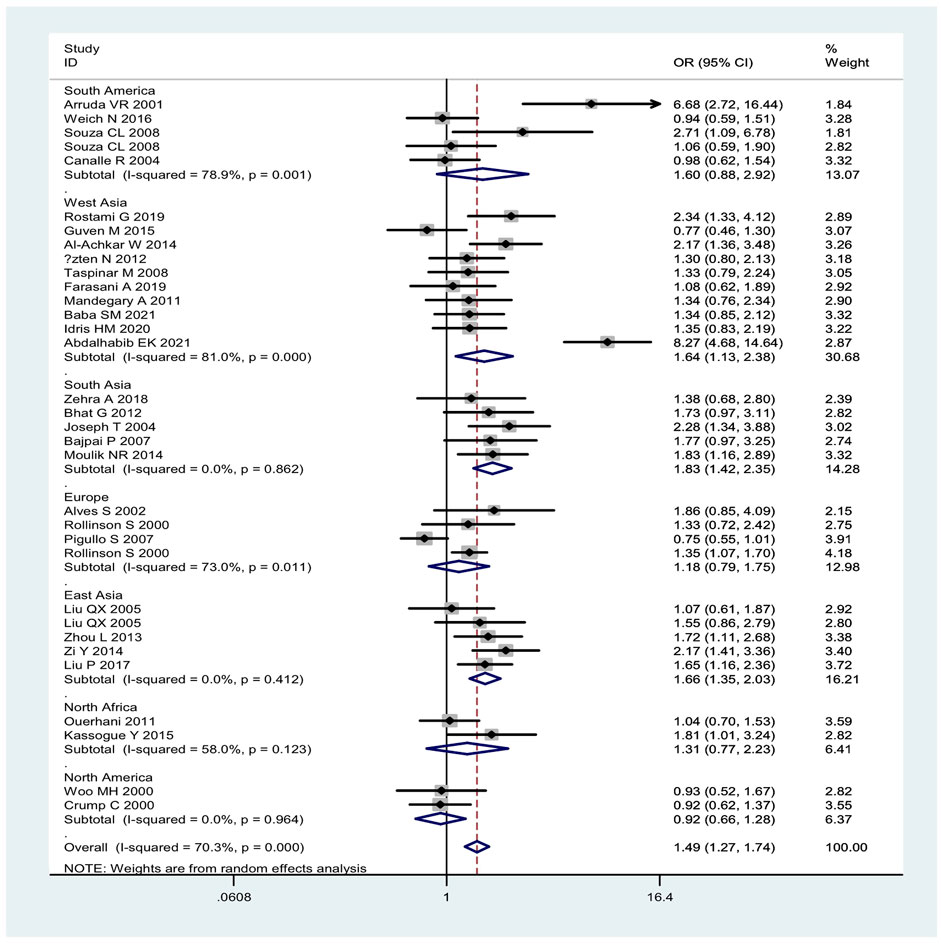
FIGURE 4. Forest plot of the combined effects of the GSTM1 and GSTT1 null genotypes with risk of leukemia in overall analysis and subgroup analysis by geographic region.
Heterogeneity and Sensitivity Analyses
Between-studies heterogeneity was observed, as shown in Tables 1, 2 and Figures 2–4. A meta-regression analysis showed that the quality score of included studies (p = 0.007) were sources of heterogeneity for the GSTM1 null genotype. For the GSTT1 null genotype and combined effects, meta-regression analyses did not find sources of heterogeneity. Moreover, we did not observe any change when one study and low-quality studies were excluded from the overall analysis.
Publication Bias
Publication bias was found for GSTM1 null genotype (p = 0.005) and the combined effects of GSTM1 and GSTT1 (p = 0.035), according to Begg’s funnel plot shape and Egger’s test in the current meta-analysis. Figures 5, 6 show the funnel plots of the nonparametric “trim and fill” method. We need to add 18 articles in the future for the GSTM1 present/null polymorphism with risk of leukemia (Figure 5). Moreover, we need to add eight studies for the combined effects of the GSTM1 present/null and GSTT1 present/null polymorphisms on the risk of leukemia (Figure 6). However, the results did not change, indicating that the current study was stable in overall analysis when the nonparametric “trim and fill” method was applied.
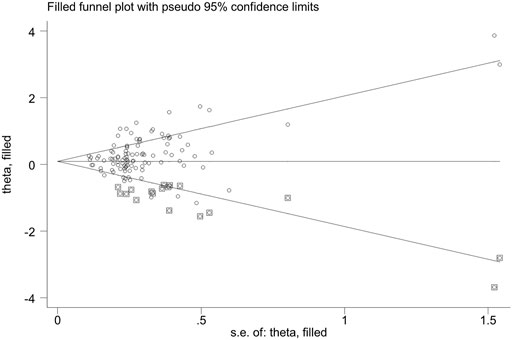
FIGURE 5. The Duval and Tweedie nonparametric “trim and fill” method’s funnel plot of the GSTM1 present/null polymorphism with risk of leukemia.
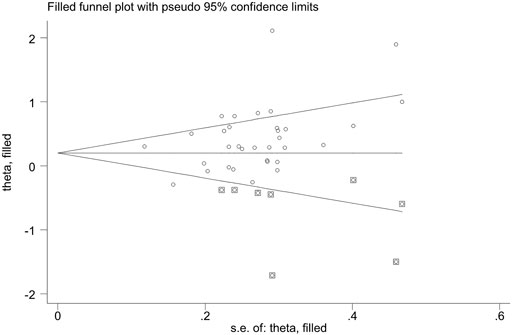
FIGURE 6. The Duval and Tweedie nonparametric “trim and fill” method’s funnel plot of the combined effects of the GSTM1 present/null and GSTTI present/null polymorphisms with risk of leukemia.
Test of Significant Associations in the Current Study
To investigate the false-positive results, FPRP and BFDP were applied. For the GSTM1 null genotype, significant associations were considered as “positive” results in overall population (FPRP < 0.001 and BFDP = 0.006), Asians (FPRP < 0.001 and BFDP < 0.001), and East Asian population (FPRP < 0.001 and BFDP = 0.002), as shown in Table 1. For the GSTT1 null genotype, significant associations were regarded as “positive” results in overall population (FPRP < 0.001 and BFDP = 0.008), AML (FPRP = 0.062 and BFDP = 0.778), Asians (FPRP = 0.001 and BFDP = 0.052), and East Asian population (FPRP = 0.009 and BFDP = 0.367), as shown in Table 2. For the combined effects of the GSTM1 and GSTT1 polymorphisms, significant associations were also considered as “positive” results in overall analysis (FPRP = 0.001 and BFDP = 0.027), Asians (FPRP = 0.005 and BFDP = 0.040), Indians (FPRP = 0.035 and BFDP = 0.095), and East Asian population (FPRP = 0.014 and BFDP = 0.040).
Discussion
Overall, the individual GSTM1 and GSTT1 null genotypes and combined effects of the two genes were associated with significantly increased leukemia risk in the overall analysis and several subgroup analyses, such as Asians, Caucasians, and so on. However, the current study applied several subgroup analyses at the expense of multiple comparisons. Therefore, FPRP and BFDP values were applied to conduct the test of false-positive results.
Glutathione S-transferases (GSTs) are a group of enzymes that play vital roles in regulating the cellular detoxification of various exogenous carcinogens (Di Pietro et al., 2010). Moreover, it is believed that GSTs can protect cells against oxidative stress and its associated DNA damage (Singh, 2015). Furthermore, it is biologically plausible that subjects carrying these null genotypes may suffer a higher risk of developing multiple malignancies because their GST proteins do not function properly. Therefore, it is widely accepted that alterations in GSTs play roles in the process associated with the etiology of cancers. Based on biochemical properties described for the GSTM1 present/null and GSTT1 present/null polymorphisms, we expected that the individual and the combined effects of the two genes were associated with the risk of leukemia in any population. However, we only found that the individual GSTM1 and GSTT1 null genotypes and combined effects of the two genes are associated with increased leukemia risk in Asians, especially in the East Asian population, and the combined effects of the two genes are also associated with increased leukemia risk in Indians when we used the FPRP and BFDP values. These results showed that the same genes may play different roles in leukemia susceptibility in different races and countries because leukemia is a complicated multigenetic disease and different genetic backgrounds and environmental factors may contribute to the discrepancy (Begg and Mazumdar, 1994). Moreover, we only found that the GSTT1 null genotype was associated with increased AML risk. The result showed that the same polymorphism also may play different roles in a different type of leukemia. Moreover, some results should be interpreted with caution, and it was necessary that a well-designed large sample study was conducted to explore the true association, such as in Southeast Asian and North African populations. Furthermore, publication bias was observed between the GSTM1 null genotype and the combined effects of the two genes on the risk of leukemia. Figures 5, 6 showed that publication bias was caused according to low-quality small-sample studies. As far as we know, random error and bias were common for the small-sample–size studies, especially in molecular epidemiological studies. Moreover, small-sample studies were easier to publish if the results were significant as they tend to yield false-positive results because they may be not rigorous and were often of low quality.
Fourteen meta-analyses (Ye and Song, 2005; Das et al., 2009; Zintzaras, 2009; Vijayakrishnan and Houlston, 2010; Tang et al., 2013; He et al., 2014a; He et al., 2014b; Ma et al., 2014; Moulik et al., 2014; Tang et al., 2014; Xu and Cao, 2014; Li et al., 2018; Zhao et al., 2018; Wang et al., 2019) reported the individual effects of the GSTM1 and GSTT1 polymorphisms with leukemia risk. Wang et al. (2019) observed that the GSTM1 and GSTT1 null genotypes were significantly associated with elevated individual susceptibility to acute lymphoblastic leukemia (ALL) and AML; the GSTT1 null genotype was also significantly associated with elevated individual susceptibility to chronic leukemia; the GSTM1 and GSTT1 null genotypes were significantly associated with elevated individual susceptibility to leukemia in Caucasians and West Asians; the GSTM1 null genotype was also significantly correlated with elevated individual susceptibility to leukemia in East Asians. Li et al. (2018) found that the GSTM1 and GSTT1 polymorphisms were both significantly correlated with hematological malignancy in Caucasians, East Asians, and West Asians, and positive results were found for the GSTM1 and GSTT1 polymorphisms in patients with certain types of acute leukemia. Zhao et al. (2018) found that the GSTM1 null genotype was associated with increased childhood ALL risk and the GSTT1 null genotype was not associated with childhood ALL risk. He et al. (2014a) revealed that the GSTM1 null genotype was associated with an increased risk of AML in East Asians and the GSTT1 null genotype in Caucasians. Tang et al. (2014) suggested that the GSTM1 and GSTT1 null genotypes might be a potential risk factor for acute leukemia in Asians. He et al. (2014b) indicated that the GSTT1 null genotype and the double-null GSTT1 and GSTM1 genotypes were associated with an increased risk of CML. Xu and Cao (2014) found that the GSTT1 null variant was significantly associated with susceptibility to childhood ALL in Asians. Tang et al. (2013) found that the GSTM1 null polymorphism was caused by childhood acute leukemia susceptibility. Ma et al. (2014) found that the GSTM1 null genotype was significantly associated with increased risk of childhood acute leukemia in the Chinese population. Vijayakrishnan and Houlston (2010) found that the GSTM1 null genotype was significantly associated with increased risk of childhood acute lymphoblastic leukemia, but should be interpreted with caution. Zintzaras (2009) suggested that no significant association was found between the GSTM1 null genotype and CML risk, while the GSTT1 null genotype was associated with increased risk of CML, especially in Indians. Das et al. (2009) indicated that significant increased risk of AML was observed with the GSTM1 null genotype, while borderline significance was seen with the GSTT1 null genotype. Ye and Song (2005) found that the GSTM1 and GSTT1 null genotypes appeared to be associated with a modest increase in the risk of ALL. Moulik et al. (2014) found that the GSTM1 null genotype was associated with increased childhood ALL risk. These results might be not credible because many original studies were not included in previously published meta-analyses. Moreover, previously published meta-analyses did not conduct the false-positive test using FPRP and BFDP values. Therefore, we performed the current study to further explore these associations.
The present study had several limitations. First, only published studies were selected. Second, the confounding factors closely related to the outcome were not controlled, such as gender, smoking, and some other factors. The current study also has several advantages over previously published meta-analyses. First, the sample size was larger. Second, we investigate the false-positive results by applying the FPRP and BFDP values.
In summary, this study strongly indicates that the individual GSTM1 and GSTT1 null genotypes and combined effects of the two genes are associated with increased leukemia risk in Asians, especially in the East Asian population; the GSTT1 null genotype is associated with increased AML risk; the combined effects of the two genes are associated with increased leukemia risk in Indians.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
TH and GZ: Research design and performance, data collection, data analysis, and paper writing. GZ and WL: Data collection. GZ and WL: Data recheck. TH: Methodology. TH: Research design and paper review.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.898937/full#supplementary-material
Abbreviations
BFDP, Bayesian false discovery probability; CIs, confidence intervals; CML, chronic myeloid leukemia; FPRP, false-positive report probabilities; GSTs, glutathione S-transferases; HWE, Hardy–Weinberg equilibrium; ORs, odds ratios.
References
Abdalhabib, E. K., Jackson, D. E., Alzahrani, B., Elfaki, E. M., Hamza, A., Alanazi, F., et al. (2021). Combined GSTT1 Null, GSTM1 Null and XPD Lys/Lys Genetic Polymorphisms and Their Association with Increased Risk of Chronic Myeloid Leukemia. Pharmgenomics Pers. Med. 14, 1661–1667. doi:10.2147/pgpm.s342625
Al-Achkar, W., Azeiz, G., Moassass, F., and Wafa, A. (2014). Influence of CYP1A1, GST Polymorphisms and Susceptibility Risk of Chronic Myeloid Leukemia in Syrian Population. Med. Oncol. 31 (5), 889. doi:10.1007/s12032-014-0889-4
Al-Eitan, L. N., Rababa'h, D. M., Alkhatib, R. Q., Khasawneh, R. H., and ALjarrah, O. A. (2016). GSTM1 and GSTP1 Genetic Polymorphisms and Their Associations with Acute Lymphoblastic Leukemia Susceptibility in a Jordanian Population. J. Pediatr. Hematol. Oncol. 38 (7), e223–e229. doi:10.1097/mph.0000000000000609
Allan, J. M., Wild, C. P., Rollinson, S., Willett, E. V., Moorman, A. V., Dovey, G. J., et al. (2001). Polymorphism in Glutathione S -transferase P1 Is Associated with Susceptibility to Chemotherapy-Induced Leukemia. Proc. Natl. Acad. Sci. U.S.A. 98 (20), 11592–11597. doi:10.1073/pnas.191211198
Alves, S., Amorim, A., Ferreira, F., Norton, L., and Prata, M. (2002). The GSTM1 and GSTT1 Genetic Polymorphisms and Susceptibility to Acute Lymphoblastic Leukemia in Children from North Portugal. Leukemia 16 (8), 1565–1567. doi:10.1038/sj.leu.2402543
Arruda, V. R., Lima, C. S. P., Grignoli, C. R. E., de Melo, M. B., Lorand-Metze, I., Alberto, F. L., et al. (2001). Increased Risk for Acute Myeloid Leukaemia in Individuals with Glutathione S -transferase Mu 1 (GSTM1) and Theta 1 (GSTT1) Gene Defects. Eur. J. Haematol. 66 (6), 383–388. doi:10.1034/j.1600-0609.2001.066006383.x
Aydin-Sayitoglu, M., Hatirnaz, O., Erensoy, N., and Ozbek, U. (2006). Role ofCYP2D6, CYP1A1, CYP2E1, GSTT1, andGSTM1 Genes in the Susceptibility to Acute Leukemias. Am. J. Hematol. 81, 162–170. doi:10.1002/ajh.20434
Baba, S. M., Pandith, A. A., Shah, Z. A., Geelani, S. A., Bhat, J. R., Gul, A., et al. (2021). GSTT1null and Rs156697 Polymorphism in GSTO2 Influence the Risk and Therapeutic Outcome of B-Acute Lymphoblastic Leukemia Patients. Front. Oncol. 11, 714421. doi:10.3389/fonc.2021.714421
Bajpai, P., Tripathi, A. K., and Agrawal, D. (2007). Increased Frequencies of Glutathione-S-Transferase (GSTM1 and GSTT1) Null Genotypes in Indian Patients with Chronic Myeloid Leukemia. Leukemia Res. 31 (10), 1359–1363. doi:10.1016/j.leukres.2007.02.003
Balta, G., Yuksek, N., Ozyurek, E., Ertem, U., Hicsonmez, G., Altay, C., et al. (2003). Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 Genotypes in Childhood Acute Leukemia. Am. J. Hematol. 73 (3), 154–160. doi:10.1002/ajh.10339
Bănescu, C., Iancu, M., Trifa, A. P., Cândea, M., Benedek Lazar, E., Moldovan, V. G., et al. (2016). From Six Gene Polymorphisms of the Antioxidant System, Only GPX Pro198Leu and GSTP1 Ile105Val Modulate the Risk of Acute Myeloid Leukemia. Oxid. Med. Cell. Longev. 2016, 2536705. doi:10.1155/2016/2536705
Bănescu, C., Trifa, A. P., Voidăzan, S., Moldovan, V. G., Macarie, I., Benedek Lazar, E., et al. (2014). CAT, GPX1, MnSOD, GSTM1, GSTT1, and GSTP1 Genetic Polymorphisms in Chronic Myeloid Leukemia: a Case-Control Study. Oxid. Med. Cell. Longev. 2014, 875861. doi:10.1155/2014/875861
Barnette, P., Scholl, R., Blandford, M., Ballard, L., Tsodikov, A., Magee, J., et al. (2004). High-Throughput Detection of Glutathione S-Transferase Polymorphic Alleles in a Pediatric Cancer Population. Cancer Epidemiol. Biomarkers Prev. 13, 304–313. doi:10.1158/1055-9965.epi-03-0178
Begg, C. B., and Mazumdar, M. (1994). Operating Characteristics of a Rank Correlation Test for Publication Bias. Biometrics 50, 1088–1101. doi:10.2307/2533446
Benjamini, Y., and Hochberg, Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. doi:10.1111/j.2517-6161.1995.tb02031.x
Bhat, G., Bhat, A., Wani, A., Sadiq, N., Jeelani, S., Kaur, R., et al. (2012). Polymorphic Variation in Glutathione-S-Transferase Genes and Risk of Chronic Myeloid Leukaemia in the Kashmiri Population. Asian Pac. J. Cancer Prev. 13 (1), 69–73. doi:10.7314/apjcp.2012.13.1.069
Bhatla, D., Gerbing, R. B., Alonzo, T. A., Mehta, P. A., Deal, K., Elliott, J., et al. (2008). DNA Repair Polymorphisms and Outcome of Chemotherapy for Acute Myelogenous Leukemia: a Report from the Children's Oncology Group. Leukemia 22, 265–272. doi:10.1038/sj.leu.2405000
Bloomfield, C. D., and Caligiury, M. (2001). “Molecular Biology of Leukemias,” in Cancer: Principles and Practice of Oncology. Editors V. T. DeVita, S. Hellman, and S. A. Rosenberg (Philadelphia: Lippincott Williams & Wilkins), 2389–2404.
Bolufer, P., Collado, M., Barragán, E., Cervera, J., Calasanz, M.-J., Colomer, D., et al. (2007). The Potential Effect of Gender in Combination with Common Genetic Polymorphisms of Drug-Metabolizing Enzymes on the Risk of Developing Acute Leukemia. Haematologica 92 (3), 308–314. doi:10.3324/haematol.10752
Canalle, R., Burim, R. V., Tone, L. G., and Takahashi, C. S. (2004). Genetic Polymorphisms and Susceptibility to Childhood Acute Lymphoblastic Leukemia. Environ. Mol. Mutagen. 43, 100–109. doi:10.1002/em.20003
Chan, J. Y.-S., Ugrasena, D. G., Lum, D. W.-K., Lu, Y., and Yeoh, A. E.-J. (2011). Xenobiotic and Folate Pathway Gene Polymorphisms and Risk of Childhood Acute Lymphoblastic Leukaemia in Javanese Children. Hematol. Oncol. 29, 116–123. doi:10.1002/hon.965
Chauhan, P. S., Ihsan, R., Mishra, A. K., Yadav, D. S., Saluja, S., Mittal, V., et al. (2012). High Order Interactions of Xenobiotic Metabolizing Genes and P53 Codon 72 Polymorphisms in Acute Leukemia. Environ. Mol. Mutagen. 53, 619–630. doi:10.1002/em.21723
Chauhan, P. S., Ihsan, R., Yadav, D. S., Mishra, A. K., Bhushan, B., Soni, A., et al. (2011). Association of Glutathione S-Transferase, EPHX, and P53 Codon 72 Gene Polymorphisms with Adult Acute Myeloid Leukemia. DNA Cell. Biol. 30, 39–46. doi:10.1089/dna.2010.1092
Chen, C. L., Liu, Q., Pui, C. H., Rivera, G. K., Sandlund, J. T., Ribeiro, R., et al. (1997). Higher Frequency of Glutathione S-Transferase Deletions in Black Children with Acute Lymphoblastic Leukemia. Blood 89 (5), 1701–1707. doi:10.1182/blood.v89.5.1701.1701_1701_1707
Chen, H. C., Hu, W. X., Liu, Q. X., Li, W. K., Chen, F. Z., Rao, Z. Z., et al. (2008). Genetic Polymorphisms of Metabolic Enzymes CYP1A1, CYP2D6, GSTM1 and GSTT1 and Leukemia Susceptibility. Eur. J. Cancer Prev. 17 (3), 251–258. doi:10.1097/cej.0b013e3282b72093
Clavel, J., Bellec, S., Rebouissou, S., Ménégaux, F., Feunteun, J., Bonaïti-Pellié, C., et al. (2005). Childhood Leukaemia, Polymorphisms of Metabolism Enzyme Genes, and Interactions with Maternal Tobacco, Coffee and Alcohol Consumption during Pregnancy. Eur. J. Cancer Prev. 14, 531–540. doi:10.1097/00008469-200512000-00007
Crump, C., Chen, C., Appelbaum, F. R., Kopecky, K. J., Schwartz, S. M., Willman, C. L., et al. (2000). Glutathione S-Transferase Theta 1 Gene Deletion and Risk of Acute Myeloid Leukemia. Cancer Epidemiol. Biomarkers Prev. 9 (5), 457–460.
Curran, J. E., Weinstein, S. R., and Griffiths, L. R. (2000). Polymorphisms of Glutathione S-Transferase Genes (GSTM1, GSTP1 and GSTT1) and Breast Cancer Susceptibility. Cancer Lett. 153, 113–120. doi:10.1016/s0304-3835(00)00361-x
D'Alò, F., Voso, M. T., Guidi, F., Massini, G., Scardocci, A., Sica, S., et al. (2004). Polymorphisms of CYP1A1 and Glutathione S-Transferase and Susceptibility to Adult Acute Myeloid Leukemia. Haematologica 89 (6), 664–670.
Das, P., Shaik, A. P., and Bammidi, V. K. (2009). Meta-analysis Study of Glutathione-S-Transferases (GSTM1,GSTP1, andGSTT1) Gene Polymorphisms and Risk of Acute Myeloid Leukemia. Leukemia Lymphoma 50 (8), 1345–1351. doi:10.1080/10428190903003236
Davies, S. M., Robison, L. L., Buckley, J. D., Radloff, G. A., Ross, J. A., and Perentesis, J. P. (2000). Glutathione S-Transferase Polymorphisms in Children with Myeloid Leukemia: a Children's Cancer Group Study. Cancer Epidemiol. Biomarkers Prev. 9, 563–566.
Davies, S. M., Bhatia, S., Ross, J. A., Kiffmeyer, W. R., Gaynon, P. S., Radloff, G. A., et al. (2002). Glutathione S-Transferase Genotypes, Genetic Susceptibility, and Outcome of Therapy in Childhood Acute Lymphoblastic Leukemia. Blood 100 (1), 67–71. doi:10.1182/blood.v100.1.67
DerSimonian, R., and Laird, N. (1986). Meta-analysis in Clinical Trials. Control. Clin. Trials 7, 177–188. doi:10.1016/0197-2456(86)90046-2
Di Pietro, G., Magno, L. A. V., and Rios-Santos, F. (2010). Glutathione S-Transferases: an Overview in Cancer Research. Expert Opin. Drug Metabolism Toxicol. 6, 153–170. doi:10.1517/17425250903427980
Dual, S., and Tweedie, R. (2000). A Nonparametric “Trim and Fill” Method of Accounting for Publication Bias in Meta-Analysis. J. Am. Stat. Assoc. 95, 89–98.
Dunna, N. R., Vure, S., Sailaja, K., Surekha, D., Raghunadharao, D., Rajappa, S., et al. (2013). Deletion of GSTM1 and T1 Genes as a Risk Factor for Development of Acute Leukemia. Asian Pac. J. Cancer Prev. 14 (4), 2221–2224. doi:10.7314/apjcp.2013.14.4.2221
Egger, M., Smith, G. D., Schneider, M., and Minder, C. (1997). Bias in Meta-Analysis Detected by a Simple, Graphical Test. Bmj 315, 629–634. doi:10.1136/bmj.315.7109.629
Eyada, T. K., El Ghonemy, E. G., El Ghoroury, E. A., El Bassyouni, S. O., and El Masry, M. K. (2007). Study of Genetic Polymorphism of Xenobiotic Enzymes in Acute Leukemia. Blood Coagul. Fibrinolysis 18, 489–495. doi:10.1097/mbc.0b013e3281eec930
Farasani, A. (2019). Genetic Variants of Glutathione S-Transferase and the Risk of Acute Myeloid Leukemia in a Saudi Population. Saudi J. Biol. Sci. 26 (7), 1525–1530. doi:10.1016/j.sjbs.2018.12.011
Feng, J. F., Wang, J., Zhang, L., Luan, B., and Wang, H. (2004). Gstm1, Gstt1 Gene Polymorphisms and Childhood Acute Myeloid Leukemia Susceptibility. Lab. Med. 19, 260–262.
Goyette, P., Sumner, J. S., Milos, R., Duncan, A. M. V., Rosenblatt, D. S., Matthews, R. G., et al. (1994). Human Methylenetetrahydrofolate Reductase: Isolation of cDNA, Mapping and Mutation Identification. Nat. Genet. 7, 195–200. doi:10.1038/ng0694-195
Gra, O. A., Glotov, A. S., Kozhekbaeva, Z., Makarova, O. V., and Nasedkina, T. V. (2008). Genetic Polymorphism in GST, NAT2, and MTRR and Susceptibility to Childhood Acute Leukemia. Mol. Biol. Mosk. 42 (2), 214–225. doi:10.1134/s0026893308020039
Guven, M., Unal, S., Erhan, D., Ozdemir, N., Baris, S., Celkan, T., et al. (2015). Role of Glutathione S-Transferase M1, T1 and P1 Gene Polymorphisms in Childhood Acute Lymphoblastic Leukemia Susceptibility in a Turkish Population. Meta Gene 5, 115–119. doi:10.1016/j.mgene.2015.06.002
Haase, D., Binder, C., Bünger, J., Fonatsch, C., Streubel, B., Schnittger, S., et al. (2002). Increased Risk for Therapy-Associated Hematologic Malignancies in Patients with Carcinoma of the Breast and Combined Homozygous Gene Deletions of Glutathione Transferases M1 and T1. Leukemia Res. 26 (3), 249–254. doi:10.1016/s0145-2126(01)00124-2
Haranatha Reddy, P., and Jamil, K. (2006). Polymorphisms in the GST (M1 andT1) Gene and Their Possible Association with Susceptibility to Childhood Acute Lymphocytic Leukemia in Indian Population. Afr. J. Biotechnol. 5, 1454–1456.
Hayes, J. D., Flanagan, J. U., and Jowsey, I. R. (2005). Glutathione Transferases. Annu. Rev. Pharmacol. Toxicol. 45, 51–88. doi:10.1146/annurev.pharmtox.45.120403.095857
Hayes, J. D., and Strange, R. C. (2000). Glutathione S-Transferase Polymorphisms and Their Biological Consequences. Pharmacology 61, 154–166. doi:10.1159/000028396
He, H. R., Zhang, X. X., Sun, J. Y., Hu, S. S., Ma, Y., Dong, Y. L., et al. (2014). Glutathione S-Transferase Gene Polymorphisms and Susceptibility to Chronic Myeloid Leukemia. Tumor Biol. 35 (6), 6119–6125. doi:10.1007/s13277-014-1810-7
He, H. R., You, H. S., Sun, J. Y., Hu, S. S., Ma, Y., Dong, Y. L., et al. (2014). Glutathione S-Transferase Gene Polymorphisms and Susceptibility to Acute Myeloid Leukemia: Meta-Analyses. Jpn. J. Clin. Oncol. 44 (11), 1070–1081. doi:10.1093/jjco/hyu121
Henderson, E. S. (1990). “Acute Leukemia: Gen-Eral Considerations,” in Hematology. Editors W. J. Williams, E. Beutler, A. J. Erslev, and M. A. Lichtman. 4 th ed (New York: McGraw-Hill), 237.
Hirschhorn, J. N., Lohmueller, K., Byrne, E., and Hirschhorn, K. (2002). A Comprehensive Review of Genetic Association Studies. Genet. Med. 4 (2), 45–61. doi:10.1097/00125817-200203000-00002
Hishida, A., Terakura, S., Emi, N., Yamamoto, K., Murata, M., Nishio, K., et al. (2005). GSTT1 and GSTM1 Deletions, NQO1 C609T Polymorphism and Risk of Chronic Myelogenous Leukemia in Japanese. Asian Pac J. Cancer Prev. 6 (3), 251–255.
Idris, H. M., Elderdery, A. Y., Khalil, H. B., and Mills, J. (2020). Genetic Polymorphism of GSTP1, GSTM1 and GSTT1 Genes and Susceptibility to Chronic Myeloid Leukaemia. Asian Pac J. Cancer Prev. 21 (2), 499–503. doi:10.31557/apjcp.2020.21.2.499
Jiang, L. J., Chen, M., and Tan, G. F. (2008). The Association between Polymorphism of Cytochrome P4501A1 and Glutathione S-Transferase M1, T1 Genes and Acute Lymphoblastic Leukemia. J Youjiang Med. Coll. Natl. 5, 721–723.
Jiang, L. J., and Tan, G. F. (2010). Association between the Polymorphisms of Cytochrome P4501a1 and Glutathione S-Transferase M1, T1 Genes and Childhood Acute Lymphocytic Leukemia. Chin. J. New Clin. Med. 3, 810–814.
Joseph, T., Kusumakumary, P., Chacko, P., Abraham, A., and Radhakrishna Pillai, M. (2004). Genetic Polymorphism ofCYP1A1,CYP2D6,GSTM1 andGSTT1 and Susceptibility to Acute Lymphoblastic Leukaemia in Indian Children. Pediatr. Blood Cancer 43, 560–567. doi:10.1002/pbc.20074
Kassogue, Y., Dehbi, H., Quachouh, M., Quessar, A., Benchekroun, S., and Nadifi, S. (2015). Association of Glutathione S-Transferase (GSTM1 and GSTT1) Genes with Chronic Myeloid Leukemia. Springerplus 4, 210. doi:10.1186/s40064-015-0966-y
Kim, H. N., Kim, N. Y., Yu, L., Tran, H. T. T., Kim, Y.-K., Lee, I.-K., et al. (2012). Association ofGSTT1polymorphism with Acute Myeloid Leukemia Risk Is Dependent on Smoking Status. Leukemia Lymphoma 53 (4), 681–687. doi:10.3109/10428194.2011.625576
Krajinovic, M., Labuda, D., Richer, C., Karimi, S., and Sinnett, D. (1999). Susceptibility to Childhood Acute Lymphoblastic Leukemia: Influence of CYP1A1, CYP2D6, GSTM1, and GSTT1 Genetic Polymorphisms. Blood 93 (5), 1496–1501. doi:10.1182/blood.v93.5.1496.405a36_1496_1501
Kreile, M., Piekuse, L., Rots, D., Dobele, Z., Kovalova, Z., and Lace, B. (2016). Analysis of Possible Genetic Risk Factors Contributing to Development of Childhood Acute Lymphoblastic Leukaemia in the Latvian Population. Aoms 3 (3), 479–485. doi:10.5114/aoms.2016.59920
Lemos, M. C., Cabrita, F. J., Silva, H. A., Vivan, M., Plácido, F., and Regateiro, F. J. (1999). Genetic Polymorphism of CYP2D6, GSTM1 and NAT2 and Susceptibility to Haematological Neoplasias. Carcinogenesis 20 (7), 1225–1229. doi:10.1093/carcin/20.7.1225
Li, M., Zheng, M., Chen, H., and Yu, H. (2018). Effects of GST Variants on the Risk Odds of Hematological Malignancy: A Meta‐analysis. J Cell. Biochem. 120, 8570–8580. doi:10.1002/jcb.28145
Li, Y. H., Wen, F. Q., Xiao, Z. H., Chen, Y. X., Zhang, Z. X., and Chen, L. L. (2012). Genetic Polymorphism of GST Gene in Children with Infectious Mononucleosis and Acute Lymphocytic Leukemia. Zhongguo Dang Dai Er Ke Za Zhi 14 (4), 260–263.
Liu, P., Zhang, M., Xie, X., Jin, J., and Holman, C. D. A. J. (2017). Green Tea Consumption and Glutathione S-Transferases Genetic Polymorphisms on the Risk of Adult Leukemia. Eur. J. Nutr. 56 (2), 603–612. doi:10.1007/s00394-015-1104-x
Liu, Q. X., Chen, H. C., Liu, X. F., Cao, Y. F., Zhang, J., and Liu, J. (2005). Study on the Relationship between Polymorphisms of Cyp1A1, GSTM1, GSTT1 Genes and the Susceptibility to Acute Leukemia in the General Population of Hunan Province. Zhonghua Liu Xing Bing Xue Za Zhi 26, 975–979.
Loffler, H., Bergmann, J., Hochhaus, A., Hehlmann, R., and Krämer, A.the German CML Study Group (2001). Reduced Risk for Chronic Myelogenous Leukemia in Individuals with the Cytochrome P-450 Gene Polymorphism CYP1A1*2A. Blood 98 (13), 3874–3875. doi:10.1182/blood.V98.13.3874
Lopes, B. A., Emerenciano, M., Gonçalves, B. A. A., Vieira, T. M., Rossini, A., and Pombo-de-Oliveira, M. S. (2015). Polymorphisms in CYP1B1, CYP3A5, GSTT1, and SULT1A1 Are Associated with Early Age Acute Leukemia. PLoS One 10 (5), e0127308. doi:10.1371/journal.pone.0127308
Lordelo, G. S., Miranda-Vilela, A. L., Akimoto, A. K., Alves, P. C. Z., Hiragi, C. O., Nonino, A., et al. (2012). Association between Methylene Tetrahydrofolate Reductase and Glutathione S-Transferase M1 Gene Polymorphisms and Chronic Myeloid Leukemia in a Brazilian Population. Genet. Mol. Res. 11 (2), 1013–1026. doi:10.4238/2012.april.19.6
Lourenco, G. J., Ortega, M. M., Nascimento, H., Teori, M. T., De Souza, C. A., Costa, F. F., et al. (2005). Polymorphisms of Glutathione S-Transferase Mu1 (GSTM1) and Theta 1 (GSTT1) Genes in Chronic Myeloid Leukaemia. Eur. J. Haematol. 75 (6), 530–531. doi:10.1111/j.1600-0609.2005.00567.x
Ma, Y., Sui, Y., Wang, L., and Li, H. (2014). Effect of GSTM1 Null Genotype on Risk of Childhood Acute Leukemia: a Meta-Analysis. Tumor Biol. 35 (1), 397–402. doi:10.1007/s13277-013-1055-x
Majumdar, S., Mondal, B. C., Ghosh, M., Dey, S., Mukhopadhyay, A., Chandra, S., et al. (2008). Association of Cytochrome P450, Glutathione S-Transferase and N-Acetyl Transferase 2 Gene Polymorphisms with Incidence of Acute Myeloid Leukemia. Eur. J. Cancer Prev. 17 (2), 125–132. doi:10.1097/cej.0b013e3282b6fd68
Mandegary, A., Rostami, S., Alimoghaddam, K., Ghavamzadeh, A., and Ghahremani, M. H. (2011). Gluthatione-S-transferase T1-Null Genotype Predisposes Adults to Acute Promyelocytic Leukemia; a Case-Control Study. Asian Pac J. Cancer Prev. 12 (5), 1279–1282.
Mantel, N., and Haenszel, W. (1959). Statistical Aspects of the Analysis of Data from Retrospective Studies of Disease. J. Natl. Cancer Inst. 22, 719–748.
Mondal, B. C., Paria, N., Majumdar, S., Chandra, S., Mukhopadhyay, A., Chaudhuri, U., et al. (2005). Glutathione S-Transferase M1 and T1 Null Genotype Frequency in Chronic Myeloid Leukaemia. Eur. J. Cancer Prev. 14 (3), 281–284. doi:10.1097/00008469-200506000-00014
Moulik, N. R., Parveen, F., Kumar, A., and Agrawal, S. (2014). Glutathione-S-transferase Polymorphism and Acute Lymphoblastic Leukemia (ALL) in North Indian Children: a Case-Control Study and Meta-Analysis. J. Hum. Genet. 59 (9), 529–535. doi:10.1038/jhg.2014.66
Muddathir, A. R. M., Abdallah, E. I., Khabour, O. F., Abdelgader, R. E., and Elgari, M. M. (2019). Age- and Gender-independent Association of Glutathione S-Transferase Null Polymorphisms with Chronic Myeloid Leukemia. Bosn. J. Basic Med. Sci. 19 (4), 350–354. doi:10.17305/bjbms.2019.4176
Müller, P., Asher, N., Heled, M., Cohen, S. B., Risch, A., and Rund, D. (2008). Polymorphisms in Transporter and Phase II Metabolism Genes as Potential Modifiers of the Predisposition to and Treatment Outcome of De Novo Acute Myeloid Leukemia in Israeli Ethnic Groups. Leukemia Res. 32, 919–929. doi:10.1016/j.leukres.2007.10.011
Naoe, T., Takeyama, K., Yokozawa, T., Kiyoi, H., Seto, M., Uike, N., et al. (2000). Analysis of Genetic Polymorphism in NQO1, GST-M1, GST-T1, and CYP3A4 in 469 Japanese Patients with Therapy-Related Leukemia/Myelodysplastic Syndrome and De Novo Acute Myeloid Leukemia. Clin. Cancer Res. 6 (10), 4091–4095.
Nasr, A., Sami, R., Ibrahim, N., and Darwish, D. (2015). Glutathione S Transferase (GSTP 1, GSTM 1, and GSTT 1) Gene Polymorphisms in Egyptian Patients with Acute Myeloid Leukemia. Indian J. Cancer 52 (4), 490–495. doi:10.4103/0019-509x.178408
Ouerhani, S., Nefzi, M. A., Menif, S., Safra, I., Douzi, K., Fouzai, C., et al. (2011). Influence of Genetic Polymorphisms of Xenobiotic Metabolizing Enzymes on the Risk of Developing Leukemia in a Tunisian Population. Bull. Cancer 98 (12), 95–106. doi:10.1684/bdc.2011.1502
Ovsepian, V. A., Vinogradova, E., and Sherstneva, E. S. (2010). Cytochrome P4501A1, Glutathione S-Transferase M1 and T1 Gene Polymorphisms in Chronic Myeloid Leukemia. Genetika 46 (10), 1360–1362.
Özten, N., Sunguroğlu, A., and Bosland, M. C. (2012). Variations in Glutathione-S-Transferase Genes Influence Risk of Chronic Myeloid Leukemia. Hematol. Oncol. 30 (3), 150–155. doi:10.1002/hon.1018
Pakakasama, S., Mukda, E., Sasanakul, W., Kadegasem, P., Udomsubpayakul, U., Thithapandha, A., et al. (2005). Polymorphisms of Drug-Metabolizing Enzymes and Risk of Childhood Acute Lymphoblastic Leukemia. Am. J. Hematol. 79, 202–205. doi:10.1002/ajh.20404
Pigullo, S., Haupt, R., Dufour, C., Di Michele, P., Valsecchi, M. G., Basso, G., et al. (2007). Are Genotypes of Glutathione S-Transferase Superfamily a Risk Factor for Childhood Acute Lymphoblastic Leukemia? Results of an Italian Case-Control Study. Leukemia 21, 1122–1124. doi:10.1038/sj.leu.2404617
Pui, C. H. (2000). Acute Lymphoblastic Leukemia in Children. Curr. Opin. Oncol. 12, 3–12. doi:10.1097/00001622-200001000-00002
Pui, M. H., and Husen, Y. A. (2000). Value of Magnetic Resonance Myelography in the Diagnosis of Disc Herniation and Spinal Stenosis. Australas Radiol. 44 (3), 281–284. doi:10.1046/j.1440-1673.2000.00813.x
Pui, C. H., Relling, M. V., and Evans, W. E. (2002). Role of Pharmacogenomics and Pharmacodynamics in the Treatment of Acute Lymphoblastic Leukaemia. Best Pract. Res. Clin. Haematol. 15 (4), 741–756. doi:10.1053/beha.2002.0225
Pui, C. H., Relling, M. W., and Evans, W. E. (2006). Is Mega Dose of Methotrexate Beneficial to Patients with Acute Lymphoblastic Leukemia?. Leuk. Lymphoma. 47 (12), 2431–2432. doi:10.1080/10428190600955837
Quintás-Cardama, A., and Cortes, J. E. (2006). Chronic Myeloid Leukemia: Diagnosis and Treatment. Mayo Clin. Proc. 81, 973–988. doi:10.4065/81.7.973
Rimando, M. G., Chua, M. N., Yuson, E. d. J., de Castro-Bernas, G., and Okamoto, T. (2008). Prevalence of GSTT1, GSTM1 and NQO1 (609C>T) in Filipino Children with ALL (Acute Lymphoblastic Leukaemia). Biosci. Rep. 28, 117–124. doi:10.1042/bsr20070010
Rollinson, S., Roddam, P., Kane, E., Roman, E., Cartwright, R., Jack, A., et al. (2000). Polymorphic Variation within the Glutathione S-Transferase Genes and Risk of Adult Acute Leukaemia. Carcinogenesis 21 (1), 43–47. doi:10.1093/carcin/21.1.43
Ross, J. A., Davies, S. M., Potter, J. D., and Robison, L. L. (1994). Epidemiology of Childhood Leukemia, with a Focus on Infants. Epidemiol. Rev. 16, 243–272. doi:10.1093/oxfordjournals.epirev.a036153
Rostami, G., Assad, D., Ghadyani, F., Hamid, M., Karami, A., Jalaeikhoo, H., et al. (2019). Influence of Glutathione S-Transferases (GSTM1, GSTT1, and GSTP1) Genetic Polymorphisms and Smoking on Susceptibility Risk of Chronic Myeloid Leukemia and Treatment Response. Mol. Genet. Genomic Med. 7 (7), e00717. doi:10.1002/mgg3.717
Saadat, I., and Saadat, M. (2000). The Glutathione S-Transferase Mu Polymorphism and Susceptibility to Acute Lymphocytic Leukemia. Cancer Lett. 158, 43–45. doi:10.1016/s0304-3835(00)00504-8
Sasai, Y., Horiike, S., Misawa, S., Kaneko, H., Kobayashi, M., Fujii, H., et al. (1999). Genotype of Glutathione S-Transferase and Other Genetic Configurations in Myelodysplasia. Leukemia Res. 23 (11), 975–981. doi:10.1016/s0145-2126(99)00119-8
Seedhouse, C., Faulkner, R., Ashraf, N., Das-Gupta, E., and Russell, N. (2004). Polymorphisms in Genes Involved in Homologous Recombination Repair Interact to Increase the Risk of Developing Acute Myeloid Leukemia. Clin. Cancer Res. 10, 2675–2680. doi:10.1158/1078-0432.ccr-03-0372
Seidegård, J., Vorachek, W. R., Pero, R. W., and Pearson, W. R. (1988). Hereditary Differences in the Expression of the Human Glutathione Transferase Active on Trans-stilbene Oxide Are Due to a Gene Deletion. Proc. Natl. Acad. Sci. U.S.A. 85, 7293–7297. doi:10.1073/pnas.85.19.7293
Singh, S. (2015). Cytoprotective and Regulatory Functions of Glutathione S-Transferases in Cancer Cell Proliferation and Cell Death. Cancer Chemother. Pharmacol. 75, 1–15. doi:10.1007/s00280-014-2566-x
Souza, C. L., Barbosa, C. G., Moura Neto, J. P. d., Barreto, J. H., Reis, M. G., and Gonçalves, M. S. (2008). Polymorphisms in the Glutathione S-Transferase Theta and Mu Genes and Susceptibility to Myeloid Leukemia in Brazilian Patients. Genet. Mol. Biol. 31 (1), 39–41. doi:10.1590/s1415-47572008000100008
Suneetha, K. J., Nancy, K. N., Rajalekshmy, K. R., Rama, R., Sagar, T. G., and Rajkumar, T. (2011). Role of Glutathione-S-Transferase and CYP1A1*2A Polymorphisms in the Therapy Outcome of South Indian Acute Lymphoblastic Leukemia Patients. Indian J. Med. Paediatr. Oncol. 32 (1), 25–29. doi:10.4103/0971-5851.81886
Suneetha, K. J., Nancy, K. N., Rajalekshmy, K. R., Sagar, T. G., and Rajkumar, T. (2008). Role of GSTM1 (Present/Null) and GSTP1 (Ile105Val) Polymorphisms in Susceptibility to Acute Lymphoblastic Leukemia Among the South Indian Population. Asian Pac J. Cancer Prev. 9 (4), 733–736.
Tang, Q., Li, J., Zhang, S., Yuan, B., Sun, H., Wu, D., et al. (2013). GSTM1 and GSTT1 Null Polymorphisms and Childhood Acute Leukemia Risk: Evidence from 26 Case-Control Studies. PLoS One 8 (10), e78810. doi:10.1371/journal.pone.0078810
Tang, Z. H., Zhang, C., Cheng, P., Sun, H. M., Jin, Y., Chen, Y. J., et al. (2014). Glutathione-S-transferase Polymorphisms (GSTM1, GSTT1 and GSTP1) and Acute Leukemia Risk in Asians: a Meta-Analysis. Asian Pac. J. Cancer Prev. 15 (5), 2075–2081. doi:10.7314/apjcp.2014.15.5.2075
Taspinar, M., Aydos, S. E., Comez, O., Elhan, A. H., Karabulut, H. G., and Sunguroglu, A. (2008). CYP1A1, GST Gene Polymorphisms and Risk of Chronic Myeloid Leukemia. Swiss Med. Wkly. 138 (1-2), 12–17.
Thakkinstian, A., McKay, G. J., McEvoy, M., Chakravarthy, U., Chakrabarti, S., Silvestri, G., et al. (2011). Systematic Review and Meta-Analysis of the Association between Complement Component 3 and Age-Related Macular Degeneration: a HuGE Review and Meta-Analysis. Am. J. Epidemiol. 173, 1365–1379. doi:10.1093/aje/kwr025
Udomsinprasert, R., Pongjaroenkit, S., Wongsantichon, J., Oakley, A. J., Prapanthadara, L.-a., Wilce, M. C. J., et al. (2005). Identification, Characterization and Structure of a New Delta Class Glutathione Transferase Isoenzyme. Biochem. J. 388, 763–771. doi:10.1042/bj20042015
Vijayakrishnan, J., and Houlston, R. S. (2010). Candidate Gene Association Studies and Risk of Childhood Acute Lymphoblastic Leukemia: a Systematic Review and Meta-Analysis. Haematologica 95 (8), 1405–1414. doi:10.3324/haematol.2010.022095
Wang, J., Zhang, L., Feng, J., Wang, H., Zhu, S., Hu, Y., et al. (2004). Genetic Polymorphisms Analysis of Glutathione S-Transferase M1 and T1 in Children with Acute Lymphoblastic Leukemia. J. Huazhong Univ. Sci. Technol. Med. Sci. 24 (3), 243–244. doi:10.1007/BF02832001
Wang, J., Wu, D., and Sun, A. (2019). Effects of GST Null Genotypes on Individual Susceptibility to Leukemia: A Meta-Analysis. Exp. Mol. Pathology 108, 137–142. doi:10.1016/j.yexmp.2019.01.004
Wang, W., Li, C., Long, Y., Zhan, C., and Xiang, C. (2002). Study on the Relationship between Glutathione S Transferase Mu Gene Deletion and Leukemia in Workers Exposed to Benzene. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 20 (1), 38–41.
Weich, N., Ferri, C., Moiraghi, B., Bengió, R., Giere, I., Pavlovsky, C., et al. (2016). GSTM1 and GSTP1 , but Not GSTT1 Genetic Polymorphisms Are Associated with Chronic Myeloid Leukemia Risk and Treatment Response. Cancer Epidemiol. 44, 16–21. doi:10.1016/j.canep.2016.07.008
Weich, N., Nuñez, M. C., Galimberti, G., Elena, G., Acevedo, S., Larripa, I., et al. (2015). Polymorphic Variants ofGSTM1,GSTT1, andGSTP1genes in Childhood Acute Leukemias: A Preliminary Study in Argentina. Hematology 20 (9), 511–516. doi:10.1179/1607845415y.0000000007
Woo, M., Shuster, J., Chen, C., Bash, R., Behm, F., Camitta, B., et al. (2000). Glutathione S-Transferase Genotypes in Children Who Develop Treatment-Related Acute Myeloid Malignancies. Leukemia 14, 232–237. doi:10.1038/sj.leu.2401660
Wu, Y. X., Gao, Y. J., Zhao, J. C., Jing, X. P., and Xia, Z. L. (2004). Preliminary Study on Polymorphism of GSTM1, CYP2E1 and NQO1 Genes and Risk Factors of Children Leukemia. Zhonghua Liu Xing Bing Xue Za Zhi 25, 819.
Xu, L. Y., and Cao, L. F. (2014). GSTT1 Genetic Polymorphism and Susceptibility to Childhood Acute Lymphoblastic Leukemia: a Meta-Analysis. Tumor Biol. 35 (2), 1433–1437. doi:10.1007/s13277-013-1197-x
Yang, L., Zhang, Y., Zhang, M. R., and Xiao, Z. J. (2005). Relationship between GSTT1, GSTM1 and NQO1 Gene Polymorphism and Acute Myeloid Leukemia and Recurrent Chromosome Translocations. Zhonghua Yi Xue Za Zhi 85, 2312–2316.
Ye, Z., and Song, H. (2005). Glutathione S-Transferase Polymorphisms (GSTM1, GSTP1 and GSTT1) and the Risk of Acute Leukaemia: a Systematic Review and Meta-Analysis. Eur. J. Cancer 41 (7), 980–989. doi:10.1016/j.ejca.2005.01.014
Yuille, M., Condie, A., Hudson, C., Kote-Jarai, Z., Stone, E., Eeles, R., et al. (2002). Relationship between Glutathione S-Transferase M1, T1, and P1 Polymorphisms and Chronic Lymphocytic Leukemia. Blood 99 (11), 4216–4218. doi:10.1182/blood.v99.11.4216
Zehra, A., Zehra, S., Ismail, M., and Azhar, A. (2018). Glutathione S-Transferase M1 and T1 Gene Deletions and Susceptibility to Acute Lymphoblastic Leukemia (ALL) in Adults. Pak J. Med. Sci. 34 (3), 666–670. doi:10.12669/pjms.343.14911
Zhang, L., Zhang, X. X., and Wang, J. (2003). Glutathione S-Transferase M1, T1 Genetic Polymorphisms in Childhood Leukemia. Zhengzhou, China: Zhengzhou University.
Zhao, T., Ma, F., and Yin, F. (2018). Role of Polymorphisms of GSTM1, GSTT1 and GSTP1 Ile105Val in Childhood Acute Lymphoblastic Leukemia Risk: an Updated Meta-Analysis. Minerva Pediatr. 70 (2), 185–196. doi:10.23736/S0026-4946.17.04657-6
Zhou, L., Zhu, Y. Y., Zhang, X. D., Li, Y., and Liu, Z. G. (2013). Risk Effects of GST Gene Polymorphisms in Patients with Acute Myeloid Leukemia: a Prospective Study. Asian Pac. J. Cancer Prev. 14 (6), 3861–3864. doi:10.7314/apjcp.2013.14.6.3861
Zi, Y., Wu, S., Ma, D., Yang, C., Yang, M., Huang, Y., et al. (2014). Association of GSTTI and GSTM1 Variants with Acute Myeloid Leukemia Risk. Genet. Mol. Res. 13 (2), 3681–3685. doi:10.4238/2014.may.9.11
Zintzaras, E. (2009). Glutathione S-Transferase M1 and T1 Genes and Susceptibility to Chronic Myeloid Leukemia: a Meta-Analysis. Genet. Test. Mol. Biomarkers 13 (6), 791–797. doi:10.1089/gtmb.2009.0079
Keywords: GSTM1, GSTT1, polymorphism, FPRP, BFDP, leukemia
Citation: Hu T, Zhou G and Li W (2022) Association Between the Individual and Combined Effects of the GSTM1 and GSTT1 Polymorphisms and Risk of Leukemia: A Meta-Analysis. Front. Genet. 13:898937. doi: 10.3389/fgene.2022.898937
Received: 18 March 2022; Accepted: 06 June 2022;
Published: 22 July 2022.
Edited by:
Juliana Mara Serpeloni, State University of Londrina, BrazilReviewed by:
Glauco Akelinghton Freire Vitiello, A.C. Camargo Cancer Center, BrazilSalvador F. Aliño, University of Valencia, Spain
Copyright © 2022 Hu, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Hu, aHV0aW5na2V5YW5AMTI2LmNvbQ==
†These authors have contributed equally to this work
 Ting Hu
Ting Hu Guozhong Zhou2†
Guozhong Zhou2†