
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 28 April 2022
Sec. Livestock Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.891204
This article is part of the Research TopicReproduction in Aquatic AnimalsView all 8 articles
 Siqi Xu1,2
Siqi Xu1,2 Shiyong Zhang1,3*
Shiyong Zhang1,3* Wenping Zhang1,2
Wenping Zhang1,2 Hongyan Liu1
Hongyan Liu1 Minghua Wang1,3
Minghua Wang1,3 Liqiang Zhong1,3
Liqiang Zhong1,3 Wenji Bian1,2,3
Wenji Bian1,2,3 Xiaohui Chen1,2,3*
Xiaohui Chen1,2,3*The Dmrt (Doublesex and Mab-3 related transcription factor) gene family is a class of crucial transcription factors characterized by a conserved DM domain related to sex determination and differentiation, which has been systematically described in various teleost fish, but less in channel catfish (Ictalurus punctatus), an important global aquaculture species in the US and China. In this study, seven Dmrt genes from channel catfish genome were identified and analyzed using bioinformatics methods. Seven IpDmrt genes were distributed unevenly across five chromosomes. Synteny analysis revealed that Dmrt1, Dmrt2a, Dmrt2b, Dmrt3, Dmrt4, and Dmrt5 were relatively conserved in teleost fish. Tissue distribution analysis showed that IpDmrt1, IpDmrt2b, IpDmrt5, and IpDmrt6 exhibited sexually dimorphic expression patterns and, among them, IpDmrt1 and IpDmrt6 had high expression levels in the testes, while IpDmrt2b and IpDmrt5 had more significant expression levels in the ovaries than in other tissues. After 17β-estradiol treatment, IpDmrt2b and IpDmrt5 were significantly up regulated, while the expression of IpDmrt1 and IpDmrt6 was significantly repressed in XY channel catfish ovaries compared with XX channel catfish ovaries. The present study provides a comprehensive insight into the Dmrt gene family of channel catfish. The results suggest that IpDmrt1 and IpDmrt6 may play an important role in testis differentiation/development, while IpDmrt2b and IpDmrt5 are critical in ovary development in this species.
Sex determination (SD) in vertebrates is a complicated physiological process, affected by both environmental and genetic factors (Shen and Wang, 2014). A diverse array of SD mechanisms, including environmental SD (ESD), and genetic SD (GSD); or a combination of both, have been identified among these animals (Ortega-Recalde et al., 2020). During the evolution of organisms from lower to higher forms, genetic factors gradually increased, while environmental factors weakened. With the evolution of mammals, the former completely replaced the impact of the latter on gonadal sex differentiation (Zhou et al., 2004). In lower vertebrates such as reptiles, amphibians, fish due to the relatively low degree of evolution, as well as the lack of an obvious difference in sex chromosomes, environmental factors, such as temperature, social status, or pH, had a certain impact on early sex differentiation, and could even induce sexual reversal and change the sex differentiation pathway (She and Yang, 2017).
The reproductive modes of teleosts are divided into gonochorism and hermaphroditism (Nagahama et al., 2021). In nature, most teleosts are gonochoristic, exhibiting two differentiated forms, namely, male and female. However, hermaphroditism is also common among teleosts, and in this mode both testes and ovaries exist either simultaneously (e.g., in coral reef fish) (Godwin et al., 2000) or sequentially (e.g., in rice field eel) (Xiao et al., 2010). The primordial gonad in gonochoristic fish can develop either as a testis or an ovary, thus the direction of gonadal sex differentiation depends on their sex-determining genes present in the genome, or on environment conditions. To date, several master sex-determining genes have been characterized in teleosts, such as Dmy from Japanese medaka (Oryzias latipes) (Matsuda et al., 2002), Gsdf from Philippine medaka (Oryzias luzonensis) (Myosho et al., 2012), Amh from Patagonian pejerrey (Odontesthes hatcheri) (Hattori et al., 2012), Amhr2 from Japanese pufferfish (Fugu rubripes) (Kamiya et al., 2012), Sox3 from brackish medaka (Oryzias dancena) (Takehana et al., 2014), and Dmrt1 from half-smooth tongue sole (Cynoglossus semilaevis) (Cui et al., 2017).
Although sex-determining genes vary in teleost fish, the Dmrt (Doublesex and Mab-3 related transcription factor) gene family is thought to be associated with sex determination and differentiation, embryo and gonadal development, as well as muscle growth, in vertebrates ranging from fish to tetrapods. This gene family belongs to the transforming growth factor-beta superfamily, which contains one or several classical DNA-binding domains. There are at least five Dmrt genes (Dmrt1, Dmrt2a, Dmrt2b, Dmrt3 and Dmrt5) in teleost fish, among which Dmrt1 is the major male-biased expression gene (Herpin and Schartl, 2011; Chen et al., 2014) participating directly in sex determination and differentiation through the activation of the gsdf-promotor and inhibition of the Cyp19a1a-promotor alone in both XY and ZZ male teleosts (Han et al., 2021).
Channel catfish (Ictalurus punctatus) is one of the most important freshwater aquaculture fish in the US and China (Chen et al., 2016). Due to its strong environmental adaptability, superior muscle quality, lower number of intermuscular spines, and easy meat processing characteristics, it is currently bred worldwide (Bao et al., 2019). The annual production of channel catfish has been stable at more than 4 × 109 kg in China (Zhong et al., 2021). At present, the Dmrt gene family has been systematically described in various teleost fish, but less so in channel catfish. The aim of this study was to provide a comprehensive insight into this important aquaculture species by identifying its Dmrt gene family members and analyzing their physical and chemical properties, gene phylogeny, as well as gene expression patterns.
The channel catfish fries used in the present study were produced at the National Genetic Breeding Center of Channel Catfish (Yangzhong, Jiangsu, China). Artificial compound feed containing 17β-estradiol (17β-E2, 60 μg/g) was selected as a vector to induce the sexual reversal of male fish by feeding catfish larvae continuously for 27 days. The dosage of 17β-E2 in the compound feed was 60 μg/g. The gonads of sex-reversed and normal female catfish at 60 dahs were identified by histological section and sex-linkage molecular marker (Zhang et al., 2019; Pan et al., 2022), respectively.
Furthermore, the ovary tissues of the two catfish types were dissected from three individuals based on the identification results. At the same time, five tissues, namely, liver, brain, kidney, head kidney, and testes/ovaries, were also dissected from three male and three female individuals. All tissue samples were preserved in the RNA Keeper Tissue Stabilizer (Vazyme, China) and were stored at -20°C until RNA extraction. During sampling, the experimental fish were anesthetized using 0.1% tricaine methanesulfonate (MS-222) (Merck, Germany). All animal handling was carried out in accordance with the ethical guidelines and protocols of the Freshwater Fisheries Research Institute of Jiangsu Province.
Candidate IpDmrt family genes were identified on the channel catfish genome using the BLAST search program based on the conserved DM domain (Pfam 00,751) with amino acids. The identified candidates and their amino acid sequences were downloaded from the channel catfish genome database stored in the NCBI database (https://www.ncbi.nlm.nih.gov/). The genes were further confirmed to contain the conserved DM domains by scanning their sequences through the Pfam (http://pfam.sanger.ac.uk/search), SMART (http://smart.embl-heidelberg.de/), and InterPro (http://www.ebi.ac.uk/interpro/) online databases.
The total RNA of sampled tissues was isolated using the RNeasy Mini Kit (Qiagen, Germany). The cDNA was subjected to reverse transcription using the PrimeScript™ first Strand cDNA Synthesis Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. According to the DNA sequences of channel catfish Dmrt genes, we designed seven pairs of primers (Supplementary Table S1) to clone their full-length cDNA sequences. The PCR conditions were as follows: 94°C for 30 s, gene-specific annealing for 30 s and elongation stage at 72°C for 60 s, a total of 34 cycles. The target products were purified using the universal DNA Purification Kit (Tiangen, Beijing, China), and then sequenced by the Sangon Biotech Co., Ltd. (Shanghai, China).
Basic information related to the IpDmrt genes, including the number of introns and exons, coding sequence length, and chromosome location, were extracted from the channel catfish genome database. The physicochemical characteristics of IpDmrt proteins, including the theoretical molecular weight (kDa), lengths, isoelectric points (pI), and grand average of hydropathicity (GRAVY), were evaluated using the online ExPASY tool (https://web.expasy.org/protparam/). The intron and exon positions, and untranslated regions were visualized using the Gene Structure Display Server 2.0 (http://gsds.gao-lab.org/).
Multiple sequence alignments of the IpDmrt proteins were performed and visualized using the T-COFFEE (tcoffee.crg.cat/apps/tcoffee/index.html) and EsPript 3.0 (https://espript.ibcp.fr/ESPript/ESPript) online software, respectively. The amino acid sequences of Dmrt proteins from human (Homo sapiens), chicken (Gallus), African clawed frog (Xenopus laevis), zebrafish (Danio rerio), large yellow croaker (Larimichthys crocea), Nile tilapia (Oreochromis niloticus), Japanese medaka, Japanese pufferfish, and channel catfish were selected to construct the phylogenetic tree in MEGA7 using the neighbor-joining method. The Jones-Taylor-Thornton (JTT) algorithm + Nearest-Neighbor-Interchange (NNI) distance were selected to automatically generate the initial tree (Default - NJ/BioNJ) model and evaluate the phylogenetic tree. The Dmrt genes were named using an “Ip” prefix and were numbered in ascending order based on the results of the phylogenetic analysis. The name of Dmrt protein sequences showed in Supplementary Table S2.
In order to examine the extent of preservation of genomic neighborhoods between IpDmrt genes and the potential counterparts of other species, such as electric eel (Electrophorus electricus), Nile tilapia, zebrafish, and Atlantic cod (Gadus morhua), the Synteny Database (http://teleost.cs.uoregon.edu/synteny_db/) and Genomicus Browser (http://www.dyogen.ens.fr/genomicus-63.01/cgi-bin/search.pl) were used as the main visualization tools. To perform the synteny analysis, the channel catfish orthologs of the Dmrt genes neighboring other teleosts’ genes were firstly examined at the chromosome level through the Synteny Database, and at the local neighborhood level using the Genomicus Browser, which provides detailed gene information related to each teleost gene’s orthologs on an individual chromosome. In the synteny analyses of Dmrt genes, the channel catfish neighboring genes as follows: Fbrsl1, Ap1b1, Fbxo21, Fbxw8, Abca2, Adamts3, Dmrt2a, Dmrt3, Dmrt1, and Adgrv1 on Chr22; Abl2, Bend5, Dmrt5, Calr, Abhd17 ab, Tmem47, Tab3, Nexn, Armh1, Multh, and Acbd6 on Chr5; Irrc40, Irrc7, Insl5a, Ak4, Jak1, Foxd3, Kank4, Cyldl, Adcyaplr1b, and Dmrt2b on Chr11; and Ndrg2, Arhgef40, Casq1b, Ipcat4, Khnyn, Drap1, Rela, Elavl2, Dmrt4, Rbpms, and Alpk1 on Chr29.
The purpose of protein interaction network analysis was to examine the relationship between Dmrt and other genes and explore their potential functions. The protein interaction networks of the IpDmrt gene family were constructed using the STRING online software (https://www.string-db.org).
Gene-specific primers (listed in Supplementary Table S3) were designed using Primer Premier 5.0 software and were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). The α-tubulin (Zhang et al., 2020) gene was employed as an internal reference gene. The qRT-PCR reaction system contained 12.5 μL of SYBR green (TaKaRa, Dalian, China), 1 μL of gene-specific primers (1.0 μM), 1 μL of cDNA, and 8.5 μL of DEPC water, reaching a total reaction volume of 25 μL. The PCR conditions were as follows: 95°C for 30 s, followed by 40 cycles of 15 s at 95°C and 30 s at 57°C; 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. Relative mRNA expression levels were determined using the 2−△△ CT method (Livak and Schmittgen, 2001). Finally, the statistical significance was calculated in R v. 4.0.5. Statistically significant differences were set at p < 0.05.
A total of seven Dmrt genes (IpDmrt1, IpDmrt2a, IpDmrt2b, IpDmrt3, IpDmrt4, IpDmrt5, and IpDmrt6) were identified in the channel catfish genome, and were named based on the nomenclature of teleost Dmrt genes and the results of phylogenetic analyses. These gene numbers were relatively consistent compared to that previously reported in other fish species, such as large yellow croaker (Wan et al., 2020), while it was less than that reported for grass carp (Ctenopharyngodon idella) (Chen et al., 2019) and rainbow trout (Oncorhynchus mykiss) (Wang et al., 2014), which experienced four rounds of whole genome duplication during evolution. The exon-intron structure analysis revealed that the IpDmrt gene family contained two to five exons; in particularly, IpDmrt1 had five exons; IpDmrt2a and IpDmrt6 had four; IpDmrt2b had three; and IpDmrt3, IpDmrt4, and IpDmrt5 had two. The length of IpDmrt proteins ranged from 282 to 507 amino acids, with an average molecular weight of 42.30062 kDa. The average pI of the IpDmrt protein family was 8.13; with the highest and lowest points reaching 9.36 and 5.91, respectively. Only one basic protein, IpDmrt3, was detected, while the others were all acidic proteins. The calculated grand average of the GRAVY values of all IpDmrt genes was -0.2835 to -0.928, indicating that they were hydrophobic in nature (Table1).
Each IpDmrt protein had a characteristic conserved DM domain (Pfam 00,751), and only protein IpDmrt1 had an additional Dmrt1 domain, while a DMA domain was detected in IpDmrt3, IpDmrt4, and IpDmrt5 (Figure 1).
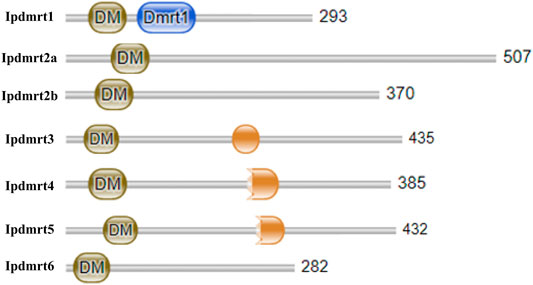
FIGURE 1. Schematic diagram of the construction of seven IpDmrt proteins. Image (a) shows the schematic diagram of seven IpDmrt protein domains; all the identified proteins present a conserved DM domain, and IpDmrt1 has an additional DMRT-1 domain.
The multiple alignment of IpDmrt protein sequences revealed the presence of a conserved DM domain at the N-terminal (Figure 2), while outside the DM domain there was an extremely lower conservation. Sequence identity analysis of DM domain showed that IpDmrt1 shared a 75% identity with its homologs from IpDmrt2a (74.58%) and IpDmrt3 (66.10%), which were located on the same chromosome (Chr 22). However, IpDmrt1 shared a higher identity with its counterparts from IpDmrt5 (82.76%). Although IpDmrt4 and IpDmrt5 were on the different chromosome, they shared the highest percentage of identity (89.66%) compared to all chromosomes (Table 2).
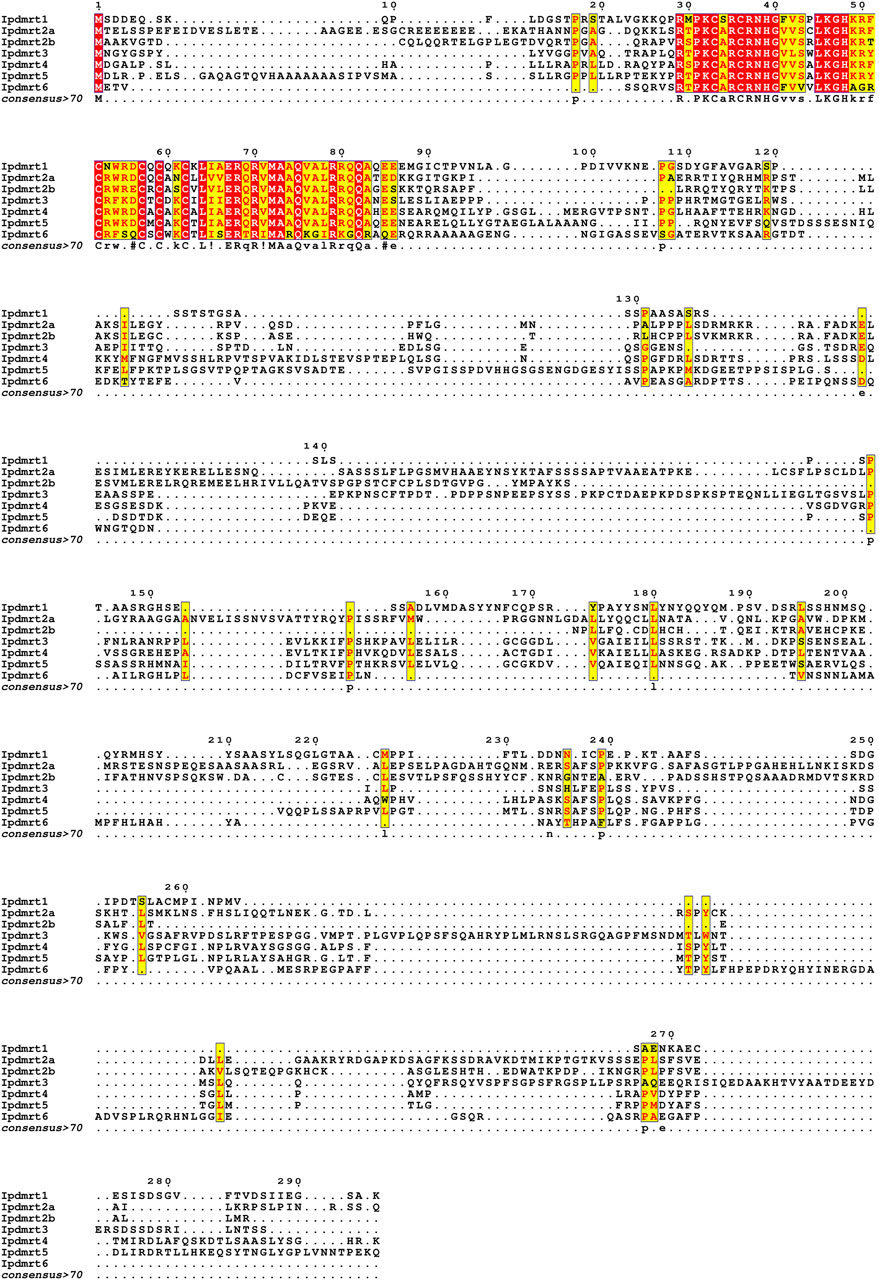
FIGURE 2. Multiple sequence alignments of Dmrt proteins. The alignments were performed using the T-COFFEE online software, and were visualized in EsPript v. 3.0 (available online). The partial protein sequence in the red box represents the DM domain, which has a higher conservation, while the protein sequence outside it has a lower level of identity.
To analyze the phylogenetic relationships among the IpDmrt gene family members, a phylogenetic tree was generated using 57 Dmrt protein sequences from various vertebrate species, ranging from fish to mammals, based on the maximum likelihood method. As shown in Figure 3, Dmrt1, Dmrt2a, Dmrt3, and Dmrt5 were found in all five teleost species. As expected, Dmrt genes were clustered into six subfamilies, which is consistent with previous studies (Dong et al., 2020; Kikkawa and Osumi, 2021), and each Dmrt gene branched along with its counterparts from different vertebrate species. Within each subfamily, the phylogenetic topology could be further divided into two subgroups: tetrapods and teleosts. Dmrt2 experienced teleost-specific whole genome duplication during evolution, and at least two copies (Dmrt2a and Dmrt2b) were observed in teleosts. However, Dmrt7 and Dmrt8 were lost in teleosts, and are only present in the mammal genome (Veith et al., 2006; Date et al., 2012).
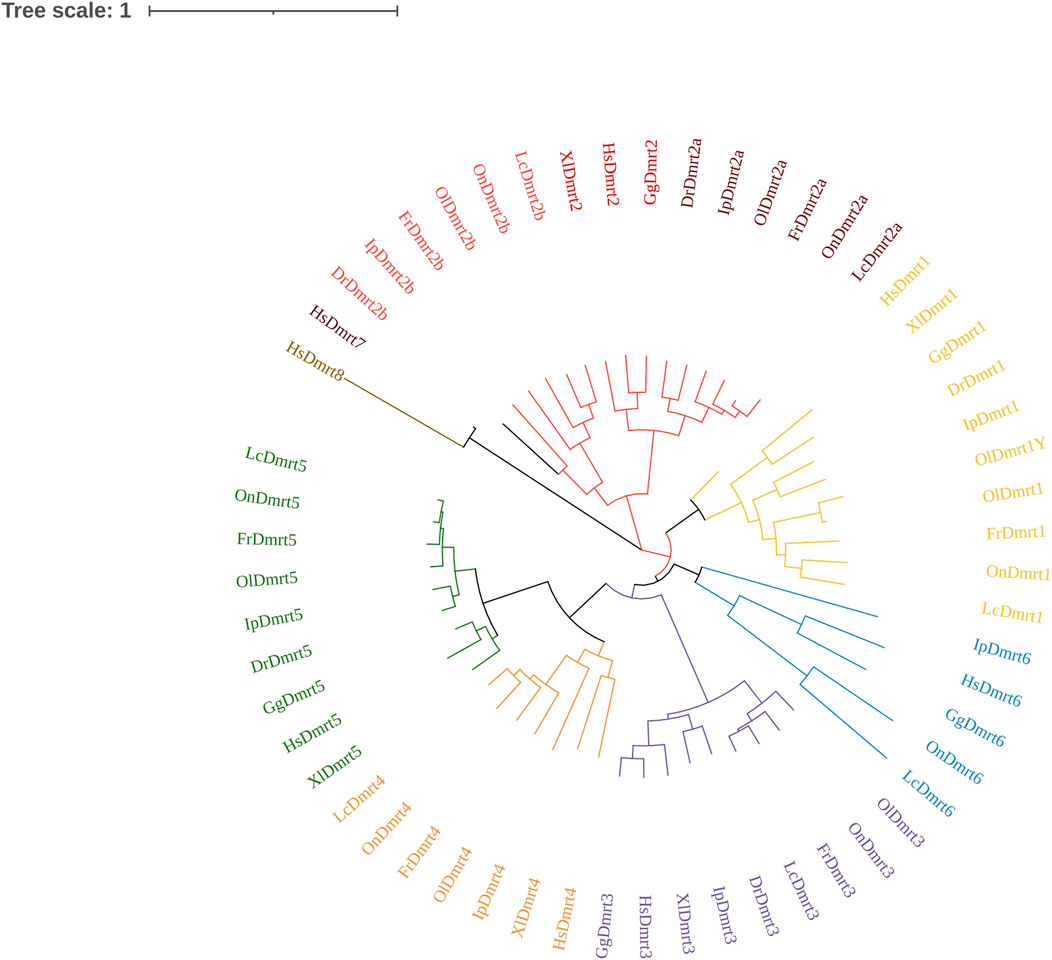
FIGURE 3. Phylogenetic tree analysis of Dmrt gene family members from human, chicken, African clawed frog, zebrafish, large yellow croaker, Nile tilapia, Japanese pufferfish, and channel catfish. The phylogenetic tree was constructed in MEGA7, using the neighbor-joining method. The GenBank accession numbers of the sequences used are listed in the Supplementary Table S1.
Gene duplication is an important process in species evolution in terms of gene expansion and functional diversity. The aim of synteny analysis is to show whether genes are conserved during evolution in teleosts. Conservative regions can be clearly identified by observing the distribution and permutation of genes on chromosomes. In this study, in order to investigate the preservation extent of the Dmrt genes among teleosts, the distribution and permutation on the corresponding chromosomes of these genes were compared, as well as their neighboring genes from channel catfish, zebrafish, electric eel, Nile tilapia, and Atlantic cod. Seven IpDmrt genes were unevenly distributed on five chromosomes by chromosomal location analysis. Specifically, IpDmrt2b, IpDmrt4, IpDmrt5, and IpDmrt6 were located on Chr11, Chr29, Chr5, and Chr7, respectively; while IpDmrt1, IpDmrt2a, and IpDmrt3 were located on the same chromosome (Chr22).
Syntenic analyses revealed that the Dmrt1-Dmrt2a-Dmrt3 syntenic block was highly conserved in teleosts (Figure 4A). Interestingly, Dmrt5 and neighboring genes were not completely conserved, showing differences in replication direction and location (Figure 4B), which indicated that this region experienced genome arrangement during evolution. On the contrary, the most conservative syntenic block, Irrc40-Irrc7-Insl5a-Ak4-Jak1-Foxd3-Kank4-Cyldl-Adcyaplr1b-Dmrt2b, was detected in channel catfish, electric eel, and zebrafish, despite the location and copy direction of these genes being different in Atlantic cod (Figure 4C). It was also found that Dmrt4 is preserved only in channel catfish, electric eel, and Atlantic cod, while it was lost in zebrafish and Nile tilapia (Figure 4D).
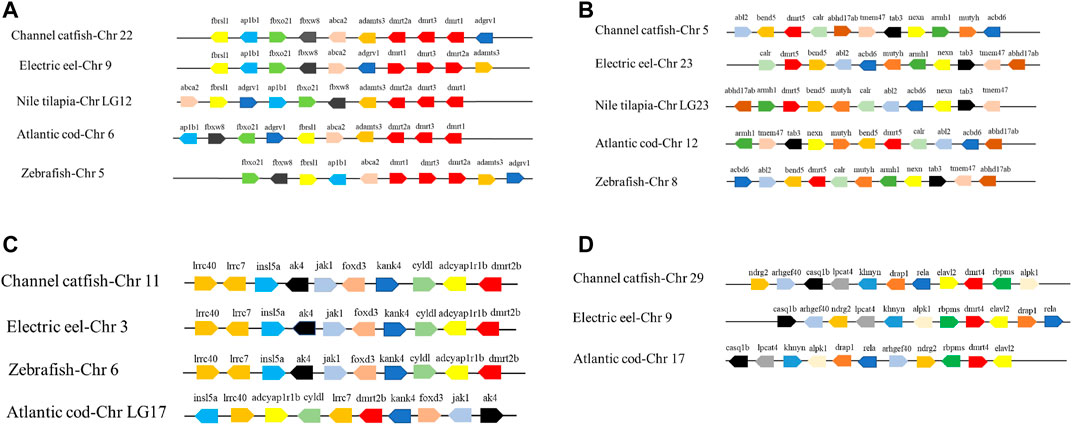
FIGURE 4. Synteny analysis of Dmrt1, Dmrt2, Dmrt3 (A), Dmrt5 (B), Dmrt2b (C), Dmrt4 (D) and their adjacent genes in electric eel, Nile tilapia, Atlantic cod, and zebrafish. Direction of the arrows indicates gene orientation.
The interaction network of channel catfish Dmrt proteins was constructed using the STRING database. Dmrt1 closely interacted not only with the Amh protein, which is thought to be associated with male sex determination, but also with Cyp19a1, a key downstream gene of female sex regulation (Figure 5A). Both Dmrt2a and Dmrt2b were related to Pxdc1 in the protein interaction network; in addition, Dmrt2a was also associated with Wnt4 (Figures 5B, C); Figures 5D–F showed that there was protein interaction among Dmrt3, Dmrt4, and Dmrt5. Moreover, Dmrt3 was demonstrated to interact with Foxl1, which was also compactly linked with Dmrt6 in the protein interaction network (Figure 5G). This analysis revealed that most genes interacting with Dmrt genes have functions in sex differentiation and gonad development, contributing to understand the mechanism/dynamics of more genes related to sex determination and differentiation in channel catfish and even in teleosts.
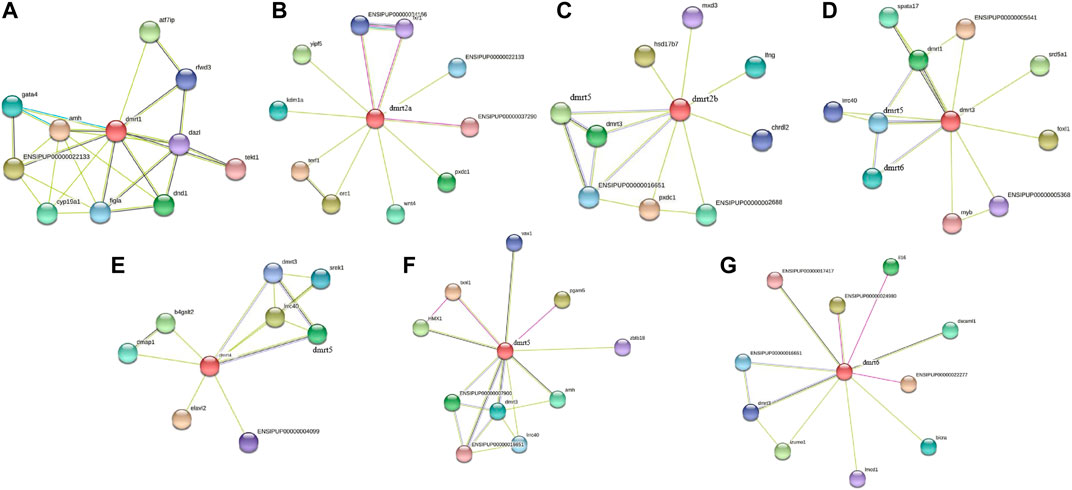
FIGURE 5. Interaction network analysis of IpDmrt proteins. Images (A), (B), (C), (D), (E), (F), and (G) show the protein interaction network of IpDmrt1, IpDmrt2a, IpDmrt2b, IpDmrt3, IpDmrt4, IpDmrt5, and IpDmrt6, respectively.
To study the spatiotemporal specific expression of IpDmrt genes, their expression profiles were investigated in five tissues obtained from female and male channel catfish, namely the brain, head kidney, kidney, liver, and ovaries or testes using qRT-PCR. Subsequently, the mRNA expression of IpDmrt genes in the ovary tissues of sex-reversed individuals were also explored. The results indicated that IpDmrt1 and IpDmrt6 exhibited a sexually dimorphic expression pattern with higher expression levels in the testes than in the ovaries (p < 0.01, Figures 6A, G). IpDmrt2a, IpDmrt3, and IpDmrt4 were highly expressed in the kidney, head kidney, and liver of male channel catfish (p < 0.05, Figures 6B, D, E). In addition, a sexually dimorphic expression pattern was also detected in IpDmrt2b and IpDmrt5, which presented more significant expression levels in the ovaries than in other tissues (p < 0.05, Figures 6C, F). Also, a relatively high expression level of Dmrt5 was also detected in the kidney, head kidney, and liver of male channel catfish (p < 0.05, Figure 6F).
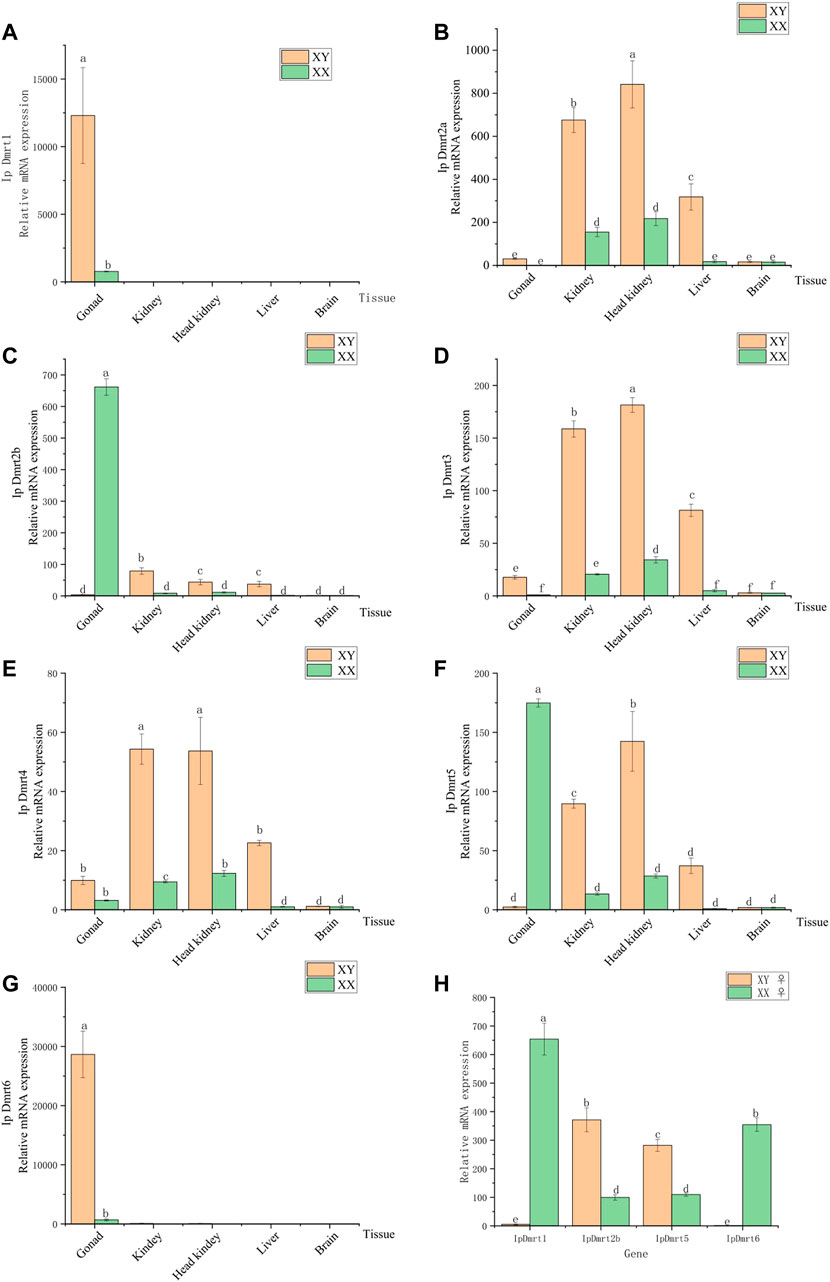
FIGURE 6. Results of the qRT-PCR assay for channel catfish Dmrt genes in testes and ovaries. The gene expression level was normalized to the α-tubulin transcript. Images (A), (B), (C), (D), (E), (F), and (G), show the Dmrt genes expression level in gonad, kidney, head kidney, liver, and brain. Images (H) show the Dmrt1, Dmrt2b, Dmrt5 and Dmrt6 genes expression level in gonad which are female (XX) and female (XY). In each image, different letters represent significant differences among lines (p < 0.05).
The qRT-PCR assay was performed to verify the response expression of IpDmrt genes during sex reversal. The expression of IpDmrt2a, IpDmrt3, and IpDmrt4 was not carried out because a sexually dimorphic expression pattern was not observed. Compared with XX channel catfish, IpDmrt2b and IpDmrt5 were significantly up-regulated in the ovaries of XY individuals, while IpDmrt1 and IpDmrt6 were significantly down-regulated. The results indicate that these Dmrt gene family members may play an important role during sex reversal in XY channel catfish.
Although it has been reported that the Dmrt family genes played vital roles in sex determination and differentiation in teleosts such as zebrafish (Graf et al., 2015; Webster et al., 2017), Chinese tongue sole (Cynoglossus semilaevis) (Cui et al., 2017; Feng et al., 2021), and spotted scat (Scatophagus argus) (Uma Farouk et al., 2018), to date, a thorough survey and research of the functions of Dmrt genes has not been conducted in channel catfish, even though its entire genome sequence has been available for several years (Chen et al., 2016; Ourth et al., 2017). In this study, a comprehensive genome-wide analysis identified a total of seven Dmrt genes in the channel catfish genome.
Previous reports have shown that the number of Dmrt genes range from five (zebrafish) to twelve (common carp), and this is correlated not only with genome size but also with genomic duplication rounds. For example, only five Dmrt genes were identified in Atlantic cod, which has a genome of 686 megabases (Mb) (Star et al., 2011; Johnsen and Andersen, 2012), while Atlantic salmon, whose genome size (2,970 Mb) is relatively large (Lien et al., 2016; Dong et al., 2020), has ten Dmrt genes. Both zebrafish and common carp (Cyprinus carpio), which belong to the order Cypriniformes, have relatively large genomes (>1,400 Mb) (Kerstin et al., 2013; De et al., 2018), but, as they underwent three and four rounds of genome replication, respectively, the former has only five Dmrt genes (Jocelyn, 2021), while the latter has 12. Compared with tetrapods, teleosts experienced three to four rounds of genome replication and seem to possess more Dmrt genes. However, Dmrt7 and Dmrt8 are present in mammals specifically, and only a couple of copy genes (Dmrt2a and Dmrt2b) are found in most 3R teleosts, which probably separated during the basal vertebrate genome duplication (2R).
The protein interaction network analysis showed that Dmrt genes have a close relationship with Wnt4, Foxl, Amh, and Cyp19a1. Amh (anti-Müllerian hormone) is considered necessary for female differentiation, because it regulates FSH to induce aromatase activity (Garg and Tal, 2016). Moreover, Amh is closely related to the development of spermatogonia by interacting with its Amhr2 (anti Müllerian hormone receptor type 2) receptor in gonads (Wu et al., 2017). The Foxl gene family members have different functions, and Foxl2 in particular is associated with ovary formation, as confirmed by numerous study (Perry et al., 2019). Another significant sex differentiation gene is Wnt4, which regulates the development of the Müllerian duct by controlling the secretion of sterol in mammals (Biason-Lauber and Konrad, 2008). Even if this structure is absent in teleosts, the genes that regulated its formation are present, and during this process they acquired a major and even more prominent role in sex determination (Adolfi et al., 2019). It has been proved that steroid hormones play crucial roles in the process of sex differentiation through indirect techniques, such as treatments with steroid hormones, steroid enzyme inhibitors, or steroid receptor antagonists (Paul-Prasanth et al., 2013). Cyp19a1 is a key gene for the balancing of the secretion levels of endogenous steroid hormones, and regulated by several upstream genes, including Dmrt1, Foxl2, Amh, and Wnt4.
In the present study, the expression levels of IpDmrt genes were detected in several tissues based on the qRT-PCR assay. Among these genes, IpDmrt1 and IpDmrt6 had a higher expression in the testes, while IpDmrt2b and IpDmrt5 had a higher expression in the ovaries, indicating that IpDmrt1/IpDmrt6 and IpDmrt2b/IpDmrt5 may have important regulatory roles in male and female sex differentiation/development, respectively. The expression levels of IpDmrt1, IpDmrt2b, IpDmrt5, and IpDmrt6 in the ovaries of normal and XY channel catfish treated with 17β-estradiol also confirmed our hypothesis; in particular, IpDmrt2b and IpDmrt5 were significantly up-regulated in XY channel catfish ovary, while IpDmrt1 and IpDmrt6 were significantly repressed. Numerous studies have proved that Dmrt1 plays a dominant role in male sex differentiation in avians, turtles, frogs, and teleosts (Bellefroid et al., 2013). However, Dmrt1 participates to antagonistic regulatory networks in association with Foxl2 to maintain the male fate in mammals. Dmrt6 is absent in most teleosts, such as zebrafish, medaka, fugu, and Atlantic salmon (Dong et al., 2020). This gene takes part in mammalian mitotic and meiotic developmental processes during spermatogenesis by repressing genes involved in spermatogonial differentiation and activating those required for meiotic prophase (Zhang T et al., 2014). It seems to have similar functions in bony fish; for example, Dmrt6 knockout in tilapia resulted in fewer spermatocytes and produced a lower level of serum 11-ketotestosterone (Zhang X et al., 2014). Tetrapods have only one Dmrt2 gene, while the majority of teleosts present at least two orthologous Dmrt2 genes, Dmrt2a and Dmrt2b, of which the former is expressed in various tissues and promotes the transition of endochondral bone formation by linking Sox9 and Runx2 (Ono et al., 2021). Dmrt2b is different from Dmrt2a, Dmrt2b is significantly expressed in teleosts’ ovaries and is critical for their development (Liu et al., 2009). Dmrt5 is required for the terminal differentiation of corticotropes and gonadotropes; this gene regulates corticotrope differentiation in the pituitary gland in a cell-autonomous manner, thereby determining gonadotrope numbers (Michaelidou et al., 2013).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
SX and SZ designed the study and carried out the analyses; XC and HL performed the technical process; SX, WZ, LZ and SZ prepared and drafted the article; MW and WB were involved in the material preparation. All authors read and approved the final article.
This research was funded by Natural Science Foundation of China (32102771); China Agriculture Research System of MOF and MARA (CARS-46); Important New Varieties Selection Project of Jiangsu Province (PZCZ201741) and Natural Science Foundation of Jiangsu Province (BK20191487).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to National Genetic Breeding Center of Channel Catfish for providing channel catfish.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.891204/full#supplementary-material
Adolfi, M. C., Nakajima, R. T., Nóbrega, R. H., and Schartl, M. (2019). Intersex, Hermaphroditism, and Gonadal Plasticity in Vertebrates: Evolution of the Müllerian Duct and Amh/Amhr2 Signaling. Annu. Rev. Anim. Biosci. 7, 149–172. doi:10.1146/annurev-animal-020518-114955
Bao, L., Tian, C., Liu, S., Zhang, Y., Elaswad, A., Yuan, Z., et al. (2019). The Y Chromosome Sequence of the Channel Catfish Suggests Novel Sex Determination Mechanisms in Teleost Fish. BMC Biol. 17 (1), 6. doi:10.1186/s12915-019-0627-7
Bellefroid, E. J., Leclère, L., Saulnier, A., Keruzore, M., Sirakov, M., Vervoort, M., et al. (2013). Expanding Roles for the Evolutionarily Conserved Dmrt Sex Transcriptional Regulators during Embryogenesis. Cell. Mol. Life Sci. 70, 3829–3845. doi:10.1007/s00018-013-1288-2
Biason-Lauber, A., and Konrad, D. (2008). WNT4 and Sex Development. Sex. Dev. 2, 210–218. doi:10.1159/000152037
Chen, L., Huang, R., Zhu, D., Yang, C., He, L., Li, Y., et al. (2019). Deep Sequencing of Small RNAs from 11 Tissues of Grass Carp Ctenopharyngodon Idella and Discovery of Sex-Related microRNAs. J. Fish. Biol. 94 (1), 132–141. doi:10.1111/jfb.13875
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole-genome Sequence of a Flatfish Provides Insights into ZW Sex Chromosome Evolution and Adaptation to a Benthic Lifestyle. Nat. Genet. 46 (3), 253–260. doi:10.1038/ng.2890
Chen, X., Zhong, L., Bian, C., Xu, P., Qiu, Y., You, X., et al. (2016). High-quality Genome Assembly of Channel Catfish, Ictalurus punctatus. GigaSci 5 (1), 39. doi:10.1186/s13742-016-0142-5
Cui, Z., Liu, Y., Wang, W., Wang, Q., Zhang, N., Lin, F., et al. (2017). Genome Editing Reveals Dmrt1 as an Essential Male Sex-Determining Gene in Chinese Tongue Sole (Cynoglossus Semilaevis). Sci. Rep. 7, 42213. doi:10.1038/srep42213
Date, S., Nozawa, O., Inoue, H., Hidema, S., and Nishimori, K. (2012). Impairment of Pachytene Spermatogenesis inDmrt7Deficient Mice, Possibly Causing Meiotic Arrest. Biosci. Biotechnol. Biochem. 76 (9), 1621–1626. doi:10.1271/bbb.120024
De Clercq, S., Keruzore, M., Desmaris, E., Pollart, C., Assimacopoulos, S., Preillon, J., et al. (2018). DMRT5 Together with DMRT3 Directly Controls hippocampus Development and Neocortical Area Map Formation. Cereb. Cortex 28 (2), 493–509. doi:10.1093/cercor/bhw384
Dong, J., Li, J., Hu, J., Sun, C., Tian, Y., Li, W., et al. (2020). Comparative Genomics Studies on the Dmrt Gene Family in Fish. Front. Genet. 11, 563947. doi:10.3389/fgene.2020.563947
Feng, B., Li, S., Wang, Q., Tang, L., Huang, F., Zhang, Z., et al. (2021). lncRNA DMRT2-AS Acts as a Transcriptional Regulator of Dmrt2 Involving in Sex Differentiation in the Chinese Tongue Sole (Cynoglossus Semilaevis). Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 253, 110542. doi:10.1016/j.cbpb.2020.110542
Garg, D., and Tal, R. (2016). The Role of AMH in the Pathophysiology of Polycystic Ovarian Syndrome. Reprod. Biomed. Online 33 (1), 15–28. doi:10.1016/j.rbmo.2016.04.007
Godwin, J., Sawby, R., Warner, R. R., Crews, D., and Grober, M. S. (2000). Hypothalamic Arginine Vasotocin mRNA Abundance Variation across Sexes and with Sex Change in a Coral Reef Fish. Brain Behav. Evol. 55 (2), 77–84. doi:10.1159/000006643
Graf, M., Teo Qi-Wen, E.-R., Sarusie, M. V., Rajaei, F., and Winkler, C. (2015). Dmrt5 Controls Corticotrope and Gonadotrope Differentiation in the Zebrafish Pituitary. Mol. Endocrinol. 29 (2), 187–199. doi:10.1210/me.2014-1176
Han, C., Wang, C., Ouyang, H., Zhu, Q., Huang, J., Han, L., et al. (2021). Characterization of Dmrts and Their Potential Role in Gonadal Development of Mandarin Fish (Siniperca chuatsi). Aquaculture Rep. 21, 2352–5134. doi:10.1016/j.aqrep.2021.100802
Hattori, R. S., Murai, Y., Oura, M., Masuda, S., Majhi, S. K., Sakamoto, T., et al. (2012). A Y-Linked Anti-müllerian Hormone Duplication Takes over a Critical Role in Sex Determination. Proc. Natl. Acad. Sci. U.S.A. 109 (8), 2955–2959. doi:10.1073/pnas.1018392109
Herpin, A., and Schartl, M. (2011). Dmrt1genes at the Crossroads: a Widespread and central Class of Sexual Development Factors in Fish. Fed. Eur. Biochem. Societies J. 278 (7), 1010–1019. doi:10.1111/j.1742-4658.2011.08030.x
Jocelyn, S. S. (2021). Characterizing the Role of the DMRT Gene Family in Zebrafish Sexual Development. Boston: University of Massachusetts Boston, 148.
Johnsen, H., and Andersen, Ø. (2012). Sex Dimorphic Expression of Five Dmrt Genes Identified in the Atlantic Cod Genome. The Fish-specific Dmrt2b Diverged from Dmrt2a before the Fish Whole-Genome Duplication. Gene 505 (2), 221–232. doi:10.1016/j.gene.2012.06.021
Kamiya, T., Kai, W., Tasumi, S., Oka, A., Matsunaga, T., Mizuno, N., et al. (2012). A Trans-species Missense SNP in Amhr2 Is Associated with Sex Determination in the Tiger Pufferfish, Takifugu rubripes (Fugu). Plos Genet. 8 (7), e1002798. doi:10.1371/journal.pgen.1002798
Kerstin, H., Matthew, D. C., Carlos, F. T., James, T., Camille, B., Matthieu, M., et al. (2013). The Zebrafish Reference Genome Sequence and its Relationship to the Human Genome. Nature 496, 498–503. doi:10.1038/nature12111
Kikkawa, T., and Osumi, N. (2021). Multiple Functions of the Dmrt Genes in the Development of the central Nervous System. Front. Neurosci. 15, 789583. doi:10.3389/fnins.2021.789583
Lien, S., Koop, B. F., Sandve, S. R., Miller, J. R., Kent, M. P., Nome, T., et al. (2016). The Atlantic salmon Genome Provides Insights into Rediploidization. Nature 533 (7602), 200–205. doi:10.1038/nature17164
Liu, S., Li, Z., and Gui, J.-F. (2009). Fish-Specific Duplicated Dmrt2b Contributes to a Divergent Function through Hedgehog Pathway and Maintains Left-Right Asymmetry Establishment Function. PLoS One 4 (9), e7261. doi:10.1371/journal.pone.0007261
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Matsuda, M., Nagahama, Y., Shinomiya, A., Sato, T., Matsuda, C., Kobayashi, T., et al. (2002). DMY Is a Y-specific DM-Domain Gene Required for Male Development in the Medaka Fish. Nature 417 (6888), 559–563. doi:10.1038/nature751
Michaelidou, K., Tzovaras, A., Missitzis, I., Ardavanis, A., and Scorilas, A. (2013). The Expression of the CEACAM19 Gene, a Novel Member of the CEA Family, Is Associated with Breast Cancer Progression. Int. J. Oncol. 42 (5), 1770–1777. doi:10.3892/ijo.2013.1860
Myosho, T., Otake, H., Masuyama, H., Matsuda, M., Kuroki, Y., Fujiyama, A., et al. (2012). Tracing the Emergence of a Novel Sex-Determining Gene in Medaka, Oryzias luzonensis. Genetics 191 (1), 163–170. doi:10.1534/genetics.111.137497
Nagahama, Y., Chakraborty, T., Paul-Prasanth, B., Ohta, K., and Nakamura, M. (2021). Sex Determination, Gonadal Sex Differentiation, and Plasticity in Vertebrate Species. Physiol. Rev. 101 (3), 1237–1308. doi:10.1152/physrev.00044.2019
Ono, K., Hata, K., Nakamura, E., Ishihara, S., Kobayashi, S., Nakanishi, M., et al. (2021). Dmrt2 Promotes Transition of Endochondral Bone Formation by Linking Sox9 and Runx2. Commun. Biol. 4 (1), 326. doi:10.1038/s42003-021-01848-1
Ortega-Recalde, O., Goikoetxea, A., Hore, T. A., Todd, E. V., and Gemmell, N. J. (2020). The Genetics and Epigenetics of Sex Change in Fish. Annu. Rev. Anim. Biosci. 8, 47–69. doi:10.1146/annurev-animal-021419-083634
Ourth, D., Marecaux, E., Raghu, D., and Peterson, B. (2017). Innate Immune Response of Channel Catfish Ictalurus punctatus Mannose-Binding Lectin to Channel Catfish Virus (CCV). Dis. Aquat. Org. 124 (2), 159–163. doi:10.3354/dao03109
Pan, N., Wang, M., Zhong, L., Bian, W., Chen, X., and Zhang, S. (2022). Identification of Male-specific SNP Markers and Development of Rapid PCR-Based Genetic Sex Identification Method in Channel Catfish (Ictalurus punctatus). Aquaculture 547, 737535. doi:10.1016/j.aquaculture.2021.737535
Paul-Prasanth, B., Bhandari, R. K., Kobayashi, T., Horiguchi, R., Kobayashi, Y., Nakamoto, M., et al. (2013). Estrogen Oversees the Maintenance of the Female Genetic Program in Terminally Differentiated Gonochorists. Sci. Rep. 3, 2862. doi:10.1038/srep02862
Perry, S., Larhammar, M., Vieillard, J., Nagaraja, C., Hilscher, M. M., Tafreshiha, A., et al. (2019). Characterization of Dmrt3-Derived Neurons Suggest a Role within Locomotor Circuits. J. Neurosci. 39 (10), 1771–1782. doi:10.1523/jneurosci.0326-18.2018
She, Z.-Y., and Yang, W.-X. (2017). Sry and SoxE Genes: How They Participate in Mammalian Sex Determination and Gonadal Development? Semin. Cel Developmental Biol. 63, 13–22. doi:10.1016/j.semcdb.2016.07.032
Shen, Z.-G., and Wang, H.-P. (2014). Molecular Players Involved in Temperature-dependent Sex Determination and Sex Differentiation in Teleost Fish. Genet. Selection Evol. 46 (1), 26. doi:10.1186/1297-9686-46-26
Star, B., Nederbragt, A. J., Jentoft, S., Grimholt, U., Malmstrøm, M., Gregers, T. F., et al. (2011). The Genome Sequence of Atlantic Cod Reveals a Unique Immune System. Nature 477 (7363), 207–210. doi:10.1038/nature10342
Takehana, Y., Matsuda, M., Myosho, T., Suster, M. L., Kawakami, K., Shin-I, T., et al. (2014). Co-option of Sox3 as the Male-Determining Factor on the Y Chromosome in the Fish Oryzias Dancena. Nat. Commun. 5, 4157. doi:10.1038/ncomms5157
Uma Farouk, M., Jiang, D. N., Liang, Z. H., Gu, H. T., Wei, Y., Chen, H. P., et al. (2018). Male-specific Dmrt1 Is a Candidate Sex Determination Gene in Spotted Scat (Scatophagus argus). Aquaculture 495, 351–358. doi:10.1016/j.aquaculture.2018.06.009
Veith, A.-M., Klattig, J., Dettai, A., Schmidt, C., Englert, C., and Volff, J.-N. (2006). Male-biased Expression of X-Chromosomal DM Domain-Less Dmrt8 Genes in the Mouse. Genomics 88 (2), 185–195. doi:10.1016/j.ygeno.2006.01.003
Wan, H.-F., Zhong, Z.-W., Jiang, Y.-H., Zou, P.-F., Zhang, Z.-P., and Wang, Y.-L. (2020). Genome-wide Investigation of Dmrt Gene Family in Large Yellow Croaker (Larimichthys Crocea). Theriogenology 156, 272–282. doi:10.1016/j.theriogenology.2020.07.010
Wang, J. H., Miao, L., Li, M. Y., Guo, X. F., Pan, N., Chen, Y. Y., et al. (2014). Cloning the Dmrt1 and DmrtA2 Genes of Ayu (Plecoglossus Altivelis) and Mapping Their Expression in Adult, Larval, and Embryonic Stages. Dongwuxue Yanjiu 35 (2), 99–107. doi:10.11813/j.issn.0254-5853.2014.2.099
Webster, K. A., Schach, U., Ordaz, A., Steinfeld, J. S., Draper, B. W., and Siegfried, K. R. (2017). Dmrt1 Is Necessary for Male Sexual Development in Zebrafish. Developmental Biol. 422 (1), 33–46. doi:10.1016/j.ydbio.2016.12.008
Wu, G.-C., Li, H.-W., Tey, W.-G., Lin, C.-J., and Chang, C.-F. (2017). Expression Profile of amh/Amh during Bi-directional Sex Change in the Protogynous orange-spotted Grouper Epinephelus coioides. PLoS One 12, e0185864. doi:10.1371/journal.pone.0185864
Xiao, Y.-M., Chen, L., Liu, J., Liu, W.-B., Chen, H.-G., Zou, L.-J., et al. (2009). Contrast Expression Patterns of JNK1 during Sex Reversal of the rice-field Eel. J. Exp. Zool. 9999B (3), a–n. doi:10.1002/jez.b.21332
Zhang, S., Li, Y., Shao, J., Liu, H., Wang, J., Wang, M., et al. (2020). Functional Identification and Characterization of IpMSTNa, a Novel Orthologous Myostatin (MSTN) Gene in Channel Catfish Ictalurus punctatus. Int. J. Biol. Macromolecules 152, 1–10. doi:10.1016/j.ijbiomac.2020.02.060
Zhang, S., Zhang, X., Chen, X., Xu, T., Wang, M., Qin, Q., et al. (2019). Construction of a High-Density Linkage Map and QTL Fine Mapping for Growth- and Sex-Related Traits in Channel Catfish (Ictalurus punctatus). Front. Genet. 10, 251. doi:10.3389/fgene.2019.00251
Zhang, T., Murphy, M. W., Gearhart, M. D., Bardwell, V. J., and Zarkower, D. (2014). The Mammalian Doublesex Homolog DMRT6 Coordinates the Transition between Mitotic and Meiotic Developmental Programs during Spermatogenesis. Development (Cambridge, England) 141 (19), 3662–3671. doi:10.1242/dev.113936
Zhang, X., Wang, H., Li, M., Cheng, Y., Jiang, D., Sun, L., et al. (2014). Isolation of Doublesex- and Mab-3-Related Transcription Factor 6 and its Involvement in Spermatogenesis in Tilapia1. Biol. Reprod. 91 (6), 136. doi:10.1095/biolreprod.114.121418
Zhong, L. Q., Wang, M. H., Chen, X. H., Zhang, S. Y., Jiang, H. C., and Bian, W. J. (2021). Current Status and Development Strategy of the Channel Catfish Industry in Jiangsu. Chin. Agric. Sci. Bull. 37 (17), 137–143.
Keywords: Dmrt gene family, sex reversal, channel catfish, sex determination, animal reproduction
Citation: Xu S, Zhang S, Zhang W, Liu H, Wang M, Zhong L, Bian W and Chen X (2022) Genome-Wide Identification, Phylogeny, and Expression Profile of the Dmrt (Doublesex and Mab-3 Related Transcription Factor) Gene Family in Channel Catfish (Ictalurus punctatus). Front. Genet. 13:891204. doi: 10.3389/fgene.2022.891204
Received: 07 March 2022; Accepted: 31 March 2022;
Published: 28 April 2022.
Edited by:
Hui Qiao, Chinese Academy of Fishery Sciences, ChinaReviewed by:
Yao Zheng, Chinese Academy of Fishery Sciences, ChinaCopyright © 2022 Xu, Zhang, Zhang, Liu, Wang, Zhong, Bian and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiyong Zhang, c2hpeW9uZ3poYW5nQGhvdG1haWwuY29t; Xiaohui Chen, Y3hpYW9odWk0MTZAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.