- 1Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education & Key Lab of Swine Genetics and Breeding of Ministry of Agriculture and Rural Affairs, Huazhong Agricultural University, Wuhan, China
- 2Institute for Animal Breeding and Genetics, University of Veterinary Medicine Hannover, Hannover, Germany
- 3Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, China
Introduction
The development of high-precision genome editing tools, such as targeted nucleases, has accelerated advances in fundamental human medicine, animal science, animal breeding, as well as disease diagnosis (Doudna and Charpentier, 2014; Kurtz et al., 2021; Rieblinger et al., 2021; Xie et al., 2021). In particular, the genome editing system known as CRISPR technology has grown rapidly since it was first reported (Jinek et al., 2012) and has become one of the most popular technologies. CRISPR/Cas9 technology can accurately identify target sequences and achieve efficient DNA cutting, thereby completing gene knock-outs/knock-ins on a genome-wide scale (Cong et al., 2013; Koike-Yusa et al., 2014).
However, due to double-strand breaks (DSBs) occurring during the editing process, this technology often introduces a large number of non-ideal InDel (insertion and deletion) mutations (Zhao et al., 2019). Subsequently, base editors (BEs), which can achieve precise editing of a single nucleotide using cytosine deaminase or adenosine deaminase without inducing DSB were developed (Gaudelli et al., 2017; Rees and Liu, 2018). Recently, prime editors (PEs) have further expanded the CRISPR-based-edit toolkit to all twelve possible base-to-base conversions, and insertion and deletion of short DNA fragments. This technology fuses reverse transcriptase and Cas9 protein, and uses a prime editing guide RNA (pegRNA) as the repair template to achieve precise gene editing (Anzalone et al., 2019). In this mini-review, we summarize and discuss recent applications of the CRISPR technology in pigs.
Gene-Edited Pigs for Human Biomedicine
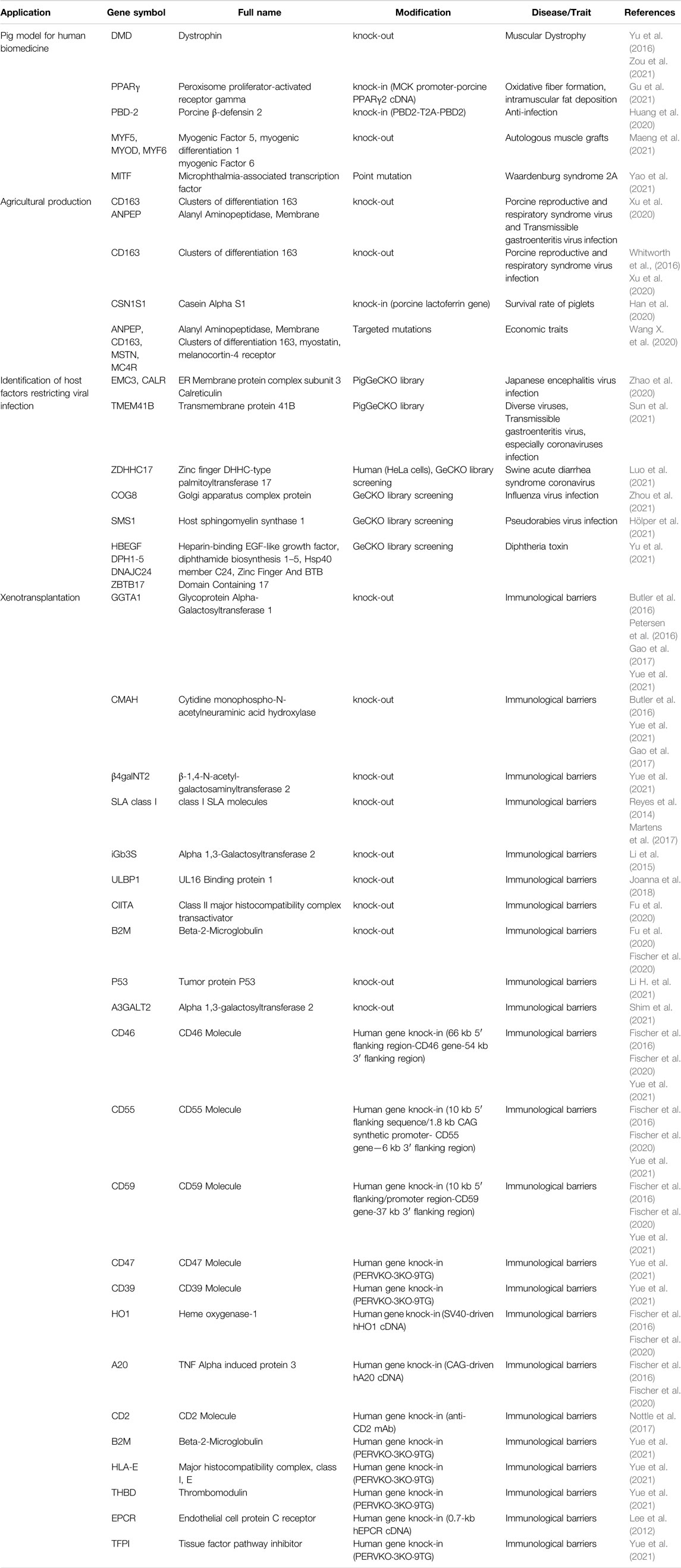
Pigs serve as an important agricultural resource and animal model in biomedical research. A variety of genetically modified pig models have been successfully generated through CRISPR-based technologies (Table 1) (Huang et al., 2020; Xu et al., 2020; Gu et al., 2021; Maeng et al., 2021; Yao et al., 2021; Yue et al., 2021; Xu et al., 2022). Duchenne muscular dystrophy (DMD) is an incurable X-linked inherited neuromuscular disorder and is caused by mutations in the dystrophin gene (DMD) (Hoffman et al., 1987). Studies in mdx (X-linked muscular dystrophy) mice, rats, dogs and monkey provided only a limited understanding of DMD disease mechanisms, as these possess different pathological manifestations from humans or cost highly (Nakamura et al., 2014; Chen et al., 2015; Nelson et al., 2016; Amoasii et al., 2018). Pigs (Sus scrofa) are closely related to humans in terms of anatomy, genetics and physiology. The generation of DMD knockout pig models using CRISPR/Cas9 technology may potentially pave the way for new treatments for patients (Yu et al., 2016; Zou et al., 2021).
Gene-Edited Pigs for Agricultural Production
CRISPR technology offers a new strategy to combat infectious diseases in pigs. Porcine reproductive and respiratory syndrome (PRRS) is one of the most economically important swine infectious diseases worldwide. CD163 was identified as the striking receptor in PRRSV entry, and by knocking it out from the genome or editing the receptor using CRISPR/Cas9, pigs fully resistant to PRRSV have been produced - a milestone in modern pig breeding (Whitworth et al., 2016; Burkard et al., 2018; Xu et al., 2020). Another study reported the construction of genome-edited pigs with marker-free site-specific knock-in of lactoferrin gene in the 3′-end of Casein alpha-s1 by CRISPR/Cas9-mediated homologous recombination (Han et al., 2020). Antibacterial activity of lactoferrin could potentially improve the survival rate of piglets in the genome-edited pigs (Han et al., 2020). There were abundant evidences that CRISPR-based technologies have great potential in human health and animal production.
CRISPR-Based Functional Genomics to Combat Infectious Diseases
CRISPR technology provides an easy way to introduce targeted mutations into mammalian cells to induce loss-of-function phenotypes (Doudna and Charpentier, 2014; Hsu et al., 2014; Ruan et al., 2017). Genome-wide CRISPR screen has now been successfully applied to identify host factors that restrict viral infections, providing a powerful tool for exploring functional genomics of virus-host interactions (Shalem et al., 2014; Hoffmann et al., 2021). To identify novel host-dependent factors, a porcine genome-scale CRISPR/Cas9 knockout (PigGeCKO) library was established and successfully used to identify several key genes (EMC3, CALR, etc.) related to Japanese encephalitis virus (JEV) infection (Zhao et al., 2020). Several reports have identified multiple host factors required for the entry of other viruses and toxins in pigs and humans by using the CRISPR screening strategy (Hölper et al., 2021; Luo et al., 2021; Sun et al., 2021; Yu et al., 2021; Zhou et al., 2021).
Emerging coronaviruses (CoVs) pose a severe threat to human and animal health worldwide. Through CRISPR screening, transmembrane protein 41B (TMEM41B) was identified as a critical host-dependency factor required for the replication of diverse viruses, especially coronaviruses (Sun et al., 2021). TMEM41B was found to be involved in the formation of SARS-CoV-2 and transmissible gastroenteritis virus (TGEV) replicative organelles (Sun et al., 2021). ZDHHC17 (zinc finger DHHC-type palmitoyltransferase 17) was identified as a potential drug target for swine acute diarrhea syndrome coronavirus (SADS-CoV) infection by genome-wide CRISPR knockout library screening in human HeLa cells (Luo et al., 2021).
Adopting the same strategy, the Golgi apparatus complex protein (COG8) was identified as a pivotal regulator of influenza virus infection (Zhou et al., 2021). Host sphingomyelin synthase 1 (SMS1) was also found to be involved in pseudorabies virus (PRV) infection when the gD-mediated entry pathway was blocked (Hölper et al., 2021). In addition, HBEGF (heparin-binding EGF-like growth factor), DPH1-5 (diphthamide biosynthesis 1–5), DNAJC24 (Hsp40 member C24), and ZBTB17 were determined as diphtheria toxin (DT) receptors (Yu et al., 2021). These are the key factors involved in the biosynthesis of diphthamide, which serves as the molecular target for DT (Yu et al., 2021). These data demonstrate that CRISPR screening strategy is a powerful tool for functional genome in livestock.
Furthermore, CRISPR technology can also be used to specifically target infectious viruses (Freije and Sabeti, 2021). African swine fever (ASF) is a highly contagious viral disease of swine, with a high mortality rate up to 100%. CRISPR/Cas9 has been successfully used to produce recombinant ASF virus (ASFV), which could help speed up vaccine production to combat the infectious virus (Abkallo et al., 2021). Indeed, the CRISPR/Cas9 in combination with Cre/Lox system has been used to develop a stable anti-pseudorabies virus (PRV) vaccine of pig (Liang et al., 2016). Vaccination and challenge experiments demonstrate that recombinant vaccine candidates generated by gene editing technology can provide immune protection in pigs (Liang et al., 2016). These studies showed that development of virus vaccine can be accelerated via CRISPR and synthetic biology technologies.
CRISPR-Based Diagnostics
The rapid detection of infectious diseases is highly needed in diagnosis and infection prevention (Pfaller, 2001; Hwang et al., 2018). CRISPR-based nucleic acid detection methods have suddenly emerged, with the potential to power the fields of genetic mutation and pathogen detection (Chen et al., 2018). This technology mainly employs Cas12, Cas13, and Cas14a, which have a target-activated trans-cleavage activity and can efficiently cleave single-stranded DNA (ssDNA) or single-stranded RNA (ssRNA) sequences (Gootenberg et al., 2017; Chen et al., 2018; Harrington et al., 2018).
To achieve point-of-care testing (POCT) of ASFV, a variety of sensitive diagnostic methods based on CRISPR technology have been established (He et al., 2020; Tao et al., 2020; Wang X. et al., 2020; Wu et al., 2020; Wei et al., 2022; Xie et al., 2022), for instance, recombinase-aided amplification (RAA)-Cas12a combined with lateral flow detection assay (Wang Y. et al., 2020), CRISPR/Cas12a based universal lateral flow biosensor assay (Wu et al., 2020), CRISPR/Cas12a enhanced fluorescence assay (Tao et al., 2020), CRISPR/Cas13 combined with lateral flow strip assay (Wei et al., 2022), as well as high-throughput and all-solution phase ASFV detection assay (He et al., 2020). Recently, to simplify the detection process, the rapid visual CRISPR assay (RAVI-CRISPR), combining a naked-eye colorimetric detection method based on CRISPR/Cas12a and a convolutional neural network was established (Xie et al., 2022). This RAVI-CRISPR/MagicEye mobile APP system is perhaps the today’s simplest platform for rapid POCT testing.
Porcine Genome Engineering for Xenotransplantation
The extreme shortage of human donor organs for the treatment of patients with end-stage organ failure is well known. Pig-to-human xenotransplantation is a most promising strategy to solve this problem, because domestic pigs are similar to humans in terms of anatomy, physiology and organ size, and are highly reproductive and low in maintenance costs (Hryhorowicz et al., 2017). However, discrepancies between pigs and humans lead to the development of immune barriers, blocking direct xenotransplantation (Vadori and Cozzi, 2015).
In the last decade, CRISPR technology accelerated the pace and extent of modifications to porcine genomes, such as knockout of major carbohydrate antigens (GGTA1, CMAH, β4galNT2) and tumor suppressor protein (p53), as well as knockin of various human complement regulatory proteins (e.g. CD46, CD55), human coagulation regulatory proteins (e.g. THBD, EPCR), human anti-inflammatory molecule (HO1), and human macrophage-inhibitory ligand (CD47), to modulate human immune response (Cooper et al., 2019; Li H. et al., 2021). These genetically modified pigs have been used in preclinical studies and greatly improved survival outcomes of xenografts of non-human primate recipients (Niu et al., 2021). In addition, multiplex CRISPR/Cas9 gene editing technology has enabled multi-fold knockouts of porcine genes in various combinations. Pigs carrying multi-fold xenoprotective transgenes and knockouts of xenoreactive antigens have been generated (Fischer et al., 2016; Zhang et al., 2018; Fischer et al., 2020; Fu et al., 2020; Shim et al., 2021; Yue et al., 2021), with great potential to completely eliminate immunological barriers. It remains a challenge, however, to effectively assess the human immune response induced by various genetic modifications and to identify the ideal gene combinations (Li P. et al., 2021). Recently, the world’s first porcine-to-human transplantation was performed at the University of Maryland Medical Center, successfully transplanting a genetically modified porcine heart into a 57-year-old man with end-stage heart disease, and the patient lived for two months after the transplant (Shah and Han, 2022). The advent of the CRISPR system has accelerated the field, bringing the successful application of xenotransplantation closer to reality.
Conclusion and Regulation of CRISPR Development
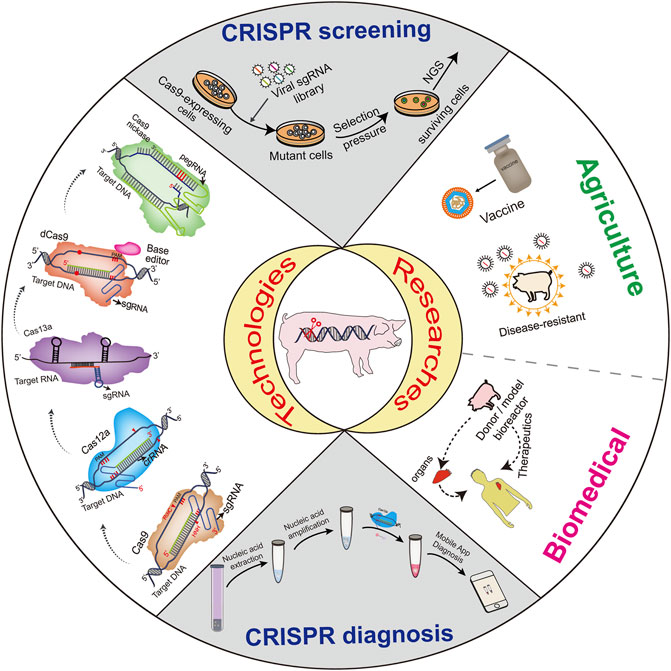
CRISPR, a sequence-specific nuclease able to edit target gene sequences, has ignited a revolution in the field of genetic engineering and site-specific editing within malfunctioning genes (Hsu et al., 2014). The system’s efficiency, robustness, and affordability allow its application to endless potential genetic targets (Figure 1). The use of CRISPR in genetic disorders, infectious diseases, defective traits and immunological barriers via gene knockout, gene knockin and gene editing has immense potential for the development of animal production, human medicine and Xenotransplantation (Doudna and Charpentier, 2014; Hsu et al., 2014; Ruan et al., 2017; Shah and Han, 2022). CRISPR technology has also been extensively employed to develop rapid point-of-care detection methods for viruses (Xie et al., 2022), with great potential in combating infectious diseases such as CoVs and ASFV. Additionally, the technology exerts important roles in clarifying the pathways of virus-host interactions, and generating recombinant viruses to speed up vaccine production. Future applications of CRISPR will enhance the quality and quantity of gene therapy and animal production, improve human health and animal welfare and will save countless lives.
Gene editing regulations for animals have not yet been globally established and vary greatly between countries. China’s regulations on genetically modified organisms (“GMOs”) mainly focus on Agricultural GMOs. In the U.S., genetically modified crops are regulated by the U.S. Department of Agriculture, which is relaxing its oversight of gene editing. While animal biotechnology is regulated by the Food and Drug Administration (FDA) under an unusual reading of the Federal Food, Drug, and Cosmetic Act of 1938, and gene editing is very strictly regulated by the FDA. In our opinion, using CRISPR technology, we can create an advanced animal that is essentially identical to the original one in all respects. Nevertheless, it is important to establish sound laws and regulations on CRISPR in the worldwide scientific community and between government agencies globally. Despite all risks, we believe that the application of CRISPR will provide benefits for everyone in the not far-distant future.
Insights and Prospects
The rapid development of life science has brought us from the “reading” stage of biological genetic information to the post-genome era, in which “rewriting” and even “new design” of genomes are gradually becoming a reality. Synthetic biology, which aims to design and create new living organisms, has developed rapidly under this background and has shown great promise for applications in biomedicine, agriculture, vaccines, manufacturing, and energy. In continuous exploration and research, gene editing technologies, especially CRISPR, have evolved from initial reliance on naturally occurring homologous recombination in cells to targeted cleavage at almost any site, and even to nucleic acid-based diagnostics. The simplicity and efficiency of its operation has greatly facilitated the genetic modification of species and disease diagnosis. Gene editing provides the means for continued modification of synthetic life and opens up more possibilities for the creation of new species through genetic modification. De novo genome synthesis and the large-scale modifications of natural genomes belong to the fields of synthetic genomics and gene editing (Xie et al., 2017), both subjects are current hot spots topics in synthetic biology research.
Since Science magazine named CRISPR technology the breakthrough of the year in 2015, this new technology has taken the gene-editing field by storm. In the past few years, CRISPR technology has rapidly swept the animal world as a popular gene editing technique. Although the research and application of gene editing technology has been developing rapidly, gene editing technology still faces challenges in terms of off-target, ethics and safety. The future development of gene editing technology needs to pay attention to the following aspects: first, strengthen planning and guidance, and attach great importance to strengthening research on basic theories and innovative methods of gene editing; second, strengthen supervision and scientific guidance, and pay attention to the applications of gene editing; third, strengthen research on ethical norms, improve the legal and policy system for gene editing supervision, and vigorously support the research and development of animal gene editing products; fourth, strengthen the popularization of science, let more people understand and accept gene editing technology, so that gene editing can better benefit mankind.
Author Contributions
SX, XZ: writing-original draft, review and editing. JR: writing-original draft. SZ: writing-review and editing. All authors have proof-read the final version.
Funding
This work was supported by the Natural Science Foundation of China (No: 32072685), the Major Project of Hubei Hongshan Laboratory (No: 2021hszd019) and the Laboratory of Lingnan Modern Agriculture Project (No: NZ2021005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abkallo, H. M., Svitek, N., Oduor, B., Awino, E., Henson, S. P., Oyola, S. O., et al. (2021). Rapid CRISPR/Cas9 Editing of Genotype IX African Swine Fever Virus Circulating in Eastern and Central Africa. Front. Genet. 12, 733674. doi:10.3389/fgene.2021.733674
Amoasii, L., Hildyard, J. C. W., Li, H., Sanchez-Ortiz, E., Mireault, A., Caballero, D., et al. (2018). Gene Editing Restores Dystrophin Expression in a Canine Model of Duchenne Muscular Dystrophy. Science 362, 86–91. doi:10.1126/science.aau1549
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 576, 149–157. doi:10.1038/s41586-019-1711-4
Burkard, C., Opriessnig, T., Mileham, A. J., Stadejek, T., Ait-Ali, T., Lillico, S. G., et al. (2018). Pigs Lacking the Scavenger Receptor Cysteine-Rich Domain 5 of CD163 Are Resistant to Porcine Reproductive and Respiratory Syndrome Virus 1 Infection. J. Virol. 92 (16), e00415–18. doi:10.1128/JVI.00415-18
Butler, J. R., Martens, G. R., Li, P., Wang, Z.-Y., Estrada, J. L., Ladowski, J. M., et al. (2016). The Fate of Human Platelets Exposed to Porcine Renal Endothelium: A Single-Pass Model of Platelet Uptake in Domestic and Genetically Modified Porcine Organs. J. Surg. Res. 200, 698–706. doi:10.1016/j.jss.2015.08.034
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., et al. (2018). CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 360, 436–439. doi:10.1126/science.aar6245
Chen, Y., Zheng, Y., Kang, Y., Yang, W., Niu, Y., Guo, X., et al. (2015). Functional Disruption of the Dystrophin Gene in Rhesus Monkey Using CRISPR/Cas9. Hum. Mol. Genet. 24, 3764–3774. doi:10.1093/hmg/ddv120
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. doi:10.1126/science.1231143
Cooper, D. K. C., Hara, H., Iwase, H., Yamamoto, T., Li, Q., Ezzelarab, M., et al. (2019). Justification of Specific Genetic Modifications in Pigs for Clinical Organ Xenotransplantation. Xenotransplantation 26, e12516. doi:10.1111/xen.12516
Doudna, J. A., and Charpentier, E. (2014). The New Frontier of Genome Engineering with CRISPR-Cas9. Science 346, 1258096. doi:10.1126/science.1258096
Fischer, K., Kraner-Scheiber, S., Petersen, B., Rieblinger, B., Buermann, A., Flisikowska, T., et al. (2016). Efficient Production of Multi-Modified Pigs for Xenotransplantation by 'combineering', Gene Stacking and Gene Editing. Sci. Rep. 6, 29081. doi:10.1038/srep29081
Fischer, K., Rieblinger, B., Hein, R., Sfriso, R., Zuber, J., Fischer, A., et al. (2020). Viable Pigs after Simultaneous Inactivation of Porcine MHC Class I and Three Xenoreactive Antigen Genes GGTA1, CMAH and B4GALNT2. Xenotransplantation 27, e12560. doi:10.1111/xen.12560
Freije, C. A., and Sabeti, P. C. (2021). Detect and Destroy: CRISPR-Based Technologies for the Response against Viruses. Cell. Host Microbe 29, 689–703. doi:10.1016/j.chom.2021.04.003
Fu, R., Fang, M., Xu, K., Ren, J., Zou, J., Su, L., et al. (2020). Generation of GGTA1−/−β2M−/−CIITA−/− Pigs Using CRISPR/Cas9 Technology to Alleviate Xenogeneic Immune Reactions. Transplantation 104, 1566–1573. doi:10.1097/TP.0000000000003205
Gao, H., Zhao, C., Xiang, X., Li, Y., Zhao, Y., Li, Z., et al. (2017). Production of α1,3-galactosyltransferase and Cytidine Monophosphate-N-Acetylneuraminic Acid Hydroxylase Gene Double-Deficient Pigs by CRISPR/Cas9 and Handmade Cloning. J. Reproduction Dev. 63, 17–26. doi:10.1262/jrd.2016-079
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable Base Editing of at to GC in Genomic DNA without DNA Cleavage. Nature 551, 464–471. doi:10.1038/nature24644
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. doi:10.1126/science.aam9321
Gu, H., Zhou, Y., Yang, J., Li, J., Peng, Y., Zhang, X., et al. (2021). Targeted Overexpression of PPARγ in Skeletal Muscle by Random Insertion and CRISPR/Cas9 Transgenic Pig Cloning Enhances Oxidative Fiber Formation and Intramuscular Fat Deposition. FASEB J. 35, e21308. doi:10.1096/fj.202001812RR
Han, X., Gao, Y., Li, G., Xiong, Y., Zhao, C., Ruan, J., et al. (2020). Enhancing the Antibacterial Activities of Sow Milk via Site-specific Knock-In of a Lactoferrin Gene in Pigs Using CRISPR/Cas9 Technology. Cell. Biosci. 10, 133. doi:10.1186/s13578-020-00496-y
Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., Witte, I. P., et al. (2018). Programmed DNA Destruction by Miniature CRISPR-Cas14 Enzymes. Science 362, 839–842. doi:10.1126/science.aav4294
He, Q., Yu, D., Bao, M., Korensky, G., Chen, J., Shin, M., et al. (2020). High-throughput and All-Solution Phase African Swine Fever Virus (ASFV) Detection Using CRISPR-Cas12a and Fluorescence Based Point-Of-Care System. Biosens. Bioelectron. 154, 112068. doi:10.1016/j.bios.2020.112068
Hoffman, E. P., Brown, R. H., and Kunkel, L. M. (1987). Dystrophin: the Protein Product of the Duchenne Muscular Dystrophy Locus. Cell. 51, 919–928. doi:10.1016/0092-8674(87)90579-4
Hoffmann, H.-H., Schneider, W. M., Rozen-Gagnon, K., Miles, L. A., Schuster, F., Razooky, B., et al. (2021). TMEM41B Is a Pan-Flavivirus Host Factor. Cell. 184, 133–148.e120. doi:10.1016/j.cell.2020.12.005
Hölper, J. E., Grey, F., Baillie, J. K., Regan, T., Parkinson, N. J., Höper, D., et al. (2021). A Genome-wide CRISPR/Cas9 Screen Reveals the Requirement of Host Sphingomyelin Synthase 1 for Infection with Pseudorabies Virus Mutant gD-Pass. Viruses 13, 1574. Viruses 13. doi:10.3390/v13081574
Hryhorowicz, M., Zeyland, J., Słomski, R., and Lipiński, D. (2017). Genetically Modified Pigs as Organ Donors for Xenotransplantation. Mol. Biotechnol. 59, 435–444. doi:10.1007/s12033-017-0024-9
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 157, 1262–1278. doi:10.1016/j.cell.2014.05.010
Huang, J., Wang, A., Huang, C., Sun, Y., Song, B., Zhou, R., et al. (2020). Generation of Marker-free Pbd-2 Knock-In Pigs Using the CRISPR/Cas9 and Cre/loxP Systems. Genes. 11, 951. doi:10.3390/genes11080951
Hwang, H., Hwang, B.-Y., and Bueno, J. (2018). Biomarkers in Infectious Diseases. Dis. Markers 2018, 1–2. doi:10.1155/2018/8509127
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821. doi:10.1126/science.1225829
Joanna, Z., Magdalena, H., Agnieszka, N.-T., Jacek, J., Ryszard, S., Zdzisław, S., et al. (2018). The Production of UL16-Binding Protein 1 Targeted Pigs Using CRISPR Technology. 3 Biotech. 8, 70. doi:10.1007/s13205-018-1107-4
Koike-Yusa, H., Li, Y., Tan, E.-P., Velasco-Herrera, M. D. C., and Yusa, K. (2014). Genome-wide Recessive Genetic Screening in Mammalian Cells with a Lentiviral CRISPR-Guide RNA Library. Nat. Biotechnol. 32, 267–273. doi:10.1038/nbt.2800
Kurtz, S., Lucas-Hahn, A., Schlegelberger, B., Göhring, G., Niemann, H., Mettenleiter, T. C., et al. (2021). Knockout of the HMG Domain of the Porcine SRY Gene Causes Sex Reversal in Gene-Edited Pigs. Proc. Natl. Acad. Sci. U.S.A. 118. doi:10.1073/pnas.2008743118
Lee, K. F. E., Lu, B., Roussel, J. C., Murray-Segal, L. J., Salvaris, E. J., Hodgkinson, S. J., et al. (2012). Protective Effects of Transgenic Human Endothelial Protein C Receptor Expression in Murine Models of Transplantation. Am. J. Transpl. 12 (9), 2363–2372. doi:10.1111/j.1600-6143.2012.04122.x
Li, H., Cheng, W., Chen, B., Pu, S., Fan, N., Zhang, X., et al. (2021). Efficient Generation of P53 Biallelic Mutations in Diannan Miniature Pigs Using RNA-Guided Base Editing. Life 11 (12), 1417. doi:10.3390/life11121417
Li, P., Estrada, J. L., Burlak, C., Montgomery, J., Butler, J. R., Santos, R. M., et al. (2015). Efficient Generation of Genetically Distinct Pigs in a Single Pregnancy Using Multiplexed Single-Guide RNA and Carbohydrate Selection. Xenotransplantation 22, 20–31. doi:10.1111/xen.12131
Li, P., Walsh, J. R., Lopez, K., Isidan, A., Zhang, W., Chen, A. M., et al. (2021). Genetic Engineering of Porcine Endothelial Cell Lines for Evaluation of Human-To-Pig Xenoreactive Immune Responses. Sci. Rep. 11 (1), 13131. doi:10.1038/s41598-021-92543-y
Liang, X., Sun, L., Yu, T., Pan, Y., Wang, D., Hu, X., et al. (2016). A CRISPR/Cas9 and Cre/Lox System-Based Express Vaccine Development Strategy against Re-emerging Pseudorabies Virus. Sci. Rep. 6, 19176. doi:10.1038/srep19176
Luo, Y., Tan, C. W., Xie, S.-Z., Chen, Y., Yao, Y.-L., Zhao, K., et al. (2021). Identification of ZDHHC17 as a Potential Drug Target for Swine Acute Diarrhea Syndrome Coronavirus Infection. mBio 12, e0234221. doi:10.1128/mBio.02342-21
Maeng, G., Das, S., Greising, S. M., Gong, W., Singh, B. N., Kren, S., et al. (2021). Humanized Skeletal Muscle in MYF5/MYOD/MYF6-null Pig Embryos. Nat. Biomed. Eng. 5, 805–814. doi:10.1038/s41551-021-00693-1
Martens, G. R., Reyes, L. M., Butler, J. R., Ladowski, J. M., Estrada, J. L., Sidner, R. A., et al. (2017). Humoral Reactivity of Renal Transplant-Waitlisted Patients to Cells from GGTA1/CMAH/B4GalNT2, and SLA Class I Knockout Pigs. Transplantation 101, e86–e92. doi:10.1097/TP.0000000000001646
Nakamura, K., Fujii, W., Tsuboi, M., Tanihata, J., Teramoto, N., Takeuchi, S., et al. (2014). Generation of Muscular Dystrophy Model Rats with a CRISPR/Cas System. Sci. Rep. 4, 5635. doi:10.1038/srep05635
Nelson, C. E., Hakim, C. H., Ousterout, D. G., Thakore, P. I., Moreb, E. A., Rivera, R. M. C., et al. (2016). In Vivo genome Editing Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Science 351, 403–407. doi:10.1126/science.aad5143
Niu, D., Ma, X., Yuan, T., Niu, Y., Xu, Y., Sun, Z., et al. (2021). Porcine Genome Engineering for Xenotransplantation. Adv. Drug Deliv. Rev. 168, 229–245. doi:10.1016/j.addr.2020.04.001
Nottle, M. B., Salvaris, E. J., Fisicaro, N., McIlfatrick, S., Vassiliev, I., Hawthorne, W. J., et al. (2017). Targeted Insertion of an Anti-CD2 Monoclonal Antibody Transgene into the GGTA1 Locus in Pigs Using FokI-dCas9. Sci. Rep. 7, 8383. doi:10.1038/s41598-017-09030-6
Petersen, B., Frenzel, A., Lucas-Hahn, A., Herrmann, D., Hassel, P., Klein, S., et al. (2016). Efficient Production of biallelicGGTA1knockout Pigsbycytoplasmic Microinjection of CRISPR/Cas9 into Zygotes. Xenotransplantation 23, 338–346. doi:10.1111/xen.12258
Pfaller, M. (2001). Molecular Approaches to Diagnosing and Managing Infectious Diseases: Practicality and Costs. Emerg. Infect. Dis. 7, 312–318. doi:10.3201/eid0702.010234
Rees, H. A., and Liu, D. R. (2018). Base Editing: Precision Chemistry on the Genome and Transcriptome of Living Cells. Nat. Rev. Genet. 19, 770–788. doi:10.1038/s41576-018-0059-1
Reyes, L. M., Estrada, J. L., Wang, Z. Y., Blosser, R. J., Smith, R. F., Sidner, R. A., et al. (2014). Creating Class I MHC-Null Pigs Using Guide RNA and the Cas9 Endonuclease. J. I. 193, 5751–5757. doi:10.4049/jimmunol.1402059
Rieblinger, B., Sid, H., Duda, D., Bozoglu, T., Klinger, R., Schlickenrieder, A., et al. (2021). Cas9-expressing Chickens and Pigs as Resources for Genome Editing in Livestock. Proc. Natl. Acad. Sci. U.S.A. 118. doi:10.1073/pnas.2022562118
Ruan, J., Xu, J., Chen-Tsai, R. Y., and Li, K. (2017). Genome Editing in Livestock: Are We Ready for a Revolution in Animal Breeding Industry? Transgenic Res. 26, 715–726. doi:10.1007/s11248-017-0049-7
Shah, A. M., and Han, J. J. (2022). First Successful Porcine to Human Heart Transplantation Performed in the United States. Artif. Organs 46 (4), 543–545. doi:10.1111/aor.14203
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelsen, T. S., et al. (2014). Genome-scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 343, 84–87. doi:10.1126/science.1247005
Shim, J., Ko, N., Kim, H.-J., Lee, Y., Lee, J.-W., Jin, D.-I., et al. (2021). Human Immune Reactivity of GGTA1/CMAH/A3GALT2 Triple Knockout Yucatan Miniature Pigs. Transgenic Res. 30 (5), 619–634. doi:10.1007/s11248-021-00271-w
Sun, L., Zhao, C., Fu, Z., Fu, Y., Su, Z., Li, Y., et al. (2021). Genome-scale CRISPR Screen Identifies TMEM41B as a Multi-Function Host Factor Required for Coronavirus Replication. PLoS Pathog. 17, e1010113. doi:10.1371/journal.ppat.1010113
Tao, D., Liu, J., Nie, X., Xu, B., Tran-Thi, T.-N., Niu, L., et al. (2020). Application of CRISPR-Cas12a Enhanced Fluorescence Assay Coupled with Nucleic Acid Amplification for the Sensitive Detection of African Swine Fever Virus. ACS Synth. Biol. 9, 2339–2350. doi:10.1021/acssynbio.0c00057
Vadori, M., and Cozzi, E. (2015). The Immunological Barriers to Xenotransplantation. Tissue Antigens 86, 239–253. doi:10.1111/tan.12669
Wang, X., Ji, P., Fan, H., Dang, L., Wan, W., Liu, S., et al. (2020). CRISPR/Cas12a Technology Combined with Immunochromatographic Strips for Portable Detection of African Swine Fever Virus. Commun. Biol. 3, 62. doi:10.1038/s42003-020-0796-5
Wang, Y., Bi, D., Qin, G., Song, R., Yao, J., Cao, C., et al. (2020). Cytosine Base Editor (hA3A-BE3-Ng)-Mediated Multiple Gene Editing for Pyramid Breeding in Pigs. Front. Genet. 11, 592623. doi:10.3389/fgene.2020.592623
Wei, N., Zheng, B., Niu, J., Chen, T., Ye, J., Si, Y., et al. (2022). Rapid Detection of Genotype II African Swine Fever Virus Using CRISPR Cas13a-Based Lateral Flow Strip. Viruses 14, 179. Viruses 14. doi:10.3390/v14020179
Whitworth, K. M., Rowland, R. R. R., Ewen, C. L., Trible, B. R., Kerrigan, M. A., Cino-Ozuna, A. G., et al. (2016). Gene-edited Pigs Are Protected from Porcine Reproductive and Respiratory Syndrome Virus. Nat. Biotechnol. 34, 20–22. doi:10.1038/nbt.3434
Wu, J., Mukama, O., Wu, W., Li, Z., Habimana, J. D. D., Zhang, Y., et al. (2020). A CRISPR/Cas12a Based Universal Lateral Flow Biosensor for the Sensitive and Specific Detection of African Swine-Fever Viruses in Whole Blood. Biosensors 10, 203. doi:10.3390/bios10120203
Xie, B., Shi, X., Li, Y., Xia, B., Zhou, J., Du, M., et al. (2021). Deficiency of ASGR1 in Pigs Recapitulates Reduced Risk Factor for Cardiovascular Disease in Humans. PLoS Genet. 17, e1009891. doi:10.1371/journal.pgen.1009891
Xie, S., Tao, D., Fu, Y., Xu, B., Tang, Y., Steinaa, L., et al. (2022). Rapid Visual CRISPR Assay: A Naked-Eye Colorimetric Detection Method for Nucleic Acids Based on CRISPR/Cas12a and a Convolutional Neural Network. ACS Synth. Biol. 11, 383–396. doi:10.1021/acssynbio.1c00474
Xie, Z. X., Li, B. Z., Mitchell, L. A., Wu, Y., Qi, X., Jin, Z., et al. (2017). "Perfect" Designer Chromosome V and Behavior of a Ring Derivative. Science 355 (6329), eaaf4704. doi:10.1126/science.aaf4704
Xu, K., Zhang, X., Liu, Z., Ruan, J., Xu, C., Che, J., et al. (2022). A Transgene-free Method for Rapid and Efficient Generation of Precisely Edited Pigs without Monoclonal Selection. Sci. China Life Sci., 1–12. doi:10.1007/s11427-021-2058-2
Xu, K., Zhou, Y., Mu, Y., Liu, Z., Hou, S., Xiong, Y., et al. (2020). CD163 and pAPN Double-Knockout Pigs Are Resistant to PRRSV and TGEV and Exhibit Decreased Susceptibility to PDCoV while Maintaining Normal Production Performance. Elife 9. doi:10.7554/eLife.57132
Yao, J., Wang, Y., Cao, C., Song, R., Bi, D., Zhang, H., et al. (2021). CRISPR/Cas9-mediated Correction of MITF Homozygous Point Mutation in a Waardenburg Syndrome 2A Pig Model. Mol. Ther. - Nucleic Acids 24, 986–999. doi:10.1016/j.omtn.2021.04.009
Yu, C., Zhong, H., Yang, X., Li, G., Wu, Z., and Yang, H. (2021). Establishment of a Pig CRISPR/Cas9 Knockout Library for Functional Gene Screening in Pig Cells. Biotechnol. J., 2100408. doi:10.1002/biot.202100408
Yu, H.-H., Zhao, H., Qing, Y.-B., Pan, W.-R., Jia, B.-Y., Zhao, H.-Y., et al. (2016). Porcine Zygote Injection with Cas9/sgRNA Results in DMD-Modified Pig with Muscle Dystrophy. Ijms 17, 1668. doi:10.3390/ijms17101668
Yue, Y., Xu, W., Kan, Y., Zhao, H.-Y., Zhou, Y., Song, X., et al. (2021). Extensive Germline Genome Engineering in Pigs. Nat. Biomed. Eng. 5, 134–143. doi:10.1038/s41551-020-00613-9
Zhang, R., Wang, Y., Chen, L., Wang, R., Li, C., Li, X., et al. (2018). Reducing Immunoreactivity of Porcine Bioprosthetic Heart Valves by Genetically-Deleting Three Major Glycan Antigens, GGTA1/β4GalNT2/CMAH. Acta Biomater. 72, 196–205. doi:10.1016/j.actbio.2018.03.055
Zhao, C., Liu, H., Xiao, T., Wang, Z., Nie, X., Li, X., et al. (2020). CRISPR Screening of Porcine sgRNA Library Identifies Host Factors Associated with Japanese Encephalitis Virus Replication. Nat. Commun. 11, 5178. doi:10.1038/s41467-020-18936-1
Zhao, C., Wang, Y., Nie, X., Han, X., Liu, H., Li, G., et al. (2019). Evaluation of the Effects of Sequence Length and Microsatellite Instability on Single-Guide RNA Activity and Specificity. Int. J. Biol. Sci. 15, 2641–2653. doi:10.7150/ijbs.37152
Zhou, A., Zhang, W., Dong, X., and Tang, B. (2021). Porcine Genome-wide CRISPR Screen Identifies the Golgi Apparatus Complex Protein COG8 as a Pivotal Regulator of Influenza Virus Infection. Crispr J. 4, 872–883. doi:10.1089/crispr.2021.0054
Keywords: CRISPR, functional genomics, pig, nucleic acid detection, human biomedical model
Citation: Ruan J, Zhang X, Zhao S and Xie S (2022) Advances in CRISPR-Based Functional Genomics and Nucleic Acid Detection in Pigs. Front. Genet. 13:891098. doi: 10.3389/fgene.2022.891098
Received: 07 March 2022; Accepted: 16 May 2022;
Published: 31 May 2022.
Edited by:
Mike McGrew, University of Edinburgh, United KingdomReviewed by:
Channabasavaiah Gurumurthy, University of Nebraska Medical Center, United StatesBjoern Petersen, Friedrich Loeffler Institute (FLI), Germany
Copyright © 2022 Ruan, Zhang, Zhao and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengsong Xie, c3N4aWVAbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jinxue Ruan
Jinxue Ruan Xuying Zhang
Xuying Zhang Shuhong Zhao
Shuhong Zhao Shengsong Xie
Shengsong Xie