- 1Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan
- 2Department of Ophthalmology, Changhua Christian Hospital, Changhua, Taiwan
- 3School of Medicine, Chung Shan Medical University, Taichung, Taiwan
- 4Department of Internal Medicine, Division of Endocrinology and Metabolism, Chung Shan Medical University Hospital, Taichung, Taiwan
- 5Department of Ophthalmology, Chung Shan Medical University Hospital, Taichung, Taiwan
- 6Department of Ophthalmology, Nobel Eye Institute, Taipei, Taiwan
- 7Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan
Long noncoding RNAs (lncRNAs) have been proven to play critical roles in diabetic retinopathy (DR). This study investigated whether the single nucleotide polymorphism (SNP) of long intergenic noncoding RNA 00673 (LINC00673) affects the clinical characteristics of diabetic retinopathy (DR). A total of three loci of LINC00673 SNPs (rs6501551, rs9914618, and rs11655237) were genotyped using TaqMan allelic discrimination in 276 and 454 individuals with and without DR, respectively. Our results revealed that LINC00673 SNP rs11655237 CT genotype (AOR: 1.592, 95% CI: 1.059–2.395, p = 0.026), CT + TT genotype (AOR: 1.255, 95% CI: 1.029–1.531, p = 0.025), and allele T (AOR: 1.185, 95% CI: 1.004–1.397, p = 0.044) yielded higher ratios in the non-proliferative diabetic retinopathy (NPDR) subgroup than in the non-DR group. Furthermore, the interval of diabetes mellitus (DM) was significantly shorter in the LINC00673 SNP rs11655237 CT + TT variant than that in the LINC00673 SNP rs11655237 wild type (10.44 ± 7.10 vs. 12.98 ± 8.34, p = 0.009). In conclusion, the LINC00673 SNP rs11655237 T allele is associated with a higher probability of NPDR development. Patients with the LINC00673 SNP rs11655237 CT + TT variant exhibited a short DM interval.
Introduction
Diabetes mellitus (DM) affects more than 9% of individuals worldwide and can lead to various vascular disorders (Zheng et al., 2018). Diabetic retinopathy (DR) is a complication of DM that is characterized by vascular damage and fluid leakage in the retina. DR can be further divided into non-proliferative diabetic retinopathy (NPDR) and proliferative diabetic retinopathy (PDR), according to the presence of retinal neovascularization (Antonetti et al., 2012). Advanced PDR can cause other ocular diseases, including tractional retinal detachment, vitreous hemorrhage, and neovascular glaucoma (Cheung et al., 2010; Sabanayagam et al., 2019). A previous study estimated that nearly 2.5 and 1.4% of the global cases of blindness and moderate-to-severe visual impairment, respectively, resulted from DR (Flaxman et al., 2017).
The etiology of DR development is multifactorial (Cheung et al., 2010; Antonetti et al., 2012). Apart from DM duration and the condition of blood sugar control (defined as the concentration of glycated hemoglobin [HbA1c] (Sabanayagam et al., 2019; Ghamdi, 2020), biomarkers can also influence the occurrence of DR (Jenkins et al., 2015). Inflammatory cytokines such as interleukins are involved in the development of DR and diabetic macular edema (Capitão and Soares, 2016). Another prominent component in the development of DR is vascular endothelial growth factor (VEGF) (Zhang et al., 2009), which is secreted by the retina during DR progression (Pe’er et al., 1996). In addition, some genes and their variants can influence DR occurrence, including the single nucleotide polymorphisms (SNPs) of VEGF and aquaporin-4 (Khan et al., 2020; Zhou et al., 2021).
Long noncoding RNAs (lncRNAs) are a group of RNAs that could contribute to angiogenesis and cell proliferation in tumor cells (Su S.-C. et al., 2018; Zhao et al., 2020; Su et al., 2021b; Ding et al., 2021). Among this group, long intergenic noncoding RNA 00673 (LINC00673) is associated with several malignancies (Zhu K. et al., 2021; Zhu Y. et al., 2021). In addition, LINC00673 SNPs can influence the expression of cancers such as oral cancer and gastric cancer (Zhao et al., 2019; Su et al., 2021a). Nevertheless, research on the relationship between the SNPs of LINC00673 and DR development is rare. Because LINC00673 is downregulated in patients with DR (Cheng et al., 2021), the SNPs of LINC00673 may also affect the clinical features of DR.
The purpose of this study was to assess the possible correlation between LINC00673 SNPs (rs6501551, rs9914618, and rs11655237) and the clinical characteristics of DR, including NPDR and PDR. The different phenotypes of LINC00673 SNP rs11655237 were also separately analyzed.
Materials and Methods
Ethical Declarations
This study, conducted in accordance with the 1964 Declaration of Helsinki and its later amendments, was approved by the Institutional Review Board of Chung Shan Medical University Hospital (project identification code: CS1-20048, approval date: 27 April 2020). Written informed consent was signed by the participants.
Patient Selection
Our case–control study was conducted at Chung Shan Medical University Hospital. A total of 730 patients diagnosed as having DM were included. Among these, 454 patients were categorized as the non-DR group and the other 276 patients were categorized as the DR group, according to fundus examination records in the ophthalmic department. The development of DR was regarded as any of the following fundus findings: dot- and flame-shaped hemorrhage, cotton-wool spot, hard exudate, venous beading, microaneurysm, or intraretinal microvascular abnormality. In the DR group, 111 patients were classified as the PDR group according to any of the following ophthalmological findings: neovascularization of the retina or optic disc, tractional retinal detachment, vitreous hemorrhage, or neovascular glaucoma. The remaining 165 patients constituted the NPDR group.
Data and Sample Collection
We reviewed the medical records of patients with DM in Chung Shan Medical University Hospital and collected basic data, including age, body mass index, sex, blood sugar level expressed as HbA1c, DM duration, blood pressure, lipid profiles, renal function, and insulin treatment regimen. To analyze LINC00673 polymorphisms, we used the methods described in our previous study (Hsieh et al., 2021). First, venous blood was drawn from each patient, and the collected blood samples were stored in separate ethylenediaminetetraacetic acid–containing tubes (Hsiao et al., 2010). Next, the blood samples were centrifuged promptly and stored in a laboratory refrigerator at approximately −80°C. Patients were excluded from the study if the genomes in their blood samples had been degraded prior to any analysis.
DNA Extraction and Determination of LINC00673 SNP Using Real-Time PCR
A total of three LINC00673 SNPs, namely, rs6501551 (A/G), rs9914618 (G/A), and rs11655237 (C/T), were selected for phenotype analyses because the minor allele frequencies of these SNPs were higher than 5% and our preceding research indicated the effect of these SNPs on other cancers (Su et al., 2021a). The procedures for DNA extraction and phenotype analyses in this study were in accordance with those of our previous study (Su S. C. et al., 2018). First, DNA was extracted from leukocytes by using QIAamp DNA kits (QIAGEN, Valencia, CA, United States), according to the manufacturer’s instruction for DNA isolation (Chung et al., 2011). Next, the DNA was preserved at approximately −20°C. Genetic polymorphisms of the LINC00673 SNPs, namely, rs6501551 (A/G) (ID: C_29084653_10), rs9914618 (G/A) (ID: C_29971800_10), and rs11655237 (C/T) (ID: C_345893_20), were then determined using the ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The final results of the LINC00673 genetic polymorphisms were analyzed using SDS version 3.0 software (Applied Biosystems).
Statistical Analysis
We used SAS version 9.4 (SAS Institute Inc., Cary, NC, United States) for all statistical analyses. Descriptive statistics such as mean, standard deviation, and percentage were used to compare demography, blood sugar status, and other laboratory data between the DR and non-DR groups. An independent t-test was used to evaluate the difference of these data between both groups. Next, multiple logistic regression models were applied to evaluate adjusted odds ratios (AORs) and 95% confidence intervals (CIs) of the LINC00673 SNP distribution and the allele distribution between the two groups. The multiple logistic regressions were adjusted for age, DM duration, insulin treatment, HbA1c, glomerular filtration rate, serum creatinine levels, and HDL cholesterol levels. The same multiple logistic regression models were used in the subgroup analyses for SNP distributions between the non-DR group and each of the NPDR and PDR subgroups. Furthermore, differences in clinicopathological characteristics for each SNP rs11655237 phenotype (i.e., CC and CT + TT) in the DR group were again analyzed using the independent t-test. A p-value less than 0.05 was considered statistically significant.
Results
Characteristics Between the Non-DR and DR Groups
The mean age of the DR group was 62.61 ± 10.72 years, which was significantly higher than that of the non-DR group (60.22 ± 11.22, p = 0.005). Parameters related to DM, including DM duration, HbA1c level, and ratio of insulin treatment, were all higher in the DR group (all p < 0.001). Serum creatinine was also higher in the DR group (p < 0.001), although the glomerular filtration rate (p < 0.001) and HDL cholesterol (p = 0.017) were lower than those of the non-DR group. The remaining parameters were not significantly different between both groups (all p > 0.05) (Table 1).
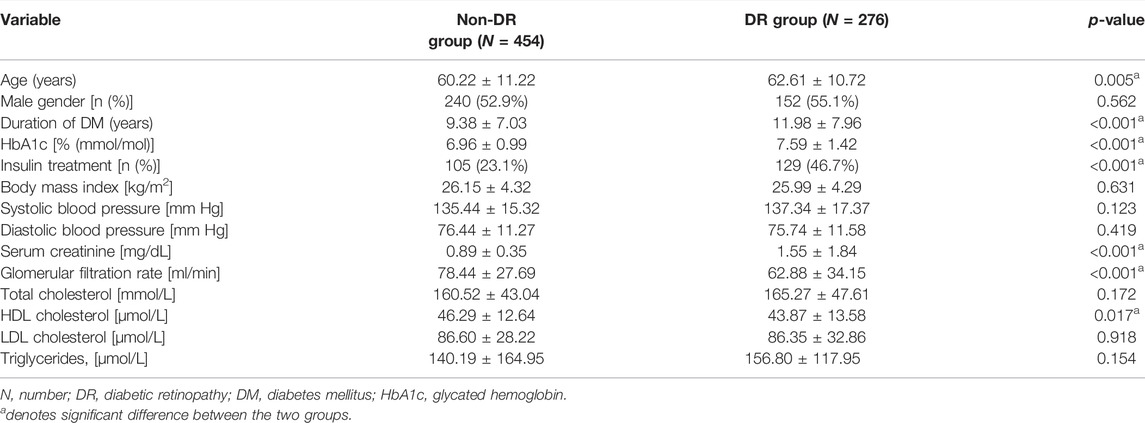
TABLE 1. Clinical and laboratory characteristics of patients with diabetic retinopathy and no diabetic retinopathy.
LINC00673 SNP Distribution Among Different DR Groups
The genotyping frequencies of each LINC00673 SNP between the DR and non-DR groups are shown in Table 2. No significant differences were observed regarding the distribution of the LINC00673 SNPs and their alleles between both groups (all p > 0.05) (Table 2). In the subgroup analyses, the LINC00673 SNP rs11655237 CT genotype (AOR: 1.592, 95% CI: 1.059–2.395, p = 0.026), LINC00673 SNP rs11655237 CT + TT genotype (AOR: 1.255, 95% CI: 1.029–1.531, p = 0.025), and LINC00673 SNP rs11655237 allele T (AOR: 1.185, 95% CI: 1.004–1.397, p = 0.044) exhibited higher ratios in the NPDR subgroup than in the non-DR group (Table 3). However, no significant differences in genotype frequencies were observed between the PDR subgroup and the non-DR group (all p > 0.05; Table 4).

TABLE 2. Adjusted odds ratio and 95% confidence intervals of diabetic retinopathy associated with LINC00673 genotypic frequencies.
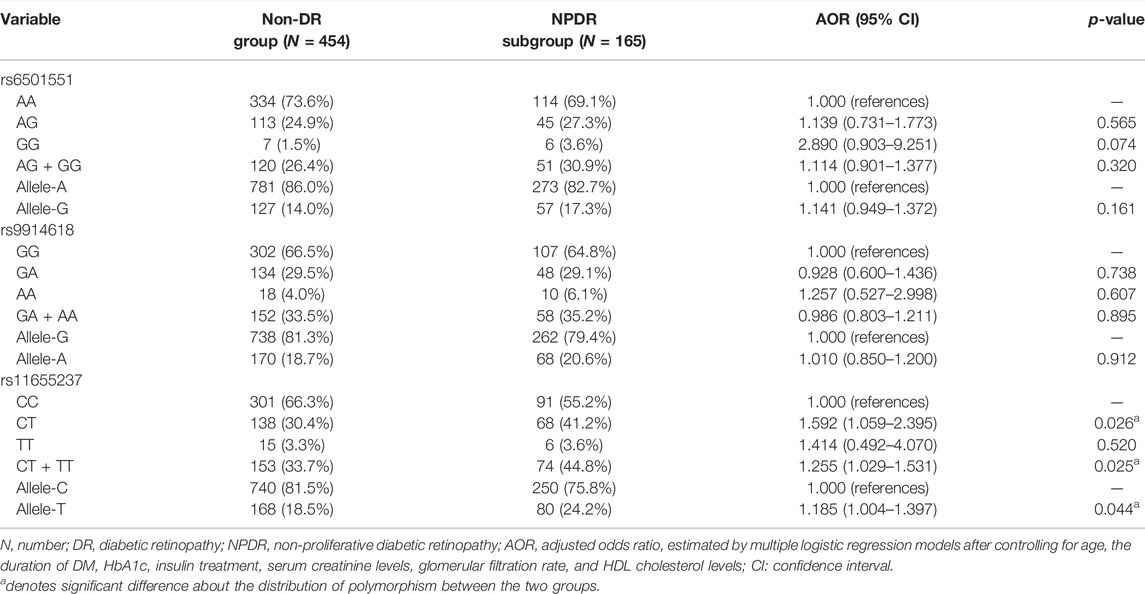
TABLE 3. Adjusted odds ratio and 95% confidence intervals of non-proliferative diabetic retinopathy associated with LINC00673 genotypic frequencies.
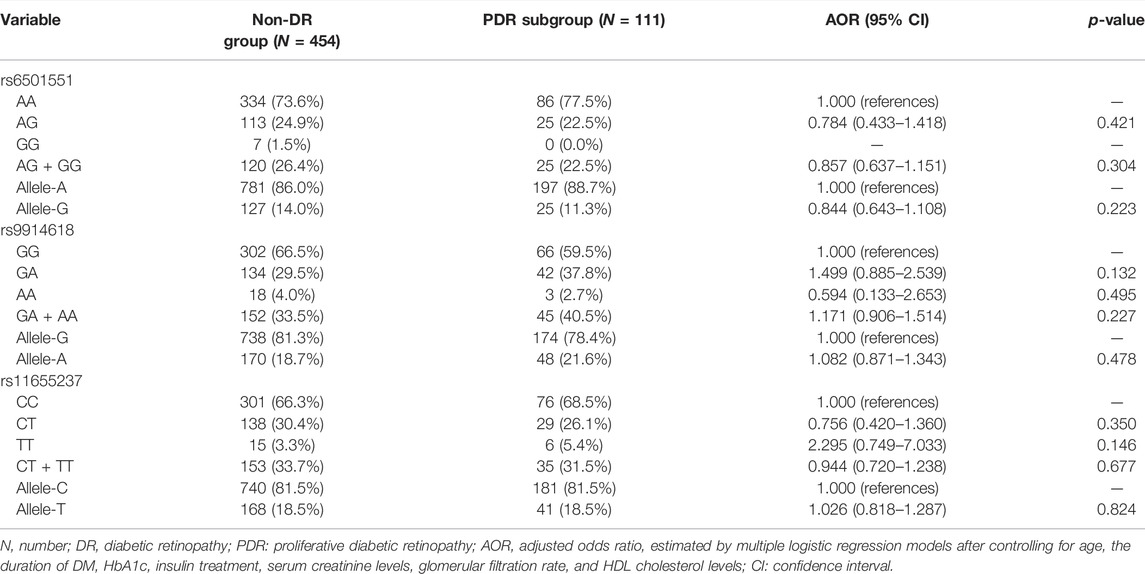
TABLE 4. Adjusted odds ratio and 95% confidence intervals of proliferative diabetic retinopathy associated with LINC00673 genotypic frequencies.
Clinical Characteristics and Distribution of LINC00673 SNP rs11655237 in the DR Group
In the DR group, DM duration was significantly shorter in the LINC00673 SNP rs11655237 CT + TT variant than in the LINC00673 SNP rs11655237 wild type (10.44 ± 7.10 versus 12.98 ± 8.34, p = 0.009). Other factors, including HbA1c, renal function, and lipid profile, produced similar values between the LINC00673 SNP rs11655237 wild type and the LINC00673 SNP rs11655237 CT + TT variant (all p > 0.05; Table 5).
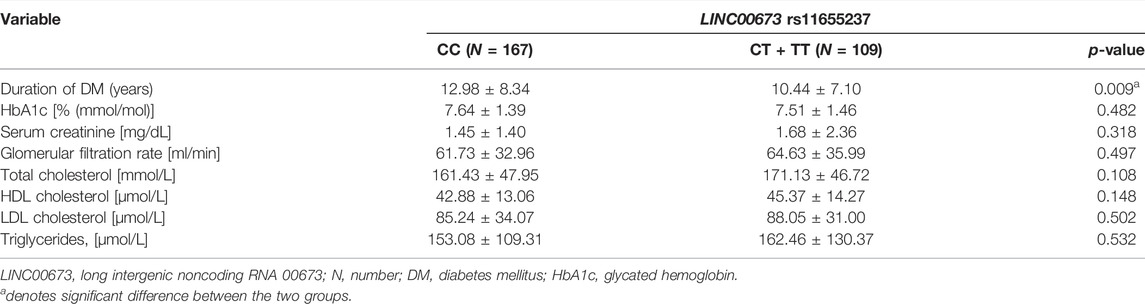
TABLE 5. Clinical characteristics of diabetic retinopathy individuals according to LINC00673 rs11655237 genotypes.
Discussion
In this study, the LINC00673 SNP rs11655237 CT and CT + TT phenotypes and the LINC00673 SNP rs11655237 T allele were more common in patients with NPDR than in the non-DR population. In addition, LINC00673 SNP rs11655237 CT + TT variants were associated with a shorter DM interval in the DR group. However, the distribution of the LINC00673 SNP rs11655237 variant was similar between the PDR and non-DR groups.
Several biochemical pathways are correlated with the occurrence of NPDR and PDR (Jenkins et al., 2015), in which inflammatory growth, and angiogenic molecules play major roles (Jenkins et al., 2015). In the development of DR, hypoxic and ischemic retinas secrete inflammatory cytokines such as interleukin-6 and monocyte chemotactic protein, resulting in a prominent inflammatory reaction and leading to vascular damage and retinal ganglion cell death (Wang and Lo, 2018). VEGF concentration is also elevated in such conditions (Huang et al., 2015). If DR progresses, the retinal concentration of VEGF further increases, and retinal neovascularization eventually develops (Osaadon et al., 2014). Anti-VEGF is the main treatment for PDR and its related complications such as vitreous hemorrhage and neovascular glaucoma (Wang and Lo, 2018). Intraocular steroid injection can also retard the progression of PDR (Iglicki et al., 2018). Regarding genetic polymorphisms, VEGF polymorphism influences the development of DR (Khan et al., 2020). In previous studies, VEGF SNP-634G > C was associated with a higher rate of DR (Qiu et al., 2013), and VEGF SNP rs2146323 was related to DR severity (Churchill et al., 2008). SNP rs11567245 of inflammatory cytokine interleukin-10 could also promote DR development (Shi et al., 2019).
SNPs of lncRNAs have been proposed to influence certain types of malignancies and angiogenic diseases (Peng et al., 2017; Yuan et al., 2019; Weng et al., 2020; Ding et al., 2021; Wang et al., 2021). In previous studies, lncRNA CCAT1 SNP rs67085638 was correlated with the development of gastric cancer (Olesiński et al., 2021), and lncRNA PCAT1 and its SNP rs2632159 were associated with a higher rate of colon cancer, which involves an angiogenesis process (Yang et al., 2019). LINC00673, a member of the lncRNA family, can enhance the development of certain neoplasms because of its chromatin modulation and transcriptional regulation functions (Lu et al., 2017; Qiao et al., 2019; Hsieh et al., 2021; Huang et al., 2021). LINC00673 SNP rs11655237 could alter the development of several cancers, including hepatocellular carcinoma, pancreatic cancer, and oral cancer (Li et al., 2020). Because LINC00673 SNP is related to cellular proliferation and lncRNA SNP is correlated with angiogenesis, LINC00673 SNP may also lead to DR development, which is characterized by both cell proliferation and angiogenesis. This hypothesis is aided by our study’s results.
Studies that have discussed the possible correlation between LINC00673 SNP and DR development are rare; only one study has evaluated the relationship between LINC00673 expression and DR (Cheng et al., 2021). In our study, the frequency of LINC00673 SNPs (i.e., rs6501551, rs9914618, and rs11655237) was not significantly different between the non-DR and DR groups. In the subgroup analyses that divided the DR group into the NPDR and PDR subgroups, the LINC00673 SNP rs11655237 CT and CT + TT variants were correlated with NPDR development, with a higher AOR. This result may support the relationship between LINC00673 SNP rs11655237 and NPDR. Conversely, the LINC00673 SNP rs11655237 TT variant was not significantly correlated with NPDR development. A total of fifteen patients had the LINC00673 SNP rs11655237 TT phenotype, of which six patients were in each of the non-DR group and NPDR subgroup, accounting for approximately 3% in each group. Such a few number of cases may lead to statistical bias. The analysis of the T allele effect on LINC00673 SNP rs11655237 revealed a significantly higher LINC00673 SNP rs11655237 T-allele distribution in the NPDR subgroup. Consequently, we speculated that the existence of the T allele in LINC00673 SNP rs11655237, whether in the homogenous or heterogeneous form, is crucial for the development of NPDR. On the contrary, the distribution of the three LINC00673 SNPs was not significantly different between the non-DR group and the PDR population. It may be possible that the effect of a single SNP is not adequate to trigger the angiogenesis process that contributes to PDR development. Whether the development of PDR is influenced by the LINC00673 SNP in combination with VEGF SNP or other biomarkers requires additional investigation.
Concerning clinicopathological characteristics, our study demonstrated that individuals who had DR with the LINC00673 SNP rs11655237 CT and TT variants exhibited a significantly shorter DM duration than that with the LINC00673 SNP rs11655237 CC phenotype. We believe this is preliminary evidence that genetic polymorphisms of LINC00673 may affect the course of DR development in patients with DM. The DM duration in patients with the LINC00673 SNP rs11655237 CT and TT variants was nearly 2.5 years shorter than that of patients with the LINC00673 SNP rs11655237 wild type and also shorter than the mean DM duration in a previous study (Gverović Antunica et al., 2019). Although the exact mechanism underlying this finding is unknown, we speculate that the LINC00673 SNP rs11655237 CT and TT variants might make the retina more vulnerable to hyperglycemic conditions, even with numerically lower HbA1c levels. This finding, along with the higher genotype frequency of LINC00673 SNP rs11655237 variants in the NPDR subgroup, may explain the prominent effect of the LINC00673 SNP rs11655237 variant on NPDR development.
Regarding demographic data and laboratory findings, patients with DR were significantly older than those without DR. Because age is an established risk factor for DR occurrence (Antonetti et al., 2012), our findings agree with those of previous studies (Antonetti et al., 2012). In addition, longer DM duration, higher serum HbA1c concentration, and higher ratio of insulin treatment were observed in the DR group, which could indicate that inadequate blood sugar control is a prominent risk factor for DR (Cheung et al., 2010). Renal function was significantly worse in the DR group. In previous studies, poorly controlled DM was associated with a higher rate of chronic kidney disease (Tziomalos and Athyros, 2015). Chronic kidney disease would in turn contribute to retinal diseases such as retinal microangiopathy and central serous chorioretinopathy (Chang et al., 2019; Kasumovic et al., 2020). It is therefore reasonable that kidney impairment and DR developed concurrently in our study. Higher HDL cholesterol levels in the DR group may have not been clinically significant because both values belonged to the low-HDL status (März et al., 2017).
Our study had several limitations. First, because our study used a case–control design, rather than a cohort design, we could not evaluate the possible causal relationship between the LINC00673 SNP and the clinical course of DR. Second, some patients may have received DR-related treatment elsewhere, which may have modified their clinical course toward NPDR and could have caused a wrong estimation of the effect of LINC00673 SNP on PDR development. Additionally, the interval between the internal medicine department visit and the ophthalmic department visit was uneven among the participants.
Conclusion
In conclusion, the existence of the LINC00673 SNP rs11655237 T allele is associated with a higher probability of NPDR development. The LINC00673 SNP rs11655237 variant is correlated with a shorter DM interval in patients with DR; consequently, genetic screening might be suggested for patients with DM. Patients with this genetic polymorphism should observe stricter blood sugar control. Further large-scale prospective studies to examine whether the LINC00673 polymorphism would interact with the VEGF polymorphism in DR are warranted.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human subjects were reviewed and approved by the Institutional Review Board of Chung Shan Medical University Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
C-CC and S-FY proposed the study concept and study design. Y-SY, EK, C-NH, M-YH, and S-FY participated in sample and data collection. C-CC, C-YL, and S-FY participated in statistical analyses. C-CC, C-YL, and S-FY wrote and edited the manuscript. All authors agree to be accountable for all aspects of the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Human Biobank of Chung Shan Medical University Hospital for providing the biological specimen and related clinical data for our research.
References
Antonetti, D. A., Klein, R., and Gardner, T. W. (2012). Diabetic Retinopathy. N. Engl. J. Med. 366, 1227–1239. doi:10.1056/nejmra1005073
Capitão, M., and Soares, R. (2016). Angiogenesis and Inflammation Crosstalk in Diabetic Retinopathy. J. Cel Biochem 117, 2443–2453.
Chang, Y.-S., Weng, S.-F., Wang, J.-J., and Jan, R.-L. (2019). Increased Risk of central Serous Chorioretinopathy Following End-Stage Renal Disease. Medicine (Baltimore) 98, e14859. doi:10.1097/md.0000000000014859
Cheng, Y., Zhu, Y., and Ma, L. (2021). LncRNA LINC00673 Is Downregulated in Diabetic Retinopathy and Regulates the Apoptosis of Retinal Pigment Epithelial Cells via Negatively Regulating P53. Dmso 14, 4233–4240. doi:10.2147/dmso.s298185
Cheung, N., Mitchell, P., and Wong, T. Y. (2010). Diabetic Retinopathy. The Lancet 376, 124–136. doi:10.1016/s0140-6736(09)62124-3
Chung, T.-T., Pan, M.-S., Kuo, C.-L., Wong, R.-H., Lin, C.-W., Chen, M.-K., et al. (2011). Impact of RECK Gene Polymorphisms and Environmental Factors on Oral Cancer Susceptibility and Clinicopathologic Characteristics in Taiwan. Carcinogenesis 32, 1063–1068. doi:10.1093/carcin/bgr083
Churchill, A. J., Carter, J. G., Ramsden, C., Turner, S. J., Yeung, A., Brenchley, P. E. C., et al. (2008). VEGF Polymorphisms Are Associated with Severity of Diabetic Retinopathy. Invest. Ophthalmol. Vis. Sci. 49, 3611–3616. doi:10.1167/iovs.07-1383
Ding, Y.-F., Wen, Y.-C., Chuang, C.-Y., Lin, C.-W., Yang, Y.-C., Liu, Y.-F., et al. (2021). Combined Impacts of Genetic Variants of Long Non-coding RNA MALAT1 and the Environmental Carcinogen on the Susceptibility to and Progression of Oral Squamous Cell Carcinoma. Front. Oncol. 11, 684941. doi:10.3389/fonc.2021.684941
Flaxman, S. R., Bourne, R. R. A., Resnikoff, S., Ackland, P., Braithwaite, T., Cicinelli, M. V., et al. (2017). Global Causes of Blindness and Distance Vision Impairment 1990-2020: a Systematic Review and Meta-Analysis. Lancet Glob. Health 5, e1221–e1234. doi:10.1016/S2214-109X(17)30393-5
Ghamdi, A. H. A. (2020). Clinical Predictors of Diabetic Retinopathy Progression; A Systematic Review. Cdr 16, 242–247. doi:10.2174/1573399815666190215120435
Gverović Antunica, A., Bućan, K., Kaštelan, S., Kaštelan, H., Ivanković, M., and Šikić, M. (2019). Prevalence of Diabetic Retinopathy in the Dubrovnik-Neretva County. Cent. Eur. J. Public Health 27, 160–164. doi:10.21101/cejph.a5213
Hsiao, P.-C., Chen, M.-K., Su, S.-C., Ueng, K.-C., Chen, Y.-C., Hsieh, Y.-H., et al. (2010). Hypoxia Inducible Factor-1α Gene Polymorphism G1790A and its Interaction with Tobacco and Alcohol Consumptions Increase Susceptibility to Hepatocellular Carcinoma. J. Surg. Oncol. 102, 163–169. doi:10.1002/jso.21539
Hsieh, M. H., Lu, H. J., Lin, C. W., Lee, C. Y., Yang, S. J., Wu, P. H., et al. (2021). Genetic Variants of lncRNA GAS5 Are Associated with the Clinicopathologic Development of Oral Cancer. J. Pers Med. 11. doi:10.3390/jpm11050348
Huang, H., He, J., Johnson, D. K., Wei, Y., Liu, Y., Wang, S., et al. (2015). Deletion of Placental Growth Factor Prevents Diabetic Retinopathy and Is Associated with Akt Activation and HIF1α-VEGF Pathway Inhibition. Diabetes 64, 200–212. doi:10.2337/db14-0016
Huang, S.-K., Ni, R.-X., Wang, W.-J., Wang, D., Zhao, M., Lei, C.-Z., et al. (2021). Overexpression of LINC00673 Promotes the Proliferation of Cervical Cancer Cells. Front. Oncol. 11, 669739. doi:10.3389/fonc.2021.669739
Iglicki, M., Zur, D., Busch, C., Okada, M., and Loewenstein, A. (2018). Progression of Diabetic Retinopathy Severity after Treatment with Dexamethasone Implant: a 24-month Cohort Study the 'DR-Pro-DEX Study'. Acta Diabetol. 55, 541–547. doi:10.1007/s00592-018-1117-z
Jenkins, A. J., Joglekar, M. V., Hardikar, A. A., Keech, A. C., O'neal, D. N., and Januszewski, A. S. (2015). Biomarkers in Diabetic Retinopathy. Rev. Diabet Stud. 12, 159–195. doi:10.1900/rds.2015.12.159
Kasumovic, A., Matoc, I., Rebic, D., Avdagic, N., and Halimic, T. (2020). Assessment of Retinal Microangiopathy in Chronic Kidney Disease Patients. Med. Arch. 74, 191–194. doi:10.5455/medarh.2020.74.191-194
Khan, S. Z., Ajmal, N., and Shaikh, R. (2020). Diabetic Retinopathy and Vascular Endothelial Growth Factor Gene Insertion/Deletion Polymorphism. Can. J. Diabetes 44, 287–291. doi:10.1016/j.jcjd.2019.08.005
Li, N., Cui, Z., Huang, D., Gao, M., Li, S., Song, M., et al. (2020). Association of LINC00673 Rs11655237 Polymorphism with Cancer Susceptibility: A Meta-Analysis Based on 23,478 Subjects. Genomics 112, 4148–4154. doi:10.1016/j.ygeno.2020.07.015
Lu, W., Zhang, H., Niu, Y., Wu, Y., Sun, W., Li, H., et al. (2017). Long Non-coding RNA Linc00673 Regulated Non-small Cell Lung Cancer Proliferation, Migration, Invasion and Epithelial Mesenchymal Transition by Sponging miR-150-5p. Mol. Cancer 16, 118. doi:10.1186/s12943-017-0685-9
März, W., Kleber, M. E., Scharnagl, H., Speer, T., Zewinger, S., Ritsch, A., et al. (2017). HDL Cholesterol: Reappraisal of its Clinical Relevance. Clin. Res. Cardiol. 106, 663–675. doi:10.1007/s00392-017-1106-1
Olesiński, T., Lutkowska, A., Balcerek, A., Sowińska, A., Piotrowski, P., Trzeciak, T., et al. (2021). Long Noncoding RNA CCAT1 Rs67085638 SNP Contribution to the Progression of Gastric Cancer in a Polish Population. Sci. Rep. 11, 15369. doi:10.1038/s41598-021-94576-9
Osaadon, P., Fagan, X. J., Lifshitz, T., and Levy, J. (2014). A Review of Anti-VEGF Agents for Proliferative Diabetic Retinopathy. Eye 28, 510–520. doi:10.1038/eye.2014.13
Pe'er, J., Folberg, R., Itin, A., Gnessin, H., Hemo, I., and Keshet, E. (1996). Upregulated Expression of Vascular Endothelial Growth Factor in Proliferative Diabetic Retinopathy. Br. J. Ophthalmol. 80, 241–245. doi:10.1136/bjo.80.3.241
Peng, W.-X., Koirala, P., and Mo, Y.-Y. (2017). LncRNA-mediated Regulation of Cell Signaling in Cancer. Oncogene 36, 5661–5667. doi:10.1038/onc.2017.184
Qiao, K., Ning, S., Wan, L., Wu, H., Wang, Q., Zhang, X., et al. (2019). LINC00673 Is Activated by YY1 and Promotes the Proliferation of Breast Cancer Cells via the miR-515-5p/MARK4/Hippo Signaling Pathway. J. Exp. Clin. Cancer Res. 38, 418. doi:10.1186/s13046-019-1421-7
Qiu, M., Xiong, W., Liao, H., and Li, F. (2013). VEGF −634G>C Polymorphism and Diabetic Retinopathy Risk: A Meta-Analysis. Gene 518, 310–315. doi:10.1016/j.gene.2013.01.018
Sabanayagam, C., Banu, R., Chee, M. L., Lee, R., Wang, Y. X., Tan, G., et al. (2019). Incidence and Progression of Diabetic Retinopathy: a Systematic Review. Lancet Diabetes Endocrinol. 7, 140–149. doi:10.1016/s2213-8587(18)30128-1
Shi, Y. L., Shi, M. Y., Yin, L. Z., Shang, J. M., and Zhuang, J. Y. (2019). IL-10 Gene Polymorphism in Diabetic Retinopathy. Eur. Rev. Med. Pharmacol. Sci. 23, 5059–5064. doi:10.26355/eurrev_201906_18169
Su, S.-C., Reiter, R. J., Hsiao, H.-Y., Chung, W.-H., and Yang, S.-F. (2018b). Functional Interaction between Melatonin Signaling and Noncoding RNAs. Trends Endocrinol. Metab. 29, 435–445. doi:10.1016/j.tem.2018.03.008
Su, S. C., Hsieh, M. J., Lin, C. W., Chuang, C. Y., Liu, Y. F., Yeh, C. M., et al. (2018a). Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J. Dent Res. 97, 717–724. doi:10.1177/0022034517749451
Su, S. C., Lin, C. W., Ju, P. C., Chang, L. C., Chuang, C. Y., Liu, Y. F., et al. (2021a). Association of LINC00673 Genetic Variants with Progression of Oral Cancer. J. Pers Med. 11. doi:10.3390/jpm11060468
Su, S. C., Yeh, C. M., Lin, C. W., Hsieh, Y. H., Chuang, C. Y., Tang, C. H., et al. (2021b). A Novel Melatonin-Regulated lncRNA Suppresses TPA-Induced Oral Cancer Cell Motility through Replenishing PRUNE2 Expression. J. Pineal Res. 71, e12760. doi:10.1111/jpi.12760
Tziomalos, K., and Athyros, V. G. (2015). Diabetic Nephropathy: New Risk Factors and Improvements in Diagnosis. Rev. Diabet Stud. 12, 110–118. doi:10.1900/rds.2015.12.110
Wang, W., and Lo, A. C. Y. (2018). Diabetic Retinopathy: Pathophysiology and Treatments. Int. J. Mol. Sci. 19. doi:10.3390/ijms19061816
Wang, Y. C., Tsao, S. M., Li, Y. T., Lee, C. Y., Tsao, T. C., Hsieh, M. J., et al. (2021). The Relationship between Long Noncoding RNA H19 Polymorphism and the Epidermal Growth Factor Receptor Phenotypes on the Clinicopathological Characteristics of Lung Adenocarcinoma. Int. J. Environ. Res. Public Health 18. doi:10.3390/ijerph18062862
Weng, W. C., Chen, C. J., Chen, P. N., Wang, S. S., Hsieh, M. J., and Yang, S. F. (2020). Impact of Gene Polymorphisms in GAS5 on Urothelial Cell Carcinoma Development and Clinical Characteristics. Diagnostics (Basel) 10. doi:10.3390/diagnostics10050260
Yang, M. L., Huang, Z., Wu, L. N., Wu, R., Ding, H. X., and Wang, B. G. (2019). lncRNA-PCAT1 Rs2632159 Polymorphism Could Be a Biomarker for Colorectal Cancer Susceptibility. Biosci. Rep. 39. doi:10.1042/BSR20190708
Yuan, L. T., Chang, J. H., Lee, H. L., Yang, Y. C., Su, S. C., Lin, C. L., et al. (2019). Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma. J. Clin. Med. 8. doi:10.3390/jcm8091406
Zhang, X., Bao, S., Hambly, B. D., and Gillies, M. C. (2009). Vascular Endothelial Growth Factor-A: a Multifunctional Molecular Player in Diabetic Retinopathy. Int. J. Biochem. Cel Biol. 41, 2368–2371. doi:10.1016/j.biocel.2009.07.011
Zhao, K., Zhang, R., Li, T., and Xiong, Z. (2019). Functional Variants of lncRNA LINC00673 and Gastric Cancer Susceptibility: a Case-Control Study in a Chinese Population. Cmar 11, 3861–3868. doi:10.2147/cmar.s187011
Zhao, Z., Sun, W., Guo, Z., Zhang, J., Yu, H., and Liu, B. (2020). Mechanisms of lncRNA/microRNA Interactions in Angiogenesis. Life Sci. 254, 116900. doi:10.1016/j.lfs.2019.116900
Zheng, Y., Ley, S. H., and Hu, F. B. (2018). Global Aetiology and Epidemiology of Type 2 Diabetes Mellitus and its Complications. Nat. Rev. Endocrinol. 14, 88–98. doi:10.1038/nrendo.2017.151
Zhou, X., Cheng, F. C., and Wang, H. L. (2021). Correlations of AQP4 Expression and Polymorphism with Diabetic Retinopathy. Eur. Rev. Med. Pharmacol. Sci. 25, 1169–1176. doi:10.26355/eurrev_202102_24819
Zhu, K., Gong, Z., Li, P., Jiang, X., Zeng, Z., Xiong, W., et al. (2021a). A Review of Linc00673 as a Novel lncRNA for Tumor Regulation. Int. J. Med. Sci. 18, 398–405. doi:10.7150/ijms.48134
Keywords: long intergenic noncoding RNA 00673, polymorphism, diabetic retinopathy, diabetes mellitus, disease duration
Citation: Chuang C-C, Yang Y-S, Kornelius E, Huang C-N, Hsu M-Y, Lee C-Y and Yang S-F (2022) Impact of Long Noncoding RNA LINC00673 Genetic Variants on Susceptibility to Diabetic Retinopathy. Front. Genet. 13:889530. doi: 10.3389/fgene.2022.889530
Received: 04 March 2022; Accepted: 24 March 2022;
Published: 25 April 2022.
Edited by:
Yen-Wei Chu, National Chung Hsing University, TaiwanReviewed by:
Leyi Wei, Shandong University, ChinaXing-Hua Liao, Wuhan University of Science and Technology, China
Copyright © 2022 Chuang, Yang, Kornelius, Huang, Hsu, Lee and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shun-Fa Yang, eXNmQGNzbXUuZWR1LnR3
 Chih-Chun Chuang1,2
Chih-Chun Chuang1,2 Chien-Ning Huang
Chien-Ning Huang Min-Yen Hsu
Min-Yen Hsu Shun-Fa Yang
Shun-Fa Yang