- 1Genome Medical Science Project, National Center for Global Health and Medicine, Toyama, Tokyo,Japan
- 2Department of Human Genetics, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
- 3Department of Microbiology, Hoshi University School of Pharmacy and Pharmaceutical Sciences, Tokyo, Japan
- 4Genome Medical Science Project, National Center for Global Health and Medicine, Ichikawa, Chiba, Japan
- 5Chulabhorn International College of Medicine, Thammasat University, Rangsit Campus, Pathum Thani, Thailand
- 6Center of Excellence in Clinical Virology, Department of Pediatrics, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
- 7Center of Excellence in Hepatitis and Liver Cancer, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
- 8Center of Excellence in Liver Diseases, King Chulalongkorn Memorial Hospital, Thai Red Cross Society, Bangkok, Thailand
- 9Liver Fibrosis and Cirrhosis Research Unit, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
- 10Department of Pediatrics, Center of Excellence for Medical Genomics, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
To identify novel host genetic variants that predispose to hepatitis B virus (HBV) persistence, we performed the first genome-wide association study in the Thai population involving 318 cases of chronic hepatitis B and 309 healthy controls after quality control measures. We detected the genome-wide significant association of the HLA class II region (HLA-DPA1/DPB1, rs7770370, p-value = 7.71 × 10−10, OR = 0.49) with HBV chronicity. Subsequent HLA allele imputation revealed HLA-DPA1*01:03 (Pc = 1.21 × 10−6, OR = 0.53), HLA-DPB1*02:01 (Pc = 2.17 × 10−3, OR = 0.50), and HLA-DQB1*06:09 (Pc = 2.17 × 10−2, OR = 0.07) as protective alleles, and HLA-DPA1*02:02 (Pc = 6.32 × 10−5, OR = 1.63), HLA-DPB1*05:01 (Pc = 1.13 × 10−4, OR = 1.72), HLA-DPB1*13:01 (Pc = 4.68 × 10−2, OR = 1.60), and HLA-DQB1*03:03 (Pc = 1.11 × 10−3, OR = 1.84) as risk alleles for HBV persistence. We also detected suggestive associations in the PLSCR1 (rs35766154), PDLIM5 (rs62321986), SGPL1 (rs144998273), and MGST1 (rs1828682) loci. Among single-nucleotide polymorphisms in the PLSCR1 locus, rs1061307 was identified as the primary functional variant by in silico/in vitro functional analysis. In addition to replicating the association of the HLA class II region, we detected novel candidate loci that provide new insights into the pathophysiology of chronic hepatitis B.
Introduction
Hepatitis B virus (HBV) causes one of the major infectious diseases in humans, and is associated with an increased risk of severe liver complications, such as hepatic decompensation, cirrhosis, and liver cancer (WHO, 2017; Polaris Observatory Collaborators, 2018). Chronic HBV infection which is defined by the presence of hepatitis B surface antigen (HBsAg) in the host for at least 6 months (WHO, 2017), is considered a polygenic and multifactorial condition (Thursz, 2001; Akcay et al., 2018; Podlaha et al., 2019). Of the 292 million people living with chronic HBV infection around the world (Polaris Observatory Collaborators., 2018), 68% reside in the African and Western Pacific regions. Additionally, among individuals who are infected with HBV in adulthood, only 1% to 5% develop chronic HBV infection (Bertoletti and Kennedy, 2015). These observations led to the speculation that the persistence of HBV infection may be dependent on the heterogeneity of the host genetic background.
Numerous studies have been conducted to detect host genetic factors that are associated with HBV persistence. Two twin studies (Lin et al., 1989; Xu et al., 2004) showed significant differences in the concordance rate of infection, clinical phenotype, and serological patterns between monozygotic twins and dizygotic twins, and between monozygotic twins and the control groups. A genome-wide linkage study (Frodsham et al., 2006) and several case-control association studies (Ben-Ari et al., 2003; Deng et al., 2004; Chang et al., 2005; Singh et al., 2007; Qiu et al., 2012; Riazalhosseini et al., 2018) showed the contribution of other genes, especially immunologically relevant genes, such as HLA, CRF2, TNFA, IFNG, CCR5, IL-6, and ESR. The first genome-wide association study (GWAS) of chronic hepatitis B (Kamatani et al., 2009) showed the significant association of 11 SNPs in a region including HLA-DPA1 and HLA-DPB1; after more samples were added to the first sample set (Mbarek et al., 2011), an independent association was detected between HLA-DQ locus and HBV persistence. Subsequent GWASs in other Asian populations (Nishida et al., 2012; Hu et al., 2013; Kim et al., 2013; Chang et al., 2014; Jiang et al., 2015; Li Y. et al., 2016; Sakaue et al., 2021; Zeng et al., 2021) replicated the significant association of the HLA class II region, and showed the association of some novel loci, such as UBE2L3, HLA-C, HLA-DQA2, HLA-DQB2, EHMT2, TCF19, CFB, NOTCH4, HLA-C, HLA-DO, CD40, GRHL2, and INTS10 (Hu et al., 2013; Kim et al., 2013; Chang et al., 2014; Jiang et al., 2015; Li Y. et al., 2016; Zeng et al., 2021). However, the genetic factors that contribute to the pathogenesis of chronic HBV infection as a complex and heterogenetic disease have not yet been fully identified.
There are more than three million patients with chronic HBV infection in Thailand, and HBsAg seroprevalence rate is 6.42% (Posuwan et al., 2019). Hepatocellular carcinoma associated with HBV infection is one of the major causes of cancer-related deaths in Thailand (Chonprasertsuk and Vilaichone, 2017). Previous case-control association studies have reported the association of several genes, such as TNFA, HLA-DRB1, IL-18, IL28B, and HLA-DP (Hirankarn et al., 2007; Kummee et al., 2007; Kamatani et al., 2009; Posuwan et al., 2014; Kimkong et al., 2015), with the chronicity of HBV infection in Thailand. To further dissect the genetic architecture of chronic HBV infection in this population, we conducted a GWAS using 627 samples, including 318 cases of chronic HBV infection and 309 healthy controls; in addition to replicating the association of HLA-DPA1/DPB1, we detected the association of some novel candidate loci.
Results
GWAS
We genotyped 647 samples, including 329 cases of chronic HBV infection and 318 healthy controls, using the Affymetrix Axiom Genome-Wide ASI 1 Array (Materials and Methods). We then conducted genotype calling and removed 1 case sample with a genotyping call rate <0.97.
An identical-by-descent (IBD) test was conducted using PLINK (v1.9) (Purcell et al., 2007), and 16 samples with PI_HAT >0.1875, including 9 cases and 7 controls, were excluded from the data. We then performed a principal component analysis (PCA) for detecting possible population outliers or population stratification. PCA revealed that the Thai samples belonged to the Asian population (Supplementary Figure S1A). Three samples, including 1 case and 2 controls, were detected as population outliers, and were excluded from the data. As there are several ethnic groups in Thailand, we also conducted a PCA on Thai samples only, and found that the genetic background of the cases and controls matched (Supplementary Figure S1B). None of the samples had discordant gender information between the clinical data and genetically determined sex. Basic characteristics of samples are provided in Supplementary Table S1.
Whole-genome imputation was conducted using IMPUTE2 software (v2.3.2) (Howie et al., 2009) with 1,000 Genomes phase 3 as the reference panel. We then applied some post-imputation quality control (QC) to exclude SNPs and short INDELs with a call rate <0.97, minor allele frequency (MAF) < 0.01, and Hardy-Weinberg equilibrium (HWE) test p-value <1 × 10−6. None of the samples had a genotype calling rate <0.97. The association analysis of whole-genome imputation data, including 318 cases, 309 controls, and 6,317,193 SNPs/short INDELs, was conducted using the 2 × 2 contingency chi-square test (Supplementary Table S2). The genomic inflation factor (λGC) was 1.036 for all tested variants, and after removal of the SNPs/short INDELs of the extended HLA region (Hg19: chr6:25,652,429-33,368,333), it decreased to 1.031. The Manhattan plot of the association analysis results is shown in Figure 1. The Q-Q plots of the observed p-values against the expected distribution of the association test statistics before and after the exclusion of SNPs from the HLA region are shown in Supplementary Figures S2A,S2B, respectively. The most significant association was detected in the HLA-DPA1/DPB1 region on chromosome 6 (rs7770370, located in the intron of the HLA-DPB1 gene, p-value = 7.71 × 10−10, OR = 0.49, 95% CI = 0.39–0.61) (Figure 2A), replicating the association of this region with chronic HBV infection that was reported in previous studies (Kamatani et al., 2009; Nishida et al., 2012; Hu et al., 2013; Kim et al., 2013; Chang et al., 2014). No other locus achieved the genome-wide level of significance (p-value < 5 × 10−8). As there are highly variable linkage disequilibrium patterns and chromosomal recombination in the HLA region (Williams, 2001), we conducted a conditional analysis to check for any other genes or loci in this region that were independently associated with chronic HBV infection, but were masked by the association signal from HLA-DPA1/DPB1. We extracted 5,590 SNPs from the HLA region, and conducted logistics regression conditioning on the lead SNP (rs7770370). However, no other locus in this region achieved the level of significance after Bonferroni correction (significance threshold: 0.05/5,590 = 8.9 × 10−6) (Figure 2B).
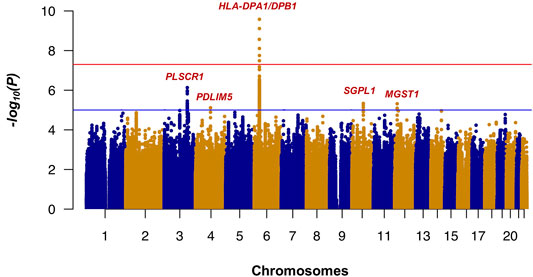
FIGURE 1. Results of the GWAS (Manhattan plot) for chronic hepatitis B in the Thai population. The HLA-DPA1/DPB1 exhibited the most significant association with chronic hepatitis B in the Thai population. p-values were calculated using the chi-squared test for 318 patients and 309 controls in the GWAS stage. The horizontal red and blue lines show the genome-wide significant threshold (p-value <5 × 10−8) and suggestive threshold (p-value <1 × 10−5), respectively.
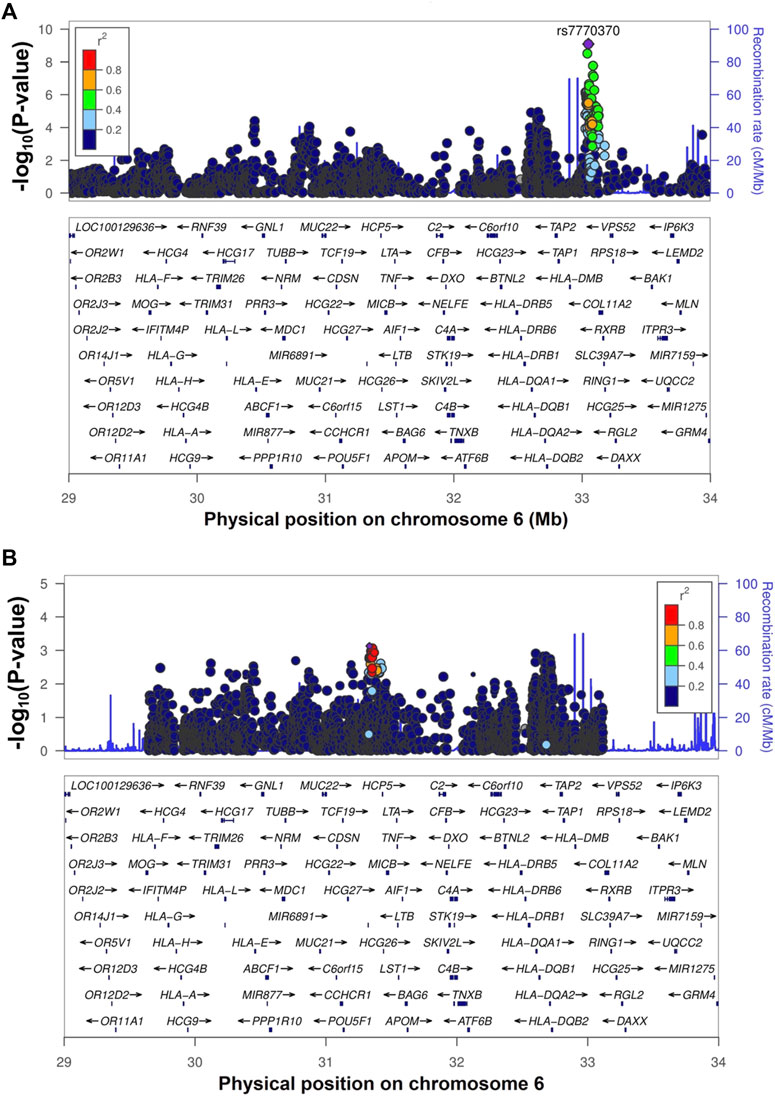
FIGURE 2. The regional association plot for the HLA-DPA1/DPB1 region. Figure (A) shows the regional association plot of the HLA region with the GWAS top SNP (rs7770370) shown by a purple circle. Figure (B) shows the plot of the HLA-DPA1/DPB1 region after conditioning on the top SNP (rs7770370).
Additionally, four other loci (Supplementary Table S2), i.e., rs35766154 located 13 kb 5′ of the PLSCR1 (Phospholipid Scramblase 1) gene on chromosome 3 (p-value = 7.39 × 10−7, OR = 0.43, 95% CI = 0.31–0.61), rs62321986 located 28 kb 5’ of the PDLIM5 (PDZ and LIM Domain 5) gene on chromosome 4 (p-value = 7.68 × 10−6, OR = 2.58, 95% CI = 1.68–3.96), rs144998273 located in the intron of SGPL1 (Sphingosine-1-Phosphate Lyase 1) gene on chromosome 10 (p-value = 4.64 × 10−6, OR = 2.52, 95% CI = 1.67–3.78), and rs1828682 located in the intron of the MGST1 (Microsomal Glutathione S-Transferase 1) gene on chromosome 12 (p-value = 4.82 × 10−6, OR = 0.32, 95% CI = 0.19–0.53), achieved the suggestive level of significance (Supplementary Figure S3). We used a TaqMan genotyping assay for experimental validation of the GWAS top SNP (rs7770370) from the HLA region, and the top genotyped and the top imputed SNPs on chromosomes 3, 4, 10, and 12 (Supplementary Table S3). All nine SNPs were successfully validated with a mean concordance rate of 98% (94% to 100%).
Association Analysis of HLA Alleles and Haplotypes With Chronic HBV Infection
In an attempt to replicate the reported association of different HLA class II alleles and discover novel alleles that predispose to chronic HBV infection in the Thai population, we conducted four-digit HLA allele imputation in the class II region (HLA-DPA1, -DPB1, and -DQB1) with the SNP data of all samples using an in-house HIBAG reference panel specific to the Thai population. As post-imputation QC, HLA alleles with a posterior probability of <0.5 were excluded from the data. To assess the accuracy of the HLA imputation, 300 samples, including 150 cases and 150 controls, were randomly chosen for HLA typing. The mean concordance rate between the imputed (after post-imputation QC) and genotyped HLA-DPA1, -DPB1, and -DQB1 genotypes was 98%, indicating the high accuracy of our in-house reference. We excluded samples with discordant genotyping information between the two methods, and HLA alleles/haplotypes with an allele frequency <0.01 in both the cases and controls. We then performed a case-control association analysis for the remaining HLA alleles/haplotypes using Pearson’s chi-squared test. p-values were corrected for the number of alleles/haplotypes tested (shown as P-corrected [Pc]). An association was considered significant if Pc < 0.05. HLA-DPA1*01:03 (Pc = 1.21 × 10−6, OR = 0.53), HLA-DPB1*02:01 (Pc = 2.17 × 10−3, OR = 0.50), and HLA-DQB1*06:09 (Pc = 2.17 × 10−2, OR = 0.07) showed a significant protective effect against chronic HBV infection, while HLA-DPA1*02:02 (Pc = 6.32 × 10−5, OR = 1.63), HLA-DPB1*05:01 (Pc = 1.13 × 10−4, OR = 1.72), HLA-DPB1*13:01 (Pc = 4.68 × 10−2, OR = 1.60), and HLA-DQB1*03:03 (Pc = 1.11 × 10−3, OR = 1.84) were significantly associated with susceptibility to chronic HBV infection (Supplementary Tables S4–S6).
To determine whether other HLA alleles had a significant association with chronic HBV infection that was masked by the most significant allele, we conducted a logistic regression analysis conditioned on the top HLA allele (HLA-DPA1*01:03); however, no other allele achieved the threshold level for significance (p-value: 0.05/18 = 2.7 × 10−3), implying that the primary HLA allele that is significantly associated with protection against HBV chronicity in the Thai population is HLA-DPA1*01:03.
We also analyzed the association of different HLA haplotypes with chronic HBV infection using Pearson’s chi-squared test. Haplotypes were estimated using the Arlequin algorithm (v3.0) (Excoffier et al., 2007). HLA haplotypes with an allele frequency <0.01 in both the cases and controls were excluded. Among two-locus HLA-DPA1-DPB1 haplotypes (Supplementary Table S7), HLA-DPA1*01:03-DPB1*02:01 (Pc = 9.5 × 10−4, OR = 0.47) showed the strongest protective association against HBV persistence, and HLA-DPA1*02:02-DPB1*05:01 (Pc = 3.74 × 10−4, OR = 1.68) showed the strongest association with susceptibility to HBV persistence. Among two-locus HLA-DPB1-DQB1 haplotypes (Supplementary Table S8), HLA-DPB1*05:01-DQB1*03:03 (Pc = 3.08 × 10−3, OR = 2.27) and HLA-DPB1*13:01-DQB1*05:02 (Pc = 9.22 × 10−3, OR = 5.42) were detected as haplotypes that were significantly associated with susceptibility to persistent HBV infection. The only significant three-locus HLA-DPA1-DPB1-DQB1 haplotype (Supplementary Table S9) was HLA-DPA1*02:02-DPB1*05:01-DQB1*03:03 (Pc = 9.68 × 10−3, OR = 2.07), which was associated with susceptibility to chronic HBV infection.
Identification of the Primary Functional Variants in the PLSCR1 Locus by in Silico/in vitro Analysis
The PLSCR1 gene was previously observed to be associated with the inhibition of HBV replication (Yang et al., 2012) in vitro and in vivo; thus, in this study, we decided to conduct a functional assay to identify the primary functional variant for this locus. Among the top 100 susceptibility SNPs in the PLSCR1 locus, rs1061307 was the only one located in genomic regions linked to histone acetylation, such as H3K27Ac and DNase I hypersensitivity, in at least one cell type in the UCSC genome browser (http://genome.ucsc.edu/index.html) (Kent et al., 2002) (Supplementary Figure S4). This SNP was selected as a potential candidate for disease susceptibility through regulation of the level of PLSCR1 gene expression. We conducted a luciferase assay (Figure 3) to evaluate the regulation of PLSCR1 gene expression by rs1061307 using the Jurkat cells (human T lymphocytes) and HepG2 cells (human liver carcinoma cells). Twenty-four hours after transfection of the pGL4.23 constructs into Jurkat cells, the luciferase activity was significantly higher with the G allele than the A allele of rs1061307 (p-value <0.05) (Figure 3B). This tendency was concordant with the e-QTL data of rs1061307 on endogenous PLSCR1 gene expression in all tissues registered in the GTEx portal database (http://gtexportal.org/home/) (Lonsdale et al., 2013), with the most significant association observed in whole blood cells (Supplementary Figure S5). This result suggested that rs1061307 was the primary functional variant for PLSCR1 locus. However, no difference in luciferase activity between the rs1061307 G and A alleles was observed in HepG2 cells (Figure 3C) which is likely due to differences in the binding of transcription factors to these alleles in different cell types.
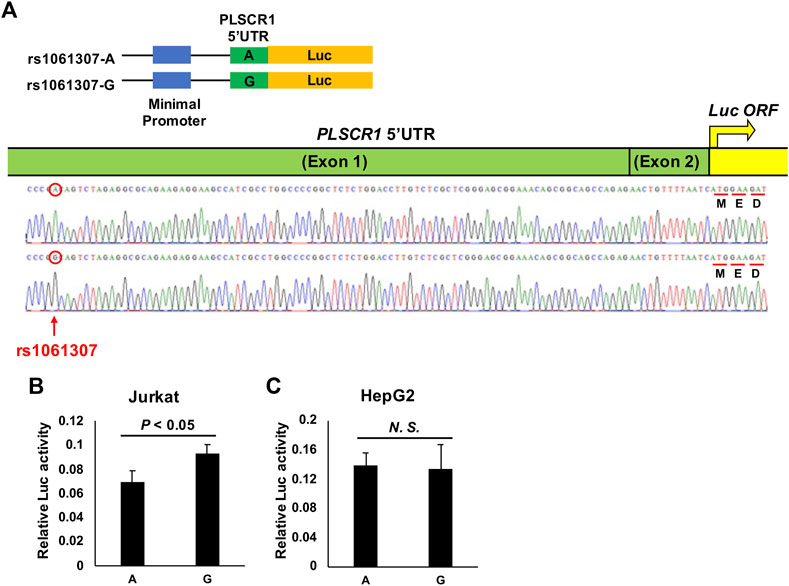
FIGURE 3. Luciferase assay for PLSCR1 rs1061307. (A) Construct of the luciferase reporter pGL4.23 (luc2/minP) vector. PLSCR1 5′UTR was conjugated with luc ORF. The transcriptional enhancing activity of these plasmid constructs was measured by assaying the luciferase (luc) activity of the transfected Jurkat (B) and HepG2 (C) cells, 24 h after transfection. The values of relative luciferase activity are shown as mean ± SD.
Discussion
In the current study, the most significant association was detected between the HLA class II region and HBV chronicity in the Thai population, replicating the results of previous studies. In addition, we observed the suggestive association of several novel candidate loci, including the PLSCR1 gene on chromosome 3, the PDLIM5 gene on chromosome 4, the SGPL1 gene on chromosome 10, and the MGST1 gene on chromosome 12. We also conducted a luciferase assay, and identified rs1061307 as the primary functional variant for the PLSCR1 gene.
In the current study, we identified the novel association of rs35766154 (PLSCR1 gene located on chromosome 3, p-value = 7.39 × 10−7, OR = 0.43, 95% CI = 0.31–0.61) with HBV chronicity. The PLSCR1 gene encodes PLSCR1, an α/β interferon (IFN)-inducible protein that has been shown to mediate antiviral activity against RNA viruses, such as vesicular stomatitis virus and encephalomyocarditis virus, through the enhancement of IFN responses (Dong et al., 2004). PLSCR1 was also observed to inhibit the replication of HBV in vivo and in vitro, as well as in a mouse model, through a significant decrease in HBV RNA and DNA intermediates and proteins (Yang et al., 2012), the activation of the Jak/Stat pathway (Yang et al., 2012), enhancement of IFN responses (Li Q. et al., 2016), and regulation of HBV-encoded X protein (HBx) stability (Yuan et al., 2015). In silico gene expression analysis of the PLSCR1 locus in the GTEx database (http://gtexportal.org/home/) (Lonsdale et al., 2013) showed that SNP rs1061307 was correlated with changes in PLSCR1 gene expression level in several tissues, with the most significant association observed in whole blood (p-value = 1.8 × 10−22). The PLSCR1 gene has several isoforms, and SNP rs1061307 is located in the promoter region of some of the isoforms. We further confirmed the significant association of rs1061307 alleles with PLSCR1 gene expression in Jurkat cells. This result suggested that rs1061307 is the primary functional variant for the PLSCR1 locus, affecting the risk of HBV chronicity in the Thai population.
There were three more suggestive associations observed on chromosomes 4, 10, and 12. Although further mapping and functional studies are needed to confirm these associations, there are some biologically plausible explanations. The SNP with the lowest p-value on chromosome 4 was rs62321986, which is located 28 kb 5’ of the PDLIM5 gene. This gene encodes Enigma Homolog 1 (ENH1, PDLIM5), a PDZ-LIM protein that interacts with and activates protein kinase C via the direct binding of its LIM domain (Kuroda et al., 1996; Maturana et al., 2011). Protein kinase C, among other protein kinases, phosphorylates HBV core protein, which modulates HBV replication at multiple stages (Kann and Gerlich, 1994; Jung et al., 2014). Another significant association was observed on chromosome 10 for SNP rs144998273 which is located in the intron of the SGPL1 gene. Based on the GTEx portal database (http://gtexportal.org/home/) (Lonsdale et al., 2013), different genotypes of rs144998273 are associated with different expression levels of the SGPL1 and PCBD1 (Pterin-4 Alpha-Carbinolamine Dehydratase 1) genes. The SGPL1 gene encodes sphingosine-1-phosphate lyase 1, which catalyzes the final degradative step in the sphingolipid metabolic pathway (Fyrst and Saba, 2008). Sphingolipids play important roles in several aspects of the HBV life cycle, and blockage of sphingolipid biosynthesis, which results in the inhibition of HBV replication, was suggested to be a potential therapeutic strategy for hepatitis B (Tatematsu et al., 2011). The PCBD1 gene encodes pterin-4 alpha-carbinolamine dehydratase, which is a dimerization cofactor for the transcription factor hepatocyte nuclear factor 1α (HNF1α), and it enhances the transcription activity of HNF1α (Sourdive et al., 1997). HNF1α downregulates the expression of HBV genes and viral replication by activating the NF-κB signaling pathway (Lin et al., 2017). The last non-HLA suggestive association in this study was observed with SNP rs1828682, which is located in the intron of the MGST1 gene on chromosome 12. This gene encodes Microsomal Glutathione S-Transferase 1, which is an important enzyme for redox control and detoxification (Morgenstern et al., 2011). It has been observed that HBV causes oxidative stress, which leads to liver carcinogenesis and cell death (Shirakata and Koike, 2003; Hu et al., 2011). Further functional studies and replication analyses in the Thai population as well as in other populations are required to confirm the associations detected in the current study.
The top SNPs of the HLA region reported by previous GWASs, such as rs3077 (HLA-DPA1) (Kamatani et al., 2009; Mbarek et al., 2011; Nishida et al., 2012), rs9277535 (HLA-DPB1) (Kamatani et al., 2009; Mbarek et al., 2011; Nishida et al., 2012), rs7453920 (HLA-DQ) (Mbarek et al., 2011), and rs2856718 (HLA-DQ) (Mbarek et al., 2011), were replicated in the current study, but not as the lead SNP. The GWAS top hit in the HLA region was rs7770370 (p-value = 7.71 × 10−10, OR = 0.49, 95% CI = 0.39–0.61). This SNP was reported as the lead SNP in two GWASs of chronic HBV infection and responsiveness to HBV vaccination (Wu et al., 2015; Huang et al., 2020). A positive correlation between the rs7770370 A allele and HLA-DPB1 protective alleles DPB1*02:01, 02:02, 03:01, 04:01, and 14:01 (r: 0.26–0.76; all p-values < 10–5) and a negative correlation between the rs7770370 A allele and HLA-DPB1 risk allele DPB1*05:01 (r: −0.76; p-values < 10–50) have been reported in a previous study (Wu et al., 2015). The high linkage between the rs7770370 A allele and HLA-DPB1 protective alleles might partly explain the protective effect of the SNP observed in the current study.
The important role of HLA class II genes in the chronicity of HBV infection has been extensively examined in different ethnicities (Singh et al., 2007; Kamatani et al., 2009; Nishida et al., 2014, 2015; Zhu et al., 2016). Here, we used a highly accurate HLA imputation method, and showed the association of several HLA alleles with chronic HBV infection in the Thai population. Our results revealed the significant association of HLA-DPA1*02:02, HLA-DPB1*05:01, HLA-DPB1*13:01, and HLA-DQB1*03:03 with susceptibility to HBV chronicity, and the significant association of HLA-DPA1*01:03, HLA-DPB1*02:01, and HLA-DQB1*06:09 with protection against HBV chronicity. HLA-DPA1*01:03 and HLA-DPB1*13:01 were previously shown to be significantly associated with chronic hepatitis B in the Thai population (Nishida et al., 2014) with protective and susceptibility effects, respectively. The HLA-DQB1*06:09 and HLA-DPA1/DPB1 alleles which we reported here, have been detected to be associated with chronic hepatitis B with similar tendencies in previous reports in different ethnic groups (Cho et al., 2008; Kamatani et al., 2009; Hu et al., 2013; Nishida et al., 2014, 2015, 2016; Li Y. et al., 2016; Zhu et al., 2016; Ou et al., 2019, 2021; Huang et al., 2020). However, HLA-DQB1*03:03 is associated with both an increased (Mbarek et al., 2011) and decreased risk of chronic hepatitis B (Xi-Lin et al., 2006; Zhang et al., 2015; Huang et al., 2016). No locus other than HLA-DPA1/DPB1 showed a significant independent association with HBV persistence after applying the regression analysis with the GWAS top hit as the covariate. However, the significant association of two alleles from the HLA-DQ locus (HLA-DQB1*03:03 and HLA-DQB1*06:09) was detected after HLA allele imputation. This shows the necessity of analyzing HLA alleles independently, because they may not be captured by GWAS arrays.
We also detected the association of two- and three-locus HLA haplotypes with HBV chronicity, including HLA-DPA1*01:03-DPB1*02:01 with protective effect, and HLA-DPA1*02:02-DPB1*05:01, HLA-DPB1*05:01-DQB1*03:03, HLA-DPB1*13:01-DQB1*05:02, and HLA-DPA1*02:02-DPB1*05:01-DQB1*03:03 with a susceptibility effect. The protective effect of HLA-DPA1*01:03-DPB1*02:01 against chronic hepatitis B was reported by Nishida et al. (Nishida et al., 2014). Although the association of HLA-DPB1*13:01 with susceptibility to HBV chronicity in Thai subjects was shown in the study by Nishida et al. (Nishida et al., 2014), no significant association of this allele was observed in any HLA-DPA1-DPB1 haplotypes in their study, which is consistent with our results. However, we detected the significant association of HLA-DPB1*13:01 in an HLA-DPB1-DQB1 haplotype (HLA-DPB1*13:01-DQB1*05:02), which again emphasizes the necessity of analyzing HLA alleles independently of HLA loci detected in GWASs. Additionally, HLA-DPB1*05:01 and three different haplotypes bearing this allele (HLA-DPA1*02:02-DPB1*05:01, HLA-DPB1*05:01-DQB1*03:03, and HLA-DPA1*02:02-DPB1*05:01-DQB1*03:03), were found to be significantly associated with susceptibility to chronic HBV infection in our study, but they were not observed to be associated with HBV chronicity in the Thai subjects in the study by Nishida et al. (Nishida et al., 2014); this discrepancy may be due to the relatively small number of healthy control subjects in their study. Two studies (Kamatani et al., 2009; Nishida et al., 2014) have reported HLA-DPA1*02:02-DPB1*05:01 as a susceptibility factor for HBV chronicity in the Japanese population. The consistency of our results with those of previous reports indicates the reliability of our findings as well as the advantage of HLA imputation for identifying disease-associated HLA alleles and loci.
There are some limitations to the current study. This study suffers from a limited sample size and lack of a replication sample set from the Thai population to validate the candidate associations detected in the GWAS stage in an independent cohort. However, we replicated the previously reported association of the HLA class II region with HBV persistence, which confirmed the reliability of our results in this cohort. In addition, the control subjects used in this study were healthy controls without previous exposure to HBV; they remain at the risk of being exposed to the virus and acquiring persistent infection, which may have limited our ability to compare the polymorphisms to those in patients with chronic HBV infection. To address these issues, larger sample sets from the Thai population and control subjects that have previously been exposed to HBV should be examined.
Conclusion
This study is the first GWAS of HBV chronicity in the Thai population. With the limitations discussed above, such as the limited sample size and lack of an independent replication sample set from the Thai population, PLSCR1 was detected as a potential genetic locus for chronic hepatitis B with plausible biological roles against HBV. Additionally, other loci, including PDLIM5 (rs62321986) and SGPL1 (rs144998273), were identified as susceptibility candidate loci for persistent HBV infection, and MGST1 (rs1828682) was identified as a protective candidate locus against persistent HBV infection. However, further research and functional studies in independent cohorts are strongly recommended. The observations of the current study indicate the necessity of conducting genomic research in diverse populations for the same disease, as differences in disease prevalence along with differences in allele frequencies and variant effect sizes in different populations may lead to detection of novel variants underlying the pathogenicity of the disease.
Materials and Methods
Samples
This study included 329 patients with chronic HBV infection who were recruited at Chulalongkorn University in Bangkok, Thailand, and 318 healthy control subjects who were blood donors at the National Blood Center, The Thai Red Cross Society, in Bangkok, Thailand. The diagnosis of chronic HBV infection was established according to the guideline for the diagnosis and treatment of chronic hepatitis B based on HBsAg seropositivity and elevated serum alanine aminotransferase levels (>1.5 times the upper limit of normal [35 IU/L]) for a period of longer than 6 months (with at least three bimonthly tests). Among the 329 patients, 100 were diagnosed with hepatocellular carcinoma, and 77 with liver cirrhosis. All healthy controls were seronegative for HBsAg and hepatitis B core antibody.
Genome-Wide SNP Genotyping and Genotype Calling
All samples were genotyped using the Axiom Genome-Wide ASI 1 Array (Thermo Fisher Scientific, Inc., Waltham, MA). Genotyping steps were performed according to the manufacturer’s protocol. The final data were produced as CEL files (intensity files). We conducted genotype calling for 600,014 SNPs using Affymetrix Genotyping Console software (v4.2.0.26). The Axiom GT1 algorithm was used for generating the genotype data. All samples had Dish QC > 0.82. One case sample with a genotype calling rate <0.97 was omitted. We then leveraged the Ps_Classification functionality of SNPolisher in Analysis Power Tools (Affymetrix, 2022) to calculate SNPs QC metrics. SNPs with good genotype quality were included in the list of recommended SNPs for downstream analysis. SNPs not included in the list were subsequently excluded from the data. SNP & Variation Suite software (Golden Helix Inc., Bozeman, MT, USA) was used to convert the results of genotype calling to input files for PLINK (v1.9) (Purcell et al., 2007).
IBD and PCA
Before conducting IBD, we performed linkage disequilibrium-based SNP pruning using PLINK (v1.9) (Purcell et al., 2007) with the option “--indep-pairwise 50 5 0.5”. We then excluded the SNPs in the prune.out output file, and conducted IBD using the options “--genome” and “--min 0.05”. One sample from each pair of samples with PI_HAT >0.1875 (halfway between the expected IBD for second- and third-degree relatives (Anderson et al., 2010)) was removed from the data.
For PCA, we downloaded the HapMap phase III data from the HapMap homepage (https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html). We merged the study data with four populations from the public HapMap data, i.e., Yoruba in Ibadan, Nigeria (YRI), Utah Residents (CEPH) with Northern and Western European Ancestry (CEU), Han Chinese in Beijing, China (CHB), and Japanese in Tokyo, Japan (JPT). QC was conducted on the data using PLINK (v1.9) (Purcell et al., 2007) with the options “--geno 0.03”, “--maf 0.01”, and “--hwe 0.000001”. Then, grm files were produced by GCTA software (Yang et al., 2011) using the option “--make-grm”. PCA was conducted by GCTA software (Yang et al., 2011) using the option “--pca 10”.
Whole-Genome Imputation
We performed pre-imputation QC and excluded SNPs with a MAF <0.01 and call rate <0.97 from all samples, and a HWE p-value <1 × 10−6 in healthy controls. The SNPs that passed the QC were pre-phased with 1,000 Genomes phase III as the reference data using SHAPEIT software (v2.17) (Delaneau et al., 2012). Whole-genome imputation of pre-phased data was conducted with 1,000 Genomes phase III as the reference panel using IMPUTE2 software (v2.3.2) (Howie et al., 2009) on 7-Mbp regions with the options “-Ne 2000”, “-k_hap 1,000”, “-k 120”, “-burnin 15”, and “-iter 50”. After imputation, SNPs with an info metric ≥0.8 were extracted.
Association Analysis
For the association analysis, PLINK (v1.9) (Purcell et al., 2007) was used with the options “--assoc”, “--adjust qq-plot”, and “--ci 0.95”.
Validation of the Association Results Using the TaqMan Assay
The TaqMan assay (Breen et al., 2000), which is a robust, accurate, cost-effective, and high-throughput technique for the discrimination of alleles differing by a single base change, was used in the current study for the validation of candidate SNPs. We conducted genotyping assays using made-to-order probes (Thermo Fisher Scientific, Waltham, MA) and the KAPA Probe Fast Universal qPCR Kit master mix (Kapa Biosystems, Boston, MA) according to the manufacturer’s instructions. The fluorescence intensity data from genotyping assays were converted into genotype calls by the Roche LightCycler 480 system (Roshe, Basal, Switzerland). The consistency rate of the genotype calls of candidate SNPs was calculated by comparison to those from the GWAS array.
HLA Typing
HLA typing was conducted using LABType SSO HLA DPA1/DPB1 kit (One Lambda, CA) and a Luminex Multi-Analyte Profiling system (xMAP; Luminex, Austin, TX) according to the manufacturer’s protocol. The genotyping results were compared to the imputation data and concordance rates were calculated.
Luciferase Assay
Specific polymerase chain reaction primers (Supplementary Table S10) were used to amplify the 5′UTR sequence of the PLSCR1 gene, including each allele of rs1061307 from human genomic DNA. Then, a NEBuilder HiFi DNA Assembly kit (New England Biolabs, Ipswich, MA) was used for the subcloning of the amplicons into the luciferase reporter pGL4.23 (luc2/minP) vector (Promega, Madison, WI) in the Nco I site (located in the translation start site of the luciferase gene). Lipofectamine 3,000 (Thermo-Fisher Scientific, Waltham, MA) was used to transfect pGL4.23 constructs of each allele (1 μg) and the pGL4.74 (hRluc/TK) vector (100 ng), which was used as an internal control, into the Jurkat and HepG2 cells. The Dual-Luciferase Reporter Assay System (Promega, Madison, WI) was used for measuring the luciferase activity. The relative luciferase activity of the major and minor alleles of rs1061307 was compared using Student’s t-test. p-values < 0.05 were regarded to be statistically significant.
Data Availability Statement
The GWAS data presented in the current study are deposited in the public database “NBDC Human Database”, accession number “hum0075.v3.Thai-gwas.v1”.
Ethics Statement
This study was approved by the Research Ethics Committees of the National Center for Global Health and Medicine, Tokyo, Japan (approval number NCGM-A-003266-02) and the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB No. 455/54 and IRB No. 431/58). All experimental procedures were conducted in accordance with the principles of the Declaration of Helsinki. All subjects provided written informed consent after receiving an explanation of the purpose of the research and the experimental protocols.
Author Contributions
KT, NN, MM, and YP conceived and designed the study. NP, PT, PK, VS, and YP collected the specimens and clinical information. SA analyzed the data. YH conducted the experiments. S-SK, YH, HS, NN, MS, MT, and YK helped to analyze the data. KT, MM, and YP guided the research process. SA wrote the manuscript. All authors reviewed the manuscript. All authors approved the final manuscript.
Funding
This work was supported by the Japan Agency for Medical Research and Development as a grant (JP20fk0210056) to NN, MS, KT, and MM; the Thailand Research Fund as a grant (RTA6280004) to PT; the National Research Council of Thailand as a grant (N34A640419) to YP; the Ratchadaphiseksomphot Endowment Fund (GRU 6105530009-1) of Chulalongkorn University to PK; the Health Systems Research Institute as a grant (64-132), and Ratchadapisek Sompoch Endowment Fund, Chulalongkorn University as a grant (764002-HE01) to VS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are deeply grateful to all the study subjects who participated in this study. We also thank the Center of Excellence in Clinical Virology for collecting the data and specimens.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.887121/full#supplementary-material
References
Affymetrix (2022). Affymetrix Website. Available at: http://www.affymetrix.com.
Akcay, I. M., Katrinli, S., Ozdil, K., Doganay, G. D., and Doganay, L. (2018). Host Genetic Factors Affecting Hepatitis B Infection Outcomes: Insights from Genome-wide Association Studies. World J. Gastroenterol. 24, 3347–3360. doi:10.3748/wjg.v24.i30.3347
Anderson, C. A., Pettersson, F. H., Clarke, G. M., Cardon, L. R., Morris, A. P., and Zondervan, K. T. (2010). Data Quality Control in Genetic Case-Control Association Studies. Nat. Protoc. 5, 1564–1573. doi:10.1038/nprot.2010.116
Ben-Ari, Z., Mor, E., Papo, O., Kfir, B., Sulkes, J., Tambur, A. R., et al. (2003). Cytokine Gene Polymorphisms in Patients Infected with Hepatitis B Virus. Am. J. Gastroenterol. 98, 144–150. doi:10.1111/j.1572-0241.2003.07179.x
Bertoletti, A., and Kennedy, P. T. (2015). The Immune Tolerant Phase of Chronic HBV Infection: New Perspectives on an Old Concept. Cell Mol. Immunol. 12, 258–263. doi:10.1038/cmi.2014.79
Breen, G., Harold, D., Ralston, S., Shaw, D., and Clair, D. St. (2000). Determining SNP Allele Frequencies in DNA Pools. BioTechniques 28, 464–470. doi:10.2144/00283st03
Chang, H.-Y., Ahn, S. H., Kim, D. Y., Shin, J.-S., Kim, Y.-S., Hong, S. P., et al. (2005). Association between CCR5 Promoter Polymorphisms and Hepatitis B Virus Infection. Korean J. Hepatol. 11, 116–124.
Chang, S.-W., Fann, C. S.-J., Su, W.-H., Wang, Y. C., Weng, C. C., Yu, C.-J., et al. (2014). A Genome-wide Association Study on Chronic HBV Infection and its Clinical Progression in Male Han-Taiwanese. PLoS One 9, e99724. doi:10.1371/journal.pone.0099724
Cho, S. W., Cheong, J. Y., Ju, Y. S., Oh, D. H., Suh, Y. J., and Lee, K. W. (2008). Human Leukocyte Antigen Class II Association with Spontaneous Recovery from Hepatitis B Virus Infection in Koreans: Analysis at the Haplotype Level. J. Korean Med. Sci. 23, 838–844. doi:10.3346/jkms.2008.23.5.838
Chonprasertsuk, S., and Vilaichone, R.-K. (2017). Epidemiology and Treatment of Hepatocellular Carcinoma in Thailand. Jpn. J. Clin. Oncol. 47, 294–297. doi:10.1093/jjco/hyw197
Delaneau, O., Marchini, J., and Zagury, J.-F. (2012). A Linear Complexity Phasing Method for Thousands of Genomes. Nat. Methods 9, 179–181. doi:10.1038/nmeth.1785
Deng, G., Zhou, G., Zhai, Y., Li, S., Li, X., Li, Y., et al. (2004). Association of Estrogen Receptor Alpha Polymorphisms with Susceptibility to Chronic Hepatitis B Virus Infection. Hepatology 40, 318–326. doi:10.1002/hep.20318
Dong, B., Zhou, Q., Zhao, J., Zhou, A., Harty, R. N., Bose, S., et al. (2004). Phospholipid Scramblase 1 Potentiates the Antiviral Activity of Interferon. J. Virol. 78, 8983–8993. doi:10.1128/JVI.78.17.8983-8993.2004
Excoffier, L., Laval, G., and Schneider, S. (2007). Arlequin (Version 3.0): An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinform Online 1, 47–50.
Frodsham, A. J., Zhang, L., Dumpis, U., Taib, N. A. M., Best, S., Durham, A., et al. (2006). Class II Cytokine Receptor Gene Cluster Is a Major Locus for Hepatitis B Persistence. PNAS 103, 9148–9153. doi:10.1073/pnas.0602800103
Fyrst, H., and Saba, J. D. (2008). Sphingosine-1-phosphate Lyase in Development and Disease: Sphingolipid Metabolism Takes Flight. Biochim. Biophys. Acta 1781, 448–458. doi:10.1016/j.bbalip.2008.05.005
Hirankarn, N., Manonom, C., Tangkijvanich, P., and Poovorawan, Y. (2007). Association of Interleukin-18 Gene Polymorphism (−607A/A Genotype) with Susceptibility to Chronic Hepatitis B Virus Infection. Tissue Antigens 70, 160–163. doi:10.1111/j.1399-0039.2007.00865.x
Howie, B. N., Donnelly, P., and Marchini, J. (2009). A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-wide Association Studies. PLoS Genet. 5, e1000529. doi:10.1371/journal.pgen.1000529
Hu, L., Chen, L., Yang, G., Li, L., Sun, H., Chang, Y., et al. (2011). HBx Sensitizes Cells to Oxidative Stress-Induced Apoptosis by Accelerating the Loss of Mcl-1 Protein via Caspase-3 Cascade. Mol. Cancer 10, 43. doi:10.1186/1476-4598-10-43
Hu, Z., Liu, Y., Zhai, X., Dai, J., Jin, G., Wang, L., et al. (2013). New Loci Associated with Chronic Hepatitis B Virus Infection in Han Chinese. Nat. Genet. 45, 1499–1503. doi:10.1038/ng.2809
Huang, J., Xiong, L., Wang, J., Liu, Y., Zhu, Q., Lei, J., et al. (2016). Association between the HLA-DQB1 Polymorphisms and the Susceptibility of Chronic Hepatitis B: A Comprehensive Meta-Analysis. Biomed. Rep. 4, 557–566. doi:10.3892/br.2016.632
Huang, Y.-H., Liao, S.-F., Khor, S.-S., Lin, Y.-J., Chen, H.-Y., Chang, Y.-H., et al. (2020). Large-scale Genome-wide Association Study Identifies HLA Class II Variants Associated with Chronic HBV Infection: a Study from Taiwan Biobank. Aliment. Pharmacol. Ther. 52, 682–691. doi:10.1111/apt.15887
Jiang, D.-K., Ma, X.-P., Yu, H., Cao, G., Ding, D.-L., Chen, H., et al. (2015). Genetic Variants in Five Novel Loci Including CFB and CD40 Predispose to Chronic Hepatitis B. Hepatology 62, 118–128. doi:10.1002/hep.27794
Jung, J., Hwang, S. G., Chwae, Y.-J., Park, S., Shin, H.-J., and Kim, K. (2014). Phosphoacceptors Threonine 162 and Serines 170 and 178 within the Carboxyl-Terminal RRRS/T Motif of the Hepatitis B Virus Core Protein Make Multiple Contributions to Hepatitis B Virus Replication. J. Virol. 88, 8754–8767. doi:10.1128/JVI.01343-14
Kamatani, Y., Wattanapokayakit, S., Ochi, H., Kawaguchi, T., Takahashi, A., Hosono, N., et al. (2009). A Genome-wide Association Study Identifies Variants in the HLA-DP Locus Associated with Chronic Hepatitis B in Asians. Nat. Genet. 41, 591–595. doi:10.1038/ng.348
Kann, M., and Gerlich, W. H. (1994). Effect of Core Protein Phosphorylation by Protein Kinase C on Encapsidation of RNA within Core Particles of Hepatitis B Virus. J. Virol. 68, 7993–8000. doi:10.1128/JVI.68.12.7993-8000.1994
Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., et al. (2002). The Human Genome Browser at UCSC. Genome Res. 12, 996–1006. doi:10.1101/gr.229102
Kim, Y. J., Kim, H. Y., Lee, J.-H., Yu, S. J., Yoon, J.-H., Lee, H.-S., et al. (2013). A Genome-wide Association Study Identified New Variants Associated with the Risk of Chronic Hepatitis B. Hum. Mol. Genet. 22, 4233–4238. doi:10.1093/hmg/ddt266
Kimkong, I., Chankaew, J., Kunanopparat, A., Hirankarn, N., and Tangkijvanich, P. (2015). Gene Polymorphisms of Interleukin 28B and the Risk to Chronic Hepatitis B Virus Infection in Thai. Tissue Antigens 85, 177–181. doi:10.1111/tan.12517
Kummee, P., Tangkijvanich, P., Poovorawan, Y., and Hirankarn, N. (2007). Association of HLA-Drb1*13 and TNF-α Gene Polymorphisms with Clearance of Chronic Hepatitis B Infection and Risk of Hepatocellular Carcinoma in Thai Population. J. Viral Hepat. 14, 841–848. doi:10.1111/j.1365-2893.2007.00880.x
Kuroda, S., Tokunaga, C., Kiyohara, Y., Higuchi, O., Konishi, H., Mizuno, K., et al. (1996). Protein-Protein Interaction of Zinc Finger LIM Domains with Protein Kinase C. J. Biol. Chem. 271, 31029–31032. doi:10.1074/jbc.271.49.31029
Li, Q., Zhang, B., Zhang, Q., Wang, X., Huo, Y., and Yang, J. (2016a). Effect of PLSCR1 on the Antiviral Activity of IFN against HBV in HepG2 Cells. Bing Du. Xue Bao 32, 747–751.
Li, Y., Si, L., Zhai, Y., Hu, Y., Hu, Z., Bei, J.-X., et al. (2016b). Genome-wide Association Study Identifies 8p21.3 Associated with Persistent Hepatitis B Virus Infection Among Chinese. Nat. Commun. 7, 11664. doi:10.1038/ncomms11664
Lin, J., Gu, C., Shen, Z., Liu, Y., Wang, W., Tao, S., et al. (2017). Hepatocyte Nuclear Factor 1α Downregulates HBV Gene Expression and Replication by Activating the NF-Κb Signaling Pathway. PLoS One 12, e0174017. doi:10.1371/journal.pone.0174017
Lin, T. M., Chen, C. J., Wu, M. M., Yang, C. S., Chen, J. S., Lin, C. C., et al. (1989). Hepatitis B Virus Markers in Chinese Twins. Anticancer Res. 9, 737–741.
Lonsdale, J., Thomas, J., Salvatore, M., Phillips, R., Lo, E., Shad, S., et al. (2013). The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 45, 580–585. doi:10.1038/ng.2653
Maturana, A. D., Nakagawa, N., Yoshimoto, N., Tatematsu, K., Hoshijima, M., Tanizawa, K., et al. (2011). LIM Domains Regulate Protein Kinase C Activity: a Novel Molecular Function. Cell Signal 23, 928–934. doi:10.1016/j.cellsig.2011.01.021
Mbarek, H., Ochi, H., Urabe, Y., Kumar, V., Kubo, M., Hosono, N., et al. (2011). A Genome-wide Association Study of Chronic Hepatitis B Identified Novel Risk Locus in a Japanese Population. Hum. Mol. Genet. 20, 3884–3892. doi:10.1093/hmg/ddr301
Morgenstern, R., Zhang, J., and Johansson, K. (2011). Microsomal Glutathione Transferase 1: Mechanism and Functional Roles. Drug Metab. Rev. 43, 300–306. doi:10.3109/03602532.2011.558511
Nishida, N., Ohashi, J., Khor, S.-S., Sugiyama, M., Tsuchiura, T., Sawai, H., et al. (2016). Understanding of HLA-Conferred Susceptibility to Chronic Hepatitis B Infection Requires HLA Genotyping-Based Association Analysis. Sci. Rep. 6, 24767. doi:10.1038/srep24767
Nishida, N., Ohashi, J., Sugiyama, M., Tsuchiura, T., Yamamoto, K., Hino, K., et al. (2015). Effects of HLA-DPB1 Genotypes on Chronic Hepatitis B Infection in Japanese Individuals. Tissue Antigens 86, 406–412. doi:10.1111/tan.12684
Nishida, N., Sawai, H., Kashiwase, K., Minami, M., Sugiyama, M., Seto, W.-K., et al. (2014). New Susceptibility and Resistance HLA-DP Alleles to HBV-Related Diseases Identified by a Trans-ethnic Association Study in Asia. PLoS One 9, e86449. doi:10.1371/journal.pone.0086449
Nishida, N., Sawai, H., Matsuura, K., Sugiyama, M., Ahn, S. H., Park, J. Y., et al. (2012). Genome-Wide Association Study Confirming Association of HLA-DP with Protection against Chronic Hepatitis B and Viral Clearance in Japanese and Korean. PLoS One 7, e39175. doi:10.1371/journal.pone.0039175
Ou, G., Liu, X., Xu, H., Ji, X., Liu, X., and Wang, J. (2021). Variation and Expression of HLA-DPB1 Gene in HBV Infection. Immunogenetics 73, 253–261. doi:10.1007/s00251-021-01213-w
Ou, G., Liu, X., Yang, L., Yu, H., Ji, X., Liu, F., et al. (2019). Relationship between HLA-DPA1 mRNA Expression and Susceptibility to Hepatitis B. J. Viral Hepat. 26, 155–161. doi:10.1111/jvh.13012
Podlaha, O., Gane, E., Brunetto, M., Fung, S., Chuang, W.-L., Pan, C. Q., et al. (2019). Large-scale Viral Genome Analysis Identifies Novel Clinical Associations between Hepatitis B Virus and Chronically Infected Patients. Sci. Rep. 9, 1–9. doi:10.1038/s41598-019-46609-7
Polaris Observatory Collaborators (2018). Global Prevalence, Treatment, and Prevention of Hepatitis B Virus Infection in 2016: a Modelling Study. Lancet Gastroenterol. Hepatol. 3, 383–403. doi:10.1016/S2468-1253(18)30056-6
Posuwan, N., Payungporn, S., Tangkijvanich, P., Ogawa, S., Murakami, S., Iijima, S., et al. (2014). Genetic Association of Human Leukocyte Antigens with Chronicity or Resolution of Hepatitis B Infection in Thai Population. PLoS One 9, e86007. doi:10.1371/journal.pone.0086007
Posuwan, N., Vuthitanachot, V., Chinchai, T., Wasitthankasem, R., Wanlapakorn, N., and Poovorawan, Y. (20192017). Serological Evidence of Hepatitis A, B, and C Virus Infection in Older Adults in Khon Kaen, Thailand and the Estimated Rates of Chronic Hepatitis B and C Virus Infection in Thais. PeerJ 7, e7492. doi:10.7717/peerj.7492
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Qiu, B., Wang, X., Zhang, P., Shi, C., Zhang, J., Qiu, W., et al. (2012). Association of TNF-α Promoter Polymorphisms with the Outcome of Persistent HBV Infection in a Northeast Chinese Han Population. Acta Biochim. Biophys. Sin. (Shanghai) 44, 712–718. doi:10.1093/abbs/gms046
Riazalhosseini, B., Mohamed, Z., Apalasamy, Y. D., Shafie, N. S., and Mohamed, R. (2018). Interleukin-6 Gene Variants Are Associated with Reduced Risk of Chronicity in Hepatitis B Virus Infection in a Malaysian Population. Biomed. Rep. 9, 213–220. doi:10.3892/br.2018.1126
Sakaue, S., Kanai, M., Tanigawa, Y., Karjalainen, J., Kurki, M., Koshiba, S., et al. (2021). A Cross-Population Atlas of Genetic Associations for 220 Human Phenotypes. Nat. Genet. 53, 1415–1424. doi:10.1038/s41588-021-00931-x
Shirakata, Y., and Koike, K. (2003). Hepatitis B Virus X Protein Induces Cell Death by Causing Loss of Mitochondrial Membrane Potential. J. Biol. Chem. 278, 22071–22078. doi:10.1074/jbc.M301606200
Singh, R., Kaul, R., Kaul, A., and Khan, K. (2007). A Comparative Review of HLA Associations with Hepatitis B and C Viral Infections across Global Populations. World J. Gastroenterol. 13, 1770–1787. doi:10.3748/wjg.v13.i12.1770
Sourdive, D. J., Transy, C., Garbay, S., and Yaniv, M. (1997). The Bifunctional DCOH Protein Binds to HNF1 Independently of its 4-Alpha-Carbinolamine Dehydratase Activity. Nucleic Acids Res. 25, 1476–1484. doi:10.1093/nar/25.8.1476
Tatematsu, K., Tanaka, Y., Sugiyama, M., Sudoh, M., and Mizokami, M. (2011). Host Sphingolipid Biosynthesis Is a Promising Therapeutic Target for the Inhibition of Hepatitis B Virus Replication. J. Med. Virol. 83, 587–593. doi:10.1002/jmv.21970
Thursz, M. (2001). Genetic Susceptibility in Chronic Viral Hepatitis. Antivir. Res. 52, 113–116. doi:10.1016/S0166-3542(01)00175-9
Williams, T. M. (2001). Human Leukocyte Antigen Gene Polymorphism and the Histocompatibility Laboratory. J. Mol. Diagn 3, 98–104. doi:10.1016/S1525-1578(10)60658-7
Wu, T.-W., Chen, C.-F., Lai, S.-K., Lin, H. H., Chu, C.-C., and Wang, L.-Y. (2015). SNP Rs7770370 in HLA-DPB1 Loci as a Major Genetic Determinant of Response to Booster Hepatitis B Vaccination: Results of a Genome-wide Association Study. J. Gastroenterol. Hepatol. 30, 891–899. doi:10.1111/jgh.12845
Xi-Lin, Z., Te, D., Jun-Hong, L., Liang-Ping, L., Xin-Hui, G., Ji-Rong, G., et al. (2006). Analysis of HLA-DQB1 Gene Polymorphisms in Asymptomatic HBV Carriers and Chronic Hepatitis B Patients in the Chinese Han Population. Int. J. Immunogenet 33, 249–254. doi:10.1111/j.1744-313X.2006.00607.x
Xu, B., Wang, Y., Deng, G., Huang, Y., Zhong, L., Liu, G., et al. (2004). The Primary Comparative Analysis between the Host Genetic Factors and Their Relationships with Clinical Phenotype of HBV Infected Twins. Zhonghua Yi Xue Za Zhi 84, 189–193. doi:10.3760/j:issn:0376-2491.2004.03.005
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). GCTA: a Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 88, 76–82. doi:10.1016/j.ajhg.2010.11.011
Yang, J., Zhu, X., Liu, J., Ding, X., Han, M., Hu, W., et al. (2012). Inhibition of Hepatitis B Virus Replication by Phospholipid Scramblase 1 In Vitro and In Vivo. Antivir. Res. 94, 9–17. doi:10.1016/j.antiviral.2012.01.010
Yuan, Y., Tian, C., Gong, Q., Shang, L., Zhang, Y., Jin, C., et al. (2015). Interactome Map Reveals Phospholipid Scramblase 1 as a Novel Regulator of Hepatitis B Virus X Protein. J. Proteome Res. 14, 154–163. doi:10.1021/pr500943x
Zeng, Z., Liu, H., Xu, H., Lu, H., Yu, Y., Xu, X., et al. (2021). Genome-wide Association Study Identifies New Loci Associated with Risk of HBV Infection and Disease Progression. BMC Med. Genomics 14, 84. doi:10.1186/s12920-021-00907-0
Zhang, Y., Zhao, F., Lan, L., Qin, Z., and Jun, L. (2015). Correlation of HLA-DQB1 Gene Polymorphism of Xinjiang Uygur with Outcome of HBV Infection. Int. J. Clin. Exp. Med. 8, 6067–6072.
Keywords: chronic hepatitis B, GWAS, SNP, HLA, allele, haplotype
Citation: Ashouri S, Khor S-S, Hitomi Y, Sawai H, Nishida N, Sugiyama M, Kawai Y, Posuwan N, Tangkijvanich P, Komolmit P, Tsuiji M, Shotelersuk V, Poovorawan Y, Mizokami M and Tokunaga K (2022) Genome-Wide Association Study for Chronic Hepatitis B Infection in the Thai Population. Front. Genet. 13:887121. doi: 10.3389/fgene.2022.887121
Received: 01 March 2022; Accepted: 16 May 2022;
Published: 13 June 2022.
Edited by:
Corrado Romano, University of Catania, ItalyReviewed by:
Martin Maiers, National Marrow Donor Program, United StatesLi Shi, Chinese Academy of Medical Sciences, China
Copyright © 2022 Ashouri, Khor, Hitomi, Sawai, Nishida, Sugiyama, Kawai, Posuwan, Tangkijvanich, Komolmit, Tsuiji, Shotelersuk, Poovorawan, Mizokami and Tokunaga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saeideh Ashouri, c2FzaG91cmlAcmkubmNnbS5nby5qcA==; Katsushi Tokunaga, a2F0b2t1bmFnYUByaS5uY2dtLmdvLmpw
 Saeideh Ashouri
Saeideh Ashouri Seik-Soon Khor1,2
Seik-Soon Khor1,2 Yuki Hitomi
Yuki Hitomi Nao Nishida
Nao Nishida Masaya Sugiyama
Masaya Sugiyama Pisit Tangkijvanich
Pisit Tangkijvanich Makoto Tsuiji
Makoto Tsuiji Vorasuk Shotelersuk
Vorasuk Shotelersuk Yong Poovorawan
Yong Poovorawan Masashi Mizokami
Masashi Mizokami Katsushi Tokunaga
Katsushi Tokunaga