
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Genet. , 12 May 2022
Sec. Genetics of Common and Rare Diseases
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.881284
This article is part of the Research Topic Unravelling the Basis of Non-Invasive Prenatal Screening Results View all 11 articles
 Giulia Bonanni1
Giulia Bonanni1 Valentina Trevisan2,3
Valentina Trevisan2,3 Marcella Zollino2,3
Marcella Zollino2,3 Marco De Santis4
Marco De Santis4 Federica Romanzi1,4
Federica Romanzi1,4 Antonio Lanzone1,4
Antonio Lanzone1,4 Elisa Bevilacqua4*
Elisa Bevilacqua4*Since the introduction of cell-free (cf) DNA analysis, Non-Invasive Prenatal Testing (NIPT) underwent a deep revolution. Pregnancies at high risk for common fetal aneuploidies can now be easily identified through the analysis of chromosome-derived components found in maternal circulation, with the highest sensitivity and specificity currently available. Consequently, the last decade has witnessed a widespread growth in cfDNA-based NIPT use, enough to be often considered an alternative method to other screening modalities. Nevertheless, the use of NIPT in clinical practice is still not devoid of discordant results. Hereby, we report a case of confined placental mosaicism (CPM) in which a NIPT false-positive result for trisomy 13 required not only amniocentesis but also cordocentesis, to rule out the fetal aneuploidy, with the additional support of molecular cytogenetics on placental DNA at delivery. Relevant aspects allowing for precision genetic diagnosis and counselling, including the number of analysed metaphases on the different fetal cells compartments and a repeated multidisciplinary evaluation, are discussed.
Cell-free (cf) DNA-based Non-Invasive Prenatal Testing (NIPT) is widely considered to be the most sensitive and specific screening option for trisomy 21, 18, and 13. However, some concerns regarding its clinical role in routine obstetric care persist. These include, inter alia, the reliability of Positive Predictive Value (PPV) estimates. According to the most recent metanalyses (Gil et al., 2015; Taylor-Philips et al., 2016; Iwarsson et al., 2017; Mackie et al., 2017), the combined False-Positive Rate (FPR) in successful tests is 0.15%. In this sense, most studies on NIPT performance still suffer from a high risk of bias, in particular, the reported FPRs are likely to be underestimated.
As it is well known, circulating cfDNA derives from both the mother and the fetal-placental unit. Consequently, the main sources of unreliability of NIPT are Confined Placental Mosaicism (CPM), maternal copy number variants, vanishing twin, and maternal cancer (Grati et al., 2014).
Despite these shortcomings, obstetric care providers are increasingly prone to prescribe cfDNA analysis as an alternative or stand-alone screening method compared to ultrasound examinations. Moreover, there is still some controversy concerning the standard protocols that would best investigate fetal anomalies during the first trimester.
With this in mind, we report a case of CPM in which a NIPT false-positive result for trisomy 13 required two further invasive diagnostic tests–an amniocentesis and a cordocentesis - to rule out the fetal aneuploidy. Molecular cytogenetics performed on placental DNA at the delivery could add relevant data for the unequivocal diagnosis of CPM.
A 31-year-old, gravida 1, para 0, Caucasian woman was referred to our hospital (Agostino Gemelli University Policlinic, Rome, RM, Italy) at 19 2/7 weeks of gestation for evaluation of suspected trisomy 13. Her previous medical and obstetric history had been unremarkable. Screening and diagnostic steps are presented in Figure 1. The first-trimester ultrasound findings were normal. At 12 2/7 weeks of gestation, she underwent the PrenatalSAFE® 5 Test (Eurofins Genoma Group Srl, Rome, Italy) through ILLUMINA VeriSeq NIPT sequencing systems, which revealed a suspected aneuploidy in chromosome 13 with a Fetal Fraction (FF) of 11% and a PPV of 92,86%. At 14 6/7 weeks of gestation, she underwent amniocentesis to confirm the positive NIPT result. By analysing 77 metaphases, we found that all but one had a normal male chromosome constitution, 46 XY. The unique cell with trisomy 13 we observed was first consistent with CPM, or, alternatively, with a very low mosaicism for trisomy 13 in the fetus.
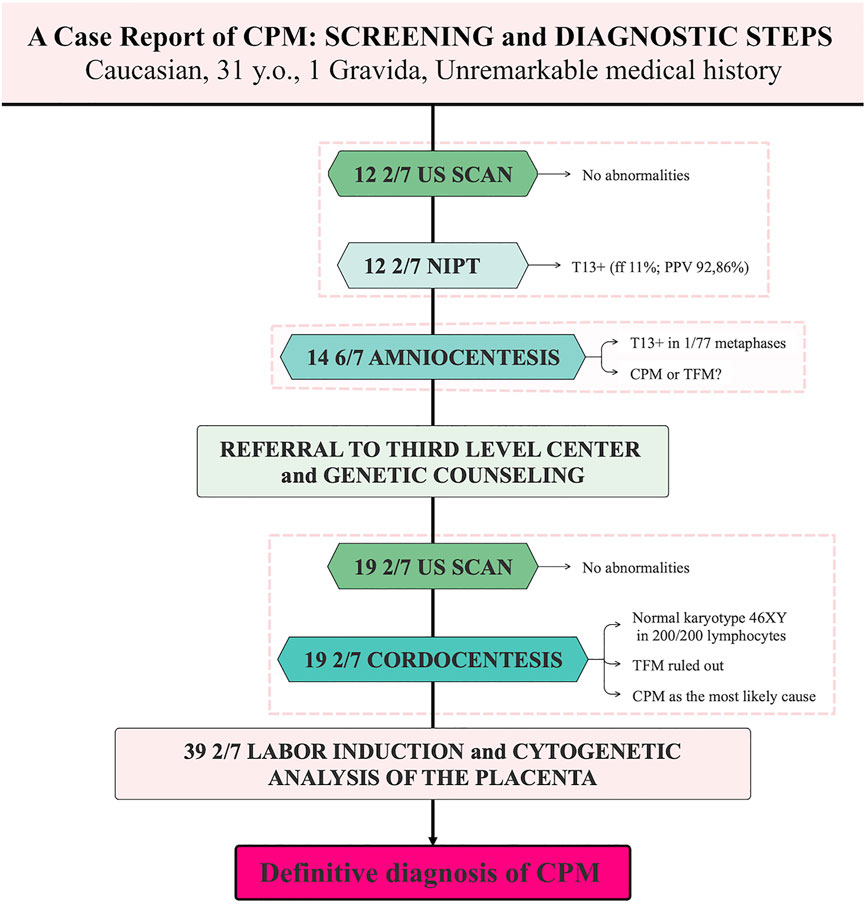
FIGURE 1. Screening and diagnostic steps in a case of Confined Placental Mosaicism. CPM, Confined Placental Mosaicism; TFM, True Fetal Mosaicism; ff, Fetal Fraction; PPV, Positive Predictive Value.
A detailed fetal ultrasound was carried out at 19 2/7 weeks of gestation with normal results.
At 19 2/7 weeks of gestation, upon genetic counselling, the couple also decided to undergo cordocentesis to rule out a True Fetal Mosaicism (TFM) for trisomy 13. The analysis of 200 fetal lymphocytes confirmed a normal male karyotype 46, XY in the totality of cells. Accordingly, CPM was considered to explain the previous results.
Subsequent ultrasound scans proved to be within normal limits.
Labor induction was performed at 39 2/7 weeks of gestation for reduction of the fetal growth trend (from 64th to 23rd percentile), that was considered to reflect the final functional dysfunction of the trisomic placenta. A male neonate with a birth weight of 3430 g and Apgar scores of 9–10 at 1–5 min, respectively, was born by vaginal delivery. Physiological newborn jaundice was present, and the infant did not present any phenotypic anomaly. Molecular cytogenetic examination (Array-CGH) of the placenta, revealed a complete trisomy of chromosome 13 in about 20% of the analysed genome, allowing to definitively establish the diagnosis of CPM. During a final genetic counselling, no risk for phenotypic abnormalities was given to the newborn, and amniocentesis for fetal chromosomal examination was suggested in subsequent pregnancies of the parents.
In a seminal paper, Lo and Wainscoat (Lo et al., 1997) described for the first time the presence of fetal DNA in maternal plasma. Since then, several studies (Sekizawa et al., 2000; Bianchi, 2004; Tjoa et al., 2006) have been carried out to investigate cfDNA mechanisms of release during pregnancy, demonstrating both the maternal and the fetal-placental unit origin. Despite the high sensitivity and specificity of currently available PCR and MPS analytical techniques for the study of cfDNA (Bianchi et al., 2012), the primary trophoblastic origin of the latter is a known driver of the relatively large number of false positive and false negative results (Grati et al., 2014). The upshot of this is the designation of NIPT as a screening - and not a diagnostic - test. In this context, the present case highlights the shortcomings of NIPT when used as an alternative to the first trimester ultrasound scan for the screening of the most common aneuploidies. We aim at evaluating some critical aspects that should be considered in protocol decision-making practices and which could best investigate fetal anomalies during the first trimester.
First, it is interesting to note that Fetal Blood Sampling (FBS) through cordocentesis can sometimes be diriment to rule out a fetal aneuploidy after a NIPT positive result. In our case, normal ultrasound findings against the background of a positive result for trisomy 13 at NIPT led initially to consider amniocentesis the best diagnostic tool to avoid erroneous results due to CPM. Despite this, cordocentesis was then deemed necessary to exclude a fetal mosaicism. This means not only exposure to all the potential risks of FBS - including bleeding from the puncture site, fetal bradycardia, pregnancy loss and vertical transmission of maternal infection (Berry et al., 2013) - but also a 47-days delay in the final response, resulting in substantial psychological stress over a long period. Such an emotional strain should be avoided since it could lead, in extremis, to an improper decision to have a first-trimester abortion for the sake of the mother’s health.
This emphasizes, above all, how important both pre- and post-test counselling are, allowing patients to understand the difference between a screening and a diagnostic test. In this sense, we believe that the best prenatal practice encompasses the interpretation of both positive and negative NIPT results in view of other screening modalities’ findings (Salomon et al., 2017). Conversely, most laboratories report the average risk in the screen-positive patient as a PPV, disregarding the prior-test risk based on age, ultrasound, prior history and screen-positive serum test. Abnormal findings at NIPT, contrasting with normal fetus development at ultrasound scan, could disclose other biological causes (Hartwig et al., 2017), such as maternal Copy Number Variations (CNVs) and Confined Placental Mosaicism (CPM) (Mardy & Wapner, 2016). In this context, even if the performance of NIPT is higher, the first trimester ultrasound scan has been proved to potentially change clinical management in almost one in 10 women if performed prior to cfDNA screening (Brown et al., 2020). This is especially the case of trisomies 18 and 13, for which a detailed ultrasound examination can detect characteristic defects.
The present case also maintains the need of a careful perinatal management when CPM is suspected. After ruling out recognized risk factors such as constitutional chromosomal abnormalities, the rate of infants with Intrauterine Growth Restriction (IUGR) associated with CPM has been estimated to be 10 times higher than in the appropriately grown controls infants (Wilkins-Haug et al., 2006). Notwithstanding this, a recent retrospective cohort study did not confirm any significant association between CPM and adverse pregnancy outcomes except for CPM for trisomy 16 (Grati et al., 2020).
In conclusion, this case provides significant clinical considerations on using NIPT in daily practice. In particular, it presents the major pitfalls of interpreting screening tests’ findings in the absence of a mutual work of integration. In recent years, we have witnessed an uncontrolled spread of cf-DNA analysis for which the economic interest of the industry has certainly contributed. In this context, a responsible integration of similar technological innovations should always be sought in clinical experience. Our clinical experience confirms that, given the trophoblastic origin of cf-DNA, NIPT cannot but be a screening test. The real benefit of cfDNA analysis lies, therefore, in its complementary use with ultrasound scan, which helps to shed light on the most likely risk of fetal aneuploidy. We suggest that, in case of cfDNA testing positive for T21, T18, and T13 during first trimester screening, in the absence of anomalies detected during the ultrasound examination, an invasive procedure by Chorionic Villus Sampling (CVS) could be recommended only for T21, since in such a case the risk of confined placental mosaicism is about 1–2%, which is comparable to the risk of mosaicism in the general population. Conversely, for T18 or T13, the best management would be to offer an amniocentesis because the risk of confined placental mosaicism is high: 3–4% for T18 and 22% for 13 (Grati et al., 2014; Grati et al., 2015; Malvestiti et al., 2015). However, based on our observation, cordocentesis can be also required to definitively rule out the fetal aneuploidy. Analysis of a larger number of metaphases from fetal blood cells, with respect to amniocytes, is recommended in these cases.
Finally, we strongly emphasize the importance of an adequate education of all obstetrical providers in order to maximize the benefit brought by cfDNA analyses. Moreover, we notice that further research is needed to examine the extent to which maternal risk factors (e.g., age, obesity, hypertension, diabetes) influence the incidence of IUGR associated to CPM.
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding author.
The written consent of the patient for the publication of the case was obtained according to the guidelines.
BG: conceptualization, data curation, writing—original draft, project administration TV: review; editing; ZM: writing—review; editing; DSM: review; editing, RF: review; editing, LA: review; editing BE: validation, writing—review; editing, supervision.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ministero della Salute - Ricerca Corrente 2022.
Berry, S. M., Stone, J., Norton, M. E., Johnson, D., and Berghella, V. (2013). Fetal Blood Sampling. Am. J. Obstetrics Gynecol. 209 (3), 170–180. doi:10.1016/j.ajog.2013.07.014
Bianchi, D. W. (2004). Circulating Fetal DNA: its Origin and Diagnostic Potential-A Review. Placenta 25 (Suppl. A), S93–S101. doi:10.1016/j.placenta.2004.01.005
Bianchi, D. W., Platt, L. D., Goldberg, J. D., Abuhamad, A. Z., Sehnert, A. J., and Rava, R. P. (2012). Genome-Wide Fetal Aneuploidy Detection by Maternal Plasma DNA Sequencing. Obstetrics Gynecol. 119 (5), 957890–957901. doi:10.1097/AOG.0b013e31824fb482
Brown, I., Fernando, S., Menezes, M., Silva Costa, F., Ramkrishna, J., Meagher, S., et al. (2020). The Importance of Ultrasound Preceding Cell‐free DNA Screening for Fetal Chromosomal Abnormalities. Prenat. Diagn. 40 (11), 1439–1446. doi:10.1002/pd.5788
Gil, M. M., Quezada, M. S., Revello, R., Akolekar, R., and Nicolaides, K. H. (2015). Analysis of Cell-free DNA in Maternal Blood in Screening for Fetal Aneuploidies: Updated Meta-Analysis. Ultrasound Obstet. Gynecol. 45 (3), 249–266. doi:10.1002/uog.14791
Grati, F. R., Ferreira, J., Benn, P., Izzi, C., Verdi, F., Vercellotti, E., et al. (2020). Outcomes in Pregnancies with a Confined Placental Mosaicism and Implications for Prenatal Screening Using Cell-free DNA. Genet. Med. 22 (2), 309–316. doi:10.1038/s41436-019-0630-y
Grati, F. R., Malvestiti, F., Ferreira, J. C. P. B., Bajaj, K., Gaetani, E., Agrati, C., et al. (2014). Fetoplacental Mosaicism: Potential Implications for False-Positive and False-Negative Noninvasive Prenatal Screening Results. Genet. Med. 16 (8), 620–624. doi:10.1038/gim.2014.3
Grati, F. R., Molina Gomes, D., Ferreira, J. C. P. B., Dupont, C., Alesi, V., Gouas, L., et al. (2015). Prevalence of Recurrent Pathogenic Microdeletions and Microduplications in over 9500 Pregnancies. Prenat. Diagn 35 (8), 801–809. doi:10.1002/pd.4613
Hartwig, T. S., Ambye, L., Sørensen, S., and Jørgensen, F. S. (2017). Discordant Non-invasive Prenatal Testing (NIPT) - a Systematic Review. Prenat. Diagn 37 (6), 527–539. doi:10.1002/pd.5049
Iwarsson, E., Jacobsson, B., Dagerhamn, J., Davidson, T., Bernabé, E., and Heibert Arnlind, M. (2017). Analysis of Cell-free Fetal DNA in Maternal Blood for Detection of Trisomy 21, 18 and 13 in a General Pregnant Population and in a High Risk Population - a Systematic Review and Meta-Analysis. Acta Obstet. Gynecol. Scand. 96 (1), 7–18. doi:10.1111/aogs.13047
Lo, Y. M. D., Corbetta, N., Chamberlain, P. F., Rai, V., Sargent, I. L., Redman, C. W., et al. (1997). Presence of Fetal DNA in Maternal Plasma and Serum. Lancet 350 (9076), 485–487. doi:10.1016/S0140-6736(97)02174-0
Mackie, F., Hemming, K., Allen, S., Morris, R., and Kilby, M. (2017). The Accuracy of Cell-free Fetal DNA-Based Non-invasive Prenatal Testing in Singleton Pregnancies: a Systematic Review and Bivariate Meta-Analysis. BJOG Int. J. Obstet. Gy 124 (1), 32–46. doi:10.1111/1471-0528.14050
Malvestiti, F., Agrati, C., Grimi, B., Pompilii, E., Izzi, C., Martinoni, L., et al. (2015). Interpreting Mosaicism in Chorionic Villi: Results of a Monocentric Series of 1001 Mosaics in Chorionic Villi with Follow-Up Amniocentesis. Prenat. Diagn 35 (11), 1117–1127. doi:10.1002/pd.4656
Mardy, A., and Wapner, R. J. (2016). Confined Placental Mosaicism and its Impact on Confirmation of NIPT Results. Am. J. Med. Genet. 172 (2), 118–122. doi:10.1002/ajmg.c.31505
Salomon, L. J., Alfirevic, Z., Audibert, F., Kagan, K. O., Paladini, D., Yeo, G., et al. (2017). ISUOG Updated Consensus Statement on the Impact of cfDNA Aneuploidy Testing on Screening Policies and Prenatal Ultrasound Practice. Ultrasound Obstet. Gynecol. 49 (6), 815–816. doi:10.1002/uog.17483
Sekizawa, A., Samura, O., Zhen, D., Falco, V., Farina, A., and Bianchi, D. W. (2000). Apoptosis in Fetal Nucleated Erythrocytes Circulating in Maternal Blood. Prenat. Diagn. 20 (11), 886–889. doi:10.1002/1097-0223(200011)20:11<886::aid-pd942>3.0.co;2-4
Taylor-Phillips, S., Freeman, K., Geppert, J., Agbebiyi, A., Uthman, O. A., Madan, J., et al. (2016). Accuracy of Non-invasive Prenatal Testing Using Cell-free DNA for Detection of Down, Edwards and Patau Syndromes: a Systematic Review and Meta-Analysis. BMJ Open 6 (1), e010002. doi:10.1136/bmjopen-2015-010002
Tjoa, M. L., Cindrova-Davies, T., Spasic-Boskovic, O., Bianchi, D. W., and Burton, G. J. (2006). Trophoblastic Oxidative Stress and the Release of Cell-free Feto-Placental DNA. Am. J. Pathology 169 (2), 400–404. doi:10.2353/ajpath.2006.060161
Keywords: NIPT, cfDNA, confined placental mosaicism, prenatal diagnosis, aneuploidies
Citation: Bonanni G, Trevisan V, Zollino M, De Santis M, Romanzi F, Lanzone A and Bevilacqua E (2022) Case Report: Challenges of Non-Invasive Prenatal Testing (NIPT): A Case Report of Confined Placental Mosaicism and Clinical Considerations. Front. Genet. 13:881284. doi: 10.3389/fgene.2022.881284
Received: 22 February 2022; Accepted: 26 April 2022;
Published: 12 May 2022.
Edited by:
Luigia De Falco, AMES, centro Polidiagnostico Strumentale, srl, ItalyReviewed by:
Balint Nagy, University of Debrecen, HungaryCopyright © 2022 Bonanni, Trevisan, Zollino, De Santis, Romanzi, Lanzone and Bevilacqua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisa Bevilacqua, ZWxpc2EuYmV2aWxhY3F1YUBwb2xpY2xpbmljb2dlbWVsbGkuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.