- 1Suzhou Polytechnic Institute of Agriculture, Suzhou, China
- 2Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, China
- 3Seed Administrative Station of Suzhou, Suzhou, China
- 4College of Agricultural and Biological Engineering (College of Tree Peony), Heze University, Heze, China
NBS-LRR genes are the largest gene family in plants conferring resistance to pathogens. At present, studies on the evolution of NBS-LRR genes in angiosperms mainly focused on monocots and eudicots, while studies on NBS-LRR genes in the basal angiosperms are limited. Euryale ferox represents an early-diverging angiosperm order, Nymphaeales, and confronts various pathogens during its lifetime, which can cause serious economic losses in terms of yield and quality. In this study, we performed a genome-wide identification and analysis of NBS-LRR genes in E. ferox. All 131 identified NBS-LRR genes could be divided into three subclasses according to different domain combinations, including 18 RNLs, 40 CNLs, and 73 TNLs. The E. ferox NBS-LRR genes are unevenly distributed on 29 chromosomes; 87 genes are clustered at 18 multigene loci, and 44 genes are singletons. Gene duplication analysis revealed that segmental duplications acted as a major mechanism for NBS-LRR gene expansions but not for RNL genes, because 18 RNL genes were scattered over 11 chromosomes without synteny loci, indicating that the expansion of RNL genes could have been caused by ectopic duplications. Ancestral gene reconciliation based on phylogenetic analysis revealed that there were at least 122 ancestral NBS-LRR lineages in the common ancestor of the three Nymphaeaceae species, suggesting that NBS-LRR genes expanded slightly during speciation in E. ferox. Transcriptome analysis showed that the majority of NBS-LRR genes were at a low level of expression without pathogen stimulation. Overall, this study characterized the profile of NBS-LRR genes in E. ferox and should serve as a valuable resource for disease resistance breeding in E. ferox.
Introduction
Euryale ferox (prickly waterlily) is an annual aquatic plant in tropical and subtropical regions of Southeast and East Asia and is the only species in the Euryale genus in the Nymphaeaceae family. It is cultivated as a nutritious food due to its high starch content; additionally it is also a common Chinese traditional medicine which can treat depression and diabetes mellitus (Ahmed et al., 2015; Wu et al., 2017; Huang et al., 2018; Liu et al., 2018). However, the productivity of E. ferox is threatened by various pests and microbial pathogens, including fungi, bacteria, and viruses. The diseases caused by these pathogens pose serious threats to not only plant growth and productivity but also the quality of the edible tissues. However, up to now, no functional disease resistance genes (R genes) have been cloned from E. ferox.
In order to cope with pathogen invasion, plants have developed sophisticated immune systems during the long-term evolutionary process. R genes in plants, which play a core role of the immune system, detect pathogens and trigger downstream resistance response, frequently accompanied by hypersensitive reactions (HR) (Cui et al., 2015; Jones et al., 2016; Wang and Chai, 2020). The nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes are the largest R gene family in plants. To date, more than 60% of the functional resistance genes cloned from angiosperms belong to the NBS-LRR gene family, which can provide resistance to bacteria, fungi, viruses, nematodes, and other pathogens (Kourelis and van der Hoorn, 2018). NBS-LRR genes originated in the common ancestor of all green plants, and diverged early into different subclasses (subfamilies) with different domain combinations along plant evolution (Xue et al., 2012; Shao et al., 2019). According to the N-terminal domain, angiosperm NBS-LRR genes can be divided into three subclasses: TIR-NBS-LRR (TNL), CC-NBS-LRR (CNL), and RPW8-NBS-LRR (RNL) (Shao et al., 2016). The majority of TNL and CNL proteins usually serve as pathogen detectors (Kourelis and van der Hoorn, 2018). NBS-LRR proteins are usually in a signaling-competent yet autoinhibited state, with LRR domain folding back onto the central NACHT domain (Hu et al., 2013). After recognizing pathogen effectors injected into the host cell, NBS-LRR proteins are activated, and NBS domain underwent conformational alterations, with exposed N-terminal domains to trigger downstream hypersensitive reactions, finally eliciting apoptosis of infected cells to suppress the pathogens transmission and proliferation (Andersen et al., 2018). The low expression of NBS-LRR genes are considered a normal stage and logically makes sense, otherwise activated HR would cause apoptosis all over plant organs and tissues. The RNL subclass can be divided into two small and ancient subclasses, namely ADR1 and NRG1, which can transduce immune signals and function downstream of ‘sensor NBS-LRR’ (sNLR) activation, so it was called ‘helper NBS-LRR’ (hNLR) (Collier et al., 2011; Shao et al., 2016; Zhong and Cheng, 2016). Furthermore, NRG1 proteins have advantages in TNL-mediated immune signaling (Peart et al., 2005; Qi et al., 2018; Castel et al., 2019; Wu et al., 2019). Recent studies indicated that as Ca2+-permeable channels, CNL and RNL proteins provoked immune response and cell death (Bi et al., 2021; Jacob et al., 2021).
Since the genome-wide study of NBS-LRR in Arabidopsis thaliana and rice early in the 21st century, NBS-LRR genes have been identified and analyzed in dozens of plant genomes (Meyers et al., 2003). These studies have found that the number of NBS-LRR genes varies greatly among different species, ranging from dozens to hundreds (Liu et al., 2021; Luo et al., 2012; Qian et al., 2017; Shao et al., 2014; Xue et al., 2020; Zhang et al., 2016; Zhou et al., 2020). These results provided abundant data resources for studying the structure, mechanism, and evolution of NBS-LRR genes. However, the current understanding of NBS-LRR gene evolution in angiosperms is mainly derived from studies of monocots and dicots, and the research on NBS-LRR genes in the basal angiosperms are limited to Amborella trichopoda (Shao et al., 2016). Recently, the genomic data of E. ferox provided not only important data resources for the study of disease resistance genes in this species but also a new research object for the in-depth study of the characteristics and evolution pattern of NBS-LRR genes in basal angiosperm species (Yang et al., 2020). In this study, NBS-LRR genes in the E. ferox genome were comprehensively identified and analyzed, which laid a foundation for the subsequent screening of disease resistance resources and the cloning of disease resistance genes and provided a theoretical basis for the evolution of NBS-LRR genes in angiosperms.
Materials and Methods
Data Source
The genome sequence and annotation files of E. ferox were downloaded from the Comparative Genome (CoGe) database (https://genomevolution.org/CoGe/GenomeInfo.pl?gid=56574). The RNA-seq data of E. ferox was downloaded from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/sra/?term=SRR9650346). The NBS-LRR genes in Nymphaeaceae colorata and N. thermarum were downloaded from the ANNA database (http://compbio.nju.edu.cn/app/ANNA/) (Liu et al., 2021).
Identification of NBS-LRR Genes
NBS-LRR genes were identified in the E. ferox genome as described previously (Shao et al., 2015). Briefly, an HMM search was first conducted for the protein sequences of E. ferox with the amino acid sequence of the NB-ARC domain (Pfam: PF00931) as a query, with a threshold expectation value of 1.0. Meanwhile, BLASTp search was performed towards the protein sequences of E. ferox using the sequence of the HMM profile of the NB-ARC domain (E-value = 1.0). Then, all hits obtained using the two methods were merged, and the redundant hits were removed. In order to confirm that blast hits contained the NBS domain, a round of HMMscan was conducted for all the obtained proteins using a more strict threshold expectation value (E-value set to 0.0001). All non-redundant candidate sequences were submitted to an online tool—the Conserved Domains Database at NCBI (CDD; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to further verify the CC, TIR, RPW8, LRR, and other integrated domains.
Chromosomal Anchoring of NBS-LRR Genes in Euryale ferox Genome
To anchor each of the NBS-LRR genes to specific positions on chromosomes of the E. ferox genome, the annotation file recording the positions of all genes was used, and the genomic locations of all the NBS-LRR genes were extracted from the file. The flanking regions of each NBS-LRR gene (a 250 kb window upstream and downstream, respectively) were searched for the presence/absence of other NBS-LRR genes. If another NBS-LRR gene was detected within the flanking regions, the two NBS-LRR genes were considered as located in the same NBS-LRR gene cluster (Ameline-Torregrosa et al., 2008), otherwise the gene initially analyzed should be considered as a singleton.
Sequence Alignment and Phylogenetic Analysis
The sequence alignment of the NBS domain and phylogenetic analysis were conducted as described by Zhang et al. (2016). First of all, amino acid sequences of the NBS domain from all the identified NBS-LRR genes were extracted and aligned by the ClustalW software integrated into MEGA 7.0 (Kumar et al., 2016), with default settings and then manually corrected to achieve a better alignment. Sequences that were too short or extremely divergent were removed to prevent interference with the alignments and subsequent phylogenetic analysis. Phylogenetic analysis was carried out with IQ-TREE using the maximum likelihood method after selecting the best-fit model using ModelFinder (Nguyen et al., 2015; Kalyaanamoorthy et al., 2017). Support values of branches were calculated by UFBoot2 (Minh et al., 2013).
Analyses of Gene Synteny and Gene Duplication Types
Paralogues in the E. ferox genomes were first examined using pair-wise all-against-all BLAST (Altschul et al., 1990), then the obtained results and the GFF annotation file were subjected to MCScanX to evaluate the microsynteny relationships between paralogues. Syntenic genes were considered as the results of whole genome duplications or chromosomal segmental duplications (Wang et al., 2012). Microsynteny relationships were displayed using TBtools (Chen et al., 2020).
Results
Genome-wide Identification of NBS-LRR Genes in Euryale ferox
Altogether 131 NBS-LRR genes (40 CNLs, 73 TNLs, and 18 RNLs) (Supplementary Table S1) were identified from the E. ferox genome, accounting for approximately 0.6% of all annotated genes. The number of NBS-LRR genes in the E. ferox genome was smaller than those in Brassicaceae, Solanaceae, Leguminosae or Poaceae as reported by previous studies (Luo et al., 2012; Shao et al., 2014; Zhang et al., 2016; Qian et al., 2017), which may have been due to a convergent contraction in NBS-LRR genes during adaptation to an aquatic lifestyle, and was consistent with the report by Liu et al. (Liu et al., 2021). Notably, of three subclasses, the number of RNL genes is higher than that in most investigated angiosperms, and TNLs account for a larger proportion, suggesting that E. ferox tends to employ TNL-mediated defense when confronting pathogens.
Diversity of NBS-LRR Gene Structure
Our previous study found that not all NBS-LRR genes possess the entire structure, and some NBS-LRR genes may lose part of their domains or fuse additional domains during duplication (Shao et al., 2014; Zhang et al., 2016). Based on the domain combinations of the translated proteins, the NBS-LRR genes in each subclass were classified into different groups (Figure 1A). The 18 RNL genes were divided into four groups according to the structure of proteins. Among them, six intact RNL genes accounted for 33.3%. Seven RNL genes encoded proteins that lacked the LRR domain, whereas four RNL genes encoded proteins that lacked both RPW8 and LRR domains. Additionally, we found an RNL gene encoding an RPW8-NBS-RPW8-NBS (RNRN) structure protein. The structures of TNL and CNL genes are both more diverse than those of RNL genes. We explored 13 different kinds of structures in 73 TNL genes and seven kinds of structures in the 40 CNL genes. Notably, these structural variations were caused not only by the loss of the typical domains of NBS-LRR genes but also by the fusing of additional domains. For example, we found four different integrated domains (IDs) in nine TNL genes. Among them, seven IDs were fused to the C-terminal domain of NBS-LRR proteins, whereas two were fused to the N-terminal domain. Moreover, transcription analysis indicated that the majority of NBS-LRR genes of E. ferox were expressed at a low level (Figure 1B; Supplementary Table S2) because they always remain silenced but highly expressed during pathogen stimulation.
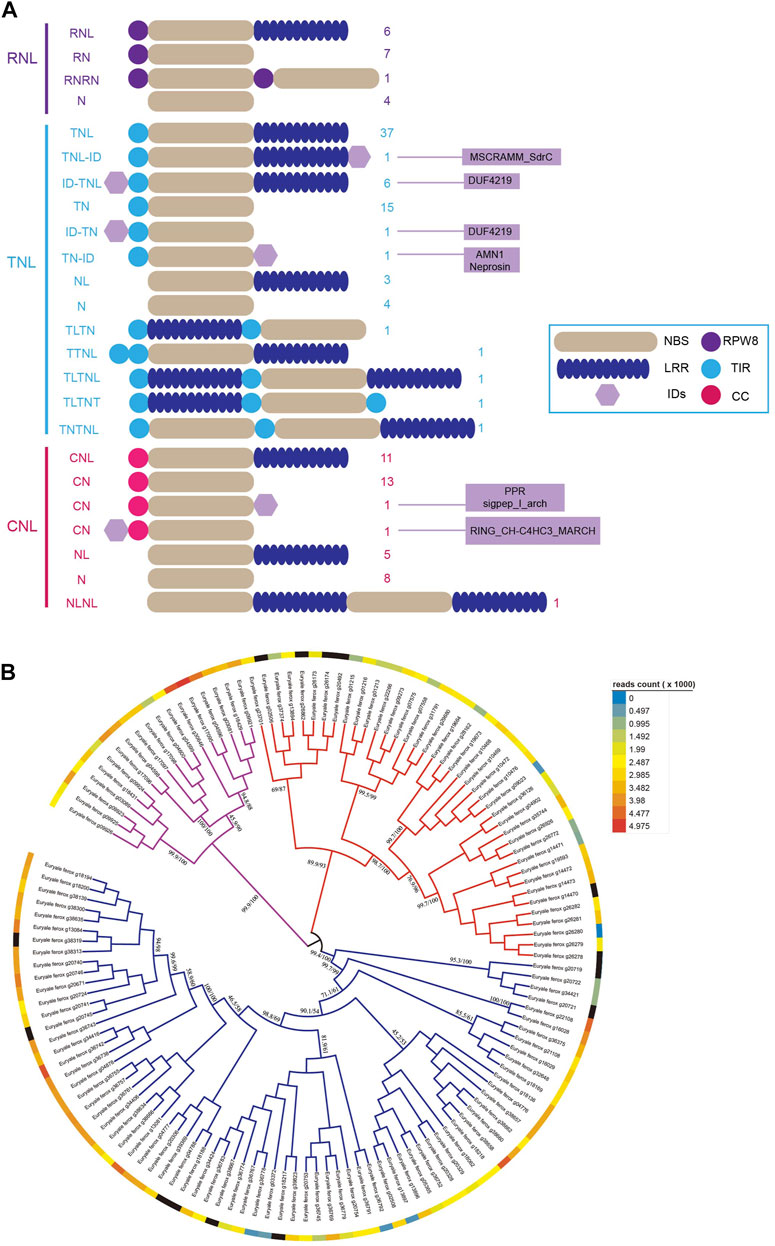
FIGURE 1. NBS-LRR genes in E. ferox and their domain combinations. (A) Twenty-four domain combinations of NBS-LRR genes in E. ferox. The numbers of NBS-LRR genes of each domain combination are listed and the IDs are also listed as a hexagon. (B) The expression profile of 131 NBS-LRR genes in E. ferox.
Chromosomal Distribution of Euryale ferox NBS-LRR Genes
The positions of NBS-LRR genes were anchored to chromosomes based on their physical location provided by the GFF3 file, and the 131 identified NBS-LRR genes were found to be scattered unevenly on 28 of 29 E. ferox chromosomes (Figure 2). Chromosomes 11, 15, and 18 had more than 10 NBS-LRR genes, whereas only one NBS-LRR gene was identified on chromosomes 1, 3, 5, 7, 17, 21, and 27. No NBS-LRR genes were detected on chromosome 22. It seems that NBS-LRR gene number is not correlated to the length of a chromosome (Supplementary Figure S1).
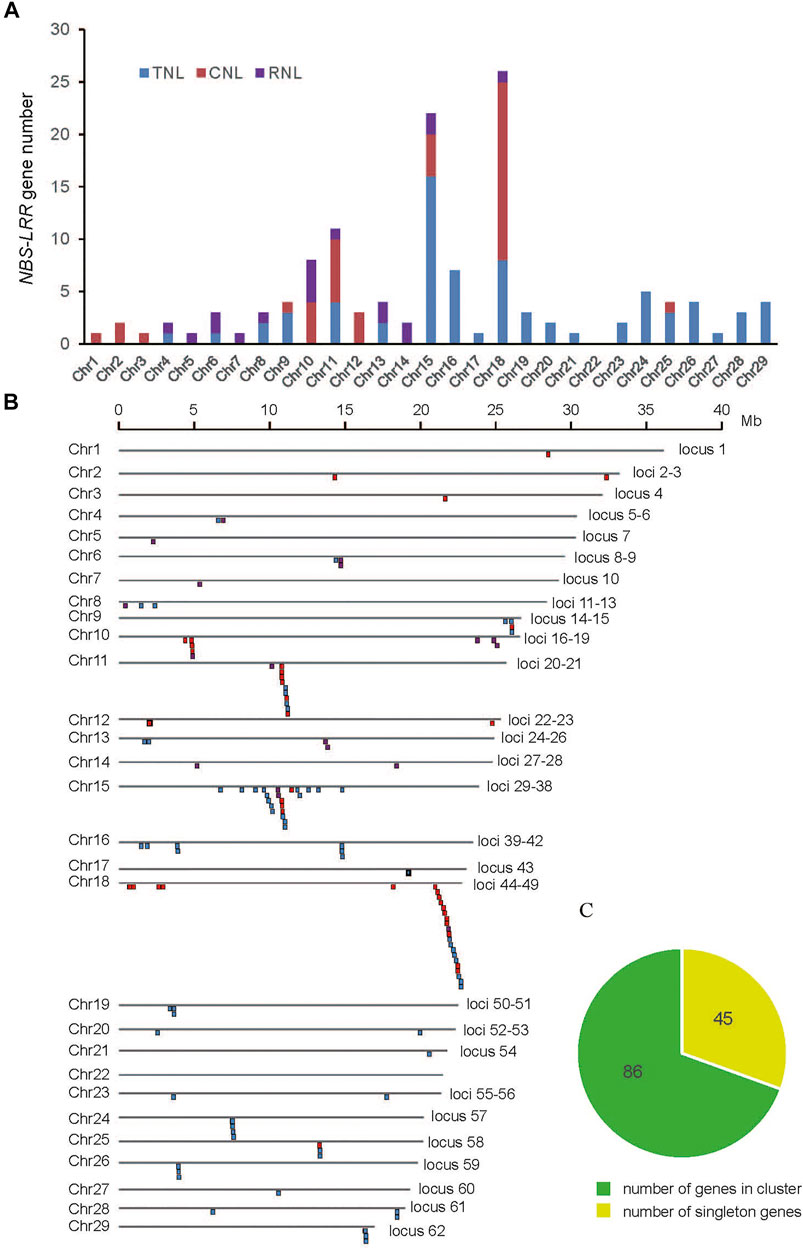
FIGURE 2. (A) The histogram shows the number of NBS-LRR genes in each chromosome. (B) Chromosomal distribution of E. ferox NBS-LRR genes. (C) The pie chart shows the proportion between singleton and clustered NBS-LRR genes.
According to the physical locations, the NBS-LRR genes on the 28 chromosomes were classified into 62 loci, including 44 singletons (one gene at one locus) and 18 multigene clusters (Figure 2). The results demonstrated that 87 NBS-LRR genes were organized into 18 clusters, which were distributed in chromosomes 11, 15, and 18. On average, there were five genes per cluster. These clusters were mainly distributed on chromosomes 11, 15, and 18, resulting in a high density of NBS-LRR genes on these chromosomes. Among the 18 clusters, the smallest one only contained two adjacent genes, while the largest one on chromosome 18 contained 10 genes.
Different Types of Duplications of Euryale ferox NBS-LRR Genes
The expansion in the NBS-LRR gene family was derived from different duplication types, among which a large proportion of NBS-LRR genes were organized into clusters due to tandem duplication. Through the analysis of the duplication types of NBS-LRR genes, we found that 28 of the 131 genes were produced via tandem duplication, 18 were produced by dispersed duplication, 15 were produced by proximal duplication, and 70 were resulted from segmental duplications or whole-genome duplications (WGD), which were distributed mainly on chromosomes 11, 15, and 18 (Figure 3; Supplementary Figure S2). For instance, gene g26862 on chromosome eight is syntenic to g20492 on chromosome 20, so these two genes were considered to be derived from a WGD or a segmental duplication.
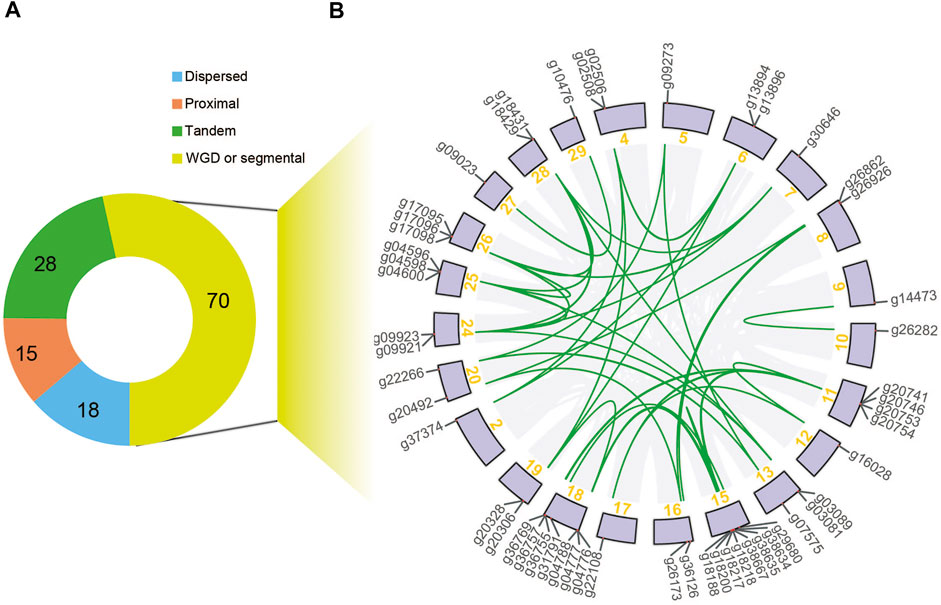
FIGURE 3. Duplication type of E. ferox NBS-LRR genes. (A) A pie chart showing NBS-LRR genes with different duplication types. (B) Syntenic relationships of the 70 segmental-duplicated NBS-LRR genes.
Phylogeny of NBS-LRR Genes in Euryale ferox
To trace the evolutionary history of NBS-LRR genes in E. ferox, a phylogenetic tree was constructed together with NBS-LRR genes from two species in Nymphaeaceae, N. colorata and N. thermarum, and Arabidopsis thaliana. The phylogenetic tree consisted of three monophyletic clades, CNL, TNL, and RNL, with high support values (Figure 4). The RNL monophyletic clade in the phylogenetic tree had a high support value (99.9%). The topology was consistent with that of other investigated angiosperms, supporting the notion that the three subclasses diverged in early ages (Xue et al., 2012; Shao et al., 2019).
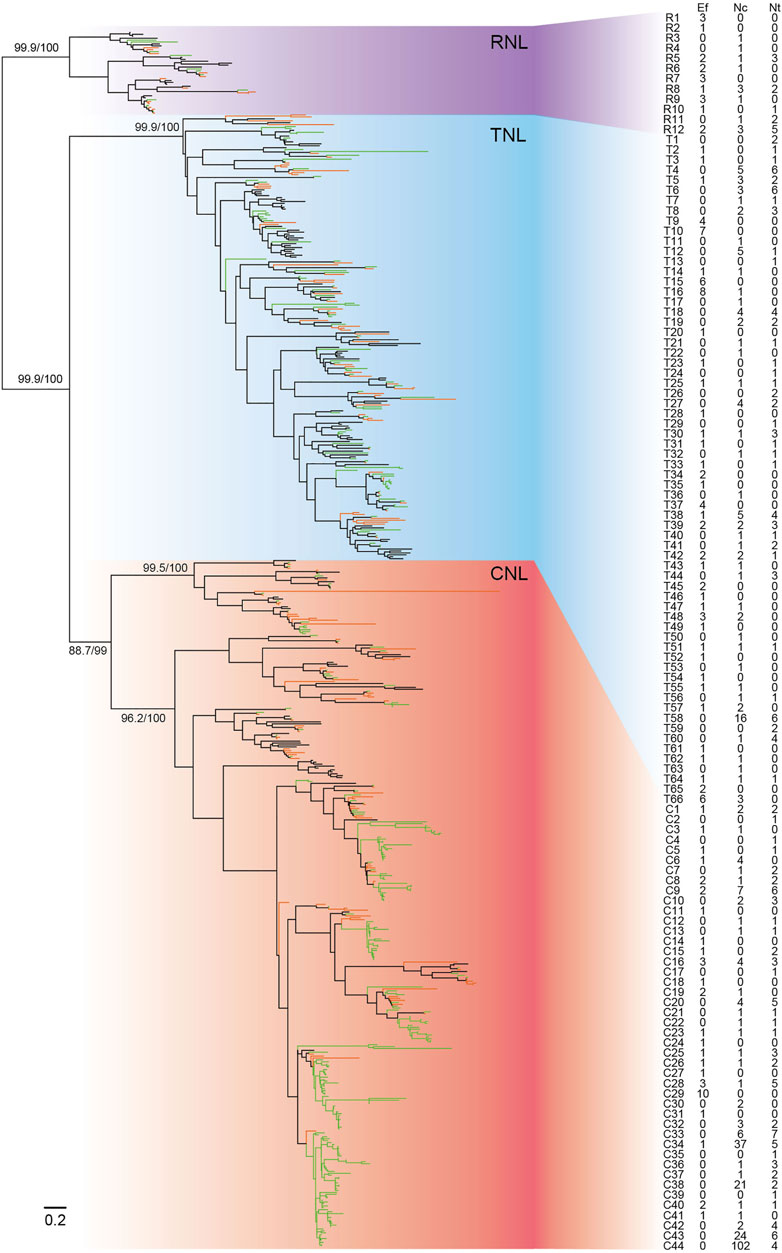
FIGURE 4. Phylogeny of NBS-LRR genes of E. ferox, N. colorata, and N. thermarum. The phylogeny was constructed based on the conserved NBS domain of NBS-LRR genes from E. ferox (Ef, green branches), N. colorata (Nc, orange branches), and N. thermarum (Nt, black branches). Branch support values obtained from a UFBoot2 test are labeled on basal nodes. Predicted ancestral lineages are labeled with numbers. The detailed phylogenetic tree is shown in Supplementary Figure S1.
Based on the reconstructed NBS-LRR gene phylogeny, 122 ancestral NBS-LRR lineages in the common ancestor of the three species (including 44 CNLs, 66 TNLs, and 12 RNLs) were recovered (Figure 4). Clusters organized by several NBS-LRR genes from E. ferox and two water lilies were detected in the phylogenetic tree, which mirrored the species-specific expansion, especially the drastic expansion of CNLs in N. colorata in the C44 lineage, with 102 CNLs. Of 122 ancestral lineages, E. ferox inherited 70 lineages, whereas there were only 131 NBS-LRR genes in the E. ferox genome, suggesting that a fairly low level of NBS-LRR gene expansion occurred in the E. ferox genome during evolution.
Discussion
NBS-LRR genes consist of hundreds of members and are the largest family among R genes. In two-tier immune systems, the NBS-LRR gene family plays important roles in effector-triggered immunity. Revealing the profile of NBS-LRR genes in a species is very important, and by exploring the evolutionary patterns of NBS-LRR genes in the species and its close relatives, we can put these conclusions into practice (Meyers et al., 2003; Zhang et al., 2016). In the past 20 years, genome-wide identification and evolutionary analysis have been conducted in many angiosperms (Liu et al., 2021; Luo et al., 2012; Qian et al., 2017; Shao et al., 2014; Xue et al., 2020; Zhang et al., 2016; Zhou et al., 2020). It is noteworthy that of the more than 300 disease resistance genes, over 60% that have been cloned from different plants belong to the NBS-LRR gene family (Kourelis and van der Hoorn, 2018). Angiosperms inherited three subclasses of NBS-LRR genes: TNLs, CNLs, and RNLs. Studies have shown that proteins encoded by CNLs and TNLs function as receptors for pathogen recognition, and these two subclasses have expanded to various degrees during the long-term ‘arms race’ with pathogens (Shao et al., 2014).
In this study, the E. ferox genome was found to harbor only 131 NBS-LRR genes, consistent with a recent report. Liu et al. (2021) proposed that a convergent contraction of NBS-LRR genes occurred during the evolution of adaptation to an aquatic lifestyle, which may be the reason for the fewer NBS-LRR genes in E. ferox compared to other angiosperms. The structures of NBS-LRR genes are relatively diverse. Among four different IDs, MARCH1, also known as membrane-associated RING finger protein 1, is a membrane-anchored E3 ubiquitin ligase that mainly expressed in cells of the immune system (Bartee et al., 2004). MARCH1 also plays a regulatory role in T cell activation during immune responses, thus, we speculated that it regulates immune responses in plants.
NBS-LRR genes can expand via different duplication methods. Many studies have shown that tandem duplication is dominant in NBS-LRR gene expansion in most angiosperms (Shao et al., 2014; Qian et al., 2017; Zhou et al., 2020). We analyzed the duplication types of NBS-LRR genes in the E. ferox genome and found that there were 28 NBS-LRR genes generated by tandem duplication in the E. ferox genome, accounting for only 21% in total. This proportion is much lower than the 70–80% in the legume family (Shao et al., 2014). Furthermore, it was documented that E. ferox experienced two rounds of WGDs (Yang et al., 2020). Thus, we thought since tandem duplications were rare in its evolutionary history, the majority of E. ferox NBS-LRR genes are derived from segmental duplications, which is different from the situation in most other angiosperms, and the fewer number of tandemly duplicated genes may also be one of the reasons for the relatively fewer NBS-LRR genes in E. ferox. Additionally, the phylogenetic analysis of NBS-LRR genes in E. ferox and two other Nymphaeaceae species revealed that there were at least 122 ancestral NBS-LRR genes in the common ancestor of the three species, whereas only 131 NBS-LRR genes were found in the E. ferox genome, which further indicated a low level of NBS-LRR gene expansion in the E. ferox genome during its long-term evolution.
The comprehensive identification of NBS-LRR genes from genomic data is not only important for revealing the evolution of this important disease resistance gene family but also serves as an important basis for discovering and utilizing functional disease resistance genes in crops. Based on the analysis of NBS-LRR genes in the rice genome, Zhang et al. (2015) used targeted cloning of NBS-LRR genes in resistant rice varieties, and verified dozens of NBS-LRR genes that play important roles in blast resistance in rice. Witek et al. (2016) successfully cloned the late blight resistance gene Rpi-AMR3I from the resistance locus of Solanum americanum by target region sequencing and third-generation sequencing based on the analysis of NBS-LRR genes in the potato genome. These studies indicate that the genome-wide identification and analysis of NBS-LRR genes promotes the cloning of functional resistance genes in the studied species and related species. E. ferox is an important functional food and medicinal plant that has been widely cultivated in China since time immemorial. However, due to its characteristics of underwater flowering and pollination, research on germplasm innovation and disease resistance breeding has lagged behind research in other species. With the breakthrough of artificial hybrid technology for E. ferox in the 1980s and the development of tissue culture technology in recent years, it is possible to breed superior varieties of E. ferox by molecular breeding. E. ferox is vulnerable to a large number of diseases, so it is particularly important to develop molecular breeding in E. ferox.
In summary, the present study identified a complete set of 131 NBS-LRR genes in the genome of E. ferox. Although the quantity is significantly lower than that of most species in Solanaceae, Leguminosae, and Poaceae, it is roughly equivalent to that of Brassicaceae crops, and higher than those of Cucurbitaceae crops (Luo et al., 2012; Lin et al., 2013; Shao et al., 2014; Morata and Puigdomènech, 2017; Qian et al., 2017). The identification of NBS-LRR genes, analysis of sequence features, and revelation of chromosomal distribution pattern will provide important references for the molecular breeding of E. ferox. In addition, Nymphaeaceae contains many kinds of important flowering plants. The comparative analysis of NBS-LRR genes in these plants will also facilitate the molecular breeding of different species in the same genus.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
L-HQ and X-QS conceived and designed the study. J-YW, YW, XZ, and G-CZ obtained and analyzed the data. J-YW and G-CZ wrote the manuscript. L-HQ and X-QS revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the earmarked fund for Jiangsu Agricultural Industry Technology System, grant number JATS (2021)118.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.880071/full#supplementary-material
References
Ahmed, D., Kumar, V., Verma, A., Shukla, G. S., and Sharma, M. (2015). Antidiabetic, Antioxidant, Antihyperlipidemic Effect of Extract of Euryale Ferox Salisb. With Enhanced Histopathology of Pancreas, Liver and Kidney in Streptozotocin Induced Diabetic Rats. SpringerPlus 4, 315. doi:10.1186/s40064-015-1059-7
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215 (3), 403–410. doi:10.1016/s0022-2836(05)80360-2
Ameline-Torregrosa, C., Wang, B.-B., O'Bleness, M. S., Deshpande, S., Zhu, H., Roe, B., et al. (2008). Identification and Characterization of Nucleotide-Binding Site-Leucine-Rich Repeat Genes in the Model Plant Medicago Truncatula. Plant Physiol. 146 (1), 5–21. doi:10.1104/pp.107.104588
Andersen, E., Ali, S., Byamukama, E., Yen, Y., and Nepal, M. (2018). Disease Resistance Mechanisms in Plants. Genes 9 (7), 339. doi:10.3390/genes9070339
Bartee, E., Mansouri, M., Hovey Nerenberg, B. T., Gouveia, K., and Früh, K. (2004). Downregulation of Major Histocompatibility Complex Class I by Human Ubiquitin Ligases Related to Viral Immune Evasion Proteins. J. Virol. 78 (3), 1109–1120. doi:10.1128/jvi.78.3.1109-1120.2004
Bi, G., Su, M., Li, N., Liang, Y., Dang, S., Xu, J., et al. (2021). The ZAR1 Resistosome Is a Calcium-Permeable Channel Triggering Plant Immune Signaling. Cell 184 (13), 3528–3541. e12. doi:10.1016/j.cell.2021.05.003
Castel, B., Ngou, P. M., Cevik, V., Redkar, A., Kim, D. S., Yang, Y., et al. (2019). Diverse NLR Immune Receptors Activate Defence via the RPW 8‐ NLR NRG 1. New Phytol. 222 (2), 966–980. doi:10.1111/nph.15659
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Collier, S. M., Hamel, L.-P., and Moffett, P. (2011). Cell Death Mediated by the N-Terminal Domains of a Unique and Highly Conserved Class of NB-LRR Protein. Mpmi 24 (8), 918–931. doi:10.1094/mpmi-03-11-0050
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered Immunity: from Pathogen Perception to Robust Defense. Annu. Rev. Plant Biol. 66, 487–511. doi:10.1146/annurev-arplant-050213-040012
Hu, Z., Yan, C., Liu, P., Huang, Z., Ma, R., Zhang, C., et al. (2013). Crystal Structure of NLRC4 Reveals its Autoinhibition Mechanism. Science 341 (6142), 172–175. doi:10.1126/science.1236381
Huang, Z., Huang, X., Wang, Q., Jiang, R., Sun, G., Xu, Y., et al. (2018). Extract of Euryale Ferox Salisb Exerts Antidepressant Effects and Regulates Autophagy through the Adenosine Monophosphate-Activated Protein Kinase-UNC-51-like Kinase 1 Pathway. IUBMB life 70 (4), 300–309. doi:10.1002/iub.1731
Jacob, P., Kim, N. H., Wu, F., El-Kasmi, F., Chi, Y., Walton, W. G., et al. (2021). Plant "helper" Immune Receptors Are Ca 2+ -permeable Nonselective Cation Channels. Science 373 (6553), 420–425. doi:10.1126/science.abg7917
Jones, J. D., Vance, R. E., and Dangl, J. L. (2016). Intracellular Innate Immune Surveillance Devices in Plants and Animals. Science 354 (6316), aaf6395. doi:10.1126/science.aaf6395
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 14 (6), 587–589. doi:10.1038/nmeth.4285
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 30 (2), 285–299. doi:10.1105/tpc.17.00579
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Lin, X., Zhang, Y., Kuang, H., and Chen, J. (2013). Frequent Loss of Lineages and Deficient Duplications Accounted for Low Copy Number of Disease Resistance Genes in Cucurbitaceae. BMC genomics 14, 335. doi:10.1186/1471-2164-14-335
Liu, X., He, Z., Yin, Y., Xu, X., Wu, W., and Li, L. (2018). Transcriptome Sequencing and Analysis during Seed Growth and Development in Euryale Ferox Salisb. BMC genomics 19 (1), 343. doi:10.1186/s12864-018-4707-9
Liu, Y., Zeng, Z., Zhang, Y.-M., Li, Q., Jiang, X.-M., Jiang, Z., et al. (2021). An Angiosperm NLR Atlas Reveals that NLR Gene Reduction Is Associated with Ecological Specialization and Signal Transduction Component Deletion. Mol. Plant 14 (12), 2015–2031. doi:10.1016/j.molp.2021.08.001
Luo, S., Zhang, Y., Hu, Q., Chen, J., Li, K., Lu, C., et al. (2012). Dynamic Nucleotide-Binding Site and Leucine-Rich Repeat-Encoding Genes in the Grass Family. Plant Physiol. 159 (1), 197–210. doi:10.1104/pp.111.192062
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-Wide Analysis of NBS-LRR-Encoding Genes in Arabidopsis[W]. The Plant cell 15 (4), 809–834. doi:10.1105/tpc.009308
Minh, B. Q., Nguyen, M. A. T., and von Haeseler, A. (2013). Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 30 (5), 1188–1195. doi:10.1093/molbev/mst024
Morata, J., and Puigdomènech, P. (2017). Variability Among Cucurbitaceae Species (Melon, Cucumber and Watermelon) in a Genomic Region Containing a Cluster of NBS-LRR Genes. BMC genomics 18 (1), 138. doi:10.1186/s12864-017-3529-5
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi:10.1093/molbev/msu300
Peart, J. R., Mestre, P., Lu, R., Malcuit, I., and Baulcombe, D. C. (2005). NRG1, a CC-NB-LRR Protein, Together with N, a TIR-NB-LRR Protein, Mediates Resistance against Tobacco Mosaic Virus. Curr. Biol. 15 (10), 968–973. doi:10.1016/j.cub.2005.04.053
Qi, T., Seong, K., Thomazella, D. P. T., Kim, J. R., Pham, J., Seo, E., et al. (2018). NRG1 Functions Downstream of EDS1 to Regulate TIR-NLR-Mediated Plant Immunity in Nicotiana Benthamiana. Proc. Natl. Acad. Sci. U S A. 115 (46), E10979–E10987. doi:10.1073/pnas.1814856115
Qian, L.-H., Zhou, G.-C., Sun, X.-Q., Lei, Z., Zhang, Y.-M., Xue, J.-Y., et al. (2017). Distinct Patterns of Gene Gain and Loss: Diverse Evolutionary Modes of NBS-Encoding Genes in Three Solanaceae Crop Species. G3 (Bethesda, Md. 7 (5), 1577–1585. doi:10.1534/g3.117.040485
Shao, Z.-Q., Xue, J.-Y., Wang, Q., Wang, B., and Chen, J.-Q. (2019). Revisiting the Origin of Plant NBS-LRR Genes. Trends Plant Science 24 (1), 9–12. doi:10.1016/j.tplants.2018.10.015
Shao, Z.-Q., Xue, J.-Y., Wu, P., Zhang, Y.-M., Wu, Y., Hang, Y.-Y., et al. (2016). Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol. 170 (4), 2095–2109. doi:10.1104/pp.15.01487
Shao, Z.-Q., Zhang, Y.-M., Hang, Y.-Y., Xue, J.-Y., Zhou, G.-C., Wu, P., et al. (2014). Long-Term Evolution of Nucleotide-Binding Site-Leucine-Rich Repeat Genes: Understanding Gained from and beyond the Legume Family. Plant Physiol. 166 (1), 217–234. doi:10.1104/pp.114.243626
Shao, Z. Q., Zhang, Y. M., Wang, B., and Chen, J. Q. (2015). Computational Identification of MicroRNA-Targeted Nucleotide-Binding Site-Leucine-Rich Repeat Genes in Plants. Bio-protocol 5, 1. doi:10.21769/bioprotoc.1637
Wang, J., and Chai, J. (2020). Molecular Actions of NLR Immune Receptors in Plants and Animals. Sci. China Life Sci. 63 (9), 1303–1316. doi:10.1007/s11427-019-1687-6
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Witek, K., Jupe, F., Witek, A. I., Baker, D., Clark, M. D., and Jones, J. D. G. (2016). Accelerated Cloning of a Potato Late Blight-Resistance Gene Using RenSeq and SMRT Sequencing. Nat. Biotechnol. 34 (6), 656–660. doi:10.1038/nbt.3540
Wu, C.-Y., Wang, H., He, X.-X., Wu, D.-W., Yue, W., and Wu, Q.-N. (2017). The Hypoglycemic and Antioxidant Effects of Polysaccharides from the Petioles and Pedicels of Euryale Ferox Salisb. On Alloxan-Induced Hyperglycemic Mice. Food Funct. 8 (10), 3803–3813. doi:10.1039/c7fo01035d
Wu, Z., Li, M., Dong, O. X., Xia, S., Liang, W., Bao, Y., et al. (2019). Differential Regulation of TNL‐mediated Immune Signaling by Redundant Helper CNLs. New Phytol. 222 (2), 938–953. doi:10.1111/nph.15665
Xue, J.-Y., Wang, Y., Wu, P., Wang, Q., Yang, L.-T., Pan, X.-H., et al. (2012). A Primary Survey on Bryophyte Species Reveals Two Novel Classes of Nucleotide-Binding Site (NBS) Genes. PloS one 7 (5), e36700. doi:10.1371/journal.pone.0036700
Xue, J.-Y., Zhao, T., Liu, Y., Liu, Y., Zhang, Y.-X., Zhang, G.-Q., et al. (2020). Genome- Wide Analysis of the Nucleotide Binding Site Leucine-Rich Repeat Genes of Four Orchids Revealed Extremely Low Numbers of Disease Resistance Genes. Front. Genet. 10, 1286. doi:10.3389/fgene.2019.01286
Yang, Y., Sun, P., Lv, L., Wang, D., Ru, D., Li, Y., et al. (2020). Prickly Waterlily and Rigid Hornwort Genomes Shed Light on Early Angiosperm Evolution. Nat. Plants 6 (3), 215–222. doi:10.1038/s41477-020-0594-6
Zhang, X., Yang, S., Wang, J., Jia, Y., Huang, J., Tan, S., et al. (2015). A Genome-wide Survey Reveals Abundant rice blastR Genes in Resistant Cultivars. Plant J. 84 (1), 20–28. doi:10.1111/tpj.12955
Zhang, Y.-M., Shao, Z.-Q., Wang, Q., Hang, Y.-Y., Xue, J.-Y., Wang, B., et al. (2016). Uncovering the Dynamic Evolution of Nucleotide-Binding Site-Leucine-Rich Repeat (NBS-LRR) Genes in Brassicaceae. J. Integr. Plant Biol. 58 (2), 165–177. doi:10.1111/jipb.12365
Zhong, Y., and Cheng, Z.-M. (2016). A Unique RPW8-Encoding Class of Genes that Originated in Early Land Plants and Evolved through Domain Fission, Fusion, and Duplication. Sci. Rep. 6, 32923. doi:10.1038/srep32923
Keywords: Euryale ferox, NBS-LRR genes, R genes, phylogenetic analysis, evolution
Citation: Qian L-H, Wu J-Y, Wang Y, Zou X, Zhou G-C and Sun X-Q (2022) Genome-Wide Analysis of NBS-LRR Genes From an Early-Diverging Angiosperm Euryale ferox. Front. Genet. 13:880071. doi: 10.3389/fgene.2022.880071
Received: 21 February 2022; Accepted: 11 April 2022;
Published: 13 May 2022.
Edited by:
Aleksandra Pekowska, Nencki Institute of Experimental Biology (PAS), PolandReviewed by:
Jinpeng Wang, Institute of Botany (CAS), ChinaJia-Yu Xue, Nanjing Agricultural University, China
Copyright © 2022 Qian, Wu, Wang, Zou, Zhou and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guang-Can Zhou, emhvdWd1YW5nY2FuQGhlemV1LmVkdS5jbg==; Xiao-Qin Sun, eGlhb3FpbnN1bkBjbmJnLm5ldA==
†These authors have contributed equally to this work
 Lan-Hua Qian1†
Lan-Hua Qian1† Guang-Can Zhou
Guang-Can Zhou Xiao-Qin Sun
Xiao-Qin Sun