
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet., 01 June 2022
Sec. Computational Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.871988
This article is part of the Research TopicGenetic and proteomic biomarkers in solid tumor detection and treatmentView all 64 articles
Accumulating evidence has revealed the vital regulatory roles of lncRNA DLX6-AS1 in various tumors at pre-transcriptional, transcriptional, and post-transcriptional levels, which makes it a potential prognosis factor and therapeutic target. In addition, the presence of lncRNA DLX6-AS1 in the exosomes of peripheral blood of patients with tumors may also contribute to it being a possible cancer-related biomarker. However, most literature studies are devoted to studying the effect of lncRNA DLX6-AS1 as a sponging molecule of miRNAs, the research of which is likely to get stuck into a dilemma. Literature studies published already have demonstrated an exciting cell malignant phenotype inhibition with the knockdown of lncRNA DLX6-AS1 in various tumor cell lines. With the comprehensive development of delivery systems, high-throughput sequencing, and aptamers, the problems of finding novel research methods and exploring the therapeutic options which are based on lncRNA DLX6-AS1 in vivo could come into a period to deal with. This review aims to summarize the research statuses of lncRNA DLX6-AS1, discuss other study methodologies and therapeutic strategies on it, which might be of help to the deep learning of lncRNA DLX6-AS1 and its application from basic to clinical research.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs with a length longer than 200 nucleotides, which lack the capability of encoding proteins and were transcriptional noises at first. lncRNAs are transcribed from the opposite strand of the protein-coding genes and overlap one-third of them. lncRNA DLX6-AS1 is usually dysregulated in many tumor tissues and cell lines. Besides, the capability of lncRNA DLX6-AS1 to bind with DNAs, RNAs, and proteins makes its regulatory roles represent at pre-transcriptional, transcriptional, and post-transcriptional levels. Increasing evidence has implied the crucial regulatory roles of lncRNA DLX6-AS1 in various tumors, demonstrating the crucial influence of lncRNA DLX6-AS1 on tumorigenesis and development and making it a candidate biomarker to assist treatment and diagnosis of tumors.
lncRNA DLX6-AS1, located at the human chromsome7q21.3 (GRCh38/hg38: chr7: 96955141-97014065) with a length of 1990 base pairs, is the reverse transcript of the upstream of the Dlx6 gene. In mouse, lncRNA DLX6-AS1 is also called Evf1 (embryonic ventral forebrain1) and is located at chromosome 6 (Kohtz and Fishell, 2004). Evf1 is transcribed from the upstream of the DLX6 gene, which comprises two exons with about a 37.5 kb intronic region between them. It was first found as one of the target genes of the sonic hedgehog in the ventral forebrain (Kohtz and Fishell, 2004). Its spliced form Evf2 (also known as DLX6OS1) is transcribed from the ultra-conserved region of the DLX5/6 ei enhancer and comprises three parts which can be divided into the Evf2 5′ unique region, the Evf2 3′ unique region, and the common region with Evf1 (Feng et al., 2006). It has been found that the regulatory roles of Evf2 involve DNA methylation, histone deacetylation, and chromosome topological changes.
Berghoff et al. (2013) found that Evf2 induced site-specific methylation of Dlx5/6 ei CpG576 and CpG757 in the E13. 5 MGE of the Evf2 transgene mice (Evf2TS/TS; R) but did not affect the expression levels of Dlx5 and Dlx6 when the expression levels of evf2 were at 0.38 x wild-type levels. Apart from regulating gene methylation, it was also revealed by Bond et al. (2009) that Evf2 could recruit MECP2 to the conserved region of DLX5/6 ei/eii which decreased the recruitment of HDAC1 in the DLX5/6eii, and thus, may be the reason for the increased expression level of Dlx5 and Dlx6. Besides, Cajigas et al. (2015) also found that Evf2 lncRNAs could form an Evf2-DLX1 ribonucleoprotein (RNP), which contained the SWI/SNF-related chromatin remodelers Brahma-related gene 1 (BRG1, SMARCA4) and Brahma-associated factor (BAF170, SMARCC2) in the DXL5/6 ei region of a developing mouse forebrain, to regulate chromatin remodeling and gene expression. They revealed that Evf2 increased BRG1 binding with key Dlx5/6 enhancers with changes in lysine acetylation of histones H3 and H4, which led to significantly reduced H3AcK9 and H3AcK18 at ei, reduction of H3AcK18 to a lesser extent at eii, and an H4AcK5 decrease at four sites, while total H4AcK decreased at three intergenic sites of Evf2TS/TS E13.5GE when compared with the decrease at those of EVF2+/+ E13.5GE (Cajigas et al., 2015) (Figure 1).
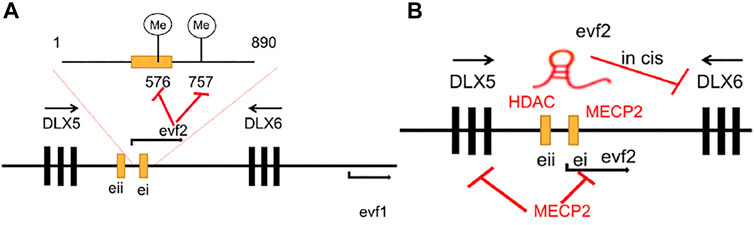
FIGURE 1. (A) Evf2 induces 576CpG and 757CpG site methylation in the ultra-conserved region of DLX5/6 ei/eii. (B) Evf2 recruits MECP2 and further absorbs HDAC1 into the DLX5/6 ei/eii region to repress DLX5 and DLX6 gene expression.
Chromosome topological changes allow the regulatory effects of enhancers to display over mega-base distances (Mir et al., 2019). Evf2, a cloud-forming DLX5/6 ultra-conserved enhancer ei lncRNA, can directly interact with its targeting genes and regulate the topology across a 27-Mb region by altering the gene–distance relationships among Dlx5/6, Umad1, and Akr1b8 genes (Cajigas et al., 2018). It was also found that evf2 could regulate the Dlx5/6UCE interactions to regulate the Rbmb8 gene repression via inducing the formation of protein pools (Sox2, Dlx, and Smarca4) (Cajigas et al., 2015).
Berghoff et al. (2013) found that evf2 can regulate evf2, DLX6, and DLX5 expression via the competitive binding of MECP2 and DLX1/2 to the DLX5/6 ei/eii region, and the repression of the DLX6 gene is via its antisense transcription. The antagonism of MECP2 and Dlx1/2 comparatively binding to the ultra-conserved region leads to three kinds of activity statuses of the DLX5/6 ei/eii region, which regulates evf2 and Dlx5 differentially: inactive (MECP2 binds two gene loci), low activity (MECP2 and DLX1/2 bind one gene locus), and high activity (Dlx1/2 occupies two gene loci) (Berghoff et al., 2013). Feng et al. (2006) also demonstrated that evf2 could serve as the coactivator of DLX2 to facilitate the expression of DLX5 and DLX6 genes.
Zhao et al. (2020) found that overexpression of lncRNA DLX6-AS1 could recruit DNA methyltransferase 1 (DNMT1) to the promoter region of its downstream target gene LARGE, which promoted the progression and lymph node metastasis of prostate cancer. It was also discovered by Zhao and Xu (2020) that lncRNA DLX6-AS1 was upregulated in endometrial cancer tissues and cell lines. It could form an RNA–DNA triplex via its triplex-forming oligonucleotide (TFO) sequence and the DLX6 triplex target site (TTS). Besides, lncRNA DLX6-AS1 also recruited a transcription factor E2F1 and histone acetyltransferase p300 to the promoter region of DLX6, which enhanced DLX6 expression and promoted proliferation, invasion, and reduced apoptosis of endometrial cancer cell lines (Zhao and Xu, 2020).
Maja Olsson and his colleagues (Olsson et al., 2016) also reported that lncRNA DLX6-AS1, one of the hyper-methylated genes in NB, was over-expressed and indicated a poorer prognosis. However, it is well known that hyper-methylation reduces transcriptional activity suppression, so the underlying molecular mechanisms need further exploration.
Lin et al. (2021) revealed that the hyper-methylated status of the promoter region of the DLX6-AS1 gene might be a novel progression-related and prognostic marker for colorectal cancer. Intriguingly, the methylation of the gene promoter region usually leads to transcriptional silencing. However, the expression level of lncRNA DLX6-AS1 in colorectal carcinoma cell lines and tissues is upregulated. Hence, there must be some other underlying mechanism that remains unknown. Besides, Zhao et al. (2021) also demonstrated that H3K4me1 can induce histone methylation around the DLX6-AS1 promoter region, which could upregulate the expression level of lncRNA DLX6-AS1 in lung squamous cell carcinoma cell lines and lead to cisplatin resistance.
LncRNA DLX6-AS1 plays key regulatory roles in regulating tumorigenesis and progress via many mechanisms. One of the most important mechanisms is formation of lncRNA DLX6-AS1-miRNAs-mRNAs ceRNAs regulatory networks, in which lncRNA DLX6-AS1 serves as an endogenous competing RNA to competitively sponge miRNAs to further up-regulate the downstream target genes (mRNAs) of miRNAs (Zeng et al., 2017; Zhang et al., 2017; An et al., 2018; Zhangmou et al., 2018; Li X. et al., 2019; Yang Q. et al., 2019; Zhang H. Y. et al., 2019; Li D. et al., 2019; Yang J. et al., 2019; Zhang N. et al., 2019; Fang et al., 2019; Huang et al., 2019; Lei et al., 2019; Sun et al., 2019; Wang et al., 2019; Zhao et al., 2019; Liu Y. et al., 2020; Du et al., 2020; Hu et al., 2020; Jia et al., 2020; Kong et al., 2020; Kong and Zhang, 2020; Liang et al., 2020; Qian et al., 2020; Wang et al., 2020; Xie et al., 2020; Yang et al., 2020; Liu et al., 2021; Wang et al., 2021; Wu et al., 2021; Zhao et al., 2021; Zheng et al., 2021; Zhu et al., 2021). Xue et al. (2021) and Feng et al. (2021) also reviewed the regulatory roles serving as a ceRNA in various tumors which influence the prognosis of patients and which may be the therapeutic and prognosis target in malignant tumors. Table 1 summarizes the microRNAs sponged by lncRNA DLX6-AS1 in various tumors.
The incidence and mortality of gastric cancer (GC) rank third and fifth, respectively (Fu et al., 2019), and is usually diagnosed at an advanced stage attributing to a lack of typical symptoms. Hence, it is imperative to find molecular biomarkers to assist early prognosis. Wu et al. (2020) found that lncRNA DLX6-AS1 could be a molecular link to stabilize mRNA levels in tumors. In gastric cancer, lncRNA DLX6-AS1 stabilizes MAP4K1 mRNA levels via regulating FUS protein expression, forming FUS-MAP4K1 protein-mRNA complexes, by which lncRNA DLX6-AS1 promotes the cell proliferation, migration, and EMT of gastric cancer.
Tian et al. (2020) made a meta-analysis between the expression levels of lncRNA DLX6-AS1 and the clinicopathology and prognosis of various cancers. They found that high expression levels of lncRNA DLX6-AS1 were associated with poor overall survival in tumor patients and overexpression of lncRNA DLX6-AS1 was associated with tumor stage (p < 0.01), tumor size (p < 0.01), lymph node metastasis (p < 0.01), and distant metastasis (p < 0.01). Hence, all the aforementioned factors have shown the vital role of research on lncRNA DLX6-AS1 in various tumors (Figure 2).
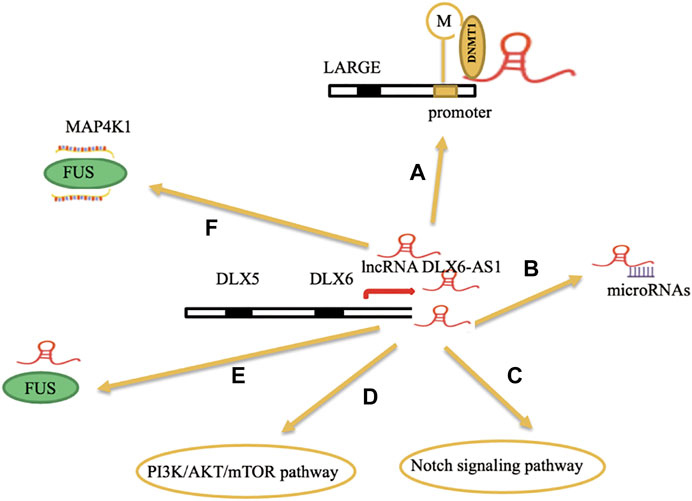
FIGURE 2. (A) Overexpression of lncRNA DLX6-AS1 could recruit DNA methyltransferase 1 (DNMT1) to the promoter region of its downstream target gene LARGE. (B) lncRNA DLX6-AS1 acts as an endogenously competing RNA to sponge miRNAs. (C,D) lncRNA DLX6-AS1 can regulate the notch signaling pathway and PI3K/AKT/mTOR pathway in epithelial ovarian cancer and colorectal cancer, respectively. (E) lncRNA DLX6-AS1 indirectly binds with MAP4K1(mRNA) via FUS(protein) to increase the stability of MAP4K1 in gastric cancer. (F) lncRNA DLX6-AS1 interacts with FUS (proteins) and regulate its expression levels in breast cancer.
Wang et al. (2021) found that lncRNA DLX6-AS1 was upregulated in serum exosome derived from patients with hepatocellular carcinoma (HCC) and the density of TAMs in cancer tissues was higher than that in adjacent tissues. They also demonstrated that HCC-exosomes could be delivered to TAMs in a tumor microenvironment by which DLX6-AS1-overexpressed TAMs could further promote the progression of HCC. In the mechanism study, they revealed that after co-culturing HCC-exosomes with monocyte THP-1 cell lines, THP-1 cells are more polarized into M2 macrophages. In addition, after co-injecting hepatocellular cell lines with primed THP-1 cells (THP-1/HCC-exosomes transferred with oe-DLX6-AS1) into the right liver lobe of the mice, they also discovered that primed THP-1 cells (THP-1/HCC-exosomes transferred with oe-DLX6-AS1) could promote HCC lung metastasis in vivo. All the above-mentioned observations show that the crosstalk between lncRNA DLX6-AS1 and tumor-associated macrophages (TAMs) could enhance the progression of HCC, which also may provide novel research clues on the regulatory roles of lncRNA DLX6-AS1 in a tumor microenvironment (TME).
Exosomes (40–100 nm) are key mediators of cell-to-cell communication by transmitting biomolecules such as mRNAs, miRNAs, and lncRNAs (Skog et al., 2008). Several research studies demonstrated that the expression level of exosome-lncRNA DLX6-AS1 in patients with cancer can be a promising prognosis biomarker in tumors.
Zhang et al. reported that the circulating exosome lncRNA DLX6-AS1 expression levels in the serum of patients with NSCLC were significantly higher than those in healthy donors, and its specificity and sensitivity were higher than that of CYFRA21-1 (Ding et al., 2021), which served as a diagnostic marker in NSCLC. They also found that the expression level of exosome lncRNA DLX6-AS1 was positively correlated with tumor differentiation, TNM stage, and lymph vascular invasion, making it a potential early diagnostic and metastatic marker for NSCLC. Ding et al. enrolled 114 patients with cervical cancer (CC), 60 patients with cervical intraepithelial neoplasia (CIN), and 110 healthy women to their study (Ding et al., 2021). They found that the exosome lncRNA DLX6-AS1 level was elevated in CC patients compared with patients with CIN and normal healthy donors. A high serum exosome lncRNA DLX6-AS1 expression level was positively associated with lymph node metastasis, histopathological differentiation, FIGO stage, and shortened survival of patients with CC (Ding et al., 2021). However, because of the limited number of enrolled patients and high false positive and negative results due to the use of serum exosome lncRNA DLX6-AS1 alone as the prognosis factor of patients with CC, they suggested that a combination with other known tumor biomarkers and clinicopathological parameters was needed to accurately predict the clinical outcome of CC.
As a tumor-promoting gene, lncRNA DLX6-AS1 is often upregulated in many tumors compared with normal tissues. Overexpression of lncRNA DLX6-AS1 could mediate the genesis and progression of many tumors, the knockdown of which could alleviate this phenomenon. Besides, high expression levels of lncRNA DLX6-AS1 are also positively associated with the clinicopathological parameters and negatively with the prognosis of patients with a tumor, which could help with the pathological diagnosis, the survival prediction, and histopathological molecular subtyping. Besides mediating the malignant phenotype of tumor cells, over-expressed lncRNA DLX6-AS1 could also be able to communicate with the tumor microenvironment (TME) such as tumor-associated macrophages (TAMs), which could make it highly expressed in cells in the TME via exosomes and further promote the progress of tumors. In addition, the exosome-lncRNA DLX6-AS1 level could be differently derived from the peripheral blood of patients with a tumor diagnosed at different stages. These features might make it a potential prognosis marker combined with other known tumor molecular markers.
lncRNA DLX6-AS1, also named evf1 in the mouse, which involves the development of the mouse’s embryonic ventral forebrain, is dysregulated in developing human telencephalon modeling autism spectrum disorder (ASD). Mariani et al. (2015) revealed that lncRNA DLX6-AS1 was one of the top ten up-regulated genes in cerebral organoids derived from members of a family with idiopathic ASD at TD11 and TD31. Wang et al. (2017) also found that DLX6-AS1 and DLX1 were two of three DEGs in CHD8+/- (CHD8 is a highly mutated gene in autism spectrum disorders) cerebral organoids, which increased ∼39- and 13-fold, respectively, and were hardly expressed in controls (Wang et al., 2017). In addition, it is well known that evf2 (the spliced form of evf1) forms a complex with DLX1 and DLX2 proteins to facilitate the enhancer activity of DLX5 and DLX6 gene expression. Hence, it supports the potential that lncRNA DLX6-AS1 cooperates with the Dlx family to regulate brain development in humans whose dysregulation is related with nervous system disorders.
lncRNA DLX6-AS1 also serves as a competitively endogenously binding RNA in preeclampsia (PE) and sepsis-induced acute kidney injury (AKI). For instance, Tan et al. (2018) found that high expression levels of lncRNA DLX6-AS1 inhibited proliferation, migration, and invasion of trophoblast JEG3 and HTR-8/SVneo cells via directly regulating the miR-376c/GADD45A axis. Interestingly, it has been proven by Liu R. et al. (2020) that the increased lncRNA DLX6-AS1 expression inhibited proliferation, invasion of trophoblast cells of the placenta, and the angiogenesis of HUVEC cells via downregulating ERP44 by sponging miR-149-5p in preeclampsia. Tan et al. (2020) reported that the knockout of lncRNA DLX6-AS1 regulated the miR-223-3p/NLRP3 axis in HK-2 cells to induce sepsis-induced acute kidney injury. In addition, Zheng et al. (2022) first demonstrated that over-expressed cAMP-response element binding protein (CREB) induces podocyte injury in diabetic nephropathy via upregulating the lncRNA DLX6-AS1 expression level in vitro and in vivo.
Most research studies on lncRNA DLX6-AS1 are related to its sponging function to microRNAs as a competitive endogenous RNA via base pairing to each other. MicroRNAs are single-stranded RNAs and generated from endogenous hairpin-shaped transcripts with a length of 19–25 nucleotides (Kim, 2005); they are predicted to regulate the activity of approximately 50% of all protein-coding genes in mammals (Krol et al., 2010). Because of its significant roles in many tumors as a suppressor gene or tumorigenic gene, therapeutics based on microRNAs or anti-microRNAs have been processed to advanced clinical trial stages. Therefore, this may be one of the important reasons for studying the regulatory function of lncRNA DLX6-AS1 in microRNAs.
Due to the biochemical approaches and high-throughput sequencing (Qian et al., 2019), the secondary structure conformations of lncRNAs have been found, and the relationship of lncRNAs between structures and functions should be considered. Hence, the studies of interactions between lncRNA DLX6-AS1 and RNAs, DNAs, or proteins may provide novel research insights into human tumors. We recommend readers to get detailed research clues and inspiration from the articles written by Qian et al. (2019) and Statello et al. (2021).
RNA interference (RNAi) is an endogenous cellular mechanism for regulating gene expression via incorporating siRNAs into RNA-induced silencing complexes to mediate target genes’ cleavage and degradation (Tabernero et al., 2013). lncRNA DLX6-AS1 is usually upregulated in many solid tumor tissues compared to adjacent normal tissues playing a tumorigenic gene role. It is reported that lncRNA DLX6-AS1 is upregulated in many tumor cell lines, and its knockdown can significantly suppress the malignant phenotype of tumor cell lines.
Novel delivery systems can successfully deliver miRNAs to tumor locations without being degraded by RNase in blood and show obvious adverse effects in patients, encapsulating si-lncRNA DLX6-AS1 in lipid nanoparticles to deliver them to tumor sites using the EPR (enhanced permeability and retention) effect on tumor locations (Matsumura et al., 1986). We can also encapsulate si-lncRNA DLX6-AS1 in specific aptamers or target motif-modified nanoparticles to deliver it to tumor locations dependent on ligand–receptor interaction. Simply inhibiting tumor cell malignant phenotypes by delivering si-lncRNA DLX6-AS1 to tumor sites may not show significant tumor inhibitory effects on patients at the advanced stages. Hence, we can encapsulate multiple siRNAs and cooperate with precise photodynamic or photo-thermal therapies to get significant therapeutic effects. However, due to the discontinuous vascular epithelium of the kidneys and spleen similar to immature vascular epithelium in tumor sites, nanoparticles also accumulate easily in these organs and thus may contribute to adverse effects. Nanoparticle drug formulations and pre-clinical trials should be rigorously designed.
Increasing evidence has implied the crucial regulatory roles of lncRNA DLX6-AS1 in various tumors which mainly represent on pre-transcriptional and post-transcriptional levels. One of the most prevalent roles is its sponging function as a ceRNA to form regulatory networks to influence cancer cells’ biological characteristics. Besides, LncRNA DLX6-AS1 can also facilitate mRNA stability to influence cell behaviors in tumors and bind with the DNA promoter region to promote methylation of DNAs which further inhibits DNA transcription. Interestingly, in tumors, the upregulation of lncRNA DLX6-AS1 mostly promotes cell proliferation, invasion, and migration, which is the opposite in preeclampsia. Wang et al. (2017) also found that DLX6-AS1 and DLX1 are dysregulated in CHD8+/- cerebral organoids, supporting its developmental regulatory role in human brain development and nervous disorders. The novel biochemical approaches and high-throughput sequencing can provide new insights into the study of the interactions between lncRNA DLX6-AS1 and RNAs, DNAs, or proteins. Along with the rapid development of delivery systems and precisely cooperated therapies, it may provide the possibility of pre-clinical therapeutics based on lncRNA DLX6-AS1 in many tumors.
This review was conceived and drafted by YZ. YZ and PL revised this article. PL supervised the study. All authors agreed to the submission of this article.
This work was supported by the Program for Innovation Research team (in Science and Technology) in Henan University (Grant No. 20IRTSTHN026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The author thanks PL for her guidance during the completion of this review about lncRNA DLX6-AS1.
An, Y., Chen, X. M., Yang, Y., Mo, F., Jiang, Y., Sun, D. L., et al. (2018). LncRNA DLX6-AS1 Promoted Cancer Cell Proliferation and Invasion by Attenuating the Endogenous Function of miR-181b in Pancreatic Cancer. Cancer Cell Int. 18 (1), 143. doi:10.1186/s12935-018-0643-7
Berghoff, E. G., Clark, M. F., Chen, S., Cajigas, I., Leib, D. E., and Kohtz, J. D. (2013). Evf2 (Dlx6as) lncRNA Regulates Ultraconserved Enhancer Methylation and the Differential Transcriptional Control of Adjacent Genes. Dev. Camb. 140 (21), 4407–4416. doi:10.1242/dev.099390
Bond, A. M., Vangompel, M. J. W., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced Gene Regulation by an Embryonic Brain ncRNA Is Critical for Adult Hippocampal GABA Circuitry. Nat. Neurosci. 12 (8), 1020–1027. doi:10.1038/nn.2371
Cajigas, I., Leib, D. E., Cochrane, J., Luo, H., Swyter, K. R., Chen, S., et al. (2015). Evf2 lncRNA/BRG1/DLX1 Interactions Reveal RNA-dependent Inhibition of Chromatin Remodeling. Development 142 (15), 2641–2652. doi:10.1242/dev.126318
Cajigas, I., Chakraborty, A., Swyter, K. R., Luo, H., Bastidas, M., Nigro, M., et al. (2018). The Evf2 Ultraconserved Enhancer lncRNA Functionally and Spatially Organizes Megabase Distant Genes in the Developing Forebrain. Mol. Cell 71 (6), 956–972. e9. doi:10.1016/j.molcel.2018.07.024
Ding, X. Z., Zhang, S. Q., Deng, X. L., and Qiang, J. H. (2021). Serum Exosomal lncRNA DLX6-AS1 Is a Promising Biomarker for Prognosis Prediction of Cervical Cancer. Technol. Cancer Res. Treat. 20 (1128), 1–6. doi:10.1177/1533033821990060
Du, C., Wang, Y., Zhang, Y., Zhang, J., Zhang, L., and Li, J. (2020). LncRNA DLX6-AS1 Contributes to Epithelial-Mesenchymal Transition and Cisplatin Resistance in Triple-Negative Breast Cancer via Modulating Mir-199b-5p/Paxillin Axis. Cell Transpl. 29, 963689720929983. doi:10.1177/0963689720929983
Fang, C., Xu, L., He, W., Dai, J., and Sun, F. (2019). Long Noncoding RNA DLX6-AS1 Promotes Cell Growth and Invasiveness in Bladder Cancer via Modulating the miR-223-Hsp90b1 axis. Cell Cycle 18 (23), 3288–3299. doi:10.1080/15384101.2019.1673633
Feng, J., Bi, C., Clark, B. S., Mady, R., Shah, P., and Kohtz, J. D. (2006). The Evf-2 Noncoding RNA Is Transcribed from the Dlx-5/6 Ultraconserved Region and Functions as a Dlx-2 Transcriptional Coactivator. Genes Dev. 20 (11), 1470–1484. doi:10.1101/gad.1416106
Feng, L., Wang, R., Wang, Y., Shen, X., Shi, Q., Lian, M., et al. (2021). Silencing Long Non‐coding RNA DLX6‐AS1 or Restoring microRNA‐193b‐3p Enhances Thyroid Carcinoma Cell Autophagy and Apoptosis via Depressing HOXA1. J. Cell Mol. Med. 25 (19), 9319–9330. doi:10.1111/jcmm.16868
Fu, X., Tian, Y., Kuang, W., Wen, S., and Guo, W. (2019). Long Non-coding RNA DLX6-AS1 Silencing Inhibits Malignant Phenotypes of Gastric Cancer Cells. Exp. Ther. Med. 17, 4715–4722. doi:10.3892/etm.2019.7521
Hu, Y., Sun, H., Hu, J., and Zhang, X. (2020). LncRNA DLX6-AS1 Promotes the Progression of Neuroblastoma by Activating STAT2 via Targeting miR-506-3p. Cancer Manag. Res. 12, 7451–7463. doi:10.2147/cmar.s252521
Huang, Y., Ni, R., Wang, J., and Liu, Y. (2019). Knockdown of lncRNA DLX6-AS1 Inhibits Cell Proliferation, Migration and Invasion while Promotes Apoptosis by Downregulating PRR11 Expression and Upregulating miR-144 in Non-small Cell Lung Cancer. Biomed. Pharmacother. 109, 1851–1859. doi:10.1016/j.biopha.2018.09.151
Jia, P., Wei, E., Liu, H., Wu, T., and Wang, H. (2020). Silencing of Long Non‐coding RNA DLX6‐AS1 Weakens Neuroblastoma Progression by the miR ‐513c‐5p/PLK4 axis. IUBMB Life 72 (12), 2627–2636. doi:10.1002/iub.2392
Kim, V. N. (2005). MicroRNA Biogenesis: Coordinated Cropping and Dicing. Nat. Rev. Mol. Cell Biol. 6 (5), 376–385. doi:10.1038/nrm1644
Kohtz, J. D., and Fishell, G. (2004). Developmental Regulation of EVF-1, a Novel Non-coding RNA Transcribed Upstream of the Mouse Dlx6 Gene. Gene Expr. Patterns 4 (4), 407–412. doi:10.1016/j.modgep.2004.01.007
Kong, L., and Zhang, C. (2020). LncRNA DLX6-AS1 Aggravates the Development of Ovarian Cancer via Modulating FHL2 by Sponging miR-195-5p. Cancer Cell Int. 20 (1), 1–12. doi:10.1186/s12935-020-01452-z
Kong, W-Q., Liang, J-J., Du, J., Ye, Z-X., Gao, P., and Liang, Y-L. (2020). Long Noncoding RNA DLX6-AS1 Regulates the Growth and Aggressiveness of Colorectal Cancer Cells via Mediating miR-26a/EZH2 Axis. Cancer Biotherapy Radiopharm. 36 (9), 753–764. doi:10.1089/cbr.2020.3589
Krol, J., Loedige, I., and Filipowicz, W. (2010). The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat. Rev. Genet. 11 (9), 597–610. doi:10.1038/nrg2843
Lei, X., Yang, S., Yang, Y., Zhang, J., Wang, Y., and Cao, M. (2019). Long Noncoding RNA DLX6-AS1 Targets miR-124-3p/CDK4 to Accelerate Ewing’s Sarcoma. Am. J. Transl. Res. 11 (10), 6569–6576.
Li, D., Tang, X., Li, M., and Zheng, Y. (2019). Long Noncoding RNA DLX6‐AS1 Promotes Liver Cancer by Increasing the Expression of WEE1 via Targeting miR‐424‐5p. J Cell. Biochem. 120 (8), 12290–12299. doi:10.1002/jcb.28493
Li, X., Zhang, H., and Wu, X. (2019). Long Noncoding RNA DLX6-AS1 Accelerates the Glioma Carcinogenesis by Competing Endogenous Sponging miR-197-5p to Relieve E2F1. Gene 686, 1–7. doi:10.1016/j.gene.2018.10.065
Liang, Y., Zhang, C.-D., Zhang, C., and Dai, D.-Q. (2020). DLX6-AS1/miR-204-5p/OCT1 Positive Feedback Loop Promotes Tumor Progression and Epithelial-Mesenchymal Transition in Gastric Cancer. Gastric Cancer 23 (2), 212–227. doi:10.1007/s10120-019-01002-1
Lin, S., Gu, S., Qian, S., Liu, Y., Sheng, J., Li, Q., et al. (2021). Genome-Wide Methylation Profiling of lncRNAs Reveals a Novel Progression-Related and Prognostic Marker for Colorectal Cancer. Front. Oncol. 11, 782077. doi:10.3389/fonc.2021.782077
Liu, R., Wang, X., and Yan, Q. (2020). The Regulatory Network of lncRNA DLX6-AS1/miR-149-5p/ERP44 Is Possibly Related to the Progression of Preeclampsia. Placenta 93, 34–42. doi:10.1016/j.placenta.2020.02.001
Liu, X., Peng, D., Cao, Y., Zhu, Y., Yin, J., Zhang, G., et al. (2021). Upregulated lncRNA DLX6-AS1 Underpins Hepatocellular Carcinoma Progression via the miR-513c/Cul4A/ANXA10 axis. Cancer Gene Ther. 28 (5), 486–501. doi:10.1038/s41417-020-00233-0
Liu, Y., Liu, X., Zhang, X., Deng, J., Zhang, J., and Xing, H. (2020). lncRNA DLX6-AS1 Promotes Proliferation of Laryngeal Cancer Cells by Targeting the miR-26a/TRPC3 Pathway. Cancer Manag. Res. 12, 2685–2695. doi:10.2147/cmar.s237181
Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., et al. (2015). FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162 (2), 375–390. doi:10.1016/j.cell.2015.06.034
Matsumura, Y., Maeda, H., and Smancs, A. (1986). A New Concept for Macromolecular Therapeutics in Cancer Chemotherapy: Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res. 46, 6387–6392.
Mir, M., Bickmore, W., Furlong, E. E. M., and Narlikar, G. (2019). Chromatin Topology, Condensates and Gene Regulation: Shifting Paradigms or Just a Phase? Development 146 (19), 1–6. doi:10.1242/dev.182766
Olsson, M., Beck, S., Kogner, P., Martinsson, T., and Carén, H. (2016). Genome-wide Methylation Profiling Identifies Novel Methylated Genes in Neuroblastoma Tumors. Epigenetics 11 (1), 74–84. doi:10.1080/15592294.2016.1138195
Qian, X., Zhao, J., Yeung, P. Y., Zhang, Q. C., and Kwok, C. K. (2019). Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem. Sci. 44 (1), 33–52. doi:10.1016/j.tibs.2018.09.012
Qian, Y., Wu, X., Wang, H., Hou, G., Han, X., and Song, W. (2020). MicroRNA-4290 Suppresses PDK1-Mediated Glycolysis to Enhance the Sensitivity of Gastric Cancer Cell to Cisplatin. Braz J. Med. Biol. Res. 53 (5), e9330–9. doi:10.1590/1414-431X20209330
Skog, J., Würdinger, T., van Rijn, S., Meijer, D. H., Gainche, L., Sena-esteves, M., et al. (2008). Glioblastoma Microvesicles Transport RNA and Proteins that Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 10 (12), 1470–1476. doi:10.1038/ncb1800
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene Regulation by Long Non-Coding RNAs and its Biological Functions. Nat. Rev. Mol. Cell Biol. [Internet] 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sun, W., Zhang, L., Yan, R., Yang, Y., and Meng, X. (2019). LncRNA DLX6-AS1 Promotes the Proliferation, Invasion, and Migration of Non-small Cell Lung Cancer Cells by Targeting the miR-27b-3p/GSPT1 axis. Onco Targets Ther. 12, 3945–3954. doi:10.2147/ott.s196865
Tabernero, J., Shapiro, G. I., LoRusso, P. M., Cervantes, A., Schwartz, G. K., Weiss, G. J., et al. (2013). First-in-humans Trial of an RNA Interference Therapeutic Targeting VEGF and KSP in Cancer Patients with Liver Involvement. Cancer Discov. 3 (4), 406–417. doi:10.1158/2159-8290.cd-12-0429
Tan, J., Fan, J., He, J., Zhao, L., and Tang, H. (2020). Knockdown of LncRNA DLX6-AS1 Inhibits HK-2 Cell Pyroptosis via Regulating miR-223-3p/NLRP3 Pathway in Lipopolysaccharide-Induced Acute Kidney Injury. J. Bioenerg. Biomembr. 52 (5), 367–376. doi:10.1007/s10863-020-09845-5
Tan, Y., Xiao, D., Xu, Y., and Wang, C. (2018). Long Non-coding RNA DLX6-AS1 Is Upregulated in Preeclampsia and Modulates Migration and Invasion of Trophoblasts through the miR-376c/GADD45A axis. Exp. Cell Res. 370 (2), 718–724. doi:10.1016/j.yexcr.2018.07.039
Tian, S., Liu, J., Kong, S., and Peng, L. (2020). LncRNA DLX6-AS1 as a Potential Molecular Biomarker in the Clinicopathology and Prognosis of Various Cancers: a Meta-Analysis. Biosci. Rep. 40 (8), BSR20193532. doi:10.1042/BSR20193532
Wang, H., Niu, X., Jiang, H., Mao, F., Zhong, B., Jiang, X., et al. (2020). Long Non-coding RNA DLX6-AS1 Facilitates Bladder Cancer Progression through Modulating miR-195-5p/VEGFA Signaling Pathway. Aging 12 (16), 16021–16034. doi:10.18632/aging.103374
Wang, L. P., Lin, J., Ma, X. Q., Xu, D. Y., Shi, C. F., Wang, W., et al. (2021). Exosomal DLX6-AS1 from Hepatocellular Carcinoma Cells Induces M2 Macrophage Polarization to Promote Migration and Invasion in Hepatocellular Carcinoma through microRNA-15a-5p/CXCL17 axis. J. Exp. Clin. Cancer Res. 40 (1), 1–16. doi:10.1186/s13046-021-01973-z
Wang, P., Mokhtari, R., Pedrosa, E., Kirschenbaum, M., Bayrak, C., Zheng, D., et al. (2017). CRISPR/Cas9-mediated Heterozygous Knockout of the Autism Gene CHD8 and Characterization of its Transcriptional Networks in Cerebral Organoids Derived from iPS Cells. Mol. Autism 8 (1), 11–17. doi:10.1186/s13229-017-0124-1
Wang, X., Lin, Y., and Liu, J. (2019). Long Non-Coding RNA DLX6-AS1 Promotes Proliferation by Acting as a ceRNA Targeting miR-199a in Cervical Cancer. Mol. Med. Rep. 19 (2), 1248–1255. doi:10.3892/mmr.2018.9729
Wu, C., Lin, W., and Fu, F. (2021). Long Non-coding RNA DLX6-AS1 Knockdown Suppresses the Tumorigenesis and Progression of Non-small Cell Lung Cancer through microRNA-16-5p/BMI1 axis. Transl. Cancer Res. TCR 10 (8), 3772–3787. doi:10.21037/tcr-21-1240
Wu, Q., Ma, J., Meng, W., and Hui, P. (2020). DLX6-AS1 Promotes Cell Proliferation, Migration and EMT of Gastric Cancer through FUS-Regulated MAP4K1. Cancer Biol. Ther. 21 (1), 17–25. doi:10.1080/15384047.2019.1647050
Xie, F., Xie, G., and Sun, Q. (2020). Long Noncoding RNA DLX6-AS1 Promotes the Progression in Cervical Cancer by Targeting miR-16-5p/ARPP19 Axis. Cancer Biotherapy Radiopharm. 35 (2), 129–136. doi:10.1089/cbr.2019.2960
Xue, C., Lv, L., Jiang, J., and Li, L. (2021). Promising Long Noncoding RNA DLX6-AS1 in Malignant Tumors. Am. J. Transl. Res. 12 (12), 7682–7692.
Yang, B., Jia, L., Ren, H., Jin, C., Ren, Q., Zhang, H., et al. (2020). LncRNA DLX6-AS1 Increases the Expression of HIF-1α and Promotes the Malignant Phenotypes of Nasopharyngeal Carcinoma Cells via Targeting MiR-199a-5p. Mol. Genet. Genomic Med. 8 (1), e1017. doi:10.1002/mgg3.1017
Yang, J., Ye, Z., Mei, D., Gu, H., and Zhang, J. (2019). Long Noncoding RNA DLX6-AS1 Promotes Tumorigenesis by Modulating miR-497-5p/FZD4/FZD6/Wnt/β-Catenin Pathway in Pancreatic cancer/FZD6/Wnt/β-Catenin Pathway in Pancreatic Cancer. Cancer Manag. Res. 11, 4209–4221. doi:10.2147/cmar.s194453
Yang, Q., Sun, J., Ma, Y., Zhao, C., and Song, J. (2019). LncRNA DLX6-AS1 promotes laryngeal squamous cell carcinoma growth and invasion through regulating miR-376c. Am. J. Transl. Res. 11 (11), 7009–7017.
Zeng, X., Hu, Z., Ke, X., Tang, H., Wu, B., Wei, X., et al. (2017). Long Noncoding RNA DLX6-AS1 Promotes Renal Cell Carcinoma Progression via miR-26a/PTEN axis. Cell Cycle 16 (22), 2212–2219. doi:10.1080/15384101.2017.1361072
Zhang, H. Y., Xing, M. Q., Guo, J., Zhao, J. C., Chen, X., Jiang, Z., et al. (2019). Long Noncoding RNA DLX6-AS1 Promotes Neuroblastoma Progression by Regulating miR-107/BDNF Pathway. Cancer Cell Int. 19 (1), 313. doi:10.1186/s12935-019-0968-x
Zhang, L., He, X., Jin, T., Gang, L., and Jin, Z. (2017). Long Non-coding RNA DLX6-AS1 Aggravates Hepatocellular Carcinoma Carcinogenesis by Modulating miR-203a/MMP-2 Pathway. Biomed. Pharmacother. 96, 884–891. doi:10.1016/j.biopha.2017.10.056
Zhang, N., Meng, X., Mei, L., Zhao, C., and Chen, W. (2019). LncRNA DLX6‐AS1 Promotes Tumor Proliferation and Metastasis in Osteosarcoma through Modulating miR‐641/HOXA9 Signaling Pathway. J Cell. Biochem. 120 (7), 11478–11489. doi:10.1002/jcb.28426
Zhangmou, R. M., Tang, T., Yu, H. M., and Yao, X. D. (2018). LncRNA DLX6-AS1/miR-129-5p/DLK1 axis Aggravates Stemness of Osteosarcoma through Wnt Signaling. Biochem. Biophys. Res. Commun. 507 (1–4), 260–266. doi:10.1016/j.bbrc.2018.11.019
Zhao, H., and Xu, Q. (2020). Long Non‐coding RNA DLX6‐AS1 Mediates Proliferation, Invasion and Apoptosis of Endometrial Cancer Cells by Recruiting p300/E2F1 in DLX6 Promoter Region. J. Cell. Mol. Med. 24 (21), 12572–12584. doi:10.1111/jcmm.15810
Zhao, P., Guan, H., Dai, Z., Ma, Y., Zhao, Y., and Liu, D. (2019). Long Noncoding RNA DLX6-AS1 Promotes Breast Cancer Progression via miR-505-3p/RUNX2 axis. Eur. J. Pharmacol. 865, 172778. doi:10.1016/j.ejphar.2019.172778
Zhao, X., Wang, J., Zhu, R., Zhang, J., and Zhang, Y. (2021). DLX6-AS1 Activated by H3K4me1 Enhanced Secondary Cisplatin Resistance of Lung Squamous Cell Carcinoma through Modulating miR-181a-5p/miR-382-5p/CELF1 axis. Sci. Rep. 11 (1), 1–13. doi:10.1038/s41598-021-99555-8
Zhao, Z., Liang, S., and Sun, F. (2020). LncRNA DLX6-AS1 Promotes Malignant Phenotype and Lymph Node Metastasis in Prostate Cancer by Inducing LARGE Methylation. Front. Oncol. 10, 1172. doi:10.3389/fonc.2020.01172
Zheng, Q., Gu, X., Yang, Q., Chu, Q., Dai, Y., and Chen, Z. (2021). DLX6-AS1 Is a Potential Biomarker and Therapeutic Target in Cancer Initiation and Progression. Clin. Chim. Acta 517 (79), 1–8. doi:10.1016/j.cca.2021.02.006
Zheng, W., Guo, J., Lu, X., Qiao, Y., Liu, D., Pan, S., et al. (2022). cAMP-Response Element Binding Protein Mediates Podocyte Injury in Diabetic Nephropathy by Targeting lncRNA DLX6-AS1. Metabolism 129, 155155. doi:10.1016/j.metabol.2022.155155
Keywords: lncRNA DLX6-AS1, regulatory mechanism, tumor, novel research methodologies, therapeutic strategies
Citation: Zhao Y and Li P (2022) Strategies of LncRNA DLX6-AS1 on Study and Therapeutics. Front. Genet. 13:871988. doi: 10.3389/fgene.2022.871988
Received: 09 February 2022; Accepted: 20 April 2022;
Published: 01 June 2022.
Edited by:
Ruowen Zhang, Jiahehongsheng (Shenzhen) Health Industry Group, ChinaReviewed by:
Dongsheng Yan, Wenzhou Medical University, ChinaCopyright © 2022 Zhao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei Li, TGlwZWlmcmVlbWFpQHp6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.