- 1College of Animal Science, Jilin University, Changchun, China
- 2Institute of Animal Husbandry and Veterinary, Jilin Academy of Agricultural Sciences, Gongzhuling, China
It is well known that Dorper (DP) is a full-bodied, fast-growing and high dressing percentage breed, while the production performance of Small-tailed Han sheep (STH) is not so excellent, in contrast to DP. Therefore, in this study, a comparative transcriptomic analysis of liver and muscle tissues from DP and STH breeds was carried out to find differentially expressed genes (DEGs) that affect their growth and meat quality traits. The results showed that the total number of DEGs was 2,188 in the two tissues. There were 950, 160 up-regulated and 1,007, 71 down-regulated genes in the liver and muscle, respectively. Several DEGs such as TGFB1, TGFB3, FABP3, LPL may be associated with growth and development in DP. Also, several GO terms were found to be associated with muscle growth and development, such as developmental growth (GO:0048589), and myofibril (GO:0030016). Further validation of eight genes (6 up-regulated, and 2 down-regulated) was performed using quantitative RT-PCR. These findings will provide valuable information for studying growth and development as well as meat quality traits in sheep.
Introduction
Since sheep were domesticated 8∼11 thousand years ago in Fertile Crescent, they have been providing mankind with excellent products such as meat, wool and milk (Lv et al., 2015). As a country with a large population, China has a huge sheep industry chain with a large consumer of sheep meat. Mutton is becoming increasingly popular with people because of its rich nutrition, high protein and low-fat characteristics. The economic value of a mutton sheep is primarily based on the quantity and quality of the meat. Meanwhile, the growth rate of the sheep is one of the main factors affecting meat yield. Therefore, one breeding objective of mutton sheep is to increase growth rate and meat yield (Seyedsharifi and Hamze Zadeh Azar, 2016). In the same environments, the genetic background becomes one of the important factors influencing meat production (Hegarty et al., 2006; Aali et al., 2017).
The Dorper (DP) is a commercial mutton breed native to South Africa, known for its excellent meat quality and outstanding growth potential (Milne, 2000; Schoeman, 2000). Meanwhile, it is an early maturing and fat depositing breed with a high average daily gain (Brand et al., 2017). Many studies on improving the growth of indigenous breeds by crossing them with Dorper have been reported (Weldeyesus, 2017; Shi et al., 2021). As a remarkable local breed in China, the Small-tailed Han sheep (STH) has a significant characteristic of high fecundity (Wang et al., 1990). Nevertheless, compared to commercial mutton sheep (e.g., Dorper, Dorset and Charollais), STH has a slower growth rate and a lower meat yield (Zhang et al., 2014; Sun et al., 2016), which seriously affects the production efficiency. Differences in the productive performance of DP and STH are mainly determined by genetic material and related regulatory factors. The wide application of RNA-seq technology has accelerated the selection and breeding of livestock. Based on transcriptome comparisons, transcripts in tissues can be well characterized, identified and quantified. Many genes affecting important traits have been identified by RNA-seq (Chen et al., 2011a; Chen et al., 2011b; Ayuso et al., 2015; Weber et al., 2016; Aali et al., 2017). To date, the comparative transcriptome analysis between DP and STH has not been described clearly and accurately.
Therefore, this study was designed to identify genes associated with growth and meat quality in liver and muscle tissues of DP and STH using RNA-seq. It will provide a theoretical basis for improving sheep breeding practices in China and may contribute to the understanding of molecular regulatory mechanisms of sheep production performance.
Materials and Methods
Experimental Animals
A total of six male individuals (three of each breed) were randomly selected from purebred SAMM population of Sheep Farm of Jilin Academy of Agricultural Sciences (Changchun, China), which with similar date of birth and weaned at around 45 days, were kept in the same environment (same diet and management practices). The animals were weighed before slaughter and analyzed using the T-test. Two comparison groups were designed for DP and STH in this study, a liver comparison group (DP_L vs. STH_L) and a muscle comparison group (DP_M vs. STH_M), and lambs were slaughtered at an approximate age of 6 months, then liver and Longissimus dorsi tissue (muscle) were collected into the tubes, which were labeled with sample number (DP_L_1∼3, STH_L_1∼3, DP_M_1∼3 and STH_M_1∼3). All samples were snap-frozen in liquid nitrogen and kept at a temperature of −80°C until RNA extraction.
RNA Isolation, Library Construction, and Sequencing
Total RNA was extracted from 12 samples using an EasyPure RNA kit (TransGen, Beijing, China) according to the manufacturer’s instructions. The purification, concentration and integrity were detected using a NanoPhotometer spectrophotometer (IMPLEN, CA, United States), the Qubit RNA Assay Kit with the Qubit 2.0 Flurometer (Life Technologies, Carlsbad, CA, United States) and the RNA Nano 6000 Assay Kit of the Bioanalyzer 2,100 system (Agilent Technologies, CA, United States), respectively. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, MA, United States) following the manufacturer’s recommendations. In brief, 12 RNA samples with high quality and concentration were used to construct the transcriptome libraries, and high-throughput sequencing was performed on the Illumina novaseq 6000 platform according to the manufacturer’s instructions, generating reads of 150 bp in length.
Data Processing
Clean data were obtained by removing reads, which contained adapters or ploy-N, and low quality reads from raw data by using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) powered by the Linux system. At the same time, Q20, Q30 and GC content of the clean data were calculated. Clean reads were then mapped to the reference genome (Oar_rambouillet_v1.0, http://asia.ensembl.org/Ovis_aries_rambouillet/) and annotated transcripts (http://ftp.esembl.org/pub/release-104/gtf/ovis_aries_rambouillet/) of Ovis aries by using the Hisat2 software (https://ccb.jhu.edu/software/hisat2/index.shtml) (Kim et al., 2015). The read numbers mapped to each gene were counted using the featureCounts software (Liao et al., 2014).
Differential Expression Analysis and Functional Annotation
Analysis of differential expressed genes (DEGs) in each group was performed by DESeq2 R package (Love et al., 2014). In order to improve accuracy, the counts calculated by the featureCounts software were filtered out if the counts were less than 5 in each sample. And the false discovery rate (FDR) of each gene in a pair-wise comparison was determined using the Benjamini-Hochberg method. Cutoffs for differential gene expression were selected as p-value and FDR <0.05, and log2fold change (log2|FC|) ≥ 1, and PCA was performed on VST values (implemented in DESeq2). To detect biological functions of DEGs, clusterProfiler R package was used to conduct the Gene Ontology (GO) functional enrichment analysis for cellular components (CC), biological processes (BP), and molecular functions (MF) (Yu et al., 2012). KOBAS.i was applied for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://kobas.cbi.pku.edu.cn/kobas3/) (Bu et al., 2021). A p-value of <0.05 (calculated using Fisher’s exact test) was set as the cutoff criteria for the GO and KEGG pathway functional enrichment analysis.
RT-qPCR Validation
RNA extraction was performed as described above and then converted to first-strand cDNA using TransScript One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China). The primers used in this study were designed by Primer 5.0 software (Supplementary Table S1) and were synthesized by Sangon Biotech (Shanghai, China). Quantitative Real-time PCR (RT-qPCR) was conducted on a Stratagene Mx3005P qPCR machine (Stratagene) with SYBR Green Technology. The cycling conditions were as follows: one cycle at 94°C for 30 s, 50 two-segment cycles (94°C for 20 s, 60°C for 1 min) and a final dissociation cycle (95°C for 1 min and progressive rise from 55°C to 95°C). Housekeeping gene ACTB was used as an internal control for quantitation, and relative expression of mRNA was calculated using 2−(ΔΔCt) method.
Results
Animal Weight and Summary of RNA-Seq Data
The average live weight was 48.27 ± 3.18 kg for DP, which was significantly higher than the weight of 40.73 ± 0.98 kg for STH (p = 0.03). To obtain a global overview of genes related to sheep growth and meat quality, we performed two pairwise comparisons: DP_L vs. STH_L, DP_M vs. STH_M. Twelve separate cDNA libraries were constructed from the liver and muscle. A total of 590,460,078 raw reads were obtained from liver and muscle tissues, the total number of bases for each of the samples varied from 6.4 to 8.4 Gb. After removing adapters, low-quality and low-complexity reads, high-quality RNA sequencing data were generated. Subsequently, 561,560,544 clean reads (84.23 Gb) were obtained, with each sample having >6.13 Gb. The Q20, Q30 and GC content of the clean data were simultaneously calculated. The Q20 (the percentage of bases with a Phred score greater than 20) and Q30 (the percentage of bases with a Phred score greater than 30) were higher than 94%, and the mean average GC content was 50%. The ratio of reads mapping uniquely to the reference sheep genome was ranged from 67.39% to 86.82% (Table 1). PCA and Spearman correlation plots showed that groups can be clearly separated from each other (Supplementary Figures S1, S2).
Differentially Expressed Genes Analyses
A total of 12,823 and 13,685 genes were found in liver tissues of DP and STH, respectively. Also, 13,061 and 12,899 genes were found in muscle tissues. The intersection analysis revealed that 11,129 genes were expressed in all 4 groups (FPKM > 0) (Supplementary Figure S3A). Then the DESeq2 R package was used to obtain DEGs. A total of 1957 DEGs were found when comparing liver transcriptomes of DP and STH sheep (FDR < 0.05) (Supplementary Table S2). Of these, 950 genes had higher expression in DP sheep compared to STH sheep and are therefore referred to as “up-regulated”, while the remaining 1,007 genes had lower expression in DP sheep and are accordingly named “down-regulated” (Figure 1A). Regarding the muscle transcriptome, 160 and 71 DEGs were detected as up-regulated and down-regulated, respectively (Supplementary Table S3) (Figure 1B). Integration analyses of DEGs revealed that 14 DEGs were up-regulated in both liver and muscle of DP, and correspondingly, the number of down-regulated in both tissues was 12 (Supplementary Figures S3B,C).
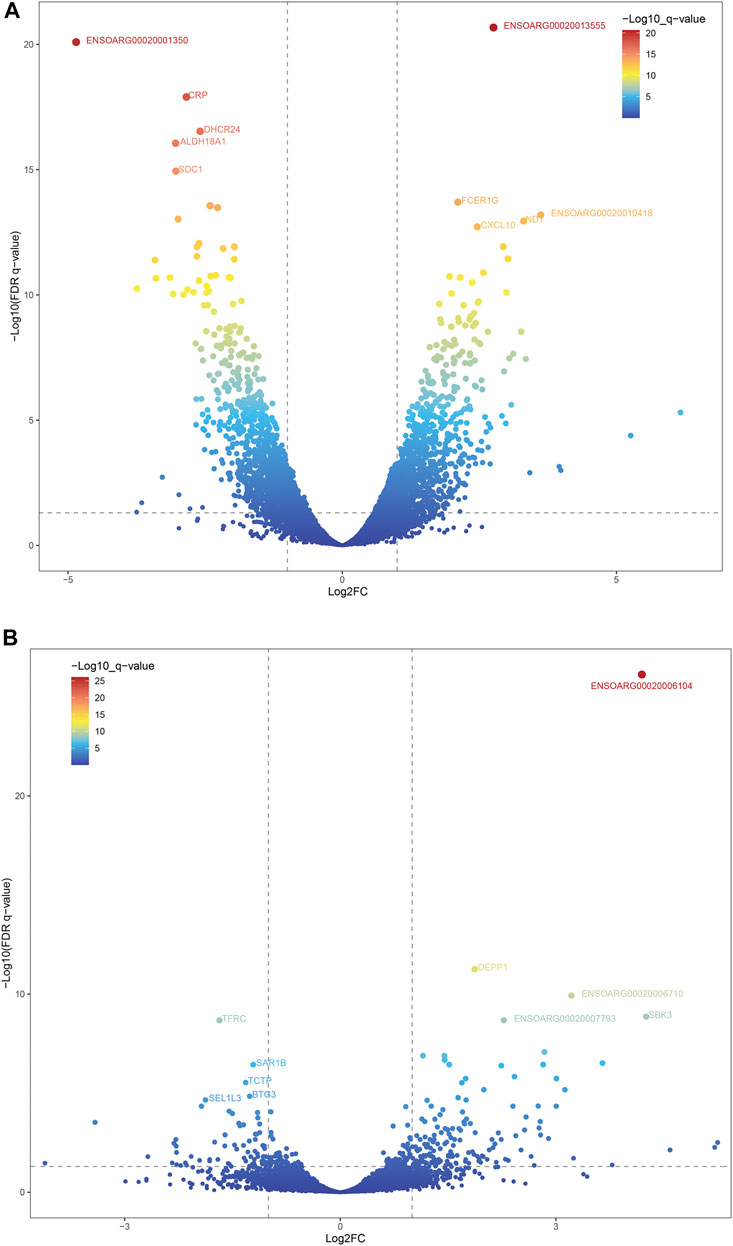
FIGURE 1. The volcano plots of DEGs of the two groups. Two vertical lines indicated the expression fold change (DP vs. STH) ≥1 and ≤−1, respectively, and the horizontal line indicated the adjusted p-value (FDR q-value) of 0.05. The color of the dot represented the FDR (q-value) levels. The top 5 DEGs were labeled on the plots. (A) DEGs of the liver; (B) DEGs of the muscle.
GO and KEGG Pathway Analyses of DEGs
GO enrichment analysis was performed to shed light on the potential function of DEGs concerned with growth and fat deposition-related traits between DP and STH. In the study, we separated the up- and down-regulated genes for GO analysis and found that many up-regulated DEGs enriched two GO terms related to growth development: developmental growth (GO:0048589) and positive regulation of developmental process (GO:0051094). And some up-regulated genes were associated with adaptability and immunity, such as adaptive immune response (GO:0002250). Down-regulated genes were enriched to a number of metabolic pathways, including small molecule metabolic process (GO:0044281), carboxylic acid metabolic process (GO:0019752), and oxoacid metabolic process (GO:0043436). Moreover, the term negative regulation of growth (GO:0045926) was discovered. In the muscle group, up-regulated genes were associated with calcium ion binding (GO:0005509), sarcomere (GO:0030017), myofibril (GO:0030016), while down-regulated genes were associated with cytochrome complex (GO:0070069), positive regulation of B cell proliferation (GO:0030890), respirasome (GO:0070469) (p < 0.05). The top 10 significant GO terms in each of the three categories were shown in (Figures 2A–F). All GO results were given in Supplementary Tables S4, S5.
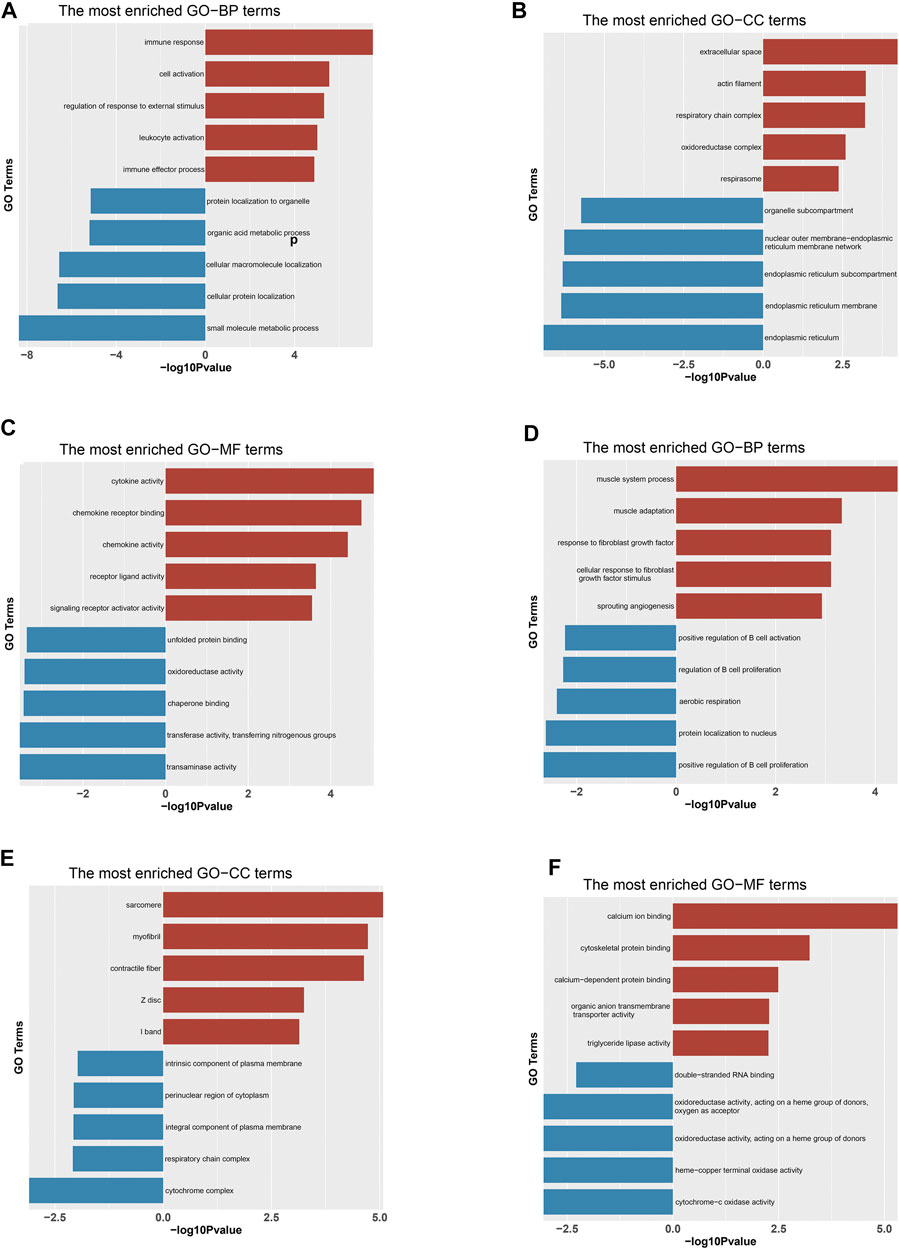
FIGURE 2. Significantly enriched Gene Ontology (GO) terms of DEGs based on their functions. Bar chart of the top 10 BP (A), CC (B) and MF (C) terms in the enrichment analysis of DEGs in the liv-er. Bar chart of the top 10 BP (D), CC (E) and MF (F) terms in the enrichment analysis of DEGs in the muscle. BP: biological process; CC: cellular component; MF: molecular function. “Red” represents for GO terms of up-regulated genes and “Blue” represents for GO terms of down-regulated genes in DP.
Based on the KEGG pathway database, we could systematically analyze gene function and molecular networks in cells. In the current study, those DEGs were assigned to 234 KEGG pathways. Of the 193 KEGG pathways in the liver group, up-regulated DEGs were annotated to development-related pathways such as Osteoclast differentiation (oas04380), immune-related pathways, including C-type lectin receptor signaling pathway (oas04625), Chemokine signaling pathway (oas04062), and T cell receptor signaling pathway (oas04660), etc. The down-regulated DEGs of the liver were mainly enriched in several metabolic pathways including Metabolic pathways (oas01100), Carbon metabolism (oas01200) and Fatty acid degradation (oas00071). In addition, signal transduction pathways such as PI3K-Akt signaling pathway (oas04151) and Hippo signaling pathway (oas04390) were also significantly enriched. A total of 41 KEGG pathways were annotated in the muscle, and the up-regulated genes were mainly associated with important signaling pathways and organismal systems such as AMPK signaling pathway (oas04152), FoxO signaling pathway (oas04068), PPAR signaling pathway (oas03320) and Insulin signaling pathway (oas04910). Meanwhile, down-regulated genes are associated with HIF-1 signaling pathway (oas04066), Metabolic pathways (oas01100) and Biosynthesis of amino acids (oas01230). The top 30 significant pathways (15 for up-regulation, 15 for down-regulation) in each group were shown in (Figures 3A–D), and all the significant KEGG results were shown in Supplementary Tables S6, S7. Statistics for GO and KEGG results for each group were available in Supplementary Table S8.
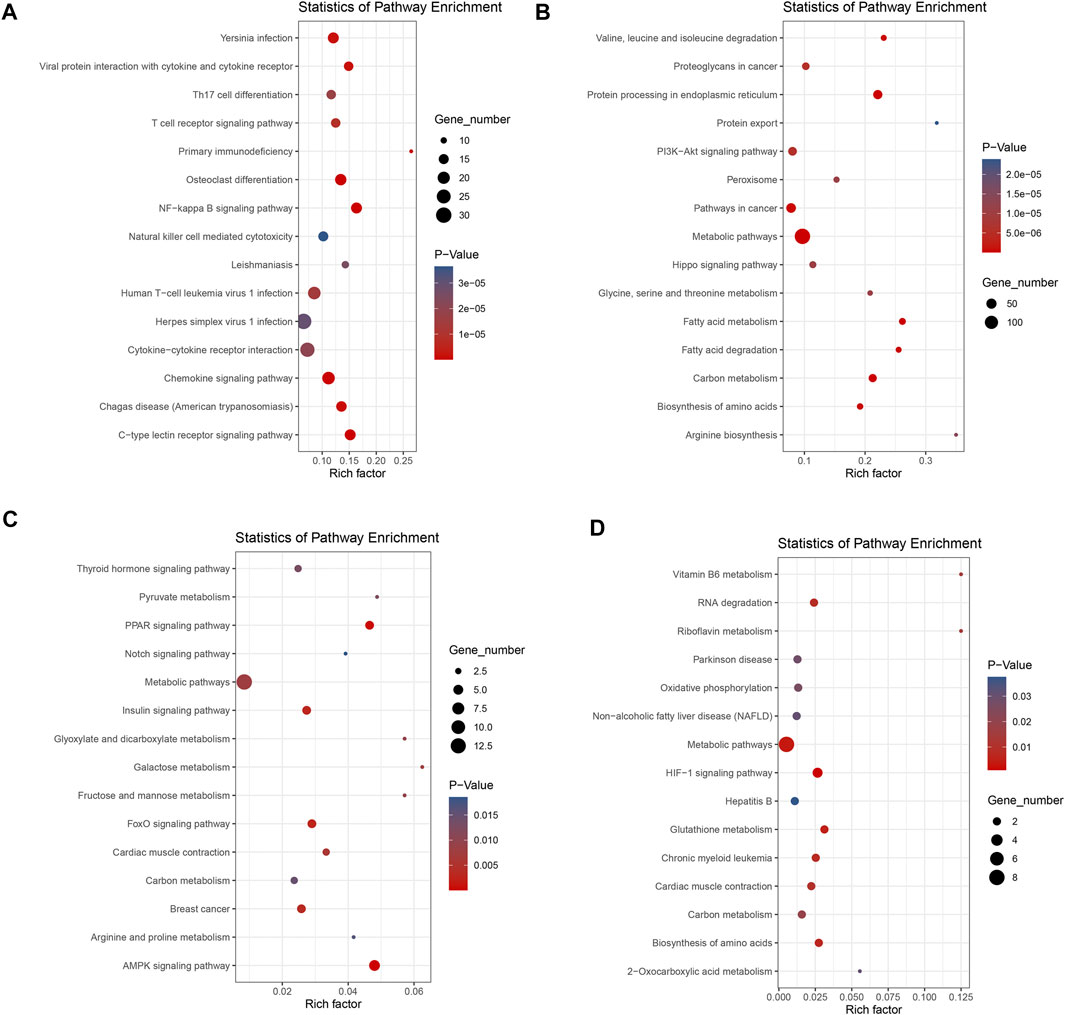
FIGURE 3. Bubble plots showing functional pathways of KEGG enrichment: The top 15 pathways shown in (A) pathways of up-regulated genes of liver; (B) pathways of down-regulated genes of liver; (C) pathways of up-regulated genes of muscle, and (D) pathways of down-regulated genes of muscle. Up- and down-regulation refers to gene expression in DP relative to STH.
RT-qPCR
In order to validate the results observed based on the RNA-seq data, the relative expression of 8 genes was quantified with RT-qPCR in the 12 samples. Six up-regulated (SOCS2, CYLD, VEGFB, FABP3, LPL, APOLD1) and 2 down-regulated genes (TGFBR2, CDKN1A) were selected. The validation results showed that the tendency of qPCR data was consistent with that of transcriptome sequencing data (Figures 4A,B).
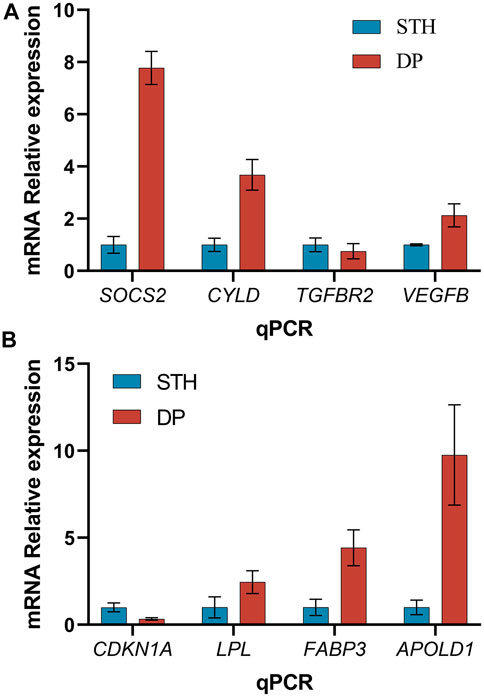
FIGURE 4. Quantitative real time PCR analysis (mean ± s.d., n = 3 biological replicates). (A) Results of 4 DEGs of liver; (B) Results of 4 DEGs of muscle.
Discussion
Mutton is a great source of food with a high content of protein and vitamins, therefore lots of people in the world could be mutton potential consumers. Because of its rapid growth and high meat production, DP is often used as a sire to improve the production performance and carcass quality of local breeds (Cheng et al., 2020). STH is widely reared in northern China due to its high reproductive capacity, imperfectly, in terms of meat production, it is not as good as the imported breeds (Di et al., 2012; Cheng et al., 2020). Many genes related to meat quality, growth and development have been identified in sheep (Di et al., 2012; Sun et al., 2016). Zhang et al. initially reported DEGs in biceps brachii tissues between DP and STH (one sample per breed) (Zhang et al., 2014). Indeed, their study provided very precious information of the DP and STH muscle transcriptomes, unfortunately, the information is limited, as a result of two individuals and inaccurate annotation of the selected reference genome. Consequently, an initial objective of this study was to identify the DEGs of the liver and muscle tissues, which may have an important role in the growth and meat yield of sheep.
In the liver, transcriptome analysis revealed that gene expression of DP and STH was significantly different. Among the DEGs, we found two GO terms namely developmental growth (GO:0048589) and positive regulation of developmental process (GO:0051094). The two terms contain many genes, which are associated with growth and development in livestock or poultry. For example, Hesx1 polymorphism is associated with average daily gain in bovine (Lai et al., 2009), MUSTN1 is thought to be involved in muscle growth and development in chickens (Li et al., 2013), and ATP2B1 is associated with growth and meat quality in pigs (Ramos et al., 2009). More importantly, we identified two members of the transforming growth factor-beta, TGFB1 and TGFB3. TGFB1 has been shown to be associated with muscle growth and pig feed utilization, and TGFB3 plays an important role in the growth and production (Long et al., 2003; Jing et al., 2015; Cao et al., 2017). SOCS2 belongs to the suppressors of cytokine signaling family, which has a central SH2 domain and a C-terminal SOCS box. The proteins of SOCS2 are multifunctional through the JAK/STAT/SOCS pathway (Metcalf et al., 2000), as modulators of cytokine and growth factor signaling. SOCS2 acts in the growth hormone (GH) signaling pathway and is therefore associated with cell growth (Metcalf et al., 2000; Horvat and Medrano, 2001; Greenhalgh et al., 2005). Low levels of SOCS2 expression inhibit several signaling pathways, including GH, prolactin, and interleukins, which negatively regulate growth hormone and insulin-like growth factor-1, whereas high levels of SOCS2 can restore or even increase the responsiveness of these growth factors (Favre et al., 1999; Pezet et al., 1999; Tannahill et al., 2005; Piessevaux et al., 2006). Further, the SOCS2 p.R96C mutation in sheep resulted in better traits such as body weight, body size, and milk production (Rupp et al., 2015). In addition, some up-regulated DEGs were significantly enriched in several immune-related terms and KEGG pathways, which makes sense as adaptability to the environment is one of the characteristics of Dorper. In our study, these genes were significantly more highly expressed in DP than in STH, and it is reasonable to assume that they have contributed to the growth of DP.
In the muscle, we identified a subset of up-regulated genes (for example, FABP3, LPL), which are probably involved in adipogenesis and fat deposition. FABP3 belongs to the fatty acid-binding proteins (FABPs) family, which, like other members of the FABPs family, is an intracellular protein involved in the transport of fatty acids from the plasma membrane to sites of ß-oxidation and triacylglycerol or phospholipid synthesis, regulating cell growth and proliferation (Chmurzyńska, 2006; Jurie et al., 2007) Many studies have shown that the FABP3 gene is involved in muscle development and meat marbling (Uemoto et al., 2007; Arora et al., 2014). Xu et al. (2018) found that FABP3 was associated with fat deposition in Yorkshire pigs. Similarly, Xu et al. (2020) reported that the expression of the FABP3 had a positive effect on the intramuscular fat (IMF) content in different muscles of Tan sheep. Lipoprotein lipase (LPL), which regulates fat in muscle and thus affects carcass quality, has been shown to significantly affect the fatty acid composition of Korean cattle muscle (Oh et al., 2013). Furthermore, both LPL and FABP3 are enriched in the PPAR signaling pathway, which is known to be a key regulatory pathway in adipogenesis (Lehrke and Lazar, 2005). In addition to this, we also found up-regulated genes including MYL2, MYL6 and TNNC1, which have been shown to correlate with meat quality (PilNam et al., 2008; Pierzchala et al., 2014; Muniz et al., 2020). In this experiment, FABP3 and LPL were significantly up-regulated in the muscle tissue of the DP, this is consistent with the actual situation, as fat deposition of DP is early (Brand et al., 2017). Hence, the increased expression level of the FABP3 and LPL may contribute to intramuscular fat deposition in Dorper.
In this study, 14 up- and 12 down-regulated DEGs were identified in both tissues of DP and STH, respectively. Among the up-regulated genes, the BICC1 gene is associated with human skeletal muscle hypertrophy and is involved in skeletal cell differentiation (Mesner et al., 2014; Seaborne et al., 2018), Moreover, this gene has been revealed as one of the selection signals for body size, meat, and growth-related trait, as well as, has been associated with birth weight in sheep (Almasi et al., 2020; Li et al., 2020), Another up-regulated gene, TFAP4 was considered to be related to chest girth in genome-wide association studies (Tao et al., 2020). These genes were significantly higher in both muscle and liver of DPs than in STH, suggesting that they are likely to be involved in body weight at slaughter in DP. Of the 12 co-expressed down-regulated genes, TFRC and MAPK6 are thought to be associated with immunity and mastitis, LARP1B and ODC1 are associated with reproductive traits, such as oestrogen and sperm quality (Park et al., 2016; Nasser et al., 2020; Zhao et al., 2020; Ghahramani et al., 2021; Zhong et al., 2021). In addition, considering that the CDKN1A gene has a negative growth regulatory role as showed in the GO enrichment analysis, the down-regulated expression of these genes may be related to the highly reproductive request, slow growth and developmental characteristics of the STH breed. However, more studies are necessary for a better understanding of the molecular networks and genes regulating economically important traits in sheep.
Conclusion
In this study, comparative transcriptomic analysis was applied to the liver and muscle of Dorper and Small-tailed Han sheep. Some DEGs, such as TGFB1, TGFB3, FABP3, LPL, and a number of GO terms may be associated with growth and fat deposition in Dorper. In addition, the CDKN1A gene was identified, which may be involved in the growth-regulating role of Small-tailed Han. The accuracy of the RNA-seq was verified using RT-qPCR. The mechanisms behind these differences need to be further investigated. Even so, we expect that our analysis will provide accessible information for future research.
Data Availability Statement
The datasets presented in this study can be found in online repositories. Accession No. PRJNA799452. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA799452/.
Ethics Statement
The animal study was reviewed and approved by Animal Welfare and Ethics Committee of the Jilin Academy of Agricultural Sciences of China (AWEC2020A05, 8 June 2020).
Author Contributions
Conceptualization, HP and MH; methodology, HP; software, HP and MH; validation, HP, MH, and WL; formal analysis, ZL and LS; investigation, ZZ; resources, HM; data curation, HP; writing—original draft preparation, HP; writing—review and editing, HP and SY; visualization, HP and MH; supervision, YL; project administration, SY and YL; funding acquisition, YL and SY. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Natural Science Foundation of Jilin Province (Nos. 20200201035JC; 20200201139JC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.868717/full#supplementary-material
References
Aali, M., Moradi-Shahrbabak, H., Moradi-Shahrbabak, M., Sadeghi, M., and Yousefi, A. R. (2017). Association of the Calpastatin Genotypes, Haplotypes, and SNPs with Meat Quality and Fatty Acid Composition in Two Iranian Fat- and Thin-Tailed Sheep Breeds. Small Ruminant Res. 149, 40–51. doi:10.1016/j.smallrumres.2016.12.026
Almasi, M., Zamani, P., Mirhoseini, S. Z., and Moradi, M. H. (2020). Genome-Wide Association Study of Weaning Traits in Lori-Bakhtiari Sheep. Ann. Anim. Sci. 20 (3), 811–824. doi:10.2478/aoas-2020-0014
Arora, R., Yadav, H. S., and Yadav, D. K. (2014). Identification of Novel Single Nucleotide Polymorphisms in Candidate Genes for Mutton Quality in Indian Sheep. Anim. Mol. Breed. 4 (4), 1–5. doi:10.5376/amb.2014.04.0001
Ayuso, M., Fernández, A., Núñez, Y., Benítez, R., Isabel, B., Barragán, C., et al. (2015). Comparative Analysis of Muscle Transcriptome between Pig Genotypes Identifies Genes and Regulatory Mechanisms Associated to Growth, Fatness and Metabolism. Plos One 10 (12), e0145162. doi:10.1371/journal.pone.0145162
Brand, T. S., Van der Westhuizen, E. J., Van der Merwe, D. A., and Hoffman, L. C. (2017). Effect of Days in Feedlot on Growth Performance and Carcass Characteristics of Merino, South African Mutton Merino and Dorper Lambs. South Afr. J. Anim. Sci. 47 (1), 26–33. doi:10.4314/sajas.v47i1.5
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 49 (W1), W317–W325. doi:10.1093/nar/gkab447
Cao, Y., Jin, H.-G., Ma, H.-H., and Zhao, Z.-H. (2017). Comparative Analysis on Genome-wide DNA Methylation in Longissimus Dorsi Muscle between Small Tailed Han and Dorper×Small Tailed Han Crossbred Sheep. Asian-Australas J. Anim. Sci. 30 (11), 1529–1539. doi:10.5713/ajas.17.0154
Chen, C., Ai, H., Ren, J., Li, W., Li, P., Qiao, R., et al. (2011a). A Global View of Porcine Transcriptome in Three Tissues from a Full-Sib Pair with Extreme Phenotypes in Growth and Fat Deposition by Paired-End RNA Sequencing. BMC Genomics 12, 448. doi:10.1186/1471-2164-12-448
Chen, Y., Gondro, C., Quinn, K., Herd, R. M., Parnell, P. F., and Vanselow, B. (2011b). Global Gene Expression Profiling Reveals Genes Expressed Differentially in Cattle with High and Low Residual Feed Intake. Anim. Genet. 42 (5), 475–490. doi:10.1111/j.1365-2052.2011.02182.x
Cheng, S., Wang, X., Wang, Q., Yang, L., Shi, J., and Zhang, Q. (2020). Comparative Analysis of Longissimus Dorsi Tissue from Two Sheep Groups Identifies Differentially Expressed Genes Related to Growth, Development and Meat Quality. Genomics 112 (5), 3322–3330. doi:10.1016/j.ygeno.2020.06.011
Chmurzyńska, A. (2006). The Multigene Family of Fatty Acid-Binding Proteins (FABPs): Function, Structure and Polymorphism. J. Appl. Genet. 47 (1), 39–48. doi:10.1007/bf03194597
Di, R., Chu, M. X., Li, Y. L., Zhang, L., Fang, L., Feng, T., et al. (2012). Predictive Potential of Microsatellite Markers on Heterosis of Fecundity in Crossbred Sheep. Mol. Biol. Rep. 39 (3), 2761–2766. doi:10.1007/s11033-011-1032-7
Favre, H., Benhamou, A., Finidori, J., Kelly, P. A., and Edery, M. (1999). Dual Effects of Suppressor of Cytokine Signaling (SOCS-2) on Growth Hormone Signal Transduction. FEBS Lett. 453 (1-2), 63–66. doi:10.1016/s0014-5793(99)00681-x
Ghahramani, N., Shodja, J., Rafat, S. A., Panahi, B., and Hasanpur, K. (2021). Integrative Systems Biology Analysis Elucidates Mastitis Disease Underlying Functional Modules in Dairy Cattle. Front. Genet. 12, 712306. doi:10.3389/fgene.2021.712306
Greenhalgh, C. J., Rico-Bautista, E., Lorentzon, M., Thaus, A. L., Morgan, P. O., Willson, T. A., et al. (2005). SOCS2 Negatively Regulates Growth Hormone Action In Vitro and In Vivo. J. Clin. Invest. 115 (2), 397–406. doi:10.1172/jci200522710
Hegarty, R. S., Warner, R. D., and Pethick, D. W. (2006). Genetic and Nutritional Regulation of Lamb Growth and Muscle Characteristics. Aust. J. Agric. Res. 57 (6), 721–730. doi:10.1071/ar06105
Horvat, S., and Medrano, J. F. (2001). Lack of Socs2 Expression Causes the High-Growth Phenotype in Mice. Genomics 72 (2), 209–212. doi:10.1006/geno.2000.6441
Jing, L., Hou, Y., Wu, H., Miao, Y., Li, X., Cao, J., et al. (2015). Transcriptome Analysis of mRNA and miRNA in Skeletal Muscle Indicates an Important Network for Differential Residual Feed Intake in Pigs. Sci. Rep. 5 (1), 1–14. doi:10.1038/srep11953
Jurie, C., Cassar-Malek, I., Bonnet, M., Leroux, C., Bauchart, D., Boulesteix, P., et al. (2007). Adipocyte Fatty Acid-Binding Protein and Mitochondrial Enzyme Activities in Muscles as Relevant Indicators of Marbling in Cattle. J. Anim. Sci. 85 (10), 2660–2669. doi:10.2527/jas.2006-837
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Lai, X., Lan, X., Chen, H., Wang, X., Wang, K., Wang, M., et al. (2009). A Novel SNP of the Hesx1 Gene in Bovine and its Associations with Average Daily Gain. Mol. Biol. Rep. 36 (7), 1677–1681. doi:10.1007/s11033-008-9368-3
Lehrke, M., and Lazar, M. A. (2005). The Many Faces of PPARγ. Cell 123 (6), 993–999. doi:10.1016/j.cell.2005.11.026
Li, J., Chen, Y., Wang, Y.-G., Zhao, X.-L., Gilbert, E., Liu, Y.-P., et al. (2013). MUSTN1 mRNA Abundance and Protein Localization Is Greatest in Muscle Tissues of Chinese Meat-Quality Chickens. Int. J. Mol. Sci. 14 (3), 5545–5559. doi:10.3390/ijms14035545
Li, X., Yang, J., Shen, M., Xie, X.-L., Liu, G.-J., Xu, Y.-X., et al. (2020). Whole-genome Resequencing of Wild and Domestic Sheep Identifies Genes Associated with Morphological and Agronomic Traits. Nat. Commun. 11 (1), 2815. doi:10.1038/s41467-020-16485-1
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 30 (7), 923–930. doi:10.1093/bioinformatics/btt656
Long, J.-R., Liu, P., Liu, Y., Lu, Y., Xiong, D., Elze, L., et al. (2003). APOE and TGF- 1 Genes Are Associated with Obesity Phenotypes. J. Med. Genet. 40 (12), 918–924. doi:10.1136/jmg.40.12.918
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Lv, F.-H., Peng, W.-F., Yang, J., Zhao, Y.-X., Li, W.-R., Liu, M.-J., et al. (2015). Mitogenomic Meta-Analysis Identifies Two Phases of Migration in the History of Eastern Eurasian Sheep. Mol. Biol. Evol. 32 (10), 2515–2533. doi:10.1093/molbev/msv139
Mesner, L. D., Ray, B., Hsu, Y.-H., Manichaikul, A., Lum, E., Bryda, E. C., et al. (2014). Bicc1 Is a Genetic Determinant of Osteoblastogenesis and Bone mineral Density. J. Clin. Invest. 124 (6), 2736–2749. doi:10.1172/jci73072
Metcalf, D., Greenhalgh, C. J., Viney, E., Willson, T. A., Starr, R., Nicola, N. A., et al. (2000). Gigantism in Mice Lacking Suppressor of Cytokine Signalling-2. Nature 405 (6790), 1069–1073. doi:10.1038/35016611
Milne, C. (2000). The History of the Dorper Sheep. Small Ruminant Res. 36 (2), 99–102. doi:10.1016/s0921-4488(99)00154-6
Muniz, M. M. M., Fonseca, L. F. S., Magalhães, A. F. B., dos Santos Silva, D. B., Canovas, A., Lam, S., et al. (2020). Use of Gene Expression Profile to Identify Potentially Relevant Transcripts to Myofibrillar Fragmentation index Trait. Funct. Integr. Genomics 20 (4), 609–619. doi:10.1007/s10142-020-00738-9
Nasser, G., Dessie, S., Michael, H., Karl, S., and Dawit, T. (2020). Transcriptome Profile and Association Study Revealed STAT3 Gene as a Potential Quality Marker of Bovine Gametes. Zygote 28 (2), 116–130. doi:10.1017/S0967199419000765
Oh, D., La, B., Lee, Y., Byun, Y., Lee, J., Yeo, G., et al. (2013). Identification of Novel Single Nucleotide Polymorphisms (SNPs) of the Lipoprotein Lipase (LPL) Gene Associated with Fatty Acid Composition in Korean Cattle. Mol. Biol. Rep. 40 (4), 3155–3163. doi:10.1007/s11033-012-2389-y
Park, H.-E., Shin, M.-K., Park, H.-T., Jung, M., Cho, Y. I., and Yoo, H. S. (2016). Gene Expression Profiles of Putative Biomarker Candidates in Mycobacterium avium subsp. Paratuberculosis-Infected Cattle. Pathog. Dis. 74 (4), ftw022. doi:10.1093/femspd/ftw022
Pezet, A., Favre, H., Kelly, P. A., and Edery, M. (1999). Inhibition and Restoration of Prolactin Signal Transduction by Suppressors of Cytokine Signaling. J. Biol. Chem. 274 (35), 24497–24502. doi:10.1074/jbc.274.35.24497
Pierzchala, M., Hoekman, A. J. W., Urbanski, P., Kruijt, L., Kristensen, L., Young, J. F., et al. (2014). Validation of Biomarkers for Loin Meat Quality (M. Longissimus ) of Pigs. J. Anim. Breed. Genet. 131 (4), 258–270. doi:10.1111/jbg.12081
Piessevaux, J., Lavens, D., Montoye, T., Wauman, J., Catteeuw, D., Vandekerckhove, J., et al. (2006). Functional Cross-Modulation between SOCS Proteins Can Stimulate Cytokine Signaling. J. Biol. Chem. 281 (44), 32953–32966. doi:10.1074/jbc.M600776200
PilNam, S., MoonSuck, K., KiBaek, K., SangHyun, H., KwangYun, S., SungSoo, L., et al. (2008). Genetic Polymorphisms of MYL2 and ADCYAP1R1 Genes and Their Association with Carcass Traits in Finished Pigs. J. Anim. Sci. Techn. 50 (3), 301–308. doi:10.5187/jast.2008.50.3.301
Ramos, A. M., Pita, R. H., Malek, M., Lopes, P. S., Guimarães, S. E. F., and Rothschild, M. F. (2009). Analysis of the Mouse High-Growth Region in Pigs. J. Anim. Breed. Genet. 126 (5), 404–412. doi:10.1111/j.1439-0388.2009.00801.x
Rupp, R., Senin, P., Sarry, J., Allain, C., Tasca, C., Ligat, L., et al. (2015). A Point Mutation in Suppressor of Cytokine Signalling 2 (Socs2) Increases the Susceptibility to Inflammation of the Mammary Gland while Associated with Higher Body Weight and Size and Higher Milk Production in a Sheep Model. Plos Genet. 11 (12), e1005629. doi:10.1371/journal.pgen.1005629
Schoeman, S. J. (2000). A Comparative Assessment of Dorper Sheep in Different Production Environments and Systems. Small Ruminant Res. 36 (2), 137–146. doi:10.1016/s0921-4488(99)00157-1
Seaborne, R. A., Strauss, J., Cocks, M., Shepherd, S., O’Brien, T. D., van Someren, K. A., et al. (2018). Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 8 (1), 1898. doi:10.1038/s41598-018-20287-3
Seyedsharifi, R., and Hamze Zadeh Azar, A. (2016). Evaluation of Slaughtered Lambs Results from Varamini Ewes Crossing with Shal, Afshar, Moghani and Varamini Rams. Iranian J. Anim. Sci. Res. 8 (1), 174–184.
Shi, J., Wang, X., Song, Y., Liu, T., Cheng, S., and Zhang, Q. (2021). Excavation of Genes Related to the Mining of Growth, Development, and Meat Quality of Two Crossbred Sheep Populations Based on Comparative Transcriptomes. Animals 11 (6), 1492. doi:10.3390/ani11061492
Sun, L., Bai, M., Xiang, L., Zhang, G., Ma, W., and Jiang, H. (2016). Comparative Transcriptome Profiling of Longissimus Muscle Tissues from Qianhua Mutton Merino and Small Tail Han Sheep. Sci. Rep. 6, 33586. doi:10.1038/srep33586
Tannahill, G. M., Elliott, J., Barry, A. C., Hibbert, L., Cacalano, N. A., and Johnston, J. A. (2005). SOCS2 Can Enhance Interleukin-2 (IL-2) and IL-3 Signaling by Accelerating SOCS3 Degradation. Mol. Cel Biol. 25 (20), 9115–9126. doi:10.1128/mcb.25.20.9115-9126.2005
Tao, L., He, X. Y., Pan, L. X., Wang, J. W., Gan, S. Q., and Chu, M. X. (2020). Genome‐wide Association Study of Body Weight and Conformation Traits in Neonatal Sheep. Anim. Genet. 51 (2), 336–340. doi:10.1111/age.12904
Uemoto, Y., Suzuki, K., Kobayashi, E., Sato, S., Shibata, T., Kadowaki, H., et al. (2007). Effects of Heart Fatty Acid-Binding Protein Genotype on Intramuscular Fat Content in Duroc Pigs Selected for Meat Production and Meat Quality Traits. Asian Australas. J. Anim. Sci. 20 (5), 622–626. doi:10.5713/ajas.2007.622
Wang, J., Li, J., and Wei, J. (1990). Selection and Improvement on Small Tail Han Sheep. China Sheep Goat Farming 1, 1–3.
Weber, K. L., Welly, B. T., Van Eenennaam, A. L., Young, A. E., Porto-Neto, L. R., Reverter, A., et al. (2016). Identification of Gene Networks for Residual Feed Intake in Angus Cattle Using Genomic Prediction and RNA-Seq. Plos One 11 (3), e0152274. doi:10.1371/journal.pone.0152274
Weldeyesus, G. (2017). Improving Live Body Weight Gain of Local Sheep through Crossbreeding with High Yielding Exotic Dorper Sheep under Smallholder Farmers. Int. J. Livest. Prod. 8 (5), 67–71. doi:10.5897/ijlp2016.0316
Xu, J., Wang, C., Jin, E., Gu, Y., Li, S., and Li, Q. (2018). Identification of Differentially Expressed Genes in Longissimus Dorsi Muscle between Wei and Yorkshire Pigs Using RNA Sequencing. Genes Genom. 40 (4), 413–421. doi:10.1007/s13258-017-0643-3
Xu, X., Liu, T., Fan, S., Ma, W., Chen, W., and Zhang, X. (2020). Effects of Fermented Caragana Korshinskii on the Intramuscular Fat Content and Expression of FABP3, UBE3C, ADRB3, LIPE, and SCD in Different Muscles of Tan Sheep. Czech J. Anim. Sci. 65 (4), 145–152. doi:10.17221/231/2019-CJAS
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Zhang, C., Wang, G., Wang, J., Ji, Z., Dong, F., and Chao, T. (2014). Analysis of Differential Gene Expression and Novel Transcript Units of Ovine Muscle Transcriptomes. PLoS One 9 (2), e89817. doi:10.1371/journal.pone.0089817
Zhao, F., Deng, T., Shi, L., Wang, W., Zhang, Q., Du, L., et al. (2020). Genomic Scan for Selection Signature Reveals Fat Deposition in Chinese Indigenous Sheep with Extreme Tail Types. Animals 10 (5), 773. doi:10.3390/ani10050773
Keywords: RNA-seq, dorper, small-tailed Han sheep, growth, mutton
Citation: Peng H, Hu M, Liu Z, Lai W, Shi L, Zhao Z, Ma H, Li Y and Yan S (2022) Transcriptome Analysis of the Liver and Muscle Tissues of Dorper and Small-Tailed Han Sheep. Front. Genet. 13:868717. doi: 10.3389/fgene.2022.868717
Received: 03 February 2022; Accepted: 21 March 2022;
Published: 11 April 2022.
Edited by:
Aline Silva Mello Cesar, University of São Paulo, BrazilReviewed by:
Bárbara Silva-Vignato, University of São Paulo, BrazilAroa Suarez Vega, Universidad de León, Spain
Copyright © 2022 Peng, Hu, Liu, Lai, Shi, Zhao, Ma, Li and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumei Li, bGlfeW1Aamx1LmVkdS5jbg==; Shouqing Yan, eWFuc3FAamx1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Hongyang Peng
Hongyang Peng Mingyue Hu1†
Mingyue Hu1†