- 1Department of Biology, Faculty of Science and Technology, University of Nairobi, Nairobi, Kenya
- 2Sub-Saharan Africa International Centre for Excellence in Malaria Research, Homa Bay, Kenya
- 3Department of Zoology, School of Physical and Biological Sciences, Maseno University, Kisumu, Kenya
- 4Department of Biochemistry, Egerton University, Nakuru, Kenya
- 5West Africa Centre for Cell Biology of Infectious Pathogen, Department of Biochemistry, Cell and Molecular Biology, University of Ghana, Accra, Ghana
- 6School of Zoological Sciences, Kenyatta University, Nairobi, Kenya
- 7Centre for Global Health Research, Kenya Medical Research Institute, Kisumu, Kenya
- 8Program in Public Health, College of Health Sciences, University of California, Irvine, Irvine, CA, United States
- 9Centre for Global Health and Diseases, Case Western Reserve University, Cleveland, OH, United States
Background: Malaria elimination and eradication efforts can be advanced by including transmission-blocking or reducing vaccines (TBVs) alongside existing interventions. Key transmission-blocking vaccine candidates, such as Pfs230 domain one and Pfs48/45 domain 3, should be genetically stable to avoid developing ineffective vaccines due to antigenic polymorphisms. We evaluated genetic polymorphism and temporal stability of Pfs230 domain one and Pfs48/45 domain three in Plasmodium falciparum parasites from western Kenya.
Methods: Dry blood spots on filter paper were collected from febrile malaria patients reporting to community health facilities in endemic areas of Homa Bay and Kisumu Counties and an epidemic-prone area of Kisii County in 2018 and 2019. Plasmodium speciation was performed using eluted DNA and real-time PCR. Amplification of the target domains of the two Pfs genes was performed on P. falciparum positive samples. We sequenced Pfs230 domain one on 156 clinical isolates and Pfs48/45 domain three on 118 clinical isolates to infer the levels of genetic variability, signatures of selection, genetic diversity indices and perform other evolutionary analyses.
Results: Pfs230 domain one had low nucleotide diversity (π = 0.15 × 10–2) with slight variation per study site. Six polymorphic sites with nonsynonymous mutations and eight haplotypes were discovered. I539T was a novel variant, whereas G605S was nearing fixation. Pfs48/45 domain three had a low π (0.063 × 10–2), high conservation index, and three segregating sites, resulting in nonsynonymous mutation and four haplotypes. Some loci of Pfs230 D1 were in positive or negative linkage disequilibrium, had negative or positive selection signatures, and others (1813, 1955) and (1813, 1983) had a history of recombination. Mutated loci pairs in Pfs48/45 domain three had negative linkage disequilibrium, and some had negative and positive Tajima’s D values with no history of recombination events.
Conclusion: The two transmission blocking vaccine candidates have low nucleotide diversity, a small number of zone-specific variants, high nucleotide conservation index, and high frequency of rare alleles. With the near fixation a polymorphic site and the proximity of mutated codons to antibody binding epitopes, it will be necessary to continue monitoring sequence modifications of these domains when designing TBVs that include Pfs230 and Pfs48/45 antigens.
Introduction
Genetic polymorphism of Plasmodium falciparum antigens has hampered efforts to develop an effective vaccine that is protective against pre-erythrocytic and asexual blood-stage parasites (Genton et al., 2002; Takala et al., 2007; Ogutu et al., 2009; Bergmann-Leitner et al., 2012; Neafsey et al., 2015; Ouattara et al., 2015). Recent efforts, however, have been made to develop vaccines that reduce and block Plasmodium falciparum transmission at the community level. Two of the existing transmission-blocking vaccine (TBV) candidates, P. falciparum surface protein 230 (Pfs230) (Sabeti et al., 2007; Lee et al., 2019, 2020; Singh et al., 2019, 2020; Tachibana et al., 2019; Huang et al., 2020; Healy et al., 2021) and P. falciparum surface protein 48/45 (Pfs48/45) (Singh et al., 2019, 2021; Lee et al., 2020) have been shown to elicit antibody responses in mice and people that block P. falciparum gametocyte fertilization in the mid-gut of the Anopheles vector.
Pfs230 is a cysteine-rich 230 kDa protein expressed by both male and female gametocytes (Rener et al., 1983; MacDonald et al., 2016). The antigen is thought to play a role in gamete fusion in the mosquito blood meal after forming a complex with another cysteine-rich protein, Pfs48/45 (Eksi et al., 2006). In comparison to antibodies elicited by immunization with other Pfs230 domains, Domain 1 (D1) has been shown to elicit transmission-blocking monoclonal antibodies with strong inhibitory activity against oocyst development in standard membrane feeding assays (Lee et al., 2019; Singh et al., 2019, 2020; Tachibana et al., 2019; Huang et al., 2020; Healy et al., 2021). Like Pfs230 D1, fusion with its counterpart Pfs48/45 D3 has good potential as a component of a TBV. The latter fused doublet antigen consists of three domains linked by disulphide bonds and contains 16 cysteine residues (Kocken et al., 1993; Lennartz et al., 2018). Unlike Pfs230, Pfs48/45 is anchored on the gamete surface membrane by glycophophatidylinositol (Kocken et al., 1993; Dijk et al., 2001; Gilson et al., 2006; Lennartz et al., 2018) and is essential for male gamete fertility. Domain 3 has been shown to elicit antibodies in the host (Graves et al., 1988; Roeffen et al., 1994; Dijk et al., 2001; Bousema et al., 2010; Jones et al., 2015; Acquah et al., 2017; Singh et al., 2019; Baptista et al., 2022). Pfs48/45 D3 is located at the C-terminus of the protein and contains binding sites for non-inhibitory and inhibitory human and mouse mAbs that reduce P. falciparum infection in mosquitoes (Vermeulen et al., 1986; Graves et al., 1988; Outchkourov et al., 2007; Chowdhury et al., 2009; Singh et al., 2017, 2019; Kundu et al., 2018; Lennartz et al., 2018; Lee et al., 2020).
Antigenic polymorphism of Pfs230 D1 and Pf48/45 D3 should be assessed in malaria endemic areas on a regular basis to support the successful development of these TBV candidates due to the fact that, if the targeted regions are genetically unstable, polymorphisms may cause critical codon changes within immunogenic epitopes, thereby reducing TBV efficacy. Several dimorphic sites on Pfs230 D1 and Pfs48/45 D3 have previously been identified (Kocken et al., 1995; Drakeley et al., 1996; Escalante et al., 1998; Conway et al., 2001; Jones et al., 2015; MacDonald et al., 2016; Kundu et al., 2018; Singh et al., 2020; Coelho et al., 2021); however, there is limited knowledge of the extent of genetic diversity, signatures of selection, and other evolutionary forces that may be shaping alleles in P. falciparum from different malaria transmission zones. We therefore performed an in-depth genetic analysis of Pfs230 D1 and Pfs48/45 D3 in parasites isolated from patients with uncomplicated falciparum malaria from three different areas in western Kenya.
Materials and Methods
Study Site and Sampling
Dry blood spots (DBS) were collected on filter paper from febrile malaria patients at health clinics in Homa Bay County (Kochia), Kisumu County (Chulaimbo), and Kisii County (Eramba) in 2018 and 2019 (Figure 1). The study site in Homa Bay is characterized by perennial transmission. Vector control consists of universal distribution of long-lasting insecticidal bed nets with annual indoor residual spraying of insecticides. The study site in Kisumu County also has perennial transmission. Vector control consists of long-lasting insecticidal bed nets (LLINs) alone. The site in Kisii County is malaria epidemic-prone with low transmission and residents use LLINs (Kapesa et al., 2018). In brief, four drops of approximately 25 µL of blood from each patient were spotted on Whatman™ Blood Stain Cards (GE Healthcare WB100014) as previously described (Coombs and Fiscus, 2009). Each card was stored individually in silica gel-containing plastic bags before being transported to the joint International Centre of Excellence for Malaria Research (ICEMR) and Tom Mboya University College Laboratory in Homa Bay town for storage at -20°C. 150 of the 372 DBS collected came from Homa Bay; 120 and 102 came from Kisumu and Kisii, respectively.
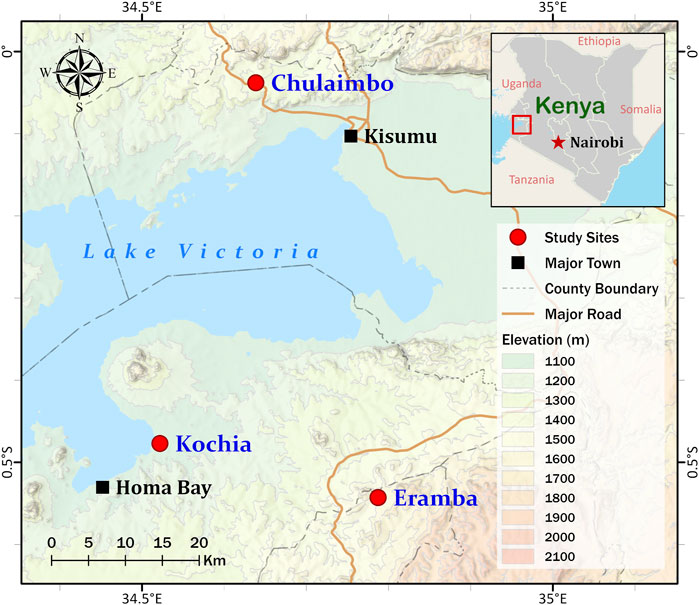
FIGURE 1. Map showing Homa Bay (Kochia), Kisumu (Chulaimbo) and Kisii (Eramba) study sites in western Kenya. The sampling points are represented by the red circles on the map. This figure was prepared with ESRI ArcGIS Pro 2.8 with field survey results and publicly available datasets. The map contains information from OpenStreetMap and OpenStreetMap 115 Foundation, which is made available under the Open Database License.
Amplification and Sequencing of Pfs230 Domain one and Pfs48/45 Domain three
Genomic DNA was extracted from filter paper using the modified Chelex resin (Chelex -100) method and stored at -20°C. As a positive control, DNA from the cultured laboratory strain NF54 was extracted and stored. Plasmodium species-specific real-time PCR targeting 18S ribosomal RNA gene was used to confirm P. falciparum positive DNA samples before amplification of specific target fragments of each gene (Ochwedo et al., 2021; Onyango et al., 2021). Primer sets were designed using Primer3 version 0.4.0 for Pfs230 D1 and Pfs48/45 D3 and in silico validation of each set was performed using the Sequence Manipulation Suite (Stothard, 2000). Among the 372 samples, 332 (89.3%) tested positive for P. falciparum DNA (n = 150, 120 and 62 from Homa Bay, Kisumu and Kisii, respectively) and were used to amplify Pfs230 D1 and Pfs48/45 D3 in a T100™ Thermal Cycler (Bio-Rad, Hercules, CA, United States ). Briefly, 3 µL of sample DNA was added to a mixture of 11.5 µL of DreamTaq Green PCR Master Mix (2X), 0.5 µL of Pfs230 D1 forward (5′-TGGTGAAGCTGTCGAAGATG-3′) and reverse primers (5′-GTGTACCACAGGGGGAAGAG-3′) targeting 514 base pairs and 7.5 µL of double-distilled water. The thermal profile was set as follows 95 °C for 3 min, 34 cycles (94 °C for 30 s, 58 °C for 30 s and 72 °C for 45 s) and final extension at 72 °C for 6 min. For Pfs48/45 D3, similar reaction volume was prepared using forward (5′-TTTTCAAGAAGGAAAAGAAAAAGC-3′) and reverse primers (5′-GCCAAAAATCCATAATATGCTGA-3′) targeting 600bp. The PCR conditions were set as follows 95°C for 3 min, 34 cycles (94°C for 30 s, 55 °C for 30 s and extension at 72°C for 45 s) final extension at 72 °C for 6 min. All the amplicons were assessed by gel electrophoresis in 1.5% w/v agarose gel before sequencing. For Pfs230 D1, 82, 39, and 35 samples from Homa Bay, Kisumu, and Kisii, respectively, were amplified. For Pfs48/45 D3, 36, 44, and 38 samples from Homa Bay, Kisumu, and Kisii, respectively, were amplified. All the PCR amplicons, together with positive controls, were purified using Exonuclease I and Shrimp Alkaline Phosphatase (ExoSAP-IT) and bi-directionally sequenced using 3730 BigDye® Terminator v3.1 Sequencing Standard kit on ABI PRISM® 3700 DNA Analyzer (Applied Biosystems, Foster City, CA, United States).
Data Analysis
All sequences were assembled using Geneious version 11.1.5 software, and multiple sequence alignment was performed using ClustalW. Polymorphic locus and codons were inferred after comparing each sequence to the respective sequence of positive control (NF54) as well as 3D7 (PF3D7_0209000 for Pfs230 and PF3D7_1346700 for Pfs48/45). DnaSP Version 6.12.03 (Rozas et al., 2017) and Arlequin version 3.5.2 (Excoffier and Lischer, 2010) were used to compute genetic diversity indices such as nucleotide diversity (π), haplotype diversity (Hd), number of haplotypes (h), number of segregating sites (S) and mean number of pairwise difference (k). Population Analysis with Reticulate Trees (Popart) version 1.7 software (Clement et al., 2000) was used to infer haplotype networks. Neutrality tests; Tajima’s D, Fu and Li’s D, Fu and Li’s F and Fu’s Fs statistics and test for the presence of Recombination events (Rm) and linkage disequilibrium (LD) were computed in DnaSP Version 6.12.03 and Arlequin version 3.5.2. Generated Tajima’s D values were plotted using GraphPad version 8.3.0. Both antigen structural delineation was done using Protein Homology/analogY Recognition Engine (PHYRE2) version 2.0 and generated models visualized and edited in UCSF Chimera version 1.15 (Pettersen et al., 2004).
Results
Analysis of Mutations Detected in Pfs230 D1 and Pfs48/45 D3
Six loci (1,616, 1813, 1955, 1964, 1967, and 1983) in Pfs230 D1 were found to be polymorphic, resulting in nonsynonymous mutations (Table 1). The mutations were skewed toward transversion, with a transversion to transition ratio (Tv: Ts) greater than 0.5. Two polymorphic sites were singletons (1964 and 1967), whereas four dimorphic sites (1,616, 1813, 1955, and 1983) were parsimony informative. These polymorphisms resulted in I539T, G605S, T652R, E655V, T656N, and K661N codon changes. Nonsynonymous alterations T652R and K661N were on separate beta (β) pleated sheets connected by a loop containing mutated codons E655V and T656N (Supplementary Figure S1). G605S was also on the loop connecting two different ß pleated sheets. In general, western Kenya parasites had a high allelic frequency of G605S (98.08%), followed by progressively lower frequencies of K661N, T652R, and I539T. E655R and T656N were each observed at a frequency of <1%. The prevalence of various alleles was almost similar across the various study sites. For example, as shown in Table 1, G605S was the most common codon change in the three study sites. Only two P. falciparum isolates from Kisumu and Homa Bay County lacked this mutation.
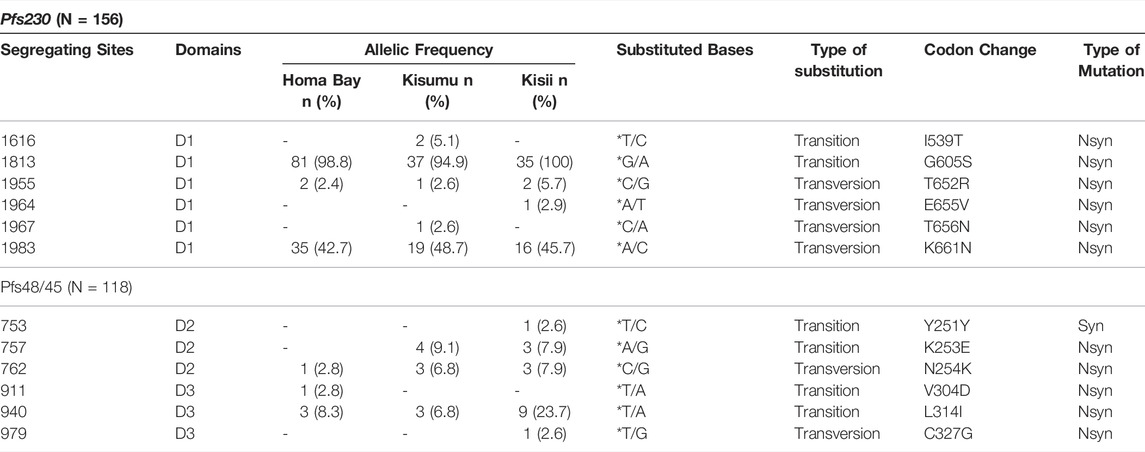
TABLE 1. Polymorphic sites on Pfs230 domain one and Pfs48/45 domain two and three from Homa Bay, Kisumu and Kisii region in western Kenya n: number of sequences harbouring mutations; *: reference (3D7and NF54) allele only; D: Domain; Nsyn: Non-synonymous mutation; Syn: Synonymous mutation; A: Adenine; C: Cytosine; T: Thymine; G: Guanine.
In contrast to the six nonsynonomous mutated sites observed in Pfs230 D1, Pfs48/45 D3 had three segregating sites (Table 1). Singleton sites were found at loci 911 and 979 in parasites isolated from patients residing in Homa Bay County and Kisii County, respectively. A low frequency polymorphism at locus 940 was observed across parasite populations in all three counties, and was parsimony-informative. These transition bias mutations at loci 911, 940, and 979 resulted in nonsynonymous mutations V304D, L314I, and C327G, respectively. The variants were in the Pfs48/45 D3 antigen loop connecting different ß pleated sheets (Supplementary Figure S1). Codon change C327G in D3 was found in only one sequence in parasites isolated from a patient in Kisii County (Table 1). The ability of the designed primer set to cover Pfs48/45 D3 also allowed for the discovery of a singleton site 753 (Y251Y) and parsimony-informative sites 757 and 762 (K253E and N254K) (Table 1; Supplementary Figure S1). Except for G605S, which was near dimorphic codons on Pfs48/45 domain 2, the superimposed structure revealed dimorphic codons of Pfs48/45 D3 antigen close to those of Pfs230 D1 (Supplementary Figure S2).
Genetic Diversity of Pfs230 and Pfs48/45 Genes in Western Kenya
Pfs230 D1 from the three sites had π of 0.15 × 10–2, k; 0.68 and haplotype diversity (Hd) of 0.57 (Table 2). The domain had a nucleotide conservation index of 98.7% with a total of eight haplotypes circulating in western Kenya (Figure 2). Kisumu had the highest π (0.18 × 10–2) followed by Kisii (0.15 × 10–2) and Homa Bay (0.12 × 10–2). The site also had the most haplotypes 6) and the highest Hd (0.63) when compared to Kisii and Homa Bay, which had four haplotypes each and Hd of 0.60 and 0.52, respectively (Figure 2). Haplotype 2 (Hap_2) with the mutated codon G605S was the most common in western Kenya and at each study site. This was followed by Hap_4 (mutated codons G605S and K661N), Hap_7 (G605S, T652R, and K661N), and Hap_8 (T652R and K661N) (Supplementary Table S1). The remaining haplotypes (Hap_1, Hap_3, Hap_6, and Hap_5) were observed at a lower frequency. Only one sequence (from Chulaimbo) in western Kenya lacked a mutated site and had 100% sequence identity to the laboratory strain PF3D7_0209000 or NF54 sequence used as a positive control (Figure 2).
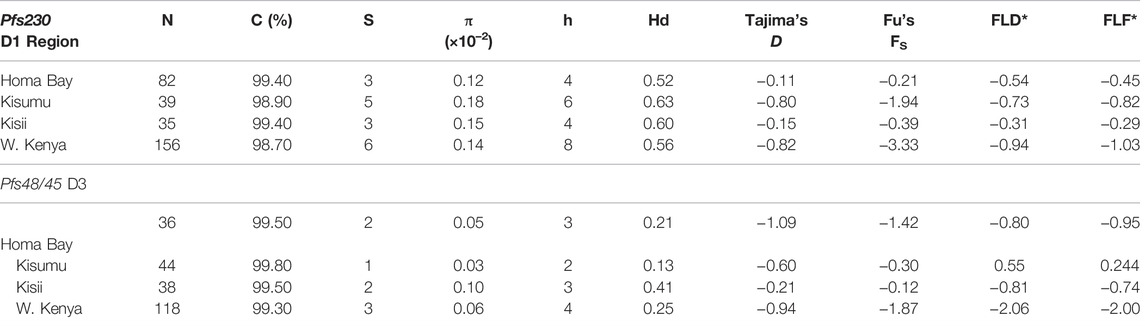
TABLE 2. Summary of genetic diversity indices for Pfs230 domain one and Pfs48/45 domain three from parasites in western Kenya N: Sample size; C: Conservation index; S: Segregating sites; π: nucleotide diversity; Vars: Variants; Hd: Haplotype diversity.
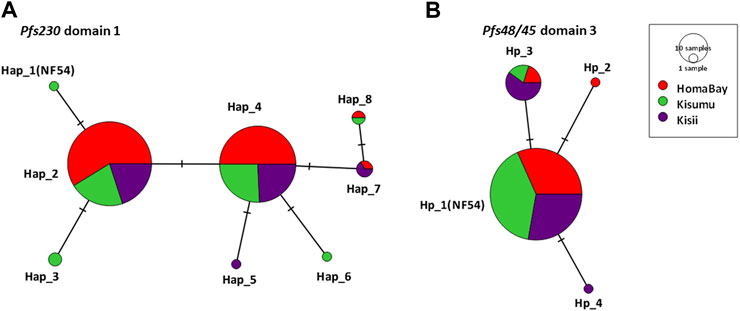
FIGURE 2. TCS-network analysis of the relationship of haplotypes based on the of Pfs230 domain one and Pfs48/45 domain three from parasites in malaria-endemic and epidemic-prone regions of western Kenya. (A). represents haplotypes based on observed sequence variation in Pfs230 D1 whereas (B) represents haplotypes in Pfs48/45 D3. The network shows the distribution of haplotype within malaria-endemic Homa Bay (red) and Kisumu (Chulaimbo) (green) as well as epidemic Kisii highland (purple). The hatch marks represent the number of mutations resulting in a specific haplotype whereas the size of the circle equates to the frequency of the observed haplotypes. Haplotypes labelled Hap_1(NF54) for Pfs230 D1 and Hp-1 (NF54) for Pfs48/45 D3 lacked mutated locus and have 100% sequence identity to laboratory strain NF54 or 3D7. Nucleotide sequences of all haplotypes were submitted to GenBank (accession: MW624857- MW625101).
The Pfs48/45 D3 from western Kenya had low π (0.06 × 10–2) and Hd (0.25) (Table 2). The domain had a conservation index of 99.3%, with four haplotypes circulating in the study area (Figure 2). Kisii parasites had the highest π (0.10 × 10–2) followed by Homa Bay (0.05 × 10–2) and Kisumu (0.03 × 10–2) (Table 2). Kisii and Homa Bay study sites each had three haplotypes in circulation, while Kisumu had four (Figure 2). The majority of haplotypes lacked a mutation (Hp_1) or had 100% sequence identity to the laboratory strain PF3D7_1346700 or the NF54 sequence used as a positive control (85.6%). This was followed by Hp_3 (mutated codon L314I), which had an overall frequency of 12.7%, while the rest (Hp_2 and Hp_4) had a frequency <1% (Supplementary Table S2).
Signatures of Selection, Linkage Disequilibrium and Recombination Events
Pfs230 D1 from P. falciparum isolates deviated from a standard neutral model. Tajima’s D (-0.8), FLD* (-0.9) and FLF* (-1.0) tests were all negative and non-significant (p > 0.05) (Table 2). However, Fu’s FS result (-3.3), was significant (p = 0.023). Tajima’s D test results were also non-significant (p > 0.05) in each site (Table 2). Despite the overall negative Tajima’s D results, there was a slight variation among individual mutated loci on Pfs230 D1. Locus 1983 (codon change K661N) had a significant (p < 0.05) positive (1.9) Tajima’s D result, whereas the rest had negative results (Figure 3). Apart from deviating from a standard neutral model, some loci pairs (1813, 1955) had positive LD (D′) results with highly significant (p < 0.001) χ 2 values of 19.7 and 12.7 among Homa Bay and Kisumu sequences, respectively (Supplementary Table S1). Among Kisii sequences, loci pairs (1813 and 1983) had positive D′ results, despite the fact that the χ 2 values was non-significant and the r2 value was low (0.03). Other loci with positive D′ results, low r2 values and non-significant (p > 0.05) χ2 values in Homa Bay and Kisumu parasites include loci pairs (1813,1983) (1955,1983) and (1967,1983). Some loci pairs on Kisumu and Kisii sequences had negative D′ results (Supplementary Table S1). On Pfs230 D1 from parasites in Homa Bay and Kisumu, recombination events were detected across loci pairs (1813, 1955) and (1813, 1983) (Supplementary Table S1).
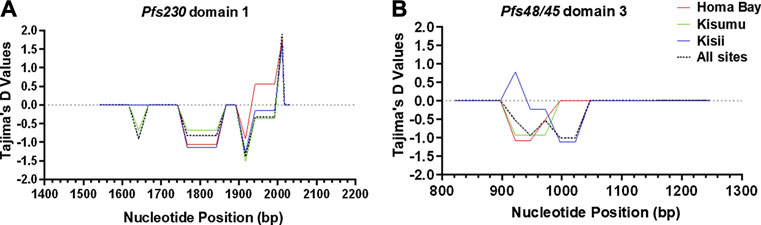
FIGURE 3. Sliding window plot of Tajima’s D values for Pfs230 domain one and Pfs48/45 domain three in western Kenya. The X-axis displays the nucleotide position (Window midpoint) whereas the Tajima’s D values are represented on the Y-axis. (A) is a representation of Pfs230 domain one whereas (B) represents of Pfs48/45 domain 3. The blue curve represents computed Tajima’s D plot for sequences of P. falciparum circulating in Kisii, the red colour is for Homa Bay, the green colour is for Kisumu (Chulaimbo) whereas the black dotted colour represents the population from the three study sites. The middle horizontal dotted line (intersecting the Y-axis at 0.0) represents a standard neutral model where the Tajima’s D value is equal to zero. Positive deviation from the grey dotted line signifies balancing selection whereas negative deviation represents purifying selection.
All of the Pfs48/45 D3 sequences had non-significant (p > 0.05) negative Tajima’s D -1.9, FLD*: 2.1 and FLF*: 2.0 results. The Fu’s FS (-1.9) result was, however, significant (p = 0.096). Locus 940 (L314I) among Kisii sequences had a significant (p > 0.05) positive Tajima’s D (0.8) results (Figure 3). There was no evidence of Rm or positive D′ results at any of the dimorphic loci within Pfs48/45 D3. However, some loci pairs from Homa Bay and Kisii had negative D′ results (Supplementary Table S2).
Discussion
The Pfs230 D1 and Pfs48/45 D3 antigens are important candidate antigens in the development of an effective TBV. Despite having a high frequency of rare alleles in western Kenya, both targets had low nucleotide diversity. Two variants, each on Pfs230 D1 and Pfs48/45 D3, were novel and private to western Kenya. The study validated five previously described polymorphic sites on Pfs230 D1 (Singh et al., 2020). In this study, G605S, one of the five mutated codons, was fixed in some study areas but not in others. Pfs230 D1 had the most mutations, while the Pfs48/45 D3 was the most conserved. Mutated loci from both domains were either under purifying or balancing selections. Other genetic forces revealed to have shaped alleles on the two genes included inbreeding and genetic drift with recombination being discovered only Pfs230 D1.
Pfs230 D1 from western Kenya had low nucleotide diversity, with significant Fu’s FS results indicating a high frequency of rare alleles. In addition to the previously reported 15 polymorphisms on Pfs230 D1 from parasites in Asian (Bangladesh, Cambodia, Laos, Myanmar, Thailand, and Vietnam) and African (Democratic Republic of the Congo, Ghana, Guinea, Malawi, Mali, Nigeria, Senegal, and Gambia) countries (MacDonald et al., 2016; Singh et al., 2020), this study discovered one additional mutation (I539T). This novel variant was identified only at the Kisumu study site along with five other polymorphisms (G605S, T652R, E655V, T656N, and K661N). These findings validate five previously described polymorphisms reported by (Singh et al., 2020). We speculate that the relatively higher nucleotide diversity index and number of haplotypes in Kisumu compared to other sites in western Kenya is related to the region’s slightly higher malaria transmission and absence of IRS activities (Oduma et al., 2021; Ochwedo et al., 2022).
Missense mutation G605S was found in parasites at a slightly higher allelic frequency (AF = 0.98) than in other geographical regions, as described by (Singh et al., 2020) (AF = 0.94) (MacDonald et al., 2016), (AF = 0.11), and (Coelho et al., 2021) (AF = 0.91). With only two clinical isolates in western Kenya lacking this mutation, G605S is almost completely fixed. This indicates the presence of selection pressure from either host antibodies, vector immune response or genetic drift (decreased variation and increasing homozygosity), may be stronger on P. falciparum populations from Kisii (low parasite population size) or Homa Bay (endemic site with declining parasite population size) compared to Kisumu (Chulaimbo) (Hancock and Di Rienzo, 2008; Honnay, 2013; Oduma et al., 2021). In contrast, the second most common polymorphism, K661N, was found in Kisumu at a higher frequency than in Kisii and Homa Bay. This reversal in the observed G605S and K661N frequencies could be attributed to factors such as recombination events (Rm), which are known to interfere with linked loci and could be effective on linked dimorphic loci pair 1813 and 1983 (responsible for G605S and K661N mutations respectively), thus increasing diversity in Kisumu (Chulaimbo) (Mejia, 2012) as opposed to Kisii and Homa Bay parasites, which also lack Rm between the two sites. Since immunogenic epitope binding light chain of transmission-blocking 4F12 monoclonal antibodies (TB 4F12 mAb) is close to the dimorphic codon G605, selection pressure from host antibodies on the epitope may be affecting the surrounding codons (MacDonald et al., 2016; Singh et al., 2020). This codon is located within a disulphide loop (from 593 to 611) that is thought to be stabilizing the epitope binding of TB 4F12 mAb (Singh et al., 2020). With near-complete fixation, the mutation may be beneficial to parasites but have a negative effect on the epitope binding affinity of TB 4F12 mAb. This needs to be looked into further by immunoassays of haplotypes with this polymorphism. Polymorphism I539T was found near codons 542–592 that contain 3G2 and 5G3 mAb binding epitopes which were previously shown to have no detectable oocyst reduction activity (Singh et al., 2020). Other polymorphisms, T652R, K661N on different ß pleated sheets, and E655V, T656N on disulphide loops linking the two loops, were distally located from the epitope that binds TB 4F12 mAb (Singh et al., 2020). When the two fusion proteins were superimposed, these four polymorphic codons were closer to mutated codon V304D, L314I, and C327G on Pfs48/45 D3, supporting the hypothesis that antibodies could be sterically interfering with protein-protein interaction (Singh et al., 2020). Plasmodium falciparum may induce these mutations in response to antibody-induced pressure in order to circumvent the blockade of fusion between Pfs230 D1 and Pfs48/45 D3, resulting in an uninterrupted gametocyte fertilization process.
The novel missense polymorphism C327G on Pfs48/45 D3 has the potential to be very important because it can interfere with one of the six cysteine residue pairings (pairing between codon C298 and C327) on the 85RF45.1 mAb epitope (Kundu et al., 2018). Other polymorphisms (Y251Y, K253E, N254K in Pfs48/45 D2 and V304D, L314I in Pfs48/45 D3) have been observed in P. falciparum populations in other malaria endemic regions (Conway et al., 2001; Jones et al., 2015; Kundu et al., 2018). However, none of these polymorphisms had been previously reported by a study conducted in the Asembo Bay area of western Kenya (Escalante et al., 1998). Though not the focus of this study, polymorphisms on codon 254 is thought to influence the type of host antibody that binds at the epitope bearing this mutation on Pfs48/45 antigen (Kocken et al., 1995). The three polymorphic codons Y251Y, K253E, and N254K on Pfs48/45 D2, are close to the disulphide loop that stabilizes the epitope binding TB 4F12 mAb on Pfs230 D1, thus suggesting steric interference from the antibodies. Pfs48/45 domain three is highly conserved, with low nucleotide and haplotype diversity when compared to Pfs230 D1. The key polymorphism based on this domain was L314I, which has a higher allelic frequency in Kisii highlands than in Homa Bay and Kisumu. Despite the presence of a high frequency of rare alleles, the majority of parasites lacked polymorphic loci on Pfs48/45 D3.
Inbreeding, recombination, and natural selection were identified as major drivers of the observed mutations in Pfs230 D1 and Pfs48/45 D3. The presence of linkage disequilibrium confirmed the history of selection pressure and inbreeding across various loci in Pfs230 D1 and Pfs48/45 D3 (Larrañaga et al., 2013). Some polymorphisms were considered intermediary because they had negative linkage disequilibrium (D′) values (Silvela et al., 1999). The negative D′ values also confirmed a history of random drift, which is decreasing the number of variants while increasing homozygosity that may play a role in the parasite’s loss of favourable mutations if it persists (Barton, 2010).
The presence of natural selection was confirmed by the Tajima’s D values. Overall negative Tajima’s D results revealed that purifying selection was affecting the majority of loci within Pfs230 D1 and Pfs48/45 D3, reducing genetic diversity (Cvijović et al., 2018). The aforementioned selection was, however, weak because the computed negative Tajima’s D values in both antigens were not significant. Individual Tajima’s D results for each codon revealed all other dimorphic codons to be under purifying selection, with the exception of K661N on Pfs230 D1 from all study sites and V304D on Pfs48/45 D3 from Kisii, which are under strong and weak balancing selection, respectively. The two mutated loci under balancing selection may play an important role within the Pfs230 D1 and Pfs48/45 D3 fusion proteins, which may explain why they are maintained in the P. falciparum population from western Kenya (Escalante et al., 1998). These findings support previous postulation (Jones et al., 2015) that selection pressure is acting on immunogenic domains of Pfs48/45.
The presence of weak purifying selection acting on dimorphic sites may impact not only host mAb binding and functional activity but also be affected by selective pressure in the mosquito vector (Lombardo and Christophides, 2016). This pressure could be exerted on individual antigens before or after complex formation. Findings in the present study support future investigations that examine functional antibody responses such as the ability of PfS230 and Pfs48/45 antibodies that activate human plasma complement and reduce mosquito infectivity in membrane feeding assays.
Conclusion
The Pfs230 D1 and Pfs48/45 D3 in P. falciparum from western Kenya have low nucleotide diversity and a high conservation index with high frequency of rare alleles. Among the observed polymorphisms in Pfs230 D1, G605S is nearly fixed in the population. Natural selection, inbreeding, and, to some extent, recombination are important driving forces in shaping these alleles in the two antigens. With the discovery of novel polymorphic sites, the two domains of the Pfs230 and Pfs48/45 from different malaria-prone regions, including areas where clinical trials have been conducted, should be monitored indefinitely. This will help track the genetic stability of the two TBV candidates.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Maseno University Ethics Review Committee (MUERC protocol No. 00456) and the University of California, Irvine Institutional Review Board (HS#2017–3512), as well as the Ministry of Health. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
KO conceptualised and designed the study, collected data, carried out the experiments, curated and analyzed the data, and drafted, edited, and revised the final manuscript. FA carried out the experiments and revised the final manuscript. WO collected data and revised the final manuscript. EM carried out the experiment and revised the final manuscript. ID revised the final manuscript. SO collected data. PO collected data, HA administration, SO administration and revised the final manuscript, AO revised the final manuscript. WM revised the final manuscript. AG revised the final manuscript. M-CL drew the map. GY developed the study’s concept and revised the final manuscript. DZ developed the concept, carried out the experiment, curated the data, and revised the final manuscript. JK developed the study’s concept and revised the final manuscript.
Funding
This research is supported by grants from the National Institutes of Health (U19 AI129326 and D43 TW001505).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.867906/full#supplementary-material
References
Acquah, F. K., Obboh, E. K., Asare, K., Boampong, J. N., Nuvor, S. V., Singh, S. K., et al. (20172017). Antibody Responses to Two New Lactococcus lactis-Produced Recombinant Pfs48/45 and Pfs230 Proteins Increase with Age in Malaria Patients Living in the Central Region of Ghana. Malar. J. 16 (1 16), 1–11. doi:10.1186/S12936-017-1955-0
Baptista, B. O., de Souza, A. B. L., Riccio, E. K. P., Bianco-Junior, C., Totino, P. R. R., Martins da Silva, J. H., et al. (2022). Naturally Acquired Antibody Response to a Plasmodium falciparum Chimeric Vaccine Candidate GMZ2.6c and its Components (MSP-3, GLURP, and Pfs48/45) in Individuals Living in Brazilian Malaria-Endemic Areas. Malar. J. 21, 6. doi:10.1186/s12936-021-04020-6
Barton, N. H. (2010). Mutation and the Evolution of Recombination. Phil. Trans. R. Soc. B 365, 1281–1294. doi:10.1098/RSTB.2009.0320
Bergmann-Leitner, E. S., Duncan, E. H., Mease, R. M., and Angov, E. (2012). Impact of Pre-existing MSP142-Allele Specific Immunity on Potency of an Erythrocytic Plasmodium falciparum Vaccine. Malar. J. 11, 315. doi:10.1186/1475-2875-11-315
Bousema, T., Roeffen, W., Meijerink, H., Mwerinde, H., Mwakalinga, S., van Gemert, G.-J., et al. (2010). The Dynamics of Naturally Acquired Immune Responses to Plasmodium falciparum Sexual Stage Antigens Pfs230 & Pfs48/45 in a Low Endemic Area in Tanzania. PLoS ONE 5, e14114. doi:10.1371/journal.pone.0014114
Chowdhury, D. R., Angov, E., Kariuki, T., and Kumar, N. (2009). A Potent Malaria Transmission Blocking Vaccine Based on Codon Harmonized Full Length Pfs48/45 Expressed in Escherichia coli. PLOS ONE 4, e6352. doi:10.1371/JOURNAL.PONE.0006352
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a Computer Program to Estimate Gene Genealogies. Mol. Ecol. 9, 1657–1659. doi:10.1046/J.1365-294X.2000.01020.X
Coelho, C. H., Tang, W. K., Burkhardt, M., Galson, J. D., Muratova, O., Salinas, N. D., et al. (20212021). A Human Monoclonal Antibody Blocks Malaria Transmission and Defines a Highly Conserved Neutralizing Epitope on Gametes. Nat. Commun. 12 (1 12), 1–12. doi:10.1038/s41467-021-21955-1
Conway, D., Machado, R. L. D., Singh, B., Dessert, P., Mikes, Z. S., Povoa, M. M., et al. (2001). Extreme Geographical Fixation of Variation in the Plasmodium falciparum Gamete Surface Protein Gene Pfs48/45 Compared with Microsatellite Loci. 115, 145–156.doi:10.1016/s0166-6851(01)00278-x
Coombs, R. W., and Fiscus, S. (2009). Processing of Dried Blood Spots Standard Operating ProcedureProcedure Title: Processing of Dried Blood Spots Standard Operating Procedure Approved by (Network): Network Name, Title Signature Date ACTG Reviewed by (Laboratory): Name, Title Signature Date Revision History Version Effective Date (Dd/mmm/yy) Comments.
Cvijović, I., Good, B. H., and Desai, M. M. (2018). The Effect of Strong Purifying Selection on Genetic Diversity. Genetics 209, 1235–1278. doi:10.1534/GENETICS.118.301058
Drakeley, C. J., Duraisingh, M. T., Póvoa, M., Conway, D. J., Targett, G. A. T., and Baker, D. A. (1996). Geographical Distribution of a Variant Epitope of Pfs4845, a Plasmodium falciparum Transmission-Blocking Vaccine Candidate. Mol. Biochem. Parasitol. 81, 253–257. doi:10.1016/0166-6851(96)02718-1
Eksi, S., Czesny, B., van Gemert, G.-J., Sauerwein, R. W., Eling, W., and Williamson, K. C. (2006). Malaria Transmission-Blocking Antigen, Pfs230, Mediates Human Red Blood Cell Binding to Exflagellating Male Parasites and Oocyst Production. Mol. Microbiol. 61, 991–998. doi:10.1111/J.1365-2958.2006.05284.X
Escalante, A. A., Lal, A. A., and Ayala, F. J. (1998). Genetic Polymorphism and Natural Selection in the Malaria Parasite Plasmodium falciparum. Genetics 149, 189–202. doi:10.1093/genetics/149.1.189
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi:10.1111/j.1755-0998.2010.02847.x
Genton, B., Betuela, I., Felger, I., Al‐Yaman, F., Anders, R. F., Saul, A., et al. (2002). A Recombinant Blood‐Stage Malaria Vaccine Reduces Plasmodium falciparum Density and Exerts Selective Pressure on Parasite Populations in a Phase 1-2b Trial in Papua New Guinea. J. Infect. Dis. 185, 820–827. doi:10.1086/339342
Gilson, P. R., Nebl, T., Vukcevic, D., Moritz, R. L., Sargeant, T., Speed, T. P., et al. (2006). Identification and Stoichiometry of Glycosylphosphatidylinositol-Anchored Membrane Proteins of the Human Malaria Parasite Plasmodium falciparum. Mol. Cell. Proteomics 5, 1286–1299. doi:10.1074/mcp.M600035-MCP200
Graves, P. M., Wirtz, R. A., Carter, R., Burkot, T. R., Looker, M., and Targett, G. A. (1988). Naturally Occurring Antibodies to an Epitope on Plasmodium falciparum Gametes Detected by Monoclonal Antibody-Based Competitive Enzyme-Linked Immunosorbent Assay. Infect. Immun. 56, 2818–2821. doi:10.1128/iai.56.11.2818-2821.1988
Hancock, A. M., and Di Rienzo, A. (2008). Detecting the Genetic Signature of Natural Selection in Human Populations: Models. Annu. Rev. Anthropol. 37, 197–217. doi:10.1146/annurev.anthro.37.081407.085141
Healy, S. A., Anderson, C., Swihart, B. J., Mwakingwe, A., Gabriel, E. E., Decederfelt, H., et al. (2021). Pfs230 Yields Higher Malaria Transmission-Blocking Vaccine Activity Than Pfs25 in Humans but Not Mice. J. Clin. Investigation 131. doi:10.1172/JCI146221
Honnay, O. (2013). Genetic Drift. Brenner’s Encyclopedia of Genetics. Second Edition, 251–253. doi:10.1016/B978-0-12-374984-0.00616-1
Huang, W. C., Deng, B., Mabrouk, M. T., Seffouh, A., Ortega, J., Long, C., et al. (2020). Particle-based, Pfs230 and Pfs25 Immunization Is Effective, but Not Improved by Duplexing at Fixed Total Antigen Dose. Malar. J. 19, 309–312. doi:10.1186/S12936-020-03368-5/FIGURES/6
Jones, S., Grignard, L., Nebie, I., Chilongola, J., Dodoo, D., Sauerwein, R., et al. (2015). Naturally Acquired Antibody Responses to Recombinant Pfs230 and Pfs48/45 Transmission Blocking Vaccine Candidates. J. Infect. 71, 117–127. doi:10.1016/j.jinf.2015.03.007
Kapesa, A., Kweka, E. J., Atieli, H., Afrane, Y. A., Kamugisha, E., Lee, M.-C., et al. (2018). The Current Malaria Morbidity and Mortality in Different Transmission Settings in Western Kenya. PLOS ONE 13, e0202031. doi:10.1371/journal.pone.0202031
Kocken, C. H. M., Jansen, J., Kaan, A. M., Beckers, P. J. A., Ponnudurai, T., Kaslow, D. C., et al. (1993). Cloning and Expression of the Gene Coding for the Transmission Blocking Target Antigen Pfs48/45 of Plasmodium falciparum. Mol. Biochem. Parasitol. 61, 59–68. doi:10.1016/0166-6851(93)90158-T
Kocken, C. H. M., Milek, R. L. B., Lensen, T. H. W., Kaslow, D. C., Schoenmakers, J. G. G., and Konings, R. N. H. (1995). Minimal Variation in the Transmission-Blocking Vaccine Candidate Pfs4845 of the Human Malaria Parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 69, 115–118. doi:10.1016/0166-6851(94)00193-Q
Kundu, P., Semesi, A., Jore, M. M., Morin, M. J., Price, V. L., Liang, A., et al. (2018). Structural Delineation of Potent Transmission-Blocking Epitope I on Malaria Antigen Pfs48/45. Nat. Commun. 9. doi:10.1038/s41467-018-06742-9
Larrañaga, N., Mejía, R. E., Hormaza, J. I., Montoya, A., Soto, A., and Fontecha, G. A. (20132013). Genetic Structure of Plasmodium falciparum Populations across the Honduras-Nicaragua Border. Malar. J. 12 (1 12), 1–10. doi:10.1186/1475-2875-12-354
Lee, S.-M., Hickey, J. M., Miura, K., Joshi, S. B., Volkin, D. B., King, C. R., et al. (2020). A C-Terminal Pfs48/45 Malaria Transmission-Blocking Vaccine Candidate Produced in the Baculovirus Expression System. Sci. Rep. 10. doi:10.1038/s41598-019-57384-w
Lee, S.-M., Wu, Y., Hickey, J. M., Miura, K., Whitaker, N., Joshi, S. B., et al. (2019). The Pfs230 N-Terminal Fragment, Pfs230D1+: Expression and Characterization of a Potential Malaria Transmission-Blocking Vaccine Candidate. Malar. J. 18, 356. doi:10.1186/s12936-019-2989-2
Lennartz, F., Brod, F., Dabbs, R., Miura, K., Mekhaiel, D., Marini, A., et al. (2018). Structural Basis for Recognition of the Malaria Vaccine Candidate Pfs48/45 by a Transmission Blocking Antibody. Nat. Commun. 9. doi:10.1038/s41467-018-06340-9
Lombardo, F., and Christophides, G. K. (2016). Novel Factors of Anopheles gambiae Haemocyte Immune Response to Plasmodium berghei Infection. Parasites Vectors 9. doi:10.1186/S13071-016-1359-Y
MacDonald, N. J., Nguyen, V., Shimp, R., Reiter, K., Herrera, R., Burkhardt, M., et al. (2016). Structural and Immunological Characterization of Recombinant 6-cysteine Domains of the Plasmodium falciparum Sexual Stage Protein Pfs230. J. Biol. Chem. 291, 19913–19922. doi:10.1074/jbc.M116.732305
Mejia, R. (2012). Natural Selection, Not Mutation: Recombination in Drosophila Increases Diversity. PLoS Biol. 10, e1001423. doi:10.1371/JOURNAL.PBIO.1001423
Neafsey, D. E., Juraska, M., Bedford, T., Benkeser, D., Valim, C., Griggs, A., et al. (2015). Genetic Diversity and Protective Efficacy of the RTS,S/AS01 Malaria Vaccine. N. Engl. J. Med. 373, 2025–2037. doi:10.1056/NEJMOA1505819
Ochwedo, K. O., Omondi, C. J., Magomere, E. O., Olumeh, J. O., Debrah, I., Onyango, S. A., et al. (20212021). Hyper-prevalence of Submicroscopic Plasmodium falciparum Infections in a Rural Area of Western Kenya with Declining Malaria Cases. Malar. J. 20 (1 20), 1–8. doi:10.1186/S12936-021-04012-6
Ochwedo, K. O., Onyango, S. A., Omondi, C. J., Orondo, P. W., Ondeto, B. M., Lee, M.-C., et al. (2022). Signatures of Selection and Drivers for Novel Mutation on Transmission-Blocking Vaccine Candidate Pfs25 Gene in Western Kenya. PLoS One 17, e0266394. doi:10.1371/journal.pone.0266394
Oduma, C. O., Ogolla, S., Atieli, H., Ondigo, B. N., Lee, M.-C., Githeko, A. K., et al. (20212021). Increased Investment in Gametocytes in Asymptomatic Plasmodium falciparum Infections in the Wet Season. BMC Infect. Dis. 21 (1 21), 1–10. doi:10.1186/S12879-020-05761-6
Ogutu, B. R., Apollo, O. J., McKinney, D., Okoth, W., Siangla, J., Dubovsky, F., et al. (2009). Blood Stage Malaria Vaccine Eliciting High Antigen-specific Antibody Concentrations Confers No Protection to Young Children in Western Kenya. PLoS ONE 4, e4708. doi:10.1371/journal.pone.0004708
Onyango, S. A., Ochwedo, K. O., Machani, M. G., Omondi, C. J., Debrah, I., Ogolla, S. O., et al. (2021). Genetic Diversity and Population Structure of the Human Malaria Parasite Plasmodium falciparum Surface Protein Pfs47 in Isolates from the Lowlands in Western Kenya. PloS one 16, e0260434. doi:10.1371/JOURNAL.PONE.0260434
Ouattara, A., Barry, A. E., Dutta, S., Remarque, E. J., Beeson, J. G., and Plowe, C. V. (2015). Designing Malaria Vaccines to Circumvent Antigen Variability. Vaccine 33, 7506–7512. doi:10.1016/j.vaccine.2015.09.110
Outchkourov, N., Vermunt, A., Jansen, J., Kaan, A., Roeffen, W., Teelen, K., et al. (2007). Epitope Analysis of the Malaria Surface Antigen Pfs48/45 Identifies a Subdomain that Elicits Transmission Blocking Antibodies. J. Biol. Chem. 282, 17148–17156. doi:10.1074/jbc.M700948200
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera?A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/JCC.20084
Rener, J., Graves, P. M., Carter, R., Williams, J. L., and Burkot, T. R. (1983). Target Antigens of Transmission-Blocking Immunity on Gametes of Plasmodium falciparum. J. Exp. Med. 158, 976–981. doi:10.1084/JEM.158.3.976
Roeffen, W., Lensen, T., Mulder, B., Teelen, K., Sauerwein, R., Eling, W., et al. (1994). Transmission Blocking Immunity as Observed in a Feeder System and Serological Reactivity to Pfs 48/45 and Pfs230 in Field Sera. Mem. Inst. Oswaldo Cruz 89 (Suppl. 2), 13–15. doi:10.1590/S0074-02761994000600004
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 34, 3299–3302. doi:10.1093/molbev/msx248
Sabeti, P. C., Varilly, P., Varilly, P., Fry, B., Lohmueller, J., Hostetter, E., et al. (2007). Genome-wide Detection and Characterization of Positive Selection in Human Populations. Nature 449, 913–918. doi:10.1038/nature06250
Silvela, L., de la Peña, I., and Gomez-Ruano, R. (19991999). Selection under Negative Linkage Disequilibrium. Random Mating versus Inbreeding. Heredity 826 82, 598–604. doi:10.1046/j.1365-2540.1999.00520.x
Singh, K., Burkhardt, M., Nakuchima, S., Herrera, R., Muratova, O., Gittis, A. G., et al. (2020). Structure and Function of a Malaria Transmission Blocking Vaccine Targeting Pfs230 and Pfs230-Pfs48/45 Proteins. Commun. Biol. 3, 1–12. doi:10.1038/s42003-020-01123-9
Singh, S. K., Plieskatt, J., Chourasia, B. K., Fabra-García, A., Garcia-Senosiain, A., Singh, V., et al. (2021). A Reproducible and Scalable Process for Manufacturing a Pfs48/45 Based Plasmodium falciparum Transmission-Blocking Vaccine. Front. Immunol. 11, 3369. doi:10.3389/FIMMU.2020.606266
Singh, S. K., Thrane, S., Chourasia, B. K., Teelen, K., Graumans, W., Stoter, R., et al. (2019). Pfs230 and Pfs48/45 Fusion Proteins Elicit Strong Transmission-Blocking Antibody Responses against Plasmodium falciparum. Front. Immunol. 10, 1256. doi:10.3389/fimmu.2019.01256
Singh, S. K., Thrane, S., Janitzek, C. M., Nielsen, M. A., Theander, T. G., Theisen, M., et al. (2017). Improving the Malaria Transmission-Blocking Activity of a Plasmodium falciparum 48/45 Based Vaccine Antigen by SpyTag/SpyCatcher Mediated Virus-like Display. Vaccine 35, 3726–3732. doi:10.1016/J.VACCINE.2017.05.054
Stothard, P. (2000). The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 28, 1102–1104. doi:10.2144/00286IR01
Tachibana, M., Miura, K., Takashima, E., Morita, M., Nagaoka, H., Zhou, L., et al. (2019). Identification of Domains within Pfs230 that Elicit Transmission Blocking Antibody Responses. Vaccine 37, 1799–1806. doi:10.1016/j.vaccine.2019.02.021
Takala, S. L., Coulibaly, D., Thera, M. A., Dicko, A., Smith, D. L., Guindo, A. B., et al. (2007). Dynamics of Polymorphism in a Malaria Vaccine Antigen at a Vaccine-Testing Site in Mali. PLoS Med. 4, e93. doi:10.1371/journal.pmed.0040093
van Dijk, M. R., Janse, C. J., Thompson, J., Waters, A. P., Braks, J. A. M., Dodemont, H. J., et al. (2001). A Central Role for P48/45 in Malaria Parasite Male Gamete Fertility. Cell. 104, 153–164. doi:10.1016/S0092-8674(01)00199-4
Vermeulen, A. N., van Deursen, J., Brakenhoff, R. H., Lensen, T. H. W., Ponnudurai, T., and Meuwissen, J. H. E. T. (1986). Characterization of Plasmodium falciparum Sexual Stage Antigens and Their Biosynthesis in Synchronised Gametocyte Cultures. Mol. Biochem. Parasitol. 20, 155–163. doi:10.1016/0166-6851(86)90027-7
Keywords: Pfs230, Pfs48/45, transmission blocking vaccines, genetic diversity, evolutionary forces
Citation: Ochwedo KO, Ariri FO, Otambo WO, Magomere EO, Debrah I, Onyango SA, Orondo PW, Atieli HE, Ogolla SO, Otieno ACA, Mukabana WR, Githeko AK, Lee M-C, Yan G, Zhong D and Kazura JW (2022) Rare Alleles and Signatures of Selection on the Immunodominant Domains of Pfs230 and Pfs48/45 in Malaria Parasites From Western Kenya. Front. Genet. 13:867906. doi: 10.3389/fgene.2022.867906
Received: 01 February 2022; Accepted: 25 April 2022;
Published: 17 May 2022.
Edited by:
Sajad Ahmad Dar, Jazan University, Saudi ArabiaReviewed by:
Muzamil Mahdi Abdel Hamid, University of Khartoum, SudanMustafa Mohammad, Guru Teg Bahadur Hospital, University of Delhi, India
Copyright © 2022 Ochwedo, Ariri, Otambo, Magomere, Debrah, Onyango, Orondo, Atieli, Ogolla, Otieno, Mukabana, Githeko, Lee, Yan, Zhong and Kazura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daibin Zhong, ZHpob25nQGhzLnVjaS5lZHU=
 Kevin O. Ochwedo
Kevin O. Ochwedo Fredrick O. Ariri
Fredrick O. Ariri Wilfred O. Otambo3,2
Wilfred O. Otambo3,2 Edwin O. Magomere
Edwin O. Magomere Wolfgang R. Mukabana
Wolfgang R. Mukabana James W. Kazura
James W. Kazura