- 1Department of Ophthalmology & Pediatrics Research Institute of Hunan Province, Hunan Children’s Hospital, Changsha, China
- 2Pediatrics Research Institute of Hunan Province, Hunan Children’s Hospital, Changsha, China
- 3Department of Ophthalmology, Hunan Children’s Hospital, Changsha, China
- 4Center for Medical Genetics & Hunan Key Laboratory of Medical Genetics, School of Life Sciences, Central South University, Changsha, China
Background: Congenital cataract is one of the most common causes of blindness in children. A rapid and accurate genetic diagnosis benefit the patients in the pediatric department. The current study aims to identify the genetic defects in a congenital cataract patient without a family history.
Case presentation: A congenital cataract patient with microphthalmia and nystagmus was recruited for this study. Trio-based whole-exome sequencing revealed a de novo variant (c.394delG, p.V132Sfs*15) in CRYGC gene. According to the American College of Medical Genetics and Genomics (ACMG) criteria, the variant could be annontated as pathogenic.
Conclusion: Our findings provide new knowledge of the variant spectrum of CRYGC gene and are essential for understanding the heterogeneity of cataracts in the Chinese population.
Background
Congenital cataract is visible at birth or during the first decade of life; it is usually diagnosed by red light reflex, ophthalmoscopy examination and ocular color doppler ultrasound. Congenital cataract is one of the most common causes of blindness in children, with an estimated prevalence of 1–6 cases per 10,000 live births (Santana and Waiswo, 2011). About 8.3%–25% of congenital cataract cases present Mendelian inheritance; autosomal dominant inheritance pattern is the most common, but autosomal recessive and X-linked patterns have also been reported (Merin and Crawford, 1971; Francois, 1982; Zhong et al., 2017).
Inherited cataracts are genetically heterogeneous. With the development of WGS techniques, more and more cataract-related genes have been mapped and identified. So far, there are at least 49 loci and 37 genes have been identified for inherited isolated forms of cataracts according to OMIM (https://www.ncbi.nlm.nih.gov/omim/). These genes can be roughly grouped into four categories: crystallins, membrane proteins, cytoskeletal proteins, and DNA/RNA-banding proteins (Shiels and Hejtmanick, 2015). Crystallins are a kind of water-soluble protein that compose about 90% of lenticular protein mass and maintain the transparency of the lens (Hoehenwarter et al., 2006). They are divided into three major classes, α-, β-, and γ-crystallins. The α-crystallins belong to the small heat shock protein (HSP20) family, accounting for up to 50% of the total soluble protein of the lens (Bhat, 2003). Furthermore, they act as chaperones by binding partially unfolded lens βγ-crystallins to prevent their aggregation and thus maintain the transparency of the lens (Bhat, 2003). The βγ-crystallins are a superfamily of proteins with a “Greek key” motif unit base. Generally, the βγ-crystallins are supposed to be the essential structural proteins of the lens, but their exact function is still not fully understood (Jaenicke and Slingsby, 2001; Bhat, 2003; Slingsby and Wistow, 2014). Human γ-crystallins include six Cryg genes (CRYGA, CRYGB, CRYGC, CRYGD, CRYGN, and CRYGS); among them, variants of CRYGC, CRYGD, CRYGS, and CRYGB have been reported to be associated with congenital cataract (Heon et al., 1999; Stephan et al., 1999; Sun et al., 2005; AlFadhli et al., 2012).
In this study, a novel 1-bp deletion (c.394delG) in CRYGC gene was detected in a congenital cataract patient by trio-based whole-exome sequencing.
Case Presentation
The patient was examined at three months old because the pupil area of both eyes was found to be white for 15 days. He had poor light tracing reactions and no family history of cataracts. An ophthalmological exam revealed bilateral phacoscotasmus (C5), shallow anterior chamber, persistent pupillary membrane, invisible fundus, and nystagmus (Figure 1A). His corneas were transparent and had a diameter of 7.5 mm. The axial lengths of his eyes were 15.13 mm (OD) and 15.05 mm (OS), respectively. The intraocular pressures (IOP) were 10.2 mmHg (OD) and 14.0 mmHg (OS). Ultrasonography showed no alterations other than the opacified lens and reduced axial lengths.
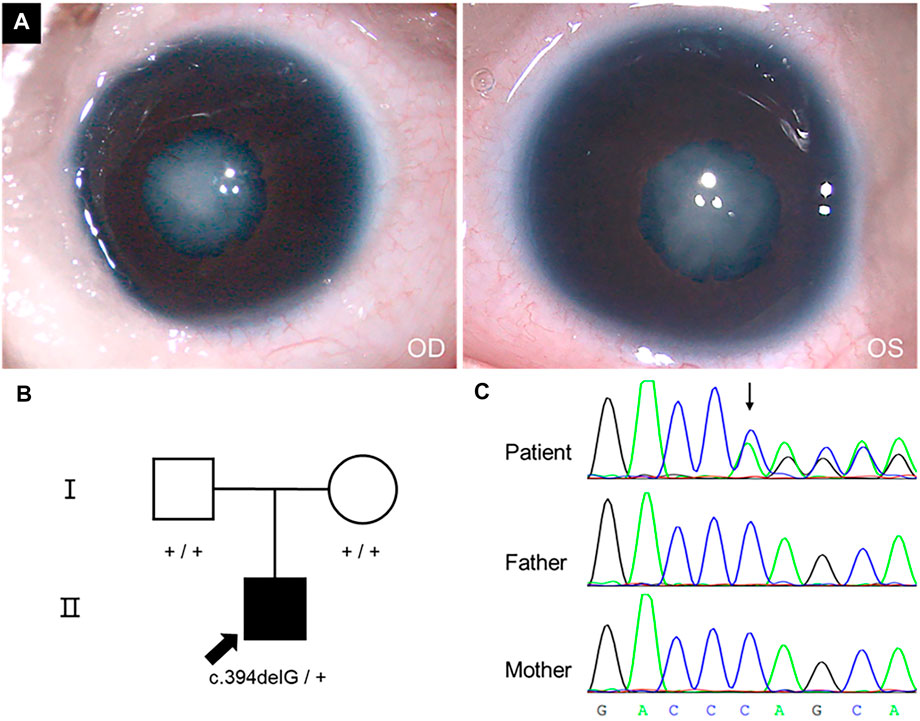
FIGURE 1. Phenotype, pedigree, and Sanger sequencing results. (A) Cataract phenotype and pupils with irregular borders of the proband were shown. (B) The pedigree of a congenital cataract trios family. (C) c.394delG variant in CRYGC gene.
A diagnosis of total cataracts and bilateral microphthalmia was made. Vitrectomy and lensectomy via anterior approach, posterior capsulorhexis, and peripheral iridectomy were performed on his both eyes. On postoperative one day, the IOP of the patient was 11 and 13 mmHg in the right and left eyes, respectively. Levofloxacin eye drops, tobramycin and dexamethasone eye drops, and tropicamide phenylephrine eye drops were used four times per day. 1 month after surgery, refractive correction in diopters (dpt) was +22.00 dpt −1.00 × 180 for the right eye and +22.00 dpt −1.00 × 180 for the left eye with spectacles. At the same time, the patient began amblyopia training under the guidance of doctors and parents.
Methods
Genomic DNA Preparation
DNA was isolated from peripheral blood using DNA Isolation Kit (Blood DNA Kit V2, CW2553). Concentrations were determined on a Qubit fluorometer (Invitrogen, Q33216) using Qubit dsDNA HS Assay Kit (Invitrogen, Q32851). Agarose gel (1%) electrophoresis was performed for quality control.
Whole-Exome Sequencing
1 μg of the isolated DNA was sheared into about 200 bp sized fragments using Bioruptor UCD-200 (Diagenode). 3 μl of the sheared DNA was electrophoresed in a 2% agarose gel to confirm the presence of fragments of the desired size range. DNA libraries were prepared with KAPA Library Preparation Kit (Kapa Biosystems, KR0453) following the manufacturer’s instructions. The libraries were estimated with Qubit dsDNA HS Assay kit (Invitrogen, Q32851). The hybridization of pooled libraries to the capture probes and remove non-hybridized library molecules were carried out by Agilent SureSelectXT2 Target Enrichment System. DNA libraries were sequenced on the Illumina Novaseq. 6000 platform (Illumina, San Diego, CA, United States) as paired-end 150-bp reads. Sample dilution, flowcell loading and sequencing were performed according to the Illumina specifications. Each sample yielded more than 10 Gb of raw data; over 95% of bases had a Phred quality score >20. The mean coverage was ×100 of the genome and the minimum coverage of ×10 was about 99%.
Data Analysis
FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) tool was used to evaluate reads quality, and our in-house script was used to filter low-quality reads. The sequenced raw reads in FastQ file format were preprocessed using Trim Galore (version 0.6.4, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove adapter-contaminated ends and low-quality bases with Phred scores < 20. Reads with > 5N bases, > 40% low-quality bases, or trimmed lengths < 30 bp were also removed. Subsequently, the quality passed reads were subsequently mapped to the human reference sequence (version: hg19) by alignment tool Burrows Wheeler Aligner (BWA, v0.7.17) (Li and Durbin, 2009). SNPs and small InDels were generated with Genome Analysis Toolkit (GATK, v3.8) (McKenna et al., 2010). The parent-child relationship was identified by King software (v2.2.7) (Manichaikul et al., 2010) to confirm the de novo variant.
Sanger Sequencing
Sanger sequencing was used to validate the variant through the filtering procedures. Primers were designed by the Primer3 program (http://frodo.wi.mit.edu/).
Result
WES yielded 14.7, 10.3, and 13.2 Gb data from genomes of proband, father, and mother, respectively. Totally, 17,823 nonsynonymous SNVs and 549 Indels were identified. Considering the patient has no family history, we checked de novo variants and recessive inherit variants at first. We identified 93 recessive inherit variants (including homozygous and compound heterozygous variants, Max MAF < 0.05), involving 54 genes. But none of these genes was associated with cataracts. In addition, there were 24 de novo variants (Max MAF < 0.005) involving 19 genes in the proband. A de novo frameshift variant c.394delG (hg19: chr2:208993058) was identified in CRYGC gene (NM_020989) through our filter pipeline. The variant would cause a frameshift from the 132nd codon and prematurely terminate at the 147th codon if a mutant protein was produced (p.V132Sfs*15). Sanger sequencing confirmed that the variant is heterozygous in the proband but absent from his parents (Figure 1C). The relationships between the three samples were confirmed (Supplementary Table S1). Furthermore, the variant was absent in the gnomAD exomes or genomes (http://gnomad.broadinstitute.org). Therefore, the c.394delG variant could be categorised as pathogenic according to the American College of Medical Genetics and Genomics (ACMG) criteria (Richards et al., 2015) (PVS1+PS2+PM2).
Discussion
To date, a total of 32 variants in CRYGC gene have been reported to be associated with congenital cataract (Heon et al., 1999; Ren et al., 2000; Santhiya et al., 2002; Gonzalez-Huerta et al., 2007; Devi et al., 2008; Yao et al., 2008; Zhang et al., 2009; Kumar et al., 2011; Guo et al., 2012; Li et al., 2012; Kondo et al., 2013; Reis et al., 2013; Gillespie et al., 2014; Prokudin et al., 2014; Li et al., 2016; Ma et al., 2016; Patel et al., 2017; Sun et al., 2017; Zhong et al., 2017; Astiazaran et al., 2018; Li et al., 2018; Zhang et al., 2019; Zhuang et al., 2019; Berry et al., 2020; Taylan Sekeroglu et al., 2020; Fernandez-Alcalde et al., 2021; Karahan et al., 2021; Rechsteiner et al., 2021), but there were few reports about the de novo mutations. In 2017, Zhong et al. reported a frameshift mutation (p.Asp65ThrfsX38) which might be de novo (Zhong et al., 2017). In 2021, Rechsteiner et al. reported a de novo mutation p.Glu107GlyfsX56, which causes cataracts and microphthalmia (Rechsteiner et al., 2021), and Fernández-Alcalde et al. reported a de novo mutation p.Leu145GlyfsX5 (Fernandez-Alcalde et al., 2021). In the present study, a de novo frameshift variant (c.394delG, p.V132Sfs*15) was identified in CRYGC gene as the cause of a patient with congenital cataract and microphthalmia.
CRYGC has a two-domain beta-structure, folded into four similar Greek key motifs (GKM); like all γ-crystallins, it has the highest intrachain symmetry (Blundell et al., 1981). The high degree of symmetry may contribute to the stability of γ-crystallins (Blundell et al., 1981). CRYGC variants in GKMs may disrupt the symmetrical structure, which changes the intra- or inter-molecular interactions, possibly leading to destabilisation and aggregation, respectively (Zhong et al., 2017). The variant p.V132Sfs*15 occurred at the beginning of GKM4 (129-171aa), leading to a frameshift and premature termination, disrupting the entire GKM4.
According to Cat-Map (Shiels et al., 2010) (https://cat-map.wustl.edu/, last updated on October 2021), the most common phenotype caused by CRYGC variants was nuclear cataracts, followed by lamellar and pulverulent cataracts. The missense variants p.F6S and p.R168W had been reported to be associated with either nuclear or lamellar cataracts (Santhiya et al., 2002; Gonzalez-Huerta et al., 2007; Devi et al., 2008; Astiazaran et al., 2018). It seems that there was no particular connection between cataract phenotypes and variant sites. Inherited cataracts could be isolated or associated with other ocular signs, including microcornea/microphthalmia, eye movement disorders (nystagmus, strabismus, amblyopia), or refractive errors. There 15 variants were reported to cause cataracts and additional ocular signs among all the 32 reported CRYGC variants. Microcornea was the most common additional ocular sign (Zhang et al., 2009; Guo et al., 2012; Reis et al., 2013; Patel et al., 2017; Sun et al., 2017; Zhong et al., 2017; Rechsteiner et al., 2021). The phenotypic heterogeneity could be due to unidentified modifier genes (Astiazaran et al., 2018) or some unknown mechanisms in which CRYGC takes part during eye development. For example, proteomics research showed that the CRYGC and some other crystallins are highly expressed in the human cornea (Subbannayya et al., 2020), indicating that these genes might involve in the cornea morphogenesis and transparency.
Next-generation DNA sequencing technologies could identify the precise genetic cause in about 45%–75% of congenital cataract families. For example, testing of WES in 11 cataract families by Kandaswamy et al. determined a genetic cause in 6 families (55%) (Kandaswamy et al., 2020). A recent study on inherited eye diseases found that WGS (through 100,000 Genomes Project) had a diagnostic yield of 44.7% (17/38) for congenital cataract families (Jackson et al., 2020). In the past few years, it has been reported that testing of a targeted gene panel (115 genes) in 36 bilateral cataracts patients identified a genetic cause in 75% of cases (Gillespie et al., 2014). However, another research using the same panel established a genetic diagnosis in 50% of congenital cataract cases (Lenassi et al., 2020). A rapid and accurate genetic diagnosis in the pediatric department helps patients understand their cause of disease, make clinical decisions, carry on the instruction for procreation, or even look for therapeutic schemes. In the current study, a genetic cause was identified in a three-month-old congenital cataract patient. He underwent cataract surgery immediately after diagnosis and had a good prognosis.
Conclusion
In conclusion, we have identified a novel frameshift variant, c.394delG, p.V132Sfs*15, within the CRYGC gene in a congenital cataract boy. Our findings provide new knowledge of the variant spectrum of CRYGC and are essential for understanding the heterogeneity of cataracts in the Chinese population.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: (BankIt2557536 BSeq#1 OM912449).
Ethics Statement
The study was approved by the Ethics Committee of Hunan Children’s Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
YL and LT: supervision and resources acquisition. YP: original manuscript writing and editing, data analysis. YZ and SZ: methodology and validation. YL: sample collection and clinical data curation and validation. ZD and YT: methodology and resources collection. YP and ZH: manuscript review and editing. All authors read and approved the final manuscript.
Funding
This work was supported by the Hunan Province Natural Science Foundation of China (Grant number: 2020JJ8076, 2020JJ8005); The Health Commission Science Research Project of Hunan Province (Grant Number: 202107021955).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors greatly thank the patient and his parents who participated in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.866246/full#supplementary-material
References
AlFadhli, S., Abdelmoaty, S., Al-Hajeri, A., Behbehani, A., and Alkuraya, F. (2012). Novel Crystallin Gamma B Mutations in a Kuwaiti Family with Autosomal Dominant Congenital Cataracts Reveal Genetic and Clinical Heterogeneity. Mol. Vis. 18, 2931–2936.
Astiazarán, M. C., García-Montaño, L. A., Sánchez-Moreno, F., Matiz-Moreno, H., and Zenteno, J. C. (2018). Next Generation Sequencing-Based Molecular Diagnosis in Familial Congenital Cataract Expands the Mutational Spectrum in Known Congenital Cataract Genes. Am. J. Med. Genet. 176, 2637–2645. doi:10.1002/ajmg.a.40524
Berry, V., Ionides, A., Pontikos, N., Georgiou, M., Yu, J., and Ocaka, L. A. (2020). The Genetic Landscape of Crystallins in Congenital Cataract. Orphanet J. Rare Dis. 15, 333. doi:10.1186/s13023-020-01613-3
Bhat, S. P. (2003). Crystallins, Genes and Cataract. Prog. Drug Res. 60, 205–262. doi:10.1007/978-3-0348-8012-1_7
Blundell, T., Lindley, P., Miller, L., Moss, D., Slingsby, C., Tickle, I., et al. (1981). The Molecular Structure and Stability of the Eye Lens: X-Ray Analysis of Gamma-Crystallin II. Nature. 289, 771–777. doi:10.1038/289771a0
Devi, R. R., Yao, W., Vijayalakshmi, P., Sergeev, Y. V., Sundaresan, P., and Hejtmancik, J. F. (2008). Crystallin Gene Mutations in Indian Families with Inherited Pediatric Cataract. Mol. Vis. 14, 1157–1170.
Fernandez-Alcalde, C., Nieves-Moreno, M., Noval, S., Peralta, J. M., Montano, V. E. F., Del Pozo, A., et al. (2021). Molecular and Genetic Mechanism of Non-syndromic Congenital Cataracts. Genes. (Basel). 12 (4), 580. doi:10.3390/genes12040580
Gillespie, R. L., O'Sullivan, J., Ashworth, J., Bhaskar, S., Williams, S., Biswas, S., et al. (2014). Personalized Diagnosis and Management of Congenital Cataract by Next-Generation Sequencing. Ophthalmology. 121, 2124–2137. e1-2. doi:10.1016/j.ophtha.2014.06.006
Gonzalez-Huerta, L. M., Messina-Baas, O. M., and Cuevas-Covarrubias, S. A. (2007). A Family with Autosomal Dominant Primary Congenital Cataract Associated with a CRYGC Mutation: Evidence of Clinical Heterogeneity. Mol. Vis. 13, 1333–1338.
Guo, Y., Su, D., Li, Q., Yang, Z., Ma, Z., Ma, X., et al. (2012). A Nonsense Mutation of CRYGC Associated with Autosomal Dominant Congenital Nuclear Cataracts and Microcornea in a Chinese Pedigree. Mol. Vis. 18, 1874–1880.
Heon, E., Priston, M., Schorderet, D. F., Billingsley, G. D., Girard, P. O., Lubsen, N., et al. (1999). The Gamma-Crystallins and Human Cataracts: a Puzzle Made Clearer. Am. J. Hum. Genet. 65, 1261–1267. doi:10.1086/302619
Hoehenwarter, W., Klose, J., and Jungblut, P. R. (2006). Eye Lens Proteomics. Amino Acids. 30, 369–389. doi:10.1007/s00726-005-0283-9
Jackson, D., Malka, S., Harding, P., Palma, J., Dunbar, H., and Moosajee, M. (2020). Molecular Diagnostic Challenges for Non-retinal Developmental Eye Disorders in the United Kingdom. Am. J. Med. Genet. C Semin. Med. Genet. 184, 578–589. doi:10.1002/ajmg.c.31837
Jaenicke, R., and Slingsby, C. (2001). Lens Crystallins and Their Microbial Homologs: Structure, Stability, and Function. Crit. Rev. Biochem. Mol. Biol. 36, 435–499. doi:10.1080/20014091074237
Kandaswamy, D. K., Prakash, M. V. S., Graw, J., Koller, S., Magyar, I., Tiwari, A., et al. (2020). Application of WES towards Molecular Investigation of Congenital Cataracts: Identification of Novel Alleles and Genes in a Hospital-Based Cohort of South India. Int. J. Mol. Sci. 21, 9569. doi:10.3390/ijms21249569
Karahan, M., Demirtas, A. A., Erdem, S., Ava, S., Tekes, S., and Keklikci, U. (2021). Crystalline Gene Mutations in Turkish Children with Congenital Cataracts. Int. Ophthalmol. 41, 2847–2852. doi:10.1007/s10792-021-01843-9
Kondo, Y., Saitsu, H., Miyamoto, T., Lee, B. J., Nishiyama, K., Nakashima, M., et al. (2013). Pathogenic Mutations in Two Families with Congenital Cataract Identified with Whole-Exome Sequencing. Mol. Vis. 19, 384–389.
Kumar, M., Agarwal, T., Khokhar, S., Kumar, M., Kaur, P., Roy, T. S., et al. (2011). Mutation Screening and Genotype Phenotype Correlation of Alpha-Crystallin, Gamma-Crystallin and GJA8 Gene in Congenital Cataract. Mol. Vis. 17, 693–707.
Lenassi, E., Clayton-Smith, J., Douzgou, S., Ramsden, S. C., Ingram, S., Hall, G., et al. (2020). Clinical Utility of Genetic Testing in 201 Preschool Children with Inherited Eye Disorders. Genet. Med. 22, 745–751. doi:10.1038/s41436-019-0722-8
Li, D., Wang, S., Ye, H., Tang, Y., Qiu, X., Fan, Q., et al. (2016). Distribution of Gene Mutations in Sporadic Congenital Cataract in a Han Chinese Population. Mol. Vis. 22, 589–598.
Li, H., and Durbin, R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760. doi:10.1093/bioinformatics/btp324
Li, J., Leng, Y., Han, S., Yan, L., Lu, C., Luo, Y., et al. (2018). Clinical and Genetic Characteristics of Chinese Patients with Familial or Sporadic Pediatric Cataract. Orphanet J. Rare Dis. 13, 94. doi:10.1186/s13023-018-0828-0
Li, X. Q., Cai, H. C., Zhou, S. Y., Yang, J. H., Xi, Y. B., Gao, X. B., et al. (2012). A Novel Mutation Impairing the Tertiary Structure and Stability of gammaC-Crystallin (CRYGC) Leads to Cataract Formation in Humans and Zebrafish Lens. Hum. Mutat. 33, 391–401. doi:10.1002/humu.21648
Ma, A. S., Grigg, J. R., Ho, G., Prokudin, I., Farnsworth, E., Holman, K., et al. (2016). Sporadic and Familial Congenital Cataracts: Mutational Spectrum and New Diagnoses Using Next-Generation Sequencing. Hum. Mutat. 37, 371–384. doi:10.1002/humu.22948
Manichaikul, A., Mychaleckyj, J. C., Rich, S. S., Daly, K., Sale, M., and Chen, W. M. (2010). Robust Relationship Inference in Genome-wide Association Studies. Bioinformatics. 26, 2867–2873. doi:10.1093/bioinformatics/btq559
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 20, 1297–1303. doi:10.1101/gr.107524.110
Merin, S., and Crawford, J. S. (1971). The Etiology of Congenital Cataracts. A Survey of 386 Cases. Can. J. Ophthalmol. 6, 178–182.
Patel, N., Anand, D., Monies, D., Maddirevula, S., Khan, A. O., Algoufi, T., et al. (2017). Novel Phenotypes and Loci Identified through Clinical Genomics Approaches to Pediatric Cataract. Hum. Genet. 136, 205–225. doi:10.1007/s00439-016-1747-6
Prokudin, I., Simons, C., Grigg, J. R., Storen, R., Kumar, V., Phua, Z. Y., et al. (2014). Exome Sequencing in Developmental Eye Disease Leads to Identification of Causal Variants in GJA8, CRYGC, PAX6 and CYP1B1. Eur. J. Hum. Genet. 22, 907–915. doi:10.1038/ejhg.2013.268
Rechsteiner, D., Issler, L., Koller, S., Lang, E., Bahr, L., Feil, S., et al. (2021). Genetic Analysis in a Swiss Cohort of Bilateral Congenital Cataract. JAMA Ophthalmol. 139, 691–700. doi:10.1001/jamaophthalmol.2021.0385
Reis, L. M., Tyler, R. C., Muheisen, S., Raggio, V., Salviati, L., Han, D. P., et al. (2013). Whole Exome Sequencing in Dominant Cataract Identifies a New Causative Factor, CRYBA2, and a Variety of Novel Alleles in Known Genes. Hum. Genet. 132, 761–770. doi:10.1007/s00439-013-1289-0
Ren, Z., Li, A., Shastry, B. S., Padma, T., Ayyagari, R., Scott, M. H., et al. (2000). A 5-base Insertion in the gammaC-Crystallin Gene Is Associated with Autosomal Dominant Variable Zonular Pulverulent Cataract. Hum. Genet. 106, 531–537. doi:10.1007/s004390000289
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and Guidelines for the Interpretation of Sequence Variants: a Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424. doi:10.1038/gim.2015.30
Santana, A., and Waiswo, M. (2011). The Genetic and Molecular Basis of Congenital Cataract. Arq. Bras. Oftalmol. 74, 136–142. doi:10.1590/s0004-27492011000200016
Santhiya, S. T., Shyam Manohar, M., Rawlley, D., Vijayalakshmi, P., Namperumalsamy, P., Gopinath, P. M., et al. (2002). Novel Mutations in the Gamma-Crystallin Genes Cause Autosomal Dominant Congenital Cataracts. J. Med. Genet. 39, 352–358. doi:10.1136/jmg.39.5.352
Shiels, A., Bennett, T. M., and Hejtmancik, J. F. (2010). Cat-Map: Putting Cataract on the Map. Mol. Vis. 16, 2007–2015.
Shiels, A., and Hejtmancik, J. F. (2015). Molecular Genetics of Cataract. Prog. Mol. Biol. Transl. Sci. 134, 203–218. doi:10.1016/bs.pmbts.2015.05.004
Slingsby, C., and Wistow, G. J. (2014). Functions of Crystallins in and Out of Lens: Roles in Elongated and Post-mitotic Cells. Prog. Biophys. Mol. Biol. 115, 52–67. doi:10.1016/j.pbiomolbio.2014.02.006
Stephan, D. A., Gillanders, E., Vanderveen, D., Freas-Lutz, D., Wistow, G., Baxevanis, A. D., et al. (1999). Progressive Juvenile-Onset Punctate Cataracts Caused by Mutation of the gammaD-Crystallin Gene. Proc. Natl. Acad. Sci. U. S. A. 96, 1008–1012. doi:10.1073/pnas.96.3.1008
Subbannayya, Y., Pinto, S. M., Mohanty, V., Dagamajalu, S., Prasad, T. S. K., and Murthy, K. R. (2020). What Makes Cornea Immunologically Unique and Privileged? Mechanistic Clues from a High-Resolution Proteomic Landscape of the Human Cornea. OMICS 24, 129–139. doi:10.1089/omi.2019.0190
Sun, H., Ma, Z., Li, Y., Liu, B., Li, Z., Ding, X., et al. (2005). Gamma-S Crystallin Gene (CRYGS) Mutation Causes Dominant Progressive Cortical Cataract in Humans. J. Med. Genet. 42, 706–710. doi:10.1136/jmg.2004.028274
Sun, Z., Zhou, Q., Li, H., Yang, L., Wu, S., and Sui, R. (2017). Mutations in Crystallin Genes Result in Congenital Cataract Associated with Other Ocular Abnormalities. Mol. Vis. 23, 977–986.
Taylan Sekeroglu, H., Karaosmanoglu, B., Taskiran, E. Z., Simsek Kiper, P. O., Alikasifoglu, M., Boduroglu, K., et al. (2020). Molecular Etiology of Isolated Congenital Cataract Using Next-Generation Sequencing: Single Center Exome Sequencing Data from Turkey. Mol. Syndromol. 11, 302–308. doi:10.1159/000510481
Yao, K., Jin, C., Zhu, N., Wang, W., Wu, R., Jiang, J., et al. (2008). A Nonsense Mutation in CRYGC Associated with Autosomal Dominant Congenital Nuclear Cataract in a Chinese Family. Mol. Vis. 14, 1272–1276.
Zhang, J., Sun, D., Wang, Y., Mu, W., Peng, Y., and Mi, D. (2019). Identification of a Novel CRYGC Mutation in a Pedigree Affected with Congenital Cataracts. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 36, 697–700. doi:10.3760/cma.j.issn.1003-9406.2019.07.010
Zhang, L., Fu, S., Ou, Y., Zhao, T., Su, Y., and Liu, P. (2009). A Novel Nonsense Mutation in CRYGC Is Associated with Autosomal Dominant Congenital Nuclear Cataracts and Microcornea. Mol. Vis. 15, 276–282.
Zhong, Z., Wu, Z., Han, L., and Chen, J. (2017). Novel Mutations in CRYGC Are Associated with Congenital Cataracts in Chinese Families. Sci. Rep. 7, 189. doi:10.1038/s41598-017-00318-1
Keywords: congenital cataract, crystallin, CRYGC, microphthalmia, whole-exome sequencing
Citation: Peng Y, Zheng Y, Deng Z, Zhang S, Tan Y, Hu Z, Tao L and Luo Y (2022) Case Report: A de novo Variant of CRYGC Gene Associated With Congenital Cataract and Microphthalmia. Front. Genet. 13:866246. doi: 10.3389/fgene.2022.866246
Received: 31 January 2022; Accepted: 11 May 2022;
Published: 27 May 2022.
Edited by:
Zi-Bing Jin, Capital Medical University, ChinaReviewed by:
Abhinav Jain, Council of Scientific and Industrial Research (CSIR), IndiaEmilia Severin, Carol Davila University of Medicine and Pharmacy, Romania
Copyright © 2022 Peng, Zheng, Deng, Zhang, Tan, Hu, Tao and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yulin Luo, bHVveXVsaW4yMDAwQDEyNi5jb20=; Lijuan Tao, aG5ldHl5MTIyMUAxNjMuY29t
†These authors have contributed equally to this work
 Yu Peng
Yu Peng Yu Zheng
Yu Zheng Zifeng Deng3
Zifeng Deng3 Shuju Zhang
Shuju Zhang Zhengmao Hu
Zhengmao Hu Yulin Luo
Yulin Luo