
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genet. , 11 May 2022
Sec. Computational Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.865371
This article is part of the Research Topic Identifying Genetics-Based Mechanisms and Treatments for Neurodevelopmental and Psychiatric Disorders Through Data Integration View all 6 articles
Human brain-related disorders, such as autism spectrum disorder (ASD), are often characterized by cell heterogeneity, as the cell atlas of brains consists of diverse cell types. There are commonality and specificity in gene expression among different cell types of brains; hence, there may also be commonality and specificity in dysregulated gene expression affected by ASD among brain cells. Moreover, as genes interact together, it is important to identify shared and cell-type-specific ASD-related gene modules for studying the cell heterogeneity of ASD. To this end, we propose integrative regularized non-negative matrix factorization (iRNMF) by imposing a new regularization based on integrative non-negative matrix factorization. Using iRNMF, we analyze gene expression data of multiple cell types of the human brain to obtain shared and cell-type-specific gene modules. Based on ASD risk genes, we identify shared and cell-type-specific ASD-associated gene modules. By analyzing these gene modules, we study the commonality and specificity among different cell types in dysregulated gene expression affected by ASD. The shared ASD-associated gene modules are mostly relevant to the functioning of synapses, while in different cell types, different kinds of gene functions may be specifically dysregulated in ASD, such as inhibitory extracellular ligand-gated ion channel activity in GABAergic interneurons and excitatory postsynaptic potential and ionotropic glutamate receptor signaling pathway in glutamatergic neurons. Our results provide new insights into the molecular mechanism and pathogenesis of ASD. The identification of shared and cell-type-specific ASD-related gene modules can facilitate the development of more targeted biomarkers and treatments for ASD.
The human brain is a highly heterogeneous organ, consisting of multiple kinds of cell types. Brain-related disorders, such as autism spectrum disorder (ASD), are often characterized by cell heterogeneity and mainly affect some specific cell types. ASD, a set of neuropsychiatric disorders, is characterized by highly genetic and phenotypic heterogeneity. To date, its actual causes and underlying mechanisms remain unclear. Although there have been hundreds of genes identified to be associated with ASD, they only account for 10–20% of ASD cases (Rylaarsdam and Guemez-Gamboa, 2019). Genes do not act alone, and what determines the manifestation of a disease in different cell types is the presence of disease-associated gene modules instead of individual genes (Kitsak et al., 2016; Guan et al., 2021). Moreover, as there are commonality and specificity in gene expression among different cell types of brains, there may also be commonality and specificity in dysregulated gene expression affected by ASD among brain cells. Therefore, based on gene expression datasets of multiple human brain cells, the detection of shared and cell-type-specific ASD-associated gene modules is of significance to study the molecular mechanism and pathogenesis of ASD.
Non-negative matrix factorization (NMF)-based methods have been developed and applied to the analyses of biological sequencing data, such as sparse NMF (sNMF) (Mairal et al., 2010) and sparse modular activity factorization (SMAF) (Cleary et al., 2017). In the context of integrating heterogeneous datasets, several methods have been proposed recently. Many of them were developed to integrate multi-modal or multi-omics data and focus on the analysis of samples, such as the joint definition of cell types of samples by taking the advantage of multiple heterogeneous datasets. For example, LIGER (Welch et al., 2019) was developed based on integrative non-negative matrix factorization (iNMF) (Yang and Michailidis, 2016) to factorize multiple datasets into a common gene-factor matrix, multiple dataset-specific gene-factor matrices, and multiple dataset-specific sample-factor matrices. Compared with the original algorithm of iNMF, LIGER adopted a novel block coordinate descent algorithm for performing iNMF, which can converge quickly. iNMF can extract consistent patterns embedded in various data sources by separating the homogeneous and heterogeneous effects among the sources, and it was mainly adopted to analyze the low-dimensional sample-factor matrices based on different kinds of data. The low-dimensional gene-factor matrices should be given more attention. The sparsity of sample representation (Yang and Michailidis, 2016) is beneficial to sample analyses, such as cell-type definition, while to perform gene module analyses, the sparsity or regularization of gene representation could be induced. Except for integrating multi-modal data, performing integrative and comparative analyses on the same type of data from multiple biological conditions, such as various cancer types or subtypes, various cell lines, and various cell types, is also valuable (Zhang and Zhang, 2019).
To depict the common and dataset-specific gene expression patterns, we proposed integrative regularized non-negative matrix factorization (iRNMF), by adopting iNMF and imposing a new regularization, to obtain a common gene-factor matrix and multiple dataset-specific gene-factor matrices. With iRNMF, we analyzed the gene expression data of multiple human brain cell types and obtained shared and cell-type-specific gene modules. Then, ASD-related risk genes were used to identify shared and cell-type-specific ASD-associated gene modules. By analyzing these gene modules, we studied the shared and cell-type-specific dysregulated gene expression patterns in ASD.
Non-negative matrix factorization can factorize a high-dimensional gene expression matrix into two low-dimensional matrices, i.e., a gene-factor matrix and a sample-factor matrix, achieving the purpose of dimension reduction. To integrate and factorize multiple gene expression datasets into a common gene-factor matrix, multiple dataset-specific gene-factor matrices, and sample-factor matrices, iNMF (Yang and Michailidis, 2016) was proposed. The optimization problem is:
where
The regularization of iNMF can make
where
until convergence. Each of the optimization subproblems mentioned previously requires solving a non-negative least-squares problem, and we used the fast block principal pivoting algorithm (Kim et al., 2014) to solve each of these subproblems.
We downloaded the single-nucleus gene expression data derived from the middle temporal gyrus (MTG) of the human cortex (Hodge et al., 2019) from the Allen Institute for Brain Science. It includes 15,928 nuclei sampled from eight human donor brains, of which 15,206 were from postmortem donors with no known neuropsychiatric or neurological conditions and 722 were from distal and normal tissues of neurosurgical donors. We preprocessed the data with R packages of scatter (McCarthy et al., 2017) and scran (Lun et al., 2016), including the quality control of nuclei and genes, and removing a minority of nuclei assigned to different cell cycle phases by the function of cyclone in scran. Nuclear and mitochondrial genes downloaded from Human MitoCarta2.0 (Calvo et al., 2016) were excluded, and protein-coding genes were retained. After removing the nuclei not assigned to any specific cell type, we obtained the expression level of 17,120 protein-coding genes in 12,246 nuclei. Then, we used scran to obtain 7,011 highly variable protein-coding genes across all nuclei, which were defined as genes with biological components that are significantly greater than zero at a false discovery rate (FDR) of 0.1. After removing the cell types containing less than 20 nuclei, we obtained the gene expression data of nuclei from glutamatergic neuron (Gluta), GABAergic interneuron (GABA), astrocyte (Ast), oligodendrocyte (Oli), and oligodendrocyte precursor cell (OPC), including 8994, 2762, 227, 112, and 133 nuclei, respectively. The gene expression of 7,011 highly variable protein-coding genes in these five cell types was used for analyses.
To determine the number of factors/gene modules m, we used the same way with LIGER, applying Kullback–Leibler (KL) divergence as a criterion. When the number of factors is too low, factors will include many genes and samples will load on many factors, with the distribution of factor loadings for a particular sample approaching a uniform distribution (Welch et al., 2019). As the number of factors approaches the true number of gene modules, each sample will generally load on only a few factors. Therefore, we calculated the KL divergence, compared to a uniform distribution, of the factor loadings for each sample and plotted the median across samples as a function of m to select the saturation point of the curve as the optimal m. We also considered the mean squared error (MSE) between
We used iRNMF to analyze gene expression datasets of multiple cell types derived from human MTG. After obtaining the cell-type-specific and shared gene module matrices
We proposed integrative regularized non-negative matrix factorization (iRNMF) to learn homogeneous and heterogeneous gene expression patterns across multiple datasets. Single-nucleus gene expression datasets of multiple cell types of human MTG (Hodge et al., 2019) were analyzed using iRNMF, involving glutamatergic neuron (Gluta), GABAergic interneuron (GABA), astrocyte (Ast), oligodendrocyte (Oli), and oligodendrocyte precursor cell (OPC) denoted by
To show the effectiveness of iRNMF, we compared iRNMF with LIGER (which only imposes regularization on
Next, we compared iRNMF with LIGER based on cell representation. For each cell type, we calculated the Pearson correlation between flatten
Lastly, we compared the gene modules obtained using LIGER and iRNMF. We calculated gene–gene correlation matrices using
Among all shared gene modules determined from W, 46 are significantly enriched with ASD genes (Supplementary Table S1). For the top ten shared ASD-associated gene modules, we list their Bonferroni-adjusted p-values, top three z-score genes (Figure 1A1), and top one significant GO term (Figure 1A2). Some top genes are ASD genes, including PDE1C and MKX in W_M42, GPC6 in W_M10, and NTNG1 in W_M26. The top one significant GO term is all related to synapses, whose dysregulation has been proven to be associated with ASD. Then, we checked which kinds of GO terms are the most common among all GO terms of all shared ASD-associated gene modules and found that the top ten common GO terms are also associated with the functioning of synapses, appearing in all shared ASD-associated gene modules (Figure 1B1). Next, we focused on the modules which have module-specific gene functions, by removing the repeated GO terms between gene modules. There are 36 shared ASD-associated gene modules with module-specific GO terms (Supplementary Table S2). The top ten modules with module-specific gene functions are also the ones shown in Figure 1A1, and their top one module-specific GO term is shown in Figure 1B2. The top three modules most significantly enriched with ASD genes, W_M78, W_M91, and W_M42, are related to cortical actin cytoskeleton, heparan sulfate proteoglycan metabolic process, and regulation of sodium ion transmembrane transport, respectively. The actin cytoskeleton has been associated with ASD and provides a strategy for ASD treatment by targeting actin regulators (Duffney et al., 2015; Hlushchenko et al., 2018). The lacking of heparin sulfate, a proteoglycan involved in a variety of neurodevelopmental processes, has been correlated with ASD (Irie et al., 2012; Pérez et al., 2016). Ion channels, including sodium, calcium, and potassium, are implicated in the etiology of ASD (Daghsni et al., 2018). It can be seen that the identified gene modules are meaningful.
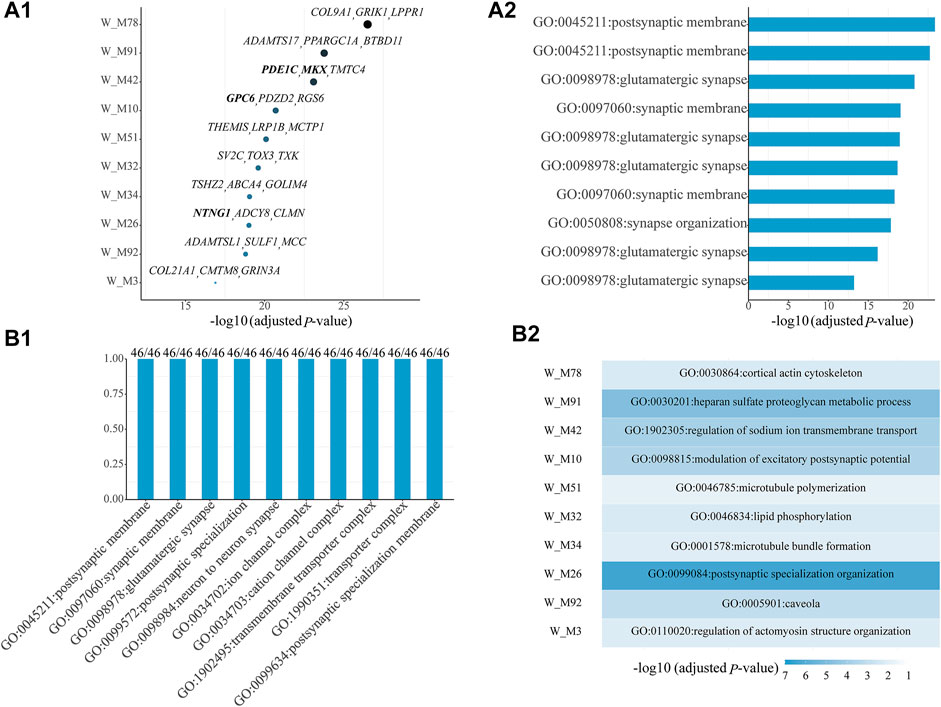
FIGURE 1. Top ten shared ASD-associated gene modules along with (A1) their Bonferroni-adjusted p-values, top three z-score genes, and (A2) top one significant GO term. (B1) Top ten common enriched GO terms among all shared ASD-associated gene modules along with the frequency of occurrence and the total number of gene modules. (B2) Module-specific top one GO term of the top ten shared ASD-associated modules. The SFARI ASD genes are bold. The Bonferroni-adjusted p-values were derived from the hypergeometric tests using module genes and ASD genes.
Among all cell-type-specific gene modules, we identified 11, 25, 29, 45, and 14 cell-type-specific ASD-associated gene modules for Ast, GABA, Gluta, Oli, and OPC, respectively (Supplementary Table S1). We list the top ten significant gene modules along with their Bonferroni-adjusted p-values, top three z-score genes, and top one significant GO term (Figure 2). Noted that for the two kinds of neurons, GABA and Gluta, the cell-type-specific ASD-associated gene modules are more significantly enriched with ASD genes and more top three genes are ASD genes, compared with glial cells. Many of the top GO terms of cell-type-specific ASD-associated gene modules are related to synapses, while different gene functions may still be dysregulated in different cell types. For instance, gamma-tubulin complex, cadherin binding, and protein tyrosine kinase activity are associated with Ast-specific ASD-associated gene modules; regulation of microtubule cytoskeleton organization, phosphatase binding, and desmosome are significant in OPC-specific ASD-associated gene modules. These may indicate that different gene functions may be dysregulated by ASD in different cells, demonstrating the cell heterogeneity of ASD.
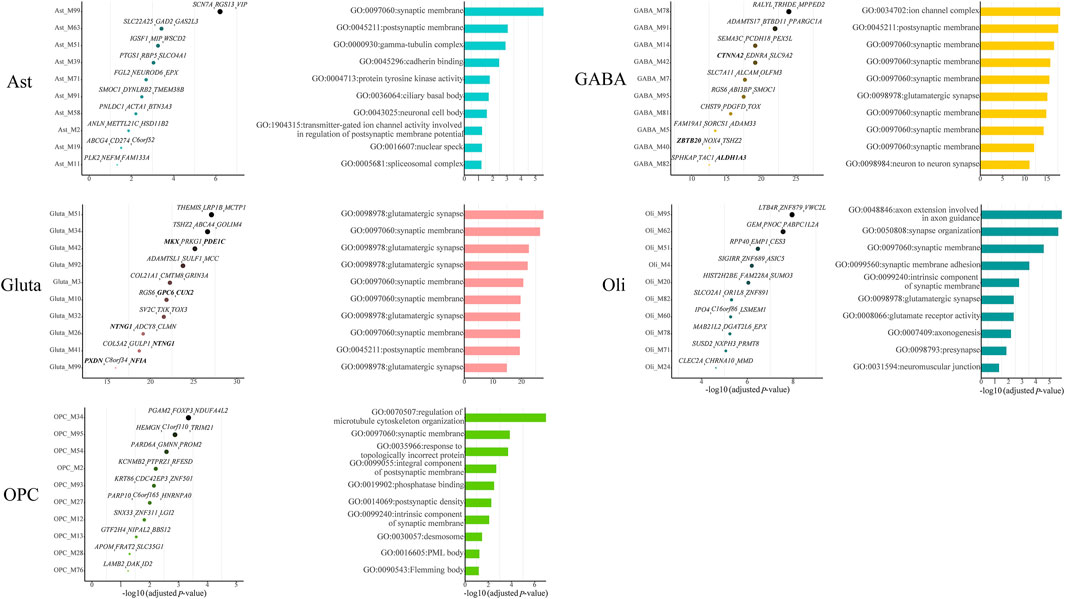
FIGURE 2. Top ten cell-type-specific ASD-associated gene modules along with their Bonferroni-adjusted hypergeometric test p-values, top three z-score genes, and top one significant GO term. The SFARI ASD genes are bold. The Bonferroni-adjusted p-values were derived from the hypergeometric tests using module genes and ASD genes.
Then, we checked which kinds of GO terms are the most common among all cell-type-specific ASD-associated gene modules in each cell type. Indeed, the functioning of synapses is important across all cell types (Figure 3A). Next, we focused on the modules which have module-specific gene functions. There are 7, 23, 24, 24, and 8 cell-type-specific ASD-associated gene modules left in Ast, GABA, Gluta, Oli, and OPC, respectively (Supplementary Table S2). We reported the top ten, along with their top three z-score genes (Figure 3B), and top one GO term (Figure 3C). In Ast, locomotory behavior, integral component of the postsynaptic membrane, and cadherin binding are functions specific to the top three modules, Ast_M99, Ast_M63, and Ast_M39. For GABA, it can be noted that inhibitory extracellular ligand-gated ion channel activity is specific to GABA_M7. On the contrary, the modulation of excitatory postsynaptic potential and ionotropic glutamate receptor signaling pathway are specific to Gluta_M10 and Gluta_M99, respectively. These gene functions are obviously associated with particular cell types. Neurons communicate with one another at synapses using two types of signals, electrical and chemical signals. At an electrical synapse, ions flow directly between cells. At a chemical synapse, neurotransmitters pass messages from the presynaptic to the postsynaptic neuron. The major excitatory and inhibitory neurotransmitters in brains are glutamate and GABA (gamma-aminobutyric acid), respectively. For Oli, regulation of dendrite morphogenesis and regulation of gliogenesis are specific to Oli_M82 and Oli_M97. For OPC, protein homooligomerization and endoplasmic reticulum unfolded protein response are specific to the top two modules, OPC_M34 and OPC_M54, respectively. The analysis of module-specific gene functions and top genes of cell-type-specific ASD-related gene modules can facilitate the development of more targeted biomarkers and treatments for ASD.
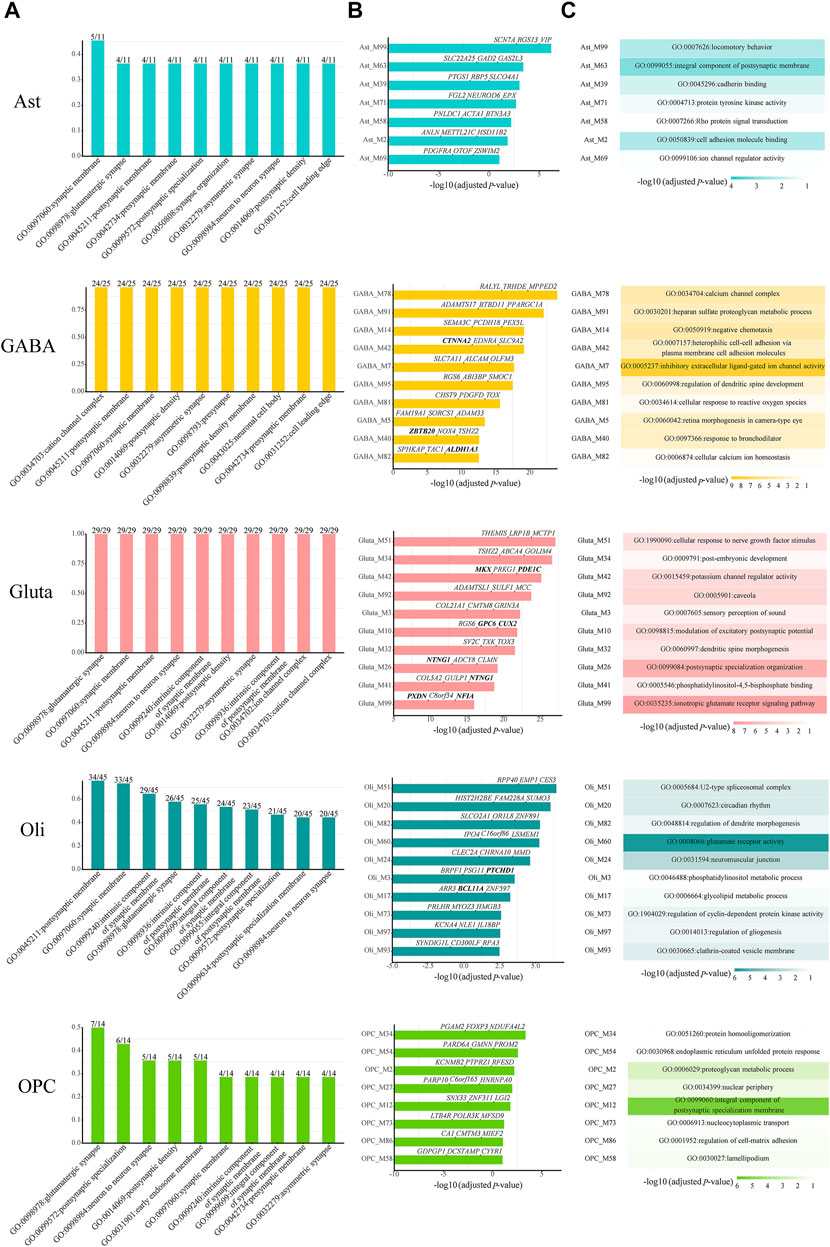
FIGURE 3. (A) Top ten common enriched GO terms among all cell-type-specific ASD-associated gene modules along with the frequency of occurrence and the total number of cell-type-specific ASD-associated gene modules. For the top ten cell-type-specific ASD-associated modules, which have module-specific GO terms, (B) their Bonferroni-adjusted hypergeometric test p-values, top three z-score genes, and (C) top one GO term are shown. The SFARI ASD genes are bold. The Bonferroni-adjusted p-values were derived from the hypergeometric tests using module genes and ASD genes.
Next, we further examined the modules with both cell-type-specific and module-specific gene functions, which are those GO terms that only appear in one module of one cell type. In GABA, Gluta, Oli, and OPC, there are 14, 18, 1, and 1 cell-type-specific ASD-associated gene modules that have both cell-type-specific and module-specific gene functions (Supplementary Table S3). It can be noted that more modules have cell type-specific and module-specific gene functions in neuronal cells, emphasizing the neurons are mainly affected by ASD. For the cell types with more than one cell-type-specific ASD-associated gene modules, we show the top ten modules along with their Bonferroni-adjusted p-values, the enriched top one GO term, and the top three z-score genes (Figure 4). Among the top three genes, CTNNA2 in GABA_M42, ZBTB2 in GABA_M40, and SLC9A9 in GABA_M75 are ASD genes. MKX and PDE1C in Gluta_M42, GPC6 and CUX2 in Gluta_M10, and PXDN and NFIA in Gluta_M99 are ASD genes. These gene modules may need more attention. We note that different kinds of gene functions are specific to ASD-associated modules of different cell types. GABA-specific ASD-associated gene modules are responsible for inhibitory extracellular ligand-gated ion channel activity and forebrain neuron differentiation, and so on. Gluta-specific ASD-associated gene modules are responsible for nerve growth factors, excitatory postsynaptic potential, and ionotropic glutamate receptor signaling pathway, and so on. Oli_M60 and OPC_M12 have a cell-type-specific and module-specific function, regulation of bone mineralization and lipid transporter activity, respectively (Supplementary Table S3). These results indicate that in different cell types, different kinds of gene functions may be specifically dysregulated in ASD, highlighting the cell heterogeneity of ASD.
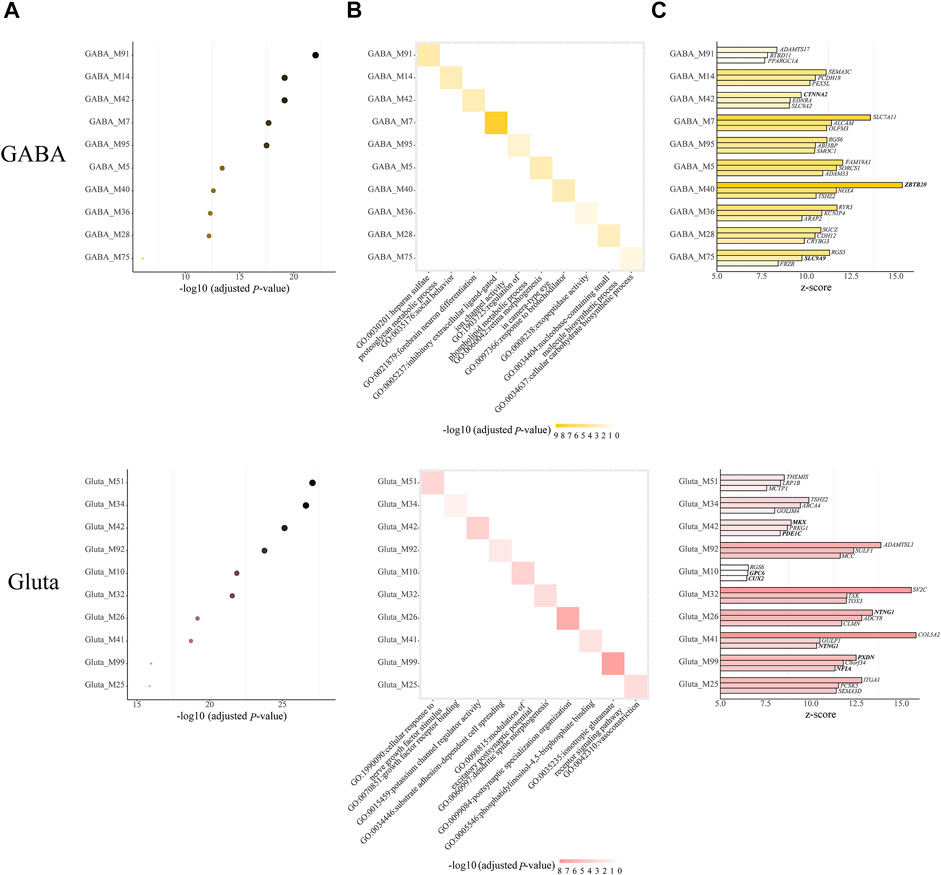
FIGURE 4. Cell-type-specific ASD-associated modules, which have both module-specific and cell type-specific GO terms, along with (A) their Bonferroni-adjusted hypergeometric test p-values, (B) top one enriched GO term, and (C) top three z-score genes. The SFARI ASD genes are bold. The Bonferroni-adjusted p-values were derived from the hypergeometric tests using module genes and ASD genes.
Brain-related diseases are often characterized by cell heterogeneity and mainly affect some specific cell types, as the brain is highly heterogeneous. To study the common and cell type-specific gene expression patterns across different brain cell types, we proposed iRNMF by adopting iNMF and imposing a further regularization. With iRNMF, we analyzed the gene expression data of multiple human brain cell types to obtain shared and cell-type-specific gene modules and cell-type-specific cell representations. By comparing iRNMF with LIGER in terms of cell representations and gene modules, it has been shown that iRNMF is effective, and the obtained low-dimensional matrices are beneficial for the downstream analyses, especially gene module analyses.
By using curated ASD candidate genes, shared and cell-type-specific ASD-associated gene modules were identified. For the shared ASD-associated gene modules, their significant gene functions are mostly relevant to the functioning of synapses, which has already been proven to be associated with ASD. Then, we identified the module-specific gene functions, including cortical actin cytoskeleton, heparan sulfate proteoglycan metabolic process, and regulation of sodium ion transmembrane transport. As to cell-type-specific ASD-associated gene modules, GABA-specific and Gluta-specific ASD-associated gene modules are more significantly enriched with ASD genes, and more top three genes are ASD genes compared with glial cells, emphasizing that the neurons are mainly affected by ASD. Many top GO terms of cell-type-specific ASD-associated gene modules are related to synapses, while different gene functions may still be specifically dysregulated by ASD in different cell types. Therefore, we focused on the functions which are specific to modules and also cell types. We noted that inhibitory extracellular ligand-gated ion channel activity and forebrain neuron differentiation are functions specifically significant in GABA; nerve growth factor, excitatory postsynaptic potential, and ionotropic glutamate receptor signaling pathway are specifically related to Gluta; lipid transporter activity is specifically significant in OPC.
By analyzing the gene functions and top important genes of shared and cell-type-specific ASD-associated gene modules, we study the shared and cell-type-specific dysregulated gene expression patterns in ASD. Moreover, we highlighted the shared ASD-associated gene modules, which have module-specific gene functions, and cell-type-specific ASD-associated gene modules, which have both module-specific and cell-type-specific gene functions. Analyzing these gene modules can facilitate the development of more targeted biomarkers and treatments for ASD. Our results provide new insights into the molecular mechanism and pathogenesis of ASD, studying the cell heterogeneity of ASD. Our method can also be used to extract homogeneous and heterogeneous patterns embedded in data from multiple biological conditions, such as various cancer types or subtypes and various cell lines.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
JG conceived and designed the study. JG, YZ, and YK conducted the analyses. JG, YZ, and GJ wrote the manuscript. All authors approved the final manuscript.
This study has been supported by the National Natural Science Foundation of China (Nos 61803320 and 61573296).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.865371/full#supplementary-material
Supplementary Figure S1 | Selection of parameter values of iRNMF and LIGER. (A) Selection of m using mean squared error (MSE) and KL divergence as criteria for iRNMF and LIGER. (B) Selection of the regularization parameter
Supplementary Figure S2 | Comparisons between iRNMF and LIGER. (A) Pearson correlation coefficient between original data
Supplementary Table S1 | Shared and cell-type-specific ASD-associated gene modules. For these modules, their Bonferroni-adjusted hypergeometric p-values, module genes sorted by z-scores, and enriched gene functions are listed.
Supplementary Table S2 | Shared and cell-type-specific ASD-associated gene modules that have module-specific gene functions. For these modules, their Bonferroni-adjusted hypergeometric p-values, module genes sorted by z-scores, and enriched module-specific gene functions are listed.
Supplementary Table S3 | Cell-type-specific ASD-associated gene modules that have both module-specific and cell type-specific gene functions. For these modules, their Bonferroni-adjusted hypergeometric p-values, module genes sorted by z-scores, and enriched module-specific and cell-type-specific gene functions are listed.
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., and Satija, R. (2018). Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol. 36, 411–420. doi:10.1038/nbt.4096
Calvo, S. E., Clauser, K. R., and Mootha, V. K. (2016). MitoCarta2.0: an Updated Inventory of Mammalian Mitochondrial Proteins. Nucleic Acids Res. 44 (D1), D1251–D1257. doi:10.1093/nar/gkv1003
Cleary, B., Cong, L., Cheung, A., Lander, E. S., and Regev, A. (2017). Efficient Generation of Transcriptomic Profiles by Random Composite Measurements. Cell 171 (6), 1424–1436. doi:10.1016/j.cell.2017.10.023
Daghsni, M., Rima, M., Fajloun, Z., Ronjat, M., Brusés, J. L., M'Rad, R., et al. (2018). Autism throughout Genetics: Perusal of the Implication of Ion Channels. Brain Behav. 8 (8), e00978. doi:10.1002/brb3.978
Duffney, L. J., Zhong, P., Wei, J., Matas, E., Cheng, J., Qin, L., et al. (2015). Autism-like Deficits in Shank3-Deficient Mice Are Rescued by Targeting Actin Regulators. Cel Rep. 11 (9), 1400–1413. doi:10.1016/j.celrep.2015.04.064
Guan, J., Lin, Y., Wang, Y., Gao, J., and Ji, G. (2021). An Analytical Method for the Identification of Cell Type-specific Disease Gene Modules. J. Transl Med. 19 (1), 20. doi:10.1186/s12967-020-02690-5
Hlushchenko, I., Khanal, P., Abouelezz, A., Paavilainen, V. O., and Hotulainen, P. (2018). ASD-associated De Novo Mutations in Five Actin Regulators Show Both Shared and Distinct Defects in Dendritic Spines and Inhibitory Synapses in Cultured Hippocampal Neurons. Front. Cel. Neurosci. 12, 217. doi:10.3389/fncel.2018.00217
Hodge, R. D., Bakken, T. E., Miller, J. A., Smith, K. A., Barkan, E. R., Graybuck, L. T., et al. (2019). Conserved Cell Types with Divergent Features in Human versus Mouse Cortex. Nature 573, 61–68. doi:10.1038/s41586-019-1506-7
Irie, F., Badie-Mahdavi, H., and Yamaguchi, Y. (2012). Autism-like Socio-Communicative Deficits and Stereotypies in Mice Lacking Heparan Sulfate. Proc. Natl. Acad. Sci. U.S.A. 109 (13), 5052–5056. doi:10.1073/pnas.1117881109
Kim, J., He, Y., and Park, H. (2014). Algorithms for Nonnegative Matrix and Tensor Factorizations: a Unified View Based on Block Coordinate Descent Framework. J. Glob. Optim 58 (2), 285–319. doi:10.1007/s10898-013-0035-4
Kitsak, M., Sharma, A., Menche, J., Guney, E., Ghiassian, S. D., Loscalzo, J., et al. (2016). Tissue Specificity of Human Disease Module. Sci. Rep. 6 (1), 35241. doi:10.1038/srep35241
Lun, A. T. L., McCarthy, D. J., and Marioni, J. C. (2016). A Step-by-step Workflow for Low-Level Analysis of Single-Cell RNA-Seq Data with Bioconductor. F1000Res 5, 2122. doi:10.12688/f1000research.9501.2
Mairal, J., Bach, F., Ponce, J., and Sapiro, G. (2010). Online Learning for Matrix Factorization and Sparse Coding. J. Machine Learn. Res. 11 (1), 19–60. doi:10.48550/arXiv.0908.0050
McCarthy, D. J., Campbell, K. R., Lun, A. T. L., and Wills, Q. F. (2017). Scater: Pre-processing, Quality Control, Normalization and Visualization of Single-Cell RNA-Seq Data in R. Bioinformatics 33 (8), btw777–1186. doi:10.1093/bioinformatics/btw777
Pérez, C., Sawmiller, D., and Tan, J. (2016). The Role of Heparan Sulfate Deficiency in Autistic Phenotype: Potential Involvement of Slit/Robo/srGAPs-Mediated Dendritic Spine Formation. Neural Dev. 11, 11. doi:10.1186/s13064-016-0066-x
Rupert, G. (2012). Simultaneous Statistical Inference. New York, NY: Springer Science & Business Media.
Rylaarsdam, L., and Guemez-Gamboa, A. (2019). Genetic Causes and Modifiers of Autism Spectrum Disorder. Front. Cel. Neurosci. 13, 385. doi:10.3389/fncel.2019.00385
Welch, J. D., Kozareva, V., Ferreira, A., Vanderburg, C., Martin, C., and Macosko, E. Z. (2019). Single-Cell Multi-Omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177 (7), 1873. doi:10.1016/j.cell.2019.05.006
Yang, Z., and Michailidis, G. (2016). A Non-negative Matrix Factorization Method for Detecting Modules in Heterogeneous Omics Multi-Modal Data. Bioinformatics 32 (1), btv544. doi:10.1093/bioinformatics/btv544
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Keywords: ASD, cell-type-specific gene module, shared gene module, gene function, integrative regularized non-negative matrix factorization
Citation: Guan J, Zhuang Y, Kang Y and Ji G (2022) Shared and Cell-Type-Specific Gene Expression Patterns Associated With Autism Revealed by Integrative Regularized Non-Negative Matrix Factorization. Front. Genet. 13:865371. doi: 10.3389/fgene.2022.865371
Received: 29 January 2022; Accepted: 11 April 2022;
Published: 11 May 2022.
Edited by:
Kaifang Pang, Baylor College of Medicine, United StatesReviewed by:
Flavia Esposito, University of Bari Aldo Moro, ItalyCopyright © 2022 Guan, Zhuang, Kang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinting Guan, anRndWFuQHhtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.