
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 04 May 2022
Sec. RNA
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.862642
This article is part of the Research Topic Genetic and epigenetic aspects of non-coding RNAs in physiology and disease View all 11 articles
Over many years, research on HIV/AIDS has advanced with the introduction of HAART. Despite these advancements, significant gaps remain with respect to aspects in HIV life cycle, with specific attention to virus-host interactions. Investigating virus-host interactions may lead to the implementation of novel therapeutic strategies against HIV/AIDS. Notably, host gene silencing can be facilitated by cellular small non-coding RNAs such as microRNAs paving the way for epigenetic anti-viral therapies. Numerous studies have elucidated the importance of microRNAs in HIV pathogenesis. Some microRNAs can either promote viral infection, while others can be detrimental to viral replication. This is accomplished by targeting the HIV-proviral genome or by regulating host genes required for viral replication and immune responses. In this review, we report on 1) the direct association of microRNAs with HIV infection; 2) the indirect association of known human genetic factors with HIV infection; 3) the regulation of human genes by microRNAs in other diseases that can be explored experimentally to determine their effect on HIV-1 infection; and 4) therapeutic interactions of microRNA against HIV infection.
The Human Immunodeficiency Virus (HIV) is a member of the lentivirus family of retroviruses that infects humans and increases susceptibility to Acquired Immunodeficiency Syndrome (AIDS). At the end of 2020, more than 38 million people were living with HIV globally (Global, 2020). While an effective vaccine remains elusive, extensive research on the inhibition of various stages of the HIV life cycle has paved the way for the development of many antiretroviral drugs (Cohen et al., 2016). Despite the progress with lifesaving, highly active antiretroviral therapy (HAART), treatment may lead to the development of drug toxicities and resistance (Pomerantz and Horn, 2003). HAART has also been implicated in the onset of adverse metabolic effects such as dyslipidaemia, elevated blood pressure, and insulin resistance (Palios et al., 2011). These compounding factors emphasise the necessity for new less toxic, more effective and additional, complementary therapeutic approaches.
Advancements in discovering and determining the function of host factors in viral biogenesis and transmission highlight the possibility of developing new therapeutic tools for preventative measures and treatment of HIV/AIDS (Hoxie and June, 2012). As such, modulating gene expression post-transcriptionally using small non-coding RNAs (sncRNAs) mediates cellular gene silencing through RNA interference (RNAi). This mode of regulation has become increasingly utilized in the development and delivery of the therapeutic anti-viral strategy (Balasubramaniam et al., 2018). Eukaryotic cells possess endogenous RNAi mechanisms, of which microRNAs (miRNAs) are the most significant family of sncRNAs (Ghildiyal and Zamore, 2009). MiRNAs are a class of small non-coding RNA molecules (21–25 nucleotides in length) that are instrumental in regulating gene expression of multiple cellular processes, including differentiation, development, apoptosis, and stress response (Felekkis et al., 2010). These molecules exert their regulatory mechanisms by mRNA degradation or translational repression (prevention of translation of target mRNAs) (Cai et al., 2009; Fabian et al., 2010; Inui et al., 2010; Subramanian and Steer, 2010). The biogenesis of miRNAs is detailed profoundly in several manuscripts, which describe the two principal pathways (canonical and non-canonical) (O'Brien et al., 2018; Ha and Kim, 2014; Macfarlane and R. Murphy, 2010; Zhao et al., 2019).
Briefly, the canonical pathway begins in the nucleus where a primary RNA (pri-miRNA), usually ∼80 nucleotides long, is transcribed from its specific gene by RNA polymerase II. The pri-miRNA is then cleaved to form a precursor miRNA (pre-miRNA), generally ∼60 nucleotides long, by the Microprocessor complex (Zhao et al., 2019). The Multiprocessor complex consists of two multiprotein units. The first is a large multiprotein unit. The second is a small multiprotein which constitutes of Drosha (RNase III enzyme) and the RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8) (Gregory et al., 2004). Once the pre-miRNA is generated, it is transported to the cytoplasm by exportin-5 and Ran-GTP, where it undergoes cleavage by Dicer (O'Brien et al., 2018). The Dicer enzyme removes the terminal loop, thus resulting in a double-stranded product that consists of the mature miRNA guide strand and a passenger strand. The mature miRNA product will be transferred onto Argonaute (AGO) protein (Macfarlane and R. Murphy, 2010). The remaining passenger strands are usually directed toward degradation. However, the guide strand is further integrated into the RNA-induced silencing complex (RISC) (O'Brien et al., 2018; Macfarlane and R. Murphy, 2010). Finally, the RISC-miRNA complex principally binds to the 3′UTR of the target mRNA. The complementarity of this binding predicts the fate of the mRNA, such that, in the event of perfect complementarity, the target mRNA is degraded. However, when this binding is incomplete, the mRNA is translationally repressed (Cai et al., 2009).
Several non-canonical pathways have been described (Annese et al., 2020). In summary, non-canonical pathways are classified into Drosha/DGCR8-independent and Dicer-independent pathways. The class of Drosha/DGCR8-independent miRNAs which originate from spliced introns are commonly known as mirtrons. These miRNAs are instantly transported to the cytoplasm via Dicer processing (Treiber et al., 2019). On the contrary, Dicer-independent miRNAs are uncommon. Drosha processes Dicer-independent miRNAs from endogenous short hairpin RNA (shRNA) transcripts, directly recognised by Ago proteins, thus making them Dicer-independent (Dai et al., 2019).
Multiple studies have linked aberrant miRNA profiles to diseases such as cancer (Croce and Calin, 2005; Calin and Croce, 2006), neurodegenerative disease (Kim et al., 2007; Wang et al., 2008), autoimmune disease (Dai et al., 2007; Stanczyk et al., 2008; Zhao et al., 2010), inflammatory diseases (Sonkoly et al., 2007), muscular disorders (Eisenberg et al., 2007), cardiovascular disorders (Carè et al., 2007; Ikeda et al., 2007), in addition to developmental abnormalities and psychiatric disorders (Lewis et al., 2003). Moreover, the five biggest infectious killers globally, including HIV/AIDS, are responsible for approximately 80% of the total contagious disease burden. About 12 million people per year succumb to these diseases, primarily in developing countries (Organization, 2020). Comparable to non-infectious conditions, miRNAs affect host and virus interactions in various ways. They are characterised as direct alteration of viral replication by influencing viral susceptibility or as indirect alteration of host genes that influence viral replication (Scaria et al., 2007; Kumar and Jeang, 2008).
MiRNAs have previously been implicated in HIV infection (Sun et al., 2016; Balasubramaniam et al., 2018; Su et al., 2018). As a field in its infancy, there is a substantial benefit in determining the impact of miRNAs on HIV infection.
This review discusses the direct alterations of miRNAs in HIV infection and the indirect alterations of known human genetic factors in HIV infection. Thereafter, we describe miRNA associations of known human genetic factors with other diseases that can be exploited to determine their specific effect on HIV infection, and the potential use of miRNAs as therapeutic interactions against HIV infection.
MiRNAs can aid or obstruct HIV infection at various stages of the viral life cycle, affecting viral replication, host immune response, and ultimately disease management (Figure 1). HIV exploits and uses cellular miRNAs to modulate its replication by directly targeting its RNA or host mRNAs that would negatively impact HIV replication. In addition, miRNAs are linked with a possible susceptibility to HIV infection in monocytes and macrophages (Wang et al., 2009; Qiuling et al., 2018). Furthermore, the viral genome may produce viral encoded miRNAs that modulate viral RNAs as well as cellular mRNAs (Cullen, 2006; Skalsky and Cullen, 2010). This suggests that HIV could potentially regulate its replication cycle and possibly program its own latency (Omoto et al., 2004; Bennasser et al., 2006; Ouellet et al., 2013; Zhang et al., 2014). Several cellular miRNAs have demonstrated the ability to modulate HIV infection, either directly or indirectly (Table 1).
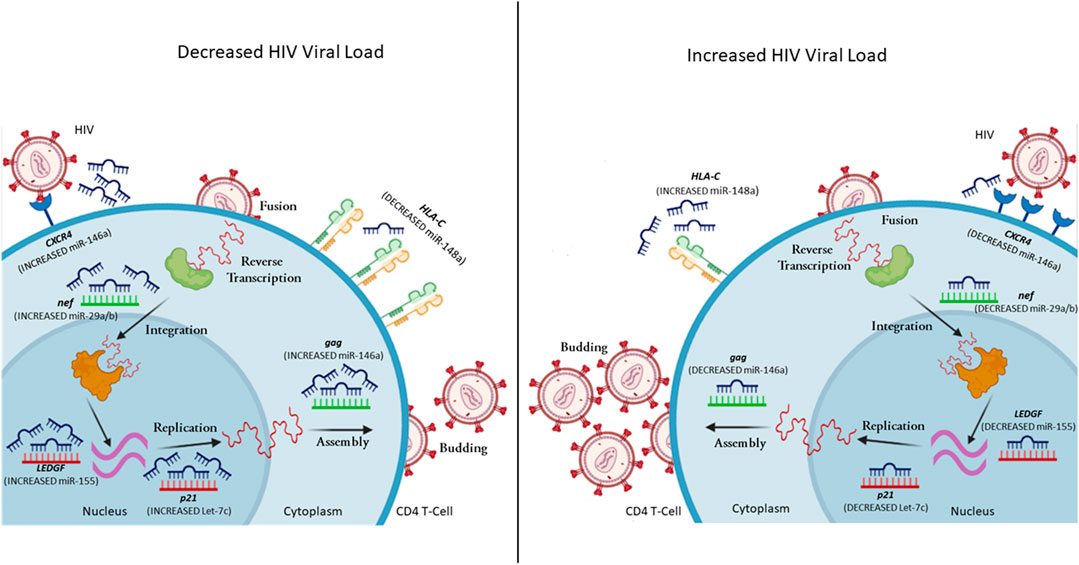
FIGURE 1. A representation of selected miRNAs that control gene expression levels, leading to variability in HIV viral load. MiRNA can regulate both host (red mRNA) and viral (green mRNA) mRNA. In the case of decreased viral load, the CD4+ T cell has increased expression of miR-146a (reduces CXCR4 and gag expression), miR-29a/b (reduces nef expression), miR-155 (reduces LEDGF expression), Let-7c (reduces p21 expression), while decreased expression of miR-148a upregulates HLA-C expression. In the case of increased viral load, the CD4+ T cell has decreased expression of miR-146a (increases CXCR4 and gag expression), miR-29a/b (increases nef expression), miR-155 (increases LEDGF expression), Let-7c (increases p21 expression), while increased expression of miR-148a down-regulates HLA-C expression (complied using BioRender).
Host derived miRNAs can bind to HIV RNA, directly regulating pathogenesis (Trobaugh and Klimstra, 2017). For instance, recent data has shown that miR-139-5p plays a role in activating latent HIV infected cells, by regulating FOX01, as well as FOS and JUN transcription factors (Okoye et al., 2021). The expression of miR-28, miR-125b, miR-150, miR-223, and miR-382 were significantly lower in activated CD4+ T cells in comparison to its resting counterpart. The same group of miRNAs may play a role in establishing viral latency by interacting with a conserved 1.2 kb fragment found in the 3′UTR of all HIV transcripts. These miRNAs can inhibit the translation of all viral proteins with the exception of nef (Huang et al., 2007). Moreover, the study showed that infected cells with established latency could be reactivated by treatment with miRNA inhibitors, suggesting that cellular miRNAs may provide a mechanistic effect towards HIV latency (Huang et al., 2007). Besides their role in promoting HIV latency, these five miRNAs play a crucial role in preventing HIV infection of monocytes and monocyte-derived macrophages (MDM). MiR-28, miR-125b, miR-150, miR-223, and miR-382 were observed at significantly higher levels in monocytes compared to MDM. These miRNAs were found to impede HIV reverse transcriptase activity in both cell types. However, the activity of HIV reverse transcriptase was dependant on the level of these miRNAs. This may explain why monocyte differentiation into macrophages is required for effective HIV infection (Wang et al., 2009).
Nef expression can also be influenced by cellular miRNAs (Ahluwalia et al., 2008; Sun et al., 2012). Ahluwalia et al. found that miR-29a and miR-29b may target HIV nef expression, which resulted in repression of nef translation and subsequent decrease in viral load (Figure 1) (Ahluwalia et al., 2008).
Moreover, in a series of refined experiments, Sun et al. demonstrated a new regulatory circuit during HIV infection (Sun et al., 2012). The downregulation of the miR-29 family could be associated with nef up-regulation and apoptosis of CD4+ cells (Sun et al., 2012). In addition, previous studies showed that miR-29 inhibited HIV replication by approximately 60%, while miR-133b, miR-138, miR-326, miR-149, and miR-92a reduced HIV viral replication by 40% (Houzet et al., 2012). In silico screening showed that these miRNAs may possibly target the 5′LTR (miR-326), env (miR-133b, miR-138), gag (miR-149), and pol (miR-92a) leading to the repression of viral replication.
Recent work by Chen et al. showed another form of miRNA regulation of HIV viruses through the interaction of miR-146a with the viral protein gag (Figure 1) (Chen et al., 2014). This interaction resulted in a viral-RNA-mediated gag assembly blockage, thereby interfering with viral budding and infectivity (Chen et al., 2014). These findings illustrate that miRNAs can alter viral gene expression via direct targeting of HIV mRNAs, with variable mechanisms of action dictated by the cell types.
MiRNAs regulate HIV infection through indirect modulation of host factor expression. One viral-dependent factor in cells is Cyclin T1, characterised as an essential part of the PTEFb complex, responsible for facilitating viral transcription (Hoque et al., 2011). The direct modulation is facilitated through the interaction with tat, which recruits the complex to HIV TAR, thereby impacting viral latency (Hoque et al., 2011). Recent work by Sung et al. described that miR-198targets and down-regulates Cyclin T1 mRNA and protein expression, which subsequently impairs the tat-mediated transcriptional activation of HIV in infected monocytes and macrophages (Sung and Rice, 2009). Over-expression of miR-198 inhibited HIV replication in macrophages, suggesting that cell type-specific mechanisms may be an effect executed by miRNAs (Sung and Rice, 2009). Additional studies identified that Cyclin T1 inhibition is exerted by cellular miRNAs (miR-27b, miR-29b, miR-150, and miR-223) in resting CD4+ T cells (Chiang et al., 2012). However, CD4+ T cell activation followed the downregulation of the miRNAs. This result was correlated with enhanced HIV susceptibility and productive replication (Chiang et al., 2012).
The viral protein tat is an essential transcriptional activator that interacts with several cellular proteins. For efficient HIV transcriptional activation, tat must be acetylated by p300-CREB binding protein associated factor (PCAF) (D'Orso and Frankel, 2009). Remarkably, miR-17/92 family of host miRNAs impedes HIV infection by downregulating PCAF (Triboulet et al., 2007). Triboulet et al. also showed that miR-17 as well as miR-20a inhibited PCAF expression at the mRNA and protein levels. In addition, HIV can actively repress miR-17-5p and miR-20a to enhance viral translation through p300/PCAF-dependant tat activation (Triboulet et al., 2007).
Another well characterised cellular factor that interacts with HIV tat to up-regulate viral transcription is the purine-rich element binding protein α (Pur-α) (Wortman et al., 2000). A collection of six cellular miRNAs (miR-15a, miR-15b, miR-16, miR-20a, miR-93, and miR-106b) enriched in monocytes were linked with the repression of Pur-α (Shen et al., 2012). Consequently, inhibition of these miRNAs in monocytes increased the expression of Pur-α, resulting in an increase in HIV infection (Shen et al., 2012).
MiR-155 has demonstrated significant effects on HIV infection through a Toll-Like receptor (TLR)-dependant mechanism (Swaminathan et al., 2012a). Swaminathan et al. showed that miR-155 is significantly up-regulated in MDMs, stimulated by TLR3 and TLR4 (Swaminathan et al., 2012a). Furthermore, up-regulation of miR-155 through TLR stimulation leads to decreased mRNA and protein expression of ADAM10, TNPO3, NUP153, and LEDGF/p75, in MDMs (Swaminathan et al., 2012a). Gene silencing of LEDGF had the most significant effect on HIV infection (Figure 1) (Swaminathan et al., 2012a). However, co-silencing of both LEDGF and ADAM10 had a more substantial impact, impairing the transport of viral pre-integration complexes (Swaminathan et al., 2012a).
The inhibition of TRIM32 by miR-155 results in post-integration latency of HIV (Ruelas et al., 2015). TRIM32 directs NF-κB to the nucleus via a tat-independent mechanism, as described by Ruelas et al. (2015). The study characterises a novel mechanism by which TRIM32 activates NF-κB. Collectively, the inhibitory effect of miR-155 on TRIM32 highlights a new tool for HIV remaining in infected reservoirs (Ruelas et al., 2015). Despite this significant study, recent studies have identified miR-155 as a potent biomarker of activated T cells and immune dysfunction in HIV-infected individuals (Jin et al., 2017a; Jin et al., 2017b; Zhang et al., 2021a).
MiRNAs can also restrict viral entry by targeting the receptors and co-receptors exploited for HIV entry. Orecchini et al. report a tat-dependant mechanism that controls CD4 receptor by up-regulating miR-222 (Orecchini et al., 2014). In addition, Lodge et al. demonstrated that miR-221 and miR-222 are up-regulated in MDMs, targeting the 3′ UTR of CD4 (Orecchini et al., 2014). The mRNA and subsequent protein expression are reduced, ultimately impairing HIV entry into MDM (Lodge et al., 2017). Labbaye et al. showed that promyelocytic leukaemia zinc finger (PLZF) could regulate miR-146a, subsequently controlling the expression of CXCR4 in vitro (Labbaye et al., 2008). Activation of resting CD4+ T cells by phytohemagglutinin results in the downregulation of miR-146a (Quaranta et al., 2015). Downregulation of miR-146a results in the overexpression of CXCR4 co-receptor promoting viral entry in CD4+ T cells (Quaranta et al., 2015).
Vpr HIV-binding protein (vprBP) is a cellular cofactor that forms part of a ubiquitin protein ligase complex. VprBP promotes HIV infection (Ma et al., 2014). Ma et al. demonstrated that miR-1236 inhibitors increased translation of vprBP in monocytes, thus facilitating HIV infection. Contrary to monocytes, miR-1236 mimics in monocyte-derived dendritic cells had supressed vprBP, which was complemented by decreased infection (Ma et al., 2014).
High surface expression of human leukocyte antigen C (HLA-C) greatly corresponded with slower disease progression via superior control of HIV viremia. Several genetic variants have been shown to disrupt miR-148a regulation of HLA-C (Kulkarni et al., 2011; Blais et al., 2012; Kulkarni et al., 2013). Disruption of miRNA binding site allows high expressing HLA-C alleles to escape miR-148a regulation (Kulkarni et al., 2011). HLA-C alleles that do not have a disrupted miR-148a binding site are tightly regulated by miR-148a and are expressed at low levels. The polymorphisms affecting HLA-C expression through disrupted miR-148a binding are rs9264942, rs67384697, and rs735316, with the variants of rs9264942 and rs67384697 being in linkage disequilibrium (Kulkarni et al., 2011; Blais et al., 2012; Kulkarni et al., 2013). All three variants are associated with control and progression of HIV infection by miR-148a-mediated post-transcriptional regulation of HLA-C.
IL-10 is a multifunctional anti-inflammatory cytokine produced by various immune cells. With regards to miRNA regulation of IL-10, the let-7 family can directly target IL10. In vitro infection with HIV elevated IL10 levels through the reduction of let-7. In addition, CD4+ T cells of chronically infected HIV-positive individuals had significantly lower let-7 levels than uninfected individuals and long-term non-progressors. (Swaminathan et al., 2012b). A single miRNA is able to regulate multiple target genes. In addition to IL10, let-7c is involved in the regulation of p21. let-7c overexpression in Jurkat cells resulted in a 1.38-fold change in p21 expression (Figure 1) (Farberov et al., 2015).
B lymphocyte-induced maturation protein-1 (Blimp-1) is a transcriptional repressor of IL-2 (an important cytokine required for T cell growth and survival). In HIV-infected individuals, BLIMP-1 may contribute to T cell dysregulation through alterations in IL-2 levels. MiR-9 inhibited BLIMP1 expression in CD4+ T cells. Chronically infected HIV-positive patients had lower miR-9 and higher BLIMP1 expression in comparison to uninfected healthy individuals and long-term non-progressors (Seddiki et al., 2013).
It is estimated that 1,254 human genes are involved in viral replication. Genome-wide RNA interference has enabled researchers to identify multiple host factors that are involved in HIV life cycle. This large array of host gene targets may be essential in the development of new therapeutic strategies against HIV. By identifying and understanding the mechanisms behind the associations of specific miRNAs and their targets, we can exploit these factors for HIV viral control. Several HIV-associated genes are shown to be under the regulation of miRNAs in other diseases.
Blocking the access of HIV into host cells is the first step in preventing the HIV proviral genome from integrating into the host’s genome. The human chemokine receptor 5 (CCR5) plays an important role in the internalization of HIV into the host cell (Lederman et al., 2006). Individuals with the 32 base pair deletion in their CCR5 gene are known to be resistant to HIV as they have lower levels of CCR5 on the surface of their CD4+ T cells. Thus, the regulation of CCR5 expression may be essential in inhibiting HIV replication. Che et al. found that miR-107 binds to the 3′UTR of CCR5 (Che et al., 2016). CCR5 proteins and gene expression were found to be significantly lower in the presence of miR-107 (Che et al., 2016). Since CCR5 is important in the HIV context, miR-107 may be of potential therapeutic value in preventing HIV infection.
Intercellular adhesion molecule 1 (ICAM-1) also plays a significant role in HIV entry. The binding of ICAM-1 with LFA-1 on the cell surface facilitates viral infectivity. ICAM-1 increases viral infectivity by directly inserting into mature HIV virions (Fortin et al., 1997; Bounou et al., 2002). Lui et al. demonstrated that ICAM1 is negatively regulated by miR-296-3p in the malignant highly metastatic M12 cell line (Liu et al., 2013). Furthermore, in prostate cancer cells there is a negative correlation between miR-296-3p and ICAM1 (Liu et al., 2013). In the context of HIV, the downregulation of ICAM1 by miR-296-3p would reduce the rate of infectivity (Liu et al., 2013).
The tripartite motif (TRIM) proteins are a family of E3 ubiquitin ligases with diverse anti-viral functions (van Gent et al., 2018). TRIM22, TRIM11, and KAP1 (TRIM28) were previously shown to have anti-HIV activity (Barr et al., 2008; Allouch et al., 2009; Yuan et al., 2016). TRIM22 inhibits the processing of viral particles and viral budding through the ubiquitylation in HIV. TRIM22 also has anti-Hepatitis C virus (HCV) activity. Tian et al. confirmed that TRIM22 was regulated by miR-215 (Tian and He, 2018). In Con1b cells, the overexpression of miR-215 facilitated HCV replication by downregulating TRIM22. Knockdown of miR-215 suppressed HCV replication through the increased expression of TRIM22 in Huh7.5.1 cells (Tian and He, 2018). In colon cancer, TRIM11 is negatively regulated by miR-24-3p, promoting cellular proliferation and inhibiting apoptosis (Yin et al., 2016). Likewise, Qi et al. demonstrated that miR-491 levels inversely corresponded with TRIM28 expression in glioblastoma multiforme (GBM) (Qi et al., 2016). Their data showed that miR-491 was reduced in GBM and indicated that the low levels of miR-491 are associated with poor prognosis (Qi et al., 2016). miR-491 inhibited TRIM28 translation in GBM cells (Qi et al., 2016).
Studies have also demonstrated a link between RAD51 expression and HIV disease (Chipitsyna et al., 2004; Cosnefroy et al., 2012; Kaminski et al., 2014; Thierry et al., 2015). Elevated expression of RAD51 promotes HIV-1 transcription (Kaminski et al., 2014). Evidence demonstrates that RAD51 may have stimulatory or inhibitory effects on specific steps of retroviral replication cycles (Thierry et al., 2015). These effects depend on RAD51 being able to recruit both transcription machinery and proteins implicated in chromatin remodelling and formulation of RAD51 stimulatory compound (Thierry et al., 2015). Findings from Gasparini et al. indicate that DNA repair is indirectly regulated by miR-155 through its interaction with RAD51 in breast cancer (Gasparini et al., 2014).
The regulation of several other HIV-associated host factors such as TRAF6, CCL4, CCL3, IRF7, RSAD2, ISG15, TLR3, SETDB1, and Rab27a by miRNAs could potentially play a role in HIV infection. Table 2 provides a list of HIV-associated host genes which should be investigated in future miRNA studies. The host’s genes and associated miRNAs described in Table 2 may provide novel therapeutic targets against HIV.
Extensive research has paved the way for developing multiple antiretroviral drugs targeting specific phases of the viral life cycle, leading to a combination of antiretroviral therapy (cART). Currently, this treatment results in controlled viral replication in many treated individuals (Cohen et al., 2016). Despite the progress with lifesaving HAART, infection with HIV remains pathogenic and incurable. In addition, these drugs lead to the development of toxicities and adverse side effects which may only be combated by changing the drug regimen. Furthermore, the increasing emergence of HIV drug resistance poses a threat to the success of the current regimens (Bertagnolio et al., 2012; Le Douce et al., 2012; Stadeli and Richman, 2013). These compounding factors highlight the importance of identifying novel and complementary treatment regimens.
RNA-based therapeutics appear ready to deliver on their promise. Significant success has been observed in several clinical trials using potential miRNA drugs in multiple infectious and non-infectious diseases, including cancer (Hatley et al., 2010; Li et al., 2010; Steele et al., 2011; Wong et al., 2012; Yamanaka et al., 2012), hepatitis C (Jopling et al., 2005; Sarasin-Filipowicz et al., 2009; Lanford et al., 2010), heart abnormalities (Thum et al., 2008; Liu et al., 2010), kidney disease, pathologic fibrosis, and even keloid formation. Interestingly, studies have also shown that dysregulated miRNA profiles play a role in HIV replication (Barr et al., 2008; Pincetic et al., 2010; Liu et al., 2011; Sirois et al., 2011; Tyagi and Kashanchi, 2012; Raposo et al., 2013; Doyle et al., 2015). The vaccine, iHIVARNA is a combination of mRNA sequences that serve as an HIV immunogen. In the first round of clinical trials, iHIVARNA is tolerated in HIV-infected patients on chronic cART (De Jong et al., 2019). Despite this progress, the application of miRNAs as diagnostic and interventional medicine remains an underexplored area of research. The clinical trial was merely a proof-of-concept trial; the stability and delivery of the mRNA are still being tested (De Jong et al., 2019).
The Achilles heel of miRNA-based viral therapy is the lack of targeted miRNA delivery systems, off-target effects, and unidentified targets of miRNAs. In addition, miRNAs are relatively unstable, which may result in insufficient circulation and poor half-life of the miRNA-based therapy. Future research should be directed towards constructing optimal miRNA delivery systems and identifying methods to prevent off-target effects. As the use of miRNAs as treatment strategies is a growing field, only a few drugs have been FDA approved (Nature Biotechnology, 2020; Zhang et al., 2021b), which highlights the potential of RNAs for therapeutic intervention. MiRNAs provide a unique, reversible approach to treating human diseases and may be our secret weapon in our fight against HIV.
MiRNAs play a significant role in regulating gene expression. While the role of miRNAs in diseases such as cancer has been thoroughly investigated, the interplay between miRNAs and HIV infection has only begun to emerge. MiRNAs have emerged as key contributors to immune dysfunction observed in HIV disease. As research develops in specific subsets and more targeted populations, the understanding of this field matures as more can be uncovered. Considering that key genes involved in the HIV life cycle are affected by differentially expressed miRNAs, there is a link between the host’s RNA interference machinery and HIV pathogenicity. Future research should focus on identifying differentially expressed miRNAs in HIV-infected donors from different population groups., which may be exploited for therapeutic benefit.
In addition, the application of specific miRNA mimics and inhibitors (Andorfer et al., 2011; He et al., 2012; De Santa et al., 2013) is an appealing avenue for future investigations. Noting that one miRNA alone may be able to target several host genetic factors, the combined effect of several miRNAs together offers the potential for a multi-targeted effect. This treatment strategy can complement current cART regimen. Furthermore, inhibition of selected miRNAs is advantageous. For instance, selectively blocking miRNAs that target anti-viral proteins or pathways could potentially enhance anti-viral responses. This approach is efficient during the onset of infection, as the anti-viral response to HIV can be improved.
Conceptualization and conceiving of idea, VR. Additional input with regards to conceptualization, RC. Writing, RC, TA, TAR, and LN. Research, RC, TA, TAR, and LN. Figure design, TA and VR. Editing of manuscript, VR. All authors contributed to the article and approved the submitted version.
VR was funded as a FLAIR Research Fellow (the Future Leader in African Independent Research (FLAIR) Fellowship Programme was a partnership between the African Academy of Sciences (AAS) and the Royal Society that was funded by the UK Government as part of the Global Challenge Research Fund (GCRF) Grant # FLAIR-FLR\R1\190204); supported by the South African Medical Research Council (SAMRC) with funds from the Department of Science and Technology (DST); and VR was also supported in part through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative (Grant # DEL-15-006) by the AAS. TA is funded by the Poliomyelitis Research Foundation (PRF) Grant # 21/49.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
In Memoriam of the late author Romona Chinniah.
Ahluwalia, J. K., Khan, S. Z., Soni, K., Rawat, P., Gupta, A., Hariharan, M., et al. (2008). Human Cellular microRNA Hsa-miR-29a Interferes with Viral Nef Protein Expression and HIV-1 Replication. Retrovirology 5, 117. doi:10.1186/1742-4690-5-117
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen Recognition and Innate Immunity. Cell 124 (4), 783–801. doi:10.1016/j.cell.2006.02.015
Allouch, A., et al. (2009). HIV-1 Acetylated Integrase Is Targeted by KAP1 (TRIM28) to Inhibit Viral Integration. Retrovirology 6 (Suppl. 2), P2. doi:10.1186/1742-4690-6-s2-p2
Amaral, A. J., Andrade, J., Foxall, R. B., Matoso, P., Matos, A. M., Soares, R. S., et al. (2017). Mi RNA Profiling of Human Naive CD 4 T Cells Links miR‐34c‐5p to Cell Activation and HIV Replication. Embo j 36 (3), 346–360. doi:10.15252/embj.201694335
Andorfer, C. A., Necela, B. M., Thompson, E. A., and Perez, E. A. (2011). MicroRNA Signatures: Clinical Biomarkers for the Diagnosis and Treatment of Breast Cancer. Trends Mol. Med. 17 (6), 313–319. doi:10.1016/j.molmed.2011.01.006
Annese, T., Tamma, R., De Giorgis, M., and Ribatti, D. (2020). microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 10, 581007. doi:10.3389/fonc.2020.581007
Balasubramaniam, M., Pandhare, J., and Dash, C. (2018). Are microRNAs Important Players in HIV-1 Infection? an Update. Viruses 10 (3), 110. doi:10.3390/v10030110
Barr, S. D., Smiley, J. R., and Bushman, F. D. (2008). The Interferon Response Inhibits HIV Particle Production by Induction of TRIM22. Plos Pathog. 4 (2), e1000007-e1000007. doi:10.1371/journal.ppat.1000007
Bennasser, Y., Bennasser, Y., Le, S. Y., Yeung, M. L., and Jeang, K. T. (2006). MicroRNAs in Human Immunodeficiency Virus-1 Infection. Methods Mol Biol342, 241–253. doi:10.1385/1-59745-123-1:241
Bertagnolio, S., De Luca, A., Vitoria, M., Essajee, S., Penazzato, M., Hong, S. Y., et al. (2012). Determinants of HIV Drug Resistance and Public Health Implications in Low- and Middle-Income Countries. Antivir. Ther. 17 (6), 941–953. doi:10.3851/imp2320
Blais, M.-E., Zhang, Y., Rostron, T., Griffin, H., Taylor, S., Xu, K., et al. (2012). High Frequency of HIV Mutations Associated with HLA-C Suggests Enhanced HLA-C-Restricted CTL Selective Pressure Associated with an AIDS-Protective Polymorphism. J.I. 188 (9), 4663–4670. doi:10.4049/jimmunol.1103472
Blanpain, C., Libert, F., Vassart, G., and Parmentier, M. (2002). CCR5 and HIV Infection. Receptors and Channels 8 (1), 19–31. doi:10.3109/10606820212135
Bounou, S., Leclerc, J. E., and Tremblay, M. J. (2002). Presence of Host ICAM-1 in Laboratory and Clinical Strains of Human Immunodeficiency Virus Type 1 Increases Virus Infectivity and CD4 + -T-Cell Depletion in Human Lymphoid Tissue, a Major Site of Replication In Vivo. J. Virol. 76 (3), 1004–1014. doi:10.1128/jvi.76.3.1004-1014.2002
Cai, Y., Yu, X., Hu, S., and Yu, J. (2009). A Brief Review on the Mechanisms of miRNA Regulation. Genomics, proteomics & bioinformatics 7 (4), 147–154. doi:10.1016/s1672-0229(08)60044-3
Calin, G. A., and Croce, C. M. (2006). MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 6 (11), 857–866. doi:10.1038/nrc1997
Carè, A., Catalucci, D., Felicetti, F., Bonci, D., Addario, A., Gallo, P., et al. (2007). MicroRNA-133 Controls Cardiac Hypertrophy. Nat. Med. 13 (5), 613–618. doi:10.1038/nm1582
Carrol, E. D., Mankhambo, L. A., Balmer, P., Nkhoma, S., Banda, D. L., Guiver, M., et al. (1999). Chemokine Responses Are Increased in HIV-Infected Malawian Children with Invasive Pneumococcal Disease. J. Acquir Immune Defic Syndr. 44 (4), 443–450. doi:10.1097/QAI.0b013e31802f8390
Chang, C., Liu, T., Huang, Y., Qin, W., Yang, H., and Chen, J. (2017). MicroRNA-134-3p Is a Novel Potential Inhibitor of Human Ovarian Cancer Stem Cells by Targeting RAB27A. Gene 605, 99–107. doi:10.1016/j.gene.2016.12.030
Che, L.-F., Shao, S.-F., and Wang, L.-X. (2016). Downregulation of CCR5 Inhibits the Proliferation and Invasion of Cervical Cancer Cells and Is Regulated by microRNA-107. Exp. Ther. Med. 11 (2), 503–509. doi:10.3892/etm.2015.2911
Chen, A. K., Sengupta, P., Waki, K., Van Engelenburg, S. B., Ochiya, T., Ablan, S. D., et al. (2014). MicroRNA Binding to the HIV-1 Gag Protein Inhibits Gag Assembly and Virus Production. Proc. Natl. Acad. Sci. U S A. 111 (26), E2676–E2683. doi:10.1073/pnas.1408037111
Chen, X.-Y., Zhang, H.-S., Wu, T.-C., Sang, W.-W., and Ruan, Z. (2013). Down-regulation of NAMPT Expression by miR-182 Is Involved in Tat-Induced HIV-1 Long Terminal Repeat (LTR) Transactivation. Int. J. Biochem. Cel Biol. 45 (2), 292–298. doi:10.1016/j.biocel.2012.11.002
Cheng, N. L., Chen, X., Kim, J., Shi, A. H., Nguyen, C., Wersto, R., et al. (2015). MicroRNA‐125b Modulates Inflammatory Chemokine CCL4 Expression in Immune Cells and its Reduction Causes CCL4 Increase with Age. Aging cell 14 (2), 200–208. doi:10.1111/acel.12294
Chiang, K., Liu, H., and Rice, A. P. (2013). miR-132 Enhances HIV-1 Replication. Virology 438 (1), 1–4. doi:10.1016/j.virol.2012.12.016
Chiang, K., Sung, T.-L., and Rice, A. P. (2012). Regulation of Cyclin T1 and HIV-1 Replication by MicroRNAs in Resting CD4 + T Lymphocytes. J. Virol. 86 (6), 3244–3252. doi:10.1128/jvi.05065-11
Chipitsyna, G., Slonina, D., Siddiqui, K., Peruzzi, F., Skorski, T., Reiss, K., et al. (2004). HIV-1 Tat Increases Cell Survival in Response to Cisplatin by Stimulating Rad51 Gene Expression. Oncogene 23 (15), 2664–2671. doi:10.1038/sj.onc.1207417
Cohen, M. S., Chen, Y. Q., McCauley, M., Gamble, T., Hosseinipour, M. C., Kumarasamy, N., et al. (2016). Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N. Engl. J. Med. 375 (9), 830–839. doi:10.1056/NEJMoa1600693
Cosnefroy, O., Tocco, A., Lesbats, P., Thierry, S., Calmels, C., Wiktorowicz, T., et al. (2012). Stimulation of the Human RAD51 Nucleofilament Restricts HIV-1 Integration In Vitro and in Infected Cells. J. Virol. 86 (1), 513–526. doi:10.1128/jvi.05425-11
Croce, C. M., and Calin, G. A. (2005). miRNAs, Cancer, and Stem Cell Division. Cell 122 (1), 6–7. doi:10.1016/j.cell.2005.06.036
D'Orso, I., and Frankel, A. D. (2009). Tat Acetylation Modulates Assembly of a Viral-Host RNA-Protein Transcription Complex. Proc. Natl. Acad. Sci. U.S.A. 106 (9), 3101–3106. doi:10.1073/pnas.0900012106
Dai, X., Kaushik, A. C., and Zhang, J. (2019). The Emerging Role of Major Regulatory RNAs in Cancer Control. Front. Oncol. 9, 920. doi:10.3389/fonc.2019.00920
Dai, Y., Huang, Y.-S., Tang, M., Lv, T.-Y., Hu, C.-X., Tan, Y.-H., et al. (2007). Microarray Analysis of microRNA Expression in Peripheral Blood Cells of Systemic Lupus Erythematosus Patients. Lupus 16 (12), 939–946. doi:10.1177/0961203307084158
De Jong, W., Aerts, J., Allard, S., Brander, C., Buyze, J., Florence, E., et al. (2019). iHIVARNA Phase IIa, a Randomized, Placebo-Controlled, Double-Blinded Trial to Evaluate the Safety and Immunogenicity of iHIVARNA-01 in Chronically HIV-Infected Patients under Stable Combined Antiretroviral Therapy. Trials 20 (1), 361–410. doi:10.1186/s13063-019-3409-1
De Santa, F., Iosue, I., and Fazi, F. (2013). microRNA Biogenesis Pathway as a Therapeutic Target for Human Disease and Cancer. Curr. Pharm. Des. 19, 745–764. doi:10.2174/138161213804581846
Dorhoi, A., Iannaccone, M., Farinacci, M., Faé, K. C., Schreiber, J., Moura-Alves, P., et al. (2013). MicroRNA-223 Controls Susceptibility to Tuberculosis by Regulating Lung Neutrophil Recruitment. J. Clin. Invest. 123 (11), 4836–4848. doi:10.1172/jci67604
Doyle, T., Goujon, C., and Malim, M. H. (2015). HIV-1 and Interferons: Who's Interfering with Whom? Nat. Rev. Microbiol. 13 (7), 403–413. doi:10.1038/nrmicro3449
Eisenberg, I., Eran, A., Nishino, I., Moggio, M., Lamperti, C., Amato, A. A., et al. (2007). Distinctive Patterns of microRNA Expression in Primary Muscular Disorders. Proc. Natl. Acad. Sci. U.S.A. 104 (43), 17016–17021. doi:10.1073/pnas.0708115104
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi:10.1146/annurev-biochem-060308-103103
Farberov, L., Herzig, E., Modai, S., Isakov, O., Hizi, A., and Shomron, N. (2015). MicroRNA-mediated Regulation of P21 and TASK1 Cellular Restriction Factors Enhances HIV-1 Infection. J. Cel Sci 128 (8), 1607–1616. doi:10.1242/jcs.167817
Felekkis, K., Touvana, E., Stefanou, C. h., and Deltas, C. (2010). microRNAs: a Newly Described Class of Encoded Molecules that Play a Role in Health and Disease. Hippokratia 14 (4), 236–240.
Fortin, J. F., Cantin, R., Lamontagne, G., and Tremblay, M. (1997). Host-derived ICAM-1 Glycoproteins Incorporated on Human Immunodeficiency Virus Type 1 Are Biologically Active and Enhance Viral Infectivity. J. Virol. 71 (5), 3588–3596. doi:10.1128/jvi.71.5.3588-3596.1997
Gasparini, P., Lovat, F., Fassan, M., Casadei, L., Cascione, L., Jacob, N. K., et al. (2014). Protective Role of miR-155 in Breast Cancer through RAD51 Targeting Impairs Homologous Recombination after Irradiation. Proc. Natl. Acad. Sci. U.S.A. 111 (12), 4536–4541. doi:10.1073/pnas.1402604111
Gerber, P. P., Cabrini, M., Jancic, C., Paoletti, L., Banchio, C., von Bilderling, C., et al. (2015). Rab27a Controls HIV-1 Assembly by Regulating Plasma Membrane Levels of Phosphatidylinositol 4,5-bisphosphate. J. Cel. Biol. 209 (3), 435–452. doi:10.1083/jcb.201409082
Ghildiyal, M., and Zamore, P. D. (2009). Small Silencing RNAs: an Expanding Universe. Nat. Rev. Genet. 10 (2), 94–108. doi:10.1038/nrg2504
Global, U. N. A. I. D. S. (2020). HIV & AIDS Statistics — Fact Sheet. Available at: https://www.unaids.org/en/resources/fact-sheet.
Gregory, R. I., Yan, K.-p., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., et al. (2004). The Microprocessor Complex Mediates the Genesis of microRNAs. Nature 432 (7014), 235–240. doi:10.1038/nature03120
Ha, M., and Kim, V. N. (2014). Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cel Biol 15 (8), 509–524. doi:10.1038/nrm3838
Hatley, M. E., Patrick, D. M., Garcia, M. R., Richardson, J. A., Bassel-Duby, R., van Rooij, E., et al. (2010). Modulation of K-ras-dependent Lung Tumorigenesis by MicroRNA-21. Cancer cell 18 (3), 282–293. doi:10.1016/j.ccr.2010.08.013
He, M.-L., Luo, M. X.-M., Lin, M. C., and Kung, H.-f. (2012). MicroRNAs: Potential Diagnostic Markers and Therapeutic Targets for EBV-Associated Nasopharyngeal Carcinoma. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1825 (1), 1–10. doi:10.1016/j.bbcan.2011.09.001
Hoque, M., Shamanna, R. A., Guan, D., Pe'ery, T., and Mathews, M. B. (2011). HIV-1 Replication and Latency Are Regulated by Translational Control of Cyclin T1. J. Mol. Biol. 410 (5), 917–932. doi:10.1016/j.jmb.2011.03.060
Houzet, L., Klase, Z., Yeung, M. L., Wu, A., Le, S.-Y., Quiñones, M., et al. (2012). The Extent of Sequence Complementarity Correlates with the Potency of Cellular miRNA-Mediated Restriction of HIV-1. Nucleic Acids Res. 40 (22), 11684–11696. doi:10.1093/nar/gks912
Hoxie, J. A., and June, C. H. (2012). Novel Cell and Gene Therapies for HIV. Cold Spring Harbor Perspect. Med. 2 (10), a007179. doi:10.1101/cshperspect.a007179
Huang, J., Wang, F., Argyris, E., Chen, K., Liang, Z., Tian, H., et al. (2007). Cellular microRNAs Contribute to HIV-1 Latency in Resting Primary CD4+ T Lymphocytes. Nat. Med. 13 (10), 1241–1247. doi:10.1038/nm1639
Ikeda, S., Kong, S. W., Lu, J., Bisping, E., Zhang, H., Allen, P. D., et al. (2007). Altered microRNA Expression in Human Heart Disease. Physiol. genomics 31 (3), 367–373. doi:10.1152/physiolgenomics.00144.2007
Inui, M., Martello, G., and Piccolo, S. (2010). MicroRNA Control of Signal Transduction. Nat. Rev. Mol. Cel Biol 11 (4), 252–263. doi:10.1038/nrm2868
Jiang, C., Zhu, W., Xu, J., Wang, B., Hou, W., Zhang, R., et al. (2014). MicroRNA-26a Negatively Regulates Toll-like Receptor 3 Expression of Rat Macrophages and Ameliorates Pristane Induced Arthritis in Rats. Arthritis Res. Ther. 16 (1), R9. doi:10.1186/ar4435
Jin, C., Cheng, L., Höxtermann, S., Xie, T., Lu, X., Wu, H., et al. (2017). MicroRNA-155 Is a Biomarker of T-Cell Activation and Immune Dysfunction in HIV-1-Infected Patients. HIV Med. 18 (5), 354–362. doi:10.1111/hiv.12470
Jin, C., Cheng, L., Lu, X., Xie, T., Wu, H., and Wu, N. (2017). Elevated Expression of miR-155 Is Associated with the Differentiation of CD8+ T Cells in Patients with HIV-1. Mol. Med. Rep. 16 (2), 1584–1589. doi:10.3892/mmr.2017.6755
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M., and Sarnow, P. (2005). Modulation of Hepatitis C Virus RNA Abundance by a Liver-specific MicroRNA. science 309 (5740), 1577–1581. doi:10.1126/science.1113329
Kaminski, R., Wollebo, H. S., Datta, P. K., White, M. K., Amini, S., and Khalili, K. (2014). Interplay of Rad51 with NF-Κb Pathway Stimulates Expression of HIV-1. PLoS ONE 9, e98304. doi:10.1371/journal.pone.0098304
Kapoor, R., Arora, S., Ponia, S. S., Kumar, B., Maddika, S., and Banerjea, A. C. (2015). The miRNA miR-34a Enhances HIV-1 Replication by Targeting PNUTS/PPP1R10, Which Negatively Regulates HIV-1 Transcriptional Complex Formation. Biochem. J. 470 (3), 293–302. doi:10.1042/bj20150700
Kim, J., Inoue, K., Ishii, J., Vanti, W. B., Voronov, S. V., Murchison, E., et al. (2007). A MicroRNA Feedback Circuit in Midbrain Dopamine Neurons. Science 317 (5842), 1220–1224. doi:10.1126/science.1140481
Kulkarni, S., Qi, Y., O’hUigin, C., Pereyra, F., Ramsuran, V., McLaren, P., et al. (2013). Genetic Interplay between HLA-C and MIR148A in HIV Control and Crohn Disease. Proc. Natl. Acad. Sci. U.S.A. 110 (51), 20705–20710. doi:10.1073/pnas.1312237110
Kulkarni, S., Savan, R., Qi, Y., Gao, X., Yuki, Y., Bass, S. E., et al. (2011). Differential microRNA Regulation of HLA-C Expression and its Association with HIV Control. Nature 472 (7344), 495–498. doi:10.1038/nature09914
Kumar, A., and Jeang, K.-T. (2008). Insights into Cellular microRNAs and Human Immunodeficiency Virus Type 1 (HIV-1). J. Cel. Physiol. 216 (2), 327–331. doi:10.1002/jcp.21488
Labbaye, C., Spinello, I., Quaranta, M. T., Pelosi, E., Pasquini, L., Petrucci, E., et al. (2008). A Three-step Pathway Comprising PLZF/miR-146a/CXCR4 Controls Megakaryopoiesis. Nat. Cel Biol 10 (7), 788–801. doi:10.1038/ncb1741
Lanford, R. E., Hildebrandt-Eriksen, E. S., Petri, A., Persson, R., Lindow, M., Munk, M. E., et al. (2010). Therapeutic Silencing of microRNA-122 in Primates with Chronic Hepatitis C Virus Infection. Science 327 (5962), 198–201. doi:10.1126/science.1178178
Le Douce, V., Janossy, A., Hallay, H., Ali, S., Riclet, R., Rohr, O., et al. (2012). Achieving a Cure for HIV Infection: Do We Have Reasons to Be Optimistic? J. Antimicrob. Chemother. 67 (5), 1063–1074. doi:10.1093/jac/dkr599
Lederman, M. M., Penn-Nicholson, A., Cho, M., and Mosier, D. (2006). Biology of CCR5 and its Role in HIV Infection and Treatment. Jama 296 (7), 815–826. doi:10.1001/jama.296.7.815
Levine, A., et al. (2009). CCL3 Genotype and Current Depression Increase Risk of HIV-Associated Dementia. Neurobehav HIV Med. Vol. 1, 1–7. doi:10.2147/nbhiv.s6820
Lewis, B. P., Shih, I.-h., Jones-Rhoades, M. W., Bartel, D. P., and Burge, C. B. (2003). Prediction of Mammalian microRNA Targets. Cell 115 (7), 787–798. doi:10.1016/s0092-8674(03)01018-3
Li, Y., Zhu, X., Gu, J., Hu, H., Dong, D., Yao, J., et al. (2010). Anti-miR-21 Oligonucleotide Enhances Chemosensitivity of Leukemic HL60 Cells to Arabinosylcytosine by Inducing Apoptosis. Hematology 15 (4), 215–221. doi:10.1179/102453310x12647083620840
Li, Z., Yin, H., Hao, S., Wang, L., Gao, J., Tan, X., et al. (2016). miR-200 Family Promotes Podocyte Differentiation through Repression of RSAD2. Sci. Rep. 6, 27105. doi:10.1038/srep27105
Liu, G., Friggeri, A., Yang, Y., Milosevic, J., Ding, Q., Thannickal, V. J., et al. (2010). miR-21 Mediates Fibrogenic Activation of Pulmonary Fibroblasts and Lung Fibrosis. J. Exp. Med. 207 (8), 1589–1597. doi:10.1084/jem.20100035
Liu, L., Oliveira, N. M., Cheney, K. M., Pade, C., Dreja, H., Bergin, A.-M. H., et al. (2011). A Whole Genome Screen for HIV Restriction Factors. Retrovirology 8, 94. doi:10.1186/1742-4690-8-94
Liu, X., Chen, Q., Yan, J., Wang, Y., Zhu, C., Chen, C., et al. (2013). MiRNA-296-3p-ICAM-1 axis Promotes Metastasis of Prostate Cancer by Possible Enhancing Survival of Natural Killer Cell-Resistant Circulating Tumour Cells. Cell Death Dis 4, e928. doi:10.1038/cddis.2013.458
Lodge, R., Ferreira Barbosa, J. A., Lombard-Vadnais, F., Gilmore, J. C., Deshiere, A., Gosselin, A., et al. (2017). Host MicroRNAs-221 and -222 Inhibit HIV-1 Entry in Macrophages by Targeting the CD4 Viral Receptor. Cel Rep. 21 (1), 141–153. doi:10.1016/j.celrep.2017.09.030
Ma, L., Shen, C.-J., Cohen, É. A., Xiong, S.-D., and Wang, J.-H. (2014). miRNA-1236 Inhibits HIV-1 Infection of Monocytes by Repressing Translation of Cellular Factor VprBP. PloS one 9 (6), e99535. doi:10.1371/journal.pone.0099535
Macfarlane, L.-A., and R. Murphy, P. (2010). MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genomics 11 (7), 537–561. doi:10.2174/138920210793175895
Martinez-Nunez, R. T., Louafi, F., Friedmann, P. S., and Sanchez-Elsner, T. (2009). MicroRNA-155 Modulates the Pathogen Binding Ability of Dendritic Cells (DCs) by Down-Regulation of DC-specific Intercellular Adhesion Molecule-3 Grabbing Non-integrin (DC-SIGN). J. Biol. Chem. 284 (24), 16334–16342. doi:10.1074/jbc.m109.011601
Modi, W. S., Lautenberger, J., An, P., Scott, K., Goedert, J. J., Kirk, G. D., et al. (2006). Genetic Variation in the CCL18-CCL3-CCL4 Chemokine Gene Cluster Influences HIV Type 1 Transmission and AIDS Disease Progression. Am. J. Hum. Genet. 79 (1), 120–128. doi:10.1086/505331
Monteleone, K., Selvaggi, C., Cacciotti, G., Falasca, F., Mezzaroma, I., D’Ettorre, G., et al. (2015). MicroRNA-29 Family Expression and its Relation to Antiviral Immune Response and Viro-Immunological Markers in HIV-1-Infected Patients. BMC Infect. Dis. 15, 51. doi:10.1186/s12879-015-0768-4
Nature Biotechnology (2020). Second RNAi Drug Approved. Nat. Biotechnol. 38, 385.doi:10.1038/s41587-020-0494-3
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 9, 402. doi:10.3389/fendo.2018.00402
Okoye, I., Xu, L., Oyegbami, O., Shahbaz, S., Pink, D., Gao, P., et al. (2021). Plasma Extracellular Vesicles Enhance HIV-1 Infection of Activated CD4+ T Cells and Promote the Activation of Latently Infected J-Lat10.6 Cells via miR-139-5p Transfer. Front. Immunol. 12, 697604. doi:10.3389/fimmu.2021.697604
Omoto, S., Ito, M., Tsutsumi, Y., Ichikawa, Y., Okuyama, H., Brisibe, E. A., et al. (2004). HIV-1 Nef Suppression by Virally Encoded microRNA. Retrovirology 1 (1), 44–12. doi:10.1186/1742-4690-1-44
Orecchini, E., Doria, M., Michienzi, A., Giuliani, E., Vassena, L., Ciafrè, S. A., et al. (2014). The HIV-1 Tat Protein Modulates CD4 Expression in Human T Cells through the Induction of miR-222. RNA Biol. 11 (4), 334–338. doi:10.4161/rna.28372
Organization, W. H. (2020). Global Health Estimates 2016: Disease burden by Cause, Age, Sex, by Country and by Region, 2000–2016, 2018. Geneva. Recuperado de 390, 10100.
Ouellet, D. L., Vigneault-Edwards, J., Létourneau, K., Gobeil, L. A., Plante, I., Burnett, J. C., et al. (2013). Regulation of Host Gene Expression by HIV-1 TAR microRNAs. Retrovirology 10 (1), 86–15. doi:10.1186/1742-4690-10-86
Palios, J., Kadoglou, N. P., and Lampropoulos, S. (2011). The Pathophysiology of HIV-/HAART-Related Metabolic Syndrome Leading to Cardiovascular Disorders: The Emerging Role of Adipokines. Experimental Diabetes Research.
Pilakka-Kanthikeel, S., Raymond, A., Atluri, V. S. R., Sagar, V., Saxena, S. K., Diaz, P., et al. (2015). Sterile Alpha Motif and Histidine/aspartic Acid Domain-Containing Protein 1 (SAMHD1)-Facilitated HIV Restriction in Astrocytes Is Regulated by miRNA-181a. J. Neuroinflammation 12, 66. doi:10.1186/s12974-015-0285-9
Pincetic, A., Kuang, Z., Seo, E. J., and Leis, J. (2010). The Interferon-Induced Gene ISG15 Blocks Retrovirus Release from Cells Late in the Budding Process. J. Virol. 84 (9), 4725–4736. doi:10.1128/jvi.02478-09
Pomerantz, R. J., and Horn, D. L. (2003). Twenty Years of Therapy for HIV-1 Infection. Nat. Med. 9 (7), 867–873. doi:10.1038/nm0703-867
Qi, Z., Cai, S., Cai, J., Chen, L., Yao, Y., Chen, L., et al. (2016). miR-491 Regulates Glioma Cells Proliferation by Targeting TRIM28 In Vitro. BMC Neurol. 16 (1), 248. doi:10.1186/s12883-016-0769-y
Qiuling, H., Chen, L., Luo, M., Lv, H., Luo, D., Li, T., et al. (2018). HIV-1-Induced miR-146a Attenuates Monocyte Migration by Targeting CCL5 in Human Primary Macrophages. AIDS Res. Hum. Retroviruses 34 (7), 580–589. doi:10.1089/AID.2017.0217
Quaranta, M. T., Olivetta, E., Sanchez, M., Spinello, I., Paolillo, R., Arenaccio, C., et al. (2015). miR-146a Controls CXCR4 Expression in a Pathway that Involves PLZF and Can Be Used to Inhibit HIV-1 Infection of CD4+ T Lymphocytes. Virology 478, 27–38. doi:10.1016/j.virol.2015.01.016
Raposo, R. A. S., Abdel-Mohsen, M., Bilska, M., Montefiori, D. C., Nixon, D. F., and Pillai, S. K. (2013). Effects of Cellular Activation on Anti-HIV-1 Restriction Factor Expression Profile in Primary Cells. J. Virol. 87 (21), 11924–11929. doi:10.1128/jvi.02128-13
Rosenberger, C. M., Podyminogin, R. L., Diercks, A. H., Treuting, P. M., Peschon, J. J., Rodriguez, D., et al. (2017). miR-144 Attenuates the Host Response to Influenza Virus by Targeting the TRAF6-IRF7 Signaling axis. Plos Pathog. 13 (4), e1006305. doi:10.1371/journal.ppat.1006305
Ruelas, D. S., Chan, J. K., Oh, E., Heidersbach, A. J., Hebbeler, A. M., Chavez, L., et al. (2015). MicroRNA-155 Reinforces HIV Latency. J. Biol. Chem. 290 (22), 13736–13748. doi:10.1074/jbc.m115.641837
Sarasin-Filipowicz, M., Krol, J., Markiewicz, I., Heim, M. H., and Filipowicz, W. (2009). Decreased Levels of microRNA miR-122 in Individuals with Hepatitis C Responding Poorly to Interferon Therapy. Nat. Med. 15 (1), 31–33. doi:10.1038/nm.1902
Scaria, V., Hariharan, M., Pillai, B., Maiti, S., and Brahmachari, S. K. (2007). Host-virus Genome Interactions: Macro Roles for microRNAs. Cell Microbiol. 9 (12), 2784–2794. doi:10.1111/j.1462-5822.2007.01050.x
Seddiki, N., Phetsouphanh, C., Swaminathan, S., Xu, Y., Rao, S., Li, J., et al. (2013). The microRNA-9/b-Lymphocyte-Induced Maturation protein-1/IL-2 axis Is Differentially Regulated in Progressive HIV Infection. Eur. J. Immunol. 43 (2), 510–520. doi:10.1002/eji.201242695
Shen, C. J., Jia, Y. H., Tian, R. R., Ding, M., Zhang, C., and Wang, J. H. (2012). Translation of Pur‐α Is Targeted by Cellular miRNAs to Modulate the Differentiation‐dependent Susceptibility of Monocytes to HIV‐1 Infection. FASEB J. 26 (11), 4755–4764. doi:10.1096/fj.12-209023
Sirois, M., Robitaille, L., Allary, R., Shah, M., Woelk, C. H., Estaquier, J., et al. (2011). TRAF6 and IRF7 Control HIV Replication in Macrophages. PLOS ONE 6 (11), e28125. doi:10.1371/journal.pone.0028125
Skalsky, R. L., and Cullen, B. R. (2010). Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 64, 123–141. doi:10.1146/annurev.micro.112408.134243
Sonkoly, E., Wei, T., Janson, P. C. J., Sääf, A., Lundeberg, L., Tengvall-Linder, M., et al. (2007). MicroRNAs: Novel Regulators Involved in the Pathogenesis of Psoriasis? PloS one 2 (7), e610. doi:10.1371/journal.pone.0000610
Stadeli, K. M., and Richman, D. D. (2013). Rates of Emergence of HIV Drug Resistance in Resource-Limited Settings: a Systematic Review. Antivir. Ther. 18 (1), 115–123. doi:10.3851/IMP2437
Stanczyk, J., Pedrioli, D. M. L., Brentano, F., Sanchez-Pernaute, O., Kolling, C., Gay, R. E., et al. (2008). Altered Expression of MicroRNA in Synovial Fibroblasts and Synovial Tissue in Rheumatoid Arthritis. Arthritis Rheum. 58 (4), 1001–1009. doi:10.1002/art.23386
Steele, C. W., Oien, K. A., McKay, C. J., and Jamieson, N. B. (2011). Clinical Potential of microRNAs in Pancreatic Ductal Adenocarcinoma. Pancreas 40 (8), 1165–1171. doi:10.1097/mpa.0b013e3182218ffb
Su, B., Fu, Y., Liu, Y., Wu, H., Ma, P., Zeng, W., et al. (2018). Potential Application of microRNA Profiling to the Diagnosis and Prognosis of HIV-1 Infection. Front. Microbiol. 9, 3185. doi:10.3389/fmicb.2018.03185
Subramanian, S., and Steer, C. J. (2010). MicroRNAs as Gatekeepers of Apoptosis. J. Cel Physiol 223 (2), 289–298. doi:10.1002/jcp.22066
Sun, B., Yang, R., and Mallardo, M. (2016). Roles of microRNAs in HIV-1 Replication and Latency. Mirna 5 (2), 120–123. doi:10.2174/2211536605666160829123118
Sun, G., Li, H., Wu, X., Covarrubias, M., Scherer, L., Meinking, K., et al. (2012). Interplay between HIV-1 Infection and Host microRNAs. Nucleic Acids Res. 40 (5), 2181–2196. doi:10.1093/nar/gkr961
Sung, T.-L., and Rice, A. P. (2009). miR-198 Inhibits HIV-1 Gene Expression and Replication in Monocytes and its Mechanism of Action Appears to Involve Repression of Cyclin T1. Plos Pathog. 5 (1), e1000263. doi:10.1371/journal.ppat.1000263
Swaminathan, G., Rossi, F., Sierra, L. J., Gupta, A., Navas-Martín, S., and Martín-García, J. (2012). A Role for microRNA-155 Modulation in the Anti-HIV-1 Effects of Toll-like Receptor 3 Stimulation in Macrophages. PLoS Pathogens 8 (9), e1002937. doi:10.1371/journal.ppat.1002937
Swaminathan, G., Rossi, F., Sierra, L.-J., Gupta, A., Navas-Martín, S., and Martín-García, J. (2012). A Role for microRNA-155 Modulation in the Anti-HIV-1 Effects of Toll-like Receptor 3 Stimulation in Macrophages. Plos Pathog. 8 (9), e1002937. doi:10.1371/journal.ppat.1002937
Swaminathan, S., Suzuki, K., Seddiki, N., Kaplan, W., Cowley, M. J., Hood, C. L., et al. (2012). Differential Regulation of the Let-7 Family of microRNAs in CD4+ T Cells Alters IL-10 Expression. J. Immunol. 188 (12), 6238–6246. doi:10.4049/jimmunol.1101196
Thierry, S., Benleulmi, M. S., Sinzelle, L., Thierry, E., Calmels, C., Chaignepain, S., et al. (2015). Dual and Opposite Effects of hRAD51 Chemical Modulation on HIV-1 Integration. Chem. Biol. 22 (6), 712–723. doi:10.1016/j.chembiol.2015.04.020
Thum, T., Gross, C., Fiedler, J., Fischer, T., Kissler, S., Bussen, M., et al. (2008). MicroRNA-21 Contributes to Myocardial Disease by Stimulating MAP Kinase Signalling in Fibroblasts. Nature 456 (7224), 980–984. doi:10.1038/nature07511
Tian, H., and He, Z. (2018). miR-215 Enhances HCV Replication by Targeting TRIM22 and Inactivating NF-Κb Signaling. Yonsei Med. J. 59 (4), 511–518. doi:10.3349/ymj.2018.59.4.511
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA Biogenesis and its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cel Biol 20 (1), 5–20. doi:10.1038/s41580-018-0059-1
Triboulet, R., Mari, B., Lin, Y.-L., Chable-Bessia, C., Bennasser, Y., Lebrigand, K., et al. (2007). Suppression of microRNA-Silencing Pathway by HIV-1 during Virus Replication. Science 315 (5818), 1579–1582. doi:10.1126/science.1136319
Trobaugh, D. W., and Klimstra, W. B. (2017). MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Molecular Medicine 23 (1), 80–93. doi:10.1016/j.molmed.2016.11.003
Tyagi, M., and Kashanchi, F. (2012). New and Novel Intrinsic Host Repressive Factors against HIV-1: PAF1 Complex, HERC5 and Others. Retrovirology 9, 19. doi:10.1186/1742-4690-9-19
van Gent, M., Sparrer, K. M. J., and Gack, M. U. (2018). TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 5, 385–405. doi:10.1146/annurev-virology-092917-043323
Wang, W.-X., Rajeev, B. W., Stromberg, A. J., Ren, N., Tang, G., Huang, Q., et al. (2008). The Expression of MicroRNA miR-107 Decreases Early in Alzheimer's Disease and May Accelerate Disease Progression through Regulation of -Site Amyloid Precursor Protein-Cleaving Enzyme 1. J. Neurosci. 28 (5), 1213–1223. doi:10.1523/jneurosci.5065-07.2008
Wang, X., Ye, L., Hou, W., Zhou, Y., Wang, Y.-J., Metzger, D. S., et al. (2009). Cellular microRNA Expression Correlates with Susceptibility of Monocytes/macrophages to HIV-1 Infection. Blood 113 (3), 671–674. doi:10.1182/blood-2008-09-175000
Wong, S. T., Zhang, X. Q., Zhuang, J. T., Chan, H. L., Li, C. H., and Leung, G. K. (2012). MicroRNA-21 Inhibition Enhances In Vitro Chemosensitivity of Temozolomide-Resistant Glioblastoma Cells. Anticancer Res. 32 (7), 2835–2841.
Wortman, M. J., Krachmarov, C. P., Kim, J. H., Gordon, R. G., Chepenik, L. G., Brady, J. N., et al. (2000). Interaction of HIV-1 Tat with Pur? in Nuclei of Human Glial Cells: Characterization of RNA-Mediated Protein-Protein Binding. J. Cel. Biochem. 77 (1), 65–74. doi:10.1002/(sici)1097-4644(20000401)77:1<65:aid-jcb7>3.0.co;2-u
Wu, M., Fan, B., Guo, Q., Li, Y., Chen, R., Lv, N., et al. (2018). Knockdown of SETDB1 Inhibits Breast Cancer Progression by miR-381-3p-Related Regulation. Biol. Res. 51 (1), 39. doi:10.1186/s40659-018-0189-0
Wu, S., He, L., Li, Y., Wang, T., Feng, L., Jiang, L., et al. (2013). miR-146a Facilitates Replication of Dengue Virus by Dampening Interferon Induction by Targeting TRAF6. J. Infect. 67 (4), 329–341. doi:10.1016/j.jinf.2013.05.003
Wu, X., Zhang, L.-L., Yin, L.-B., Fu, Y.-J., Jiang, Y.-J., Ding, H.-B., et al. (2017). Deregulated MicroRNA-21 Expression in Monocytes from HIV-Infected Patients Contributes to Elevated IP-10 Secretion in HIV Infection. Front. Immunol. 8, 1122. doi:10.3389/fimmu.2017.01122
Yamanaka, S., Olaru, A. V., An, F., Luvsanjav, D., Jin, Z., Agarwal, R., et al. (2012). MicroRNA-21 Inhibits Serpini1, a Gene with Novel Tumour Suppressive Effects in Gastric Cancer. Dig. Liver Dis. 44 (7), 589–596. doi:10.1016/j.dld.2012.02.016
Yang, F., Xu, Z., Duan, S., and Luo, M. (2016). MicroRNA-541 Promotes the Proliferation of Vascular Smooth Muscle Cells by Targeting IRF7. Am. J. Transl Res. 8 (2), 506–515.
Yin, Y., Zhong, J., Li, S.-W., Li, J.-Z., Zhou, M., Chen, Y., et al. (2016). TRIM11, a Direct Target of miR-24-3p, Promotes Cell Proliferation and Inhibits Apoptosis in colon Cancer. Oncotarget 7 (52), 86755–86765. doi:10.18632/oncotarget.13550
Yuan, T., Yao, W., Tokunaga, K., Yang, R., and Sun, B. (2016). An HIV-1 Capsid Binding Protein TRIM11 Accelerates Viral Uncoating. Retrovirology 13 (1), 72. doi:10.1186/s12977-016-0306-5
Zhang, H.-S., Chen, X.-Y., Wu, T.-C., Sang, W.-W., and Ruan, Z. (2012). MiR-34a Is Involved in Tat-Induced HIV-1 Long Terminal Repeat (LTR) Transactivation through the SIRT1/NFκB Pathway. FEBS Lett. 586 (23), 4203–4207. doi:10.1016/j.febslet.2012.10.023
Zhang, H.-S., Wu, T.-C., Sang, W.-W., and Ruan, Z. (2012). MiR-217 Is Involved in Tat-Induced HIV-1 Long Terminal Repeat (LTR) Transactivation by Down-Regulation of SIRT1. Biochim. Biophys. Acta. 1823 (5), 1017–1023. doi:10.1016/j.bbamcr.2012.02.014
Zhang, M. M., Bahal, R., Rasmussen, T. P., Manautou, J. E., and Zhong, X.-b. (2021). The Growth of siRNA-Based Therapeutics: Updated Clinical Studies. Biochem. Pharmacol. 189, 114432. doi:10.1016/j.bcp.2021.114432
Zhang, Q., He, Y., Nie, M., and Cai, W. (2017). Roles of miR-138 and ISG15 in Oral Squamous Cell Carcinoma. Exp. Ther. Med. 14 (3), 2329–2334. doi:10.3892/etm.2017.4720
Zhang, Y., Fan, M., Geng, G., Liu, B., Huang, Z., Luo, H., et al. (2014). A Novel HIV-1-Encoded microRNA Enhances its Viral Replication by Targeting the TATA Box Region. Retrovirology 11 (1), 23–15. doi:10.1186/1742-4690-11-23
Zhang, Z., Wu, Y., Chen, J., Hu, F., Chen, X., and Xu, W. (2021). Expression of microRNA-155 in Circulating T Cells is an Indicator of Immune Activation Levels in HIV-1 Infected Patients. HIV Res. Clin. Pract. 22 (3), 71–77.
Zhao, C., Sun, X., and Li, L. (2019). Biogenesis and Function of Extracellular miRNAs. ExRNA 1 (1), 1–9. doi:10.1186/s41544-019-0039-4
Keywords: microRNA, HIV, host-genetics, epigenetics, miRNA
Citation: Chinniah R, Adimulam T, Nandlal L, Arumugam T and Ramsuran V (2022) The Effect of miRNA Gene Regulation on HIV Disease. Front. Genet. 13:862642. doi: 10.3389/fgene.2022.862642
Received: 26 January 2022; Accepted: 13 April 2022;
Published: 04 May 2022.
Edited by:
Gabriel Adelman Cipolla, Federal University of Paraná, BrazilReviewed by:
Weizhong Chang, National Cancer Institute at Frederick (NIH), United StatesCopyright © 2022 Chinniah, Adimulam, Nandlal, Arumugam and Ramsuran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veron Ramsuran, cmFtc3VyYW52QHVrem4uYWMuemE=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.