- 1Key Laboratory of Preservation of Human Genetic Resources and Disease Control in China, Harbin Medical University, Ministry of Education, Harbin, China
- 2Laboratory of Medical Genetics, Harbin Medical University, Harbin, China
- 3The Second Hospital of Heilongjiang Province, Harbin, China
- 4College of Basic Medicine, Harbin Medical University-Daqing, Daqing, China
Background: Some studies have shown that the base excision repair (BER) pathway has an effect on HIV-1 replication. APEX1 and XRCC1 as key BER genes may affect DNA repair capacity. However, the roles of single nucleotide polymorphisms (SNPs) in APEX1 and XRCC1 and their impact on HIV-1 infection and AIDS progression remain unclear.
Methods: A custom-designed 48-Plex SNPscan Kit was used for detection of single nucleotide polymorphisms. 601 HIV-1-infected men who have sex with men (MSM) and 624 age-matched healthy individuals were recruited in northern China. Four SNPs (rs1130409, rs1760944, rs2307486 and rs3136817) in APEX1 gene and three SNPs (rs1001581, rs25487 and rs25489) in XRCC1 gene were genotyped. The generalized multifactor dimension reduction (GMDR) method was used to identify the SNP-SNP interactions.
Results: In this study, rs1130409 G allele, rs1001581 C allele and rs25487 C allele were associated with a higher risk of HIV-1 infection susceptibility (p = 0.020, p = 0.007 and p = 0.032, respectively). The frequencies of APEX1 haplotype TT and XRCC1 haplotype CT showed significant differences between cases and controls (p = 0.0372 and p = 0.0189, respectively). Interestingly, stratified analysis showed that the frequency of rs1001581 C allele was significantly higher in AIDS patients with the CD4+ T-lymphocyte count <200 cells/μl than those with >200 cells/μl (p = 0.022). Moreover, significant gene-gene interactions among rs1130409, rs1001581 and rs25487 were identified by GMDR (p = 0.0107). Specially, individuals with five to six risk alleles have a higher susceptibility to HIV-1 infection than those with zero to two risk alleles (p < 0.001).
Conclusion: APEX1 and XRCC1 gene polymorphisms were associated with the susceptibility to HIV-1 infection and AIDS progression in MSM populations in northern China.
Introduction
The spread of HIV-1 infection around the world has become a major global public health problem. HIV-1 virus is the pathogen of AIDS, which can infect human immune cells, causing immune system dysfunction and making the human body susceptible to a variety of diseases (Bandera et al., 2019). Several reports have shown that different heterogeneities of host genetics and host immune response contribute to different chronic complications and AIDS progression (O’Brien and Nelson, 2004; Speelmon et al., 2006).
DNA damage triggers genome instability, thereby burdening cells with dangerously disadvantageous mutations (Whitaker et al., 2017). Activated powerful DNA damage response (DDR) pathways through the DNA repair system, cell cycle checkpoints and cell death pathways to minimize the harmful effects of DNA damage. Base excision repair (BER), one of the primary DNA repair pathways, repairs the endogenous DNA damages including deaminations, depurinations, alkylations and a plethora of oxidative damages, and can enhance DNA single-strand break repair (SSBR) (Nemec et al., 2010; Hegde et al., 2012). Based on the multiple functions of DNA glycosylase, there are two classic BER pathways: short patch BER and long patch BER. APEX1 and XRCC1 are two key enzymes in short patch BER.
APEX1 is a 2.21 kb gene located on chromosome 14q11.2-q12. It encodes an apurinic endonuclease, which recognizes and cleaves phosphodiester bonds through a hydrolysis mechanism, and specifically activates DNA damage repair (Parsons et al., 2004). APEX1 polymorphism has been reported to be associated with the efficacy of platinum-based adjuvant chemotherapy on cervical cancer (Chung et al., 2006) and non-small cell lung cancer (Pérez-Ramírez et al., 2019). In addition, it may increase the risk of other cancers, such as breast cancer (Mitra et al., 2008), colorectal cancer (Wang L. et al., 2016) and pancreatic cancer (Chen et al., 2019).
The XRCC1 gene is located on chromosome 19q13.2-13.3 and is about 33 kb long. In DNA damage repair, the encoded protein functions as a scaffold protein for specific repair enzymes and plays a role in the subsequent enzymatic steps (Caldecott, 2003; Caldecott, 2008). Polymorphisms of XRCC1 may be involved in the pathogenesis of many diseases, including head and neck cancer (Nanda et al., 2018), diabetes (Wang et al., 2019) and Alzheimer’s disease (Qian et al., 2010). It has long been speculated that insertion of HIV-1 cDNA leads to the creation of DSBs and leaves single-stranded gaps connecting the viral and host DNA (Skalka and Katz, 2005). Effective repair of these lesions is critical to host cell integrity and subsequent transcription of the viral genome, and BER may play this role (Mbonye and Karn, 2017). Functional SNPs in the BER gene can affect gene expression or function, affect DNA repair ability, and eventually lead to cell dysfunction and mutagenesis (Hung et al., 2005).
In addition, it has been reported that BER may influence the sequence preference of HIV-1 integration site (Bennett et al., 2014). However, the association between BER gene polymorphisms and risk of HIV-1 infection has not been fully evaluated. Our previous works have confirmed that polymorphisms in mismatch repair gene MSH6 and homologous recombination gene MRE11 may play an important role in the development of AIDS (Liu C. et al., 2019; Wang C. et al., 2016). In this study, we investigated whether seven potentially functional SNPs in the XRCC1 and APEX1 genes are associated with susceptibility to HIV-1 infection and the AIDS progression in men who have sex with men (MSM) populations in northern China.
Methods
Subjects
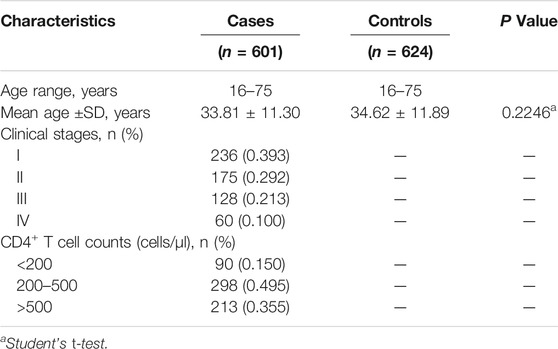
In this case-control study, 601 HIV-1 positive males were recruited from the Center for Disease Control and Prevention of Heilongjiang Province in northern China. All patients had acquired HIV-1 infection through male and male homosexual transmission. We recruited 624 healthy men diagnosed as HIV-1 seronegative by comprehensive medical examination at the Second Affiliated Hospital of Harbin Medical University, as a control group. Based on age, 1:1 frequency matching was performed at 10-year intervals. The detailed characteristics of samples are shown in Table 1. The study was approved by the local ethics review committee, and all subjects signed written consent prior to the study.
SNPs Selection and Genotyping
Potential functional polymorphisms in the XRCC1 and APEX1 were identified through the dbSNP database (http://www.ncbi.nlm.nih.gov/) and HaploReg v4.1 (http://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). With minor allele frequency (MAF) > 0.1 in CHB (Han Chinese in Beijing), four SNPs (rs1130409 T > G, rs1760944 T > G, rs2307486A > G and rs3136817 T > C) in APEX1 gene and three SNPs (rs1001581 C > T, rs25487 T > C, rs25489 C > T) in XRCC1 gene were finally selected. The candidate SNPs are located in the untranslated regions (5′-UTR, intron) and exon of genes, and there is no significant linkage disequilibrium (R2 < 0.8) among these SNPs.
Following the manufacturer’s instructions, genomic DNA was extracted from 200 μl whole blood of each sample using the QIAamp blood kit (Qiagen, Hilden, Germany). Genotyping was performed using a customized 48-plex SNPscan kit (Genesky Biotechnologies Inc., Shanghai, China), which used double ligation and multiplex fluorescent PCR. For quality control, 5% of random control samples were genotyped twice to verify that the accuracy and repeatability of genotyping was 100%.
Statistical Analysis
The Hardy-Weinberg equilibrium (HWE) of the control group was confirmed for each polymorphism by χ2 goodness-of-fit test. Two-sided chi-square test (adjusted chi-square test, yates for 1 ≤ T < 5) was used to compare allele and genotype frequencies between HIV-1 infected individuals and healthy controls. In association tests, odds ratios (ORs) and 95% confidence intervals (95% CI) were estimated as the relative risk associated with SNPs. The linkage disequilibrium (LD) and haplotype analysis was constructed by Haploview v.4.2 (http://sourceforge.net/projects/haploview/). Comparison of the CD4+ T-lymphocyte count in different genetic models was performed using Wilcoxon rank–sum test. Generalized multifactor dimensionality reduction (GMDR) (http://www.ssg.uab.edu/gmdr/) was used to analyze the gene-gene interaction models, and the best model was selected based on the consistency and accuracy test results of cross-validation. Association between the number of risk alleles and AIDS was calculated using logistic regression. Bonferroni correction was implemented to correct for multiple testing. Statistical analyses were performed using SPSS v.22.0 (IBM-SPSS, Inc., Chicago, IL, United States) and R statistical software (v3.6.3). P value <0.05 was considered statistically significant.
Results
Characteristics of Participants
A total of 601 AIDS patients and 624 healthy controls living in Heilongjiang Province were recruited in this study. Table 1 shows the basic characteristics of participants based on the CD4+ T-lymphocyte count and clinical staging. The mean ages of the patients and control subjects were 33.81 ± 11.30 and 34.62 ± 11.89 years, respectively. There was no significant difference in the distribution of age and gender between the two groups. The genotype distributions of candidate SNPs in control samples were in agreement with HWE (p > 0.05) (Table 2).
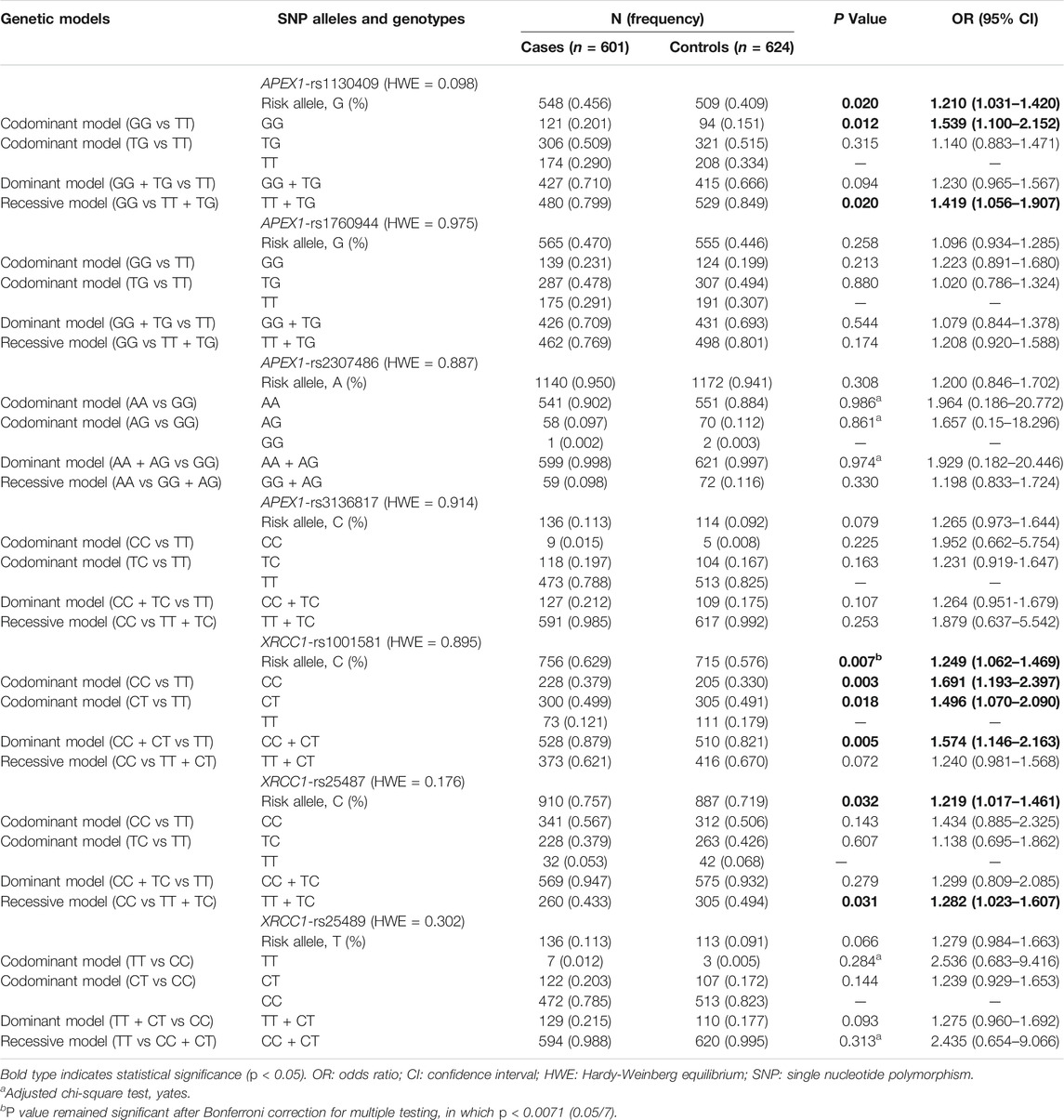
TABLE 2. Associations between APEX1 and XRCC1 gene polymorphisms and susceptibility to HIV-1 infection.
Association Between APEX1 and XRCC1 Gene Polymorphisms and Susceptibility to HIV-1 Infection
As shown in Table 2, the allele frequencies of APEX1 rs1130409 and XRCC1 rs1001581 and rs25487 were significantly different between the case and control groups (p < 0.05). Specially, the rs1130409 G (OR = 1.210, 95% CI = 1.031-1.420, p = 0.020), rs1001581 C (OR = 1.249, 95% CI = 1.062-1.469, p = 0.007) and rs25487 C (OR = 1.219, 95% CI = 1.017–1.461, p = 0.032) were associated with a higher risk of susceptibility to HIV-1 infection. In addition, the distribution of rs1001581 allele still showed significant differences between cases and controls after Bonferroni correction for multiple testing, in which p < 0.0071 (0.05/7) was set as statistically significant.
For the rs1130409, rs1001581 and rs25487 polymorphisms, there were significant differences in genotype distribution between HIV-1 infected group and control group under multiple genetic models (Table 2). In the analysis of APEX1 rs1130409, genotype GG was associated with increased susceptibility to HIV-1 infection (recessive model: GG vs TT + TG: OR = 1.419, 95% CI = 1.056–1.907, p = 0.020; codominant model: GG vs TT: OR = 1.539, 95% CI = 1.100–2.152, p = 0.012). Additionally, XRCC1 rs1001581 genotypes (CC + CT) were associated with increased susceptibility to HIV-1 infection (dominant model: CC + CT vs TT: OR = 1.574, 95% CI = 1.146-2.163, p = 0.005; codominant model: CC vs TT: OR = 1.691, 95% CI = 1.193–2.397, p = 0.003; CT vs TT: OR = 1.496, 95% CI = 1.070–2.090, p = 0.018). Moreover, the rs25487 CC genotype was also associated with increased susceptibility to HIV-1 infection (recessive model: CC vs TT + TC: OR = 1.282, 95% CI = 1.023–1.607, p = 0.031). However, after Bonferroni correction for multiple tests with p < 0.0018 (0.05/28), all these associations were no longer statistically significant.
Haplotype Analysis
Haploview software was used for linkage disequilibrium (LD) analysis. Two haplotype blocks were detected in APEX1 and XRCC1. APEX1 haplotype block1 is composed of rs3136817 and rs1130409 (Figure 1A), and XRCC1 haplotype block1 is composed of rs25489 and rs1001581 (Figure 1B). After the P value calculated by multiple tests of 10,000 permutations, the frequencies of APEX1 haplotype TT and XRCC1 haplotype CT showed significant differences between cases and controls (p = 0.0372 and p = 0.0189, respectively) (Supplementary Table S1).
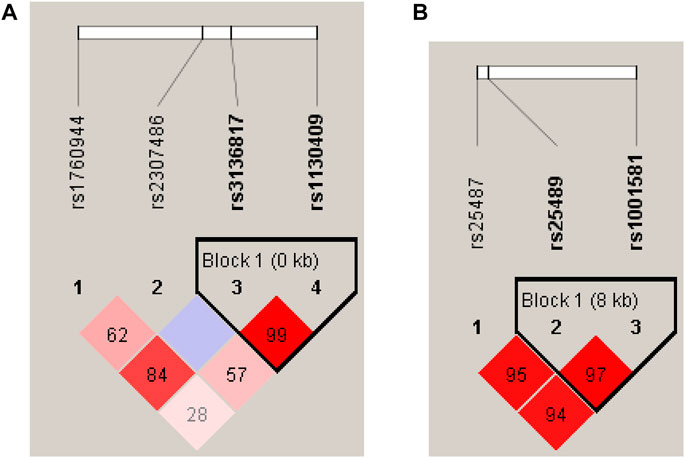
FIGURE 1. The results of LD analysis. (A) Haplotype block map for 4 SNPs of the APEX1 gene, (B) Haplotype block map for 3 SNPs of the XRCC1 gene. The haplotypes were constructed according to the prevalence of SNPs and LD among them. Numbers in squares indicate D’ values. The white cell indicates D’ < 1 and LOD (log of the likelihood odds ratio) < 2; The blue cell indicates D’ = 1 and LOD <2; the pink or red cell indicates D’ < 1 and LOD ≥2; The bright red cell indicates D’ = 1 and LOD ≥2.
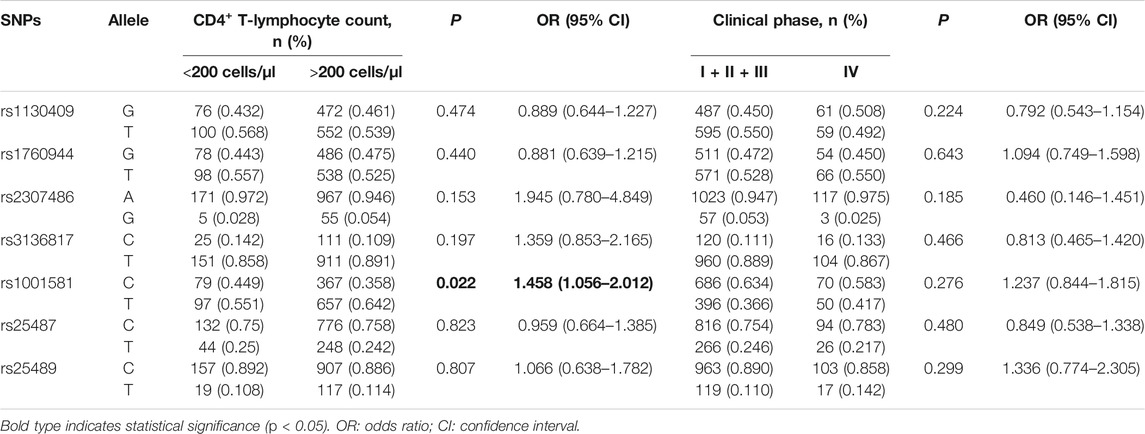
Association Between APEX1 and XRCC1 Polymorphisms and AIDS Progressions
As the number of CD4+ T lymphocytes decreases, the immunity of HIV-1 infected patients will be severely impaired, leading to complications such as pneumonia and tumors. The CD4+ T-lymphocyte count was used as a surrogate marker for AIDS progression in our study. The frequency of rs1001581 C was significantly higher in the cases with the CD4+ T-lymphocyte count <200 cells/μl than those with >200 cells/μl (OR = 1.458, 95% CI = 1.056–2.012, p = 0.022) (Table 3). However, after Bonferroni correction for multiple tests with p < 0.0071 (0.05/7), all these associations were no longer statistically significant. We further found that patients with rs25487 CC and TC genotypes have a lower CD4+ T-lymphocyte count than those with TT genotype in the dominant model (p = 0.027) (Figure 2A), while there was no difference in CD4+ T-lymphocyte count in the recessive genetic model (Figure 2B).
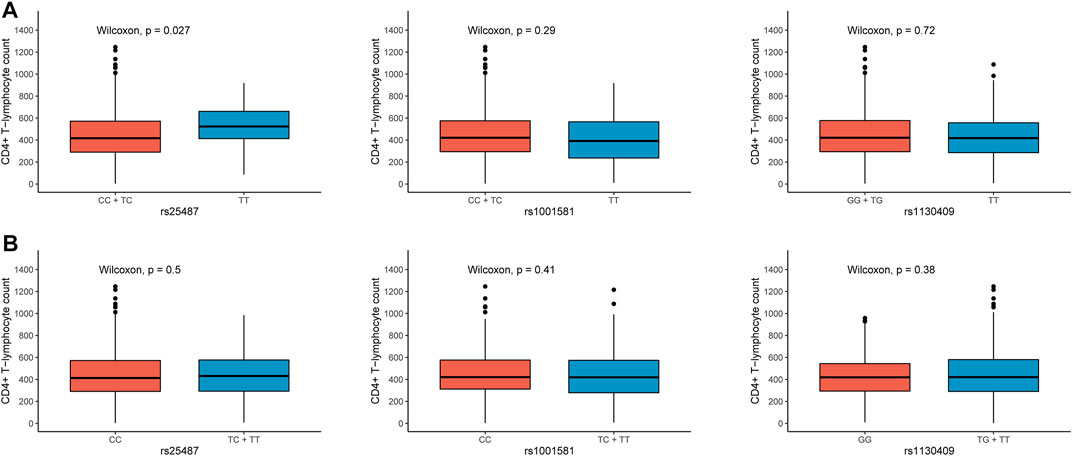
FIGURE 2. Differential analysis of CD4+ T-lymphocyte count in the different genetic models of rs25487, rs1001581 and rs1130409. (A) Dominant model, (B) Recessive model.
Gene-gene Interactions Analysis
The GMDR model was employed to find the best SNP-SNP interaction combination among seven SNPs in APEX1 and XRCC1 genes. The results showed a significant two-locus model involving rs1130409 and rs1001581 (p = 0.0107) and a significant three-locus model involving rs1130409, rs1001581 and rs25487 (p = 0.0107) (Supplementary Table S2). The susceptibility to HIV-1 infection may be altered by the three genetic variants. We further assessed the association between the sum of risk alleles of rs1130409 G, rs1001581 C and rs25487 C and susceptibility to HIV-1 infection. Figure 3 A shows the numbers of patients and healthy individuals with each risk allele in cases and controls, and we observed an additive effect of risk alleles on the presence of AIDS (Figure 3B). The OR for AIDS was 1.10 (95% CI = 0.835–1.462) in patients with three to four risk alleles, and the corresponding OR in those with five to six risk alleles was 1.703 (95% CI = 1.242–2.335, p < 0.001).
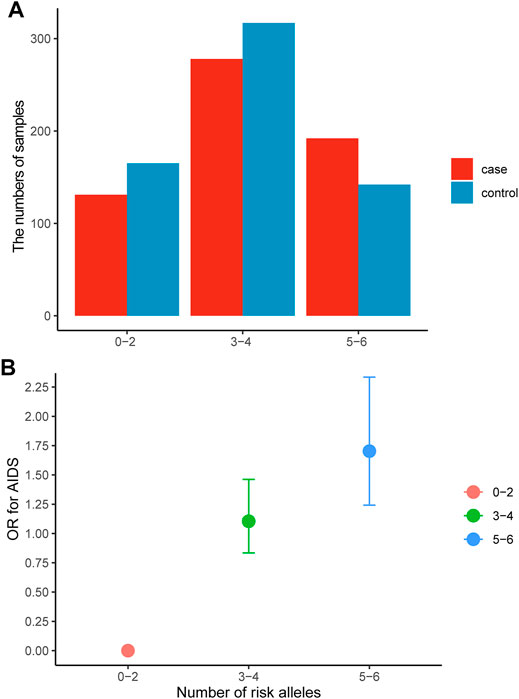
FIGURE 3. Association between the number of risk alleles in rs1130409, rs1001581 and rs25487 and AIDS. (A) the numbers of patients and healthy individuals with each risk allele in cases and controls, (B) The risk of AIDS by the number of risk allele categories. Data are crude ORs and 95% CIs. The 0–2 group was the reference. OR = odds ratio.
Discussion
It has been reported that the genetic background of the host will affect the susceptibility to HIV-1 infection and disease progression (Juno et al., 2012; Vidyant et al., 2017; de Silva et al., 2018). The interruption or dysregulation of SSBR can lead to genome instability, thereby promoting the integration of viral cDNA into the host genome (Sancar et al., 2004). One study also showed that the BER pathway is essential for effective lentiviral integration (Yoder et al., 2011). These findings suggest that the BER pathway may play a role in regulating HIV infection and integration. APEX1 and XRCC1 play a critical role in maintaining the integrity of the BER system. During the short patch BER process, APEX1 endonuclease cuts the 5′phosphodiester bond into an abasic site and recruits Polβ to fill the single nucleoside gap linked by the enzyme complex including LIG3 and XRCC1 (Izumi et al., 2000; Chatterjee and Walker, 2017). Therefore, this study aims to investigate the role of functional polymorphisms of XRCC1 and APEX1 genes in the susceptibility and clinical progression of HIV-1 infection, and to provide a theoretical basis for the prevention and treatment of the disease.
In our study, homozygous genotype GG and G allele of APEX1 rs1130409 were found to be significantly associated with increased susceptibility to HIV-1 infection. The rs1130409 G allele has been reported to increase the risk of other diseases. Mitra et al. confirmed a statistical association between the rs1130409 G allele and breast cancer risk in north Indian women (Mitra et al., 2008). Recently, a study conducted by Usategui-Martín et al. showed a statistically significant association between Paget’s disease of bone and the rs1130409 G allele in Spanish (Usategui-Martín et al., 2018). rs1130409 in the APEX1 exon 5 leads to the conversion of the residues at the carboxyl end of Asp148Glu, which may affect the endonuclease activity of APEX1. In addition, it has been reported that HIV-1 Rev protein interacts with APEX1 confirmed by in vitro binding experiments in HeLa cells (Naji et al., 2012). Yan et al. also demonstrated that siRNA-mediated knockdown of APEX1 significantly reduced the level of HIV-1 cDNA integration and virus production in HeLa-CD4 cells, thereby inhibiting HIV-1 infection (Yan et al., 2009). The above functional studies on APEX1 and rs1130409 can explain, to a certain extent, that individuals carrying rs1130409 G allele and GG genotype are more likely to be infected with HIV-1.
We also observed that homozygous genotype CC and C allele of XRCC1 rs25487 were significantly associated with susceptibility to HIV-1 infection. Several studies found that the T allele or TT genotype were associated with the risk of a variety of cancers. For example, Liu et al. observed that rs25487 T allele significantly increased the risk of cervical cancer in the female population of southwest China (Liu G. C. et al., 2019). A study also showed that T allele of rs25487 was associated with poor prognosis of HBV-related hepatocellular carcinoma patients after hepatectomy (Yu et al., 2016). Mahmoud et al. suggested that rs25487 TT genotype could be regarded as possible genotypic risk factor for the development of HCV-related hepatocellular carcinoma (Mahmoud et al., 2019). Another study pointed out that patients with rs25487 genotype TT + CT had a higher risk of endometrial cancer in northern China (Chen et al., 2016). The results of the above association studies are inconsistent, which may be caused by the different disease types and populations used in the different studies. Moreover, we further found that patients with rs25487 CC and TC genotypes have a lower CD4+ T-lymphocyte count than those with TT genotype. rs25487 is a non-synonymous SNP, which changes the glutamine of XRCC1 exon 10 into arginine, thereby reducing the affinity between XRCC1 protein and DNA repair complex and affecting the ability of DNA repair.
In addition, our results showed that homozygous genotype CC, heterozygous genotype CT and allele C of XRCC1 rs1001581 were significantly associated with increased susceptibility to HIV-1 infection. The rs1001581 allele C still showed a significant association with susceptibility to HIV-1 infection, even after Bonferroni correction for multiple testing. Interestingly, there was a significant association between rs1001581 and CD4+ T-lymphocyte count, and the C allele could significantly accelerate the disease progression of AIDS. Gu et al. proved that rs1001581 C to T variation was significantly related to the protective effect of the Han population against HAV infection (Gu et al., 2018). Another study showed that rs1001581 heterozygous genotype CT was associated with the risk of advanced non-small cell lung cancer in Koreans (Kim et al., 2010). These results indicated that XRCC1 rs1001581 may serve as an important biomarker for a variety of diseases and be beneficial for future evaluation of individual prognosis of AIDS. Although rs1001581 is located in XRCC1 intron, it may affect the function of XRCC1 encoding protein by altering its transcriptional activity. A functional study also reported that siRNA-mediated knockdown of APEX1 and XRCC1 can inhibit HIV infection in HeLa P4/ R5 cells (Espeseth et al., 2011).
AIDS is a consequence of the interaction between the individual’s genetic background and the environmental factors faced by the individual, in which gene-gene interaction may play a significant role. According to our GMDR model analysis, there was a significant SNP-SNP interaction between rs1130409, rs1001581 and rs25487. We observed a trend toward increased AIDS risk as the number of rs1130409 G, rs1001581 C and rs25487 C alleles increased, suggesting a cumulative effect of genetic variants on AIDS risk. Moreover, the association of haplotypes were analyzed to evaluate the synergy of SNPs. The frequencies of TT haplotype of APEX1 and CT haplotype of XRCC1 differed between cases and controls, and these associations were statistically significant. These results indicated that the interaction of rs1130409, rs1001581 and rs25487 significantly increased the risk of susceptibility to HIV-1 infection.
In the current study, we applied the Bonferroni correction to reduce the number of false positives in multiple significance testing. Although only the rs1001581 allele passed the calibration criteria after P-value was adjusted, the positive results of other SNPs should not be ignored. Undoubtedly, there are still some limitations in this study. First, there is a lack of information about critical factors in cases, including history of injection drug use, clinical data of viral loads and other clinical manifestations. Second, cases and controls were not exposed to the same conditions, and we did not collect blood samples from healthy MSM controls due to privacy regulations.
Conclusion
In conclusion, the present study suggests that the polymorphisms of APEX1 and XRCC1 may increase the risk of HIV-1 susceptibility and the clinical progression of AIDS in MSM populations in northern China. In order to further understand the effect of BER gene polymorphism on AIDS, it is necessary to carry out functional studies and genetic association studies in different ethnic groups with larger samples.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the protocols used in this study were evaluated and approved by the Ethics Committee of Harbin Medical University (No. HMUIRB20180019) and written informed consent was obtained from all participants. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
BL, KW, and JW performed the experiment and computational analysis. BL wrote the entire manuscript. YH, XY, LX WS, XJ, JW, and SF compiled the patient’s medical history and laboratory data. YQ and XZ contributed to the interpretation of the clinical manifestations and revised the manuscript for critical intellectual content. All authors have read and approved the manuscript.
Funding
This work was funded by the Program for the Postdoctoral Foundation of Heilongjiang Province (LRB08-340) of YQ and the National Natural Science Foundation of China (81373220) of XZ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge the numerous sample donors for making this work possible.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.861355/full#supplementary-material
References
Bandera, A., Gori, A., Clerici, M., and Sironi, M. (2019). Phylogenies in ART: HIV Reservoirs, HIV Latency and Drug Resistance. Curr. Opin. Pharmacol. 48, 24–32. doi:10.1016/j.coph.2019.03.003
Bennett, G. R., Peters, R., Wang, X.-H., Hanne, J., Sobol, R. W., Bundschuh, R., et al. (2014). Repair of Oxidative DNA Base Damage in the Host Genome Influences the HIV Integration Site Sequence Preference. PLoS One 9 (7), e103164. doi:10.1371/journal.pone.0103164
Caldecott, K. W. (2008). Single-Strand Break Repair and Genetic Disease. Nat. Rev. Genet. 9 (8), 619–631. doi:10.1038/nrg2380
Caldecott, K. W. (2003). XRCC1 and DNA Strand Break Repair. DNA Repair 2 (9), 955–969. doi:10.1016/s1568-7864(03)00118-6
Chatterjee, N., and Walker, G. C. (2017). Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagen. 58 (5), 235–263. doi:10.1002/em.22087
Chen, J., Wang, H., and Li, Z. (2019). Association between Polymorphisms of X-Ray Repair Cross Complementing Group 1 Gene and Pancreatic Cancer Risk: A Systematic Review with Meta-Analysis. Pathol. Oncol. Res. 25 (3), 897–904. doi:10.1007/s12253-017-0364-6
Chen, L., Liu, M.-M., Liu, H., Lu, D., Zhao, X.-D., and Yang, X.-J. (2016). ERCC1 and XRCC1 but Not XPA Single Nucleotide Polymorphisms Correlate with Response to Chemotherapy in Endometrial Carcinoma. Onco Targets Ther. 9, 7019–7028. doi:10.2147/OTT.S110976
Chung, H. H., Kim, M.-K., Kim, J. W., Park, N.-H., Song, Y.-S., Kang, S.-B., et al. (2006). XRCC1 R399Q Polymorphism Is Associated with Response to Platinum-Based Neoadjuvant Chemotherapy in Bulky Cervical Cancer. Gynecol. Oncol. 103 (3), 1031–1037. doi:10.1016/j.ygyno.2006.06.016
de Silva, T. I., Leligdowicz, A., Carlson, J., Garcia-Knight, M., Onyango, C., Miller, N., et al. (2018). HLA-Associated Polymorphisms in the HIV-2 Capsid Highlight Key Differences between HIV-1 and HIV-2 Immune Adaptation. AIDS 32 (6), 709–714. doi:10.1097/QAD.0000000000001753
Espeseth, A. S., Fishel, R., Hazuda, D., Huang, Q., Xu, M., Yoder, K., et al. (2011). siRNA Screening of a Targeted Library of DNA Repair Factors in HIV Infection Reveals a Role for Base Excision Repair in HIV Integration. PLoS One 6 (3), e17612. doi:10.1371/journal.pone.0017612
Gu, M., Qiu, J., Guo, D., Xu, Y., Liu, X., Shen, C., et al. (2018). Evaluation of Candidate Genes Associated with Hepatitis A and E Virus Infection in Chinese Han Population. Virol. J. 15 (1), 47. doi:10.1186/s12985-018-0962-2
Hegde, M. L., Izumi, T., and Mitra, S. (2012). Oxidized Base Damage and Single-Strand Break Repair in Mammalian Genomes: Role of Disordered Regions and Posttranslational Modifications in Early Enzymes. Prog. Mol. Biol. Transl Sci. 110, 123–153. doi:10.1016/B978-0-12-387665-2.00006-7
Hung, R. J., Hall, J., Brennan, P., and Boffetta, P. (2005). Genetic Polymorphisms in the Base Excision Repair Pathway and Cancer Risk: a HuGE Review. Am. J. Epidemiol. 162 (10), 925–942. doi:10.1093/aje/kwi318
Izumi, T., Hazra, T. K., Boldogh, I., Tomkinson, A. E., Park, M. S., Ikeda, S., et al. (2000). Requirement for Human AP Endonuclease 1 for Repair of 3′-blocking Damage at DNA Single-Strand Breaks Induced by Reactive Oxygen Species. Carcinogenesis 21 (7), 1329–1334. doi:10.1093/carcin/21.7.1329
Juno, J., Tuff, J., Choi, R., Card, C., Kimani, J., Wachihi, C., et al. (2012). The Role of G Protein Gene GNB3 C825TPolymorphism in HIV-1 Acquisition, Progression and Immune Activation. Retrovirology 9, 1. doi:10.1186/1742-4690-9-1
Kim, I.-S., Lee, G.-W., Kim, D. C., Kim, H.-G., Kim, S., Oh, S. Y., et al. (2010). Polymorphisms and Haplotypes in the XRCC1 Gene and the Risk of Advanced Non-small Cell Lung Cancer. J. Thorac. Oncol. 5 (12), 1912–1921. doi:10.1097/JTO.0b013e3181f46708
Liu, C., Qiao, Y., Xu, L., Wu, J., Mei, Q., Zhang, X., et al. (2019). Association between Polymorphisms in MRE11 and HIV-1 Susceptibility and AIDS Progression in a Northern Chinese MSM Population. J. Antimicrob. Chemother. 74 (7), 2009–2018. doi:10.1093/jac/dkz132
Liu, G. C., Zhou, Y.-F., Su, X.-C., and Zhang, J. (2019). Interaction between TP53 and XRCC1 Increases Susceptibility to Cervical Cancer Development: a Case Control Study. BMC Cancer 19 (1), 24. doi:10.1186/s12885-018-5149-0
Mahmoud, A. A., Hassan, M. H., Ghweil, A. A., Abdelrahman, A., Mohammad, A. N., and Ameen, H. H. (2019). Urinary 8-hydroxydeoxyguanosine in Relation to XRCC1 Rs25487 G/A (Arg399Gln) and OGG1 Rs1052133 C/G (Ser326Cys) DNA Repair Genes Polymorphisms in Patients with Chronic Hepatitis C and Related Hepatocellular Carcinoma. Cancer Manag. Res. 11, 5343–5351. doi:10.2147/CMAR.S209112
Mbonye, U., and Karn, J. (2017). The Molecular Basis for Human Immunodeficiency Virus Latency. Annu. Rev. Virol. 4 (1), 261–285. doi:10.1146/annurev-virology-101416-041646
Mitra, A. K., Singh, N., Singh, A., Garg, V. K., Agarwal, A., Sharma, M., et al. (2008). Association of Polymorphisms in Base Excision Repair Genes with the Risk of Breast Cancer: A Case-Control Study in North Indian Women. Oncol. Res. 17 (3), 127–135. doi:10.3727/096504008785055567
Naji, S., Ambrus, G., Cimermančič, P., Reyes, J. R., Johnson, J. R., Filbrandt, R., et al. (2012). Host Cell Interactome of HIV-1 Rev Includes RNA Helicases Involved in Multiple Facets of Virus Production. Mol. Cell Proteomics 11 (4), M111.015313. doi:10.1074/mcp.M111.015313
Nanda, S. S., Gandhi, A. K., Rastogi, M., Khurana, R., Hadi, R., Sahni, K., et al. (2018). Evaluation of XRCC1 Gene Polymorphism as a Biomarker in Head and Neck Cancer Patients Undergoing Chemoradiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 101 (3), 593–601. doi:10.1016/j.ijrobp.2018.03.039
Nemec, A. A., Wallace, S. S., and Sweasy, J. B. (2010). Variant Base Excision Repair Proteins: Contributors to Genomic Instability. Semin. Cancer Biol. 20 (5), 320–328. doi:10.1016/j.semcancer.2010.10.010
O'Brien, S. J., and Nelson, G. W. (2004). Human Genes that Limit AIDS. Nat. Genet. 36 (6), 565–574. doi:10.1038/ng1369
Parsons, J. L., Dianova, I. I., and Dianov, G. L. (2004). APE1 Is the Major 3'-Phosphoglycolate Activity in Human Cell Extracts. Nucleic Acids Res. 32 (12), 3531–3536. doi:10.1093/nar/gkh676
Pérez-Ramírez, C., Cañadas-Garre, M., Alnatsha, A., Villar, E., Valdivia-Bautista, J., Faus-Dáder, M. J., et al. (2019). Pharmacogenetics of Platinum-Based Chemotherapy: Impact of DNA Repair and Folate Metabolism Gene Polymorphisms on Prognosis of Non-Small Cell Lung Cancer Patients. Pharmacogenomics J. 19 (2), 164–177. doi:10.1038/s41397-018-0014-8
Qian, Y., Chen, W., Wu, J., Tao, T., Bi, L., Xu, W., et al. (2010). Association of Polymorphism of DNA Repair Gene XRCC1 with Sporadic Late-Onset Alzheimer's Disease and Age of Onset in Elderly Han Chinese. J. Neurol. Sci. 295 (1-2), 62–65. doi:10.1016/j.jns.2010.05.002
Sancar, A., Lindsey-Boltz, L. A., Ünsal-Kaçmaz, K., and Linn, S. (2004). Molecular Mechanisms of Mammalian DNA Repair and the DNA Damage Checkpoints. Annu. Rev. Biochem. 73, 39–85. doi:10.1146/annurev.biochem.73.011303.073723
Skalka, A. M., and Katz, R. A. (2005). Retroviral DNA Integration and the DNA Damage Response. Cell Death Differ 12 (Suppl. 1), 971–978. doi:10.1038/sj.cdd.4401573
Speelmon, E. C., Livingston-Rosanoff, D., Li, S. S., Vu, Q., Bui, J., Geraghty, D. E., et al. (2006). Genetic Association of the Antiviral Restriction Factor TRIM5α with Human Immunodeficiency Virus Type 1 Infection. J. Virol. 80 (5), 2463–2471. doi:10.1128/JVI.80.5.2463-2471.2006
Usategui-Martín, R., Gutiérrez-Cerrajero, C., Jiménez-Vázquez, S., Calero-Paniagua, I., García-Aparicio, J., Corral-Gudino, L., et al. (2018). Polymorphisms in Genes Implicated in Base Excision Repair (BER) Pathway Are Associated with Susceptibility to Paget's Disease of Bone. Bone 112, 19–23. doi:10.1016/j.bone.2018.04.003
Vidyant, S., Chatterjee, A., Agarwal, V., and Dhole, T. N. (2017). Susceptibility to HIV-1 Infection Is Influenced by Toll Like Receptor-2 (−196 to −174) Polymorphism in a north Indian Population. J. Gene Med. 19 (8), e2971. doi:10.1002/jgm.2971
Wang, C., Zhao, C., Zhang, X., Xu, L., Jia, X., Sun, H., et al. (2016). The Polymorphisms of MSH6 Gene Are Associated with AIDS Progression in a Northern Chinese Population. Infect. Genet. Evol. 42, 9–13. doi:10.1016/j.meegid.2016.04.016
Wang, L., Qian, J., Ying, C., Zhuang, Y., Shang, X., and Xu, F. (2016). X-ray Cross-Complementing Groups 1 Rs1799782 C>T Polymorphisms and Colorectal Cancer Susceptibility: A Meta-Analysis Based on Chinese Han Population. J. Cancer Res. Ther. 12 (Supplement), C264–C267. doi:10.4103/0973-1482.200753
Wang, Y., Wang, C., Qi, S., Liu, Z., Su, G., and Zheng, Y. (2019). Interaction between XRCC 1 Gene Polymorphisms and Diabetes on Susceptibility to Primary Open-Angle Glaucoma. Exp. Biol. Med. (Maywood) 244 (7), 588–592. doi:10.1177/1535370219842791
Whitaker, A. M., Schaich, M. A., Smith, M. R., Flynn, T. S., and Freudenthal, B. D. (2017). Base Excision Repair of Oxidative DNA Damage: From Mechanism to Disease. Front. Biosci. 22, 1493–1522. doi:10.2741/4555
Yan, N., Cherepanov, P., Daigle, J. E., Engelman, A., and Lieberman, J. (2009). The SET Complex Acts as a Barrier to Autointegration of HIV-1. Plos Pathog. 5 (3), e1000327. doi:10.1371/journal.ppat.1000327
Yoder, K. E., Espeseth, A., Wang, X.-H., Fang, Q., Russo, M. T., Lloyd, R. S., et al. (2011). The Base Excision Repair Pathway Is Required for Efficient Lentivirus Integration. PLoS ONE 6 (3), e17862. doi:10.1371/journal.pone.0017862
Yu, L., Liu, X., Han, C., Lu, S., Zhu, G., Su, H., et al. (2016). XRCC1 Rs25487 Genetic Variant and TP53 Mutation at Codon 249 Predict Clinical Outcomes of Hepatitis B Virus-Related Hepatocellular Carcinoma after Hepatectomy: A Cohort Study for 10 years' Follow up. Hepatol. Res. 46 (8), 765–774. doi:10.1111/hepr.12611
Keywords: human immunodeficiency virus, acquired immune deficiency syndrome, base excision repair, single nucleotide polymorphism, male and male homosexual transmission
Citation: Liu B, Wang K, Wu J, Hu Y, Yang X, Xu L, Sun W, Jia X, Wu J, Fu S, Qiao Y and Zhang X (2022) Association of APEX1 and XRCC1 Gene Polymorphisms With HIV-1 Infection Susceptibility and AIDS Progression in a Northern Chinese MSM Population. Front. Genet. 13:861355. doi: 10.3389/fgene.2022.861355
Received: 24 January 2022; Accepted: 21 February 2022;
Published: 16 March 2022.
Edited by:
Zhaowen Qiu, Northeast Forestry University, ChinaReviewed by:
Qing Zhan, King Abdullah University of Science and Technology, Saudi ArabiaJiayang Guo, Xiamen University, China
Copyright © 2022 Liu, Wang, Wu, Hu, Yang, Xu, Sun, Jia, Wu, Fu, Qiao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuandong Qiao, cWlhb3l1YW5kb25nQGhyYm11LmVkdS5jbg==; Xuelong Zhang, emhhbmd4dWVsb25nQGhyYm11LmVkdS5jbg==
†These authors have contributed equally to this work
 Bangquan Liu1,2†
Bangquan Liu1,2† Jiawei Wu
Jiawei Wu Wenjing Sun
Wenjing Sun Yuandong Qiao
Yuandong Qiao