- 1Posttranscriptional Gene Regulation, University Medical Centre Mainz, Mainz, Germany
- 2Institute for Clinical Chemistry and Laboratory Medicine, University Medical Centre Mainz, Mainz, Germany
- 3Centre for Thrombosis and Hemostasis (CTH), University Medical Centre Mainz, Mainz, Germany
- 4Institute of Molecular Biology (IMB), Mainz, Germany
- 5Institute of Developmental Biology and Neurobiology (IDN), Johannes Gutenberg University, Mainz, Germany
- 6German Centre for Cardiovascular Research (DZHK), Berlin, Germany
- 7Centre for Healthy Aging (CHA) Mainz, Mainz, Germany
The integrity of the genome is governed by multiple processes to ensure optimal survival and to prevent the inheritance of deleterious traits. While significant progress has been made to characterize components involved in the DNA Damage Response (DDR), little is known about the interplay between RNA processing and the maintenance of genome stability. Here, we describe the emerging picture of an intricate bidirectional coupling between RNA processing and genome integrity in an integrative manner. By employing insights from a recent large-scale RNAi screening involving the depletion of more than 170 components that direct (alternative) polyadenylation, we provide evidence of bidirectional crosstalk between co-transcriptional RNA 3′end processing and the DDR in a manner that optimizes genomic integrity. We provide instructive examples illustrating the wiring between the two processes and show how perturbations at one end are either compensated by buffering mechanisms at the other end, or even propel the initial insult and thereby become disease-eliciting as evidenced by various disorders.
DNA Damage Regulation
The exposure of cells to exogenous or endogenous stresses (such as UV-light, radiation, certain pharmaceuticals, oxidative agents as well as replication or transcription errors) can result in structural alterations of the DNA. To maintain the integrity of the genome, cells are equipped with intricate (and sometimes redundant) molecular networks that are ready to both detect and correct DNA damage. These networks take part in a protective cellular program known as DNA damage response (DDR) that, beyond merely detecting the damage, is also responsible for activating cellular checkpoints, regulating gene expression, repairing the lesions, and inducing apoptosis, in case the damage exceeds the cell-intrinsic repair capacity. The coordinated sequence of these events involves dedicated components, including DNA damage sensors (such as the apical kinases ATM or ATR and the DNA-coating counterparts MRE11-RAD50-NBS1 complex and RPA) as well as effector proteins (including the checkpoint kinases Chk1 and Chk2), which in turn arrest the cell cycle until the damage is repaired. Although there is still much to be learned, the main processes underlying the DDR are understood in great detail (Zhou and Elledge, 2000; Ciccia and Elledge, 2010; Pilié et al., 2019). It is becoming increasingly apparent however, that RNA regulation is adding a layer of complexity to the DDR in terms of RNA transcription, RNA turnover, and post-transcriptional modifications. Indeed, RNA metabolism is emerging as a critical contributor to genome integrity, starting from early observations that steady-state transcripts levels are decreased upon DNA damage (Lenzken et al., 2013). Several layers of transcriptome regulation are involved in this effect. For example, initiation of transcription has been reported to be repressed in response to UV light-induced damage due to sequestration of the TATA-binding protein from the preinitiation complex (Vichi et al., 1997) and depletion of the initiating hypophosphorylated form of RNA polymerase II (RNAPII) (Rockx et al., 2000). UV light has also been shown to increase phosphorylation of the C-terminal domain (CTD) of RNAPII, leading to a decrease in elongation rates along with changes in alternative splicing (Muñoz et al., 2010). But also pre-mRNA 3′end processing has been linked to mRNA stability during DDR (Kleiman and Manley, 2001). Processing of the RNA 3′end is a crucial feature of most genes and impinges on translocation of the mRNA from the nucleus to the cytoplasm, transcript stability, and ultimately protein output (Danckwardt et al., 2008). It is therefore not surprising that communication between the 3′end processing machinery and the DDR exists to ensure optimal RNA processing and hence genome stability.
The present work focuses on the bidirectional connection between DDR and 3′end processing of genes, i.e. how the DDR affects 3′ end processing and how this, in turn, impinges on the DDR. Apart from illustrating a few well-known examples that connect DDR with 3′end processing, we are also showcasing insights from a recently performed large-scale screening (Ogorodnikov et al., 2018) suggesting that alterations of 3′end processing that normally play a role in damage repair may, under certain circumstances, also propel the DNA damage.
DDR Alterations Affect 3′End Processing Events
In addition to capping and splicing, almost all transcripts in eukaryotes undergo further processing at the RNA 3′end. For most genes, this involves endonucleolytic cleavage and non-templated polyadenylation (CPA) before the mature RNA can be exported into the cytoplasm (Danckwardt et al., 2008) (and refs. therein). As CPA controls almost all genes, regulation of CPA has evolved as an important layer of gene expression. Under damaging conditions, CPA plays a role in RNA surveillance, which prevents the “release” of inappropriate and potentially deleterious transcripts (Nourse et al., 2020). CPA is carried out by a multi-subunit complex involving over 80 trans-acting proteins organized in four core protein subcomplexes, the cleavage and polyadenylation specificity factor (CPSF), cleavage factor I (CFI), cleavage factor II (CFII), and the cleavage stimulation factor (CstF) (Proudfoot, 2016). The recruitment of these multimeric complexes to dedicated, but largely poorly conserved, processing sites (Gruber et al., 2016) ensures that 3′end processing of the nascent transcript occurs in a timely manner and at the right position (Danckwardt et al., 2008). 3′end processing involves an intricate interaction between these complexes (and subcomponents) and RNA polymerase II (RNAP II) (Proudfoot, 2016). Surprisingly, the CPA machinery does not exclusively associate with components involved in the processing of the pre-mRNA but also interacts with certain DDR proteins (Figure 1A). For example, CstF-50, a CstF subunit, interacts with BARD1 and BRCA1 (Kleiman and Manley, 2001), both of which have established functions in DNA repair and checkpoint controls. Upon UV irradiation, levels of CstF-50/BARD1/BRCA1 complexes have been shown to increase (Kleiman and Manley, 2001) leading to a reduction of RNA 3′ endonucleolytic cleavage by direct interaction between CstF-50 and the BARD1 subunit of the BRCA1/BARD1 heterodimer (Kleiman and Manley, 1999). Moreover, BRCA/BARD1 targets the hyperphosphorylated RNAP II for ubiquitination and ultimately proteasomal degradation, inhibiting the coupled transcription-RNA processing machinery and facilitating repair (Kleiman et al., 2005). Consistent with this bridging function of CstF between CPA and DDR, depletion of CstF proved to enhance UV sensitivity, prevent DNA repair, and lead to cell-cycle arrest and apoptosis in B Cells and HeLa cells (Takagaki and Manley, 1998; Mirkin et al., 2008). Similarly to the BRCA1-BARD1-CstF-50 complex, CstF-50 was also shown to bind to the p53-BARD1 complex in-vitro, which results in an inhibition of the 3′end cleavage (Nazeer et al., 2011). Thus, modulating CPA appears to be an important target of the DDR, which could increase survival in multiple ways: i) in preventing damaged transcripts from producing toxic proteins and ii) to promote efficient repair of the genomic lesion (further detailed below). In addition, shifting the balance between polyadenylation and deadenylation can also control the total level of (preexistent) mature RNAs in response to DNA damage, as illustrated by the interaction between CstF-50 and Poly(A)-Specific Ribonuclease (PARN) (Cevher et al., 2010). Here, the Nuclear cap-binding protein subunit 1 (CBP80), which inhibits deadenylation through PARN binding, dissociates from PARN and thereby activates cap-dependent deadenylation resulting in reduced levels of total mRNAs (Balatsos et al., 2006). Additionally, PARN can directly decrease the stability of the p53 mRNA in non-stress conditions through ARE sequence present in the 3′UTR of p53 mRNA (Devany et al., 2013). Under DNA damage conditions, p53 protein accumulation allows its association to, and activation of, PARN ultimately decreasing the expression of target mRNAs in the p53-dependent DDR pathway.
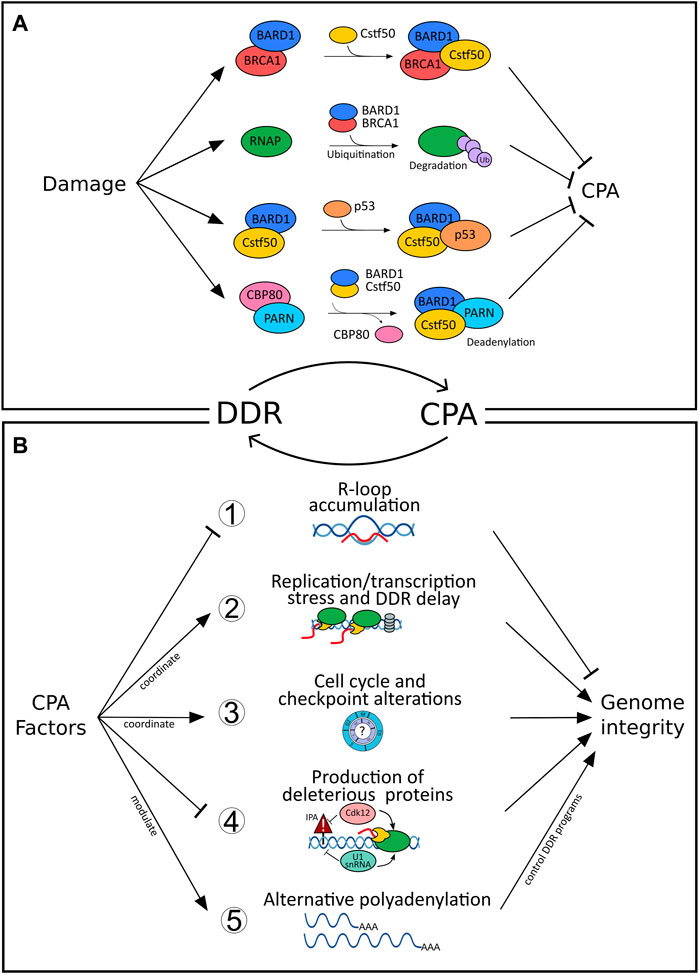
FIGURE 1. Bidirectional connections between DNA damage response (DDR) and cleavage and polyadenylation (CPA). (A) Inhibition of cleavage and polyadenylation (CPA) as a response to DNA damage fosters repair of the genomic lesions and prevents the release of defective transcripts. For example, CstF-50/BARD1/BRCA1-complex formation after UV irradiation blocks RNA 3′ endonucleolytic cleavage (Kleiman and Manley, 2001) and results in RNAP II ubiquitination and proteasomal degradation (Kleiman et al., 2005). Similarly, binding of the p53-BARD1 complex inhibits the 3′end cleavage activity of CstF-50 (Nazeer et al., 2011). The interaction between CstF-50 and PARN shifts the balance between polyadenylation and deadenylation, ultimately controlling the level of mature RNAs in response to DNA damage (Cevher et al., 2010). CBP80, which inhibits deadenylation through PARN binding, dissociates from PARN upon damage and activates cap-dependent deadenylation resulting in reduced levels of total mRNAs (Balatsos et al., 2006). In contrast, (B) CPA can impair genome integrity when deregulated (further details see text, Supplementary Table S1 and Figure 2). For example, (1) loss of function of select CPA components (such as PCF11, CLP1, FIP1L1, CFT2, WDR33) impairs genome integrity by resulting in R-loop formation (Stirling et al., 2012; Teloni et al., 2019). (2) Transcription and replication are coordinated to prevent collision between both machineries and prevent genomic instability (Gaillard and Aguilera, 2014). CFIm depletion impairs transcription termination, which interferes with replication and results in the delay of DDR (Gaillard and Aguilera, 2014) as well as increased sensitivity to UV light (Mirkin et al., 2008). (3) Alterations of CPA components result in cell cycle and checkpoint alterations; for example, CstF64 deficiency results in G0/G1 arrest (Takagaki and Manley, 1998) or loss of symplekin in a G2/M arrest (Ruepp et al., 2011). (4) Cdk12 suppresses intronic polyadenylation (IPA) and thereby fosters the production of full-length gene products, which for example affects many homologous recombination genes. Intronic alternative polyadenylation resulting from U1snRNA downregulation after UV (Devany et al., 2016) alters IPA of genes involved in the UV-response (such as POLR2A and CDKN1A). (5) >70% of all genes are affected by alternative polyadenylation (APA) resulting in functional diversity (Nourse et al., 2020). CPA factors pervasively control APA (Marini et al., 2021) and thereby drive programs involved in the DDR (Figure 2; Supplementary Table S1).
Taken together, these mechanisms involving core CPA factors during DDR evidence an important role of 3′end processing in minimizing the risk of generating inadequate polyadenylated mRNA isoforms resulting from damaging conditions.
Additional Factors Modulating CPA During DDR
Although evidence linking 3′end processing and DDR exists, the underlying molecular connections are still poorly defined. Insights may emerge through the realization that several factors engaged in the DDR are also a part of the 3′end processing complex (Shi et al., 2009). This is indeed the case for the serine/threonine-protein kinase complex DNA-PKcs-Ku70-Ku86, although its direct role in the phosphorylation of CPA components has not yet been fully clarified (Ariumi et al., 1999). DNA-PK can phosphorylate the Poly (ADP-ribose) polymerase 1 (PARP1), which is both functionally associated with CPA and involved in the DDR. RBBP6, an E3 ubiquitin ligase initially discovered to interact with p53 (Simons et al., 1997), has also been found as part of the 3′end processing machinery. Mpe1, the yeast homolog of the E3 ubiquitin-protein ligase RBBP6, is able to ubiquitinate the poly(A) polymerase PAP1 (Lee and Moore, 2014), suggesting that a possible connection between ubiquitination and CPA for mammal cells may also exist. Corroborating this idea, some RBBP6 protein isoforms alter polyadenylation signal selection and promote alternative polyadenylation at distal signals throughout the genome (Di Giammartino et al., 2014). Furthermore, the tumor suppressors CSR1 (cellular stress response 1) and CDC73 have been found to modulate 3′end processing via inducing CPSF3 translocation from the nucleus to the cytoplasm and consequently inhibiting CPA or facilitating association of 3′end processing factors with actively-transcribed chromatin, respectively (Rozenblatt-Rosen et al., 2009; Zhu et al., 2009).
Selective 3′End Processing of DDR Genes
The efficient response to DNA damage requires the expression of multiple DDR genes for the repair process to begin. DDR expression must occur, despite the above-described global inhibition of transcription and 3′UTR processing (Figure 1A). Due to this paradoxical scenario, compensatory mechanisms must be in place to allow selective transcription and escape from the global 3′UTR processing inhibition following DNA damage. In fact, increased transcription for some DDR-responsive genes has been observed (Christmann and Kaina, 2013) and it is conceivable that their upregulation compensates for the inhibition of processing. However, specific regulatory features have also been found to facilitate the 3′end processing of DDR genes. The intrinsic strength of polyA signals (PAS), downstream sequence elements (DSE), and upstream sequence elements (USE) appear to be enhanced in DDR genes fostering effective 3′end processing even in conditions favoring low-levels of CPA complex formation. RNA secondary structures are involved, as exemplified by the tumor suppressor p53 (Decorsière et al., 2011). The p53 pre-mRNA is capable of bypassing the general 3′processing inhibition upon UV-C stimulation via interaction with the ribonucleoproteins hnRNP H/F after an RNA helicase, DHX36, unwinds a G-quadruplex structure downstream of a p53 polyadenylation site to allow access of hnRNP H/F promoting effective 3′end processing (Newman et al., 2017).
Apart from processes acting in cis, 3′end processing of DDR genes can also be facilitated through determinants in trans. For instance, select genes that belong to the p53 pathway bypass the requirement for RNAP II phosphorylation on Ser2, which is usually required for proper 3′end processing (Gomes et al., 2006). However, other early response genes, such as c-fos and junB, are dependent on Ser2 phosphorylation, suggesting that different compensatory mechanisms might exist for different stress stimuli (Fujita et al., 2008). Other types of RNA polymerases can also participate in DDR regulation. This is the case of the non-canonical Star-PAP and its ability to bind directly the pre mRNA (Laishram and Anderson, 2010). Upon stress induction, Star-PAP can regulate 3′end processing in a gene- and condition-specific manner. As an example, etoposide treatment induces Star-PAP binding to the proapoptotic gene Bcl-2 interacting killer (BIK) mRNA (Li et al., 2012) while other stressors (oxidative stress) induce Star-PAP regulation of the cytoprotective heme oxygenase HO-1 (Mellman et al., 2008) and quinone oxidoreductase NQO1 (Gonzales et al., 2008). In addition, Star-PAP can also promote alternative polyadenylation (APA), a pervasive mechanism of transcriptome diversification (Ogorodnikov et al., 2016) (further detailed below), of the tumor suppressor PTEN (Li et al., 2017) mediating increase of PTEN protein upon DNA damage. This further supports the involvement of CPA in the regulation of damage-responsive mRNAs. But also factors not belonging to the core CPA machinery are known to modulate APA after DNA damage. The cyclin-dependent kinase CDK12, beyond maintaining RNAP II processivity through its phosphorylation, is also implicated in various pre-mRNA processing mechanisms. Transcript isoforms of the DNA damage response activator ATM are regulated via modulation of alternative last exon (ALE) inclusion by CDK12 (Tien et al., 2017). Further, CDK12 is known to protect from premature CPA and loss of expression of long genes, including those participating in the DDR (Krajewska et al., 2019). p38 MAPK, another kinase known to promote cell survival under damaging conditions (Phong et al., 2010), modulates polyadenylation after (genotoxic) stress in prothrombin and several other cancer-associated genes including BCL2L2, OTUD7B, PDCD10, PDGFRA as well as KIN with a role in DNA replication and the cellular response to DNA damage (Danckwardt et al., 2007; Danckwardt et al., 2011). Here, select trans-acting RNA-binding proteins (U2AF35, U2AF65, FBP2, FBP3) bind to defined RNA elements (USEs) in a phosphorylation-dependent, antagonistic manner to confer specificity of stress-dependent CPA modulation (Danckwardt et al., 2011). This example illustrates how determinants in cis and trans can co-cooperate resulting in “dual authentication” of condition-dependent selective 3′end RNA processing.
Ultimately, regulation of DDR genes can also occur through mRNA decay mechanisms, which are usually strongly determined by the 3′UTR architecture. One such example is provided by the recruitment of RNA-binding proteins (RBPs) to AU-rich elements (ARE) typically residing in this region. These interactions can either stabilize or destabilize transcripts. For instance, the ARE-binding protein AUF1 can destabilize the growth arrest and DNA damage-inducible gene GADD45a under healthy conditions (Lal et al., 2006). Here, AUF1 competes with the Poly(A) binding protein PABP (Sagliocco et al., 2006) for binding to the polyA tail and promotes recruitment of exosome and proteasomal degradation. Upon genotoxic stress, however, AUF1 dissociates resulting in prolonged GADD45α mRNA half-life. Similarly, interaction with the RBPs TTP and KSRP, known to relay signals of genotoxic stress (Briata et al., 2016; Lee et al., 2020), leads to rapid degradation of ARE-containing target mRNAs through recruitment of deadenylases (Winzen et al., 2007; Clement et al., 2011). Conversely, binding of HuR fosters stabilization of transcripts involved in carcinogenesis, cell proliferation and survival, and oxidative and genotoxic cellular response (Westmark et al., 2005; Li et al., 2018). Collectively, these findings document that the processing of DDR genes can be regulated on various layers to likely accommodate the specific needs under conditions of genotoxic stress.
CPA Factors can Modulate the DDR
Although mounting evidence suggests that 3′end processing may be a key regulator taking part in genome stability and the damage response, the reciprocal role of CPA components in modulating the DDR remains elusive (see above). Evidence for a critical function of CPA for the maintenance of genome integrity, independent from a primary genotoxic insult, is provided by R-loops (Figure 1B). R-loops are three-stranded structures composed of an RNA-DNA hybrid and a displaced single-stranded DNA (ssDNA) (Crossley et al., 2019; García-Muse and Aguilera, 2019). When R-loops are not removed in a timely manner, they can lead to compromised genome integrity. R-loops are abundant in the proximity of polyadenylation sites and correlate with efficient termination of transcription (Sanz et al., 2016). The coordinated co-transcriptional processing and packaging of the nascent transcript into “inert” ribonucleoprotein particles (RNPs) ensures that R-loops occur in a scheduled manner preventing the formation of otherwise deleterious RNA-DNA hybrid structures (Danckwardt et al., 2008). Events that perturb the coordinated co-transcriptional processing result in unscheduled R-loop accumulation, which can lead to replication-associated DNA damage. A loss-of-function screening study performed in budding yeast highlighted seven essential protein components of 3′end processing machinery (including PCF11, CLP1, FIP1L1, and CFT2) to maintain genome integrity by suppressing R-loop formation (Stirling et al., 2012). In another study, deregulation of WDR33, a component of the core CPA machinery, impaired cleavage of nascent pre-mRNA leading to R-loop accumulation and slowing of the replication fork during the S phase (Teloni et al., 2019). Additional factors, localized at the RNA 3′end and cooperating in transcription termination demonstrate the same functional outcome, as in the case of loss of function of Rtt103, yeast homolog of RPRD1B, (Stirling et al., 2012), Sen1 (senataxin) (Mischo et al., 2011) and XRN2 (Morales et al., 2016). Taken together, the evidence is consistent with findings regarding the harmful effects of (unscheduled) R-loops (beyond their physiological role in transcription termination) and supports a novel contribution of CPA in promoting genome integrity (Figure 1B).
CPA regulated replication stress can however also take other forms (beyond R-loops defects) and different players come into action. For instance, CFIm depletion mediates a delayed activation of DNA damage checkpoint signaling and RNA polymerase II degradation following DNA damage (Gaillard and Aguilera, 2014) as well as increased sensitivity to UV light (Mirkin et al., 2008). Similarly, depletion of CstF64, leads to destabilization of the entire Cstf complex, rendering cells deficient in recovery from UV treatment and in the repair of the UV-induced DNA lesions (Mirkin et al., 2008). CstF64 deficient cells accumulate in the G0/G1 stage of cell cycle (Takagaki and Manley, 1998). Alterations in cell cycle progression are also seen upon depletion of symplekin (Ruepp et al., 2011), which may affect repair pathway decisions. Similarly, depletion of CPSF73 and CPSF100, with resulting effects on symplekin expression, affected histone pre-mRNA processing (Sullivan et al., 2009). Finally, CstF2tau has recently been shown to control the abundance of snRNAs (through alternative oligoadenylation) resulting in alternative splicing of several RNAs including the histone deacetylase HDAC2 (Kargapolova et al., 2017), a critical component in the maintenance of genome stability (Miller et al., 2010; Dovey et al., 2013).
The role of CPA factors in safeguarding genome integrity has also been documented beyond their function in RNA processing. After ubiquitin-mediated degradation of RNAP II, chromatin must be made accessible to allow the repair machinery to access the lesion. Here, co-recruitment of CstF-50 with the ubiquitin escort factor p97 contributes to the displacement of ubiquitinated histones H2A, H2B, and nucleosome remodeling (Fonseca et al., 2018). Moreover, Cdk12 has recently been shown to globally suppress intronic polyadenylation thereby fostering the production of full-length gene products (Figure 1B). This also affects many homologous recombination genes, and accordingly Cdk12 loss of function mutations frequently found in tumors globally impair genome integrity (Dubbury et al., 2018).
These findings indicate that individual CPA components (and associated factors) can likely impair central processes that are directly involved in the suppression or the propagation of genotoxic stress. However, what is the evidence that CPA components can be more globally linked to the surveillance of genome integrity?
Global Insights Linking DDR With Alternative Cleavage and Polyadenylation
Early reports on UV-irradiated sun-damaged fibroblasts suggested a link between the usage of different polyA signals (PAS) and genotoxic stress (Schwartz et al., 1998). The regulation of PAS choice in response to damaging agents seems to depend on the trigger as well as on the cell type. Intronic alternative polyadenylation, mediated by U1snRNA downregulation was observed upon UV treatment of mammalian cells (Devany et al., 2016). Topoisomerase inhibitors, on the other hand, promoted alternative last exons (ALE) through APA (alternative polyadenylation) (Dutertre et al., 2014). In yeast, the same UV treatment led to a global lengthening of transcripts (Graber et al., 2013), also observed on cells after treatment with anisomycin (Hollerer et al., 2016). In contrast, the damaging agent arsenite led to transcript shortening and preferential degradation of species with long 3′ends (Zheng et al., 2018).
Recently, diversification of the transcriptome at the 3′end by APA emerged as a pervasive and evolutionarily conserved layer of gene regulation (Derti et al., 2012). It affects more than 70% of all genes, resulting in transcript isoforms with distinct 3′end termini. APA thereby considerably expands the diversity of the transcriptome 3′end (TREND). This leads to mRNA isoforms with profoundly different physiological effects, by affecting protein output, production of distinct protein isoforms, or modulating protein localization (Mayr, 2017). As APA is globally regulated in various conditions, including developmental and adaptive programs [with perturbations resulting in numerous disorders (Nourse et al., 2020)], it is tempting to speculate that APA may also modulate, respond and contribute to perturbations of DNA damage and its resolution. Interrogating the dynamic APA landscape may thus provide further insights into the functional connection between the CPA machinery and DNA damage. A recent large scale RNAi screening based on the depletion of >170 CPA components and associated factors involved in numerous facets of RNA metabolism showed, on a genome-wide level, how individual (CPA) factors affect the APA landscape (Ogorodnikov et al., 2018), including the resulting effects on gene ontologies (Marini et al., 2021). While CPA components pervasively regulate APA [with key components, namely NUDT21, CPSF6 and PCF11, affecting the largest proportion of genes (Marini et al., 2021)], a significant proportion of APA is controlled by components involved in transcription and other co- and post-transcriptional events (e.g., splicing and RNA turnover) or epigenetic modification (Ogorodnikov et al., 2018). Interestingly, this screening also identified APA regulation to be caused by factors involved in genome surveillance or known to drive tumor-suppressive programs (e.g., TP53), as well as other processes involved in the coupling between oncogenic signals and 3′end processing (such as BARD1, see above) (Marini et al., 2021). Corroborating the role of CPA factors in DDR, alterations of expression levels of CPA components have already been evidenced in cell cycle processes and cancer biology. For instance, NUDT21 levels were found to be downregulated in glioblastoma tumors, where the resulting 3′UTR shortening causally led to enhanced cellular proliferation and tumorigenicity, probably through the upregulation of growth-promoting factors, such as cyclin D1 (Masamha et al., 2014). In agreement, PAK1, which recently emerged as a component of the DDR (Pérez-Yépez et al., 2018), was found to be a downstream target of NUDT21, serving as a predictive prognostic marker for glioblastoma patients (Chu et al., 2019). Contrary to NUDT21 downregulation, CPSF6 or CSTF2 were shown to be upregulated in hepatocellular carcinoma and urothelial carcinoma respectively (Chen et al., 2018; Tan et al., 2021). This in turn correlated with the upregulation of NQO1 and RAC1 through the favorable usage of its proximal 3′UTR poly(A) promoting tumor formation and progression. Further, PCF11 has previously been shown to control pathways (such as EIF2 and IGF1) converging on WNT-signaling (Ogorodnikov et al., 2018), functionally associated with DNA damage (Karimaian et al., 2017; Zhao et al., 2018; Pasadi and Muniyappa, 2020). Interestingly PCF11 constitutes a central component shaping transcriptome 3′end diversity. At the same time, it also represents a developmental switch that directly links RNA 3′end processing with aberrant development and tumor formation (Ogorodnikov et al., 2018). Persistently high levels of PCF11 expression in the postnatal period arrest neuronal precursors in an immature state and ultimately give rise to neuroblastomas, the most common solid tumor in children (Maris, 2010). While high-level PCF11 determines a fatal disease progression, low levels of PCF11 instead associate with favorable outcome and spontaneous tumor regression (Ogorodnikov et al., 2018). The contribution of PCF11 to APA-mediated DDR is seemingly noticeable when taking a closer look into which biological processes are modulated in a PCF11-dependent manner (http://shiny.imbei.uni-mainz.de:3838/trend-db/) (Marini et al., 2021) (Figure 2A). The DNA metabolism cluster, defined by 83 significantly APA-affected genes, falls into various enriched subsets of DNA metabolism including DNA repair and cell responses to DNA damage. Accordingly, the vast majority of all those genes are associated with various disorders (mainly developmental) but also with other entities including cancer. This corresponds to the previously made observations indicating a central role of PCF11 for differentiation/dedifferentiation and developmental programs (Ogorodnikov et al., 2018; Kamieniarz-Gdula et al., 2019).
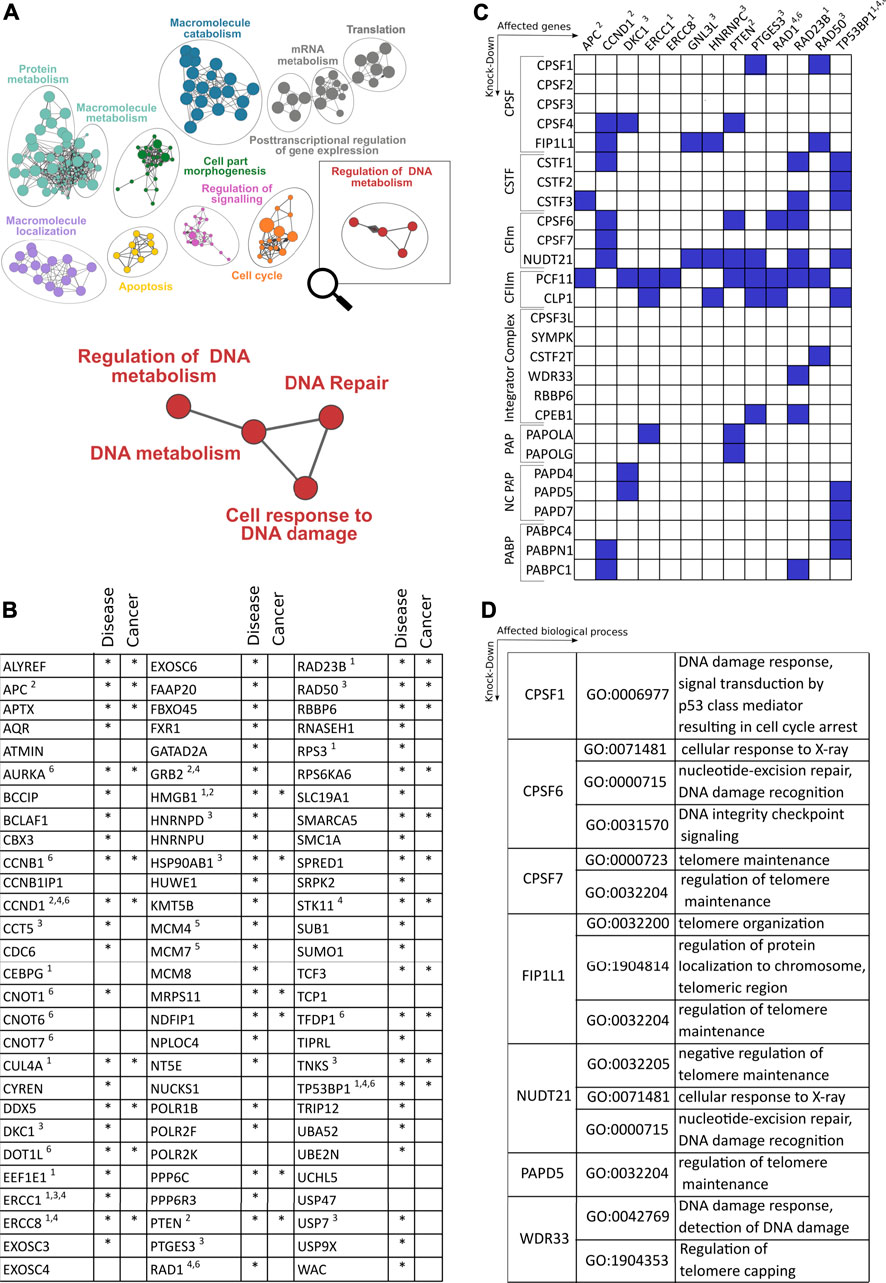
FIGURE 2. Role of cleavage and polyadenylation factors (CPA) on genes and pathways involved in DNA damage response. (A) Enriched GO terms based on genes that show alterations of polyadenylation after depletion of the CFIm component PCF11 (Ogorodnikov et al., 2018; Marini et al., 2021). (B) Close-up of nodes with functional enrichment centering around regulation of DNA metabolism based on 83 genes regulated upon depletion of PCF11. The table depicts the identity of altered genes and their involvement in disease including cancer (selected entities of DDR response mechanisms highlighted are: 1. DNA repair/nucleotide excision, 2. ATM-dependent DNA response, 3. Telomer regulation, 4. Response to X-ray, 5. Double strand breaks, 6. DNA damage checkpoint response). (C) Matrix of selected genes (x-axis; functional assignment see legend in (B)) involved in DDR with significant alterations of polyadenylation after depletion of canonical and non-canonical 3′end processing factors (y-axis; blue boxes indicate significant changes; data obtained from TREND-DB (Marini et al., 2021) covering a large scale RNAi screening (Ogorodnikov et al., 2018) coupled to transcriptome-wide interrogation of alterations in polyadenylation by TRENDseq (Ogorodnikov and Danckwardt, 2021); further details see text). (D) Gene Ontology (GO) enrichment of processes centering around DDR upon depletion of CPA factors indicated (Supplementary Table S1 provides further details on DDR genes affected by alternative polyadenylation).
As one would expect based on the pervasive function of several CPA components for APA modulation (Marini et al., 2021) and the resulting functional impact on protein output (Ogorodnikov et al., 2018), PCF11 does not only control APA of critical genes involved in the DDR (encompassing a wide variety of functional categories, including DNA damage checkpoint responses, DNA repair, nucleotide excision, ATM-dependent DNA response, telomere function, response to X-ray and double-strand breaks). In fact, albeit to variable extent, depletion of almost all CPA components results in the regulation 3′end processing of critical regulators involved in several aspects of DDR (Figure 2C). While 3′end processing of some of these DDR components appears to be selectively regulated (e.g., the CSA Ubiquitin Ligase Complex Subunit ERCC Excision Repair 8 (ERCC8), ERCC1, or the tumor suppressor adenomatosis polyposis coli (APC)), others tend to be more broadly controlled by several factors of the CPA machinery. Interestingly this includes central genes with known functions in nucleotide excision, DNA damage checkpoint- and ATM-dependent DNA damage response (CCND1, PTEN, RAD23B, TP53BP1). This suggests that modulation of RNA 3′processing can directly influence several components involved in the maintenance of genome integrity. Accordingly, beyond PCF11 (Figures 2A,B), the functional enrichment upon depletion of further CPA components (CPSF1, CPSF6, CPSF7, FIP1L1, NUDT21, PAPD5 and WDR33) also reveals significantly enriched GO terms that are associated with various aspects of DNA damage control and repair (Figure 2D). This suggests that mechanisms have evolved that directly link co-transcriptional processing of pre-mRNAs to genome integrity and support a critical function of APA as a pervasive and evolutionarily conserved layer of gene regulation in this context (Figure 2; Supplementary Table S1).
An interesting, yet to be explored, hypothesis arising from these findings is that such a coupling could serve as a buffering system conferring robustness to biological systems. This would ensure that the otherwise detrimental consequences arising from aberrant RNA processing (such as R-loop formation) result in the induction of compensatory mechanisms governing genome integrity (Ngo et al., 2021); Figure 1B), and vice versa, or result in apoptosis of affected cells when a certain threshold is exceeded. Alternatively, aberrant processing could also initiate (and propagate) a failure of genome surveillance, and thereby aggravate a phenotype resulting from initially (minor) perturbations of 3′end processing. As such, the frequently observed perturbations of APA in cancer deserve attention (Nourse et al., 2020). They can act as non-genomic drivers of cancer (Ogorodnikov et al., 2018) and may also result in consecutive downstream perturbations of mechanisms involved in the surveillance of genome integrity (such as WNT-, EIF2- or IGF1-signaling (Ogorodnikov et al., 2018)) with established roles in genome surveillance (Wek et al., 2006; Vermeij et al., 2016; Pasadi and Muniyappa, 2020), eventually perpetuating and even aggravating the initial disease phenotype. Aberrant 3′end processing could thereby contribute to the clonal evolution of tumor lesions (Hugo et al., 2015). Vice versa, APA alterations could also reflect the consequences of impaired DDR with potentially both ends, either aggravating the resulting effects further or resulting in responses that ultimately help the cell to (sense and) repair the mutation and thereby resolve the genomic instability (Figures 1A,B).
Collectively, CPA components that mediate RNA 3′processing and shape the transcriptome diversity are thus emerging as important pillars in the maintenance of genome integrity and vice versa. In view of the emerging opportunities to interfere with CPA (and APA) in a targeted manner (Nourse et al., 2020) further functional characterization of the bidirectional link between both processes may also open hitherto novel untapped therapeutic avenues.
Outlook
Tools capable of elucidating polyadenylation and involved components in a transcriptome-wide manner (Kargapolova et al., 2017; Ogorodnikov and Danckwardt, 2021) bear great potential to decipher the resulting functional implications including the coupling of 3′end processing with genome integrity. Combined with technologies that enable the genome-wide interrogation of intermediates of impaired DNA damage repair inflict [such as R-loops; (Ginno et al., 2012; Yan et al., 2019; Wang et al., 2021)] or the mapping of the resulting consequences [dsDNA breaks etc., (Singh et al., 1988; Zhu et al., 2019)], complemented by loss-of-function studies, will help to further unwind the intricate reciprocal coupling between CPA and DDR. These studies are urgently needed to better understand the sequence of these events, their contribution to common pathologies, and to uncover novel potentially druggable driver lesions in devastating disorders such as cancer.
Author Contributions
SS, BL, and SD viewed the literature and prepared the manuscript.
Funding
Work in the Danckwardt lab is kindly supported by the German Research Foundation Priority Program SPP 1935, German Research Foundation grants DA 1189/2-1 and DA 1189/5-1, Germany Research Foundation Major Research Instrumentation Program INST 371/33-1, the German Research Foundation graduate school GRK 1591, Federal Ministry of Education and Research (BMBF01EO1003), German Society of Clinical and Laboratory Medicine and the Hella Bühler Award for Cancer Research. SS is supported by the EU Horizon 2020 Innovative Training Network, Thromboinflammation in Cardiovascular Research (TICARDIO). Work in the Luke lab was supported by the German Research Foundation Heisenberg program LU 1709/2-1.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to express their gratitude to current and former members of the Danckwardt lab.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.854907/full#supplementary-material
Supplementary Table S1 | Examples of DDR genes that are affected by alternative polyadenylation (APA) and DDR components affecting cleavage and polyadenylation.
References
Akman, B. H., Can, T., and Erson-Bensan, A. E. (2012). Estrogen-induced Upregulation and 3′-UTR Shortening of CDC6. Nucleic Acids Res. 40, 10679–10688. doi:10.1093/nar/gks855
Ariumi, Y., Masutani, M., Copeland, T. D., Mimori, T., Sugimura, T., Shimotohno, K., et al. (1999). Suppression of the poly(ADP-Ribose) Polymerase Activity by DNA-dependent Protein Kinase In Vitro. Oncogene 18, 4616–4625. doi:10.1038/sj.onc.1202823
Balatsos, N. A. A., Nilsson, P., Mazza, C., Cusack, S., and Virtanen, A. (2006). Inhibition of mRNA Deadenylation by the Nuclear Cap Binding Complex (CBC). J. Biol. Chem. 281, 4517–4522. doi:10.1074/jbc.m508590200
Batista, L., Bourachot, B., Mateescu, B., Reyal, F., and Mechta-Grigoriou, F. (2016). Regulation of miR-200c/141 Expression by Intergenic DNA-Looping and Transcriptional Read-Through. Nat. Commun. 7, 8959. doi:10.1038/ncomms9959
Briata, P., Bordo, D., Puppo, M., Gorlero, F., Rossi, M., Perrone‐Bizzozero, N., et al. (2016). Diverse Roles of the Nucleic Acid-Binding Protein KHSRP in Cell Differentiation and Disease, 7. Wiley Interdiscip Rev RNA, 227–240. doi:10.1002/wrna.1327Diverse Roles of the Nucleic Acid‐binding Protein KHSRP in Cell Differentiation and DiseaseWIREs RNA
Cevher, M. A., Zhang, X., Fernandez, S., Kim, S., Baquero, J., Nilsson, P., et al. (2010). Nuclear Deadenylation/polyadenylation Factors Regulate 3′ Processing in Response to DNA Damage. Embo j 29, 1674–1687. doi:10.1038/emboj.2010.59
Chen, X., Zhang, J. X., Luo, J. H., Wu, S., Yuan, G. J., Ma, N. F., et al. (2018). CSTF2-Induced Shortening of the RAC1 3'UTR Promotes the Pathogenesis of Urothelial Carcinoma of the Bladder. Cancer Res. 78, 5848–5862. doi:10.1158/0008-5472.CAN-18-0822
Chirackal Manavalan, A. P., Pilarova, K., Kluge, M., Bartholomeeusen, K., Rajecky, M., Oppelt, J., et al. (2019). CDK12 Controls G1/S Progression by Regulating RNAPII Processivity at Core DNA Replication Genes. EMBO Rep. 20, e47592. doi:10.15252/embr.201847592
Christmann, M., and Kaina, B. (2013). Transcriptional Regulation of Human DNA Repair Genes Following Genotoxic Stress: Trigger Mechanisms, Inducible Responses and Genotoxic Adaptation. Nucleic Acids Res. 41, 8403–8420. doi:10.1093/nar/gkt635
Chu, Y., Elrod, N., Wang, C., Li, L., Chen, T., Routh, A., et al. (2019). Nudt21 Regulates the Alternative Polyadenylation of Pak1 and Is Predictive in the Prognosis of Glioblastoma Patients. Oncogene 38, 4154–4168. doi:10.1038/s41388-019-0714-9
Ciccia, A., and Elledge, S. J. (2010). The DNA Damage Response: Making it Safe to Play with Knives. Mol. Cel 40, 179–204. doi:10.1016/j.molcel.2010.09.019
Clement, S. L., Scheckel, C., Stoecklin, G., and Lykke-Andersen, J. (2011). Phosphorylation of Tristetraprolin by MK2 Impairs AU-Rich Element mRNA Decay by Preventing Deadenylase Recruitment. Mol. Cel Biol 31, 256–266. doi:10.1128/mcb.00717-10
Crossley, M. P., Bocek, M., and Cimprich, K. A. (2019). R-loops as Cellular Regulators and Genomic Threats. Mol. Cel 73, 398–411. doi:10.1016/j.molcel.2019.01.024
Danckwardt, S., Gantzert, A.-S., Macher-Goeppinger, S., Probst, H. C., Gentzel, M., Wilm, M., et al. (2011). p38 MAPK Controls Prothrombin Expression by Regulated RNA 3′ End Processing. Mol. Cel 41, 298–310. doi:10.1016/j.molcel.2010.12.032
Danckwardt, S., Hentze, M. W., and Kulozik, A. E. (2008). 3′ End mRNA Processing: Molecular Mechanisms and Implications for Health and Disease. Embo j 27, 482–498. doi:10.1038/sj.emboj.7601932
Danckwardt, S., Kaufmann, I., Gentzel, M., Foerstner, K. U., Gantzert, A.-S., Gehring, N. H., et al. (2007). Splicing Factors Stimulate Polyadenylation via USEs at Non-canonical 3′ End Formation Signals. Embo j 26, 2658–2669. doi:10.1038/sj.emboj.7601699
Decorsière, A., Cayrel, A., Vagner, S., and Millevoi, S. (2011). Essential Role for the Interaction between hnRNP H/F and a G Quadruplex in Maintaining P53 Pre-mRNA 3'-end Processing and Function during DNA Damage. Genes Dev. 25, 220–225. doi:10.1101/gad.607011
Derti, A., Garrett-Engele, P., Macisaac, K. D., Stevens, R. C., Sriram, S., Chen, R., et al. (2012). A Quantitative Atlas of Polyadenylation in Five Mammals. Genome Res. 22, 1173–1183. doi:10.1101/gr.132563.111
Devany, E., Park, J. Y., Murphy, M. R., Zakusilo, G., Baquero, J., Zhang, X., et al. (2016). Intronic Cleavage and Polyadenylation Regulates Gene Expression during DNA Damage Response through U1 snRNA. Cell Discov 2, 16013. doi:10.1038/celldisc.2016.13
Devany, E., Zhang, X., Park, J. Y., Tian, B., and Kleiman, F. E. (2013). Positive and Negative Feedback Loops in the P53 and mRNA 3′ Processing Pathways. Proc. Natl. Acad. Sci. U.S.A. 110, 3351–3356. doi:10.1073/pnas.1212533110
Di Giammartino, D. C., Li, W., Ogami, K., Yashinskie, J. J., Hoque, M., Tian, B., et al. (2014). RBBP6 Isoforms Regulate the Human Polyadenylation Machinery and Modulate Expression of mRNAs with AU-Rich 3′ UTRs. Genes Dev. 28, 2248–2260. doi:10.1101/gad.245787.114
Dovey, O. M., Foster, C. T., Conte, N., Edwards, S. A., Edwards, J. M., Singh, R., et al. (2013). Histone Deacetylase 1 and 2 Are Essential for normal T-Cell Development and Genomic Stability in Mice. Blood 121, 1335–1344. doi:10.1182/blood-2012-07-441949
Dubbury, S. J., Boutz, P. L., and Sharp, P. A. (2018). CDK12 Regulates DNA Repair Genes by Suppressing Intronic Polyadenylation. Nature 564, 141–145. doi:10.1038/s41586-018-0758-y
Dutertre, M., Chakrama, F. Z., Combe, E., Desmet, F.-O., Mortada, H., Polay Espinoza, M., et al. (2014). A Recently Evolved Class of Alternative 3′-terminal Exons Involved in Cell Cycle Regulation by Topoisomerase Inhibitors. Nat. Commun. 5, 3395. doi:10.1038/ncomms4395
Fan, Z., Devlin, J. R., Hogg, S. J., Doyle, M. A., Harrison, P. F., Todorovski, I., et al. (2020). CDK13 Cooperates with CDK12 to Control Global RNA Polymerase II Processivity. Sci. Adv. 6. doi:10.1126/sciadv.aaz5041
Fonseca, D., Baquero, J., Murphy, M. R., Aruggoda, G., Varriano, S., Sapienza, C., et al. (2018). mRNA Processing Factor CstF-50 and Ubiquitin Escort Factor P97 Are BRCA1/BARD1 Cofactors Involved in Chromatin Remodeling during the DNA Damage Response. Mol. Cel Biol 38. doi:10.1128/MCB.00364-17
Fontana, G. A., Rigamonti, A., Lenzken, S. C., Filosa, G., Alvarez, R., Calogero, R., et al. (2017). Oxidative Stress Controls the Choice of Alternative Last Exons via a Brahma-BRCA1-CstF Pathway. Nucleic Acids Res. 45, 902–914. doi:10.1093/nar/gkw780
Fujita, T., Ryser, S., Piuz, I., and Schlegel, W. (2008). Up-regulation of P-TEFb by the MEK1-Extracellular Signal-Regulated Kinase Signaling Pathway Contributes to Stimulated Transcription Elongation of Immediate Early Genes in Neuroendocrine Cells. Mol. Cel Biol 28, 1630–1643. doi:10.1128/mcb.01767-07
Gaillard, H., and Aguilera, A. (2014). Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in Saccharomyces cerevisiae. Plos Genet. 10, e1004203. doi:10.1371/journal.pgen.1004203
García-Muse, T., and Aguilera, A. (2019). R Loops: From Physiological to Pathological Roles. Cell 179, 604–618.
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I., and Chédin, F. (2012). R-loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Mol. Cel 45, 814–825. doi:10.1016/j.molcel.2012.01.017
Gomes, N. P., Bjerke, G., Llorente, B., Szostek, S. A., Emerson, B. M., and Espinosa, J. M. (2006). Gene-specific Requirement for P-TEFb Activity and RNA Polymerase II Phosphorylation within the P53 Transcriptional Program. Genes Dev. 20, 601–612. doi:10.1101/gad.1398206
Gonzales, M. L., Mellman, D. L., and Anderson, R. A. (2008). CKIα Is Associated with and Phosphorylates Star-PAP and Is Also Required for Expression of Select Star-PAP Target Messenger RNAs. J. Biol. Chem. 283, 12665–12673. doi:10.1074/jbc.m800656200
Graber, J. H., Nazeer, F. I., Yeh, P.-c., Kuehner, J. N., Borikar, S., Hoskinson, D., et al. (2013). DNA Damage Induces Targeted, Genome-wide Variation of Poly(A) Sites in Budding Yeast. Genome Res. 23, 1690–1703. doi:10.1101/gr.144964.112
Gruber, A. J., Schmidt, R., Gruber, A. R., Martin, G., Ghosh, S., Belmadani, M., et al. (2016). A Comprehensive Analysis of 3′ End Sequencing Data Sets Reveals Novel Polyadenylation Signals and the Repressive Role of Heterogeneous Ribonucleoprotein C on Cleavage and Polyadenylation. Genome Res. 26, 1145–1159. doi:10.1101/gr.202432.115
Hollerer, I., Curk, T., Haase, B., Benes, V., Hauer, C., Neu-Yilik, G., et al. (2016). The Differential Expression of Alternatively Polyadenylated Transcripts Is a Common Stress-Induced Response Mechanism that Modulates Mammalian mRNA Expression in a Quantitative and Qualitative Fashion. Rna 22, 1441–1453. doi:10.1261/rna.055657.115
Hugo, W., Shi, H., Sun, L., Piva, M., Song, C., Kong, X., et al. (2015). Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 162, 1271–1285. doi:10.1016/j.cell.2015.07.061
Kamieniarz-Gdula, K., Gdula, M. R., Panser, K., Nojima, T., Monks, J., Wiśniewski, J. R., et al. (2019). Selective Roles of Vertebrate PCF11 in Premature and Full-Length Transcript Termination. Mol. Cel 74, 158–172. e159. doi:10.1016/j.molcel.2019.01.027
Kargapolova, Y., Levin, M., Lackner, K., and Danckwardt, S. (2017). sCLIP-an Integrated Platform to Study RNA-Protein Interactomes in Biomedical Research: Identification of CSTF2tau in Alternative Processing of Small Nuclear RNAs. Nucleic Acids Res. 45, 6074–6086. doi:10.1093/nar/gkx152
Karimaian, A., Majidinia, M., Bannazadeh Baghi, H., and Yousefi, B. (2017). The Crosstalk between Wnt/β-Catenin Signaling Pathway with DNA Damage Response and Oxidative Stress: Implications in Cancer Therapy. DNA Repair 51, 14–19. doi:10.1016/j.dnarep.2017.01.003
Kleiman, F. E., and Manley, J. L. (1999). Functional Interaction of BRCA1-Associated BARD1 with Polyadenylation Factor CstF-50. Science 285, 1576–1579. doi:10.1126/science.285.5433.1576
Kleiman, F. E., and Manley, J. L. (2001). The BARD1-CstF-50 Interaction Links mRNA 3′ End Formation to DNA Damage and Tumor Suppression. Cell 104, 743–753. doi:10.1016/s0092-8674(01)00270-7
Kleiman, F. E., Wu-Baer, F., Fonseca, D., Kaneko, S., Baer, R., and Manley, J. L. (2005). BRCA1/BARD1 Inhibition of mRNA 3′ Processing Involves Targeted Degradation of RNA Polymerase II. Genes Dev. 19, 1227–1237. doi:10.1101/gad.1309505
Krajewska, M., Dries, R., Grassetti, A. V., Dust, S., Gao, Y., Huang, H., et al. (2019). CDK12 Loss in Cancer Cells Affects DNA Damage Response Genes through Premature Cleavage and Polyadenylation. Nat. Commun. 10, 1757. doi:10.1038/s41467-019-09703-y
Kunisky, A. K., Anyaeche, V. I., Herron, R. S., Park, C. Y., and Hwang, H.-W. (2021). Shift in MSL1 Alternative Polyadenylation in Response to DNA Damage Protects Cancer Cells from Chemotherapeutic Agent-Induced Apoptosis. Cel Rep. 37, 109815. doi:10.1016/j.celrep.2021.109815
Laishram, R. S., and Anderson, R. A. (2010). The Poly A Polymerase Star-PAP Controls 3′-end Cleavage by Promoting CPSF Interaction and Specificity toward the Pre-mRNA. Embo j 29, 4132–4145. doi:10.1038/emboj.2010.287
Lal, A., Abdelmohsen, K., Pullmann, R., Kawai, T., Galban, S., Yang, X., et al. (2006). Posttranscriptional Derepression of GADD45α by Genotoxic Stress. Mol. Cel 22, 117–128. doi:10.1016/j.molcel.2006.03.016
Lee, J.-H., Kang, B.-H., Jang, H., Kim, T. W., Choi, J., Kwak, S., et al. (2015). AKT Phosphorylates H3-Threonine 45 to Facilitate Termination of Gene Transcription in Response to DNA Damage. Nucleic Acids Res. 43, 4505–4516. doi:10.1093/nar/gkv176
Lee, S. D., and Moore, C. L. (2014). Efficient mRNA Polyadenylation Requires a Ubiquitin-like Domain, a Zinc Knuckle, and a RING finger Domain, All Contained in the Mpe1 Protein. Mol. Cel Biol 34, 3955–3967. doi:10.1128/mcb.00077-14
Lee, T.-H., Choi, J. Y., Park, J.-M., and Kang, T.-H. (2020). Posttranscriptional Control of the Replication Stress Response via TTP-Mediated Claspin mRNA Stabilization. Oncogene 39, 3245–3257. doi:10.1038/s41388-020-1220-9
Lenzken, S. C., Loffreda, A., and Barabino, S. M. (20132013). RNA Splicing: a New Player in the DNA Damage Response. Int. J. Cel Biol 2013, 153634. doi:10.1155/2013/153634
Li, W., Laishram, R. S., Ji, Z., Barlow, C. A., Tian, B., and Anderson, R. A. (2012). Star-PAP Control of BIK Expression and Apoptosis Is Regulated by Nuclear PIPKIα and PKCδ Signaling. Mol. Cel 45, 25–37. doi:10.1016/j.molcel.2011.11.017
Li, W., Li, W., Laishram, R. S., Hoque, M., Ji, Z., Tian, B., et al. (2017). Distinct Regulation of Alternative Polyadenylation and Gene Expression by Nuclear Poly(A) Polymerases. Nucleic Acids Res. 45, 8930–8942. doi:10.1093/nar/gkx560
Li, Y., Estep, J. A., and Karginov, F. V. (2018). Transcriptome-wide Identification and Validation of Interactions between the miRNA Machinery and HuR on mRNA Targets. J. Mol. Biol. 430, 285–296. doi:10.1016/j.jmb.2017.12.006
Lin, Y., Li, Z., Ozsolak, F., Kim, S. W., Arango-Argoty, G., Liu, T. T., et al. (2012). An In-Depth Map of Polyadenylation Sites in Cancer. Nucleic Acids Res. 40, 8460–8471. doi:10.1093/nar/gks637
Marini, F., Scherzinger, D., and Danckwardt, S. (2021). TREND-DB-a Transcriptome-wide Atlas of the Dynamic Landscape of Alternative Polyadenylation. Nucleic Acids Res. 49, D243–d253. doi:10.1093/nar/gkaa722
Maris, J. M. (2010). Recent Advances in Neuroblastoma. N. Engl. J. Med. 362, 2202–2211. doi:10.1056/nejmra0804577
Masamha, C. P., Xia, Z., Yang, J., Albrecht, T. R., Li, M., Shyu, A.-B., et al. (2014). CFIm25 Links Alternative Polyadenylation to Glioblastoma Tumour Suppression. Nature 510, 412–416. doi:10.1038/nature13261
Mayr, C. (2017). Regulation by 3′-Untranslated Regions. Annu. Rev. Genet. 51, 171–194. doi:10.1146/annurev-genet-120116-024704
Mellman, D. L., Gonzales, M. L., Song, C., Barlow, C. A., Wang, P., Kendziorski, C., et al. (2008). A PtdIns4,5P2-Regulated Nuclear Poly(A) Polymerase Controls Expression of Select mRNAs. Nature 451, 1013–1017. doi:10.1038/nature06666
Miller, K. M., Tjeertes, J. V., Coates, J., Legube, G., Polo, S. E., Britton, S., et al. (2010). Human HDAC1 and HDAC2 Function in the DNA-Damage Response to Promote DNA Nonhomologous End-Joining. Nat. Struct. Mol. Biol. 17, 1144–1151. doi:10.1038/nsmb.1899
Mirkin, N., Fonseca, D., Mohammed, S., Cevher, M. A., Manley, J. L., and Kleiman, F. E. (2008). The 3' Processing Factor CstF Functions in the DNA Repair Response. Nucleic Acids Res. 36, 1792–1804. doi:10.1093/nar/gkn005
Mischo, H. E., Gómez-González, B., Grzechnik, P., Rondón, A. G., Wei, W., Steinmetz, L., et al. (2011). Yeast Sen1 Helicase Protects the Genome from Transcription-Associated Instability. Mol. Cel 41, 21–32. doi:10.1016/j.molcel.2010.12.007
Morales, J. C., Richard, P., Patidar, P. L., Motea, E. A., Dang, T. T., Manley, J. L., et al. (2016). XRN2 Links Transcription Termination to DNA Damage and Replication Stress. Plos Genet. 12, e1006107. doi:10.1371/journal.pgen.1006107
Muñoz, M. J., De La Mata, M., and Kornblihtt, A. R. (2010). The Carboxy Terminal Domain of RNA Polymerase II and Alternative Splicing. Trends Biochem. Sci. 35, 497–504.
Nazeer, F. I., Devany, E., Mohammed, S., Fonseca, D., Akukwe, B., Taveras, C., et al. (2011). p53 Inhibits mRNA 3′ Processing through its Interaction with the CstF/BARD1 Complex. Oncogene 30, 3073–3083. doi:10.1038/onc.2011.29
Newman, M., Sfaxi, R., Saha, A., Monchaud, D., Teulade-Fichou, M.-P., and Vagner, S. (2017). The G-quadruplex-specific RNA Helicase DHX36 Regulates P53 Pre-mRNA 3′-End Processing Following UV-Induced DNA Damage. J. Mol. Biol. 429, 3121–3131. doi:10.1016/j.jmb.2016.11.033
Ngo, G. H. P., Grimstead, J. W., and Baird, D. M. (2021). UPF1 Promotes the Formation of R Loops to Stimulate DNA Double-Strand Break Repair. Nat. Commun. 12, 3849. doi:10.1038/s41467-021-24201-w
Nilsson, K., Wu, C., Kajitani, N., Yu, H., Tsimtsirakis, E., Gong, L., et al. (2018). The DNA Damage Response Activates HPV16 Late Gene Expression at the Level of RNA Processing. Nucleic Acids Res. 46, 5029–5049. doi:10.1093/nar/gky227
Nourse, J., Spada, S., and Danckwardt, S. (2020). Emerging Roles of RNA 3'-end Cleavage and Polyadenylation in Pathogenesis, Diagnosis and Therapy of Human Disorders. Biomolecules 10. doi:10.3390/biom10060915
Oegema, R., Baillat, D., Schot, R., Van Unen, L. M., Brooks, A., Kia, S. K., et al. (2017). Human Mutations in Integrator Complex Subunits Link Transcriptome Integrity to Brain Development. Plos Genet. 13, e1006809. doi:10.1371/journal.pgen.1006809
Ogorodnikov, A., and Danckwardt, S. (2021). TRENDseq-A Highly Multiplexed High Throughput RNA 3′ End Sequencing for Mapping Alternative Polyadenylation. Methods Enzymol. 655, 37–72. doi:10.1016/bs.mie.2021.03.022
Ogorodnikov, A., Kargapolova, Y., and Danckwardt, S. (2016). Processing and Transcriptome Expansion at the mRNA 3′ End in Health and Disease: Finding the Right End. Pflugers Arch. - Eur. J. Physiol. 468, 993–1012. doi:10.1007/s00424-016-1828-3
Ogorodnikov, A., Levin, M., Tattikota, S., Tokalov, S., Hoque, M., Scherzinger, D., et al. (2018). Transcriptome 3′end Organization by PCF11 Links Alternative Polyadenylation to Formation and Neuronal Differentiation of Neuroblastoma. Nat. Commun. 9, 5331. doi:10.1038/s41467-018-07580-5
Okada, N., Lin, C.-P., Ribeiro, M. C., Biton, A., Lai, G., He, X., et al. (2014). A Positive Feedback between P53 and miR-34 miRNAs Mediates Tumor Suppression. Genes Dev. 28, 438–450. doi:10.1101/gad.233585.113
Pasadi, S., and Muniyappa, K. (2020). Evidence for Functional and Regulatory Cross-Talk between Wnt/β-Catenin Signalling and Mre11-Rad50-Nbs1 Complex in the Repair of Cisplatin-Induced DNA Cross-Links. Oncotarget 11, 4028–4044. doi:10.18632/oncotarget.27777
Passacantilli, I., Panzeri, V., Bielli, P., Farini, D., Pilozzi, E., Fave, G. D., et al. (2017). Alternative Polyadenylation of ZEB1 Promotes its Translation during Genotoxic Stress in Pancreatic Cancer Cells. Cell Death Dis 8, e3168. doi:10.1038/cddis.2017.562
Pérez-Yépez, E. A., Saldívar-Cerón, H. I., Villamar-Cruz, O., Pérez-Plasencia, C., and Arias-Romero, L. E. (2018). p21 Activated Kinase 1: Nuclear Activity and its Role during DNA Damage Repair. DNA Repair (Amst) 65, 42–46.
Phong, M. S., Van Horn, R. D., Li, S., Tucker-Kellogg, G., Surana, U., and Ye, X. S. (2010). p38 Mitogen-Activated Protein Kinase Promotes Cell Survival in Response to DNA Damage but Is Not Required for the G 2 DNA Damage Checkpoint in Human Cancer Cells. Mol. Cel Biol 30, 3816–3826. doi:10.1128/mcb.00949-09
Pilié, P. G., Tang, C., Mills, G. B., and Yap, T. A. (2019). State-of-the-art Strategies for Targeting the DNA Damage Response in Cancer. Nat. Rev. Clin. Oncol. 16, 81–104. doi:10.1038/s41571-018-0114-z
Proudfoot, N. J. (2016). Transcriptional Termination in Mammals: Stopping the RNA Polymerase II Juggernaut. Science 352, aad9926. doi:10.1126/science.aad9926
Proudfoot, N. J. (2011). Ending the Message: Poly(A) Signals Then and Now. Genes Dev. 25, 1770–1782. doi:10.1101/gad.17268411
Quereda, V., Bayle, S., Vena, F., Frydman, S. M., Monastyrskyi, A., Roush, W. R., et al. (2019). Therapeutic Targeting of CDK12/CDK13 in Triple-Negative Breast Cancer. Cancer Cell 36, 545–558. e547. doi:10.1016/j.ccell.2019.09.004
Raptis, V., Georgianos, P. I., Sarafdis, P. A., Sioulis, A., Makedou, K., Makedou, A., et al. (2013). Elevated Asymmetric Dimethylarginine Is Associated with Oxidant Stress Aggravation in Patients with Early Stage Autosomal Dominant Polycystic Kidney Disease, 38. Basel: Kidney Blood Press Res, 72–82. doi:10.1159/000355756Elevated Asymmetric Dimethylarginine Is Associated with Oxidant Stress Aggravation in Patients with Early Stage Autosomal Dominant Polycystic Kidney DiseaseKidney Blood Press. Res.
Riaz, M., Raz, Y., Van Putten, M., Paniagua-Soriano, G., Krom, Y. D., Florea, B. I., et al. (2016). PABPN1-Dependent mRNA Processing Induces Muscle Wasting. Plos Genet. 12, e1006031. doi:10.1371/journal.pgen.1006031
Richard, P., Trollet, C., Stojkovic, T., De Becdelievre, A., Perie, S., Pouget, J., et al. (2017). Correlation between PABPN1 Genotype and Disease Severity in Oculopharyngeal Muscular Dystrophy. Neurology 88, 359–365. doi:10.1212/wnl.0000000000003554
Rockx, D. A. P., Mason, R., Van Hoffen, A., Barton, M. C., Citterio, E., Bregman, D. B., et al. (2000). UV-induced Inhibition of Transcription Involves Repression of Transcription Initiation and Phosphorylation of RNA Polymerase II. Proc. Natl. Acad. Sci. U.S.A. 97, 10503–10508. doi:10.1073/pnas.180169797
Rozenblatt-Rosen, O., Nagaike, T., Francis, J. M., Kaneko, S., Glatt, K. A., Hughes, C. M., et al. (2009). The Tumor Suppressor Cdc73 Functionally Associates with CPSF and CstF 3′ mRNA Processing Factors. Proc. Natl. Acad. Sci. U.S.A. 106, 755–760. doi:10.1073/pnas.0812023106
Ruepp, M.-D., Schweingruber, C., Kleinschmidt, N., and Schümperli, D. (2011). Interactions of CstF-64, CstF-77, and Symplekin: Implications on Localisation and Function. MBoC 22, 91–104. doi:10.1091/mbc.e10-06-0543
Sagliocco, F., Laloo, B., Cosson, B., Laborde, L., Castroviejo, M., Rosenbaum, J., et al. (2006). The ARE-Associated Factor AUF1 Binds Poly(A) In Vitro in Competition with PABP. Biochem. J. 400, 337–347. doi:10.1042/bj20060328
Sanz, L. A., Hartono, S. R., Lim, Y. W., Steyaert, S., Rajpurkar, A., Ginno, P. A., et al. (2016). Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cel 63, 167–178. doi:10.1016/j.molcel.2016.05.032
Schwartz, E., Gelfand, J. M., Mauch, J. C., and Kligman, L. H. (1998). Generation of a Tropoelastin mRNA Variant by Alternative Polyadenylation Site Selection in Sun-Damaged Human Skin and Ultraviolet B-Irradiated Fibroblasts. Biochem. Biophysical Res. Commun. 246, 217–221. doi:10.1006/bbrc.1998.8601
Shi, Y., Di Giammartino, D. C., Taylor, D., Sarkeshik, A., Rice, W. J., Yates, J. R., et al. (2009). Molecular Architecture of the Human Pre-mRNA 3′ Processing Complex. Mol. Cel 33, 365–376. doi:10.1016/j.molcel.2008.12.028
Simons, A., Melamed-Bessudo, C., Wolkowicz, R., Sperling, J., Sperling, R., Eisenbach, L., et al. (1997). PACT: Cloning and Characterization of a Cellular P53 Binding Protein that Interacts with Rb. Oncogene 14, 145–155. doi:10.1038/sj.onc.1200825
Singh, N. P., Mccoy, M. T., Tice, R. R., and Schneider, E. L. (1988). A Simple Technique for Quantitation of Low Levels of DNA Damage in Individual Cells. Exp. Cel Res. 175, 184–191. doi:10.1016/0014-4827(88)90265-0
Stirling, P. C., Chan, Y. A., Minaker, S. W., Aristizabal, M. J., Barrett, I., Sipahimalani, P., et al. (2012). R-loop-mediated Genome Instability in mRNA Cleavage and Polyadenylation Mutants. Genes Dev. 26, 163–175. doi:10.1101/gad.179721.111
Sullivan, K. D., Steiniger, M., and Marzluff, W. F. (2009). A Core Complex of CPSF73, CPSF100, and Symplekin May Form Two Different Cleavage Factors for Processing of Poly(A) and Histone mRNAs. Mol. Cel 34, 322–332. doi:10.1016/j.molcel.2009.04.024
Sun, M., Ding, J., Li, D., Yang, G., Cheng, Z., and Zhu, Q. (2017). NUDT21 Regulates 3′-UTR Length and microRNA-Mediated Gene Silencing in Hepatocellular Carcinoma. Cancer Lett. 410, 158–168. doi:10.1016/j.canlet.2017.09.026
Takagaki, Y., and Manley, J. L. (1998). Levels of Polyadenylation Factor CstF-64 Control IgM Heavy Chain mRNA Accumulation and Other Events Associated with B Cell Differentiation. Mol. Cel 2, 761–771. doi:10.1016/s1097-2765(00)80291-9
Tan, S., Zhang, M., Shi, X., Ding, K., Zhao, Q., Guo, Q., et al. (2021). CPSF6 Links Alternative Polyadenylation to Metabolism Adaption in Hepatocellular Carcinoma Progression. J. Exp. Clin. Cancer Res. 40, 85. doi:10.1186/s13046-021-01884-z
Teloni, F., Michelena, J., Lezaja, A., Kilic, S., Ambrosi, C., Menon, S., et al. (2019). Efficient Pre-mRNA Cleavage Prevents Replication-Stress-Associated Genome Instability. Mol. Cel 73, 670–683. e612. doi:10.1016/j.molcel.2018.11.036
Tian, B., and Manley, J. L. (2017). Alternative Polyadenylation of mRNA Precursors. Nat. Rev. Mol. Cel Biol 18, 18–30. doi:10.1038/nrm.2016.116
Tien, J. F., Mazloomian, A., Cheng, S.-W. G., Hughes, C. S., Chow, C. C. T., Canapi, L. T., et al. (2017). CDK12 Regulates Alternative Last Exon mRNA Splicing and Promotes Breast Cancer Cell Invasion. Nucleic Acids Res. 45, 6698–6716. doi:10.1093/nar/gkx187
Tummala, H., Walne, A., Collopy, L., Cardoso, S., De La Fuente, J., Lawson, S., et al. (2015). Poly(A)-specific Ribonuclease Deficiency Impacts Telomere Biology and Causes Dyskeratosis Congenita. J. Clin. Invest. 125, 2151–2160. doi:10.1172/jci78963
Vermeij, W. P., Hoeijmakers, J. H. J., and Pothof, J. (2016). Genome Integrity in Aging: Human Syndromes, Mouse Models, and Therapeutic Options. Annu. Rev. Pharmacol. Toxicol. 56, 427–445. doi:10.1146/annurev-pharmtox-010814-124316
Vichi, P., Coin, F., Renaud, J. P., Vermeulen, W., Hoeijmakers, J. H., Moras, D., et al. (1997). Cisplatin- and UV-Damaged DNA Lure the Basal Transcription Factor TFIID/TBP. Embo j 16, 7444–7456. doi:10.1093/emboj/16.24.7444
Wang, K., Wang, H., Li, C., Yin, Z., Xiao, R., Li, Q., et al. (2021). Genomic Profiling of Native R Loops with a DNA-RNA Hybrid Recognition Sensor. Sci. Adv. 7. doi:10.1126/sciadv.abe3516
Wang, L., Lang, G.-T., Xue, M.-Z., Yang, L., Chen, L., Yao, L., et al. (2020). Dissecting the Heterogeneity of the Alternative Polyadenylation Profiles in Triple-Negative Breast Cancers. Theranostics 10, 10531–10547. doi:10.7150/thno.40944
Wang, Q., He, G., Hou, M., Chen, L., Chen, S., Xu, A., et al. (2018). Cell Cycle Regulation by Alternative Polyadenylation of CCND1. Sci. Rep. 8, 6824. doi:10.1038/s41598-018-25141-0
Wek, R. C., Jiang, H.-Y., and Anthony, T. G. (2006). Coping with Stress: eIF2 Kinases and Translational Control. Biochem. Soc. Trans. 34, 7–11. doi:10.1042/bst0340007
Westmark, C. J., Bartleson, V. B., and Malter, J. S. (2005). RhoB mRNA Is Stabilized by HuR after UV Light. Oncogene 24, 502–511. doi:10.1038/sj.onc.1208224
Williamson, L., Saponaro, M., Boeing, S., East, P., Mitter, R., Kantidakis, T., et al. (2017). UV Irradiation Induces a Non-coding RNA that Functionally Opposes the Protein Encoded by the Same Gene. Cell 168, 843–855. e813. doi:10.1016/j.cell.2017.01.019
Wilson, W. C., Hornig-Do, H.-T., Bruni, F., Chang, J. H., Jourdain, A. A., Martinou, J.-C., et al. (2014). A Human Mitochondrial Poly(A) Polymerase Mutation Reveals the Complexities of post-transcriptional Mitochondrial Gene Expression. Hum. Mol. Genet. 23, 6345–6355. doi:10.1093/hmg/ddu352
Winzen, R., Thakur, B. K., Dittrich-Breiholz, O., Shah, M., Redich, N., Dhamija, S., et al. (2007). Functional Analysis of KSRP Interaction with the AU-Rich Element of Interleukin-8 and Identification of Inflammatory mRNA Targets. Mol. Cel Biol 27, 8388–8400. doi:10.1128/mcb.01493-07
Yan, Q., Shields, E. J., Bonasio, R., and Sarma, K. (2019). Mapping Native R-Loops Genome-wide Using a Targeted Nuclease Approach. Cel Rep. 29, 1369–1380. e1365. doi:10.1016/j.celrep.2019.09.052
Yang, S. W., Li, L., Connelly, J. P., Porter, S. N., Kodali, K., Gan, H., et al. (2020). A Cancer-specific Ubiquitin Ligase Drives mRNA Alternative Polyadenylation by Ubiquitinating the mRNA 3′ End Processing Complex. Mol. Cel 77, 1206–1221. e1207. doi:10.1016/j.molcel.2019.12.022
Zhang, J., Sun, W., Ren, C., Kong, X., Yan, W., and Chen, X. (2019). A PolH Transcript with a Short 3′UTR Enhances PolH Expression and Mediates Cisplatin Resistance. Cancer Res. 79, 3714–3724. doi:10.1158/0008-5472.can-18-3928
Zhao, Y., Tao, L., Yi, J., Song, H., and Chen, L. (2018). The Role of Canonical Wnt Signaling in Regulating Radioresistance. Cell Physiol Biochem 48, 419–432. doi:10.1159/000491774
Zheng, D., Wang, R., Ding, Q., Wang, T., Xie, B., Wei, L., et al. (2018). Cellular Stress Alters 3′UTR Landscape through Alternative Polyadenylation and Isoform-specific Degradation. Nat. Commun. 9, 2268. doi:10.1038/s41467-018-04730-7
Zhou, B.-B. S., and Elledge, S. J. (2000). The DNA Damage Response: Putting Checkpoints in Perspective. Nature 408, 433–439. doi:10.1038/35044005
Zhu, Y., Biernacka, A., Pardo, B., Dojer, N., Forey, R., Skrzypczak, M., et al. (2019). qDSB-Seq Is a General Method for Genome-wide Quantification of DNA Double-Strand Breaks Using Sequencing. Nat. Commun. 10, 2313. doi:10.1038/s41467-019-10332-8
Keywords: cleavage and polyadenylation, DNA damage response, genome integrity, cancer, alternative polyadenylation, systematic screening, resillience, aging
Citation: Spada S, Luke B and Danckwardt S (2022) The Bidirectional Link Between RNA Cleavage and Polyadenylation and Genome Stability: Recent Insights From a Systematic Screen. Front. Genet. 13:854907. doi: 10.3389/fgene.2022.854907
Received: 14 January 2022; Accepted: 28 March 2022;
Published: 28 April 2022.
Edited by:
Iwona Rzeszutek, University of Rzeszow, PolandReviewed by:
Shalini Sharma, University of Arizona, United StatesYutaka Saito, National Institute of Advanced Industrial Science and Technology (AIST), Japan
De-Li Shi, Sorbonne University, France
Copyright © 2022 Spada, Luke and Danckwardt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sven Danckwardt, U3Zlbi5EYW5ja3dhcmR0QHVuaW1lZGl6aW4tbWFpbnouZGU=
 Stefano Spada
Stefano Spada Brian Luke4,5
Brian Luke4,5 Sven Danckwardt
Sven Danckwardt