- Department of Oral Pathology, Xiangya Stomatological Hospital & Hunan Key Laboratory of Oral Health Research & Hunan 3D Printing Engineering Research Center of Oral Care, Central South University, Changsha, China
For decades, the desire for tissue regeneration has never been quenched. Dental-derived mesenchymal stem cells (DMSCs), with the potential of self-renewal and multi-directional differentiation, have attracted much attention in this topic. Growing evidence suggests that non-coding RNAs (ncRNAs) can activate various regulatory processes. Even with a slight decrease or increase in expression, ncRNAs can weaken or even subvert cellular fate. Therefore, a systematic interpretation of ncRNAs that guide the differentiation of DMSCs into cells of other tissue types is urgently needed. In this review, we introduce the roles of ncRNAs in the differentiation of DMSCs, such as osteogenic differentiation, odontogenic differentiation, neurogenic differentiation, angiogenic differentiation and myogenic differentiation. Additionally, we illustrate the regulatory mechanisms of ncRNAs in the differentiation of DMSCs, such as epigenetic regulation, transcriptional regulation, mRNA modulation, miRNA sponges and signalling. Finally, we summarize the types and mechanisms of ncRNAs in the differentiation of DMSCs, such as let-7 family, miR-17∼92 family, miR-21, lncRNA H19, lncRNA ANCR, lncRNA MEG3, circRNA CDR1as and CircRNA SIPA1L1. If revealing the intricate relationship between ncRNAs and pluripotency of DMSCs 1 day, the application of DMSCs in regenerative medicine and tissue engineering will be improved. Our work could be an important stepping stone towards this future.
Introduction
DMSCs have been widely studied because of their ready availability, easy accessibility and lack of complex ethical issues. Recently, numerous types of DMSCs have been isolated and characterized, including dental pulp stem cells (DPSCs), periodontal ligament stem cells (PDLSCs), stem cells from exfoliated deciduous teeth (SHEDs), dental follicle stem cells (DFPCs), stem cells from apical papilla (SCAPs) and gingival mesenchymal stem cells (GMSCs) (Yin et al., 2021). Similar to other stem cells, DMSCs have multiple differentiation potential and can differentiate into a variety of tissue-like cells under specific induction conditions.
NcRNAs are an abundant class of RNAs that do not encode proteins. Only a few years ago these transcripts were considered “dark matter” in the genome, but now they play leading roles in the regulation of biological processes (Cai et al., 2021). There are many types of ncRNAs, and the main classes of functional ncRNAs that are not translated into proteins include microRNA (miRNA), long ncRNA (lncRNA) and circular RNA (circRNA). MiRNAs are small (∼22 nucleotides), single-stranded, non-coding RNAs encoded by endogenous genes. MiRNA biogenesis often begins with primary miRNA (pri-miRNAs) in the nucleus. Following processing by the Drosha and DRCG8, precursor miRNA (pre-miRNA) is exported from the nucleus by exportin 5. Then, it undergoes further processing to generate mature miRNA. LncRNAs are large non-coding RNAs >200 nucleotides in length, transcribed by RNA polymerase II, and its biogenesis process is similar to miRNA. CircRNAs differ from linear RNAs in that they can form covalently closed continuous loops with their 3′ heads and 5′ tails bound together and their unique closed-loop structure makes them more resistant to RNAseR digestion (Sangere et al., 1976; Danan et al., 2012). At the same time, it has highly conserved characteristics and specificity in expression patterns between cells, tissues and developmental stages (Mumtaz et al., 2020). In the present review, we summarize the important role of miRNA, lncRNA and circRNA in the multidirectional differentiation of DMSCs.
ncRNAs and the Differentiation of DMSCs
Many miRNAs, lncRNAs and circRNAs are associated with the differentiation of DMSCs (Figure 1). NcRNAs provide an additional and promising possibility of the regulation of the differentiation of DMSCs that has not been fully elucidated to date. With increasing numbers of ncRNAs discovered in this process, it has become possible to use these ncRNA-related therapeutic methods in the field of regenerative medicine and tissue engineering.
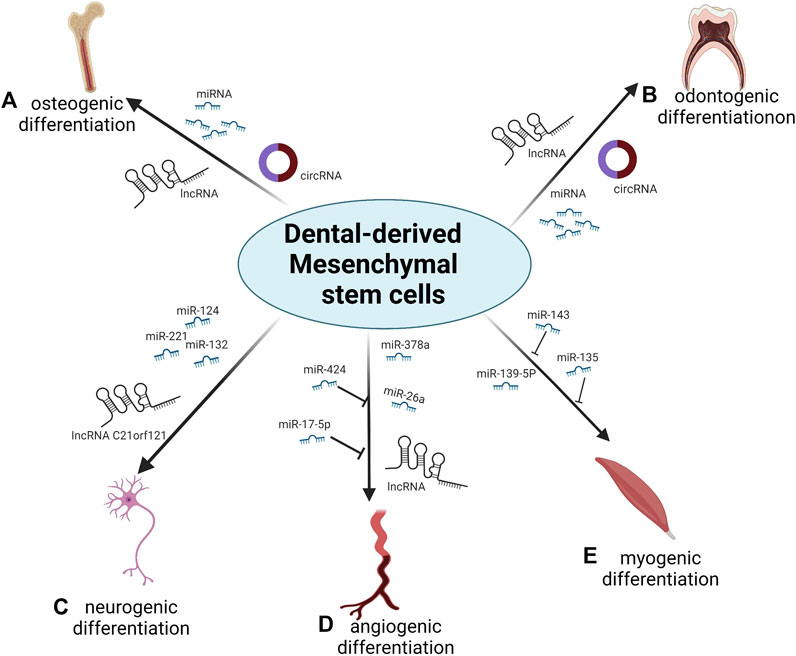
FIGURE 1. Overview of the role of ncRNAs during differentiation of DMSCs. (A) Numerous miRNAs, lncRNAs and circRNAs regulate the osteogenic differentiation of DMSCs. (B) Numerous miRNAs, lncRNAs and circRNAs regulate the odontogenic differentiation of DMSCs. (C) miR-132, miR-124, miR-221and lncRNA C21orf121 promote the neurogenic differentiation of DMSCs. (D) miR-378a and miR-26a promote the angiogenic differentiation of DMSCs; miR-424 and miR-17-5p inhibit the angiogenic differentiation of DMSCs; numerous lncRNAs regulate the angiogenic differentiation of DMSCs. (E) miR-139-5p promotes the myogenic differentiation of DMSCs; miR-143 and miR-135 inhibit the myogenic differentiation of DMSCs.
ncRNAs and the Osteogenic Differentiation of DMSCs
Hao used (Hao et al., 2017) a miRNA microarray to detect the different expression profiles of miRNAs in PDLSCs during the osteogenic differentiation process. The results indicated a significant change in the expression level of 116 miRNAs(30 upregulated and 86 downregulated) in PDLSCs after 14d of osteogenic induction. (Zheng et al., 2018) used RNA sequencing to detect the different expression profiles of lncRNAs in PDLSCs at different time points during osteogenic differentiation. The results showed that 48 lncRNAs had significant changes on days 3, 7 and 14, of which 17 lncRNAs were upregulated and 31 were downregulated in PDLSCs. (Gu et al., 2017) compared the lncRNA profiles and circRNAs profiles of PDLSCs on the seventh day with or without osteogenic differentiation medium using RNA sequencing. The results showed that 17 lncRNAs were upregulated and 31 were downregulated while 766 circRNAs were significantly upregulated and 690 circRNAs were downregulated during osteogenic differentiation in PDLSCs. These results suggest an important regulatory role that ncRNAs might play in the osteogenic differentiation of PDLSCs.
ncRNAs and the Odontogenic Differentiation of DMSCs
A microarray analysis revealed that (Gong et al., 2012) 12 upregulated miRNAs and 10 downregulated miRNAs to be differentially expressed in human DPSCs during odontogenic differentiation process. Further bioinformatic analysis showed that the target genes of these miRNAs are related to the mitogen-activated protein kinase (MAPK) and the Wnt signalling pathways; both pathways are of particular interest to odontogenesis. (Zheng and Jia, 2016) used RNA sequencing to compare the lncRNA profiles between freshly-isolated and cultured dental mesenchymal cells. The analysis indicated that there were a total of 144 lncRNAs (among which 108 were upregulated and 36 were downregulated) that participated in odontogenic differentiation. They also constructed 54 coexpression relationships in the odontogenic process, as well as an lncRNA-mRNA coexpression network. There are 139 differentially expressed genes between odontogenic induced and undifferentiated human DPSCs, with downstream pathways implicating cell cycle, extracellular matrix receptor interaction, and transforming growth factor (TGF)-β signaling pathways (Chen et al., 2016). Microarray research conducted by Chen et al. (2020) showed that 187 circRNAs are differentially expressed after odontogenic induction of human DPSCs. Moreover, hsa_circRNA_104101 was highly assumed to enhance the odontogenic differentiation of human DPSCs. These results indicate that ncRNAs might play crucial roles in this process and regulate odontogenesis-related pathways.
ncRNAs and the Neurogenic Differentiation of DMSCs
Retinal diseases are the leading cause of irreversible visual impairment and blindness in developed countries. In the process of transdifferentiation of hPDLSCs into retinal ganglion-like cells, 71 human miRNAs were differentially expressed (44 were up-regulated and 27 were down-regulated), among which miR-132 was a key regulator of retinal fate determination of hPDLSCs (Ng et al., 2015). In the past, auditory neuropathy (loss or degeneration of spiral ganglion neurons [SGN]) was considered a serious problem with no alternative therapy. MiR-124 was a key regulator of neuronal development in the inner ear and was expressed at high levels in auditory and vestibular neurons. Some scholars transfected hDPSCs with miR-124 to observe the changes of neural progenitor cells and protein levels, and found that the expression of neural marker Nestin could be changed, which opened up broad prospects for the treatment and replacement of SGN (Mehri-Ghahfarrokhi et al., 2019). Chromatin unwinding DNA-binding protein 8(CHD8) inhibited the onset of autism spectrum disorder (ASD), and the upregulation of miR-221 activated the Wnt/β-catenin pathway by binding to CHD8, which facilitating the differentiation from SHEDs to neurons (Wen et al., 2020). Autism is a childhood neurodevelopmental disorder with complex genetic origin. A study in a rat model of autism found that lncRNA C21orf121 promotes the differentiation of SHEDs into neurons by targeting miR-140-5p to upregulate BMP2 (Liu J. et al., 2018).
ncRNAs and the Angiogenic Differentiation of DMSCs
Growing evidence reveals that miRNAs play a critical role in angiogenic processes (Hernández-Romero et al., 2019). When cultured in a medium supplemented with bFGF and VEGF-165, DPSCs were induced toward endothelial differentiation, during which miR-424 was downregulated gradually, revealing that miR-424 negatively regulated the endothelial differentiation of DPSCs (Liu et al., 2014). Extracellular vesicles secreted by periodontitis-compromised DPSCs (P-EVs) carrying miR-378a promoted endothelial cells angiogenesis by downregulating Sufu to activate the Hedgehog/Gli1 signalling pathway (Zhou et al., 2021). In addition, overexpression of miR-17-5p blocked the pro-angiogenic ability of inflamed PDLSCs (Zhang Z. et al., 2020). SHED aggregate exosomes shuttled miR-26a promote angiogenesis both in SHED and Human umbilical vein endothelial cells (HUVECs) via TGF-β/SMAD2/3 signalling (Wu et al., 2021). An lncRNA microarray analysis showed that (Li X. et al., 2020) 376 lncRNAs were significantly increased and 426 lncRNAs were down-regulated among DPSCs cultured in vascular induction medium, suggesting that lncRNAs were also essential for angiogenic processes.
ncRNAs and the Myogenic Differentiation of DMSCs
MiRNAs also play a crucial role in the induction of myogenic differentiation. MiR-139-5p elevated the skeletal myogenic differentiation of hDPSCs by interacting with the Wnt/β-catenin signaling pathway (Xie and Shen, 2018). The expression of miR-143 and miR-135 were significantly downregulated in 5-Aza-CdR-induced myoblast DPSCs. Interestingly, the addition of miR-143 or miR-135 inhibitors to the culture medium stimulates the myogenic properties of DPSCs, which eventually fuses to form myotube (Li et al., 2015).
Regulatory Mechanisms of ncRNAs in the Differentiation of DMSCs
ncRNAs can regulate the differentiation of DMSCs by diverse mechanisms (Figure 2).
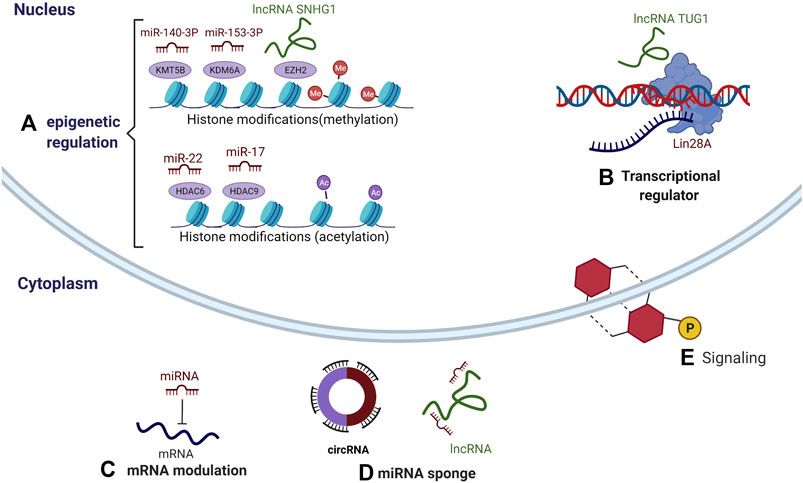
FIGURE 2. Multiple mechanisms of ncRNAs regulating the differentiation of DMSCs. (A) Epigenetic regulation: miR-140-3p interacts with KMT5B, miR-153-3p interacts with KDM6A, and lncRNA SNHG1 interacts with EZH2, resulting in abnormal methylation. miR-22 interacts with HDAC6, and miR-17 interacts with HDAC9, resulting in abnormal acetylation. (B) gene transcription: lncRNA TUG1 interacts with RBP family member-Lin28A, affecting the transcriptional activation of downstream genes. (C) mRNA modulation: miRNA targets the 3′ untranslated region (UTR) of mRNA. (D) miRNA sponges: lncRNAs and circular RNAs target miRNAs. (E) signaling: all three ncRNAs can participate in signaling pathways.
Epigenetic Regulation
Common epigenetic regulation mainly involves multifaceted dynamics, including DNA methylation, histone modifications. It has been reported that mature miRNAs presence in the nucleus, which can activate or silence genes through mechanisms of epigenetic pathways. MiRNAs involved in epigenetic modifications are referred to as epi-miRNAs, such as miR-140-3p, miR-153-3p, miR-22, miR-17. Lysine methyltransferase 5B(KMT5B) belongs to a group of histone methyltransferases (Rodas-Junco et al., 2017; Shen et al., 2019). (Zheng et al., 2021) found that miR-140-3p enhances the osteo/odontogenic differentiation of DPSCs by inhibiting KMT5B under the hypoxia condition simulated by cobalt chloride (CoCl2). Lysine demethylase 6A (KDM6A) belongs to a group of histone demethylases. (Jiang and Jia, 2021) revealed that miR-153-3p inhibits osteogenic differentiation of PDLSCs through KDM6A-induced demethylation of H3K27me3 (trimethylated histone H3 Lysine 27). HDACs can transfer acetyl groups from the histone, resulting in histone hypoacetylation and chromatin accumulation. MiR-22 promoted osteogenic differentiation of human PDLSCs by targeting HDAC6 (Yan et al., 2017). (Li L. et al., 2018) discovered that miR-17 and HDAC9 formed an inhibitory loop under chronic inflammatory conditions, which negatively affected each other. There were HDAC9-enriched promoter regions in miR-17, while miR-17 induced PDLSCS osteogenesis by inhibiting HDAC9.
In the nucleus, lncRNAs regulate the gene expression in various ways. These include epigenetic modifications by the recruitment of chromatin modifiers. Enhancer of the zeste homolog 2 (EZH2) is a histone methyltransferase, specifically in charge of methylation of H3K27me3. IncRNA SNHG1 inhibited the osteogenic differentiation of PDLSCs through EZH2-mediated H3K27me3 methylation of Kruppel-like factor 2(KLF2) promotor (Li X. et al., 2020).
Transcriptional Regulation
LncRNAs can specifically bind with a specific family of proteins called RNA binding proteins (RBPs) to regulate the biological functions of these proteins and then affect the transcriptional activation of downstream genes. As a novel protein type, RBPs have been demonstrated to interact with lncRNAs and have attracted increasing attention from researchers (Kim et al., 2017; Kishore et al., 2011; Wu et al., 2016). He et al. (2018) demonstrated that lin-28 homologue A (Lin28A, a member of the RBP family) contains multiple binding sites of lncRNA-taurine upregulated gene 1 (TUG1) and lncRNA-TUG1 facilitates osteogenic differentiation of PDLSCs via interacting with Lin28A.
mRNA Modulation
In the cytoplasm, binding of mature miRNAs to the 3′ untranslated region (UTR) of specific mRNA targets leads to gene repression by undermining mRNA stability or reducing translation. For example, miRNAs negatively regulate the osteogenesis of DMSCs by targeting the 3′-UTR of osteogenic genes, such as alkaline phosphatase (ALP), osteocalcin (OCN), Runt-related transcription factor 2 (RUNX2), etc. MiR-584-5p inhibits the osteogenic differentiation of human PDLCs by binding to ALPL mRNA and reducing the expression of ALPL protein (Wang et al., 2021). MiR-218 has been confirmed to target Runx2 and play important inhibitory roles in the osteogenic differentiation of PDLSCs, DPSCs and GSCs (Gay et al., 2014). MiR-204 negatively regulates the osteogenic differentiation of human DFPCs (hDPCs) by targeting Runx2 and ALP (Ito et al., 2020).
miRNA Sponges
Some lncRNAs and circular RNAs regulate the activity of miRNAs because they have binding sites that retain them, thus modulating the activity of miRNAs. LncRNAs and circular RNAs, which present this mechanism, are considered miRNA sponges, and they are part of a complex interaction network in the transcriptome or also called the theory of competitive endogenous RNAs(ceRNAs) (Rincon-Riveros et al., 2021).
LINC00968 promotes osteogenic differentiation of DPSCs via regulation of miR-3658/RUNX2 (Liao et al., 2020). LncRNA FER1L4 promotes osteogenic differentiation of human PDLSCs via targeting miR-874-3p (Huang et al., 2020). LncRNA X-Inactive Specific Transcript (XIST) promotes osteogenic differentiation of PDLSCs by sponging miR-214-3p (Feng et al., 2020).
Moreover, circAKT3 positively regulates osteogenic differentiation of human DPSCs via miR-206/CX43 axis (Zhang B. et al., 2020). CircRNA hsa_circ_0026827 has been reported to promote osteoblast differentiation of human DPSCs via Beclin1 and the RUNX1 signaling pathways by sponging miR-188-3p (Ji et al., 2020b). Furthermore, role of exosomes in osteogenic differentiation of DPSCs has been investigated and it was found that circLPAR1 adsorbs to hsa-miR-31 to avoid its inhibitory effects on osteogenesis of DPSCs (Xie et al., 2020).
Based on above, it is well established that the common mechanisms of lncRNAs and circRNAs in DMSCs are competently binding with miRNAs to deliver a promoting effect on osteogenic differentiation.
ncRNA and Signaling
From a molecular point of view, DMSCs differentiation is mediated by several signaling pathways. Notably, all three ncRNAs can become key participants in signaling pathways through various interactions with other biomolecules (Figure 3).
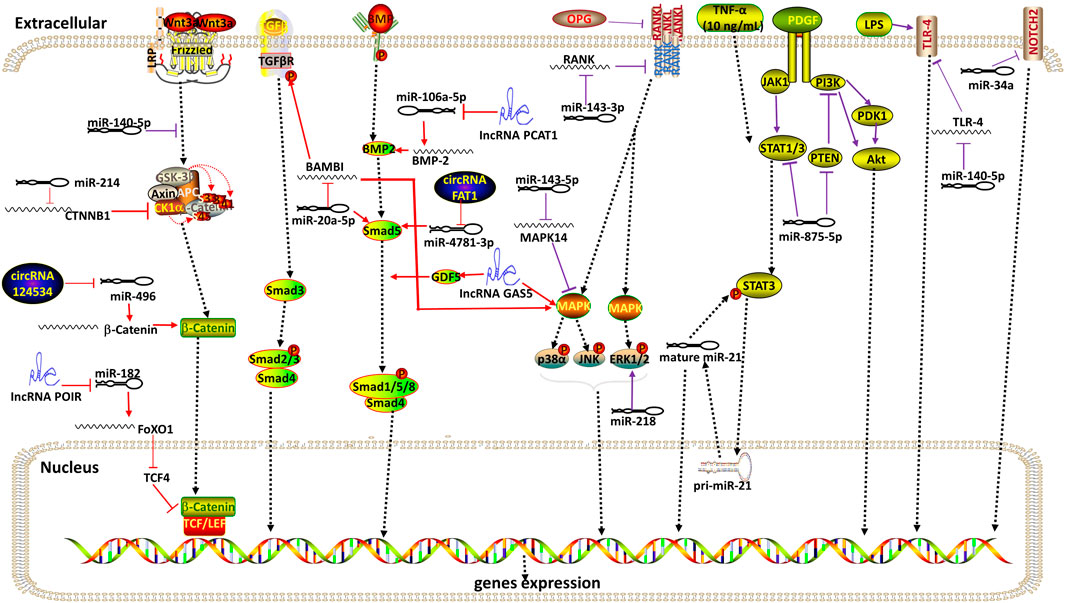
FIGURE 3. Overview of signaling pathways involved by ncRNAs during the differentiation of DMSCs. Red indicates signaling pathways associated with osteogenic differentiation. Purple indicates signaling pathways associated with odontogenic differentiation. Arrow indicates promotion and T indicates inhibition. Collectively, ncRNAs related to Wnt/β-catenin signaling pathway are miR-140-5p, miR-214, circRNA124534 and lncRNA POIR. MiR-20a-5p is involved in TGF-β, BMP and MAPK signaling pathway. NcRNAs associated with the BMP signaling pathway are lncPCAT1 and circRNA FAT1. LncRNA GAS5 is involved in BMP and MAPK signaling pathway. MiR-143-3p inhibits OPG-RANKL signaling pathway. NcRNAs related to the MAPK pathway are miR-143-5p, miR-218. MiR-875-5P is related to PDGF signaling pathway. There is a reciprocal positive feedback loop between miR-21 and STAT3 at low concentrations of TNF-α (1–10 ng/ml). MiR-140-5p inhibits the TLR-4 pathway under LPS stimulation. MiR-34a inhibits Notch signaling pathway.
Wnt Signaling Pathway
The Wnt/β-catenin signaling pathway plays an important role in regulating DMSCs differentiation, which is a key signaling pathway. Canonical Wnt signaling is activated by the Wnt1/3a protein when it binds to the Frizzled (FZD), and the low-density LRP5 or LRP6 complex at the cell surface. In the absence of Wnt, β-catenin is phosphorylated by a complex that contains GSK3. β-catenin accumulates in the cytosol and translocates in the nucleus to bind TCF/LEF transcription factor, thereby regulating target gene transcription, such as Runx2, OSX, Hoxc-8 et al. MiR-214 promotes PDLSCs osteoblastic differentiation by directly interacting with the 3′-UTR of the β-catenin gene CTNNB1, and suppressing Wnt/β-catenin signaling through the inhibition of β-catenin (Cao et al., 2017). MiR-140-5p regulates the odontoblastic differentiation of DPSCs via the Wnt1/β-catenin signaling pathway (Lu et al., 2019). LncRNA-POIR and miR-182 suppress each other and form a network to regulate FoxO1. FoxO1 increased bone formation of pPDLSCs by competing with TCF-4 for β-catenin and inhibiting the canonical Wnt pathway (Wang et al., 2016). CircRNA124534, acting as a miRNA sponge, directly interacts with miR-496 and consequently regulates β-catenin, which in turn exerts osteogenesis of DPSCs (Ji et al., 2020a).
MAPK Signaling Pathway
Mitogen-activated protein kinases (MAPKs) are a family of evolutionarily conserved serine/threonine kinases which have three main subfamilies, namely, extracellular signal-regulated kinase (ERK), c jun N-terminal kinase (JNK), and p38. Downregulation of miR-143-5p induces the expression of MAPK14 and odontoblastic differentiation markers (ALP, DSPP, and OCN), which in turn promotes odontogenic differentiation of hDPSCs by activating the p38 MAPK signaling pathway (Wang et al., 2019). MiR-218 plays a negative role in the odontogenesis of DPSCs by promoting ERK1/2 pathway (Chang et al., 2019).
TGF-β Signaling Pathway
The TGF-β superfamily consists of over 40 members, of which the best known are TGF-βs and Bone morphogenetic proteins (BMPs). In the Smad-dependent signaling pathway, Smad1, Smad5, and Smad8 are BMP activated, whereas Smad 2 and 3 are TGF-β activated. R-Smads form a complex with common-mediator Smad (Co-Smad), Smad4, and enter the nucleus to modulate osteogenic genes expression. Activin membrane binding inhibitor (BAMBI), a TGF-β pseudoreceptor, could interact with TGF-β receptors to inhibit the formation of receptor complexes and then prevent downstream signaling (Onichtchouk et al., 1999). MiR-20a-5p contributes to osteogenic differentiation of hDPSCs through regulating BAMBI and activating the phosphorylation of Smad5 and p38 (Cen et al., 2021). Growth differentiation factor 5 (GDF5) is part of the BMP family. lncRNA growth arrest specific transcript 5 (GAS5) promotes osteogenic differentiation of hPDLSCs by regulating GDF5 and p38/JNK Signaling Pathway (Yang et al., 2020). lncRNA prostate cancer-associated ncRNA transcript-1 (lncPCAT1) promotes the osteogenic differentiation of PDLSCs by sponging miR-106a-5p to upregulate miR-106a-5p-targeted gene BMP2 which activates BMP signaling pathway (Jia et al., 2019). CircRNA FAT1 promotes osteoblastic differentiation of PDLSCs through miR-4781-3p/SMAD5 pathway (Ye et al., 2021).
RANKL/RANK/OPG Signaling Pathway
RANKL/RANK/OPG system consists of three essential signaling molecules: the cytokine receptor activator of nuclear factor (NF)-kB-ligand (RANKL), the receptor activator of NF-kB (RANK), and the soluble decoy receptor osteoprotegerin (OPG). Binding between RANKL and RANK induces receptor trimerization, which triggers the activation of downstream signaling pathways (such as NF-κB, and the MAPK). (Yang et al., 2020) indicated that down-regulated miR-143-3p resulted in up-regulated RANK and activation of the OPG–RANKL signalling pathway to promote odontoblast differentiation of hDPSCs.
PDGF Signaling Pathway
Platelet-derived growth factor (PDGF) activates multiple signaling pathways including PI3-kinase pathways. Signal transducer and activator of transcription (STAT) pathway has been shown to interact with PI3-kinase pathways and STATs are regulated by PDGF signaling (Simon et al., 2000). Funada et al. (2020) transfected mimic miR875-5p into mouse DPSCs and found that cell migration toward dental epithelial cells was significantly induced by miR875-5p via the PDGF signaling pathway. Xu et al. (2018) observed that miR-21 promotes the odontogenic differentiation of DPSCs in low concentration (1–10 ng/ml) of TNF-α for that miR-21 and STAT3 have a positive reciprocal feedback loop.
Notch Signaling Pathway
The Notch signaling pathway is critical for development and cell differentiation. Notch signaling has been confirmed to inhibit odontoblastic differentiation of hDPSCs. (Sun et al., 2014) found that crosstalk between miR-34a and Notch signaling promotes odontoblastic differentiation in SCAPs.
TLR-4 Signaling Pathway
A certain concentration of Lipopolysaccharides (LPS) promotes the odontoblastic differentiation of DPSCs through the Toll-like receptor (TLR-4) signaling pathway (Zhou et al., 2015). A study (Sun et al., 2017) showed that the expression of miR-140-5p is decreased during LPS-induced odontoblastic differentiation of DPSCs in vitro and miR-140-5p suppresses the odontogenic differentiation of DPSCs by binding to the 3′UTR of TLR-4 mRNA.
Types and Mechanisms of ncRNAs in the Differentiation of DMSCs
Various ncRNAs can affect the differentiation of DMSCs through multiple molecular pathways. The various molecular mechanisms through which ncRNAs regulate the differentiation of DMSC are summarized below.
Let-7 Family
MicroRNA let-7 family acts as the key regulator of the differentiation of DMSCs. Ma et al. (2016) found that the Insulin-like growth factor-1 (IGF-1)/IGF-1 receptor (IGF-1R)/hsa-let-7c axis can control the odonto/osteogenic differentiation of IGF-1-treated SCAPs via the regulation of JNK and p38 MAPK signaling pathways. Liu G.-X. et al. (2018) demonstrated that overexpression of let-7c significantly inhibited the expression of IGF-1R and downregulated the osteo/odontogenic differentiation of DPSCs. Furthermore, the ERK, JNK, and P38 MAPK pathways were significantly inhibited following the overexpression of hsa-let-7c in DPSCs. Hsa-let-7b, another member of the let-7 family, can repress the osteogenic differentiation of PDLSCs by regulating CTHRC1 (Fu et al., 2021). Hsa-let-7b also suppresses the odonto/osteogenic differentiation capacity of SCAPs by targeting matrix metalloproteinase 1 (MMP1) (Wang et al., 2018). As mentioned above, the let-7 family may be an inhibitor of odonto/osteogenic differentiation of DMSCs.
MiR-17∼92a Family
MiR17-92a family was first described in mammals in 2001, and miR-17 is a core member of the miR-17-92 family. (Liu et al., 2013) found that miR-17 regulated the osteogenic differentiation of PDLSCs by reducing the expression of transcriptional factor 3 (TCF3) and inhibiting the Wnt signalling pathway. In contrast, (Liu et al., 2011) demonstrated that Under normal conditions, miR-17 negatively regulates osteogenic differentiation of PDLSCs, but in the TNF-α-induced chronic inflammatory microenvironment, miR-17 induces osteogenic differentiation by binding to the Smad ubiquitin regulatory factor one (Smurf1, an inhibitor of osteoblast differentiation)3′-UTR. Totally, the function of miR-17 changed depending on the microenvironment (Figure 4).
MiR-21
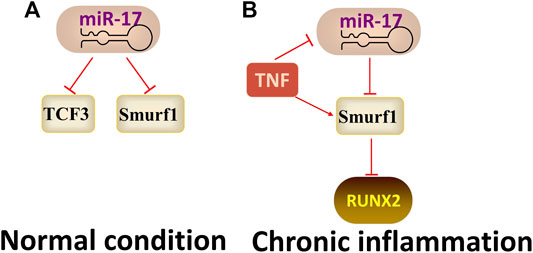
FIGURE 4. The regulatory mechanism of miR-17. Arrow indicates promotion and T indicates inhibition. (A) miR-17 inhibits osteogenic differentiation by targeting TCF3 or Smurf1 when under normal condition. (B) miR-17 promotes osteogenic differentiation by binding to Smurf1 when under the TNF-α-induced inflammatory microenvironment.
The function of miR-21 also influenced by the microenvironment. A study (Yang et al., 2017) showed that the expression of miR-21 was up-regulated during the osteogenic differentiation of PDLSCs Under normal conditions, while the expression of miR-21 was down-regulated in the TNF-α-induced inflammatory microenvironment. Meanwhile, TNF-α suppresses osteogenic differentiation of human PDLSCs by inhibiting miR-21/Spry1 functional axis. In yet another study, miR-21 inhibits the osteogenic differentiation of PDLSCs by targeting the Smad5 (Wei et al., 2017). Interestingly, when exposed to mechanical stretch, miR-21 can target activin receptor type IIB (ACVR2B, a negative regulator of osteogenic differentiation, involved in the TGF-β pathway) to promote the osteogenic differentiation of PDLSCs (Wei et al., 2015). Therefore, miR-21 is a mechanosensitive gene that functions through self-maintenance under different physiological and pathological conditions (Figure 5).
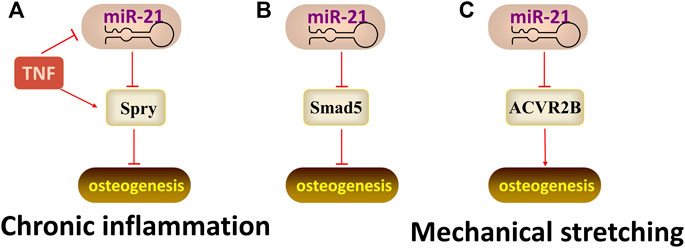
FIGURE 5. The regulatory mechanism of miR-21. Arrow indicates promotion and T indicates inhibition. (A) MiR-21 inhibits osteogenic differentiation by targeting Spry1 when under the TNF-α-induced inflammatory microenvironment. (B) MiR-21 inhibits osteogenic differentiation by targeting Smad5. (C) MiR-21 promotes osteogenic differentiation by targeting ACVR2B when exposed to mechanical stretch.
IncRNA H19
The lncRNA H19 is transcribed from the H19/IGF2 gene located on human chromosome 11p15.5 and has a molecular weight of 2.3 kilobase. Recent studies have shown that H19 can participate in the differentiation of DMSCs by adsorbing and inhibiting the expression of miRNAs (acting as a ceRNA). (Li et al., 2019) revealed that H19 promotes the osteo/odontogenic differentiation of SCAPs and “H19/miR-141/SPAG9/MAPK” positive feedback loop plays paramount role. Mechanistically, H19, as a ceRNA, serves as a miRNA sponge for miR-141. H19 competitively bound to miR-141 and prevented SPAG9 from miRNA-mediated degradation, thus significantly elevating phosphorylated levels of p38 and JNK and facilitating the osteo/odontogenic differentiation of SCAPs. Zhong et al. (2020) found that H19 plays a positive regulatory role in odontoblastic differentiation of hDPSCs through H19/miR-140-5p/BMP-2/FGF9 axis. H19 is both epigenetically regulated and utilizes epigenetic mechanisms to regulate odontogenic differentiation of hDPSCs. Zeng et al. (2018) demonstrated that the overexpression of H19 reduced the expression level of S-adenosylhomocysteine hydrolase (SAHH), which can block the methylation activity of DNA methyltransferases (DNMTs). Besides, upregulated H19 expression significantly repressed SAHH and DNMT3B activity, which then enhanced the DLX3 expression by inhibiting DNMT3B-mediated methylation of DLX3. Therefore, the H19/SAHH axis epigenetically promoted odontogenic differentiation of hDPSCs. (Du et al., 2021) revealed that H19 inhibited LATS1 to promote the production of odontoblasts. The effects of H19 on hDPSCs were achieved by repressing LATS1 through EZH2-induced trimethylation of H327me3 (Figure 6).
IncRNA ANCR
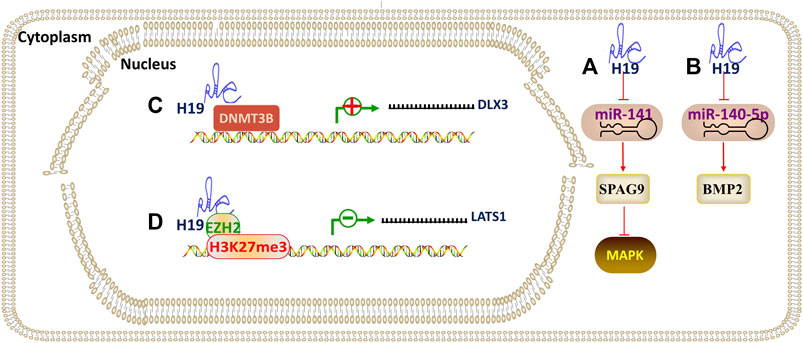
FIGURE 6. The regulatory mechanism of lncRNA H19. Arrow indicates promotion and T indicates inhibition. (A) H19 promotes osteo/odontogenic differentiation through H19/miR-141/SPAG9/MAPK axis. (B) H19 promotes odontoblastic differentiation through H19/miR-140-5p/BMP-2/FGF9 axis. (C) H19 epigenetically promotes odontoblastic differentiation by inhibiting DNMT3B-mediated methylation of DLX3. (D) H19 epigenetically promotes odontoblastic differentiation by repressing LATS1 through EZH2-induced trimethylation of H327me3.
Anti-differentiation non-coding RNA (ANCR, subsequently named differentiation-antagonizing non-protein coding RNA (DANCR)), was identified in 2012, which function was mainly to inhibit differentiation and to enhance the undifferentiated state of somatic progenitor populations (Kretz et al., 2012). lncRNA DANCR suppresses odontoblast-like differentiation of human DPSCs by inhibiting wnt/β-catenin pathway (Chen et al., 2016). (Jia et al., 2015) found that down-regulated lncRNA-ANCR promoted osteogenic differentiation of PDLSCs and the regulating effect of lncRNA-ANCR was associated with the canonical WNT signalling pathway. Peng et al. (2018) showed that lncRNA-ANCR suppresses bone formation of PDLSCs via sponging miRNA-758. Furthermore, downregulation of lncRNA-ANCR promoted the osteogenic, adipogenic and neurogenic differentiation of DMSCs (DPSCs, PDLSCs and SCAPs Jia et al., 2016). These data provide an insight effect of lncRNA-ANCR on DMSCs and indicate that ANCR is a vital regulatory factor in DMSCs differentiation.
lncRNA MEG3
Maternally expressed gene 3 (MEG3), also known as gene trap locus 2 (Gtl2), has been considered as a lncRNA for tumor suppression. MEG3 is known to hinder osteogenic differentiation of PDLSCs by competing with BMP2 mRNA for heterogeneous nuclear ribonucleoprotein I (hnRNPs, an RNA-binding protein), leading to repression of BMP2 (Liu et al., 2019b). Another study (Zhao et al., 2020) demonstrated that MEG3 increases the Smurf1 by serving as a ceRNA to sequester miR-543 in human DPSCs. MEG3 also promotes the osteogenic differentiation of PDLSCs by targeting miR-27a-3p for the upregulation of IGF1 and activation of PI3K/Akt signaling (Liu et al., 2019a). Downregulation of MEG3 resulted in enhancement of osteogenic differentiation of human DFPCs through epigenetically regulating the Wnt/β-catenin pathway, and ChIP analysis showed that these effects were due to the EZH2 regulation of H3K27me3 level on the Wnt genes promotors (Deng et al., 2018). As mentioned above, MEG3 coordinates osteogenic differentiation in DMSCs by targeting multiple molecules (Figure 7).
CircRNA CDR1as
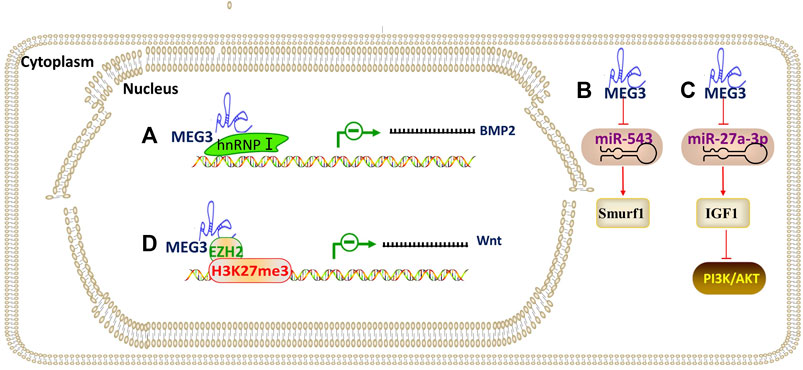
FIGURE 7. The regulatory mechanism of lncRNA MEG3. Arrow indicates promotion and T indicates inhibition. (A) MEG3 leads to repression of BMP2 by competing with BMP2 mRNA for hnRNPs (an RNA-binding protein), which in turn inhibits osteogenic differentiation. (B) MEG3 promotes osteogenic differentiation through MEG3/miR-543/Smurf1 axis. (C) MEG3 promotes osteogenic differentiation through MEG3/miR-27a-3p/IGF1/PI3K/Akt axis. (D) MEG3 epigenetically regulating Wnt through the EZH2 regulation of H3K27me3 level on the Wnt genes promotors.
Antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as), also known as CIRS-7, has approximately 70 conserved miR-7 binding sites and could therefore act as a miR-7 “sponge”. (Gu et al., 2021) demonstrated that CDR1as promotes the pluripotent state of PDLSCs by inhibiting miR-7-mediated suppression of KLF4 expression, and hnRNPM can promote the expression of CDR1as in PDLSCs. CDR1as was also reported (Li X. et al., 2018) to regulate the osteoblastic differentiation of PDLSCs by triggering the activation of Smad and p38 MAPK signaling pathway, as well as upregulation of GDF5. The circRNA-CDR1as/miR-7/GDF5/SMAD and p38 MAPK signaling pathway may be involved in PDLSCs upon the periodontitis burst.
CircRNA SIPA1L1
CircSIPA1L1 is produced by a transcript encoding circSIPA1L1 on human chromosome 14. The expression of circSIPA1L1 in the mineralization induction group was about 8 times that of the control group, suggesting vital functions of circSIPA1L1 in osteogenesis. CircSIPA1L1 was found to regulate miR-617/Smad3 axis to potentiate osteogenic differentiation of DPSCs (Ge et al., 2020). In yet another study, circSIPA1L1 regulated osteoblastic differentiation of SCAPs via miR-204-5p/ALPL pathway (Li Y. et al., 2020).
Conclusion
In this review, we summarized the functions and mechanisms of ncRNAs, which played important roles in the differentiation of DMSCs. DMSCs are currently considered as valid biological sources for use in tissue regeneration by tissue engineering approaches. Therefore, deep knowledge of the mechanisms which govern DMSCs differentiation is of pivotal relevance. During the differentiation of DMSCs, all three ncRNAs are regulated by multiple signalling pathways. MiRNAs regulate gene expression by targeting the 3′UTR of target genes, while they are influenced by the epigenetics and microenvironment (inflammation, mechanical stretching), forming a complex and intertwined regulatory network. LncRNAs have broad functional versatility, because they contribute to the regulation of gene expression at different levels in the nucleus or cytoplasm and at transcriptional and post-transcriptional levels. Both circRNAs and lncRNAs function as ceRNAs, and there may be a competitive lncRNA/circRNA-miRNA-mRNA regulatory network system. With increasing numbers of miRNAs, lncRNAs and circRNAs discovered in this process, it has become possible to use these ncRNA-related therapeutic methods in the field of tissue repair and regeneration.
However, the current studies on the regulation of multi-directional differentiation of DMSCs by ncRNAs mostly focus on the following aspects: odontogenic differentiation, osteogenic differentiation, neurogenic differentiation, angiogenic differentiation and myogenic differentiation. DMSCs also have the ability to differentiate into liver tissue-like cells and islet-like cells, and reports on the regulation of ncRNAs in these tissue-like cells are still lacking. Therefore, further studies on the differentiation of other tissue-like cells are encouraged. Moreover, the related markers, signaling pathways (Wnt, TGFβ, MAPK), and ncRNAs (hsa-let-7c, H19) partially overlap during the osteogenic and odontogenic differentiation of DMSCs. There should be more reports in the future explaining the difference and connection of ncRNAs in their differentiation. Besides, most studies are primarily performed in vitro, and such in vitro differentiation is often limited and may not represent true differentiation of the cells themselves. Therefore, further studies on ncRNAs are needed, including in vivo experiments and animal disease models. Finally, different types of DMSCs have the ability to differentiate into different lineages. Among all subpopulations of DMSCs, DPSCs and PDLSCs are most extensively studied. Future studies should emphasize the effective combination of ncRNAs with various types of DMSCs to provide a potential scheme for tissue regeneration. Overall, whereas interest and investigation in the contribution of ncRNAs to the differentiation of DMSCs have increased considerably, the field is still a long way from understanding the full extent of the contribution of ncRNAs and the mechanisms by which ncRNAs exert their potential effects in this field.
Author Contributions
BZ: conceptualization, writing—original draft, visualization, writing—review and editing. JH: funding acquisition, and supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Clinical Medical Technology Innovation Guidance Project of Hunan Province in China, 2020SK53501.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Figures 1,2 created with BioRender.com.
References
Cai, J., Qi, H., Yao, K., Yao, Y., Jing, D., Liao, W., et al. (2021). Non-Coding RNAs Steering the Senescence-Related Progress, Properties, and Application of Mesenchymal Stem Cells. Front. Cel Dev. Biol. 9, 650431. doi:10.3389/fcell.2021.650431
Cao, F., Zhan, J., Chen, X., Zhang, K., Lai, R., and Feng, Z. (2017). miR-214 Promotes Periodontal Ligament Stem Cell Osteoblastic Differentiation by Modulating Wnt/β-Catenin Signaling. Mol. Med. Rep. 16 (6), 9301–9308. doi:10.3892/mmr.2017.7821
Cen, X., Pan, X., Zhang, B., Huang, W., Pei, F., Luo, T., et al. (2021). miR-20a-5p Contributes to Osteogenic Differentiation of Human Dental Pulp Stem Cells by Regulating BAMBI and Activating the Phosphorylation of Smad5 and P38. Stem Cel Res Ther 12 (1), 421. doi:10.1186/s13287-021-02501-8
Chang, K., Chen, R.-S., Chang, F.-H., and Chen, M.-H. (2019). Promoting Dentinogenesis of DPSCs through Inhibiting microRNA-218 by Using Magnetic Nanocarrier Delivery. J. Formos. Med. Assoc. 118 (6), 1005–1013. doi:10.1016/j.jfma.2018.10.018
Chen, L., Song, Z., Huang, S., Wang, R., Qin, W., Guo, J., et al. (2016). lncRNA DANCR Suppresses Odontoblast-like Differentiation of Human Dental Pulp Cells by Inhibiting Wnt/β-Catenin Pathway. Cell Tissue Res 364 (2), 309–318. doi:10.1007/s00441-015-2333-2
Chen, M., Yang, Y., Zeng, J., Deng, Z., and Wu, B. (2020). circRNA Expression Profile in Dental Pulp Stem Cells during Odontogenic Differentiation. Stem Cell Int. 2020, 1–19. doi:10.1155/2020/5405931
Danan, M., Schwartz, S., Edelheit, S., and Sorek, R. (2012). Transcriptome-wide Discovery of Circular RNAs in Archaea. Nucleic Acids Res. 40 (7), 3131–3142. doi:10.1093/nar/gkr1009
Deng, L., Hong, H., Zhang, X., Chen, D., Chen, Z., Ling, J., et al. (2018). Down-regulated lncRNA MEG3 Promotes Osteogenic Differentiation of Human Dental Follicle Stem Cells by Epigenetically Regulating Wnt Pathway. Biochem. Biophysical Res. Commun. 503 (3), 2061–2067. doi:10.1016/j.bbrc.2018.07.160
Du, Z., Shi, X., and Guan, A. (2021). lncRNA H19 Facilitates the Proliferation and Differentiation of Human Dental Pulp Stem Cells via EZH2-dependent LATS1 Methylation. Mol. Ther. - Nucleic Acids 25, 116–126. doi:10.1016/j.omtn.2021.04.017
Feng, Y., Wan, P., and Yin, L. (2020). Long Noncoding RNA X-Inactive Specific Transcript (XIST) Promotes Osteogenic Differentiation of Periodontal Ligament Stem Cells by Sponging MicroRNA-214-3p. Med. Sci. Monit. 26, e918932. doi:10.12659/msm.918932
Fu, L., Li, N., Ye, Y., Ye, X., Xiao, T., Wu, X., et al. (2021). MicroRNA Hsa-Let-7b Regulates the Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Targeting CTHRC1. Stem Cell Int. 2021, 1–15. doi:10.1155/2021/5791181
Funada, K., Yoshizaki, K., MIyazaki, K., Han, X., Yuta, T., Tian, T., et al. (2020). microRNA-875-5p Plays Critical Role for Mesenchymal Condensation in Epithelial-Mesenchymal Interaction during Tooth Development. Sci. Rep. 10 (1), 4918. doi:10.1038/s41598-020-61693-w
Gay, I., Cavender, A., Peto, D., Sun, Z., Speer, A., Cao, H., et al. (2014). Differentiation of Human Dental Stem Cells Reveals a Role for microRNA-218. J. Periodont Res. 49 (1), 110–120. doi:10.1111/jre.12086
Ge, X., Li, Z., Zhou, Z., Xia, Y., Bian, M., and Yu, J. (2020). Circular RNA SIPA1L1 Promotes Osteogenesis via Regulating the miR-617/Smad3 axis in Dental Pulp Stem Cells. Stem Cel Res Ther 11 (1), 364. doi:10.1186/s13287-020-01877-3
Gong, Q., Wang, R., Jiang, H., Lin, Z., and Ling, J. (2012). Alteration of microRNA Expression of Human Dental Pulp Cells during Odontogenic Differentiation. J. Endodontics 38 (10), 1348–1354. doi:10.1016/j.joen.2012.06.016
Gu, X., Li, M., Jin, Y., Liu, D., and Wei, F. (2017). Identification and Integrated Analysis of Differentially Expressed lncRNAs and circRNAs Reveal the Potential ceRNA Networks during PDLSC Osteogenic Differentiation. BMC Genet. 18 (1), 100. doi:10.1186/s12863-017-0569-4
Gu, X., Li, X., Jin, Y., Zhang, Z., Li, M., Liu, D., et al. (2021). CDR1as Regulated by hnRNPM Maintains Stemness of Periodontal Ligament Stem Cells via miR‐7/KLF4. J. Cel Mol Med 25 (9), 4501–4515. doi:10.1111/jcmm.16541
Hao, Y., Ge, Y., Li, J., Hu, Y., Wu, B., and Fang, F. (2017). Identification of MicroRNAs by Microarray Analysis and Prediction of Target Genes Involved in Osteogenic Differentiation of Human Periodontal Ligament Stem Cells. J. Periodontol. 88 (10), 1105–1113. doi:10.1902/jop.2017.170079
He, Q., Yang, S., Gu, X., Li, M., Wang, C., and Wei, F. (2018). Long Noncoding RNA TUG1 Facilitates Osteogenic Differentiation of Periodontal Ligament Stem Cells via Interacting with Lin28A. Cell Death Dis 9 (5), 455. doi:10.1038/s41419-018-0484-2
Hernández-Romero, I. A., Guerra-Calderas, L., Salgado-Albarrán, M., Maldonado-Huerta, T., and Soto-Reyes, E. (2019). The Regulatory Roles of Non-coding RNAs in Angiogenesis and Neovascularization from an Epigenetic Perspective. Front. Oncol. 9, 1091. doi:10.3389/fonc.2019.01091
Huang, Y., Han, Y., Guo, R., Liu, H., Li, X., Jia, L., et al. (2020). Long Non-coding RNA FER1L4 Promotes Osteogenic Differentiation of Human Periodontal Ligament Stromal Cells via miR-874-3p and Vascular Endothelial Growth Factor A. Stem Cel Res Ther 11 (1), 5. doi:10.1186/s13287-019-1519-z
Ito, K., Tomoki, R., Ogura, N., Takahashi, K., Eda, T., Yamazaki, F., et al. (2020). MicroRNA-204 Regulates Osteogenic Induction in Dental Follicle Cells. J. Dental Sci. 15 (4), 457–465. doi:10.1016/j.jds.2019.11.004
Ji, F., Pan, J., Shen, Z., Yang, Z., Wang, J., Bai, X., et al. (2020a). The Circular RNA circRNA124534 Promotes Osteogenic Differentiation of Human Dental Pulp Stem Cells through Modulation of the miR-496/β-Catenin Pathway. Front. Cel Dev. Biol. 8, 230. doi:10.3389/fcell.2020.00230
Ji, F., Zhu, L., Pan, J., Shen, Z., Yang, Z., Wang, J., et al. (2020b). hsa_circ_0026827 Promotes Osteoblast Differentiation of Human Dental Pulp Stem Cells through the Beclin1 and RUNX1 Signaling Pathways by Sponging miR-188-3p. Front. Cel Dev. Biol. 8, 470. doi:10.3389/fcell.2020.00470
Jia, B., Qiu, X., Chen, J., Sun, X., Zheng, X., Zhao, J., et al. (2019). A Feed‐forward Regulatory Network lncPCAT1/miR‐106a‐5p/E2F5 Regulates the Osteogenic Differentiation of Periodontal Ligament Stem Cells. J. Cel Physiol 234 (11), 19523–19538. doi:10.1002/jcp.28550
Jia, Q., Chen, X., Jiang, W., Wang, W., Guo, B., and Ni, L. (2016). The Regulatory Effects of Long Noncoding RNA-ANCRon Dental Tissue-Derived Stem Cells. Stem Cell Int. 2016, 1–12. doi:10.1155/2016/3146805
Jia, Q., Jiang, W., and Ni, L. (2015). Down-regulated Non-coding RNA (lncRNA-ANCR) Promotes Osteogenic Differentiation of Periodontal Ligament Stem Cells. Arch. Oral Biol. 60 (2), 234–241. doi:10.1016/j.archoralbio.2014.10.007
Jiang, H., and Jia, P. (2021). MiR‐153‐3p Inhibits Osteogenic Differentiation of Periodontal Ligament Stem Cells through KDM6A‐induced Demethylation of H3K27me3. J. Periodont Res. 56 (2), 379–387. doi:10.1111/jre.12830
Kim, C., Kang, D., Lee, E. K., and Lee, J.-S. (20172017). Long Noncoding RNAs and RNA-Binding Proteins in Oxidative Stress, Cellular Senescence, and Age-Related Diseases. Oxidative Med. Cell Longevity 2017, 1–21. doi:10.1155/2017/2062384
Kishore, S., Jaskiewicz, L., Burger, L., Hausser, J., Khorshid, M., and Zavolan, M. (2011). A Quantitative Analysis of CLIP Methods for Identifying Binding Sites of RNA-Binding Proteins. Nat. Methods 8 (7), 559–564. doi:10.1038/nmeth.1608
Kretz, M., Webster, D. E., Flockhart, R. J., Lee, C. S., Zehnder, A., Lopez-Pajares, V., et al. (2012). Suppression of Progenitor Differentiation Requires the Long Noncoding RNA ANCR. Genes Dev. 26 (4), 338–343. doi:10.1101/gad.182121.111
Li, D., Deng, T., Li, H., and Li, Y. (2015). MiR-143 and miR-135 Inhibitors Treatment Induces Skeletal Myogenic Differentiation of Human Adult Dental Pulp Stem Cells. Arch. Oral Biol. 60 (11), 1613–1617. doi:10.1016/j.archoralbio.2015.08.010
Li, L., Liu, W., Wang, H., Yang, Q., Zhang, L., Jin, F., et al. (2018). Mutual Inhibition between HDAC9 and miR-17 Regulates Osteogenesis of Human Periodontal Ligament Stem Cells in Inflammatory Conditions. Cel Death Dis 9 (5), 480. doi:10.1038/s41419-018-0480-6
Li, X., Xu, W., Wu, J., Lin, X., Chen, Y., Wen, J., et al. (2020). Differential Expression of Long Noncoding RNAs from Dental Pulp Stem Cells in the Microenvironment of the Angiogenesis. Arch. Oral Biol. 113, 104691. doi:10.1016/j.archoralbio.2020.104691
Li, X., Zheng, Y., Zheng, Y., Huang, Y., Zhang, Y., Jia, L., et al. (2018). Circular RNA CDR1as Regulates Osteoblastic Differentiation of Periodontal Ligament Stem Cells via the miR-7/GDF5/SMAD and P38 MAPK Signaling Pathway. Stem Cel Res Ther 9 (1), 232. doi:10.1186/s13287-018-0976-0
Li, Y., Bian, M., Zhou, Z., Wu, X., Ge, X., Xiao, T., et al. (2020). Circular RNA SIPA1L1 Regulates Osteoblastic Differentiation of Stem Cells from Apical Papilla via miR-204-5p/ALPL Pathway. Stem Cel Res Ther 11 (1), 461. doi:10.1186/s13287-020-01970-7
Li, Z., Guo, X., and Wu, S. (2020). Epigenetic Silencing of KLF2 by Long Non-coding RNA SNHG1 Inhibits Periodontal Ligament Stem Cell Osteogenesis Differentiation. Stem Cel Res Ther 11 (1), 435. doi:10.1186/s13287-020-01953-8
Li, Z., Yan, M., Yu, Y., Wang, Y., Lei, G., Pan, Y., et al. (2019). LncRNA H19 Promotes the Committed Differentiation of Stem Cells from Apical Papilla via miR-141/SPAG9 Pathway. Cel Death Dis 10 (2), 130. doi:10.1038/s41419-019-1337-3
Liao, C., Zhou, Y., Li, M., Xia, Y., and Peng, W. (2020). LINC00968 Promotes Osteogenic Differentiation In Vitro and Bone Formation In Vivo via Regulation of miR-3658/RUNX2. Differentiation 116, 1–8. doi:10.1016/j.diff.2020.09.005
Liu, G.-X., Ma, S., Li, Y., Yu, Y., Zhou, Y.-X., Lu, Y.-D., et al. (2018). Hsa-let-7c Controls the Committed Differentiation of IGF-1-Treated Mesenchymal Stem Cells Derived from Dental Pulps by Targeting IGF-1R via the MAPK Pathways. Exp. Mol. Med. 50 (4), 1–14. doi:10.1038/s12276-018-0048-7
Liu, J., Zhang, Z. Y., Yu, H., Yang, A. P., Hu, P. F., Liu, Z., et al. (2018). Long Noncoding RNA C21orf121/bone Morphogenetic Protein 2/microRNA‐140‐5p Gene Network Promotes Directed Differentiation of Stem Cells from Human Exfoliated Deciduous Teeth to Neuronal Cells. J. Cel Biochem 120, 1464–1476. doi:10.1002/jcb.27313
Liu, W., Gong, Q., Ling, J., Zhang, W., Liu, Z., and Quan, J. (2014). Role of miR-424 on Angiogenic Potential in Human Dental Pulp Cells. J. Endodontics 40 (1), 76–82. doi:10.1016/j.joen.2013.09.035
Liu, W., Liu, Y., Guo, T., Hu, C., Luo, H., Zhang, L., et al. (2013). TCF3, a Novel Positive Regulator of Osteogenesis, Plays a Crucial Role in miR-17 Modulating the Diverse Effect of Canonical Wnt Signaling in Different Microenvironments. Cel Death Dis 4 (3), e539. doi:10.1038/cddis.2013.65
Liu, Y., Liu, C., Zhang, A., Yin, S., Wang, T., Wang, Y., et al. (2019a). Down-regulation of Long Non-coding RNA MEG3 Suppresses Osteogenic Differentiation of Periodontal Ligament Stem Cells (PDLSCs) through miR-27a-3p/IGF1 axis in Periodontitis. Aging 11 (15), 5334–5350. doi:10.18632/aging.102105
Liu, Y., Liu, W., Hu, C., Xue, Z., Wang, G., Ding, B., et al. (2011). MiR-17 Modulates Osteogenic Differentiation through a Coherent Feed-Forward Loop in Mesenchymal Stem Cells Isolated from Periodontal Ligaments of Patients with Periodontitis. Stem Cells 29 (11), 1804–1816. doi:10.1002/stem.728
Liu, Y., Zeng, X., Miao, J., Liu, C., Wei, F., Liu, D., et al. (2019b). Upregulation of Long Noncoding RNA MEG3 Inhibits the Osteogenic Differentiation of Periodontal Ligament Cells. J. Cel Physiol 234 (4), 4617–4626. doi:10.1002/jcp.27248
Lu, X., Chen, X., Xing, J., Lian, M., Huang, D., Lu, Y., et al. (2019). miR-140-5p Regulates the Odontoblastic Differentiation of Dental Pulp Stem Cells via the Wnt1/β-Catenin Signaling Pathway. Stem Cel Res Ther 10 (1), 226. doi:10.1186/s13287-019-1344-4
Ma, S., Liu, G., Jin, L., Pang, X., Wang, Y., Wang, Z., et al. (2016). IGF-1/IGF-1R/hsa-let-7c axis Regulates the Committed Differentiation of Stem Cells from Apical Papilla. Sci. Rep. 6, 36922. doi:10.1038/srep36922
Mehri-Ghahfarrokhi, A., Pourteymourfard-Tabrizi, Z., Farrokhi, E., Chaleshtori, M. H., and Jami, M.-S. (2019). Increased Levels of miR-124 in Human Dental Pulp Stem Cells Alter the Expression of Neural Markers. J. Otology 14 (4), 121–127. doi:10.1016/j.joto.2019.04.001
Mumtaz, P. T., Taban, Q., Dar, M. A., Mir, S., Haq, Z. U., Zargar, S. M., et al. (2020). Deep Insights in Circular RNAs: from Biogenesis to Therapeutics. Biol. Proced. Online 22, 10. doi:10.1186/s12575-020-00122-8
Ng, T. K., Yung, J. S. Y., Choy, K. W., Cao, D., Leung, C. K. S., Cheung, H. S., et al. (2015). Transdifferentiation of Periodontal Ligament-Derived Stem Cells into Retinal Ganglion-like Cells and its microRNA Signature. Sci. Rep. 5, 16429. doi:10.1038/srep16429
Onichtchouk, D., Chen, Y.-G., Dosch, R., Gawantka, V., Delius, H., Massague´, J., et al. (1999). Silencing of TGF-β Signalling by the Pseudoreceptor BAMBI. Nature 401 (6752), 480–485. doi:10.1038/46794
Peng, W., Deng, W., Zhang, J., Pei, G., Rong, Q., and Zhu, S. (2018). Long Noncoding RNA ANCR Suppresses Bone Formation of Periodontal Ligament Stem Cells via Sponging miRNA-758. Biochem. Biophysical Res. Commun. 503 (2), 815–821. doi:10.1016/j.bbrc.2018.06.081
Rincón-Riveros, A., Morales, D., Rodríguez, J. A., Villegas, V. E., and López-Kleine, L. (2021). Bioinformatic Tools for the Analysis and Prediction of ncRNA Interactions. Ijms 22 (21), 11397. doi:10.3390/ijms222111397
Rodas-Junco, B. A., Canul-Chan, M., Rojas-Herrera, R. A., De-la-Peña, C., and Nic-Can, G. I. (2017). Stem Cells from Dental Pulp: What Epigenetics Can Do with Your Tooth. Front. Physiol. 8, 999. doi:10.3389/fphys.2017.00999
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. U.S.A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Shen, W.-C., Lai, Y.-C., Li, L.-H., Liao, K., Lai, H.-C., Kao, S.-Y., et al. (2019). Methylation and PTEN Activation in Dental Pulp Mesenchymal Stem Cells Promotes Osteogenesis and Reduces Oncogenesis. Nat. Commun. 10 (1), 2226. doi:10.1038/s41467-019-10197-x
Simon, A. R., Vikis, H. G., Stewart, S., Fanburg, B. L., Cochran, B. H., and Guan, K.-L. (2000). Regulation of STAT3 by Direct Binding to the Rac1 GTPase. Science 290 (5489), 144–147. doi:10.1126/science.290.5489.144
Sun, D.-g., Xin, B.-c., Wu, D., Zhou, L., Wu, H.-b., Gong, W., et al. (2017). miR-140-5p-mediated Regulation of the Proliferation and Differentiation of Human Dental Pulp Stem Cells Occurs through the Lipopolysaccharide/toll-like Receptor 4 Signaling Pathway. Eur. J. Oral Sci. 125 (6), 419–425. doi:10.1111/eos.12384
Sun, F., Wan, M., Xu, X., Gao, B., Zhou, Y., Sun, J., et al. (2014). Crosstalk between miR-34a and Notch Signaling Promotes Differentiation in Apical Papilla Stem Cells (SCAPs). J. Dent Res. 93 (6), 589–595. doi:10.1177/0022034514531146
Wang, B. L., Wang, Z., Nan, X., Zhang, Q. C., and Liu, W. (2019). Downregulation of microRNA‐143‐5p Is Required for the Promotion of Odontoblasts Differentiation of Human Dental Pulp Stem Cells through the Activation of the Mitogen‐activated Protein Kinases 14‐dependent P38 Mitogen‐activated Protein Kinases Signaling Pathway. J. Cel Physiol 234 (4), 4840–4850. doi:10.1002/jcp.27282
Wang, C., Dong, L., Wang, Y., Jiang, Z., Zhang, J., and Yang, G. (2021). Bioinformatics Analysis Identified miR-584-5p and Key miRNA-mRNA Networks Involved in the Osteogenic Differentiation of Human Periodontal Ligament Stem Cells. Front. Genet. 12, 750827. doi:10.3389/fgene.2021.750827
Wang, L., Wu, F., Song, Y., Li, X., Wu, Q., Duan, Y., et al. (2016). Long Noncoding RNA Related to Periodontitis Interacts with miR-182 to Upregulate Osteogenic Differentiation in Periodontal Mesenchymal Stem Cells of Periodontitis Patients. Cel Death Dis 7 (8), e2327. doi:10.1038/cddis.2016.125
Wang, Y., Pang, X., Wu, J., Jin, L., Yu, Y., Gobin, R., et al. (2018). MicroRNA Hsa ‐ Let ‐7 B Suppresses the Odonto/osteogenic Differentiation Capacity of Stem Cells from Apical Papilla by Targeting MMP1. J. Cel. Biochem. 119 (8), 6545–6554. doi:10.1002/jcb.26737
Wei, F., Liu, D., Feng, C., Zhang, F., Yang, S., Hu, Y., et al. (2015). microRNA-21 Mediates Stretch-Induced Osteogenic Differentiation in Human Periodontal Ligament Stem Cells. Stem Cell Dev. 24 (3), 312–319. doi:10.1089/scd.2014.0191
Wei, F., Yang, S., Guo, Q., Zhang, X., Ren, D., Lv, T., et al. (2017). MicroRNA-21 Regulates Osteogenic Differentiation of Periodontal Ligament Stem Cells by Targeting Smad5. Sci. Rep. 7 (1), 16608. doi:10.1038/s41598-017-16720-8
Wen, B., He, C., Zhang, Q., Zhang, F., Li, N., Pan, Y., et al. (2020). Overexpression of microRNA-221 Promotes the Differentiation of Stem Cells from Human Exfoliated Deciduous Teeth to Neurons through Activation of Wnt/β-Catenin Pathway via Inhibition of CHD8. Cell Cycle 19 (23), 3231–3248. doi:10.1080/15384101.2020.1816308
Wu, H., Yin, Q.-F., Luo, Z., Yao, R.-W., Zheng, C.-C., Zhang, J., et al. (2016). Unusual Processing Generates SPA LncRNAs that Sequester Multiple RNA Binding Proteins. Mol. Cel 64 (3), 534–548. doi:10.1016/j.molcel.2016.10.007
Wu, M., Liu, X., Li, Z., Huang, X., Guo, H., Guo, X., et al. (2021). SHED Aggregate Exosomes Shuttled miR‐26a Promote Angiogenesis in Pulp Regeneration via TGF‐β/SMAD2/3 Signalling. Cell Prolif 54 (7), e13074. doi:10.1111/cpr.13074
Xie, L., Guan, Z., Zhang, M., Lyu, S., Thuaksuban, N., Kamolmattayakul, S., et al. (2020). Exosomal circLPAR1 Promoted Osteogenic Differentiation of Homotypic Dental Pulp Stem Cells by Competitively Binding to Hsa-miR-31. Biomed. Res. Int. 2020, 1–13. doi:10.1155/2020/6319395
Xie, Y., and Shen, G. (2018). MicroRNA-139-5p E-levates S-keletal M-yogenic D-ifferentiation of H-uman A-dult D-ental P-ulp S-tem C-ells through Wnt/β-catenin S-ignaling P-athway. Exp. Ther. Med. 16 (4), 2835–2842. doi:10.3892/etm.2018.6585
Xu, K., Xiao, J., Zheng, K., Feng, X., Zhang, J., Song, D., et al. (2018). MiR-21/STAT3 Signal Is Involved in Odontoblast Differentiation of Human Dental Pulp Stem Cells Mediated by TNF-α. Cell Reprogramming 20 (2), 107–116. doi:10.1089/cell.2017.0042
Yan, G.-Q., Wang, X., Yang, F., Yang, M.-L., Zhang, G.-R., Wang, G.-K., et al. (2017). MicroRNA-22 Promoted Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Targeting HDAC6. J. Cel. Biochem. 118 (7), 1653–1658. doi:10.1002/jcb.25931
Yang, C., Jia, R., Zuo, Q., Zheng, Y., Wu, Q., Luo, B., et al. (2020). microRNA‐143‐3p Regulates Odontogenic Differentiation of Human Dental Pulp Stem Cells through Regulation of the Osteoprotegerin-RANK Ligand Pathway by Targeting RANK. Exp. Physiol. 105 (5), 876–885. doi:10.1113/ep087992
Yang, N., Li, Y., Wang, G., Ding, Y., Jin, Y., and Xu, Y. (2017). Tumor Necrosis Factor-α Suppresses Adipogenic and Osteogenic Differentiation of Human Periodontal Ligament Stem Cell by Inhibiting miR-21/Spry1 Functional axis. Differentiation 97, 33–43. doi:10.1016/j.diff.2017.08.004
Yang, Q., Han, Y., Liu, P., Huang, Y., Li, X., Jia, L., et al. (2020). Long Noncoding RNA GAS5 Promotes Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Regulating GDF5 and P38/JNK Signaling Pathway. Front. Pharmacol. 11, 701. doi:10.3389/fphar.2020.00701
Ye, Y., Ke, Y., Liu, L., Xiao, T., and Yu, J. (2021). CircRNA FAT1 Regulates Osteoblastic Differentiation of Periodontal Ligament Stem Cells via miR-4781-3p/SMAD5 Pathway. Stem Cell Int. 2021, 1–16. doi:10.1155/2021/5177488
Yin, J.-Y., Luo, X.-H., Feng, W.-Q., Miao, S.-H., Ning, T.-T., Lei, Q., et al. (2021). Multidifferentiation Potential of Dental-Derived Stem Cells. Wjsc 13 (5), 342–365. doi:10.4252/wjsc.v13.i5.342
Zeng, L., Sun, S., Han, D., Liu, Y., Liu, H., Feng, H., et al. (2018). Long Non-coding RNA H19/SAHH axis Epigenetically Regulates Odontogenic Differentiation of Human Dental Pulp Stem Cells. Cell Signal. 52, 65–73. doi:10.1016/j.cellsig.2018.08.015
Zhang, B., Huo, S., Cen, X., Pan, X., Huang, X., and Zhao, Z. (2020). circAKT3 Positively Regulates Osteogenic Differentiation of Human Dental Pulp Stromal Cells via miR-206/CX43 axis. Stem Cel Res Ther 11 (1), 531. doi:10.1186/s13287-020-02058-y
Zhang, Z., Shuai, Y., Zhou, F., Yin, J., Hu, J., Guo, S., et al. (2020). PDLSCs Regulate Angiogenesis of Periodontal Ligaments via VEGF Transferred by Exosomes in Periodontitis. Int. J. Med. Sci. 17 (5), 558–567. doi:10.7150/ijms.40918
Zhao, L.-D., Xu, W.-C., Cui, J., Liang, Y.-C., Cheng, W.-Q., Xin, B.-C., et al. (2020). Long Non-coding RNA Maternally Expressed Gene 3 Inhibits Osteogenic Differentiation of Human Dental Pulp Stem Cells via microRNA-543/smad Ubiquitin Regulatory Factor 1/runt-Related Transcription Factor 2 axis. Arch. Oral Biol. 118, 104838. doi:10.1016/j.archoralbio.2020.104838
Zheng, H., Wang, N., Li, L., Ge, L., Jia, H., and Fan, Z. (2021). miR-140-3p Enhanced the Osteo/odontogenic Differentiation of DPSCs via Inhibiting KMT5B under Hypoxia Condition. Int. J. Oral Sci. 13 (1), 41. doi:10.1038/s41368-021-00148-y
Zheng, Y., and Jia, L. (2016). Long Noncoding RNAs Related to the Odontogenic Potential of Dental Mesenchymal Cells in Mice. Arch. Oral Biol. 67, 1–8. doi:10.1016/j.archoralbio.2016.03.001
Zheng, Y., Li, X., Huang, Y., Jia, L., and Li, W. (2018). Time Series Clustering of mRNA and lncRNA Expression during Osteogenic Differentiation of Periodontal Ligament Stem Cells. PeerJ 6, e5214. doi:10.7717/peerj.5214
Zhong, J., Tu, X., Kong, Y., Guo, L., Li, B., Zhong, W., et al. (2020). LncRNA H19 Promotes Odontoblastic Differentiation of Human Dental Pulp Stem Cells by Regulating miR-140-5p and BMP-2/FGF9. Stem Cel Res Ther 11 (1), 202. doi:10.1186/s13287-020-01698-4
Zhou, H., Li, X., Wu, R. X., He, X. T., An, Y., Xu, X. Y., et al. (2021). Periodontitis‐compromised Dental Pulp Stem Cells Secrete Extracellular Vesicles Carrying miRNA‐378a Promote Local Angiogenesis by Targeting Sufu to Activate the Hedgehog/Gli1 Signalling. Cel Prolif 54 (5), e13026. doi:10.1111/cpr.13026
Keywords: dental-derived mesenchymal stem cells, non-coding RNAs, multi-directional differentiation, osteogenic differentiation, odontogenic differentiation
Citation: Zeng B and Huang J (2022) Progress in the Study of Non-Coding RNAs in Multidifferentiation Potential of Dental-Derived Mesenchymal Stem Cells. Front. Genet. 13:854285. doi: 10.3389/fgene.2022.854285
Received: 13 January 2022; Accepted: 17 March 2022;
Published: 05 April 2022.
Edited by:
Ping Hu, Shanghai Institute of Biochemistry and Cell Biology (CAS), ChinaReviewed by:
Alejandro Correa, Carlos Chagas Institute (ICC), BrazilZhekai Hu, The University of Hong Kong, Hong Kong SAR, China
Wei Qiu, Nanfang Hospital, Southern Medical University, China
Kannan Thirumulu Ponnuraj, Universiti Sains Malaysia Health Campus, Malaysia
Copyright © 2022 Zeng and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junhui Huang, ODA4MDAzQGNzdS5lZHUuY24=
 Biyun Zeng
Biyun Zeng Junhui Huang
Junhui Huang