- Institute of Cotton Research of Chinese Academy of Agricultural Science/State Key Laboratory of Cotton Biology, Anyang, China
Growth-regulating factors-interacting factors (GIFs) are a type of transcription co-activators in plants, playing crucial roles in plants’ growth, development, and stress adaptation. Here, a total of 35 GIF genes were identified and clustered into two groups by phylogenetic analysis in four cotton genus. The gene structure and conserved domain analysis proved the conservative characteristics of GIF genes in cotton. The function of GIF genes was evaluated in two cotton accessions, Ji A-1-7 (33xi) and King, which have larger and smaller lateral root numbers, respectively. The results showed that the expression of GhGIF4 in Ji A-1-7 (33xi) was higher than that in King. The enzyme activity and microstructure assay showed a higher POD activity, lower MDA content, and more giant cells of the lateral root emergence part phenotype in Ji A-1-7 (33xi) than in King. A mild waterlogging assay showed the GIF genes were down-regulated in the waterlogged seedling. Further confirmation of the suppression of GhGIF4 in cotton plants further confirmed that GhGIF4 could reduce the lateral root numbers in cotton. This study could provide a basis for future studies of the role of GIF genes in upland cotton.
1 Introduction
Growth-regulating factors-interacting factors (GIFs) are a class of transcription co-activators of growth-regulating factors (GRFs). GIFs usually interact with GRFs to form a plant-specific transcriptional complex (Kim and Tsukaya, 2015). The AtGIF gene family was first described by Kim in Arabidopsis in 2004 (Kim and Kende, 2004). It was shown that the GIF1 gene could act as a functional homolog of the human SYT transcription coactivator. Subsequently, GIF genes were associated with plant growth and development (Horiguchi et al., 2005; Vercruyssen et al., 2014).
Previous research showed that GIF1 positively regulated cell proliferation in lateral organs like leaves and petals of Arabidopsis. A narrower and smaller leaf phenotype (than control), which was caused by reduced cell numbers along the leaf-width axis, was observed in the gif1 mutant and transgenic plants, indicating that AtGIF1 may function as a transcriptional co-activator that is involved in regulating the growth and shape of plant leaves and petals (Kim and Kende, 2004). AtGIF1 was also called AN3 (ANGUSTIFOLIA3) in Arabidopsis. Another study confirmed the above result by revealing that an3 and grf5 mutations exhibited a narrow-leaf phenotype due to reduced of cell numbers, while overexpression of AN3/GRF5 increased leaf size with standard shape (Horiguchi et al., 2005). AN3 could interact with the SWI/SNF chromatin remodeling complexes to regulate leaf development during the transition period from cell proliferation to cell differentiation (Vercruyssen et al., 2014). It was also found GIF regulates the formation of the pistil in Arabidopsis and single gif mutant lines showed a similar phenotype to control, but the gif triple mutant gif1/gif2/gif3 exhibited an abnormal pistil (Liang et al., 2014). A recent research (Ercoli et al., 2018) revealed that the GIFs are necessary to maintain the precise expression patterns of key developmental factors, and the GIF transcriptional coregulators could control QC (the quiescent center) organization and the size of the meristem in Arabidopsis (Ercoli et al., 2018). GIF2 and GIF3 are two additional proteins involved in cell proliferation and lateral organ growth development (Lee et al., 2009). The gif2 and gif3 mutated plants accommodate a smaller lateral organ than wild type plant species, caused by the synergistic reduction in cell numbers (Lee et al., 2009). Recent data shows that GRFs alone or in chimeras with GIFs could dramatically boost regeneration from tissue cultures in a broad range of plant species which is significant for plant transformation and gene editing (Debernardi et al., 2020; Kong et al., 2020; Luo and Palmgren, 2020).
The role of GIFs has also been in the regulation of plant growth and the development of crops. OsGIF1/2/3 with OsGRF4 controls the grain size and weight in rice, resulting from increased and heavier grain yield (Duan et al., 2015). GIF1 was also to be involved in the regulation of shoot architecture and meristem determinacy in maize (Zhang et al., 2018). Four SlGIF genes were identified to interact with various SlGRF proteins to affect the development of tomatoes (Ahiakpa et al., 2020). In tea (Camellia sinensis), the GIF and GRF gene families could control the tea plant’s leaves by affecting the developmental process and hormonal stimuli responses (Wu et al., 2017). One gene, CsGIF1, showed a different expression pattern under abiotic stresses (Wang et al., 2017).
While GIFs have been studied in some plant species, this gene family also requires investigation in cotton. Cotton is an important industrial crop that provides natural fibre for textile industries and seed-soil for animal and human consumption. Recent advances in cotton genomics provide an opportunity to characterize the GIF gene family in Gossypium. Multiple high-quality genome data sets were available for several cotton species, such as G. arboreum (Li et al., 2014; Du et al., 2018), G. hirsutum (Li et al., 2015), G. barbadense (Wang et al., 2019), and G. raimondii (Paterson et al., 2012; Wang et al., 2012). In this study, we identified and characterized the GIF gene family in four cotton species by analyzing their gene structures, conserved motifs, cis-elements, and expression patterns. We further explored the role of GIF genes in response to waterlogging stress, and verified the function of GhGIF4 in lateral root development by using a virus-induced gene silencing (VIGS) assay. The findings of this study will provide essential and valuable information for future studies on the functional characterization of the GIF gene family in cotton.
2 Materials and Methods
2.1 Identification of GRF1-Interacting Factor Homologs in Gossypium
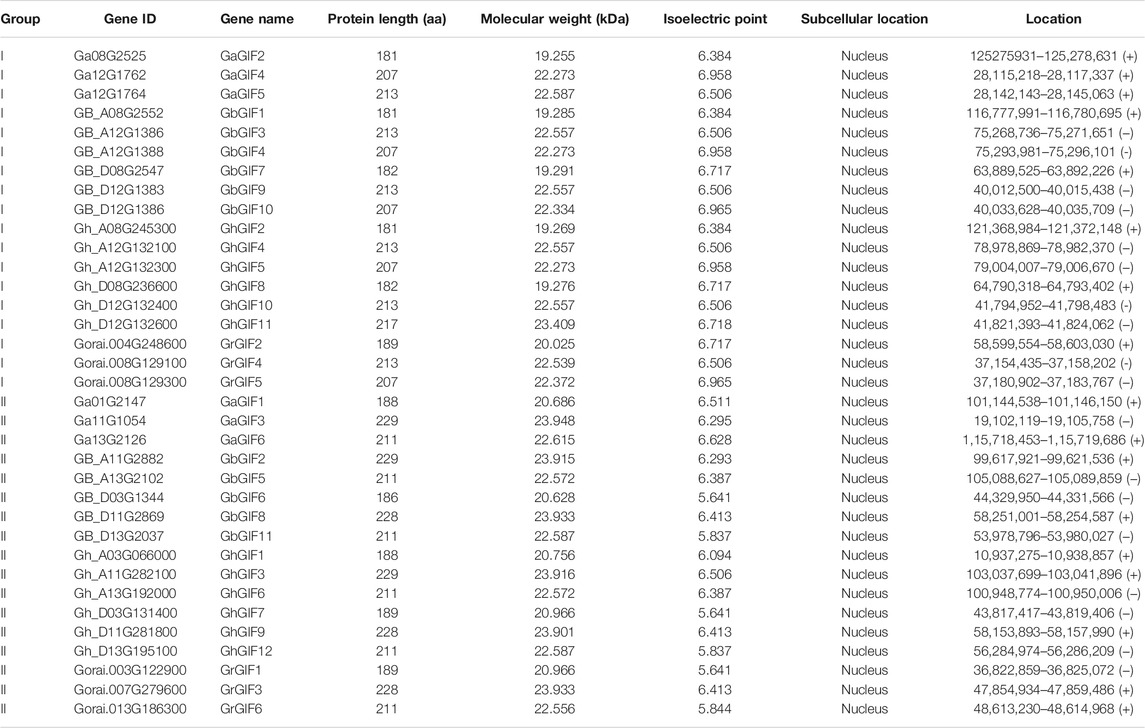
Four Gossypium species (G. arboreum, G. hirsutum, G.barbadense, and G. raimondii) were used in this study. The genome and protein sequence files of those Gossypium species were downloaded from the cotton functional genome database (CottonFGD, https://cottonfgd.org/) (Zhu et al., 2017). The hidden Markov model (HMM) profile of the conserved GIF domain SSXT (PF05030) was obtained from PFAM (http://pfam.xfam.org). HMMER 3.0 (http://hmmer.org/) was used to search the four Gossypium genomes for GIF candidate genes. The redundant candidate genes were eliminated, and the remaining genes were further confirmed by SMART (http://smart.emblheidelberg.de/). Both of the protein physicochemical properties (including the number of amino acids, molecular weight (MW), and theoretical isoelectric point (pI) of GIF proteins) and chromosomal gene positions were obtained from CottonFGD (https://cottonfgd.org/).
2.2 Characterization and Sequence Analysis of Related GIF Genes
The chromosomal location of all identified GIF genes was visualized by TBtools (v1.098661) (Chen et al., 2020). The identified GIF protein sequences of cotton were aligned using the ClustarW program in MEGA5 software (Tamura et al., 2011). Afterward, the neighbor-joining (NJ) tree was constructed using MEGA5 by running 1,000 bootstrap replicates. The gene structure of GIF genes was analyzed by the gene structure display server (GSDS2.0, http://gsds.gao-lab.org/). The conserved structural motifs of GIF genes were analyzed with the MEME online software (http://meme-suite.org/tools/meme).
2.3 Prediction of Cis-Elements of GIF Genes
The 2000 bp upstream of the translation initiation site of each of GIF gene was extracted by TBtools (v1.098661) (Chen et al., 2020), and the GIF sequences were submitted to the PlantCARE online server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict their cis-elements.
2.4 Gene Expression Analysis of GhGIFs
TPM (Transcripts Per Kilobase of exon model per Million mapped reads) expression values of GhGIFs were obtained from CottonFGD’s online website (https://cottonfgd.org/). We analyzed the expression differences of GhGIFs under different tissues (root, stem, leaf, torus, pental, anther, bract, filament, pistil, and sepal) and different stress treatments (cold, drought, and salt treatments). The average TPM values for each sample were calculated and were visualized by Heatmap Illustrator HemI 1.0.3.7 (http://hemi.biocuckoo.org/). Genes with a TPM value fold change (treatment/control) > 2 were considered up-regulated genes, and fold change (treatment/control) < 0.5 were termed as down-regulated genes.
2.5 Plant Cultivation and Phenotypic Investigation
Two G. hirsutum accessions, Ji A-1-7 (33xi) and King, were selected to be cultured and investigated in this study. Cotton seeds were provided by the National Medium-term Gene Bank of Cotton in China. All the seeds were first soaked in water for 1 day and germinated in potting sand for 3 days. Then, the resulting seedlings were transferred into hydroponic containers with a half-strength of Hoagland nutrient solution. All the seedlings were maintained in controlled conditions at 26°C during the day and 23°C at night, with a 16 h light-to-8 h dark photoperiod. The root phenotype was observed. The primary root length and lateral root numbers were counted on the 8-day after sowing. The lateral root samples were collected and stored at −80°C for further qRT-PCR analysis.
2.6 Lateral Root Initiation Observation
We noticed that the lateral roots emerged at 96 h old. To better understand the lateral root initiation mechanism, we dissected the initiation region of the lateral root at 96 h after sowing. A 1.5 cm section of root samples were collected and mixed in FAA fixative for 24 h, then embedded in paraffin and stained with toluidine blue staining solution. The structures of cells for the lateral root initiation region were observed under a Leica upright microscope, Leica Dm6B.
2.7 CAT, MDA, and POD Measurements
The lateral root samples of 8 day-old seedlings were utilized to measure the Micro Catalase (CAT), Malondialdehyde (MDA), and Peroxidase (POD) content. Fresh cotton lateral root samples (0.1 g) were weighed into a 2 ml centrifuge tube with two steel balls and 1 ml of extract buffer, ground in an automatic sample cryo-grinder (Shanghai Jingxin Industrial Development Co., Ltd., China, Shanghai) at 4°C, centrifuged at 4°C × 12,000 rpm for 10 min, and the supernatant taken for further enzyme measurement. Then, the CAT, MDA, and POD activities were tested by their Kit (Micro Catalase (CAT) Assay Kit (Suzhou Grace Biotechnology Co., Ltd., China, Suzhou), Malondialdehyde (MDA) Assay Kit (Suzhou Grace Biotechnology Co., Ltd., China, Suzhou), and Peroxidase (POD) assay Kit (Suzhou Grace Biotechnology Co., Ltd., China, Suzhou)) according to the manual instructions.
2.8 qRT-PCR
The total RNA of cotton lateral roots and leaves were isolated by RNAprep Pure Plant kit (Tiangen, China) according to the manufacturer’s protocol. Approximately 1 μg of RNA was reversely synthesized into cDNA using the Prime Script TM RT reagent kit with g DNA Eraser kit (TaKaRa). For quantitative real-time PCR (qRT-PCR), we used Primer5 software to design primers for selected candidate genes (Supplementary Table S3), and those PCR products were checked by agarose gel electrophoresis. Later, the qRT-PCR was performed by the Roche LightCycler 480 II real-time PCR system using the PerfectStart Green qPCR SuperMix (Transgen Biotech Co.,Ltd.,). The Histone3 gene was set as an internal control, and the relative expression of GhGIFs genes was calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001).
2.9 Waterlogging Experiment
In order to clarify whether the GhGIF genes are affected by waterlogging, we conducted a waterlogging assay on the two representative cotton cultivars mentioned. The experiment used the sand culture method to grow seedlings. Seeds of Ji A-1-7 (33xi) and King were planted in a medium-size plastic box. Each box contained eight seeds, and four similar seedlings were selected and kept in each box on the ninth day. Both the two cotton cultivars contained 24 boxes, and half of them were used for waterlogging treatment. When cotton seedlings were grown to the period of the two-leaf and one-heart seedling stage, the treated boxes were added with water every day, so that the water surface was always about 1 cm higher than the sand surface. The control plants were watered according to the expected standard. Four weeks later, we observed the phenotype of all the plants and sampled leaves to detect the expression of three GhGIF genes (GhGIF4, GhGIF5, and GhGIF8) by qRT-PCR.
2.10 VIGS
A virus-induced gene silencing (VIGS) experiment was performed to verify whether GhGIF genes affect lateral root development in cotton. The VIGS primers are designed using https://crm.vazyme.com/cetool/multifragment.html (Supplementary Table S3). Briefly, a 300 bp fragment of GhGIF4 was cloned into the EcoRⅠ/BamHⅠ sites of the pYL156 vector to construct the TRV: GhGIF4 recombinant plasmid. The TRV: GhGIF4 vector was transferred into Agrobacterium tumefaciens LBA4404. Seedlings of Ji A-1-7 (33xi) were cultured with a half-strength of Hoagland nutrient solution and were used as VIGS experimental plants. The A. tumefaciens strain LBA4404 was cultured overnight (14–18 h) in LB with rifampicin (30 μg/ml) and kanamycin (50 μg/ml) at 28°C with pYL156, pYL156: CLA1, pYL192, and transformed pYL156: GhGIF4. We adjusted the OD600 of the cell suspension to about 0.8 and let it continue to incubate in the dark for 3–4 h. Then, the A. tumefaciens carrying pYL156, pYL156: CLA1, and pYL156: GhGIF4 was mixed with A. tumefaciens harboring pYL192 at 1:1, respectively. The A. tumefaciens mixed suspensions were infiltrated into the leaves of the Ji A-1-7 (33xi) on 5 days-old seedlings using a needleless syringe. Those plants were placed in a dark place for 24 h and then grew in the greenhouse at 26°C days and 23°C nights with a 16 hour-to-8 h dark cycle. About 2 weeks later, the CLA1 plants were bleaching, and we observed the root morphology for all plants. The lateral root was collected for further analysis of qRT-PCR and enzyme activity (MDA, POD).
3 Results
3.1 GIF Genes Identification and Chromosomal Locations in Four Gossypium Species
Combining the results of CottonFGD (https://cottonfgd.org/), HMMER3.1 (http://hmmer.org/), and SMART (http://smart.embl-heidelberg.de/), we identified a total of 12, 11, 6, and 6 full-length putative GIF genes in G. hirsutum, G. barbadense, G. arboretum, and G. raimondii, respectively. For convenience, the family GIF members of the four cotton species were renamed GhGIF1 to GhGIF12; GbGIF1 to GbGIF11; GaGIF1 to GaGIF6; and GrGIF1 to GrGIF6, respectively.
As shown in Figure 1, the GIF genes were unevenly distributed on different Gossypium chromosomes. In G. arboretum (Figure 1A), 2 GaGIF genes (GaGIF4 and GaGIF5) were allocated on Chr12, while the other four GaGIF genes were distributed on Chr01, Chr08, Chr11, and Chr13, respectively. Similarly, two GrGIF genes (GrGIF4 and GrGIF5) were observed on Chr08 in G. raimondii, and the other four GrGIF genes were observed on Chr03, Chr04, Chr07, and Chr13, respectively (Figure 1B). The GIF genes of G. hirsutum were distributed on ten chromosomes, five in subgroup At and five in subgroup Dt (Figure 1C). Furthermore, two adjacent GIF genes were distributed on the Chr12 chromosomes of the At and Dt subgroups both in G. hirsutum and G. barbadense. A total of 11 GbGIF genes were observed on nine chromosomes in G. barbadense (Figure 1D). Five of them were distributed among four At chromosomes (A08, A11, A12, A13), and six were distributed among five Dt chromosomes (D03, D08, D11, D12, D13).
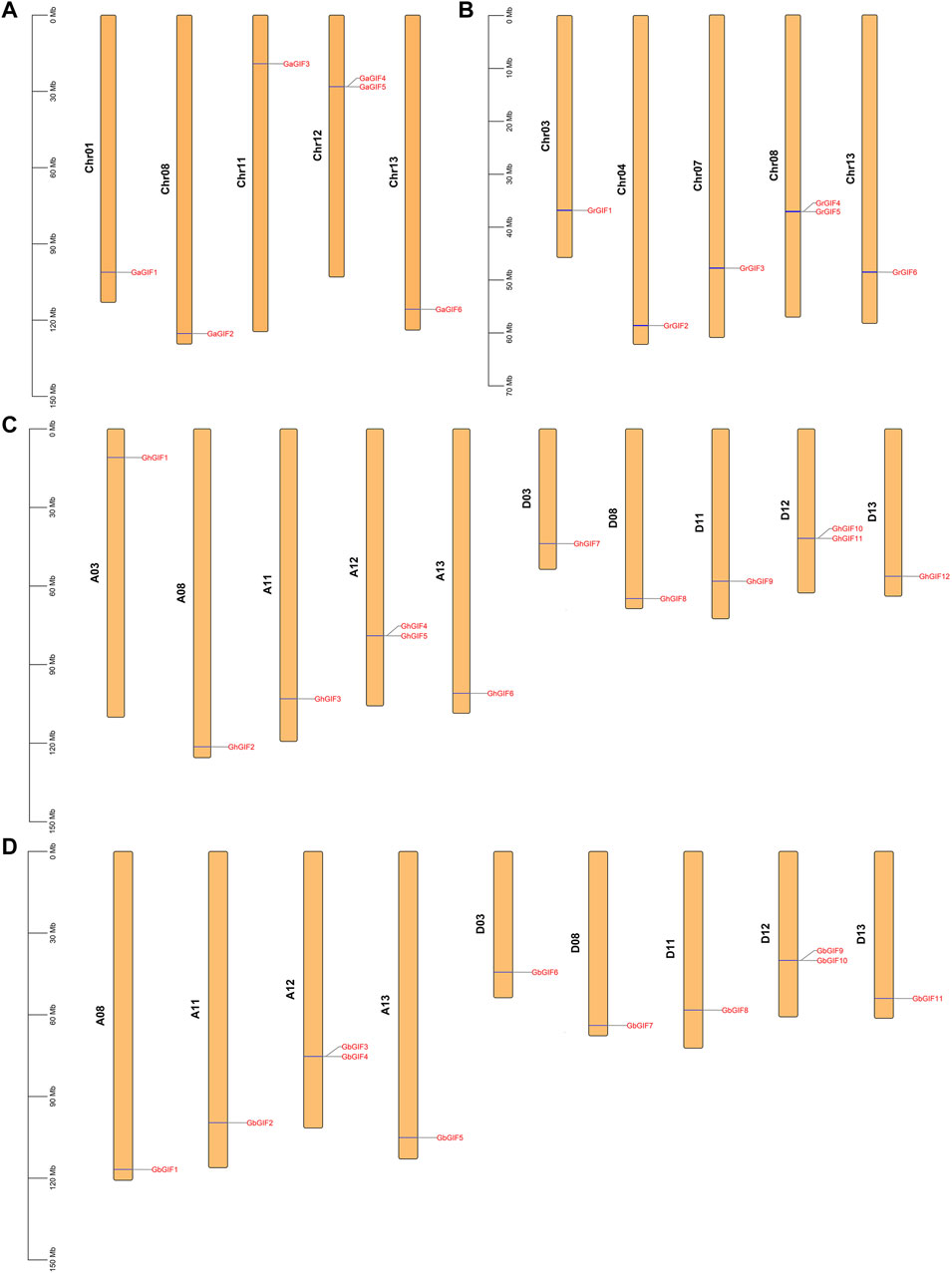
FIGURE 1. Chromosome distribution of GIF genes in four Gossypium species. (A) G. arboreum (B) G. raimondii (C) G. hirsutum (D) G. barbadense.
Protein sequence analysis showed that all GIF proteins could encode 181–229 amino acids with a molecular weight (Mw) ranging from 19.255 to 23.948 kDa, and an isoelectric point (pI) ranging from 5.641 to 6.965 (Table 1, Supplementary Table S1). Subcellular mapping prediction indicated that all 35 GIF genes were located in the nucleus (Table 1).
3.2 GIF Gene Phylogenetic Tree, Conserved Motif, and Gene Structure Analysis
A phylogenetic tree was constructed using 35 GIF protein sequences from four Gossypium species to understand the evolutionary relationship between GIF genes better. Subsequent phylogenetic analysis revealed that GIF genes could be classified into two subfamilies (Group I and II). Group I contains 18 GIF genes, while Group II has 17 GIF genes (Figure 2A). Further, we identified fifteen conserved motifs among 35 cotton GIF genes using a MEME program (Figure 2B). The number of conserved motifs in GIF genes was different from 7 to 11. Motifs 1, 3, 4, and 5, were detected in all the GIF genes. All Group I members include Motif 2 and 8, and two-thirds of Group I members include Motif 6, 7, and 11. Moreover, all the Group II members include Motif 6 and 11, and 70.59% of Group II members include Motif 2, 9, and 10 (Figure 2B).
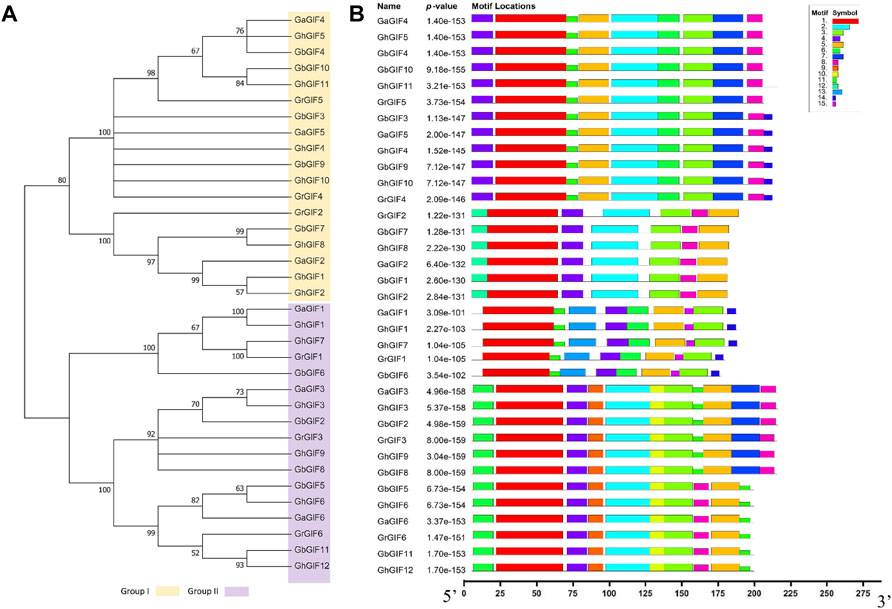
FIGURE 2. Phylogenetic and conserved motif analysis of GIF genes in Gossypium. (A) phylogenetic tree (B) The 35 GIF genes have 15 conserved motifs.
The structural gene variation of the GIF genes was then compared, yielding a comprehensive illustration of their evolutionary relationship. A variable structural pattern of exon-intron was observed in GIF family genes. As shown in Supplementary Figure S1, all the GIF genes have four CDS sequences, with three shorter and one longer. All of the GIF genes were identified with three introns. However, in Group I, the intron lengths of all GIF genes were similar; and in Group II, the intron lengths of GaGIF3, GhGIF3, GbGIF2, GrGIF3, GhGIF8, and GhGIF9 are significantly longer than those of other GIF genes.
3.3 Cis-Element Analysis of GIF Promoters
Cis-acting elements of promoters may play a crucial role in regulating of gene expression. In this study, we extracted the 2000 bp promoter sequence that was upstream of the start codon of each GIF gene and their cis-elements identification was due by using the PlantCare tool. We also screened out the stress and hormone-responsive elements for further analysis. As shown in Figure 3, the stress-related responsive mainly included anaerobic, anoxic, defense/stress, drought, low temperature and wound, and their cis-acting elements were ARE, GC-motif, TC-rich repeats, MBS, LTR, and WUN-motif respectively (Figure 3A, Supplementary Table S2). The majority were anaerobic responsive, and the representative GIF genes were GhGIF4, GhGIF5, GhGIF3, etc. The cis-elements of hormone-responsive mainly exhibited abscisic acid (ABRE), auxin (TGA-element), gibberellin (TATC-box, P-box, GARE-motif), MeJA (TGACG-motif, CGTCA-motif), and salicylic acid (TCA-element). The number of gibberellin-related cis-acting elements was the largest, and the GIF genes in G.arboretum occupied a larger ratio among them (Figure 3B; Supplementary Table S2).
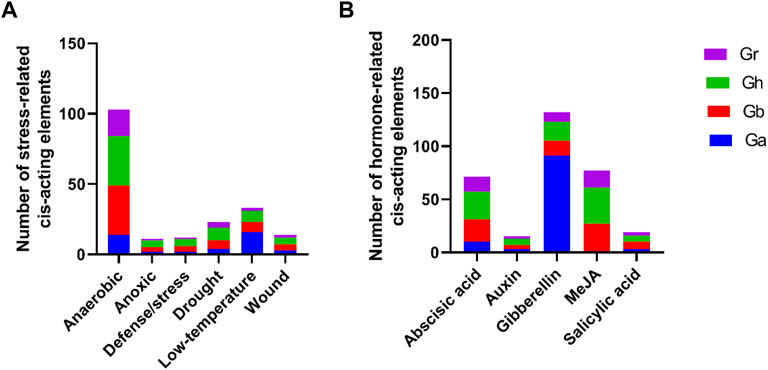
FIGURE 3. Cis-element analysis of GIF promoters in four Gossypium species. (A) analysis of the stress-related cis-acting elements (B) analysis of hormone-related cis-acting elements.
3.4 Expression Analysis of GhGIF Genes
As shown in Figure 4, we compared the transcriptional patterns of GhGIF genes using multiple tissues and stresses of TM-1. Tissue expression pattern analysis showed that eight GhGIF genes (GhGIF2, GhGIF3, GhGIF4, GhGIF5, GhGIF8, GhGIF9, GhGIF10, and GhGIF11) exhibited different expression levels in various tissues, indicating that those GIF genes have complete biological functions in the growth and development of cotton (Figure 4A). Among the above GIF genes, four GhGIFs (GhGIF4, GhGIF5, GhGIF10, and GhGIF11) were highly expressed in root tissues, suggesting that these genes may play an essential role in root development. GhGIF5 and GhGIF8 are highly expressed in leaf tissues, which indicates that GhGIF5 and GhGIF8 may be involved in leaf development. At the same time, we also analyzed the expression level of GhGIF genes under cold, drought, and salt stress. Under cold stress treatment, the expression levels of GhGIF2, GhGIF4, and GhGIF10 genes were down-regulated, and the longer the cold treatment, the greater the degree of down-regulation. In contrast, the expression of GhGIF5 would gradually increase after cold treatment (Figure 4B). Under drought stress, the expression of GhGIF4 would gradually increase after 6 h of treatment and reach its maximum at 12 h. Similarly, the expression of GhGIF10 was up-regulated during the three periods of drought (3, 6, and 12 h), and reached its maximum at 12h, but we detected a down-regulation of this gene at 24 h. In addition, three genes of GhGIF2, GhGIF5, and GhGIF11 also exhibited different expression levels in drought stress, indicating that these genes may be involved in drought stress (Figure 4C). Five genes of GhGIF2, GhGIF4, GhGIF5, GhGIF10 and GhGIF11, exhibited a varied expression in different salt treatments, which indicated that those genes might play an important role in salt stress (Figure 4D).
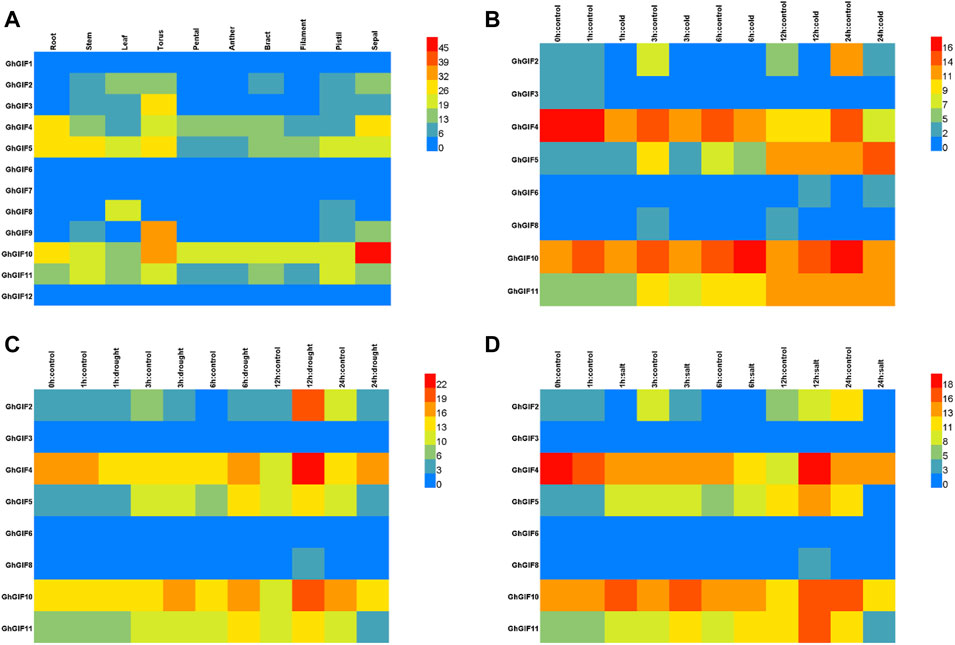
FIGURE 4. TPM expression of GhGIF genes in different organizations and different stresses. (A) TPM expression of GhGIF genes in different organizations in TM-1 (B) TPM expression of GhGIF genes under cold stress (C) TPM expression of GhGIF genes under drought stress (D) TPM expression of GhGIF genes under salt stress.
3.5 Evaluation of the Lateral Root Phenotype in Ji A-1-7 (33xi) and King
Ji A-1-7 (33xi) showed significantly larger lateral roots than King (Figures 5A,B). Further qRT-PCR expression results showed that three genes of GhGIF4, GhGIF6, and GhGIF10, expressed significantly different expression levels in lateral roots between Ji A-1-7 (33xi) and King with the highest expression value recorded for GhGIF4 (Figure 5C). The POD, CAT, and MDA results showed no difference in the CAT activity between the above two accessions. However, a significant difference was detected for POD activity and MDA content. The lateral root’s POD activity is higher in Ji A-1-7 (33xi), but King’s MDA content is higher (Figures 5D–F). The paraffin sectioning assay indicated that Ji A-1-7 (33xi) had more giant cells than King with the average cell area in Ji A-1-7 (33xi) being significantly higher than King on the lateral root emergence part (Figures 5G,H).
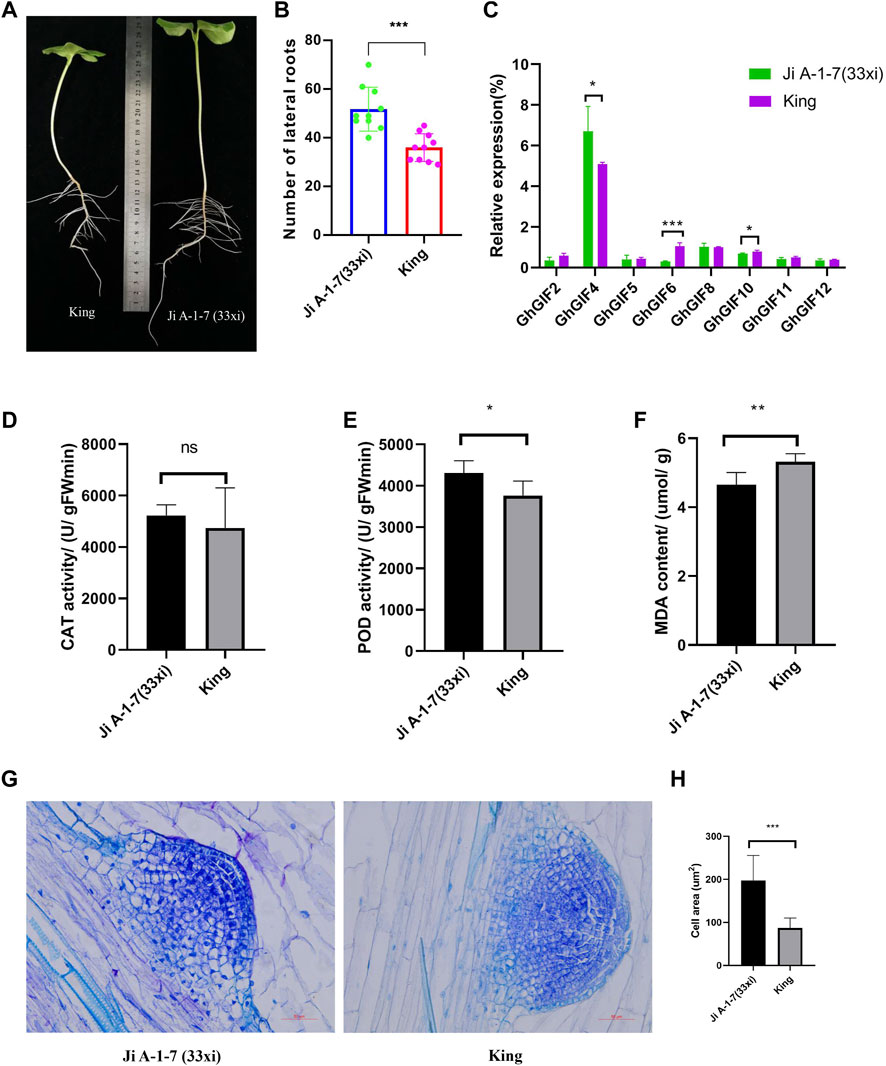
FIGURE 5. Comparison of lateral roots between Ji A-1-7 (33xi) and King. (A) Phenotype of Ji A-1-7 (33xi) and King on the 8-days after sowing (B) Comparison of lateral root numbers (C) Relative expression of eight GhGIF genes in lateral roots (D) CAT activity of lateral roots (E) POD activity of lateral roots (F) MDA content of lateral roots (G) microstructure of the lateral root emergence part in Ji A-1-7 (33xi) and King (H) Comparison of cell area between Ji A-1-7 (33xi) and King.
3.6 Waterlogging Stress Experiment
Surprisingly, we found that the waterlogged plants grew more prosperous than the control plants. The plant height, leaf numbers and areas (last second leave) were significantly higher than those of control plants (Figures 6A,B). Unfortunately, we failed to count the lateral root numbers due to their easy breakage when washing their surface sand soil. GhGIF4, GhGIF5, and GhGIF8 expression levels in the leaf of Ji A-1-7 (33xi) showed significantly higher in the waterlogging plants than in the control (Figure 6C). Expression levels of those genes in King also showed a similar pattern but did not reach a significant level (Figure 6C).
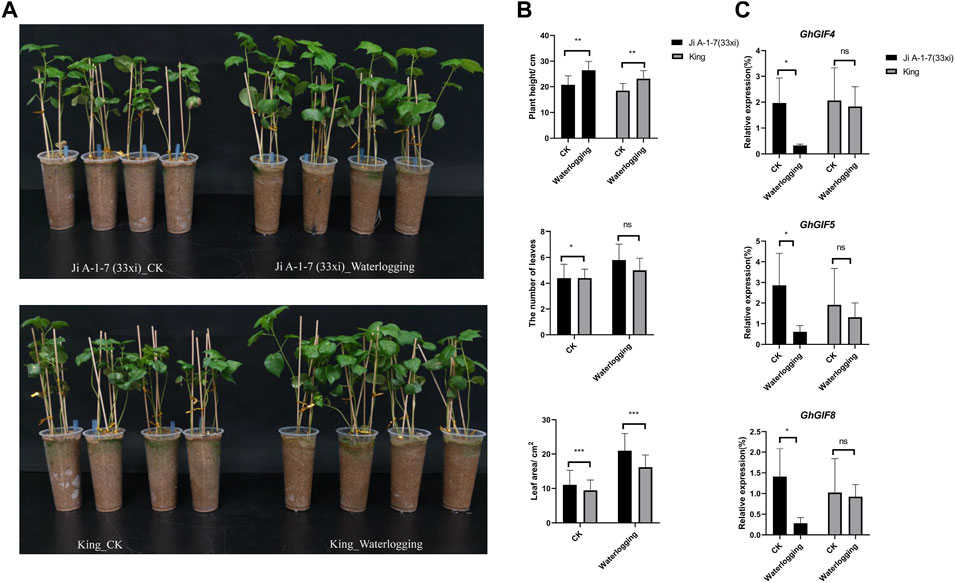
FIGURE 6. Response of GhGIF genes to waterlogging. (A) The phenotype of Ji A-1-7 (33xi) and King under waterlogging stress (B) Count of plant height, lateral root numbers, and the leaf areas (C) Relative expression of GhGIF4, GhGIF5, and GhGIF8 in leaves of waterlogging and its control (CK) seedlings.
3.7 GhGIF4 Silencing in Ji A-1-7 (33xi)
There were three GIF genes with high expression levels related to lateral development, while three GIF genes varied in response to waterlogging stress. Among the GIF genes, the common key gene is GhGIF4, resulting in that we further evaluate the function of this gene on lateral root development in cotton by VIGS. We observed the silenced seedlings and their controls when the positive control plants that injected the recombinant vector pYL156:CLA1 showed an albino phenotype (Figure 7A). The expression of GhGIF4 in silenced plants was significantly lower than the negative (CK) and positive (pYL156:00) controls. Moreover, the silencing efficiency of VIGS is about 40–60% (Figure 7B). The number of lateral roots in silenced plants was also significantly lower than in their controls (Figure 7C). Later, we also compared the lateral roots’ POD activity and MDA content between the silenced plants and their controls. A similar result was observed for the POD activity and MDA content (Figures 7D,E). Therefore, we speculate that GhGIF4 may positively regulate cotton lateral root numbers but not be influenced by the POD activity and MDA content.
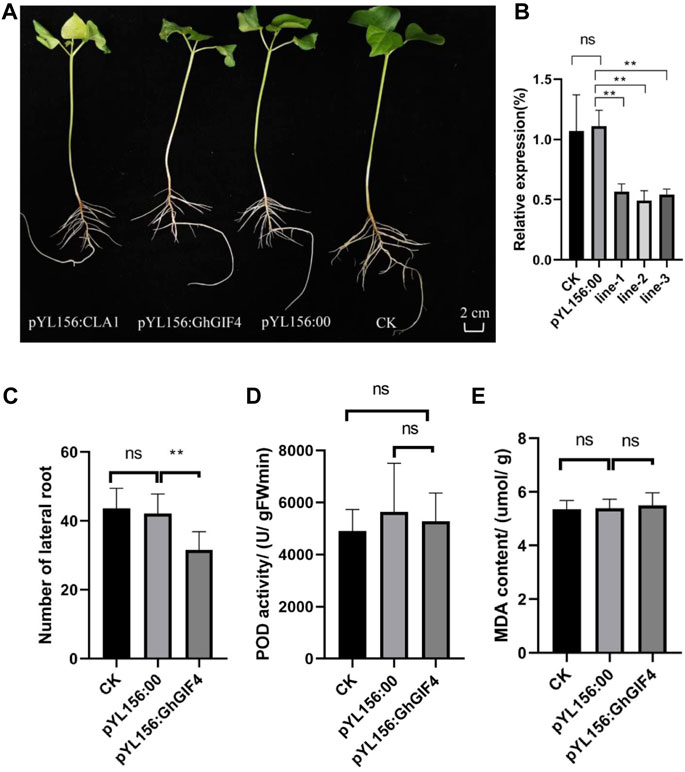
FIGURE 7. Virus-induced Gene Silencing (VIGS) of GhGIF4 in cotton. (A) The phenotype of the gene silencing plants (B) Relative expression of GhGIF4 in silenced and its control plants (C) The number of lateral roots (D) POD activity (E) MDA content. Note, all values represented the mean ± s.d. (n≥3 replicates), ns represent not significant, ∗ represent significance at p < 0.05 level.
4 Discussion
Recently, several high-quality genomes of Gossypium including G. arboreum, G. raimondii, G. hirsutum and G. barbadense have been released, which would provide valuable information for us to study the GIF family in cotton. Previous studies mainly reported the GIF genes were involved in plant growth and development, such as controlling the leaf, seed, and root meristem homeostasis in Arabidopsis (Rodriguez et al., 2010; Debernardi et al., 2014; Liang et al., 2014; Ercoli et al., 2018); modulating the tissue and organ size in rice (Song et al., 2011; Duan et al., 2015; Li et al., 2016); and regulating the shoot architecture and meristem determinacy in maize (Zhang et al., 2018). Here, we identified a total of 35 GIF genes and analyzed their structures and phylogenetic relationships in four Gossypium species. We further performed preliminary studies to verify the response of GIF genes to adversity stress and the effect of GhGIF4 on cotton lateral root development. Our results have been provided new insights into GIF proteins for cotton.
Previous reports well described that G. arboretum could be seen as the ancestor of G. hirsutum by providing an A subgenome (Kebede et al., 2007). Intriguingly, in this study, we found that the number of GIF genes identified in G. hirsutum was twice as much as that in G. arboreum. Their distributions were consistent with the cotton allotetraploid evolutionary process (Flagel and Wendel, 2009). So, we speculated that the GIF genes were multiplied by the polyploidy event of subfamily At and Dt during the formation of G. hirsutum. Furthermore, this speculation was also suited to the GIF genes identified in G. raimondii and G. barbadense. However, only 11 GbGIF genes were identified in G.barbadense, with five on four At chromosomes (A08, A11, A12, A13) and six on five Dt chromosomes (D03, D08, D11, D12, D13). It seems that there should be one GbGIF gene on the A03 chromosome. We have not identified any GIF genes on the A03 in G. barbadense, and the reason may be due to the occurrence of gene loss events. Based on the phylogenetic analysis, those 35 GIF protein sequences identified in the four cotton species can be classified into two groups: Group I and Group II. We further analyzed the conserved motif and gene structures of those GIF proteins, and the result showed that both the conserved motifs and their gene structures are relatively conservative. Four motifs of motif_1, motif_3, motif_4, and motif_5 were all detected in all the 35 GIF proteins.
Moreover, all the GIF genes in Gossypium have four CDS sequences and three introns. However, the intron length was different between Groups I and II. The different structures of those GIF genes between Group I and Group II implied that the varied functions of GIF genes might be closely related to evolution.
Currently, upland cotton is the largest natural fiber crop globally. However, during its vegetative and reproductive periods, its yield and fiber quality are adversely affected by abiotic stresses such as high temperatures, cold, drought, waterlogging, and salt stress. Recently, the GIF genes were also involved in abiotic stresses. For example, Wang et al. (2017) cloned a CsGIF1 gene in tea (Camellia sinensis) and found the CsGIF1 had different expression patterns under different abiotic stresses (Wang et al., 2017). In this study, our results mainly found that GhGIF2, GhGIF4 and GhGIF10 were always down-regulated in the cold stress treatment, and those genes may be closely regulated in response to cold stress. GhGIF4 and GhGIF10 may also respond to drought stress due to their specific expression patterns. Five genes, GhGIF2, GhGIF4, GhGIF5, GhGIF10, and GhGIF11, may respond to salt stress. In the past 2 years, floods have frequently occurred in Henan, China, which has caused severe yield and quality losses to the local cotton. However, experiments with waterlogging are rare in cotton. We tried a mild waterlogging treatment on two representative cotton accessions in this study. Surprisingly, we found cotton seedlings treated with waterlogging grew more vigorously than control plants, with higher plant height and more vigorous leaves. Of course, in the actual field, experienced growers know that vigorous leaves and higher plant heights are not conducive to the final yield of cotton. From this perspective, our results are reasonable, and we speculated that severe floods would cause the hypoxic death of seedlings and further severely affect the yield of cotton. We also detected the expression levels of GhGIF4, GhGIF5, and GhGIF8, and the result demonstrated that those genes were down-regulated in the waterlogged seedlings, especially for Ji A-1-7 (xi). Therefore, we hypothesized that the GIF genes of GhGIF4, GhGIF5, and GhGIF8 may respond to waterlogging in upland cotton.
A recent study showed that AN3/GIFs might be involved in different pathways that control the quiescent centre (QC) organization and the meristem size in Arabidopsis (Ercoli et al., 2018). In this study, we found that the gene for GhGIF4 may affect the lateral root numbers in upland cotton. The relative GIF gene expressions, enzyme activity, and microstructure of the emergence part of the lateral roots of Ji A-1-7 (33xi) and King were also compared. Among the eight selected GhGIF genes, GhGIF4 was expressed the highest and showed a significant difference between the two representative accessions. The lateral root’s POD activity in Ji A-1-7 (33xi) was higher, while the MDA content in King was higher. Moreover, the microstructure assay showed that the lateral root of Ji A-1-7 (33xi) had more giant cells than King on the lateral root emergence part. Furthermore, a further VIGS assay for GhGIF4 showed smaller lateral root numbers than control, which emphasized the vital role of GhGIF4 in lateral root development in upland cotton.
5 Conclusion
In this study, we identified 35 GIF genes in four cotton species. We analyzed their gene structures, phylogenetic relationships, conserved domains, cis-elements, and expression patterns in various organizations and different stresses. The qRT-PCR of GIF genes in lateral roots showed that the GhGIF4 expressed higher and significantly differed between Ji A-1-7 (33xi) and King, which had larger and smaller later root numbers, respectively. The enzyme activity and microstructure of the emergence part of the lateral roots in those two cotton accessions were also compared. The result showed that Ji A-1-7 (33xi) had a higher POD activity and lowered MDA content and more giant cells of the lateral root phenotype than King. A mild waterlogging assay showed that the waterlogging-seedlings grew a more vigorous phenotype than control, and three selected GIF genes were all down-regulated. A VIGS assay of GhGIF4 further verified the vital role of GhGIF4 in lateral root development in the cotton. Together, our findings provide a foundation for further functional studies of the GIF genes in cotton plant growth, lateral root development, and stress tolerance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
DH, XD, and YJ conceived and designed the experiment. YG conducted experiments, DH analyzed the data and wrote the manuscript. YJ, TM, and ZI revised the grammar, and all the other authors revised the manuscript. All authors reviewed and approved the final manuscript.
Funding
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund (No.Y2021YJ06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YY declared a shared affiliation with the authors to the handling editor at the time of review.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The National Medium-term Gene Bank of Cotton in China provided us with cotton seeds, and we are grateful for their kindly helps.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.851343/full#supplementary-material
References
Ai, G., Zhang, D., Huang, R., Zhang, S., Li, W., Ahiakpa, J. K., et al. (2020). Genome-wide Identification and Molecular Characterization of the Growth-Regulating Factors-Interacting Factor Gene Family in Tomato. Genes (Basel) 11, 1435. doi:10.3390/genes11121435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional Control ofGRFtranscription Factors by microRNA miR396 and GIF Co-activator Affects Leaf Size and Longevity. Plant J. 79, 413–426. doi:10.1111/tpj.12567
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF-GIF Chimeric Protein Improves the Regeneration Efficiency of Transgenic Plants. Nat. Biotechnol. 38, 1274–1279. doi:10.1038/s41587-020-0703-0
Du, X., Huang, G., He, S., Yang, Z., Sun, G., Ma, X., et al. (2018). Resequencing of 243 Diploid Cotton Accessions Based on an Updated A Genome Identifies the Genetic Basis of Key Agronomic Traits. Nat. Genet. 50, 796–802. doi:10.1038/s41588-018-0116-x
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 Controls Grain Size and Yield in rice. Nat. Plants 2, 15203. doi:10.1038/nplants.2015.203
Ercoli, M. F., Ferela, A., Debernardi, J. M., Perrone, A. P., Rodriguez, R. E., and Palatnik, J. F. (2018). GIF Transcriptional Coregulators Control Root Meristem Homeostasis. Plant Cell 30, 347–359. doi:10.1105/tpc.17.00856
Flagel, L. E., and Wendel, J. F. (2009). Evolutionary Rate Variation, Genomic Dominance and Duplicate Gene Expression Evolution during Allotetraploid Cotton Speciation. New Phytol. 186, 184–193. doi:10.1111/j.1469-8137.2009.03107.x
Hoe Kim, J., and Tsukaya, H. (2015). Regulation of Plant Growth and Development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR Duo. Exbotj 66, 6093–6107. doi:10.1093/jxb/erv349
Horiguchi, G., Kim, G.-T., and Tsukaya, H. (2005). The Transcription Factor AtGRF5 and the Transcription Coactivator AN3 Regulate Cell Proliferation in Leaf Primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi:10.1111/j.1365-313x.2005.02429.x
Kebede, H., Burow, G., Dani, R. G., and Allen, R. D. (2007). A-genome Cotton as a Source of Genetic Variability for Upland Cotton (Gossypium Hirsutum). Genet. Resour. Crop Evol. 54, 885–895. doi:10.1007/s10722-006-9157-6
Kim, J. H., and Kende, H. (2004). A Transcriptional Coactivator, AtGIF1, Is Involved in Regulating Leaf Growth and Morphology in Arabidopsis. Proc. Natl. Acad. Sci. 101, 13374–13379. doi:10.1073/pnas.0405450101
Kong, J., Martin-Ortigosa, S., Finer, J., Orchard, N., Gunadi, A., Batts, L. A., et al. (2020). Overexpression of the Transcription Factor GROWTH-REGULATING FACTOR5 Improves Transformation of Dicot and Monocot Species. Front. Plant Sci. 11, 572319. doi:10.3389/fpls.2020.572319
Lee, B. H., Ko, J.-H., Lee, S., Lee, Y., Pak, J.-H., and Kim, J. H. (2009). The ArabidopsisGRF-INTERACTING FACTORGene Family Performs an Overlapping Function in Determining Organ Size as Well as Multiple Developmental Properties. Plant Physiol. 151, 655–668. doi:10.1104/pp.109.141838
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome Sequence of Cultivated Upland Cotton (Gossypium Hirsutum TM-1) Provides Insights into Genome Evolution. Nat. Biotechnol. 33, 524–530. doi:10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome Sequence of the Cultivated Cotton Gossypium Arboreum. Nat. Genet. 46, 567–572. doi:10.1038/ng.2987
Li, S., Gao, F., Xie, K., Zeng, X., Cao, Y., Zeng, J., et al. (2016). The OsmiR396c‐OsGRF4‐OsGIF1 Regulatory Module Determines Grain Size and Yield in rice. Plant Biotechnol. J. 14, 2134–2146. doi:10.1111/pbi.12569
Liang, G., He, H., Li, Y., Wang, F., and Yu, D. (2014). Molecular Mechanism of microRNA396 Mediating Pistil Development in Arabidopsis. Plant Physiol. 164, 249–258. doi:10.1104/pp.113.225144
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Luo, G., and Palmgren, M. (2021). GRF-GIF Chimeras Boost Plant Regeneration. Trends Plant Sci. 26, 201–204. doi:10.1016/j.tplants.2020.12.001
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated Polyploidization of Gossypium Genomes and the Evolution of Spinnable Cotton Fibres. Nature 492, 423–427. doi:10.1038/nature11798
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., and Palatnik, J. F. (2010). Control of Cell Proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi:10.1242/dev.043067
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739. doi:10.1093/molbev/msr121
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S.-K., et al. (2014). ANGUSTIFOLIA3 Binds to SWI/SNF Chromatin Remodeling Complexes to Regulate Transcription during Arabidopsis Leaf Development. The Plant Cell 26, 210–229. doi:10.1105/tpc.113.115907
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The Draft Genome of a Diploid Cotton Gossypium Raimondii. Nat. Genet. 44, 1098–1103. doi:10.1038/ng.2371
Wang, M., Tu, L., Yuan, D., ZhuShen, D., Shen, C., Li, J., et al. (2019). Reference Genome Sequences of Two Cultivated Allotetraploid Cottons, Gossypium Hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi:10.1038/s41588-018-0282-x
Wang, Y., Wang, Y., Wang, W., Wu, Z., and Zhuang, J. (2017). Cloning and Expression Analysis of CsGIF1 Gene under Abiotic Stress in Camellia Sinensis. Acta Botanica Boreali-Occidentalia Sinica 37, 2363–2369. doi:10.7606/j.issn.1000-4025.2017.12.2363
Wu, Z.-J., Wang, W.-L., and Zhuang, J. (2017). Developmental Processes and Responses to Hormonal Stimuli in tea Plant (Camellia Sinensis) Leaves Are Controlled by GRF and GIF Gene Families. Funct. Integr. Genomics 17, 503–512. doi:10.1007/s10142-017-0553-0
Yan, S., Zou, G., Li, S., Wang, H., Liu, H., Zhai, G., et al. (2011). Seed Size Is Determined by the Combinations of the Genes Controlling Different Seed Characteristics in rice. Theor. Appl. Genet. 123, 1173–1181. doi:10.1007/s00122-011-1657-x
Zhang, D., Sun, W., Singh, R., Zheng, Y., Cao, Z., Li, M., et al. (2018). GRF-interacting Factor1 Regulates Shoot Architecture and Meristem Determinacy in maize. Plant Cell 30, 360–374. doi:10.1105/tpc.17.00791
Keywords: cotton, GIF, lateral root, waterlogging, VIGS
Citation: Hu D, Ge Y, Jia Y, He S, Geng X, Wang L, Pan Z, Iqbal Z, Mahmood T, Li H, Chen B, Wang X, Pang B and Du X (2022) Identification and Characterization of the Growth-Regulating Factors-Interacting Factors in Cotton. Front. Genet. 13:851343. doi: 10.3389/fgene.2022.851343
Received: 09 January 2022; Accepted: 23 February 2022;
Published: 14 March 2022.
Edited by:
Yuan-Ming Zhang, Huazhong Agricultural University, ChinaReviewed by:
Dawei Xue, Hangzhou Normal University, ChinaYoulu Yuan, Cotton Research Institute (CAAS), China
Xingchun Wang, Shanxi Agricultural University, China
Copyright © 2022 Hu, Ge, Jia, He, Geng, Wang, Pan, Iqbal, Mahmood, Li, Chen, Wang, Pang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiongming Du, ZHVqZWZmcmV5ODg0OEBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Daowu Hu†
Daowu Hu† Shoupu He
Shoupu He Zhaoe Pan
Zhaoe Pan Zubair Iqbal
Zubair Iqbal Tahir Mahmood
Tahir Mahmood Hongge Li
Hongge Li Xiongming Du
Xiongming Du