
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 12 May 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.849357
This article is part of the Research Topic Biotechnological and Genomic Approaches for Enhancing Agronomic Performance of Crops View all 19 articles
Parts of this article's content have been modified or rectified in:
Erratum: Heterologous expression of Arabidopsis AtARA6 in soybean enhances salt tolerance
 Zhipeng Hong1†
Zhipeng Hong1† Yang Li1†
Yang Li1† Yang Zhao1†
Yang Zhao1† Mingyu Yang1
Mingyu Yang1 Xiaoming Zhang1
Xiaoming Zhang1 Yuhan Teng1
Yuhan Teng1 Linjie Jing1
Linjie Jing1 Danxun Kong1
Danxun Kong1 Tongxin Liu1
Tongxin Liu1 Shuanglin Li1
Shuanglin Li1 Fanli Meng1*
Fanli Meng1* Qi Wang2*
Qi Wang2* Ling Zhang3*
Ling Zhang3*Salt damage is an important abiotic stress affecting the agronomic traits of soybean. Soybeans rapidly sense and transmit adverse signals when salt-damaged, inducing a set of response mechanisms to resist salt stress. AtARA6 encodes a small GTPase, which plays an important role in Arabidopsis vesicle transport and salt tolerance. In this study, we transformed the Arabidopsis gene AtARA6 into the cultivated soybean Shen Nong 9 (SN9). To investigate the salt tolerance pathways affected by AtARA6 in soybean, we performed transcriptome sequencing using transgenic soybean and wild-type (SN9) under salt treatment and water treatment. Our results suggest that AtARA6 is involved in the regulation of soybean SNARE complexes in the vesicle transport pathway, which may directly strengthen salt tolerance. In addition, we comprehensively analyzed the RNA-seq data of transgenic soybean and SN9 under different treatments and obtained 935 DEGs. GO analysis showed that these DEGs were significantly enriched in transcription factor activity, sequence-specific DNA binding, and the inositol catabolic process. Three salt-responsive negative regulator transcription factors, namely MYC2, WRKY6, and WRKY86, were found to be significantly downregulated after salt treatment in transgenic soybeans. Moreover, four genes encoding inositol oxygenase were significantly enriched in the inositol catabolic process pathway, which could improve the salt tolerance of transgenic soybeans by reducing their reactive oxygen species content. These are unique salt tolerance effects produced by transgenic soybeans. Our results provide basic insights into the function of AtARA6 in soybeans and its role in abiotic stress processes in plants.
Soybean (Glycine max [L.] Merr.) is an important oilseed crop that provides high-quality nutrients and is rich in proteins, unsaturated fatty acids, and many bioactive substances, such as isoflavones. Soybean oil is not only used as food but also as a fuel (Qi and Lee, 2014). Due to the diversity of uses, the demand for soybeans is continuously increasing. However, soybean yields are chronically compromised by abiotic stresses, such as drought and salt stress (Phang et al., 2008; Manavalan et al., 2009). Soil salinity affects 20% of the world’s arable land and severely affects soybean yield and quality (Hamwieh and Xu, 2008; Wang et al., 2010). Currently, more than 100 countries worldwide are facing serious soil salinization (Tester and Davenport, 2003; Munns and Tester, 2008). To address this problem, the search for salt-tolerant genes and the breeding of salt-tolerant soybean varieties have become a research priority for breeders.
A variety of techniques have been used to breed salt-tolerant soybean. While traditional breeding methods have limitations due to long cycle times, transgenic techniques can quickly yield soybean germplasm with good agronomic traits and genetic stability, through overexpression or downregulation of target genes. With the advent of advanced molecular biology techniques, salt tolerance genes and salt tolerance mechanisms have been widely studied. Many genes can improve salt tolerance in soybeans by coordinating ion transport. For example, overexpression of the Arabidopsis AtNHX (Na+, K+/H+ exchangers) antiporter gene, AtNHX5, enhances transgenic soybean salt resistance by increasing the concentration of Na+ in leaves and roots (Wu et al., 2016).
Drought and salt damage are the most common stressors that have a significant impact on plant growth and development, causing significant losses in crop yield (Beeson et al., 2007; Tester and Langridge, 2010; Agarwal et al., 2013). Plants have multiple ways to deal with abiotic stresses, such as vesicular transport. Substantial evidence suggests that vesicular trafficking ensures the efficient transport of stress-related ions to vacuoles. For example, the involvement of SchRabGDI1 in endocytic transport and tolerance to salt stress in Solanum chilense has been reported (Martin-Davison et al., 2017).
In eukaryotic organisms, membrane transport is a conserved cellular process that moves materials between organelles. This process occurs in three steps: vesicle budding, shuttling between organelles, and fusion. Small extracellular molecules are first screened by clathrin, then the donor cell membrane sags to form vesicles through endocytosis. The vesicles first bud from the donor membrane and then move along the cytoskeleton to the target membrane, where they undergo docking and tethering processes, among others. Finally, the fusion of vesicles and target membranes mediated by SNARE proteins complete material transport. As a house-keeping process, it is tightly regulated. Membrane transport mediated by vesicles/tubules is an important component of substance transport in plants. RAB-GTPase, a small GTPase with a molecular size of 20–25 kDa, is involved in vesicular transport and functions as a molecular switch in the cycling between GDP and GTP (Bucci and Chiariello, 2006; Schwartz et al., 2007). RAB5 belongs to the RAB family. Arabidopsis has three RAB5 homolog genes, AtARA7 (RABF2b), AtRHA1 (RABF2a), and AtARA6 (RABF1) (Ueda et al., 2001). Both AtARA7 and AtRHA1 are localized in the perivacuolar compartment (PVC), while AtARA6 is localized at the plasma membrane, in the Golgi network (TGN), and in multivesicular endosomes (MVEs), suggesting that it may be involved in intracellular endosomal transport (Hoepflinger et al., 2013). In the TGN, AtARA6 affects SNARE complexes (soluble N-ethylmaleimide-sensitive factor attachment protein receptor), which are crucial for the fusion step, by regulating the R-SNARE/VAMP727 complex on the endosome and the Q-SNARE/SYP121 complex on the plasma membrane (Ebine et al., 2011).
The importance of the SNARE pathway has been investigated under salt stress conditions (Singh et al., 2016). In Arabidopsis, the AtARA6 knockout line has a moderately salt-sensitive phenotype, while the AtARA6-overexpression line has enhanced salt tolerance (Ebine et al., 2011). SYP121 functions in drought and ABA signaling pathways in tomato (Leyman et al., 1999). Disruption of SYP121 decreases the water permeability of the cell membrane (Besserer et al., 2012). In Arabidopsis, the SYP121 mutant, AtSYP121, shows suppressed stomatal opening, indicating that the gene may be involved in the drought response (Eisenach et al., 2012). Recently, VAMP727 was found to physically interact with BRI1, a brassinosteroid receptor (Zhang et al., 2019b), thus responding to broad stress stimuli. Another RAB family member, Rab7, has been shown to negatively affect salt tolerance in Arabidopsis (Mazel et al., 2004).
AtARA6 is different from other RAB5 members because of its unique N-terminal myristoylation and palmitoylation, but it has a common activator, VPS9a (Grosshans et al., 2006). To date, AtARA6 homologues have been found in all sequenced plants, except for single-celled green algae, and have not been found in animals (Ebine et al., 2012). AtARA6 exhibits a variety of biological functions, including endosomal transport, signal transduction, stress response, and growth regulation. In this study, we heterologously expressed the AtARA6 gene in soybean and found that salt tolerance was enhanced (Supplementary Figure S1). We performed RNA-seq to uncover potential pathways affected by the AtARA6 transgene. Our study provides insights into the function of AtARA6 and lays a foundation for breeding new germplasms of transgenic salt-tolerant soybean.
Col-0 wild-type Arabidopsis was incubated in a normal growth chamber with a light/dark cycle of 16 h/8 h at 22°C/16°C. Transgenic soybean and SN9 were cultured in a greenhouse environment with a light/dark cycle of 16 h/8 h at 28°C/25°C.
Total RNA was extracted from 3 week-old wild-type Arabidopsis leaves and was reverse transcribed into cDNA using the PrimeScript RT kit (Takara, Shiga, Japan). The 609 bp CDS of Arabidopsis AtARA6 (AT3G54840) was amplified with primers (5′-GAAGAGAAGAAGCACATCCCAT-3′ 5′- ATGGGATGTGCTTCTTCTCTTC-3′) designed using Primer 5.0. The amplification products were recovered using a DNA Gel Extraction Kit (Tiangen Biotechnology Co. Ltd., Beijing, China) and sent for sequencing. The AtARA6 CDS was cloned into the pTF101 vector via Spe1 and Sac1 sites. The plasmid was transformed into Agrobacterium tumefaciens strain EHA101. Cultivated soybean SN9 was infected with the strain by the cotyledon nodulation method. Regenerated plants were obtained after exosome infiltration, clump shoot induction, clump shoot screening, and rooting culture (Paz et al., 2006; Zhang et al., 2019a). The bar gene driven by the CaMV35S promoter in the PTF101 vector was used for the positive progeny screen.
The transgenic progeny was first tested using LibertyLink test strips, according to the manufacturer’s instructions (EnviroLogix Inc., Portland, ME, United States). Genomic DNA from test strip-positive soybean leaves was extracted using a DNA extraction kit (Kangwei Century Biotechnology Co., Ltd., Beijing, China), and primers were designed according to the CDS sequences of AtARA6 and Bar genes. The soybean actin gene was used as a control (Supplementary Table S1).
The transgenic soybeans and recipient soybean seeds were sowed in vermiculite and watered with 0, 100 mM, or 200 mM NaCl solutions (Wang et al., 2018). Root length was measured after 1 week. For the salt stress treatment at emergence stages, the NaCl concentrations chosen followed the method of Li et al. (2017), with a slight modification: we first sowed the seeds in vermiculite and treated them with water for 10 days until the main leaves fully unfolded, then they were treated with 200 mM NaCl. The salt solution was changed every 3 days. Soybean phenotypes were recorded at weeks 1, 2, and 3 after treatment.
We measured representative physiological indicators of salt tolerance in transgenic lines and SN9, including superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) activities after 3 weeks of salt treatment. SOD activity was determined based on the photochemical reduction capacity of p-Nitro-Blue tetrazolium chloride, and the absorbance value was read at 560 nm (Beauchamp and Fridovich, 1971). POD activity was determined based on the amount of reaction per unit time using guaiacol as a substrate (Reuveni, 1992). During the disappearance of H2O2, the activity of CAT is detected by monitoring the change in absorbance at 240 nm (Yang et al., 2008). Malondialdehyde (MDA) reflects the level of peroxidation of plant cell membrane lipids, and we measured MDA contents to assess the degree of damage to transgenic lines and WT plants (Landi, 2017). Chlorophyll was extracted in a mixture of acetone and ethanol, and the absorbance was measured at 645, 652, and 663 nm (Arnon, 1949). The determination of proline was based on the method described by Irigoyen et al. (1992).
The transgenic positive line2 and WT, after salt stress treatment for 5 days, were named S1 and S2, respectively. The transgenic positive line2 and WT, after water treatment for 5 days, were used as controls, and named H1 and H2, respectively. Total leaf RNA was extracted using the OminiPlant RNA Kit (Dnase I) (Kangwei Century Biotechnology Co., Ltd., Beijing, China), according to the manufacturer’s instructions. Transcriptome sequencing was performed at Biomarker Biotechnology (Beijing, China). The mRNA was enriched with magnetic beads with Oligo (dT), and cDNA was synthesized using random primers. The purified double-stranded cDNA was end-repaired, A-tail was added, sequencing connectors were ligated, and the cDNA library was obtained by PCR. The RNA-seq library was constructed using Illumina novaseq 6000.
Leaves collected 5 days after salt treatment were used for qPCR validation. Total plant RNA was extracted using a Plant RNA Extraction Kit (Kangwei Century Biotechnology Co., Ltd., Beijing, China), and gDNA Remover Mix was used to remove genomic DNA contamination. An ABScript II RT Mix (ABClonal technology, Wuhan, China) was used for the reverse transcription reaction, according to the manufacturer’s instructions. To investigate the expression of AtARA6 at different treatment times and the expression of genes with which AtARA6 may have a reciprocal relationship, gene primers and GmActin (NM_001289231) primers were designed using the NCBI online website (Supplementary Table S2). The cDNA was amplified using SYBR Green Fast qRT-PCR Mix (ABClonal Technology Co. Ltd., Wuhan, China) using the chimeric fluorescence method. Each treatment had three replicates. Two internal reference primers of GmActin were used to ensure the accuracy of qPCR. qPCR experiments were performed using the LightCycler 480 II machine. Relative expression of GmActin was calculated using the 2−ΔΔCT method.
Raw data generated by sequencing were processed using FastQC and Trim_galore software (the quality control threshold was 25, reads with a length lower than 50 were discarded, the allowable error rate was 0.1, and the minimum number of bases overlapping with the adapter sequence was 3) to obtain high-quality data for mapping (reads with a ratio of N greater than 10% and reads (Q ≤ 10%) whose bases accounted for more than 50% of the entire reads were removed). The Williams 82 reference genome was constructed using HISAT2 software (version 2.1.0) (Parameter; hisat2-build -p 4 W82_ref. fa -snp W82_ref.snp -ss W82_ref. ss -exon W82_ref. exon W82_hisat2_index) (Kim et al., 2015), and unique reads were compared. Reads on pairs were assembled and quantified by DESeq2 (the test method was “Wald significance tests” and the fit type was “parametric”) using the StringTie comparison (the TPM value for the minimum allowed input transcript was 0.1 and the minimum isoform fraction was 0.1) (Love et al., 2014; Pertea et al., 2015). StringTie used FPKM (transcript per million fragments mapped) as a measure of transcript or gene expression level. Upregulated and downregulated genes between transgenic soybean and SN9 under salt treatments were determined using the edgeR package in R (Robinson et al., 2010), and all analyzed False Discovery Rate (FDR) thresholds were less than 0.05 (Fold Change ≥ 2). All raw data have been uploaded to the SRA database in NCBI (ID: PRJNA789350).
Differentially expressed genes (DEGs) generated between each treatment were analyzed for gene ontology (GO) gene function clustering and enrichment via online websites (http://www.geneontology.org and http://bioinfo.cau.edu.cn/agriGO) with an FDR threshold set at 0.05 (Huang da et al., 2009; Du et al., 2010). Gene ontology was based on the National Center for Biotechnology Information (NCBI) non-redundant protein sequences (Nr), Swiss-Prot, protein family, and clusters of orthologous groups of proteins (KOG/COG) database. KEGG [Kyoto Encyclopedia of Genes and Genomes (KEGG) and Ortholog database (KO)] pathway enrichment analysis of DEGs was performed using the (http://www.genome.jp/kegg) website (Kanehisa et al., 2021), and we screened for pathways with p values less than 0.05.
All data were analyzed using two-way Analysis of Variance to evaluate significant differences at *p < 0.05, **p < 0.01, and ***p < 0.001. All statistical analyses were carried out IBM SPSS Statistics 22 (IBM Corp., Armonk, NY, United States). All DEGs and pathways used for GO analysis and KEGG analysis can be found in Supplementary Table S3 and Supplementary File S1.
Cultivated soybean, strain SN9, was transformed by agrobacterium harboring AtARA6, and a total of six transformation lines, named Line1, Line2, Line3, Line4, Line5, and Line6, were obtained in the T0 generation (Figure 1). PCR was used to detect the presence of AtARA6 and Bar genes separately. Five positive transgenic lines were identified, consistent with the results of the LibertyLink test strips (Figures 2A–C). The positive lines were propagated to T3 to ensure stable transgene inheritance. qRT-PCR results demonstrated that AtARA6 showed the highest expression level at 5 days after salt treatment (Figures 2D,E).
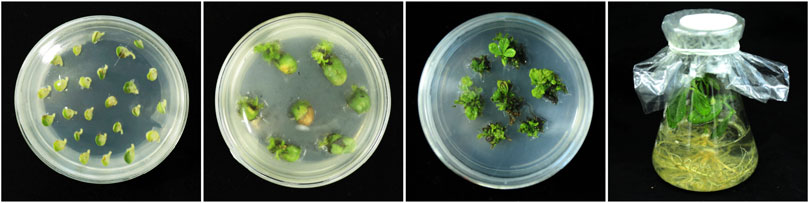
FIGURE 1. Genetic transformation of AtARA6. Agrobacterium tumefaciens EHA101-mediated genetic transformation of soybean, including exosome infiltration, clump shoot induction, clump shoot screening, and rooting culture.
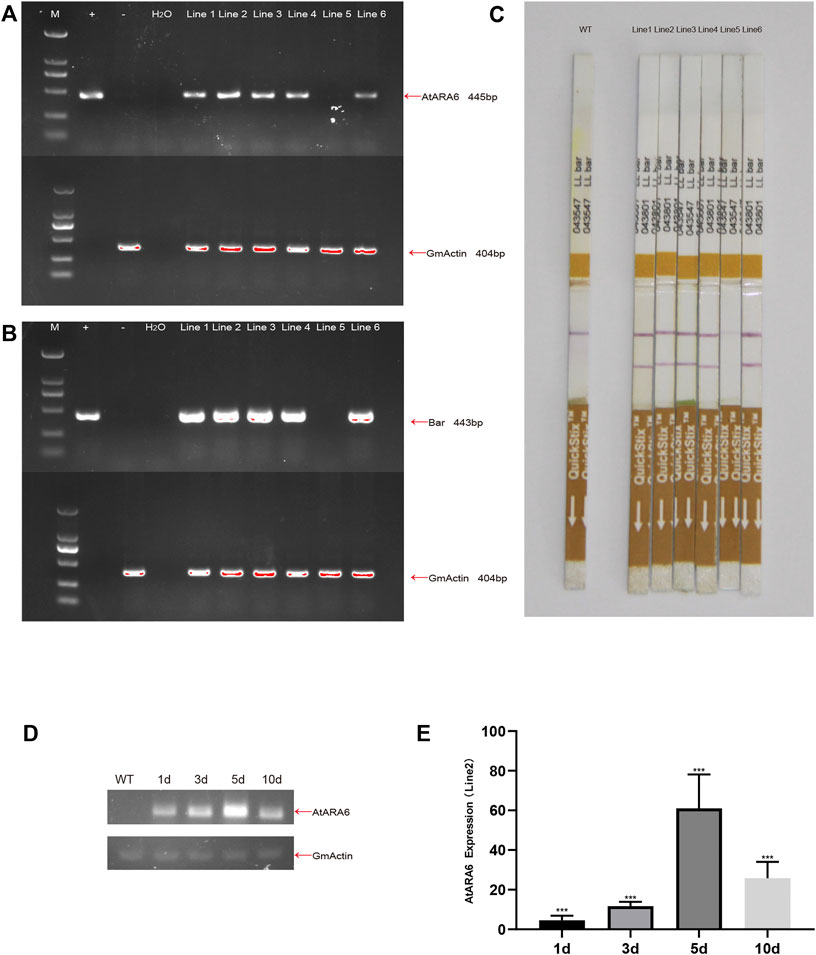
FIGURE 2. Confirmation of AtARA6 transgenesis. (A,B) Positive transgenic soybean progeny Lines1-6: 404 bp of GmActin was used to detect plant genomic DNA, AtARA6 445 bp, Bar 443=bp (M, Trans2K Plus II DNA marker; +, AtARA6 expression vector plasmid DNA; −, WT genomic DNA). (C) LibertyLink test strips for genetically transformed soybeans. (D) RT-PCR analysis of AtARA6 gene expression at different times under 200 mM NaCl treatment. (E) qRT-PCR analysis of the relative expression of AtARA6 in different durations under salt treatment.
We assessed the salt tolerance of transgenic soybean and WT plants in the germination and emergence stages separately. Three positive T3 generation transgenic lines and WT were sown in vermiculite treated with different concentrations of salt solution. The length of the roots was compared after 7 days of treatment. As shown in Figures 3A,B, there was no significant difference in root length between transgenic soybean and WT plants in the water treatment group. However, transgenic soybean roots were longer than WT in both 100 mM and 200 mM NaCl solutions (Supplementary Table S4), suggesting that transgenic soybean has stronger germination ability and adaptability under salt stress.
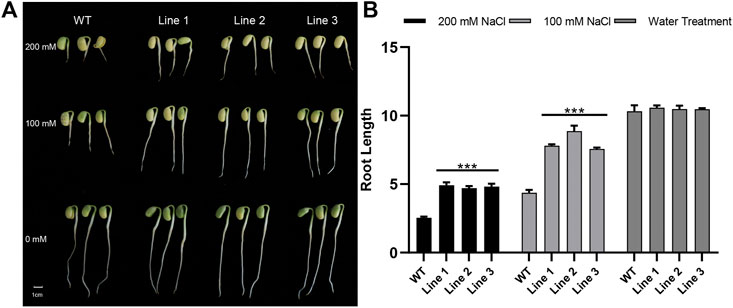
FIGURE 3. Salt treatment phenotypes of transgenic soybean and WT during germination. (A) Phenotypes of transgenic soybean Lines1-3 and WT treated with 200 mM NaCl, 100 mM NaCl, or water for 7 days at the germination stage. (B) Root length phenotypes of WT and transgenic soybean under salt stress during germination (p < 0.05).
We then treated transgenic soybean with unfolded opposite true leaves in 200 mM NaCl solution WT leaves started to turn yellow and dry at 1 week, and the true leaves wilted and fell off. There were no significant changes in the transgenic lines Line1 and Line3, although the true leaves of Line2 turned slightly yellow (Figure 4A). After 2 weeks of salt treatment, most of the lower leaves of WT plants lost their green color, appeared dehydrated, the petioles withered and fell off, the upper leaves turned yellow and appeared dehydrated, and only a few of the apical ternate leaves were unaffected. In contrast, only 1-2 true leaves of transgenic soybean Line1 and Line2 turned yellow, and the upper part of the plants stayed upright and grew normally (Figure 4B). After 3 weeks, the entire WT plant was withered, the growth point was necrotic, and the leaves were severely shed. The true leaves of Line1 and Line2 transgenic soybean did not fall off, and Line3 only had true leaves that turned yellow. The degree of salt damage in transgenic soybean was thus significantly lower than that in WT (Figure 4C). These results showed that the transformation using the AtARA6 gene significantly improved the salt tolerance pathway of SN9 and enhanced soybean resistance to salt stress.

FIGURE 4. Salt treatment phenotypes of transgenic soybean and WT at emergence stages. (A–C) Phenotypes of transgenic soybean Lines1-3 and WT (left) treated with 200 mM NaCl at emergence stages for 1, 2, and 3 weeks.
To assess the salt tolerance level of transgenic soybean, we measured the SOD, POD, and CAT activities of transgenic soybean and WT plants under salt stress. All these activities in both transgenic soybean and WT increased in the 200 mM NaCl treatment group. However, the increase in these activities in transgenic soybean was significantly greater than that in WT, suggesting that transgenic soybean has a stronger tolerance capacity under salt stress (Figures 5A–C). Similarly, the increased proline content in the WT was much lower than that in the transgenic line (Figure 5D). MDA content reflects the degree of damage to plant membrane lipids. Both transgenic soybean and WT MDA contents increased under salt stress, but the transgenic line increased less than SN9 (Figure 5E). The chlorophyll content of the soybeans decreased under salt treatment, but that of the transgenic line was over 2-fold higher than that of the WT plants (Figure 5F). Under stress, the plant’s defense system responds by generating protective enzymes such as SOD, POD, and CAT, which can reduce ROS levels in cells. Proline, an osmoregulatory substance, can regulate the osmotic potential of plant cells. Protective enzyme activity and proline content increased in both transgenic soybean and WT under salt stress. However, the elevation was much higher in transgenic soybean than in WT. In addition, the transgenic soybean plasma membrane was less damaged than the WT, and chlorophyll levels were higher than in WT. Such differences suggest that AtARA6 can aid soybean resistance to salt stress and reduce the degree of damage to the organism under such conditions.
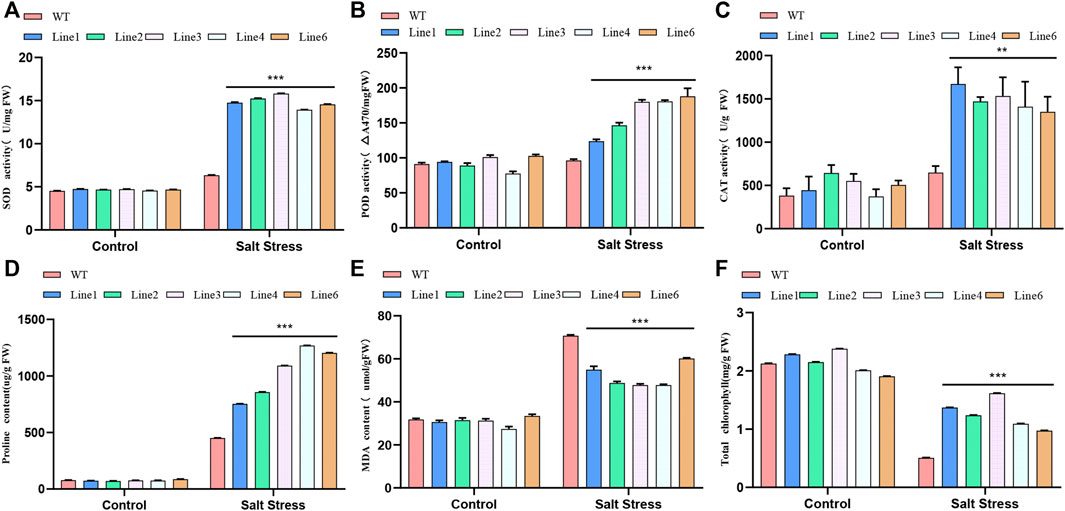
FIGURE 5. Determination of physiological parameters of transgenic soybean and WT treated with salt stress. (A) Superoxide dismutase (SOD) activity. (B) Peroxidase (POD) activity. (C) Catalase (CAT) activity. (D) Proline content in leaves (µg/g). (E) Malondialdehyde content in leaves (µmol/g). (F) Chlorophyll content of leaves after salt treatment (mg/g). All values were measured after 21 days of salt treatment. Statistical analysis was performed using one-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001, p > 0.05 indicates the difference between transgenic line and WT was not significant.
To investigate the potential pathways by which AtARA6 affects salt tolerance, we performed RNA-seq analysis using Line2 and WT in the true leaf stage under salt stress for 5 days, named S1 and S2, respectively. The same plant materials were mock-treated with water, named H1 and H2. The clean data obtained from each of these samples was over 5.84 Gb, and the percentage of Q30 bases was above 92.13%. The clean data of each sample were compared with the Williams 82 reference genome separately, and the comparison efficiency ranged from 89.56% to 94.08% (Table 1). Between H1 and S1, we detected 3,916 DEGs, including 2,399 downregulated DEGs and 1,517 upregulated DEGs in transgenic soybean under salt treatment. In the H2 versus S2 group, we detected 5,027 DEGs, including 1,729 downregulated DEGs and 3,298 upregulated DEGs in SN9 under salt treatment (Figure 6). Furthermore, we compared the DEGs in the H2-S2 and H1-S1 groups, which shared only 246 upregulated DEGs (∼5.4%) and 310 downregulated DEGs (∼8.1%) (Figure 6). These results indicate that overexpression of the AtARA6 gene in soybean resulted in significant alterations in gene transcription in SN9, which may be the reason for improved salt tolerance.
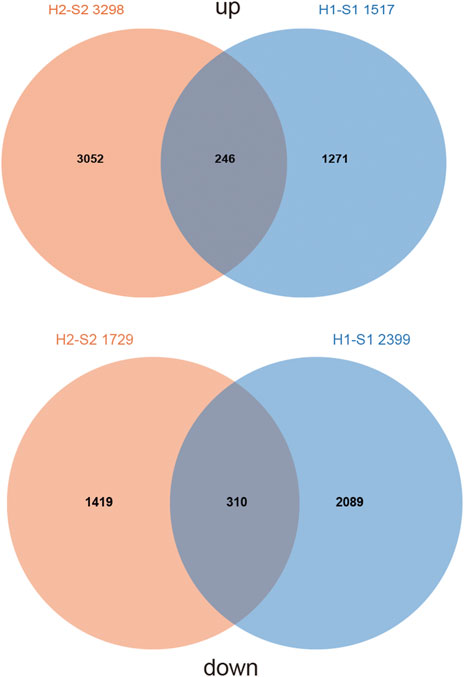
FIGURE 6. Comparison of DEGs between transgenic positive lines and SN9 under salt treatment and water treatment.
In the vesicular transport pathway, SNARE complexes are responsible for the transport of substances from endosomes to the plasma membrane. AtARA6 regulates the formation of SNARE complexes containing VAMP727 and SYP121 (Ebine et al., 2011). In S1 and S2 DEGs, we found two homologs of SYP121 in soybean (Glyma.02G069700 and Glyma.16G151200). The KEGG results showed that they were significantly enriched in the SNARE pathway (Supplementary Table S5). VAMP727 encodes a vesicle-associated membrane protein 727 on the target membrane and plays a role in the fusion of vesicles with the target membrane. We found two homologous genes of VAMP727 in soybean (Glyma.01G179300 and Glyma.11G062900), which were significantly upregulated in transgenic soybean after salt treatment. KEGG enrichment results showed that these two genes are in the endocytosis pathway (Supplementary Table S5). We verified 4 upregulated genes using qRT-PCR (Figure 7). Our results show that AtARA6 upregulates the SNARE genes SYP121 and VAMP727.
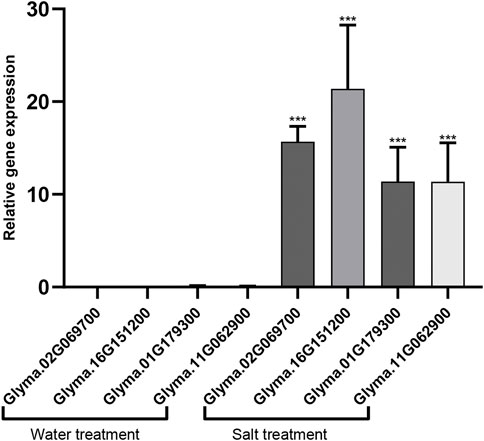
FIGURE 7. qRT_PCR analysis of homologous genes of SYP121 and VAMP727. The homologous genes of SYP121, Glyma.02G069700 and Glyma.16G151200. The homologous genes of VAMP727, 01G179300 and Glyma.11G062900. They were significantly up-regulated under in transgenic soybean after salt treatment. Three biological replicates were used. Data was calculated using the 2−ΔΔCT method, Statistical analysis was performed using ANOVA (p < 0.05), *p < 0.05, **p < 0.01, and ***p < 0.001.
To investigate the functions of DEGs, we performed GO analysis of DEGs in transgenic soybean and WT plants under salt and water treatments. We focused on 1,271 upregulated DEGs and 2,089 downregulated genes in the transgenic line under salt treatment (Figure 6). The expression of some of these genes may be perturbed by the AtARA6 transgene. For 1,271 upregulated DEGs, significant GO terms in RNA-related processes were enriched, including RNA metabolic processes (GO:0016070, p = 1.10E-05), RNA biosynthetic processes (GO:0032774, p = 4.00E-05), regulation of transcription (GO:0006355, p = 1.10E-05), and regulation of gene expression (GO:0010468, p = 1.60E-05) (Table 2). We hypothesize that such upregulated RNA-related pathways may help to explain the stronger salt resistance of the transgenic line. It is well-known that plants use a wide array of mechanisms, including transcriptional regulation and posttranscriptional mechanisms, to survive under diverse stress conditions. At the post-transcriptional level, many studies have demonstrated that Ca2+-dependent transcriptional reprogramming is important for the stress response (Galon et al., 2008; Yuan et al., 2018a; Yuan et al., 2018b). In addition, RNA modification has been shown to be related to stress responses, such as splicing, capping, polyadenylation, and translocation. Overexpression of the Arabidopsis RNA binding protein AtRGGA enhances salt tolerance (Ambrosone et al., 2015). Similarly, overexpression of Beta vulgaris RNA binding protein, salt tolerant 3 (BvSATO3), in Arabidopsis increased plant salt resistance (Rosa Téllez et al., 2020).
For 2,089 downregulated DEGs in the transgenic line under salt treatment, significant GO terms in metabolism-related processes, (GO:0008152, p = 1.50E-27), primary metabolic processes (GO:0044238, p = 0.0037), cellular metabolism (GO:0044237, p = 2.50E-08), macromolecule metabolism (GO:0043170, p = 0.019), protein metabolism (GO:0019538, p = 0.0019), carbohydrate metabolism (GO:0044262, p = 0.00077), and glucose metabolism (GO:0006006, p = 1.60E-05) (Table 3). We believe that the reduction of the expression of certain metabolism-related genes may indirectly reduce the pressure to maintain cellular homeostasis, which is perturbed by salt stress. A similar result was found for gene expression changes in the sugar beet under salt stress (Rosa Téllez et al., 2020).
In addition, we analyzed the direct mechanism via which AtARA6 improves salt tolerance in soybean. Between the S1 and S2 groups, we identified 6,572 DEGs that represent unique processes carried out by transgenic soybean under salt treatment. Among the GO terms, Rho guanyl-nucleotide exchange factor activity (GO:0005089, p = 0.0045), Ras guanyl-nucleotide exchange factor activity (GO:0005088, p = 0.0045), GTPase regulator activity (GO:0030695, p = 0.0093), and GTPase activity (GO:0003924, p = 0.044) pathways were significantly enriched in the molecular function module in upregulated DEGs in transgenic soybean under salt treatment (Supplementary Figure S2). Rho-like GTPases are plant-specific molecular switches that play a critical role in plant survival under abiotic stresses (Miao et al., 2018). The expression levels of DEGs enriched in these pathways under different treatments were found to be higher in transgenic soybean than in WT (Figure 8). Therefore, we suggest that the upregulation of these four pathways by the AtARA6 gene acted as a direct factor to improve the salt tolerance of transgenic soybean.
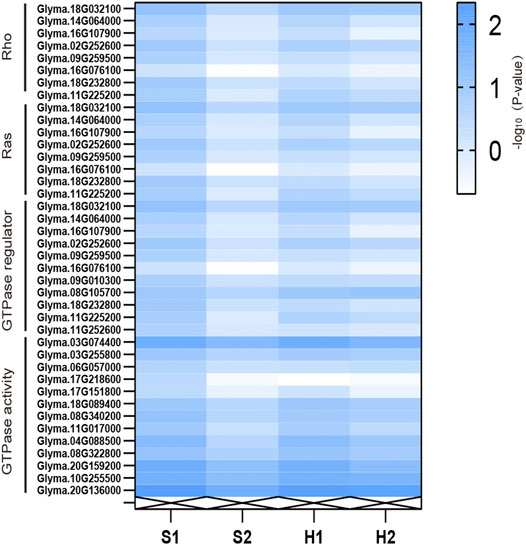
FIGURE 8. RNA-seq reveals the expression levels of DEGs enriched in the Rho/Ras/GTPase regulator/GTPase activity pathways under different treatments.
After the GO analysis of DEGs between different treatments, we concluded that the transgenic soybean showed significant enrichment in terms of transcription level and substance metabolism. In order to explore the changes caused by the transformation of AtARA6 in soybean, we combined S1, S2 and H1, H2 for analysis. We first found 3,815 DEGs between H1 vs. S1 and 4,933 DEGs between H2 vs. S2. Among these DEGs, 1,344 DEGs were shared and could be considered as changes in soybean caused by salt stress. At the same time, we identified 6,456 DEGs between S1 vs. S2 as a result of transfection of the AtARA6 gene. We found 935 DEGs between 1,344 and 6,456 DEGs, which represented the unique effects of AtARA6 gene transformation under salt stress (Supplementary Figure S3). GO analysis of 935 DEGs showed that they had the highest levels of transcription factor activity, sequence-specific DNA binding (GO:0003700, p = 5.2e-11, MF), and inositol catabolic process (GO:0019310, p = 4.0e-10, BP), and the difference was significant (Figure 9). Furthermore, we identified multiple salt stress-responsive transcription factors. MYC2 is a negative regulator of proline synthesis and improves salt tolerance by regulating the proline content in Arabidopsis (Verma et al., 2020). In addition, WRKY6 and WRKY86 have been reported to negatively regulate plant salt tolerance (Li et al., 2019; Fang et al., 2021). In our RNA-seq results, the expression levels of MYC2, WRKY6, and WRKY86 were all downregulated (Supplementary Table S5), indicating that they are involved in the salt tolerance pathway of transgenic soybean. Moreover, the phospholipase signaling pathway is an important pathway for plant salt tolerance. The N-terminus of phospholipase D contains a domain that binds to phosphoinositide. At the same time, PLDα can bind to the heterotrimeric protein subunit to regulate GTPase activity and control stomatal movement and loss of water (Zhao and Wang, 2004). GO results showed that transgenic soybeans were significantly enriched in the inositol catabolic process and inositol oxygenase activity (GO:0050113, p = 2.7e-9) pathways. Four DEGs were found to be enriched in this pathway (Glyma.08G199300, Glyma.07G126600, Glyma.05G224500, and Glyma.07G013900), all of which functioned to encode inositol oxygenase. In rice, inositol oxygenase has been reported to scavenge reactive oxygen species (Duan et al., 2012). In our study, these four genes were strongly upregulated after transgenic soybean salt treatment, indicating that transgenic soybeans could regulate the inositol catabolic process to improve salt tolerance.
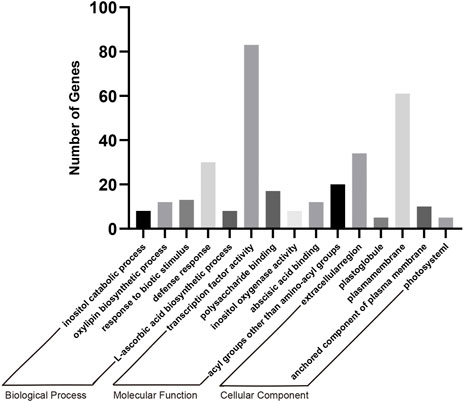
FIGURE 9. GO enrichment analysis of 935 DEGs including BP, CC and MF. p-values < 0.01 for all GOs. Transcription factor activity, sequence-specific DNA binding (GO:0003700, p = 5.2e-11, MF) and inositol catabolic process (GO:0019310, p = 4.0e-10, BP) are the most significant.
To further investigate the mechanisms for the increased salt tolerance of transgenic soybean, we performed KEGG analysis on up and downregulated DEGs under salt stress in transgenic soybean compared to WT (Figure 10A). Among the upregulated DEGs, there was enrichment in DNA replication (ko03030, p = 6.04e-80), mismatch repair (ko03430, p = 7.22e-25), pyrimidine metabolism (ko00240, p = 5.26e-43), homologous recombination (ko03440, p = 8.41e-32), base excision repair (ko03410, p = 1.17e-26), nucleotide excision repair (ko03420, p = 7.24e-32), and starch and sucrose metabolism (ko00500, p = 1.68e-98) (Supplementary Table S7). Previous studies have shown that salt stress disrupts genome stability in plants (Roy et al., 2013). Exposure to high salinity increases the risk of DNA double-strand breaks (Roy et al., 2013). Our transgenic soybean showed upregulated genes related to DNA replication and mismatch repair pathways, possibly partially underlying its stronger ability to cope with salt stress.
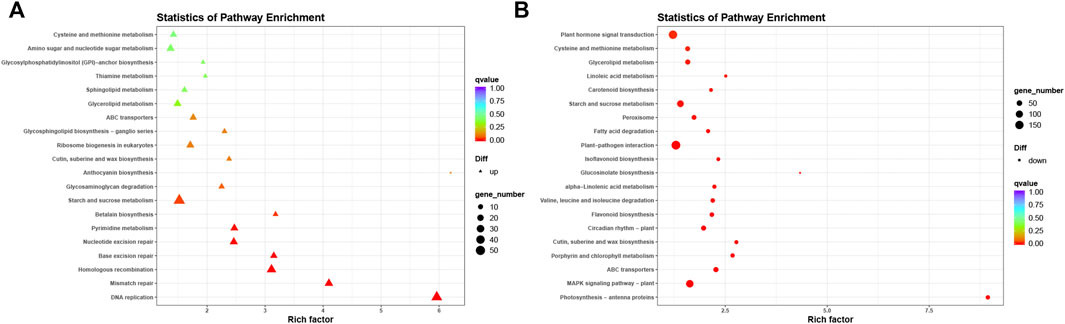
FIGURE 10. Kyoto Encyclopedia of Genes and Genomes analysis of up-regulation and down-regulation of DEGs. (A,B) KEGG analysis on up-regulated (left) and down-regulated (right) DEGs under salt stress in transgenic soybean compared to WT.
In the downregulated DEGs, significantly enriched pathways included photosynthesis-antenna proteins (ko00196, p = 4.53e-51), porphyrin and chlorophyll metabolism (ko00860, p = 2.64e-41), cutin suberine and wax biosynthesis (ko00073, p = 1.10e-48), flavonoid biosynthesis (ko00941, p = 2.80e-43), cysteine and methionine metabolism (ko00270, p = 8.57e-88), and linoleic acid metabolism (ko00591, p = 1.84e-37; Figure 10B) (Supplementary Table S8). Many downregulated pathways were concentrated in biosynthesis and metabolism processes, which were in line with our hypothesis based on GO analysis that reduction of certain metabolism-related gene expression may indirectly reduce the pressure to maintain cellular homeostasis, which is perturbed by salt stress. The downregulated pathways in our study were consistent with those of previous studies. For example, cysteine and methionine levels decreased significantly in broccoli after salt treatment (Lopez-Berenguer et al., 2008). In peanuts, linoleic acid synthesis genes were also downregulated after salt treatment (Sui et al., 2018).
Salt stress is an important factor that affects soybean yield and growth. Transgene breeding has proven to be a promising method for obtaining salt-tolerant cultivars. For example, soybean GmNHX1 can accelerate the transport of Na+ from the cytoplasm to the vesicles. Salt tolerance was improved in both soybean and Arabidopsis by overexpression of GmNHX1 (Sun et al., 2019). In rice, overexpression of OsRab7 increases endocytosis and leads to increased salt tolerance (Peng et al., 2014).
In this study, an Arabidopsis key vesicle trafficking gene, AtARA6, was transformed into soybean and stable expression lines were confirmed by PCR and qRT-PCR. Some stress-related indicators, such as protective enzymes, proline, and MDA, were found to be upregulated, perhaps underlying the ability of transgenic soybean to withstand adversity. Our results showed that the transgenic soybean grew normally at the germination and emergence stages under salt treatment, compared to WT, which showed growth stagnation and eventual death.
AtARA6 belongs to the RAB5 family. Its N-terminal myristoylation and palmitoylation sites distinguish it from other RAB5 members. The relationship between these sites and salt tolerance has been demonstrated in both AtARA6 and other genes. For example, overexpression of AtARA6 in Arabidopsis under salt treatment yields longer roots compared to WT, indicating stronger salt resistance. However, overexpression of AtARA6 with a 29 amino acid truncation at the N-terminus shows a similar root length to WT, indicating that the N-terminus is important for salt resistance (Yin et al., 2017). Similarly, the Arabidopsis AtSOS3 gene plays a role in the salt response. The rescue of the salt-hypersensitive phenotype of AtSOS3-1 knockout plants required the full-length AtSOS3 gene, but the AtSOS3 gene with an N-terminal mutant did not rescue the phenotype (Ishitani et al., 2000).
In Arabidopsis, SNARE is a pathway in which AtARA6 is directly involved, and AtARA6 has been shown to enhance salt tolerance in Arabidopsis through vesicular transport. In this study, we focused on SYP121 and VAMP727 genes. qRT-PCR results showed that their expression was upregulated under salt treatment, more in transgenic plants than in WT plants, suggesting that the transformation of the AtARA6 gene accelerated vesicle movement in soybean and that the improvement in salt tolerance in soybean is due to this, as it is in Arabidopsis. Many functions of AtARA6 in Arabidopsis have been reported: for example, the AtARA6 overexpression line is more tolerant to dark-induced senescence (DIS) (Yin et al., 2017). Recent studies have shown that AtARA6 can affect the expression of the starch gene QQS in Arabidopsis, thus participating in the dynamic balance between starch and soluble sugars in Arabidopsis. A comparison of wild-type and AtARA6 mutants revealed that AtARA6 inhibited the proliferation of pathogenic bacteria (Tsutsui et al., 2015). We explored the function of AtARA6 in transgenic soybeans. Here, we performed RNA-seq on transgenic soybean and SN9 under different treatments using leaves but not roots. Although leaves are not the first organ to respond to salt stress, considering the function of AtARA6, it has been reported that Qc-SNAREs transfer excess Na+ to vacuoles, thus increasing salt tolerance in Arabidopsis (Liu et al., 2007). As the vacuoles in the primary roots were immature, leaves became the first choice. In addition, plants can also regulate body water through the opening and closing of stomata. SYP121 and VAMP71 have been reported to be related to stomata closure (Grefen et al., 2015; Salinas-Cornejo et al., 2019). Leaves contain a larger number of stomata compared to roots; hence, they may be an important factor affecting the salt tolerance of soybeans. We identified significant enrichment of RNA-related processes and metabolic processes such as protein metabolism (GO:0019538), suggesting that transgenic plants can adapt to high salt stress through transcriptional regulation and metabolic regulation. At the same time, we comprehensively analyzed the DEGs of transgenic soybean and SN9 under different treatments and found that inositol catabolic process and transcription factor activity and sequence-specific DNA binding were significantly enriched in transgenic soybean under stress conditions, indicating that the two pathways are used for salt tolerance improvement. We have summarized the salt tolerance pathway of the AtARA6 transgenic soybean in a map (Figure 11). Reactive oxygen species scavenging mechanisms are one of the important pathways for plants to cope with abiotic stress. In our study, four genes encoding inositol oxygenases were found to be significantly upregulated in transgenic soybeans after salt treatment, which was consistent with our physiological indicator measurements. In addition, we identified multiple transcription factors that respond to salt stress, which can serve as candidate genes for studying salt tolerance in transgenic soybeans. Under drought and salt stress, WRKY6 negatively regulates the ability of Arabidopsis and cotton to tolerate abiotic stress; WRKY86 can bind to maize W-box, thereby increasing maize catalase activity, reducing malondialdehyde content, and improving salt tolerance. Moreover, the transcription factor MYC2 of the bHLH family enhances salt tolerance in Arabidopsis by regulating proline content. They provided the basis for the changes in physiological indexes of transgenic soybean under salt stress. These findings suggest that transgenic soybeans have salt tolerance pathways other than the SNARE pathway, in contrast to Arabidopsis.
Meanwhile, terms like Rho guanyl-nucleotide exchange factor activity, Ras guanyl-nucleotide exchange factor activity, GTPase regulator activity, and GTPase activity were found to be significantly enriched. To demonstrate that the enrichment of these pathways is not caused by AtARA6 homologous genes in soybean, we investigated the expression of five AtARA6 homologous genes. It was found that the expression levels of these five genes were not significantly different between SN9 and transgenic soybean under salt treatments, indicating that the upregulation of these pathways is caused by the AtARA6 gene itself.
Sometimes, plants need to cope with diverse stress threats simultaneously. On one hand, combined stress conditions require a trade-off between stress adaptation and growth. Starch and sucrose metabolism was one of the most significant upregulated terms in our KEGG analysis. Starch has been proved to be a key player in response to water deficit, high salinity, and extreme temperatures. Under these adverse environmental conditions, chloroplasts and photosynthesis may be compromised, as shown in our study. Starch metabolism and remobilization provide extra energy and carbon for the plant to survive. The correlation between starch metabolism and diverse stress resistance has been widely proved. For example, in the model plant, Arabidopsis, salt stress induces rapid carbohydrate metabolism and increases soluble sugars (Kempa et al., 2008). Similarly, in sugar beet roots, upregulated starch and sucrose metabolism was found by RNA-seq of the plants with higher salt resistance levels (Liu et al., 2020).
Previous studies have shown that salt stress affects genome stability and induces DNA double-strand breaks in plants (Roy et al., 2013). The notable enriched upregulated KEGG terms in our transgenic soybean, mismatch repair and DNA replication, may be associated with salt tolerance by maintaining genome stability. Although the connection is not fully understood, some evidence has linked genome stability and stress resistance. For example, the eukaryotic pre-replicative complex (PreRC), including heterohexameric minichromosome maintenance proteins (MCM), is responsible for making sure the DNA is replicated only once per cell division cycle. Overexpression of a MCM family member, MCM6, in tobacco confers higher salinity tolerance (Tuteja et al., 2011). Moreover, topoisomerase is the specific enzyme that can remove or add DNA supercoils and untangle snarled DNA. Enhanced salt tolerance was found in transgenic tobacco overexpressing NtTopoIIα-1, a topoisomerase gene (John et al., 2016).
On the other hand, prioritization of different stress adaptations is also critical. In Arabidopsis, salt treatment induces decreased resistance to pathogens such as Pseudomonas syringae, Alternaria brassicicola, and Botrytis cinerea (Haller et al., 2020). Similarly, Matthias et al. found that ABA, a stress plant hormone, can repress immunity against a bacterial strain, Pto hrcC−, in old leaves (Berens et al., 2019). In cotton, overexpression of the GhMKK1 gene enhances salt tolerance, as well as the susceptibility to the pathogen Ralstonia solanacearum (Lu et al., 2013). In our study, we found overexpression of an Arabidopsis gene, AtARA6, enhances soybean salt resistance. Through KEGG analysis, we found that downregulated genes were significantly enriched in plant-pathogen pathways. Our data suggest that heterologous expression of the AtARA6 gene in soybean may prioritize the salt tolerance response over the pathogen response.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA789350.
FM and ZH designed the experiment and wrote the manuscript; ZH, YL and YZ carried out experiments; MY, YT, LJ and DK carried out the salt treatment experiment; TL and SL carried out the germination experiment; FM and ZH carried out the statistical analysis.
This study was financially supported by the Basic Research Funds of JAAS (KYJF2021JQ001), the Chinese National Natural Science Foundation (32172032), China National Novel Transgenic Organisms Breeding Project (2016ZX08004-004-006) and the national project (CARS-04-PS04), the Major Project of New Varieties of Genetically Modified Organism of China (2016ZX08004-002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.849357/full#supplementary-material
Agarwal, P. K., Shukla, P. S., Gupta, K., and Jha, B. (2013). Bioengineering for Salinity Tolerance in Plants: State of the Art. Mol. Biotechnol. 54 (1), 102–123. doi:10.1007/s12033-012-9538-3
Ambrosone, A., Batelli, G., Nurcato, R., Aurilia, V., Punzo, P., Bangarusamy, D. K., et al. (2015). The Arabidopsis RNA-Binding Protein AtRGGA Regulates Tolerance to Salt and Drought Stress. Plant Physiol. 168 (1), 292–306. doi:10.1104/pp.114.255802
Arnon, D. I. (1949). Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 24 (1), 1–15. doi:10.1104/pp.24.1.1
Beauchamp, C., and Fridovich, I. (1971). Superoxide Dismutase: Improved Assays and an Assay Applicable to Acrylamide Gels. Anal. Biochem. 44 (1), 276–287. doi:10.1016/0003-2697(71)90370-8
Beeson, T. D., Mastracchio, A., Hong, J.-B., Ashton, K., and MacMillan, D. W. C. (2007). Enantioselective Organocatalysis Using SOMO Activation. Science 316 (5824), 582–585. doi:10.1126/science.1142696
Berens, M. L., Wolinska, K. W., Spaepen, S., Ziegler, J., Nobori, T., Nair, A., et al. (2019). Balancing Trade-Offs between Biotic and Abiotic Stress Responses through Leaf Age-dependent Variation in Stress Hormone Cross-Talk. Proc. Natl. Acad. Sci. U S A. 116 (6), 2364–2373. doi:10.1073/pnas.1817233116
Besserer, A., Burnotte, E., Bienert, G. P., Chevalier, A. S., Errachid, A., Grefen, C., et al. (2012). Selective Regulation of maize Plasma Membrane Aquaporin Trafficking and Activity by the SNARE SYP121. Plant Cell 24 (8), 3463–3481. doi:10.1105/tpc.112.101758
Bucci, C., and Chiariello, M. (2006). Signal Transduction gRABs Attention. Cell Signal 18 (1), 1–8. doi:10.1016/j.cellsig.2005.07.001
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: a GO Analysis Toolkit for the Agricultural Community. Nucleic Acids Res. 38 (Web Server issue), W64–W70. doi:10.1093/nar/gkq310
Duan, J., Zhang, M., Zhang, H., Xiong, H., Liu, P., Ali, J., et al. (2012). OsMIOX, a Myo-Inositol Oxygenase Gene, Improves Drought Tolerance through Scavenging of Reactive Oxygen Species in rice (Oryza Sativa L.). Plant Sci. 196, 143–151. doi:10.1016/j.plantsci.2012.08.003
Ebine, K., Fujimoto, M., Okatani, Y., Nishiyama, T., Goh, T., Ito, E., et al. (2011). A Membrane Trafficking Pathway Regulated by the Plant-specific RAB GTPase ARA6. Nat. Cel Biol. 13 (7), 853–859. doi:10.1038/ncb2270
Ebine, K., Miyakawa, N., Fujimoto, M., Uemura, T., Nakano, A., and Ueda, T. (2012). Endosomal Trafficking Pathway Regulated by ARA6, a RAB5 GTPase Unique to Plants. Small GTPases 3 (1), 23–27. doi:10.4161/sgtp.18299
Eisenach, C., Chen, Z. H., Grefen, C., and Blatt, M. R. (2012). The Trafficking Protein SYP121 of Arabidopsis Connects Programmed Stomatal Closure and K(+) Channel Activity with Vegetative Growth. Plant J. 69 (2), 241–251. doi:10.1111/j.1365-313X.2011.04786.x
Fang, X., Li, W., Yuan, H., Chen, H., Bo, C., Ma, Q., et al. (2021). Mutation of ZmWRKY86 Confers Enhanced Salt Stress Tolerance in maize. Plant Physiol. Biochem. 167, 840–850. doi:10.1016/j.plaphy.2021.09.010
Galon, Y., Nave, R., Boyce, J. M., Nachmias, D., Knight, M. R., and Fromm, H. (2008). Calmodulin-binding Transcription Activator (CAMTA) 3 Mediates Biotic Defense Responses in Arabidopsis. FEBS Lett. 582 (6), 943–948. doi:10.1016/j.febslet.2008.02.037
Grefen, C., Karnik, R., Larson, E., Lefoulon, C., Wang, Y., Waghmare, S., et al. (2015). A Vesicle-Trafficking Protein Commandeers Kv Channel Voltage Sensors for Voltage-dependent Secretion. Nat. Plants 1, 15108. doi:10.1038/nplants.2015.108
Grosshans, B. L., Ortiz, D., and Novick, P. (2006). Rabs and Their Effectors: Achieving Specificity in Membrane Traffic. Proc. Natl. Acad. Sci. U. S. A. 103 (32), 11821–11827. doi:10.1073/pnas.0601617103
Haller, E., Iven, T., Feussner, I., Stahl, M., Frohlich, K., Loffelhardt, B., et al. (2020). ABA-Dependent Salt Stress Tolerance Attenuates Botrytis Immunity in Arabidopsis. Front. Plant Sci. 11, 594827. doi:10.3389/fpls.2020.594827
Hamwieh, A., and Xu, D. (2008). Conserved Salt Tolerance Quantitative Trait Locus (QTL) in Wild and Cultivated Soybeans. Breed. Sci. 58 (4), 355–359. doi:10.1270/jsbbs.58.355
Hoepflinger, M. C., Geretschlaeger, A., Sommer, A., Hoeftberger, M., Nishiyama, T., Sakayama, H., et al. (2013). Molecular and Biochemical Analysis of the First ARA6 Homologue, a RAB5 GTPase, from green Algae. J. Exp. Bot. 64 (18), 5553–5568. doi:10.1093/jxb/ert322
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4 (1), 44–57. doi:10.1038/nprot.2008.211
Irigoyen, J. J., Einerich, D. W., and Sánchez-Díaz, M. (1992). Water Stress Induced Changes in Concentrations of Proline and Total Soluble Sugars in Nodulated Alfalfa (Medicago Sativd) Plants. Physiol. Plantarum 84, 55–60. doi:10.1111/j.1399-3054.1992.tb08764.x
Ishitani, M., Liu, J., Halfter, U., Kim, C. S., Shi, W., and Zhu, J. K. (2000). SOS3 Function in Plant Salt Tolerance Requires N-Myristoylation and Calcium Binding. Plant cell 12 (9), 1667–1678. doi:10.1105/tpc.12.9.166710.2307/3871181
John, R., Ganeshan, U., Singh, B. N., Kaul, T., Reddy, M. K., Sopory, S. K., et al. (2016). Over-expression of Topoisomerase II Enhances Salt Stress Tolerance in Tobacco. Front. Plant Sci. 7, 1280. doi:10.3389/fpls.2016.01280
Kanehisa, M., Sato, Y., and Kawashima, M. (2021). KEGG Mapping Tools for Uncovering Hidden Features in Biological Data. Protein Sci. 31 (1), 47–53. doi:10.1002/pro.4172
Kempa, S., Krasensky, J., Dal Santo, S., Kopka, J., and Jonak, C. (2008). A central Role of Abscisic Acid in Stress-Regulated Carbohydrate Metabolism. PLoS One 3 (12), e3935. doi:10.1371/journal.pone.0003935
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Landi, M. (2017). Commentary to: "Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds" by Hodges et al., Planta (1999) 207:604-611. Planta 245 (6), 1067. doi:10.1007/s00425-017-2699-3
Leyman, B., Geelen, D., Quintero, F. J., and Blatt, M. R. (1999). A Tobacco Syntaxin with a Role in Hormonal Control of Guard Cell Ion Channels. Science (New York, N.Y.) 283 (5401), 537–540. doi:10.1126/science.283.5401.537
Li, Y., Chen, Q., Nan, H., Li, X., Lu, S., Zhao, X., et al. (2017). Overexpression of GmFDL19 Enhances Tolerance to Drought and Salt Stresses in Soybean. PLoS One 12 (6), e0179554. doi:10.1371/journal.pone.0179554
Li, Z., Li, L., Zhou, K., Zhang, Y., Han, X., Din, Y., et al. (2019). GhWRKY6 Acts as a Negative Regulator in Both Transgenic Arabidopsis and Cotton During Drought and Salt Stress. Front. Genet. 10, 392. doi:10.3389/fgene.2019.00392
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007). Salt Stress Responses in Arabidopsis Utilize a Signal Transduction Pathway Related to Endoplasmic Reticulum Stress Signaling. Plant J. 51 (5), 897–909. doi:10.1111/j.1365-313X.2007.03195.x
Liu, L., Wang, B., Liu, D., Zou, C., Wu, P., Wang, Z., et al. (2020). Transcriptomic and Metabolomic Analyses Reveal Mechanisms of Adaptation to Salinity in Which Carbon and Nitrogen Metabolism Is Altered in Sugar Beet Roots. BMC Plant Biol. 20 (1), 138. doi:10.1186/s12870-020-02349-9
Lopez-Berenguer, C., Martinez-Ballesta, M. C., Garcia-Viguera, C., and Carvajal, M. (2008). Leaf Water Balance Mediated by Aquaporins under Salt Stress and Associated Glucosinolate Synthesis in Broccoli. Plant Sci. 174 (3), 321–328. doi:10.1016/j.plantsci.2007.11.012
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Lu, W., Chu, X., Li, Y., Wang, C., and Guo, X. (2013). Cotton GhMKK1 Induces the Tolerance of Salt and Drought Stress, and Mediates Defence Responses to Pathogen Infection in Transgenic Nicotiana Benthamiana. PLoS One 8 (7), e68503. doi:10.1371/journal.pone.0068503
Manavalan, L. P., Guttikonda, S. K., Tran, L. S., and Nguyen, H. T. (2009). Physiological and Molecular Approaches to Improve Drought Resistance in Soybean. Plant Cel. Physiol. 50 (7), 1260–1276. doi:10.1093/pcp/pcp082
Martin-Davison, A. S., Perez-Diaz, R., Soto, F., Madrid-Espinoza, J., Gonzalez-Villanueva, E., Pizarro, L., et al. (2017). Involvement of SchRabGDI1 from Solanum Chilense in Endocytic Trafficking and Tolerance to Salt Stress. Plant Sci. 263, 1–11. doi:10.1016/j.plantsci.2017.06.007
Mazel, A., Leshem, Y., Tiwari, B. S., and Levine, A. (2004). Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol. 134 (1), 118–128. doi:10.1104/pp.103.025379
Miao, H., Sun, P., Liu, J., Wang, J., Xu, B., and Jin, Z. (2018). Overexpression of a Novel ROP Gene from the Banana (MaROP5g) Confers Increased Salt Stress Tolerance. Int. J. Mol. Sci. 19 (10), 3108. doi:10.3390/ijms19103108
Munns, R., and Tester, M. (2008). Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi:10.1146/annurev.arplant.59.032607.092911
Paz, M. M., Martinez, J. C., Kalvig, A. B., Fonger, T. M., and Wang, K. (2006). Improved Cotyledonary Node Method Using an Alternative Explant Derived from Mature Seed for Efficient Agrobacterium-Mediated Soybean Transformation. Plant Cel. Rep. 25 (3), 206–213. doi:10.1007/s00299-005-0048-7
Peng, X., Ding, X., Chang, T., Wang, Z., Liu, R., Zeng, X., et al. (2014). Overexpression of a Vesicle Trafficking Gene, OsRab7, Enhances Salt Tolerance in rice. ScientificWorldJournal 2014, 483526. doi:10.1155/2014/483526
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 33 (3), 290–295. doi:10.1038/nbt.3122
Phang, T. H., Shao, G., and Lam, H. M. (2008). Salt Tolerance in Soybean. J. Integr. Plant Biol. 50 (10), 1196–1212. doi:10.1111/j.1744-7909.2008.00760.x
Qi, D. H., and Lee, C. F. (2014). Influence of Soybean Biodiesel Content on Basic Properties of Biodiesel-Diesel Blends. J. Taiwan Inst. Chem. Eng. 45 (2), 504–507. doi:10.1016/j.jtice.2013.06.021
Reuveni, R. (1992). Peroxidase Activity as a Biochemical Marker for Resistance of Muskmelon (Cucumis Melo) to Pseudoperonospora Cubensis. Phytopathology 82, 749–753. doi:10.1094/phyto-82-749
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26 (1), 139–140. doi:10.1093/bioinformatics/btp616
Rosa Téllez, S., Kanhonou, R., Castellote Bellés, C., Serrano, R., Alepuz, P., and Ros, R. (2020). RNA-Binding Proteins as Targets to Improve Salt Stress Tolerance in Crops. Agronomy 10 (2). doi:10.3390/agronomy10020250
Roy, S., Choudhury, S. R., Sengupta, D. N., and Das, K. P. (2013). Involvement of AtPol Lambda in the Repair of High Salt- and DNA Cross-Linking Agent-Induced Double Strand Breaks in Arabidopsis. Plant Physiol. 162 (2), 1195–1210. doi:10.1104/pp.113.219022
Salinas-Cornejo, J., Madrid-Espinoza, J., and Ruiz-Lara, S. (2019). Identification and Transcriptional Analysis of SNARE Vesicle Fusion Regulators in Tomato (Solanum lycopersicum) during Plant Development and a Comparative Analysis of the Response to Salt Stress with Wild Relatives. J. Plant Physiol. 242, 153018. doi:10.1016/j.jplph.2019.153018
Schwartz, S. L., Cao, C., Pylypenko, O., Rak, A., and Wandinger-Ness, A. (2007). Rab GTPases at a Glance. J. Cel. Sci. 120 (Pt 22), 3905–3910. doi:10.1242/jcs.015909
Singh, D., Yadav, N. S., Tiwari, V., Agarwal, P. K., and Jha, B. (2016). A SNARE-Like Superfamily Protein SbSLSP from the Halophyte Salicornia Brachiata Confers Salt and Drought Tolerance by Maintaining Membrane Stability, K+/Na+ Ratio, and Antioxidant Machinery. Front. Plant Sci. 7, 737. doi:10.3389/fpls.2016.00737
Sui, N., Wang, Y., Liu, S., Yang, Z., Wang, F., and Wan, S. (2018). Transcriptomic and Physiological Evidence for the Relationship between Unsaturated Fatty Acid and Salt Stress in Peanut. Front. Plant Sci. 9, 7. doi:10.3389/fpls.2018.00007
Sun, T. J., Fan, L., Yang, J., Cao, R. Z., Yang, C. Y., Zhang, J., et al. (2019). A Glycine max Sodium/hydrogen Exchanger Enhances Salt Tolerance through Maintaining Higher Na+ Efflux Rate and K+/Na+ Ratio in Arabidopsis. BMC Plant Biol. 19 (1), 469. doi:10.1186/s12870-019-2084-4
Tester, M., and Davenport, R. (2003). Na+ Tolerance and Na+ Transport in Higher Plants. Ann. Bot. 91 (5), 503–527. doi:10.1093/aob/mcg058
Tester, M., and Langridge, P. (2010). Breeding Technologies to Increase Crop Production in a Changing World. Science 327 (5967), 818–822. doi:10.1126/science.1183700
Tsutsui, T., Nakano, A., and Ueda, T. (2015). The Plant-Specific RAB5 GTPase ARA6 Is Required for Starch and Sugar Homeostasis in Arabidopsis thaliana. Plant Cel Physiol. 56 (6), 1073–1083. doi:10.1093/pcp/pcv029
Tuteja, N., Tran, N. Q., Dang, H. Q., and Tuteja, R. (2011). Plant MCM Proteins: Role in DNA Replication and beyond. Plant Mol. Biol. 77 (6), 537–545. doi:10.1007/s11103-011-9836-3
Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a Plant-Unique Novel Type Rab GTPase, Functions in the Endocytic Pathway of Arabidopsis thaliana. EMBO J. 20 (17), 4730–4741. doi:10.1093/emboj/20.17.4730
Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., and Sinha, A. K. (2020). A bHLH Transcription Factor, MYC2, Imparts Salt Intolerance by Regulating Proline Biosynthesis in Arabidopsis. FEBS J. 287 (12), 2560–2576. doi:10.1111/febs.15157
Wang, Z., Wang, J., Bao, Y., Wu, Y., and Zhang, H. (2010). Quantitative Trait Loci Controlling rice Seed Germination under Salt Stress. Euphytica 178 (3), 297–307. doi:10.1007/s10681-010-0287-8
Wang, Y., Jiang, L., Chen, J., Tao, L., An, Y., Cai, H., et al. (2018). Overexpression of the Alfalfa WRKY11 Gene Enhances Salt Tolerance in Soybean. PLoS One 13 (2), e0192382. doi:10.1371/journal.pone.0192382
Wu, X. X., Li, J., Wu, X. D., Liu, Q., Wang, Z. K., Liu, S. S., et al. (2016). Ectopic Expression of Arabidopsis thaliana Na+(K+)/H+ Antiporter Gene, AtNHX5, Enhances Soybean Salt Tolerance. Genet. Mol. Res. 15 (2). doi:10.4238/gmr.15027483
Yang, L., Tang, R., Zhu, J., Liu, H., Mueller-Roeber, B., Xia, H., et al. (2008). Enhancement of Stress Tolerance in Transgenic Tobacco Plants Constitutively Expressing AtIpk2beta, an Inositol Polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol. Biol. 66 (4), 329–343. doi:10.1007/s11103-007-9267-3
Yin, C., Karim, S., Zhang, H., and Aronsson, H. (2017). Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS). Int. J. Mol. Sci. 18 (2), 309. doi:10.3390/ijms18020309
Yuan, P., Du, L., and Poovaiah, B. W. (2018a). Ca2+/Calmodulin-Dependent AtSR1/CAMTA3 Plays Critical Roles in Balancing Plant Growth and Immunity. Int. J. Mol. Sci. 19 (6), 1764. doi:10.3390/ijms19061764
Yuan, P., Tanaka, K., Du, L., and Poovaiah, B. W. (2018b). Calcium Signaling in Plant Autoimmunity: A Guard Model for AtSR1/CAMTA3-Mediated Immune Response. Mol. Plant 11 (5), 637–639. doi:10.1016/j.molp.2018.02.014
Zhang, L., Li, T., Wang, Y., Zhang, Y., and Dong, Y. S. (2019a). FvC5SD Overexpression Enhances Drought Tolerance in Soybean by Reactive Oxygen Species Scavenging and Modulating Stress-Responsive Gene Expression. Plant Cel Rep. 38 (9), 1039–1051. doi:10.1007/s00299-019-02424-y
Zhang, L., Liu, Y., Zhu, X. F., Jung, J. H., Sun, Q., Li, T. Y., et al. (2019b). SYP22 and VAMP727 Regulate BRI1 Plasma Membrane Targeting to Control Plant Growth in Arabidopsis. New Phytol. 223 (3), 1059–1065. doi:10.1111/nph.15759
Keywords: soybean, AtARA6, salt tolerance, RAB GTPase, RAB5, SNARE pathway
Citation: Hong Z, Li Y, Zhao Y, Yang M, Zhang X, Teng Y, Jing L, Kong D, Liu T, Li S, Meng F, Wang Q and Zhang L (2022) Heterologous Expression of Arabidopsis AtARA6 in Soybean Enhances Salt Tolerance. Front. Genet. 13:849357. doi: 10.3389/fgene.2022.849357
Received: 06 January 2022; Accepted: 23 March 2022;
Published: 12 May 2022.
Edited by:
Sreepriya Pramod, Altria, United StatesReviewed by:
Chathurani Ranathunge, Eastern Virginia Medical School, United StatesCopyright © 2022 Hong, Li, Zhao, Yang, Zhang, Teng, Jing, Kong, Liu, Li, Meng, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fanli Meng, bWVuZ2ZhbmxpQG5lYXUuZWR1LmNu; Qi Wang, bmVhdXdxQDE2My5jb20=; Ling Zhang, emx5X2phYXNAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.