- 1School of Pharmacy, Heilongjiang University of Chinese Medicine, Harbin, China
- 2Faculty of Electrical Engineering and Information Technology, Technical University of Chemnitz, Chemnitz, Germany
- 3School of Forestry, Northeast Forestry University, Harbin, China
- 4Yichun Branch of Heilongjiang Academy of Forestry, Yichun, China
- 5Jiangsu Kanion Pharmaceutical Co. Ltd., Lianyungang, China
- 6State Key Laboratory of New-tech for Chinese Medicine Pharmaceutical Process, Lianyungang, China
- 7Key Laboratory of Basic and Application Research of Beiyao (Heilongjiang University of Chinese Medicine), Ministry of Education, Harbin, China
Acer ukurunduense refers to a deciduous tree distributed in Northeast Asia and is a widely used landscaping tree species. Although several studies have been conducted on the species’ ecological and economic significance, limited information is available on its phylo-genomics. Our study newly constitutes the complete chloroplast genome of A. ukurunduense into a 156,645-bp circular DNA, which displayed a typical quadripartite structure. In addition, 133 genes were identified, containing 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes. In total, 107 simple sequence repeats and 49 repetitive sequences were observed. Thirty-two codons indicated that biased usages were estimated across 20 protein-coding genes (CDS) in A. ukurunduense. Four hotspot regions (trnK-UUU/rps16, ndhF/rpl32, rpl32/trnL-UAG, and ycf1) were detected among the five analyzed Acer species. Those hotspot regions may be useful molecular markers and contribute to future population genetics studies. The phylogenetic analysis demonstrated that A. ukurunduense is most closely associated with the species of Sect. Palmata. A. ukurunduense and A. pubipetiolatum var. pingpienense diverged in 22.11 Mya. We selected one of the hypervariable regions (trnK-UUU/rps16) to develop a new molecular marker and designed primers and confirmed that the molecular markers could accurately discriminate five Acer species through Sanger sequencing. By sequencing the cp genome of A. ukurunduense and comparing it with the relative species of Acer, we can effectively address the phylogenetic problems of Acer at the species level and provide insights into future research on population genetics and genetic diversity.
Introduction
The Acer L. genus comprises approximately 200 species and has a dominant role in the forests of the Northern Hemisphere (Stace and Hind, 1997; Bi et al., 2016; Gao et al., 2020). Acer L. species have been used for several thousand years in China, have gained high economic value, and are now among the most important timber trees (Wu et al., 2008; Saeki and Murakami 2009; Dormontt et al., 2020). Flavonoids, tannins, terpenoids, sterols, alkaloids, diarylheptanoids, diarylheptanoids, and others have been isolated from Acer, possessing high bioactivity and exerting pharmacological effects (Tang et al., 2012; Ren and Feng, 2018; Yang et al., 2018; Hou et al., 2019; Liang et al., 2019; Piao et al., 2020; Kausar et al., 2021). Additionally, in Chinese folk medicine, Acer has also been widely used to treat angina pectoris, coronary artery cirrhosis, and cardiac–cerebral vascular disease. A. ukurunduense mainly grows in broadleaf forests at altitudes of 300–1,500 m, with distribution in northeast China, North Korea, and the Russian Far-East. It is an important autumn leaf-viewing tree in northeast China and can be used as a street tree or in a landscape garden for landscaping. Despite its importance, the phylogenetic relationship of the genus remains unclear (Tian et al., 2002).
The whole chloroplast (cp) genome of tobacco was first described in 1986 (Shinozaki et al., 1986; Daniell et al., 2016). The cp genome comprises multi-functional organelles playing a significant role, such as photosynthesis and the carbon cycle (Singh et al., 2009; Schmidt et al., 2020; Zupok et al., 2021). Generally, the cp genome is more conserved than the nuclear genome, a circular molecule of double-stranded DNA. It usually has a quadripartite configuration consisting of a pair of inverted repeat (IR) regions separated by a large single copy (LSC) region and a small single copy (SSC) region (Eid 2009; Wicke et al., 2011; Shahzadi et al., 2020). The cp genome of land plants ranges from 120 to 170 kb in size and gene order and can thus provide plenty of genetic information for investigating evolutionary divergence and the inter-specific relationships of plants (Palmer, 1985; Wolfe et al., 1987; Daniell et al., 2016). Cp information has been extensively applied in analyses of plant genetic diversity, conservation genetics, and molecular marker development (Moore et al., 2010; Li et al., 2016; Gao et al., 2018; Cui et al., 2019).
Studies have indicated that in many forests throughout the Northern Hemisphere, Acer is a dominant genus in terms of abundance or total basal area but that it may also function as a foundation genus (Ellison et al., 2019). Qiao et al. (2020) used data to identify a suite of candidate foundation species in Chinese forests. A. ukurunduense and A. barbinerve were the only two species that met the criteria as candidate foundation species. Acer is the foundation species in many terrestrial ecosystems of Northern China. In a previous study, these five Acer species (A. mandshuricum, A. tataricum, A. pictum, A. tegmentosum, and A. ukurunduense) are widely distributed in the Xiaoxing’an Mountains of China and play important roles in different forest types (Hou and Wang, 2013; Wen, 2021). Currently, cp genome data have been obtained for numerous species of Acer (Zhao et al., 2018; Fu et al., 2020; Xia et al., 2020; Dong et al., 2021; Xia et al., 2022). However, no cp genome data were reported for A. ukurunduense, which is endemic to Northeast China. The evolutionary history and phylogenetic relationship of the genus Acer and the family Aceraceae have not been reported.
In the present study, we assembled a complete cp genome sequence for A. ukurunduense using the Illumina sequencing platform. The objective was to study the cp genome structure features of A. ukurunduense, determine its effectiveness in establishing species phylogeny, assess its structural efficacy in taxon delimitation, and divergence time was estimated based on the cp genome. In addition, we are interested in identifying molecular markers in the cp genomes of five Acer species distributed in the Xiaoxing’an Mountains of China. The present study aimed to reveal the phylogenetic position of A. ukurunduense and provide insights into Acer evolution.
Materials and Methods
Supplementary Figure S1 provides a technology roadmap to help the reader better understand the study.
Plant Material and DNA Sequencing
Whole and fresh green leaves of A. ukurunduense were collected in Yichun, Heilongjiang Province, China. The voucher herbarium was deposited in the Pharmacy College of the Heilongjiang University of Chinese Medicine under the voucher number YCL20210606001. Total genomic DNA was isolated from leaves (100 mg) according to the modified CTAB method (Porebski et al., 1997). The remaining samples and DNA extraction were stored at −80°C. In addition, we selected the available complete cp genomes of four other Acer species from GenBank, including A. mandshuricum (NC_049164), A. tataricum (MK479223), A. pictum (NC_049127), and A. tegmentosum (NC_056233).
Cp Genome Assembly and Annotation
We produced a 150-bp paired-end library and sequenced it on the Illumina sequencing platform (Benagen, Wuhan, China). The clean reads were filtered using the SOAPnuke v1.3.0 pipeline to obtain plastid-like reads. Filter raw data parameters as follows: 1) remove sequences with N content exceeding 10% of the read length base number, 2) the number of low-quality bases exceeds 50% of the number of bases in the sequence, and 3) contain sequences with unremoved joints, and finally obtain high-quality clean data. We assembled the filtered reads based on the GetOrganelle v1.7.5 (Jin et al., 2020) program with published cp genomes as a reference (Xia et al., 2022). Regarding the genome annotation, we annotated the cp genome of A. ukurunduense by CPGAVAS2 (http://47.96.249.172:16019/analyzer/annotate) (Shi et al., 2019) and then manually adjusted and annotated genes. The complete cp genome sequence and gene annotation of A. ukurunduense were submitted to the public GenBank database (MZ736145). Then, we used Chloroplot (https://irscope.shinyapps.io/Chloroplot/) to generate a circular cp genome map (Zheng et al., 2020).
Comparative Cp Genome Analyses
The mVISTA program (https://genome.lbl.gov/vista/mvista/submit.shtml) (Frazer et al., 2004), which applied the Shuffle-LAGAN model (Brudno et al., 2003), was used to align, annotate, and visually compare the cp genome of A. ukurunduense with the cp genomes of four Acer species (A. mandshuricum, A. tataricum, A. pictum, and A. tegmentosum). We chose A. mandshuricum as the reference.
IR/SC Boundary Analysis, Codon Usage, and Genetic Divergence Analyses
The sizes and junction sites of the LSC/IRb/SSC/IRa regions of the five Acer species were compared using the IRscope online program (https://irscope.shinyapps.io/irapp/) (Amiryousefi et al., 2018). In the genome structural rearrangement analysis, the five species’ cp genomes were aligned using the MAUVE program (Darling et al., 2004). Condon usage, synonymous codon usage, and RSCU were determined using CodonW v1.4.2. Nucleotide divergence of Acer species was calculated based on nucleotide diversity values by employing DnaSP v6 (Rozas et al., 2017) software. Before analysis, we excluded gaps and missing data.
Repeat Sequences and SSR Analysis
Reverse, forward, complement, and palindromic repeats were then identified within the five species’ cp genomes by the REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputer) (Kurtz et al., 2001) program, with parameters set as follows: 1) the minimal repeat sequence size was 20 bp, 2) the maximum computed repeat was 50, 3) the identity was not less than 90%, and 4) the hamming distance was 3. The SSR loci of A. ukurunduense cp genomes were located using MISA (v2.1) software (http://pgrc.ipk-gatersleben.de/misa/) (Beier et al., 2017). The minimum numbers were: eight repeats for mononucleotide types; four repeats for dinucleotide types; four repeats for trinucleotide types; and three repeats for tetranucleotide types, pentanucleotide types, and hexanucleotide types. The shortest size between any two SSR loci was more than 100 bp.
Phylogenetic Relationship Analyses and Estimation of the Divergence Time
To parse the phylogenetic status of Acer species within the Aceraceae family, multiple sequence alignments were generated using the whole cp genome sequences of 105 Acer species and downloaded from the NCBI database (Supplementary Table S1), with Sapindus mukorossi and Arabidopsis thaliana as outgroups. The cp genome sequence alignment analysis of the 107 species was generated using MAFFT (v7.481) (Katoh et al., 2019) with default parameters. The maximum likelihood (ML) tree (with default parameter settings and 1,000 bootstrap repeats) was performed using the IQ-TREE (v2.1.3) (Lanfear et al., 2020) software with the best-fitting model TVM + F + R5. A Bayesian inference (BI) analysis was performed using the Markov Chain Monte Carlo (MCMC) algorithm in Mr. Bayes, using 1,000,000 generations, a sampling frequency of 100, and default values for other parameters. Visualizing and refining the output tree was implemented in MEGA X.
A timetree was inferred using the ReltimeML-option in MEGA X. The reference-node age was obtained by the divergence time of A. mandshuricum–A. fabri (20.9–32.5 million years ago) and A. acuminatum–A. saccharum (11.7–31.9 million years ago) (http://www.timetree.org/, accessed on June 8, 2022).
Validation of Molecular Markers
Variable intergenic regions can be used to develop molecular markers to discriminate among the five species. We selected the trnK-UUU/rps16 hypervariable region as an amplified fragment. The primer was designed using the Primer 3 input online software (https://primer3.ut.ee/), and the primer is shown in Supplementary Table S2. The PCR reaction system is as follows: 12.5 µl 2×Taq PCR Master Mix, 1 µl for each of the 10 µM forward and reverse primers, 2 µl total DNA, and 8.5 µl ddH2O. PCR amplification procedures with the following conditions: denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, 68°C for 1 min, and a final extension at 68°C for 7 min. PCR products (5 µl) were visualized on 1% agarose gels and then subjected to Sanger sequencing via the ABI3730XL Sequencing platform (Comate Bioscience Co., Ltd. Changchun, China).
Results
Chloroplast Genome Features of A. ukurunduense
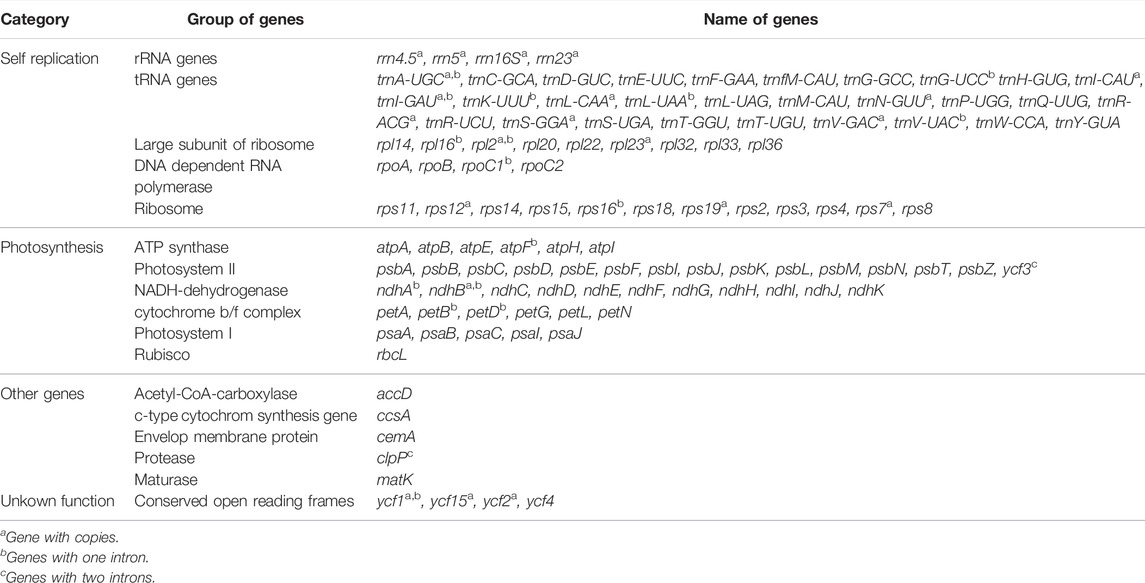
The A. ukurunduense cp genome is 156,645 bp, including the LSC region of 85,061 bp, the SSC region of 18,114 bp, and a pair of inverted repeat regions (IRa and IRb) of 53,470 bp (Figure 1). The average GC content ratio is 37.9%, and the average AT content ratio is 62.1%. In terms of gene content and composition, the features of the A. ukurunduense cp genome are similar to those of other previously researched Acer species. One-hundred thirty three genes were encoded, including 88 code protein genes, eight rRNAs and 37 tRNAs. We can put them into four classes according to their function, including genes for photosynthesis, self-replication, other genes, and unknown (Table 1). We detected a total of 20 genes are repeated in the IR regions, among which four ribosomal RNA genes (rrn4.5, rrn5, rrn16S, and rrn23), seven transfer RNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and nine PCGs (ndhB, rpl2, rpl23, rps12, rps19, rps7, ycf1, ycf2, and ycf15). Twenty-two genes contain introns. There are two genes (ycf3 and clpP) containing two introns, and the other 20 genes contain one intron (Supplementary Table S3).
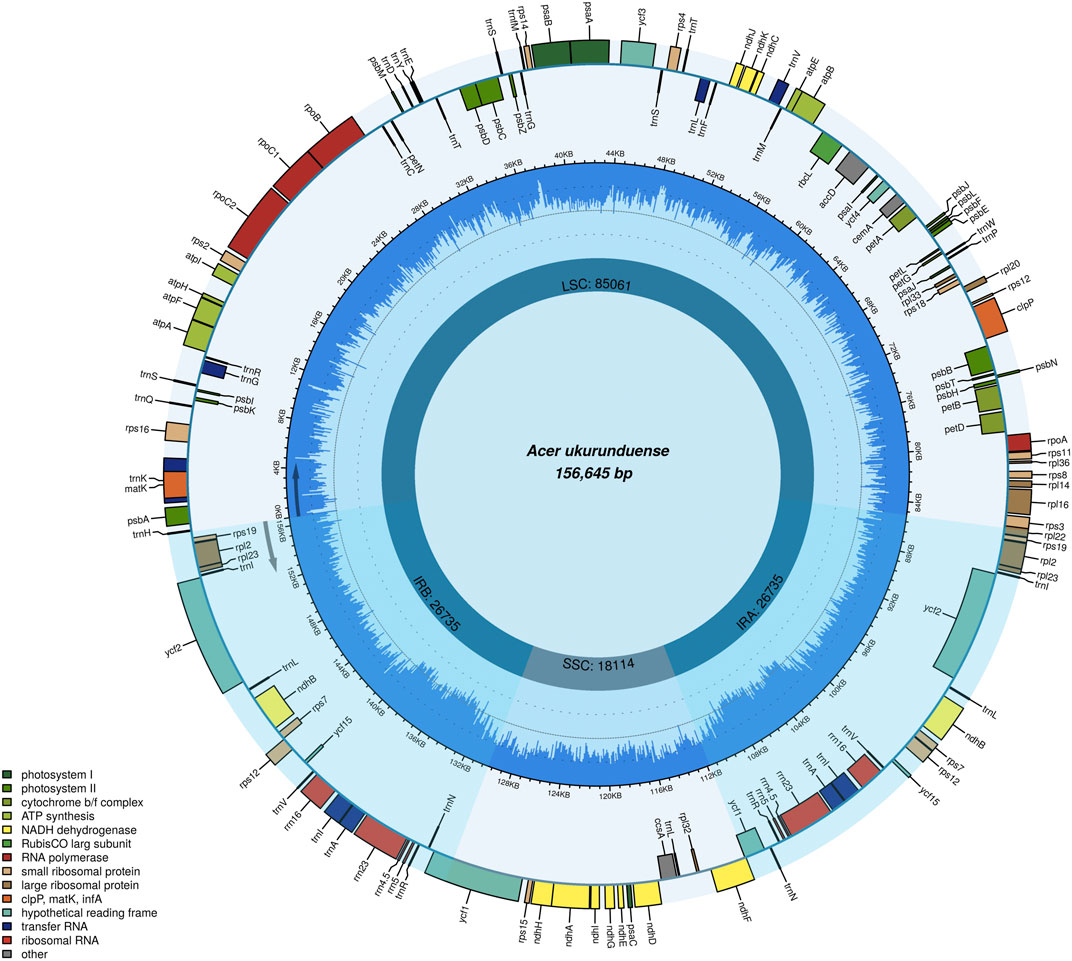
FIGURE 1. Gene map of the A. ukurunduense complete cp genome. Genes drawn within and out of the circle are transcribed in the clockwise and anticlockwise directions, respectively. Genes are color-filled, which represents different functions.
Codon Usage Analysis
The relative synonymous codon usage (RSCU) frequency was estimated in the five Acer species (Figure 2). A total of 20 amino acids were detected for protein biosynthesis by tRNA molecules in the cp genomes of the five species, and the relative synonymous codon usage (RSCU) values are listed in Supplementary Table S4. The codon numbers in all combined sequences (CDS) were A. mandshuricum (26,677), A. tataricum (26,660), A. pictum (26,682), A. tegmentosum (26,716), and A. ukurunduense (26,885), respectively. Among all codons, the most abundant codons encoded were leucine (AUU) (1,091–1,114), and the least abundant codons encoded were L-cysteine (UGC) (82–84). We were surprised to find that the RSCU values of 30 codons and one stop codon (UAA) were greater than 1 after the statistics of the codon RSCU values of five species. For A. pictum, there are 27 codons that are end-rich in A or U, and in the other four species, there are 28 codons that are end-rich in A or U, which among all codons whose RSCU value was greater than 1 did not contain a stop codon (UAA). These results indicate that the cp genomes of five Acer genus species tend to end with A or U.
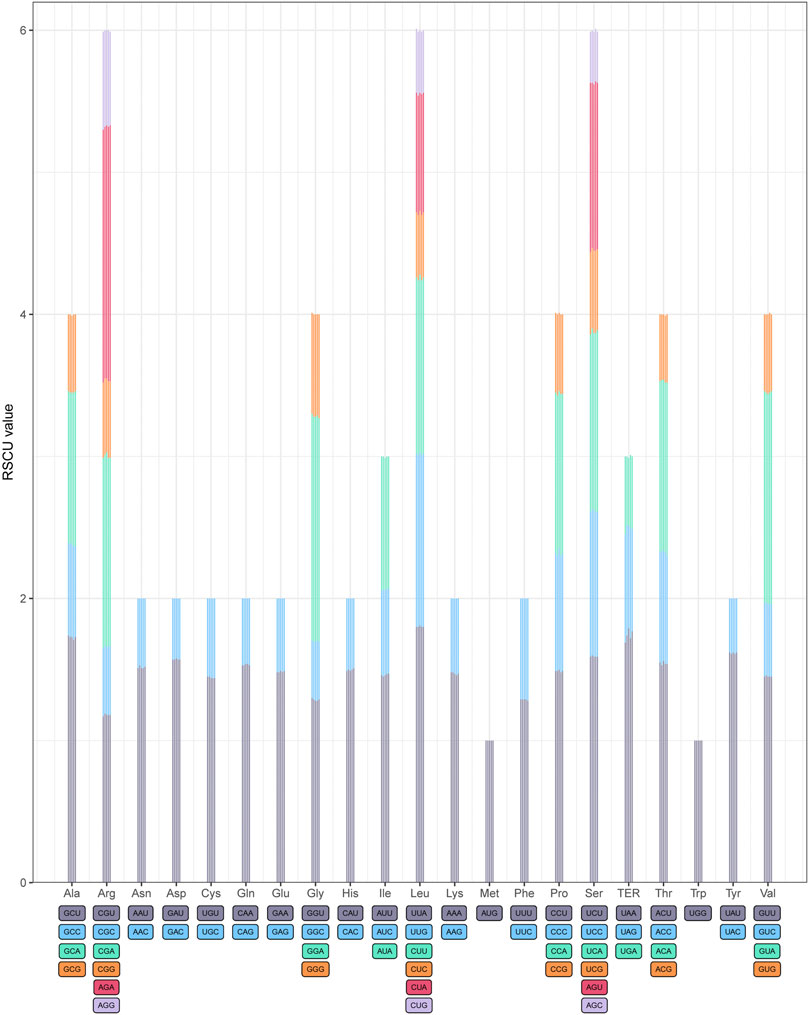
FIGURE 2. Codon content of twenty amino acids in all protein-coding genes of five Acer species cp genomes. The histogram of each amino acid indicated codon usage within Acer (from left to right: A. mandshuricum, A. pictum, A. tataricum, A. tegmentosum, and A. ukurunduense).
Long-Repeat and Simple Sequence Repeat (SSR) Analysis
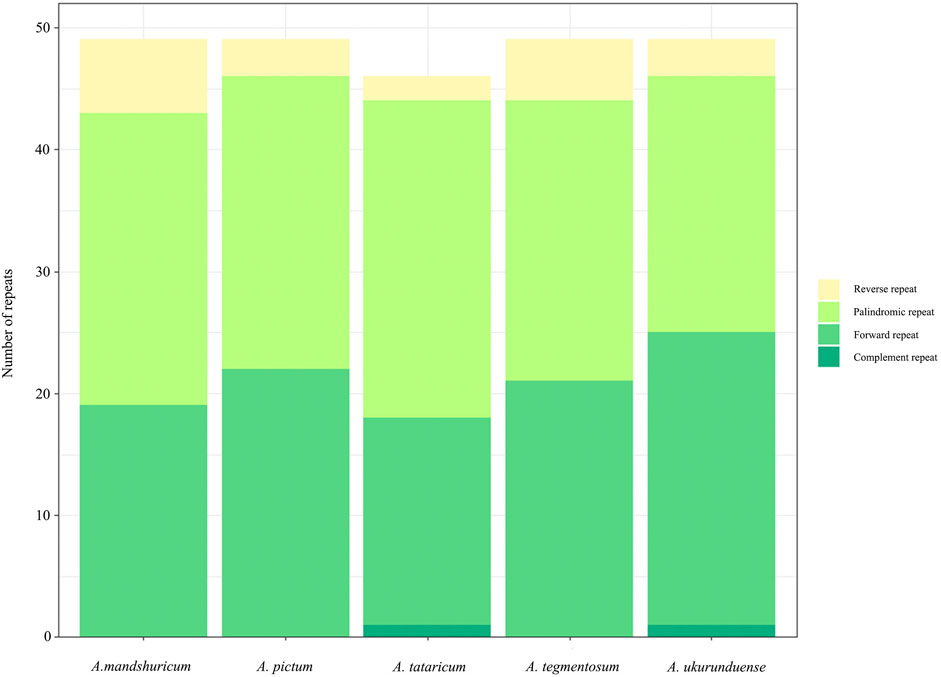
Four species of tandem repeats were presented in this study, including forward, reverse, complement, and palindrome. The detailed number of tandem repeats for each species is illustrated in Figure 3. Expect for A. tataricum, the number of tandem repeats was 49. Only A. tataricum and A. ukurunduense were included in a complement repeat. In addition, the other numbers of three repeats are also different. There were 46 (two reverse, 26 palindromic, 17 forward, and one complement), 49 (six reverse, 24 palindromic, and 19 forward), and 49 (three reverse, 24 palindromic, and 22 forward), 49 (five reverse, 23 palindromic, and 21 forward), 49 (three reverse, 21 palindromic, 24 forward, and one complement) long repeats in A. tataricum, A. mandshuricum, A. pictum, A. tegmentosum, and A. ukurunduense, respectively (Supplementary Table S5).
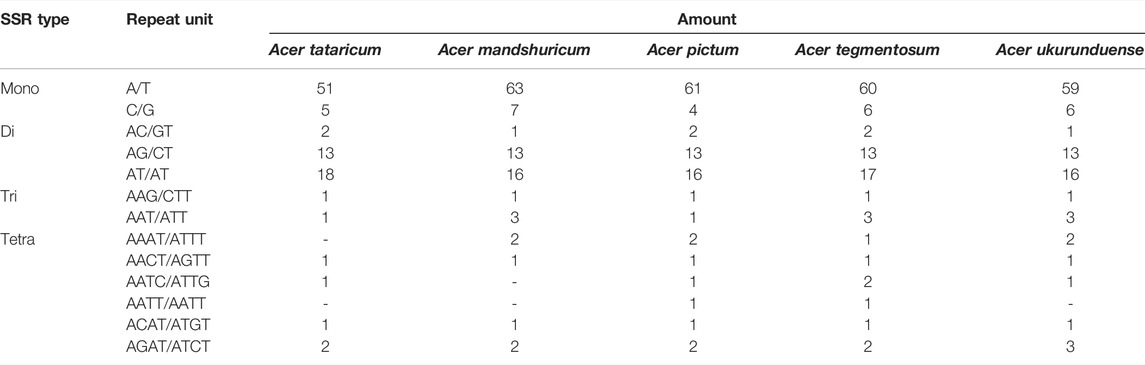
The analysis of SSRs exhibited four types of SSRs (Table 2). The total number of SSRs was 96 in A. tataricum, 109 in A. mandshuricum, 106 in A. pictum, 110 in A. tegmentosum, and 107 in A. ukurunduense. The types and content of motifs are different between species. In five species, mono-nucleotide has the largest proportion (58.33%–64.22%), followed by di-nucleotide (27.52%–34.38%), tetra-nucleotide (5.21%–7.55%), and tir-nucleotide (1.89%–3.74%). A mononucleotide accounted for 86.44%–93.84%. The majority of SSRs in all species were A/T mononucleotides. In the cp genome, the LSC region has the most SSRs, ranging from 57 to 71 in the five species analyzed here. However, the number of SSRs in IR regions from 22 to 24 is the lowest, from 13 to 19 in the SSC region, and from 57 to 71 in the LSC region (Figure 4B). The number of SSRs is the largest in the intergenic and the number of SSRs is the lowest in the intron (Figure 4C).
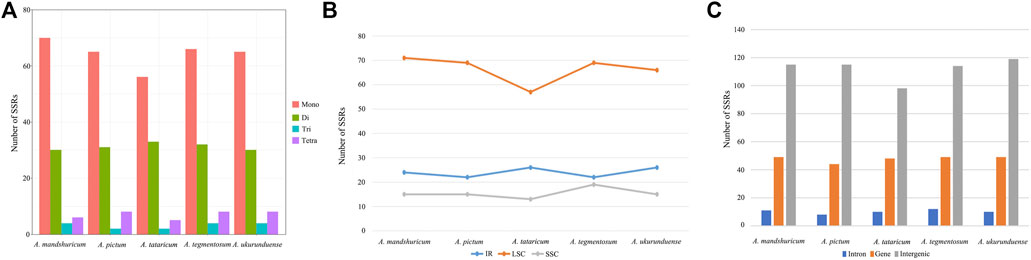
FIGURE 4. Number of different types of SSRs from five Acer species cp genomes. (A) Number of different SSR types detected in five cp genomes. (B) Number of SSRs in LSC, SSC, and IR regions. (C) Number of SSRs in the intergenic regions, genes, and introns.
Border Region Expansion and Contraction
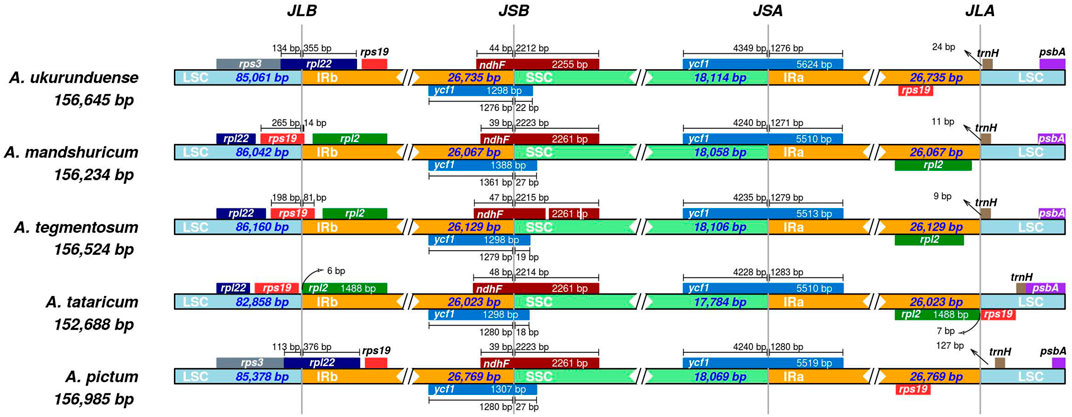
Differences in the size of the cp genome are mainly affected by the expansion and contraction of the border regions. In the current study, LSC/IRs and IRs/SSC borders and the genes located near junctions of different regions were compared and analyzed from the five Acer species cp genomes (Figure 5). The result indicated that the rps19 gene was based on the junction of LSC/IRb in two of five species, while the rpl22 gene was utterly situated at the junction of LSC/IRb in the two species of A. ukurunduense and A. pictum. The junction of IRb/SSC was located in the overlap region of the ycf1 gene, and the gene was relatively conserved. The ycf1 gene at the SSC/IRa junction and trnH gene at the right of the IRa/LSC junction in five species and rpl2 gene in A. ukurunduense, A. tataricum, and A. pictum were compared. Deletion of the rps19 gene occurred in A. mandshuricum and A. tegmentosum. The structure and gene content experienced no significant expansion or contraction of IR regions of A. ukurunduense and A. pictum, differing from the other three, which have similar gene boundary structures. These genomes presented obvious variances at the junctions, the gene sequence in LSC/IRb junction from 5 to 3′ is rps3, rpl22, and rps19 in A. ukurunduense and A. pictum, while the other three species are rpl22, rps19, and rpl2. Furthermore, a comparison of the border regions was made among Acer species. During this analysis, the adjacent genes and some remarkable variations were discovered. The rpl2 gene was absent from the regions of IRa and IRb in A. ukurunduense and A. pictum.
Analysis of Nucleotide Diversity
The Pi values ranged from 0 to 0.029 in the five Acer species (Figure 6 and Supplementary Table S6). This result showed that the species in the order Acer could undergo rapid nucleotide substitution. The IR regions had unobvious nucleotide variability compared with the SSC and LSC regions. There are four variable regions (trnK-UUU/rps16, ndhF/rpl32, rpl32/trnL-UAG, and ycf1) with Pi values exceeding 0.02, and all divergent regions reveal higher nucleotide diversity, which is observed in the intergenic region.
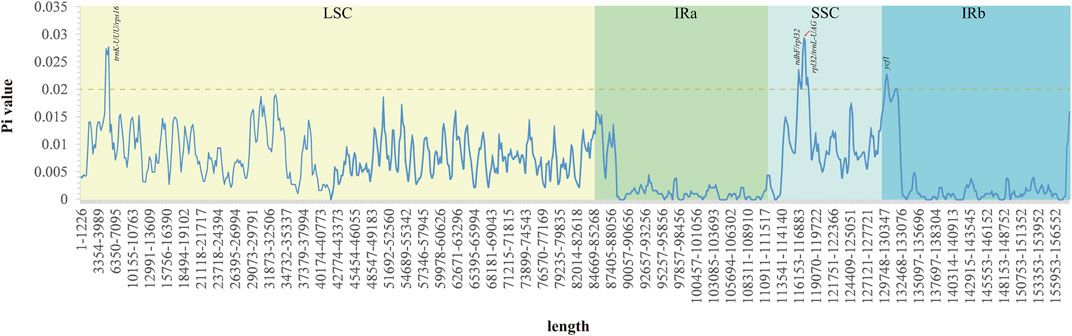
FIGURE 6. Nucleotide variability across the cp genomes of the five Acer species studied. The highest values were annotated based on the localization of predicted genes of the A. ukurunduense cp genome.
Comparative Genome Analysis
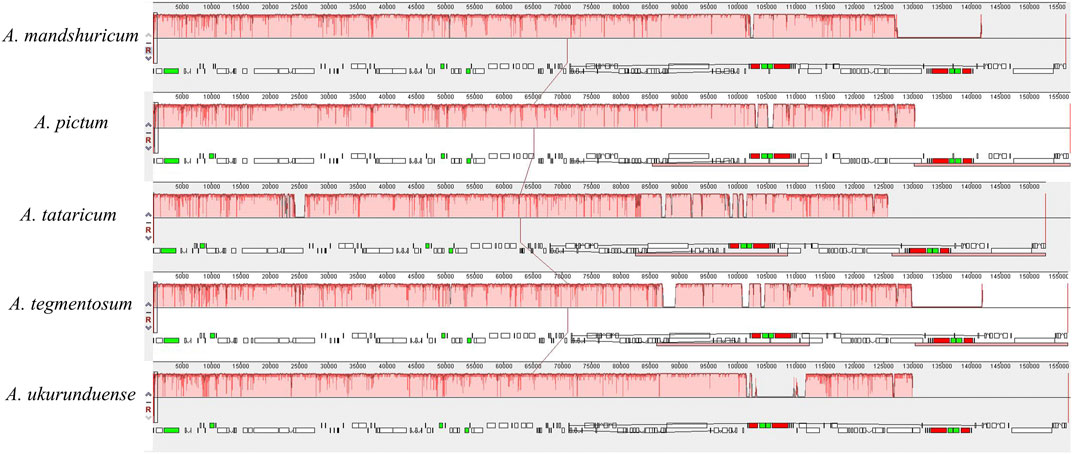
Comparative genomic analysis and the available DNA sequences make it possible to have a comprehensive view of the Acer genus. The cp genome sequences of A. ukurunduense were compared to A. mandshuricum, A. tataricum, A. pictum, and A. tegmentosum based on the MAUVE program (Figure 7). The results show that the structures of the cp genomes of the five Acer species are extremely similar and have good collinearity. By conducting the mVISTA program analysis (Figure 8), some of the high divergent regions were monitored, including matk/rps16, trnQ-UUG/rps16, trnQ-UUG/psbI, trnS-GCU/atpA, atpH/atpI, psbZ/trnG-GCC, ycf3/trnS-GGA, trnT-UGU/trnL-UAA, trnF-GAA/ndhJ, petA/psbJ, ndhF/trnL-UAG, and ycf1.
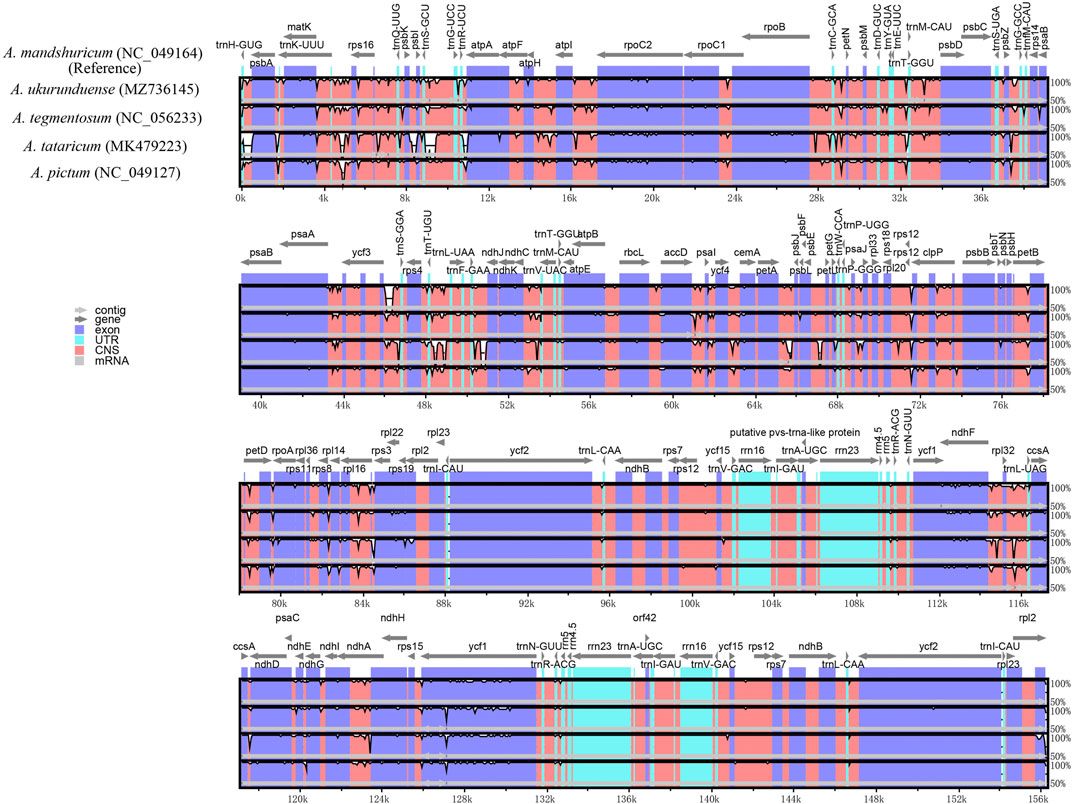
FIGURE 8. Visualized alignments of five cp genomes of Acer using mVISTA, with A. mandshuricum as the reference. The horizontal axis indicates the five species’ cp genome of the alignment. The vertical scale represents the percentage identity, ranging from 50 to 100%.
Phylogenetic Analysis and Divergence Time Estimation
A total of 105 Acer species cp genomes and two species (Sapindus mukorossi and Arabidopsis thaliana) as outgroups were constructed from 15 Sect. using the ML tree and BI tree, respectively (Figures 9A,B). The two datasets (ML and BI) topologies generated similar structures. Fourteen species of Sect. Platanoidea were clustered together. Eleven species of Sect. Macrantha were clustered together. The ML and BI trees show that Sect. Platanoidea and Sect. Macrantha were able to gather into single bands. Sect. Macrantha is closely related to Sect. Platanoidea, followed by Sect. Glabra. Seven species of Sect. Pentaphylla were clustered together. Five species of Sect. Trifoliata were not clustered into monophyly. Four species of Sect. Trifoliata were clustered together, and A. sutchuenense was separated into a single clade individually. Thirteen species of Sect. Acer were clustered together. The Sect. Pentaphylla is closely related to the Sect. Trifoliata, followed by the Sect. Acer. Sect. Palmata and Sect. Lithocarpa are closely related. As one of the only two species in Sect. Spicata, A. ukurunduense is more closely related to Sect. Palmata. The species of Sect. Negundo are closely related to Sect. Glabra, Sect. Trifoliata, and Sect. Ginnala, respectively. There are relatively close relationships among Scet. Parviflora, Sect. Indivisa, Sect. Rubra, and Sect. Hyptiocarpa.
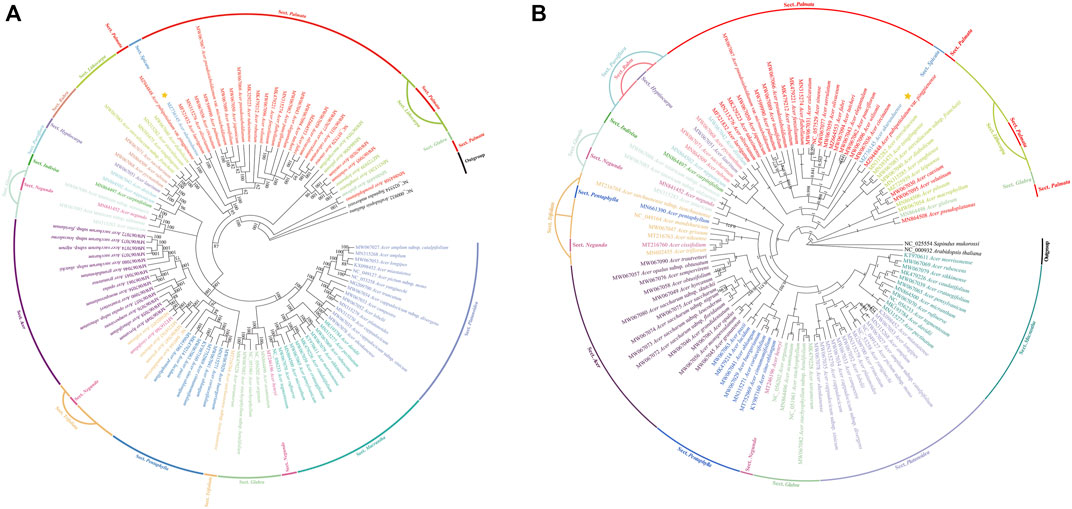
FIGURE 9. Phylogenetic trees based on 107 species of the whole-cp genome sequences using maximum likelihood (ML) (A) and Bayesian inference (BI) (B) analyses. Numbers at nodes are values for bootstrap support. Sequences from Sapindus mukorossi and Arabidopsis thaliana served as outgroups. The position of A. ukurunduense is indicated by a yellow star.
The result of the divergence time estimation of 107 species based on chloroplast sequences is shown in Supplementary Figure S2. The age of the crown of the Acer genus was estimated to be in the upper Oligocene period, approximately 28.97 million years ago (Mya). The separation between Sect. Platanoidea and Sect. Macrantha occurred at 24.98 Mya. The divergence between Sect. Glabra, Sect. Pentaphylla, Sect. Trifoliata, and Sect. Acer was dated to 24.6 Mya. The estimated divergence time for Sect. Palmata with the most species was 25.66 Mya. A. ukurunduense and A. pubipetiolatum var. pingpienense diverged at 22.11 Mya.
Molecular Markers Based on the Hypervariable Regions of Acer Cp Genomes
This study developed a specific molecular marker for accurate identification based on the hypervariable regions of the cp genomes of five Acer species from northeast China. Agarose gel electrophoresis detection showed that the size of the amplified fragment was the same as that of the target fragment (Supplementary Figure S3). Sequencing of PCR products showed six SNP and two insertion-deletion mutations (Indel) locus in the sequences of the five species. In addition, surprisingly, we discovered that three SNP loci and an Indel loci in the amplified sequence accurately identified the five species (A. mandshuricum, A. tataricum, A. pictum, A. tegmentosum, and A. ukurunduense) (Figure 10).
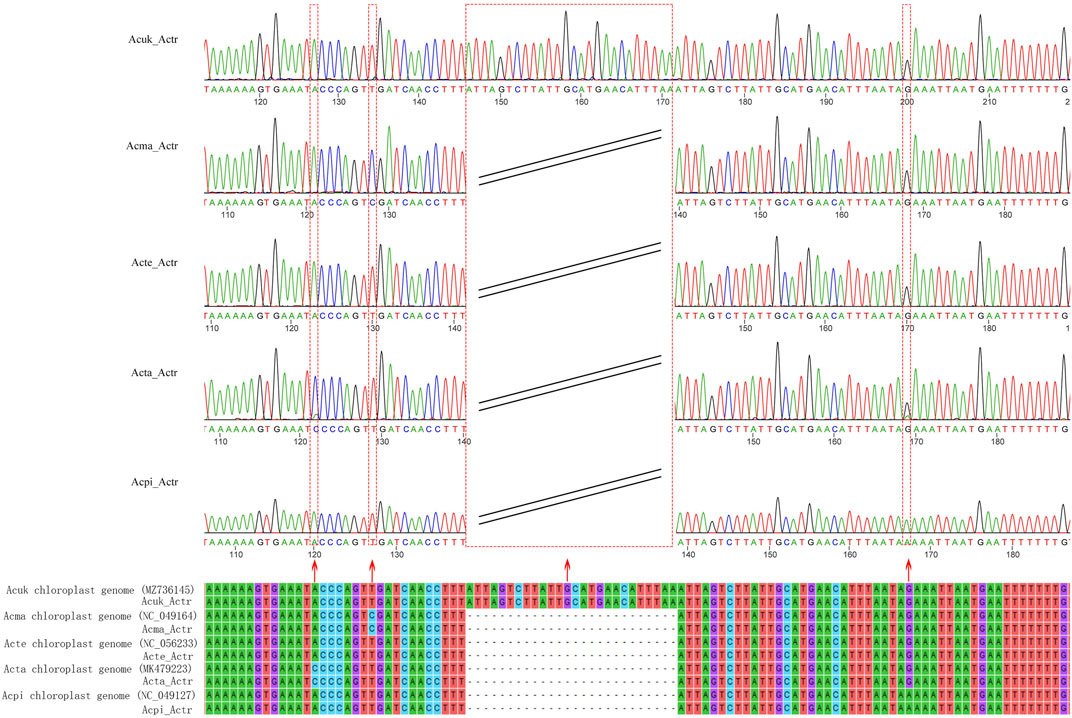
FIGURE 10. Alignment of the sequences of the PCR products amplified using the primer of AcerTS and the reference sequences. The red arrows indicate SNPs, Indel loci, and their corresponding peaks. Acuk: A. ukurunduense; Acma: A. mandshuricum; Acte: A. tegmentosum; Acta: A. tataricum; Acpi: A. pictum.
Discussion
Cp Genome Structure
There are some genes with unknown functions in this genome, such as ycf1, ycf2, ycf4, and ycf15. In most plants, the IRb and SSC boundary lie on the ycf1 gene, which may be associated with the expansion of the IRb/SSC boundary (Kumar et al., 2018). In the cp genome of A. ukurunduense, we also observed the ycf15 gene, which exists in most angiosperms but whose function remains unknown (Shi et al., 2013; Jung et al., 2021). Compared with the LSC and SSC regions, GC content in the IR region was the highest, mainly due to four rRNA genes occupying a greater area.
Comparative Genomics
Codon usage bias is often used at unequal frequencies in all genomes and is well documented across species from all three domains of life (Duret, 2002; Chen et al., 2004). Selection operates on synonymous mutations because they influence various biological processes, some of which are linked to the expression of proteins and nucleic acid composition (Plotkin and Kudla, 2011; Zhou et al., 2016). In addition, amino acid hydrophilicity, tRNA abundance, gene position, gene expression rate, and base group composition are prime factors affecting codon usage. The choice of codon biases directly impacted nonuniform ribosome decoding rates on mRNA levels, which in turn influenced the protein construction that directly affects protein function in diverse biological processes. Hence, codon usage is a critical factor and, through translation, affects the mRNA secondary structure. Thus, codon usage is closely related to the efficiency of translation and posttranscriptional processes (Yi Liu et al., 2021). In the genetic code, except for tryptophan and methionine, which have only one codon, all other amino acids have more than one degenerate codon. Therefore, there is a certain bias in selecting degenerate codons in the coding process. When the RSCU value is greater than 1, the codon is preferred. This finding was unexpected and suggests that nearly all amino acid codons in A. ukurunduense, cp genomes have preferences. The main reason is the transcription process error prevented by amino acid activity. In the cp genome, the natural selection and evolution processes dominated a very significant determinant in influencing codon usage bias, which is that the use of codons also affects the structure and function of coding proteins. The results showed that the cp genomes of A. ukurunduense contain some codons that encode necessary amino acids.
SSRs are molecular markers as a major tool with high variation within the same species that are used in investigating genomic polymorphism and polymorphism studies. Different species have different types and content of motifs. The majority of SSRs in all species were A/T mononucleotides. The presence of SSRs in the cp genome is conducive to resolving genetic diversity between related genera and increasing the power of interspecific studies, possibly in combination with other informative nuclear genome SSRs (Lohne and Borsch 2005).
Comparative Genomics
Almost all land plants have relatively conserved cp genomes. However, the arrangement of genes in the junctions of cp genomes differs from one species to another. A large IR expansion has also been reported in angiosperm lineages containing Erodium (Blazier et al., 2016), Passiflora (Rabah et al., 2019), and Pelargonium (Zhu et al., 2016). The IR regions will expand and contract, causing some genes distributed to be not fixed in the IR region or single copy region, generating the difference in genome size. The phenomenon contributes to resolving the process of genome evolution (Dugas et al., 2015). Our analyses reveal that the IR boundary expands in A. ukurunduense and A. pictum compared to the other three species. In A. ukurunduense and A. pictum, the rpl2 gene disappears, while the rps19 gene replicates, and the LSC/IRb boundary moves to the rpl22 gene position. The results of the correlational analysis show that the expansion and contraction of the IR regions may be the determining factor that led to the size and gene locus changes of cp genomes.
There existed no significant rearrangements and no large fragment inversion among the compared cp genomes, and the sequence of genes was consistent and highly conserved. In general, the alignment sequence divergence across assemblies reveals that the cp genome in Acer is relatively conservative, especially in the gene coding regions when compared with its noncoding counterparts. By analyzing highly variable regions, ten highly divergent regions may be unique barcodes applied to species identification, also providing phylogenetic information. We also analyzed the disparity of cp genomes. This result showed that the intergenic regions had more variations than the gene coding regions, which is due to most fragments in IR regions having a relatively low Pi value. Some of these regions have been previously studied in other species’ cp genomes. The trnK-UUU/rps16 has been successfully used to confirm the evolutionary relationship and complete Latin name changes of the Sium alliance within the Apiaceae tribe Oenantheae (Spalik et al., 2009). The phylogenetic analysis based on sequencing data from different molecular markers ndhF/rpl32 and rpl32/trnL-UAG is also separate in the families Aristidoideae and Eragrostideae, and in the genuses Fagopyrum and Dolomiaea, respectively (Shen et al., 2020; Fan et al., 2021; Kuan Liu et al., 2021; Guo et al., 2022). The ycf1 has been employed to assist in inferring the evolution of Pinus, Hoya, and Curcuma (Liang et al., 2020; Odago et al., 2021; Zeb et al., 2022). Those highly variable regions can be crucial molecular markers for closely related plants and contribute to generally phylogenetic relationship analysis research in the Acer genus.
Phylogenetic Analysis and Estimated Divergence Time
Based on the development of next-generation sequencing technologies, cp genomes have been widely used to clearly explain plant phylogenetic relationships and evolution (Burke et al., 2016; Du et al., 2017). The phylogenetic tree was constructed based on the maximum likelihood (ML) approach with high bootstrap support values. The BI tree reveals the evolutionary position of A. ukurunduense and its relationship with other Acer species. ML and BI trees are broadly similar in the evolutionary classification of Acer. The results showed that Acer, Macrantha, and Platanoidea are monophyly, similar to previous studies (Figures 9A,B) (Gao et al., 2017; Areces-Berazain et al., 2021). The genetic relationships of most species are very stable, which helps to further study the classification of Acer. However, we found that different analysis methods can lead to changes in the relationship between Sect. The relationship between Sect. Trifoliata and Sect. Pentaphylla is closer to Sect. Acer in the ML evolutionary tree, whereas in the BI tree, the three Sects. are all related. The cut-off value for the condensed tree in the ML tree is 50%. If the parameter is increased to 60%, the relationship results will be consistent with those in the BI tree. Li et al. (2019) further inferred that Sect. Trifoliata may have evolved from Sect. Pentaphylla and should be incorporated into Sect. Pentaphylla is based on the bioinformatics analysis of 500 nuclear sites. However, Xia et al. (2022) did not support merging Sect. Trifoliata into Sect. Pentaphyll. Therefore, we suggest that further studies on the genetic relationship between Sect. Trifoliata and Sect. Pentaphylla can be performed based on nDNA or even whole gene data. In addition, the previous study indicated A. ukurunduense was closely related to Sect. Palmata based on plastomes and nuclear sequences for Acer species (Areces-Berazain et al., 2021). Our results strongly supported the conclusion. Acer pictum subsp. mono is traditionally considered a sister to A. yangjuechi. Yu et al. (2020) proposed A. pictum subsp. mono and A. yangjuechi as sister species according to the “local varieties”. Compared with previous research results, this study, for the first time, confirmed the phylogenetic relationship of A. ukurunduense in Acer, and the phylogenetic relationship of other species was consistent with the findings of Xia et al. (2022). The comparison between the ML and BI trees is sufficient to confirm the close phylogenetic relationships of the Sect. Glabra, Sect. Platanoidea, and Sect. Macrantha. Sect. Acer, Sect. Pentaphylla, and Sect. Trifoliata were also closely related, which is consistent with the results of recent studies (Areces-Berazain et al., 2020). The divergence time of the Acer genus basically coincides with the previous study by Renner et al. (2008). Furthermore, our result is in conformity with the previous study that found Acer diversified at a constant rate during the Oligocene and that diversification may not have been affected by the climatic cooling (Areces-Berazain et al., 2021). This phylogenetic data and divergence time could be useful for resolving the phylogenetic evolutionary relationships of Aceraceae and for the rapid and accurate classification of valuable germplasm resources.
trnK-UUU/rps16 as a DNA Marker Used for Five Acer Species Identification
DNA molecular markers are short and relatively conserved fragment sequences in the plasmid sequence and distinguish species by stable mutation sites and insertion deletions. DNA molecular markers are widely used in traditional plant identification, accurate identification of medicinal plants, and invasive alien species. The hypervariable regions can be used as potential molecular genetic markers for identifying and evolving closely related species (Turktas, 2012; Park et al., 2017; Li et al., 2018; Chen et al., 2019). Through the previous analysis of ten sequences with highly variable regions, we preliminarily inferred that only one highly variable region (trnK-UUU/rps16) could be used to distinguish five species. This was proved by a sequencing comparison of trnK-UUU/rps16 amplified fragments; the result fully verified the accuracy of the primers developed based on the trnK-UUU/rps16 high variable region in identifying and providing a new method for discriminating Acer species. This method can also identify new and unique identification markers for other species and related species.
Conclusion
In the present study, the cp genome of A. ukurunduense was investigated. By comparing and analyzing cp genome sequences of A. ukurunduense with those of other four species of Acer distributed in the forests of the Xiaoxing’an Mountains, China, our analyses reveal that A. ukurunduense was similar in gene size, content, and order, which has a typical quadripartite structure of most land angiosperm cp genomes. SSR analysis can exploit valuable information to develop a higher rate of mutation DNA markers for genetic diversity surveys, distinguish different germplasms, and conduct other ecological and evolutionary studies of A. ukurunduense. The contraction and expansion that occurred in the boundary regions indicated the size variation of the genome compared to the selected Aceraceae species. Moreover, phylogenetic evolution of the 105 cp genomes in Acer strongly supports the close relationship between different species. Additionally, experimental verification of the newly developed molecular marker shows that the trnK-UUU/rps16 molecular marker can accurately identify five Acer species, which is of great significance to the taxonomic study of Acer species. These conclusions give valuable information for developing phylogenetic and genetic tools for further research on Aceraceae plants.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Formal analysis was performed by WR and CL; drafting the manuscript and preparation of figures and tables were performed by WR, CL, SY, and ZW; revision and manuscript editing were performed by ZJ, TW, MZ, ML, and JS; conceptualization and supervision were performed by WM; resources and funding acquisition were provided by HG. All the authors read and approved the submission of the final manuscript.
Funding
This work was supported by grants from the Key Project at the central government level, the ability to establish sustainable use for valuable Chinese medicine resources (2060302), Open Fund of State Key Laboratory of New-tech for Chinese Medicine Pharmaceutical Process (SKL 2020M0302), and Heilongjiang Touyan Innovation Team Program (HLJTYTP2019001).
Conflict of Interest
WM was employed by Jiangsu Kanion Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Authors sincerely thank Yuan Jiang for providing helpful suggestions and support related to the analysis performed. Authors also thank Prof. Aihua Zhang for her useful comments and precious remarks on the article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.849182/full#supplementary-material
Supplementary Figure S1 | Technology roadmap.
Supplementary Figure S2 | Divergence time analysis based on the chloroplast genome.
Supplementary Figure S3 | Agarose gel electrophoresis of trnK-UUU/rps16 fragments amplified in five species.
Supplementary Table S1 | NCBI accession number of the sequence used in phylogenetic analysis.
Supplementary Table S2 | Pair of primers for amplifying DNA barcodes.
Supplementary Table S3 | Location and length of cp genes containing introns in A. ukurunduense.
Supplementary Table S4 | Codon usage of five Acer species’ cp genomes.
Supplementary Table S5 | Simple sequence repeats of five Acer species’ cp genomes.
Supplementary Table S6 | Nucleotide diversity analysis data.
References
Amiryousefi, A., Hyvönen, J., Poczai, P., and Hancock, J. (2018). IRscope: an Online Program to Visualize the Junction Sites of Chloroplast Genomes. Bioinformatics 34, 3030–3031. doi:10.1093/bioinformatics/bty220
Areces-Berazain, F., Wang, Y., Hinsinger, D. D., and Strijk, J. S. (2020). Plastome Comparative Genomics in Maples Resolves the Infrageneric Backbone Relationships. PeerJ 8, e9483. doi:10.7717/peerj.9483
Areces-Berazain, F., Hinsinger, D. D., and Strijk, J. S. (2021). Genome-wide Supermatrix Analyses of Maples (Acer, Sapindaceae) Reveal Recurring Inter-continental Migration, Mass Extinction, and Rapid Lineage Divergence. Genomics 113, 681–692. doi:10.1016/j.ygeno.2021.01.014
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: a Web Server for Microsatellite Prediction. Bioinformatics 33, 2583–2585. doi:10.1093/bioinformatics/btx198
Bi, W., Gao, Y., Shen, J., He, C., Liu, H., Peng, Y., et al. (2016). Traditional Uses, Phytochemistry, and Pharmacology of the Genus Acer (Maple): A Review. J. Ethnopharmacol. 189, 31–60. doi:10.1016/j.jep.2016.04.021
Blazier, J. C., Jansen, R. K., Mower, J. P., Govindu, M., Zhang, J., Weng, M.-L., et al. (2016). Variable Presence of the Inverted Repeat and Plastome Stability inErodium. Ann. Bot. 117, 1209–1220. doi:10.1093/aob/mcw065
Brudno, M., Malde, S., Poliakov, A., Do, C. B., Couronne, O., Dubchak, I., et al. (2003). Glocal Alignment: Finding Rearrangements during Alignment. Bioinformatics 19 (Suppl. 1), i54–i62. doi:10.1093/bioinformatics/btg1005
Burke, S. V., Lin, C.-S., Wysocki, W. P., Clark, L. G., and Duvall, M. R. (2016). Phylogenomics and Plastome Evolution of Tropical Forest Grasses (Leptaspis, Streptochaeta: Poaceae). Front. Plant Sci. 7, 1993. doi:10.3389/fpls.2016.01993
Chen, Q., Wu, X., and Zhang, D. (2019). Phylogenetic Analysis of Fritillaria Cirrhosa D. Don and its Closely Related Species Based on Complete Chloroplast Genomes. PeerJ 7, e7480. doi:10.7717/peerj.7480
Chen, S. L., Lee, W., Hottes, A. K., Shapiro, L., and McAdams, H. H. (2004). Codon Usage between Genomes Is Constrained by Genome-wide Mutational Processes. Proc. Natl. Acad. Sci. U.S.A. 101, 3480–3485. doi:10.1073/pnas.0307827100
Cui, Y., Nie, L., Sun, W., Xu, Z., Wang, Y., Yu, J., et al. (2019). Comparative and Phylogenetic Analyses of Ginger (Zingiber Officinale) in the Family Zingiberaceae Based on the Complete Chloroplast Genome. Plants 8, 283. doi:10.3390/plants8080283
Daniell, H., Lin, C.-S., Yu, M., and Chang, W.-J. (2016). Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 17, 134. doi:10.1186/s13059-016-1004-2
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 14, 1394–1403. doi:10.1101/gr.2289704
Dong, P.-B., Wang, R.-N., Afzal, N., Liu, M.-L., Yue, M., Liu, J.-N., et al. (2021). Phylogenetic Relationships and Molecular Evolution of Woody Forest Tree Family Aceraceae Based on Plastid Phylogenomics and Nuclear Gene Variations. Genomics 113, 2365–2376. doi:10.1016/j.ygeno.2021.03.037
Dormontt, E. E., Jardine, D. I., van Dijk, K.-J., Dunker, B. F., Dixon, R. R. M., Hipkins, V. D., et al. (2020). Forensic Validation of a SNP and INDEL Panel for Individualisation of Timber from Bigleaf Maple (Acer macrophyllum Pursch). Forensic Sci. Int. Genet. 46, 102252. doi:10.1016/j.fsigen.2020.102252
Du, Y.-p., Bi, Y., Yang, F.-p., Zhang, M.-f., Chen, X.-q., Xue, J., et al. (2017). Complete Chloroplast Genome Sequences of Lilium: Insights into Evolutionary Dynamics and Phylogenetic Analyses. Sci. Rep. 7, 5751. doi:10.1038/s41598-017-06210-2
Dugas, D. V., Hernandez, D., Koenen, E. J. M., Schwarz, E., Straub, S., Hughes, C. E., et al. (2015). Mimosoid Legume Plastome Evolution: IR Expansion, Tandem Repeat Expansions and Accelerated Rate of Evolution in clpP. Sci. Rep. 5, 16958. doi:10.1038/srep16958
Duret, L. (2002). Evolution of Synonymous Codon Usage in Metazoans. Curr. Opin. Genet. Dev. 12, 640–649. doi:10.1016/S0959-437X(02)00353-2
Eid, J. (2009). Real-Time DNA Sequencing from Single Polymerase Molecules. Sci. (New York, N.Y.) 323, 133–138. doi:10.1126/science.1162986
Ellison, A., Buckley, H., Case, B., Cardenas, D., Duque, Á., Lutz, J., et al. (2019). Species Diversity Associated with Foundation Species in Temperate and Tropical Forests. Forests 10, 128. doi:10.3390/f10020128
Fan, Y., Jin, Y. n., Ding, M., Tang, Y., Cheng, J., Zhang, K., et al. (2021). The Complete Chloroplast Genome Sequences of Eight Fagopyrum Species: Insights into Genome Evolution and Phylogenetic Relationships. Front. Plant Sci. 12, 799904. doi:10.3389/fpls.2021.799904
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 32, W273–W279. doi:10.1093/nar/gkh458
Fu, Q., Yu, X., Xia, X., Zheng, Y., and Zhang, C. (2020). Complete Chloroplast Genome Sequence of Acer Nikoense (Sapindaceae). Mitochondrial DNA Part B 5, 3118–3119. doi:10.1080/23802359.2020.1797574
Gao, J., Liao, P.-C., Meng, W.-H., Du, F. K., and Li, J.-Q. (2017). Application of DNA Barcodes for Testing Hypotheses on the Role of Trait Conservatism and Adaptive Plasticity in Acer L. Section Palmata Pax (Sapindaceae). Braz. J. Bot. 40, 993–1005. doi:10.1007/s40415-017-0404-1
Gao, C., Deng, Y., and Wang, J. (2018). The Complete Chloroplast Genomes of Echinacanthus Species (Acanthaceae): Phylogenetic Relationships, Adaptive Evolution, and Screening of Molecular Markers. Front. Plant Sci. 9, 1989. doi:10.3389/fpls.2018.01989
Gao, J., Liao, P.-C., Huang, B.-H., Yu, T., Zhang, Y.-Y., and Li, J.-Q. (2020). Historical Biogeography of Acer L. (Sapindaceae): Genetic Evidence for Out-Of-Asia Hypothesis with Multiple Dispersals to North America and Europe. Sci. Rep. 10, 1–10. doi:10.1038/s41598-020-78145-0
Guo, X.-X., Qu, X.-J., Zhang, X.-J., and Fan, S.-J. (2022). Comparative and Phylogenetic Analysis of Complete Plastomes Among Aristidoideae Species (Poaceae). Biology 11, 63. doi:10.3390/biology11010063
Hou, H. Y., and Wang, L. H. (2013). Species Composition and Main Populations Spatial Distribution Pattern in Korean Pine Broadleaved Forest in Xiaoxing' an Mountains of Northeast China. Ying Yong Sheng Tai Xue Bao 24, 3043–3049. doi:10.13287/j.1001-9332.2013.0526
Hou, Y., Jin, C., An, R., Yin, X., Piao, Y., Yin, X., et al. (2019). A New Flavonoid from the Stem Bark of Acer Tegmentosum. Biochem. Syst. Ecol. 83, 1–3. doi:10.1016/j.bse.2018.12.006
Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., dePamphilis, C. W., Yi, T.-S., et al. (2020). GetOrganelle: a Fast and Versatile Toolkit for Accurate De Novo Assembly of Organelle Genomes. Genome Biol. 21, 241. doi:10.1186/s13059-020-02154-5
Jung, J., Kim, C., and Kim, J.-H. (2021). Insights into Phylogenetic Relationships and Genome Evolution of Subfamily Commelinoideae (Commelinaceae Mirb.) Inferred from Complete Chloroplast Genomes. BMC Genomics 22, 231. doi:10.1186/s12864-021-07541-1
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform 20, 1160–1166. doi:10.1093/bib/bbx108
Kausar, F., Farooqi, M.-A., Farooqi, H.-M. -U., Salih, A.-R. -C., Khalil, A.-A. -K., Kang, C.-w., et al. (2021). Phytochemical Investigation, Antimicrobial, Antioxidant and Anticancer Activities of Acer Cappadocicum Gled. Life 11, 656. doi:10.3390/life11070656
Kuan Liu, K., Wang, R., Guo, X.-X., Zhang, X.-J., Qu, X.-J., and Fan, S.-J. (2021). Comparative and Phylogenetic Analysis of Complete Chloroplast Genomes in Eragrostideae (Chloridoideae, Poaceae). Plants 10, 109. doi:10.3390/plants10010109
Kumar, S., Mu, X., Wang, P., Du, J., Gao, Y. G., and Zhang, J. (2018). The Chloroplast Genome of Cerasus Humilis: Genomic Characterization and Phylogenetic Analysis. Plos One 13, e0196473. doi:10.1371/journal.pone.0196473
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic acids Res. 29, 4633–4642. doi:10.1093/nar/29.22.4633
Li, B., Li, Y., Cai, Q., Lin, F., Huang, P., and Zheng, Y. (2016). Development of Chloroplast Genomic Resources for Akebia Quinata (Lardizabalaceae). Conserv. Genet. Resour. 8, 447–449. doi:10.1007/s12686-016-0593-0
Li, Y., Zhang, Z., Yang, J., and Lv, G. (2018). Complete Chloroplast Genome of Seven Fritillaria Species, Variable DNA Markers Identification and Phylogenetic Relationships within the Genus. PLoS One 13, e0194613. doi:10.1371/journal.pone.0194613
Li, J., Stukel, M., Bussies, P., Skinner, K., Lemmon, A. R., Lemmon, E. M., et al. (2019). Maple Phylogeny and Biogeography Inferred from Phylogenomic Data. Jnl Sytematics Evol. 57, 594–606. doi:10.1111/jse.12535
Liang, Q., Wang, W., Yuan, F., Liu, X., Li, D., and Yang, K. Q. (2019). Characterization of Yuanbaofeng (Acer Truncatum Bunge) Samaras: Oil, Fatty Acid, and Phytosterol Content. Industrial Crops Prod. 135, 344–351. doi:10.1016/j.indcrop.2019.04.032
Liang, H., Zhang, Y., Deng, J., Gao, G., Ding, C., Zhang, L., et al. (2020). The Complete Chloroplast Genome Sequences of 14 Curcuma Species: Insights into Genome Evolution and Phylogenetic Relationships within Zingiberales. Front. Genet. 11, 802. doi:10.3389/fgene.2020.00802
Löhne, C., and Borsch, T. (2005). Molecular Evolution and Phylogenetic Utility of the petD Group II Intron: a Case Study in Basal Angiosperms. Mol. Biol. Evol. 22, 317–332. doi:10.1093/molbev/msi019
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 37, 1530–1534. doi:10.1093/molbev/msaa015
Moore, M. J., Soltis, P. S., Bell, C. D., Burleigh, J. G., and Soltis, D. E. (2010). Phylogenetic Analysis of 83 Plastid Genes Further Resolves the Early Diversification of Eudicots. Proc. Natl. Acad. Sci. U.S.A. 107, 4623–4628. doi:10.1073/pnas.0907801107
Odago, W. O., Waswa, E. N., Nanjala, C., Mutinda, E. S., Wanga, V. O., Mkala, E. M., et al. (2021). Analysis of the Complete Plastomes of 31 Species of Hoya Group: Insights into Their Comparative Genomics and Phylogenetic Relationships. Front. Plant Sci. 12, 814833. doi:10.3389/fpls.2021.814833
Palmer, J. D. (1985). Comparative Organization of Chloroplast Genomes. Annu. Rev. Genet. 19, 325–354. doi:10.1146/annurev.ge.19.120185.001545
Park, I., Kim, W., Yeo, S.-M., Choi, G., Kang, Y.-M., Piao, R., et al. (2017). The Complete Chloroplast Genome Sequences of Fritillaria Ussuriensis Maxim. And Fritillaria Cirrhosa D. Don, and Comparative Analysis with Other Fritillaria Species. Molecules 22, 982. doi:10.3390/molecules22060982
Piao, Y., Zhang, C., Ni, J., Yin, X., An, R., and Jin, L. (2020). Chemical Constituents from the Stem Bark of Acer Tegmentosum. Biochem. Syst. Ecol. 89, 103982. doi:10.1016/j.bse.2019.103982
Plotkin, J. B., and Kudla, G. (2011). Synonymous but Not the Same: the Causes and Consequences of Codon Bias. Nat. Rev. Genet. 12, 32–42. doi:10.1038/nrg2899
Porebski, S., Bailey, L. G., and Baum, B. R. (1997). Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Mol. Biol. Rep. 15, 8–15. doi:10.1007/BF02772108
Qiao, X., Zhang, J., Xu, Y., Mi, X., Cao, M., Ye, W., et al. (2020). Foundation Species across a Latitudinal Gradient in Chnia. Ecol. Ecol. Soc. Am. 102, e03234. doi:10.1101/2020.03.15.986182
Rabah, S. O., Shrestha, B., Hajrah, N. H., Sabir, M. J., Alharby, H. F., Sabir, M. J., et al. (2019). Passifloraplastome Sequencing Reveals Widespread Genomic Rearrangements. Jnl Sytematics Evol. 57, 1–14. doi:10.1111/jse.12425
Ren, H., and Feng, Z. (2018). Comparison and Correlation Analysis of Flavonoids and Chlorogenic Acid Contents in Different Strains of Acer Truncatum. Food Sci. Technol. 38, 319–326. doi:10.1590/fst.23317
Renner, S. S., Grimm, G. W., Schneeweiss, G. M., Stuessy, T. F., and Ricklefs, R. E. (2008). Rooting and Dating Maples (Acer) with an Uncorrelated-Rates Molecular Clock: Implications for North American/Asian Disjunctions. Syst. Biol. 57, 795–808. doi:10.1080/10635150802422282
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 34, 3299–3302. doi:10.1093/molbev/msx248
Saeki, I., and Murakami, N. (2009). Chloroplast DNA Phylogeography of the Endangered Japanese Red Maple (Acer Pycnanthum): the Spatial Configuration of Wetlands Shapes Genetic Diversity. Divers. Distributions 15, 917–927. doi:10.1111/j.1472-4642.2009.00609.x
Schmidt, S. B., Eisenhut, M., and Schneider, A. (2020). Chloroplast Transition Metal Regulation for Efficient Photosynthesis. Trends Plant Sci. 25, 817–828. doi:10.1016/j.tplants.2020.03.003
Shahzadi, I., Abdullah, Mehmood, F., Ali, Z., Ahmed, I., and Mirza, B. (2020). Chloroplast Genome Sequences of Artemisia Maritima and Artemisia Absinthium: Comparative Analyses, Mutational Hotspots in Genus Artemisia and Phylogeny in Family Asteraceae. Genomics 112, 1454–1463. doi:10.1016/j.ygeno.2019.08.016
Shen, J., Zhang, X., Landis, J. B., Zhang, H., Deng, T., Sun, H., et al. (2020). Plastome Evolution in Dolomiaea (Asteraceae, Cardueae) Using Phylogenomic and Comparative Analyses. Front. Plant Sci. 11, 376. doi:10.3389/fpls.2020.00376
Shi, C., Liu, Y., Huang, H., Xia, E.-H., Zhang, H.-B., and Gao, L.-Z. (2013). Contradiction between Plastid Gene Transcription and Function Due to Complex Posttranscriptional Splicing: an Exemplary Study of Ycf15 Function and Evolution in Angiosperms. PLoS One 8, e59620. doi:10.1371/journal.pone.0059620
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 47, W65–W73. doi:10.1093/nar/gkz345
Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., et al. (1986). The Complete Nucleotide Sequence of the Tobacco Chloroplast Genome: its Gene Organization and Expression. EMBO J. 5, 2043–2049. doi:10.1002/j.1460-2075.1986.tb04464.x
Singh, N. D., Ding, Y., and Daniell, H. (2009). Chloroplast-derived Vaccine Antigens and Biopharmaceuticals: Protocols for Expression, Purification, or Oral Delivery and Functional Evaluation. Methods Mol. Biol. 483, 163–192. doi:10.1007/978-1-59745-407-0_10
Spalik, K., Downie, S. R., and Watson, M. F. (2009). Generic Delimitations within the Sium Alliance (Apiaceae Tribe Oenantheae) Inferred from cpDNA Rps16 -5′trnK (UUU) and nrDNA ITS Sequences. Taxon 58, 735–748. doi:10.1002/tax.583004
Stace, C., and Hind, N. (1997). Maples of the World; Dandelions of Great Britain and Ireland. Botanical J. Linn. Soc. 125, 367–368. Oxford University Press. doi:10.1111/j.1095-8339.1997.tb02266.x
Tang, W., Wang, J.-j., Xu, J.-x., Wang, L., Huang, J., and Chen, Y. (2012). Advances of Chemical Composition of Medicinal Plants in Aceraceae. North. Hortic. 36, 194–200. doi:10.1148/radiographics.7.5.3331209
Tian, X., Guo, Z. H., and De-Zhu, L. I. (2002). Phylogeny of Aceraceae Based on ITS and Trn L-F Data Sets. Acta bot. sin. 44, 714–724. doi:10.1127/0340-269X/2002/0032-0317
Turktas, M., Aslay, M., Kaya, E., and Ertuğrul, F. (2012). Molecular Characterization of Phylogeneticrelationships in Fritillaria Species Inferred from Chloroplast trnL-trnF Sequences. Turk J. Biol. 36, 552–560. doi:10.3906/biy-1201-30
Wen, H. (2021). Characteristics and Diversity of Plant and Fungi Resources in Greater Khingan Mountains and Lesser Khingan Mountains. Harbin: Northeast Foreatry University.
Wicke, S., Schneeweiss, G. M., dePamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The Evolution of the Plastid Chromosome in Land Plants: Gene Content, Gene Order, Gene Function. Plant Mol. Biol. 76, 273–297. doi:10.1007/s11103-011-9762-4
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of Nucleotide Substitution Vary Greatly Among Plant Mitochondrial, Chloroplast, and Nuclear DNAs. Proc. Natl. Acad. Sci. U. S. A. 84, 9054–9058. doi:10.2307/30764
Wu, Z. Y., Raven, P. H., Hong, D. Y., Wu, Z. Y., Raven, P. H., and Hong, D. Y. (2008). Flora of China, Vol. 7. Beijing: Menispermaceae through Capparaceae.
Xia, X., Yu, X., Fu, Q., Zheng, Y., and Zhang, C. (2020). Complete Chloroplast Genome Sequence of the Three-Flowered Maple, Acer Triflorum (Sapindaceae). Mitochondrial DNA Part B 5, 1859–1860. doi:10.1080/23802359.2020.1751000
Xia, X., Yu, X., Fu, Q., Zhao, Y., Zheng, Y., Wu, Y., et al. (2022). Comparison of Chloroplast Genomes of Compound-Leaved Maples and Phylogenetic Inference with Other Acer Species. Tree Genet. Genomes 18. doi:10.1007/s11295-022-01541-2
Yang, L., Yin, P., Ho, C.-T., Yu, M., Sun, L., and Liu, Y. (2018). Effects of Thermal Treatments on 10 Major Phenolics and Their Antioxidant Contributions in Acer Truncatum Leaves and Flowers. R. Soc. open Sci. 5, 180364–364. doi:10.1098/rsos.180364
Yi Liu, Y., Yang, Q., and Zhao, F. (2021). Synonymous but Not Silent: the Codon Usage Code for Gene Expression and Protein Folding. Annu. Rev. Biochem. 90, 375–401. doi:10.1146/annurev-biochem-071320-112701
Yu, T., Gao, J., Huang, B.-H., Dayananda, B., Ma, W.-B., Zhang, Y.-Y., et al. (2020). Comparative Plastome Analyses and Phylogenetic Applications of the Acer Section Platanoidea. Forests 11, 462. doi:10.3390/f11040462
Zeb, U., Wang, X., AzizUllah, A., Fiaz, S., Khan, H., Ullah, S., et al. (2022). Comparative Genome Sequence and Phylogenetic Analysis of Chloroplast for Evolutionary Relationship Among Pinus Species. Saudi J. Biol. Sci. 29, 1618–1627. doi:10.1016/j.sjbs.2021.10.070
Zhao, J., Xu, Y., Xi, L., Yang, J., Chen, H., and Zhang, J. (2018). Characterization of the Chloroplast Genome Sequence of Acer Miaotaiense: Comparative and Phylogenetic Analyses. Molecules 23, 1740. doi:10.3390/molecules23071740
Zheng, S., Poczai, P., Hyvönen, J., Tang, J., and Amiryousefi, A. (2020). Chloroplot: an Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 11, 1123. doi:10.3389/fgene.2020.576124
Zhou, Z., Dang, Y., Zhou, M., Li, L., Yu, C.-h., Fu, J., et al. (2016). Codon Usage Is an Important Determinant of Gene Expression Levels Largely through its Effects on Transcription. Proc. Natl. Acad. Sci. U.S.A. 113, E6117–E6125. doi:10.1073/pnas.1606724113
Zhu, A., Guo, W., Gupta, S., Fan, W., and Mower, J. P. (2016). Evolutionary Dynamics of the Plastid Inverted Repeat: the Effects of Expansion, Contraction, and Loss on Substitution Rates. New Phytol. 209, 1747–1756. doi:10.1111/nph.13743
Keywords: Acer ukurunduense, Aceraceae, chloroplast genome, phylogenetic analysis, species identification
Citation: Ren W, Liu C, Yan S, Jiang Z, Wang T, Wang Z, Zhang M, Liu M, Sun J, Gao J and Ma W (2022) Structural Characterization of the Acer ukurunduense Chloroplast Genome Relative to Related Species in the Acer Genus. Front. Genet. 13:849182. doi: 10.3389/fgene.2022.849182
Received: 21 April 2022; Accepted: 09 June 2022;
Published: 14 July 2022.
Edited by:
Milind B. Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaReviewed by:
Anuj Kumar, Dalhousie University, CanadaJiaojun Yu, Huanggang Normal University, China
Sampath Perumal, University of Saskatchewan, Canada
Copyright © 2022 Ren, Liu, Yan, Jiang, Wang, Wang, Zhang, Liu, Sun, Gao and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhui Gao, emhsNzM3NjAzQDE2My5jb20=; Wei Ma, bWF3ZWlobGp1Y21AMTI2LmNvbQ==
†These authors have contributed equally to this work
 Weichao Ren
Weichao Ren Chi Liu2†
Chi Liu2† Zhen Wang
Zhen Wang Meiqi Liu
Meiqi Liu