- 1School of Pharmacy, Jiangxi University of Chinese Medicine, Nanchang, China
- 2Affiliated Hospital of Inner Mongolia Minzu University, Inner Mongolia Minzu University, Tongliao, China
Citrus fruit contains rich nutrients which is edible and of officinal value. Citrus flavanones are widely used in the treatment of cardiovascular and other diseases, and they are a foundational material of Chinese medicine. The chalcone-flavanone isomerase (CHI) plays a key role in flavanone synthesis. Therefore, we comprehensively analyzed CHI genes in Citrus species. Here, thirty CHI genes were identified for the first time in six Citrus species, which were divided into CHI and FAP groups. Evolutionary analysis showed that CHI gene members were highly conserved and were an ancient family. All CsCHI genes showed the highest expression level after the second physiological fruit-falling period in C. sinensis. CsCHI1 and CsCHI3 were highly expressed at 50 days after the flowering (DAF) stage in albedo. The expression of CsFAP2 and CsCHI3 genes at the 50 DAF stage was 16.5 and 24.3 times higher than that at the 220 DAF stage, respectively. The expression of CsCHI1, CsCHI3, and CsFAP2 genes in the peel was higher than that in the pulp, especially in common sweet orange. The CsCHI3 gene maintained a high expression level in the epicarp and juice sac at all periods. The members of CHIs interacted with chalcone synthase (CHS), flavonol synthase/flavanone 3-hydroxylase (FLS) and naringenin, and 2-oxoglutarate 3-dioxygenase (F3H) to form heterodimers, which might together play a regulatory role and participate in the flavonoid pathway. This study will provide the basis for the selection of flavonoids in plant tissues and periods and fundamental information for further functional studies.
Introduction
The Citrus fruit belongs to the Rutaceae family and is one of the most widely cultivated fruit crops worldwide (Wu et al., 2018). Many researchers believe that citrus originated in Southeast Asia and began to be cultivated 4,000 years ago (Scora, 1975; Gmitter and Hu, 1990; Xu et al., 2013). Globally, the annual output of citrus fruit is more than 120 million tons (FAO statistics, see URLs), which is not only a nutritional source for human health but also rich in medicinal ingredients (Tocmo et al., 2020; Zhao et al., 2020). Vitamin C, as an important part of human nutrition, mainly comes from Citrus fruits (Xu et al., 2013). Moreover, phytochemical studies report that the Citrus plant has constituents including flavonoids, limonoids, and carotenoids (Zibaee et al., 2020; Addi et al., 2022). Citrus extracts were widely used in the treatment of cardiovascular, gastrointestinal, and other diseases and have anti-oxidant, anti-inflammatory, and nerve-protective effects (Szczepaniak et al., 2020; Zibaee et al., 2020; Rao et al., 2021).
Flavonoids are a kind of important secondary metabolites in plants, mainly in the form of glycosides (Winkel-Shirley, 2001; Kumar and Pandey, 2013). Phe and malonyl-coenzyme A form flavonoids through the fatty acid pathway and flavonoids constitute a diversified aromatic molecular family (Winkel-Shirley, 2001). The characteristic of fruit flavanones is that a disaccharidic moiety is connected to the 7 position of aglycone. Narirutin and naringin in grapefruit, hesperidin and narirutin in orange, and eriocitrin in lemon are the most representative flavanones (Peterson et al., 2006; Chanet et al., 2012; Kumar and Pandey, 2013). Citrus flavanones positively influence the cardio-metabolic system and prevent cardiovascular disease (Dauchet et al., 2005; Dauchet et al., 2006; He et al., 2006). For example, the positive effects of Citrus flavanones on the cardiovascular system are mainly manifested in the reduction of endothelial dysfunction, improvement of vascular function, and lipid level reduction (He et al., 2006; Testai and Calderone, 2017). The beneficial mechanism of Citrus flavanones on the cardiovascular system is mainly manifested in the vasodilator activity, anti-ischemic activity, glucose tolerance, and anti-oxidant and anti-inflammatory actions (Testai and Calderone, 2017). In addition, flavanones also have other pharmacological properties, such as anti-aging and anti-tumor activities, anti-oxidation, and immunity regulation (Yin et al., 2019).
Many studies have shown that naringenin plays an important role in the synthesis of flavanones (Yin et al., 2019). Meanwhile, CHI is a key enzyme in the synthesis of naringenin by the isomerization of 4, 2′, 4′, 6′-tetrahydroxychalcone (Shirley et al., 1992). In 1986, the first CHI gene was successfully cloned in Pisum sativum L. (Mehdy and Lamb, 1987). So far, more than 3,000 nucleotide sequences have been registered on the National Center for Biotechnology Information (NCBI) GenBank, involving 290 species in 71 families (Yin et al., 2019). Among all these species, medicinal plants account for a large proportion, such as Glycyrrhiza uralensis, Ginkgo biloba L., and Mirabilis himalaica (Zhang et al., 2009; Zhao et al., 2012; Lan et al., 2016). Arabidopsis studies have shown the function of CHI as a unique enhancer in the flavone pathway (Jiang et al., 2015). The CHI gene promotes fruit yellowing in fresh-cut Chinese water-chestnut (He and Pan, 2017). In addition, the study also found that the expression of the CHI gene was positively correlated with flavonoid accumulation in plants (Wang et al., 2010; Guan et al., 2014; Guo et al., 2015).
At present, the regulation of flavonoids in the genus Citrus mainly focuses on the identification and function of key transcription factors and some enzymes. However, the systematic analysis of the CHI gene family in genus Citrus has not been reported. Recently, a type IV CHI gene was identified in Citrus reticulata cv. Suavissima, which enhances the accumulation of flavanones and flavones (Zhao et al., 2021). This study further supports the importance of comprehensively identifying and analyzing the CHI gene family in Citrus, including gene structure, molecular characterizations, molecular evolution, and genes expression patterns. To conclude, this study improves the understanding of Citrus CHI genes and provides a reference for selecting tissue and period for flavonoid extraction. This study will be of great significance for further understanding the mechanism of flavonoid synthesis in Citrus species.
Materials and Methods
Identification and Characterization of Putative CHI Proteins in Citrus
The Citrus genome and genome annotation files were obtained from the Citrus Pan-genome to Breeding Database (CPBD) (Xu et al., 2013; Wang X. et al., 2017; Wang et al., 2018; Huang et al., 2021), including Citrus clementina, Citrus grandis, Citrus reticulate, Citrus media, Citrus ichangensis, and Citrus sinensis. The Arabidopsis and rice genome and genome annotation files were downloaded from The Arabidopsis Information Resource (TAIR) and Rice Genome Annotation Project (RGAP), respectively (Kawahara et al., 2013; Berardini et al., 2015). Selaginella moellendorffii and Physcomitrium patens genome files were obtained from Phytozome (Rensing Stefan et al., 2008; Banks et al., 2011; Goodstein et al., 2012).
The chalcone domain proteins were identified from six Citrus species using the HMMER software according to the chalcone domain, with a threshold of e-value < e−5 (Johnson et al., 2010). The domain of chalcone-flavanone isomerase (CHI) proteins (PF02431) was downloaded from the Pfam database and confirmed by the Swiss-Prot database (El-Gebali et al., 2019; UniProt, 2019). We validated candidate protein sequences again using the SMART database, and removed protein sequences with obvious errors, of length smaller than 150 aa, and/or ≥95% identity (Sun et al., 2015; Letunic and Bork, 2018). The molecular weight (MW) and isoelectric point (pI) of chalcone domain proteins were calculated by using the ExPASy online tool (Bjellqvist et al., 1993). The subcellular localization of chalcone domain proteins was predicted by using the Protein Subcellular Localization Prediction Tool (PSORT) (Peabody et al., 2020).
Gene Structure and Motif Analyses
The sequences and annotation information of CHI genes were obtained from the genome database. We identified the gene structures, including exon, intron, and 3′ UTR and 5′ UTR regions, according to the genome annotation file using the TBtool software (Chen et al., 2020). We retrieved motifs of CHI protein sequences using the Multiple Em for Motif Elicitation (MEME) tool with the following parameters: the motif width was set to 6–50, the motif number was set to 12, and any number of repetitions (Bailey et al., 2015).
Phylogenetic and Synteny Analyses
The phylogenetic trees were conducted according to neighbor-joining (NJ) and maximum likelihood (ML) methods, respectively (Saitou and Nei, 1987; Jones et al., 1992). To categorize CHIs, six referential A. thaliana CHIs were used: AtCHI1 (AT3G55120), AtCHI2 (AT5G66220), AtCHI3 (AT5G05270), AtFAP1 (AT5G66230), AtFAP2 (AT2G26310), and AtFAP3 (AT1G53520) (Berardini et al., 2015). The phylogenetic trees of the NJ method were conducted using MEGA 7.0 with the following parameters: 1,000 bootstrap resampling, pairwise deletion option, and the Jones–Taylor–Thornton (JTT) model. The phylogenetic trees of the ML method were conducted using MEGA 7.0 with the following parameters: 1,000 bootstrap resampling, complete deletion option, and the Jones–Taylor–Thornton (JTT) model (Kumar et al., 2016).
For the purpose of identifying the synteny of CHI genes, the genome sequence of C. grandis and C. sinensis was downloaded on a local server. First, we merged the genomic data corresponding to the two species. The protein sequences were aligned using the BLAST software with the following parameters: e-value ≤ 1e−5 and number threads = 10 (Ye et al., 2006). We analyzed the genome-wide synteny using the MCScanX software with alignment significance (E-value < 1e−5) (Wang et al., 2012). The gene pairs of synteny were extracted from the collinearity and tandem files. We visualized synteny gene pairs at the whole chromosome level using the R package circlize.
Expression Pattern Analysis of CHI Genes
RNA-seq data for C. sinensis were obtained from the NCBI GEO DataSets under accession numbers PRJNA689213 and PRJNA517400 (Feng et al., 2021; Huang et al., 2021). The transcriptome data of pulp and peel contained two types of sweet oranges (Valencia orange and common sweet orange), involving six varieties. The Valencia orange included “Rohde Red Valencia,” “Delta Valencia,” and “Cutter Valencia” oranges. The common sweet orange included “Xianfeng,” “Jincheng,” and “Taoye” oranges (Huang et al., 2021). Compared with the common sweet orange, Valencia orange belonged to late-ripening sweet oranges which had poor mastication traits (Wu et al., 2020). The transcriptome data of fruit development included four tissues (albedo, epicarp, juice sac, and segment membrane), involving four periods (the second physiological fruit-falling period, the expansion period, the coloring period, and the full-ripening period), which were divided into six time points (Feng et al., 2021). We extracted the expression of CHI genes and analyzed the expression pattern using the R package pheatmap.
To further verify the specific expression of genes, we detected the relative expression of CsCHI genes in fruit tissue by real-time quantitative PCR. The isolation of total RNA and the construction of the cDNA library were carried out using the TaKaRa kit (Code No. 9767 and Code No. RR047A). Specific primers of CsCHI genes for qRT-PCR were designed using Primer3Plus tools (Supplementary Table S1). We calculated the relative expression of CsCHI genes using the delta–delta CT method with the actin gene from sweet orange as the reference gene.
CHI Genes Involved in Flavonoid Metabolism Analysis
Based on the genome sequence and annotation file, the protein sequence and annotation information of CHI proteins were extracted by TBtool software (Chen et al., 2020). We predicted protein interactions in the STRING database by homologous sequence alignment (Szklarczyk et al., 2017). Molecular regulatory pathways were analyzed by the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000). The chemical molecular structure of matter was visualized using the MolView tool (Smith, 1995).
Results
Characterization of Chalcone Domain Proteins in Citrus Species
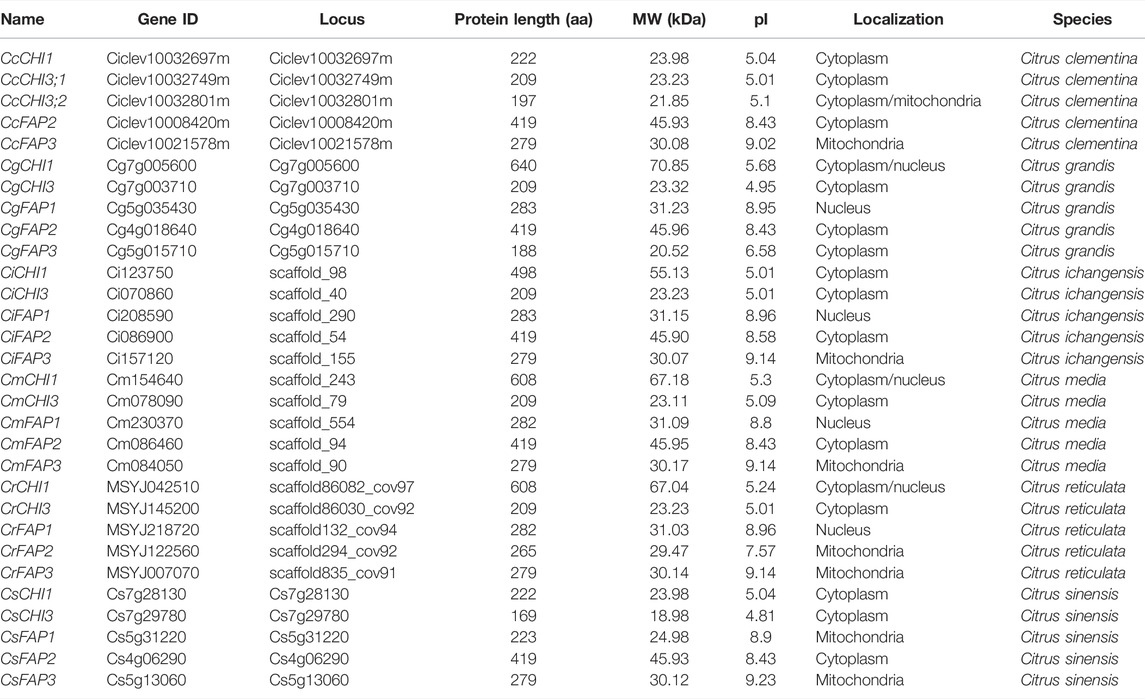
By combining BLAST and HMM searches, a total of 30 chalcone domain proteins were identified across six Citrus species (Table 1). Then, each putative protein was assigned to their closest Arabidopsis orthologous proteins and named (Supplementary Figure S1; Table 1). In total, this Citrus chalcone domain included 13 CHI and 17 fatty acid-binding protein (FAP) genes (Table 1). We obtained five CHI genes in each species, but C. clementina contained two CHI3 (CcCHI3;1 and CcCHI3;2) genes without FAP1 genes. The sequence lengths varied between 169 and 640 amino acids (aa), the isoelectric point (pI) ranged from 4.81 to 9.23, and the molecular weight (MW) varied from 18.98 to 70.85 kDa (Table 1). Subcellular localization prediction results showed that all CHI subfamily members were predicted to be targeted to the cytoplasm, whereas, CrCHI1, CmCHI1, and CgCHI1 were predicted to be located in the cytoplasm and nucleus, and CcCHI3; 2 was predicted to be located in the cytoplasm and mitochondria (Table 1).
We showed the relationships among the 30 CHI genes in the phylogenetic tree (Figure 1A). These proteins were clustered into five groups, which were similar to the groups of Arabidopsis CHI (Supplementary Figure S1). A cluster analysis again verified the differences between C. clementina CHI members and other species. The motif analysis showed that 12 conserved motifs were identified in 30 CHI/FAP proteins, and the length of the 12 motifs ranged from 21 to 50 aa (Figure 1B). Motifs 1, 3, and 5 together spread over the chalcone domain of CHI proteins (Figure 1B). Even though CHIs and FAPs had the chalcone domain, their amino acid sequences were not completely consistent. Although most of the FAP2 members shared the 11 conserved motifs, CrFAP2 lacked motifs 6, 10, and 12 (Figure 1B). We found that the members of each group had similar motif characteristics. For example, motifs 5, 3, 1, and 2 joined together and appeared in the CHI3 group, whereas motifs 9, 3, 1, 5, 4, and 2 joined together and appeared in the CHI1 group (Figure 1B). Citrus CHI genes had a rather loose gene structure, including introns ranging from 3 to 10 (Figure 1C). Except for CsCHI1 carrying three introns, most of the CHI1 genes had six introns in their genomic DNA. In the FAP2, all the genes possessed numerous introns, with 10 or 11 introns (Figure 1C). Combined with the phylogenetic tree, it was found that the CHI genes closely related to evolution had similar exon and intron structures in terms of intron number, location, and exon length (Figure 1).
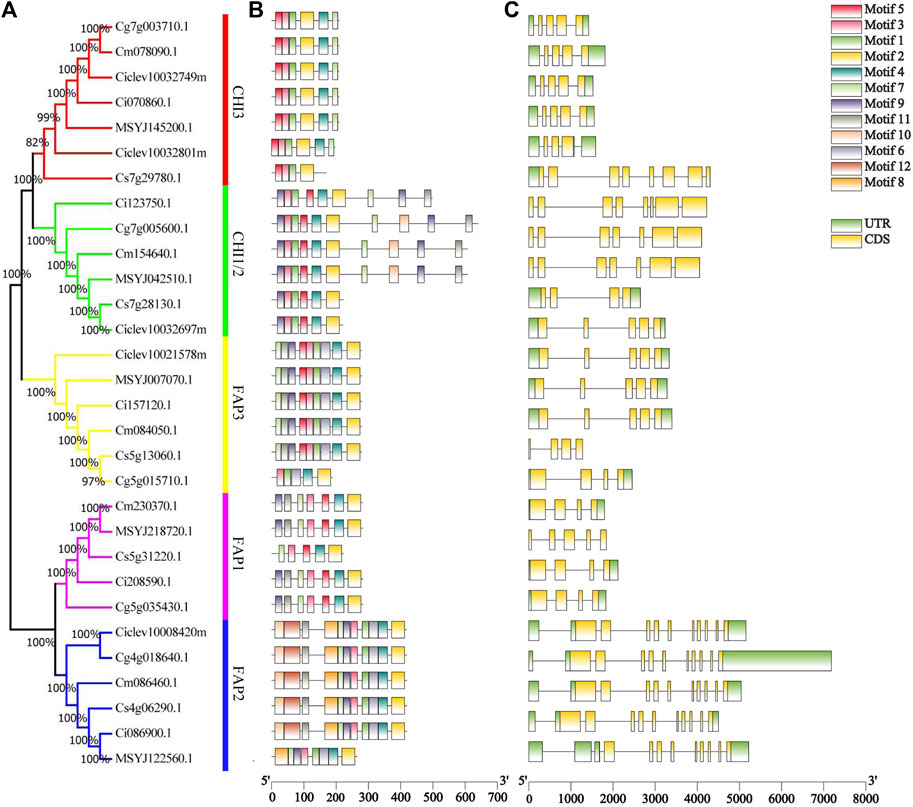
FIGURE 1. Characterization of 30 Citrus chalcone domain proteins. (A) Phylogenetic tree of 30 chalcone domain protein sequences. (B) Conserved motifs of chalcone domain proteins. Different colored boxes represent different motifs. (C) Gene structures of chalcone domain protein genes. Grey lines represent introns, yellow boxes represent exons, and green boxes represent UTR.
Phylogenetic Analysis of CHI Genes
The evolutionary relationships of CHI genes were further explored by phylogenetic and syntenic analyses. Fifty two CHI genes were obtained in 10 species, including seven dicotyledons (C. clementina, C. grandis, C. reticulata, C. media, C. reticulata, C. sinensis, and A. thaliana), monocotyledons (O. sativa), lycophyte (S. moellendorffii), and moss (P. patens) (Figure 2). Here, we identified four, four, and seven CHI genes in P. patens, S. moellendorffii, and O. sativa, respectively. The phylogenetic tree showed five clades, including CHI1/2, CHI3, FAP1, FAP2, and FAP3, whereas, the phylogenetic tree of seven dicotyledons strongly supported these subclades (Supplementary Figure S1). Four S. moellendorffii CHI genes were distributed to clades CHI1/2, CHI3, FAP2, and FAP3, respectively (Figure 2). Four P. patens CHI genes were distributed to clades CHI3, FAP2, and FAP3, respectively (Figure 2). The CHI gene might retain more ancient genetic information in plant evolution. We found that the CHI genes of angiosperms had a closer relationship in the subclades. Among them, the CHI genes of Citrus species were more closely related to A. thaliana than O. sativa. In general, CHI genes were a good gene resource for studying plant evolution. Although each plum plant contains five CHI genes, the CcFAP1 gene was missing in C. clementina, but two CHI3 genes (CcCHI3;1 and CcCHI3;2) were added in the CHI3 clades, which might be related to the expansion/contraction event of the gene family in the process of C. clementina evolution.
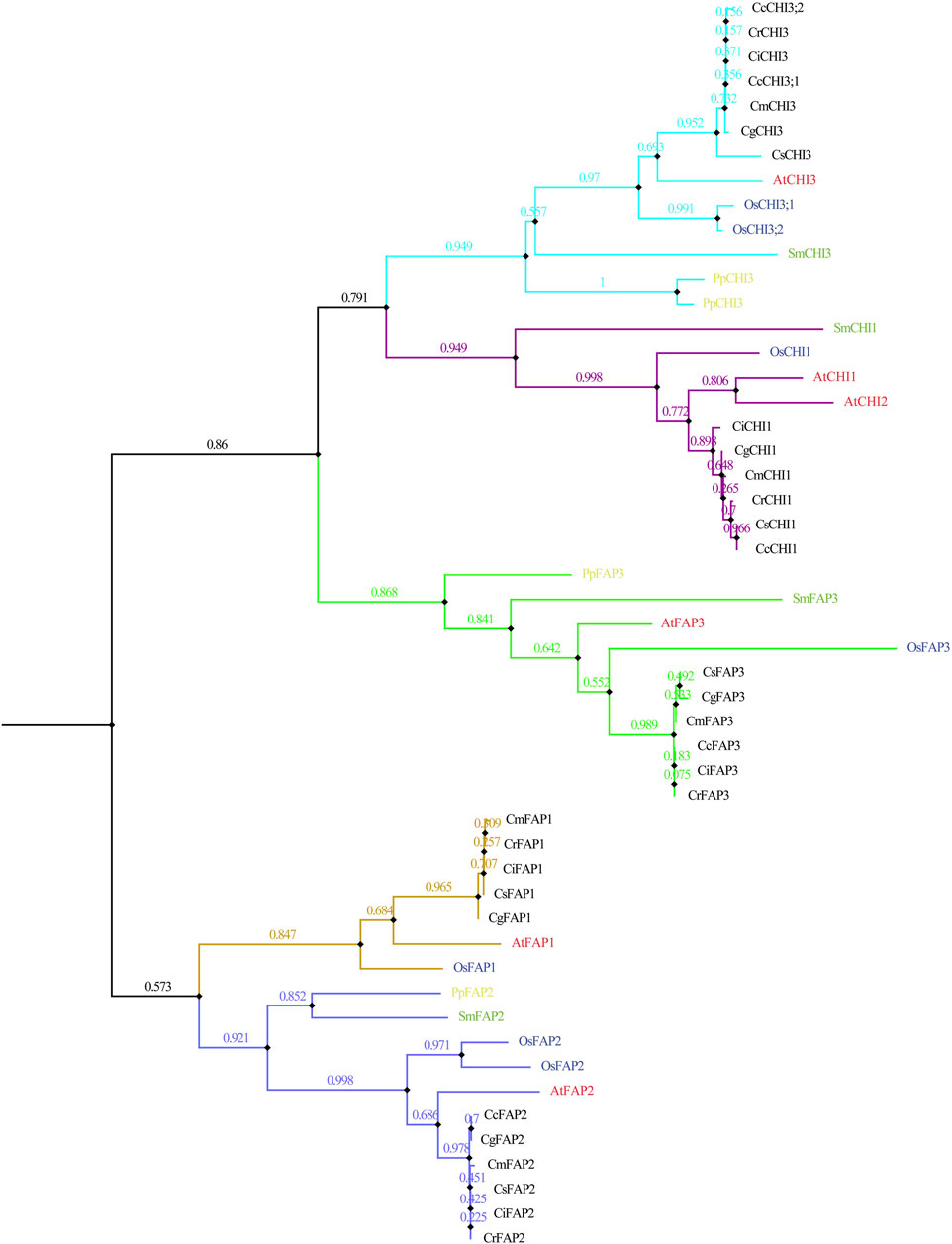
FIGURE 2. Phylogeny of representative CHI genes from the 10 plant species based on the protein sequences. Sm represents S. moellendorffii, Pp represents P. patens, Os represents O. sativa, At represents A. thaliana, Cc represents C. clementina, Cg represents C. grandis, Cr represents C. reticulata, Cm represents C. media, Cr represents C. reticulata, and Cs represents C. sinensis.
Synteny Analysis
Based on the genome at the chromosome level of C. grandis and C. sinensis, we carried out a syntenic analysis of the CHI gene. CgCHI and CsCHI genes were located on chromosomes 4, 5, and 7, respectively. The collinear blocks 11, 22, and 18 containing 4,251 gene pairs were identified on chromosomes 4, 5, and 7, respectively (Figure 3A, Supplementary Table S2). The distribution of the syntenic genes across chromosomes showed that there was an obvious correlation between the chromosomes (Cg4g vs. Cs4g, Cg5g vs. Cs5g, and Cg7g vs. Cs7g). The checking gene collinearity within a genome showed that 80% of CHI genes (CsCHI3 and CgCHI3, CsFAP3 and CgFAP3, CsFAP2 and CgFAP2, and CsFAP1 and CgFAP1) were located in collinear blocks for C. grandis and C. sinensis (Supplementary Table S3). One tandem duplication was detected for CHI genes among C. grandis (CgFAP2 and CgFAP2t), which were located on chromosome 4 [(Figure 3B, (Supplementary Table S3)]. These results further proved the close relationship between C. grandis and C. sinensis.
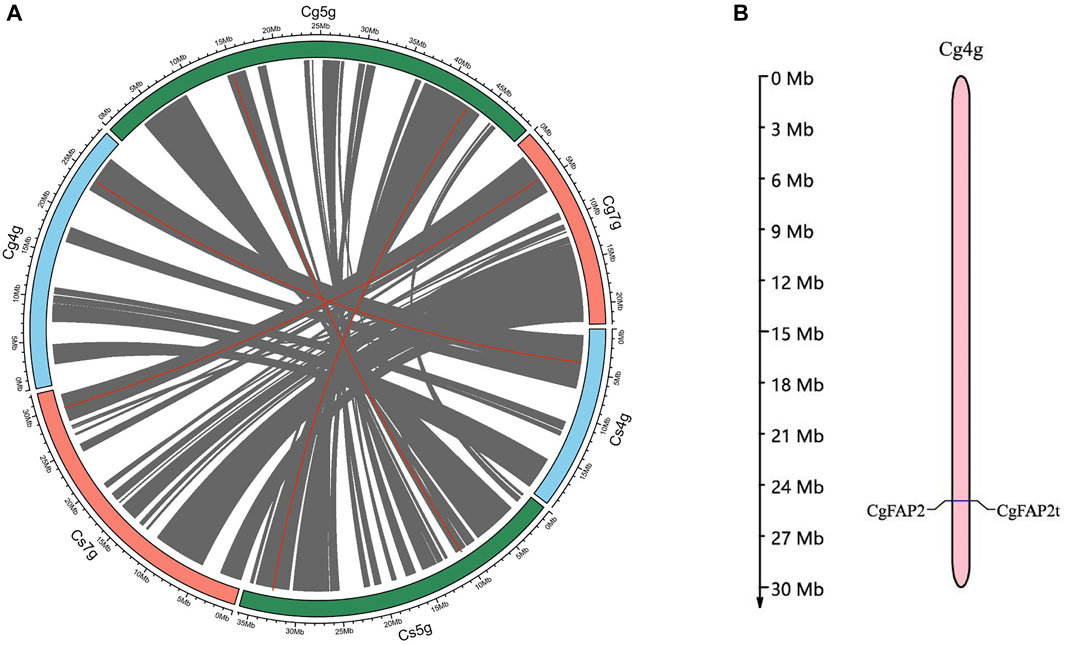
FIGURE 3. Syntenic analyses of CHI genes in C. grandis and C. sinensis. (A) Collinear genes of C. grandis and C. sinensis. Grey lines represent all collinear genes, red lines represent CHI genes between C. grandis and C. sinensis. (B)Tandem genes of C. grandis CgFAP2 genes. Expression pattern analysis of CHI genes during the fruit development
Expression Pattern Analysis of CHI Genes During the Fruit Development
We collected the expression profiles of C. sinensis (‘Fengjie 72-1′) albedo, epicarp, juice sac, and segment membrane from six fruit development stages, including 50 days after flowering (the second physiological fruit-falling period); 80, 120, and 155 days after flowering (the expansion period); 180 days after flowering (the coloring period), and 220 days after flowering (the full-ripening period) (Feng et al., 2021). The average expression of all CsCHI genes was the highest in the 50 DAF stage, then decreased rapidly, and increased slightly in the 155 DAF stage (Supplementary Table S4). As shown in Figure 4A, CsCHI1 and CsCHI3 were highly expressed at the 50 DAF stage in albedo. After that, with the continuous growth and development of fruits, the expression level dropped during the 80 to 220 DAF stage. Surprisingly, the expression level of the CsFAP2 gene was very high in the 50 DAF stage, which was 12.2 times higher than that in the 220 DAF stage. The CsFAP3 gene maintained a high expression level in the 80 to 180 DAF stage. As shown in Figure 4B, the expression of CHI genes was higher in the epicarp than in albedo. The expression of CsCHI1 was upregulated from the 155 DAF stage, and the expression of the CsFAP2 gene was relatively low at the 80 DAF stage. As shown in Figure 4C, all genes had the highest expression at the 50 DAF stage in the juice sac, and then the expression began to decline, except the CsFAP1 gene. The expression of the CsFAP2 gene at the 50 DAF stage was 16.5 times higher than that at the 220 DAF stage. The expressions of CsFAP2 and CsCHI3 genes at the 50 DAF stage were 16.5 and 24.3 times higher than that at the 220 DAF stage, respectively. As shown in Figure 4D, five CsCHI genes had the highest expression at the 50 DAF stage in the segment membrane, and then the expression began to decline. We found that the expression of the CsFAP1 gene increased slightly at the 155 and 180 DAF stages. The analysis of gene expression patterns showed that CsCHI genes played a major regulatory role at the 50 DAF stage.
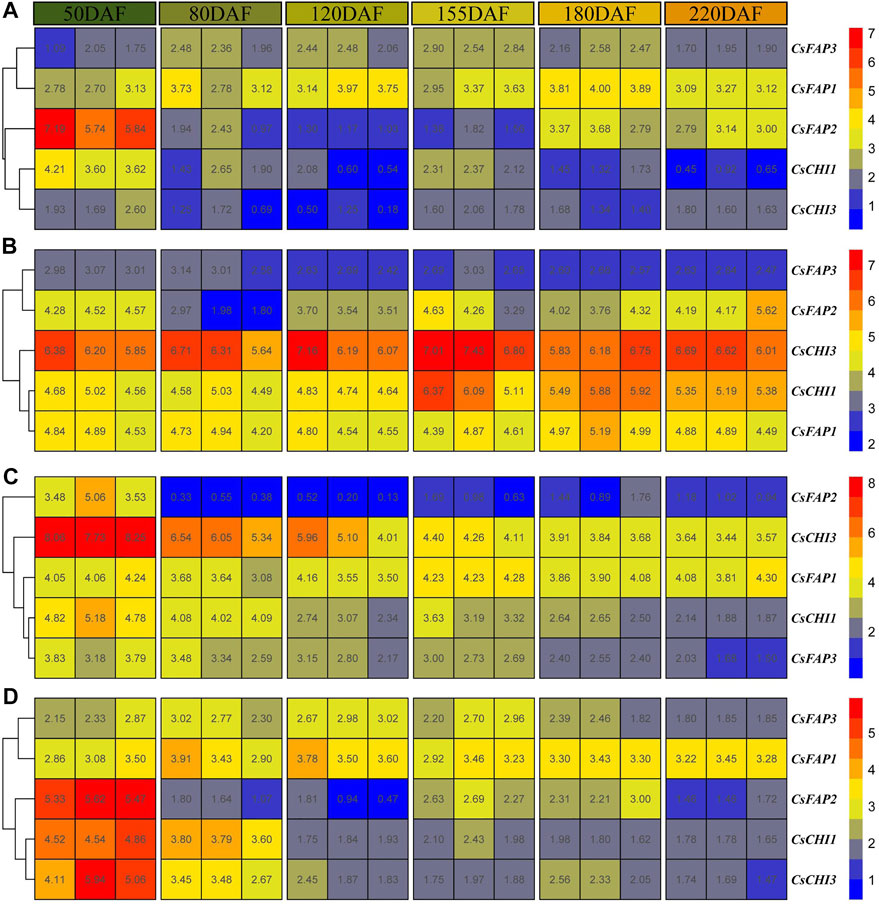
FIGURE 4. Expression pattern of CHI genes during the fruit development. (A–D) shows the expression patterns of CHI genes in the albedo, epicarp, juice sac, and segment membrane, respectively. 50, 80, 120, 155, 180, and 220 DAF represent 50, 80, 120, 155, 180, and 200 DAF, respectively.
Expression Pattern Analysis of CHI Genes in Different Tissues
To investigate the expression pattern of CsCHI genes, 36 samples were used for the expression patterns analysis, including peel and pulp from six varieties based on the available transcriptome data (Huang et al., 2021). The six varieties were selected from two categories: common sweet orange and Valencia orange. The Valencia orange ripened later than the common sweet orange and had a poor mastication trait (Wu et al., 2020). We found that five CsCHI genes showed specific expression in the pulp and peel of different varieties (Figure 5, Supplementary Table S5). On the whole, these genes showed higher expression levels in the peel, and the lowest expression levels in the common sweet orange pulp. CsCHI1 and CsFAP2 genes with similar expression patterns were clustered into the same subset and specifically expressed in the peel (Figure 5). Compared with pulp, the average expression of CsCHI1 and CsFAP2 genes were upregulated by 2.6 and 4.5 times in the peel, respectively. We found that the CsFAP1 gene was highly expressed in the peel and pulp, and only the CsFAP1 gene was downregulated in the peel, but the difference was not significant (Figure 5). These results showed that the CsFAP1 gene had no tissue-specific expression pattern in the peel and pulp. Interestingly, the CsFAP3 gene was more likely to be upregulated in the three Valencia orange varieties. Meanwhile, the expression of CsCHI1, CsFAP2 and CsFAP3 genes was hardly detected in the common sweet orange pulp (Figure 5).
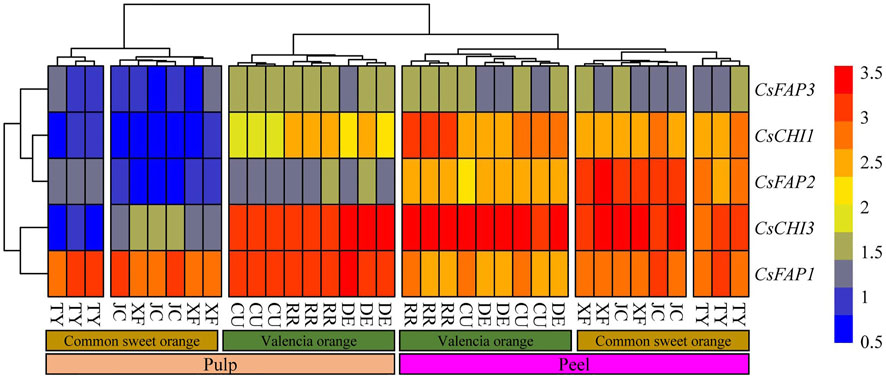
FIGURE 5. Expression pattern of CHI genes in pulp and peel. RR represents “Rohde Red Valencia,” DE represents “Delta Valencia,” CU represents “Cutter Valencia,”, XF represents “Xianfeng,” JC represents “Jincheng,” and TY represents “Taoye” oranges.
To further examine the expression level of CsCHI genes in different fruit tissues, we compared the expression levels of these genes in the albedo, epicarp, juice sac, and segment membrane (Figure 6; Supplementary Figure S2). We observed that CsCHI genes were mainly expressed in the epicarp and juice sac. Interestingly, the expression pattern of the CsCHI gene in the epicarp and juice sac showed a negative correlation during fruit development. The CsCHI3 gene was highly expressed in the epicarp and juice sac, followed by the segment membrane. The expression of the CsCHI3 gene was hardly detected in the albedo at any developmental stage of the fruit. CsCHI1 remained highly expressed after the expansion period in the epicarp. Meanwhile, the expression of CsCHI1 was detected in the albedo at the early stage of fruit development. The expression level of CsFAP3 was relatively low in four tissues, and there was no obvious tissue-specific expression pattern. CsFAP1 and CsFAP2 genes were mainly expressed in the epicarp, especially in the late stage of fruit development.
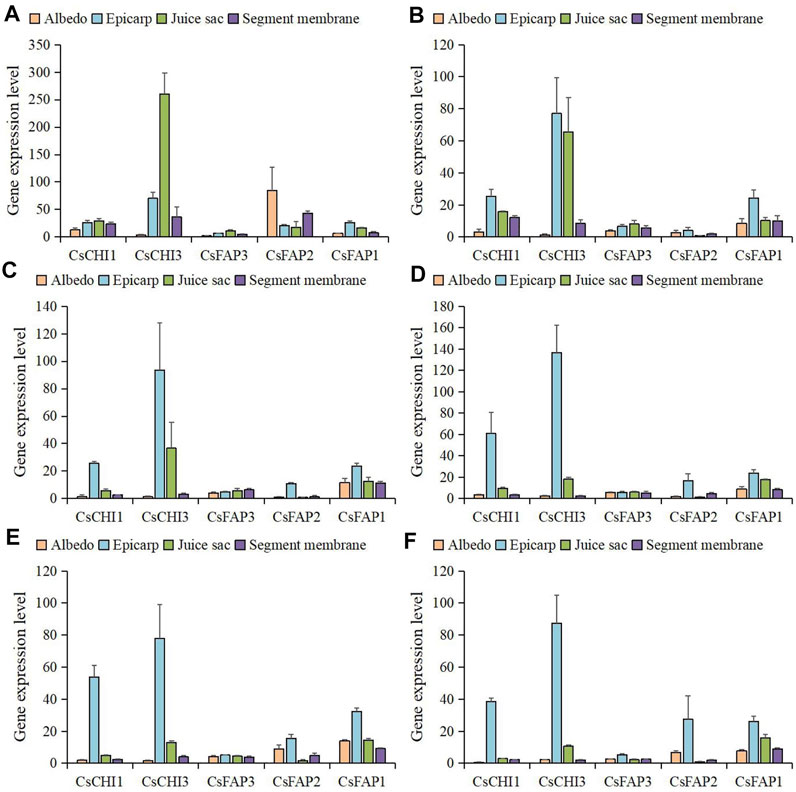
FIGURE 6. Expression level of CHI genes in albedo, epicarp, juice sac, and segment membrane. (A–F) shows the expression patterns of CHI genes at the 50, 80, 120, 155, 180, and 220 DAF stages.
Analysis of CHI Genes Involved in Flavonoid Metabolism
To understand the role of CHI genes in the anthocyanin synthesis pathway, we combined the KEGG and STRING databases and previous studies to analyze the molecular regulation mechanism of CHI genes. We found that five CHI genes had similar gene expression patterns, and there was direct interaction between their expressed proteins (Figure 7A, Supplementary Figure S3; Supplementary Table S6). These results indicated that the part members of CHI genes might play a regulatory role together and participated in the regulation of life activities. In addition, the CHIs and TTs, F3H, and FLSs also had strong interactions, and were the key proteins in this interaction network (Figure 7B). TT4, belonging to the chalcone and stilbene synthase family, encoded chalcone synthase (CHS). CHS could catalyze p-coumaroyl-CoA to form naringenin chalcone, which was a key enzyme involved in the biosynthesis of flavonoids. Naringenin chalcone further formed naringenin under the catalysis of CHI. Naringin formed apigenin and dihydrokaempferol under the action of flavonol synthase/flavanone 3-hydroxylase (FLS) and naringenin, 2-oxoglutarate 3-dioxygenase (F3H), respectively (Figure 7C; Supplementary Figure S4). FLS catalyzed the oxidation of both enantiomers of naringenin to give both cis- and trans-dihydrokaempferols. F3H catalyzed the 3-beta-hydroxylation of 2S-flavanones to 2R, 3R-dihydroflavonols which were intermediates in the biosynthesis of flavonols, anthocyanidins, catechins, and proanthocyanidins in plants. The results showed that CHIs played a key role in flavonoid metabolism and were an essential substrate for naringin synthesis.
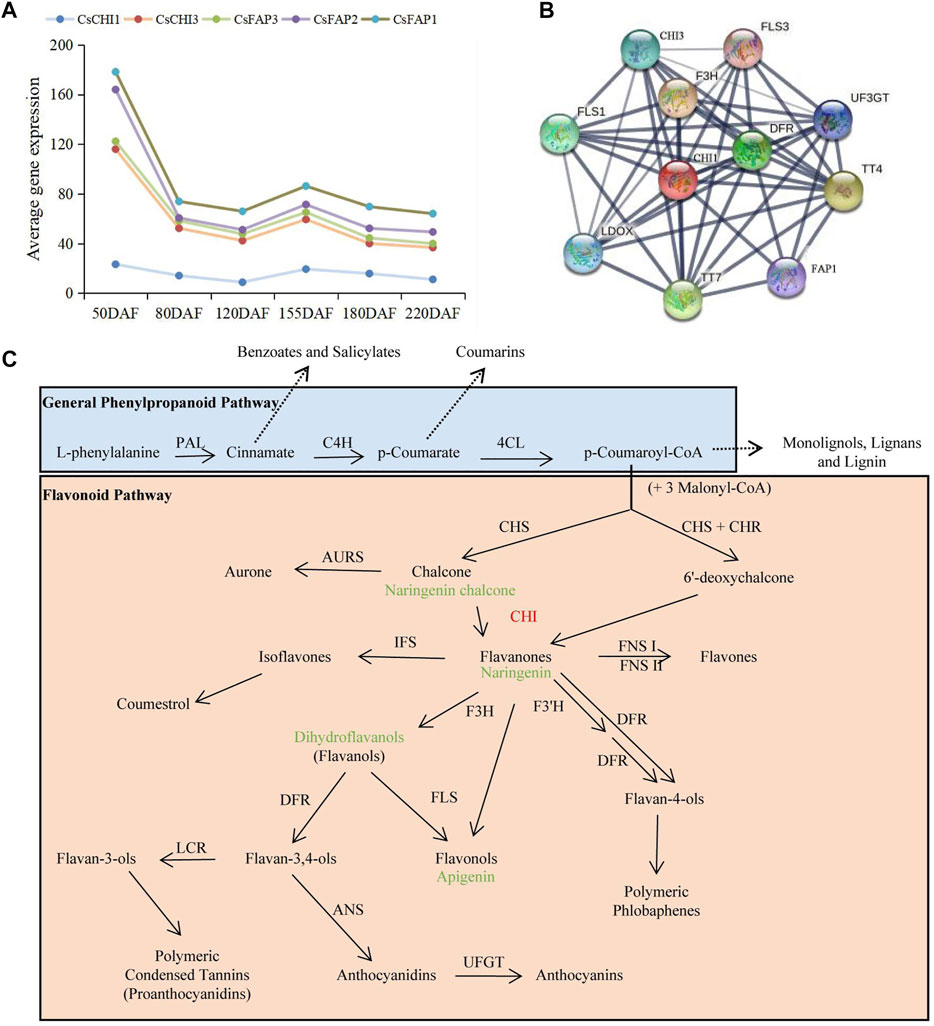
FIGURE 7. molecular regulation mechanism of CHI genes. (A) Expression trend of CHI genes during the fruit development. (B) Interaction networks of the CHI based on their ortholog proteins to A. thaliana. (C) Flavonoid biosynthetic pathway.
Discussion
Flavonoids are composed of many metabolites with different structures, which play a key role in plant growth and development and have important medicinal value (Moore et al., 2014; Testai and Calderone, 2017; Brunetti et al., 2018). Naringenin, an aglycone called naringenin according to its chemical structure, belongs to flavonoids (Szczepaniak et al., 2020). Naringenin produced by CHI is an important precursor of other flavonoids. The Arabidopsis genome contains six CHI proteins with the chalcone domain, which are AtCHI1 (AT3G55120), AtCHI2 (AT5G66220), AtCHI3 (AT5G05270), AtFAP1 (AT5G66230), AtFAP2 (AT2G26310), and AtFAP3 (AT1G53520) (UniProt, 2019). In our study, 30 CHI proteins were identified in six Citrus species, with five proteins in each species. The CHI2 protein homologous to Arabidopsis was missing in all Citrus species. AtCHI2 catalyzes the intramolecular cyclization of bicyclic chalcone to tricyclic (s)-flavanone, which has the same function as AtCHI1 (UniProt, 2019). AtCHI1 and AtCHI2 are considered to be two proteins, and the identity of the sequence is only 63.6%. However, the study of chalcone-flavanone isomerase protein genes in Arabidopsis shows that AtCHI2 is a pseudogene (Ngaki et al., 2012). The CHI genes of six Citrus plants showed that the pseudogene Citrus CHI2 could not be retained in the process of evolution.
In previous studies, CHI proteins are divided into four types (Type I, II, III, and IV) according to characteristics of the sequence structure (Zhao et al., 2021). Only the CHI proteins of types I and II have enzymatic cyclization activity (Ralston et al., 2005). The CHI of type III (FAP) is the prototype of other CHI-fold proteins, but it has a fatty acid-binding ability (Ngaki et al., 2012). The secondary structure of the CHI of type IV is similar to CHI of type I and II, while the key residues are substituted (Ngaki et al., 2012). The CHI protein of Arabidopsis contains three CHI and three FAP proteins (Ngaki et al., 2012). At the same time, the CHI proteins of six Citrus species are evenly divided into five subgroups according to the homologues of Arabidopsis. Each subgroup had a similar gene structure and conserved motif distribution. These results also further confirmed the functional differences of different types of CHI proteins.
CHI is obviously a lack of related proteins in the primary metabolism of the flavanone pathway (Jez et al., 2000; Jez et al., 2002), which makes the origin of CHI difficult to understand. In 2012, Micheline et al. showed that CHI originated from FAP3 through phylogenetic analysis (Ngaki et al., 2012). Similar results were also confirmed in our CHI phylogenetic tree. The chalcone-binding site of bona fide CHI and key catalytic residues lacked in CHI-like homologues of bacteria and fungi (Gensheimer and Mushegian, 2004). At the same time, some studies showed that CHI genes are restricted to vascular plants (Ngaki et al., 2012). The difference in the number of CHI genes between non-seed plants and Citrus may be the loss or increase of genes caused by species divergence events. The phylogenetic differentiation of CHI is not significant between seed and non-seed plants (P. patens and S. moellendorffii). The results show that CHI genes belong to an ancient family and the CHI gene study of soybean has the same conclusion (Ralston et al., 2005).
The level of gene expression directly affects the content of transcripts and the regulation of genes. GmCHI genes show root-specific expression in soybean and differential expression by nodulation signals (Ralston et al., 2005). In addition, LjCHI genes show differential expression under fungal elicitor treatment in Lasianthus japonicas (Shimada et al., 2003). The expression of the CitCHIL1 gene in flower tissue was higher than that in roots, stems, and leaves in Citrus reticulata cv. Suavissima (Zhao et al., 2021). This study showed that the expression of CsCHI1, CsCHI3, and CsFAP2 genes in peel was higher than that in pulp, especially in the common sweet orange. The CsCHI3 gene maintained a high expression level in the epicarp and juice sac at all periods. The developing tissue of Arabidopsis, including roots, seeds, embryos, cotyledons, tapetum, macrospores, preanthesis, and young seedlings, shows high expression of FAPs (Ngaki et al., 2012). The expression of the AtFAP2 gene can be detected in the whole life cycle. However, AtFAP1 and AtFAP3 genes are only expressed in developing and reproductive tissues. These genes have a maximal expression in seeds at 6 DAF (Ngaki et al., 2012). The expression of CitCHI1, CitCHIL1/2, and CitFAP1/3 is the highest at 30 DAF, while CitFAP2 reaches the peak at 120 DAF (Zhao et al., 2021). In this study, all CHI genes maintained a high expression level at 50 DAF, and then showed a down-expression trend during fruit development. The expression processing of CHI genes in plants is dynamic and characterized by spatio-temporal specificity.
In plants, CHI proteins play essential roles in flavonoid biosynthesis (Mehdy and Lamb, 1987; Winkel-Shirley, 2001; Gensheimer and Mushegian, 2004). The expression pattern of the CitCHIL1 gene was highly positively correlated with the accumulation of flavonoids, and was highly synchronized with the expression of CitCHI, CitCHS1, and CitCHS2 genes (Zhao et al., 2021). The content of flavonoids in the peel is higher than that in pulp (Wang Y. et al., 2017), which is consistent with the expression of CHI genes in this study. These results also mean that flavonoid content can be evaluated by detecting the expression of CHI and CHS genes. The beneficial mechanism of Citrus flavanone on the cardiovascular system is mainly manifested in vasodilator activity, anti-ischemic activity, glucose tolerance, and anti-oxidant and anti-inflammatory action (Testai and Calderone, 2017). In addition, flavanones also have other pharmacological properties, such as anti-aging and anti-tumor activities, anti-oxidation, and immunity regulation (Jung et al., 2003; Yamamoto et al., 2008; Testai, 2015; Da Pozzo et al., 2017; Yin et al., 2019). Flavanone compounds are unevenly distributed in fruits, mainly in the albedo and in the membranes separating cloves (Testai and Calderone, 2017). The albedo and membranous parts of Citrus fruits are usually discarded in the present processing and eating process. The rational use of Citrus resources can not only produce more valuable products for human beings, but also reduce environmental pollution.
Conclusion
In conclusion, we comprehensively analyzed the molecular characteristics, gene structure, evolutionary history, expression pattern, and molecular mechanism of CHI genes in six Citrus species. Thirty CHI genes were identified among six Citrus species. Citrus CHI gene members were highly conserved and are an ancient family. All CsCHI genes showed the highest expression level after the second physiological fruit-falling period. CsCHI1 and CsCHI3 were highly expressed at the 50 DAF stage in the albedo. The expression of CsCHI1, CsCHI3, and CsFAP2 genes in peel was higher than that in the pulp. The CsCHI3 gene maintained a high expression level in the epicarp and juice sac at all periods. The expression patterns of CsCHI genes were analyzed, which provided the basis for the selection of flavonoids in plant tissues and periods. Our study deepens the understanding of the structure and functions of CHIs and extends the knowledge on the transcriptional regulation of flavanones.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA689213; https://www.ncbi.nlm.nih.gov/, PRJNA517400.
Author Contributions
Conceptualization, QW and JZ; methodology, QW, TB, ML, and YL; formal analysis, QW and YX; investigation, TB, ML, and YL; writing—original draft preparation, QW and JZ; writing—review and editing, QW, TB, ML, YX, and TZ; supervision, JZ; and funding acquisition, QW. All authors approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China in 2020 (82060724) and the Natural Science Foundation of Inner Mongolia Autonomous Region in 2019 (2019MS08031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.848141/full#supplementary-material
References
Addi, M., Elbouzidi, A., Abid, M., Tungmunnithum, D., Elamrani, A., and Hano, C. (2022). An Overview of Bioactive Flavonoids from Citrus Fruits. Appl. Sci. 12 (1), 29. doi:10.3390/app12010029
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43 (W1), W39–W49. doi:10.1093/nar/gkv416
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., dePamphilis, C., et al. (2011). The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 332 (6032), 960–963. doi:10.1126/science.1203810
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The Arabidopsis Information Resource: Making and Mining the "gold Standard" Annotated Reference Plant Genome. genesis 53 (8), 474–485. doi:10.1002/dvg.22877
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J.-C., et al. (1993). The Focusing Positions of Polypeptides in Immobilized pH Gradients Can Be Predicted from Their Amino Acid Sequences. Electrophoresis 14 (1), 1023–1031. doi:10.1002/elps.11501401163
Brunetti, C., Fini, A., Sebastiani, F., Gori, A., and Tattini, M. (2018). Modulation of Phytohormone Signaling: A Primary Function of Flavonoids in Plant-Environment Interactions. Front. Plant Sci. 9, 1042. doi:10.3389/fpls.2018.01042
Chanet, A., Milenkovic, D., Manach, C., Mazur, A., and Morand, C. (2012). Citrus Flavanones: What Is Their Role in Cardiovascular Protection? J. Agric. Food Chem. 60 (36), 8809–8822. doi:10.1021/jf300669s
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Da Pozzo, E., Costa, B., Cavallini, C., Testai, L., Martelli, A., Calderone, V., et al. (2017). The Citrus Flavanone Naringenin Protects Myocardial Cells against Age-Associated Damage. Oxidative Med. Cell. longevity 2017, 1–12. doi:10.1155/2017/9536148
Dauchet, L., Amouyel, P., and Dallongeville, J. (2005). Fruit and Vegetable Consumption and Risk of Stroke: A Meta-Analysis of Cohort Studies. Neurology 65 (8), 1193–1197. doi:10.1212/01.wnl.0000180600.09719.53
Dauchet, L., Amouyel, P., Hercberg, S., and Dallongeville, J. (2006). Fruit and Vegetable Consumption and Risk of Coronary Heart Disease: A Meta-Analysis of Cohort Studies. J. Nutr. 136 (10), 2588–2593. doi:10.1093/jn/136.10.2588
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam Protein Families Database in 2019. Nucleic Acids Res. 47 (D1), D427–D432. doi:10.1093/nar/gky995
Feng, G., Wu, J., Xu, Y., Lu, L., and Yi, H. (2021). High‐spatiotemporal‐resolution Transcriptomes Provide Insights into Fruit Development and Ripening in Citrus Sinensis. Plant Biotechnol. J. 19 (7), 1337–1353. doi:10.1111/pbi.13549
Gensheimer, M., and Mushegian, A. (2004). Chalcone Isomerase Family and Fold: No Longer Unique to Plants. Protein Sci. 13 (2), 540–544. doi:10.1110/ps.03395404
Gmitter, F. G., and Hu, X. (1990). The Possible Role of Yunnan, China, in the Origin of Contemporary Citrus Species (Rutaceae). Econ. Bot. 44 (2), 267–277. doi:10.1007/BF02860491
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a Comparative Platform for green Plant Genomics. Nucleic Acids Res. 40 (Database issue), D1178–D1186. doi:10.1093/nar/gkr944
Guan, C., Song, X., Ji, J., Li, X., Jin, C., Guan, W., et al. (2014). Salicylic Acid Treatment Enhances Expression of Chalcone Isomerase Gene and Accumulation of Corresponding Flavonoids during Fruit Maturation of Lycium Chinense. Eur. Food Res. Technol. 239 (5), 857–865. doi:10.1007/s00217-014-2282-0
Guo, J., Zhou, W., Lu, Z., Li, H., Li, H., and Gao, F. (2015). Isolation and Functional Analysis of Chalcone Isomerase Gene from Purple-Fleshed Sweet Potato. Plant Mol. Biol. Rep. 33 (5), 1451–1463. doi:10.1007/s11105-014-0842-x
He, F. J., Nowson, C. A., and MacGregor, G. A. (2006). Fruit and Vegetable Consumption and Stroke: Meta-Analysis of Cohort Studies. The Lancet 367 (9507), 320–326. doi:10.1016/S0140-6736(06)68069-0
He, F., and Pan, Y. (2017). Purification and Characterization of Chalcone Isomerase from Fresh-Cut Chinese Water-Chestnut. LWT - Food Sci. Tech. 79, 402–409. doi:10.1016/j.lwt.2017.01.034
Huang, Y., Xu, Y., Jiang, X., Yu, H., Jia, H., Tan, C., et al. (2021). Genome of a Citrus Rootstock and Global DNA Demethylation Caused by Heterografting. Hortic. Res. 8 (1), 69. doi:10.1038/s41438-021-00505-2
Jez, J. M., Bowman, M. E., and Noel, J. P. (2002). Role of Hydrogen Bonds in the Reaction Mechanism of Chalcone Isomerase. Biochemistry 41 (16), 5168–5176. doi:10.1021/bi0255266
Jiang, W., Yin, Q., Wu, R., Zheng, G., Liu, J., Dixon, R. A., et al. (2015). Role of a Chalcone Isomerase-like Protein in Flavonoid Biosynthesis inArabidopsis Thaliana. Exbotj 66 (22), 7165–7179. doi:10.1093/jxb/erv413
Johnson, L. S., Eddy, S. R., and Portugaly, E. (2010). Hidden Markov Model Speed Heuristic and Iterative HMM Search Procedure. BMC bioinformatics 11, 431. doi:10.1186/1471-2105-11-431
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 8 (3), 275–282. doi:10.1093/bioinformatics/8.3.275
Jung, U. J., Kim, H. J., Lee, J. S., Lee, M. K., Kim, H. O., Park, E. J., et al. (2003). Naringin Supplementation Lowers Plasma Lipids and Enhances Erythrocyte Antioxidant Enzyme Activities in Hypercholesterolemic Subjects. Clin. Nutr. 22 (6), 561–568. doi:10.1016/S0261-5614(03)00059-1
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28 (1), 27–30. doi:10.1093/nar/28.1.27
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza Sativa Nipponbare Reference Genome Using Next Generation Sequence and Optical Map Data. Rice 6 (1), 4. doi:10.1186/1939-8433-6-4
Kumar, S., and Pandey, A. K. (2013). Chemistry and Biological Activities of Flavonoids: An Overview. Scientific World J. 2013, 1–16. doi:10.1155/2013/162750
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Lan, X., Quan, H., Xia, X., Yin, W., and Zheng, W. (2016). Molecular Cloning and Transgenic Characterization of the Genes Encoding Chalcone Synthase and Chalcone Isomerase from the Tibetan Herbal plantMirabilis Himalaica. Biotechnol. Appl. Biochem. 63 (3), 419–426. doi:10.1002/bab.1376
Letunic, I., and Bork, P. (2018). 20 Years of the SMART Protein Domain Annotation Resource. Nucleic Acids Res. 46 (D1), D493–D496. doi:10.1093/nar/gkx922
Mehdy, M. C., and Lamb, C. J. (1987). Chalcone Isomerase cDNA Cloning and mRNA Induction by Fungal Elicitor, Wounding and Infection. EMBO J. 6 (6), 1527–1533. doi:10.1002/j.1460-2075.1987.tb02396.x
Moore, B. D., Andrew, R. L., Külheim, C., and Foley, W. J. (2014). Explaining Intraspecific Diversity in Plant Secondary Metabolites in an Ecological Context. New Phytol. 201 (3), 733–750. doi:10.1111/nph.12526
Ngaki, M. N., Louie, G. V., Philippe, R. N., Manning, G., Pojer, F., Bowman, M. E., et al. (2012). Evolution of the Chalcone-Isomerase Fold from Fatty-Acid Binding to Stereospecific Catalysis. Nature 485 (7399), 530–533. doi:10.1038/nature11009
Noel, J. P., Jez, J. M., Bowman, M. E., and Dixon, R. A. (2000). Structure and Mechanism of the Evolutionarily Unique Plant Enzyme Chalcone Isomerase. Nat. Struct. Biol. 7 (9), 786–791. doi:10.1038/79025
Peabody, M. A., Lau, W. Y. V., Hoad, G. R., Jia, B., Maguire, F., Gray, K. L., et al. (2020). PSORTm: a Bacterial and Archaeal Protein Subcellular Localization Prediction Tool for Metagenomics Data. Bioinformatics (Oxford, England) 36 (10), 3043–3048. doi:10.1093/bioinformatics/btaa136
Peterson, J. J., Dwyer, J. T., Beecher, G. R., Bhagwat, S. A., Gebhardt, S. E., Haytowitz, D. B., et al. (2006). Flavanones in Oranges, Tangerines (Mandarins), Tangors, and Tangelos: a Compilation and Review of the Data from the Analytical Literature. J. Food Compost. Anal. 19, S66–S73. doi:10.1016/j.jfca.2005.12.006
Ralston, L., Subramanian, S., Matsuno, M., and Yu, O. (2005). Partial Reconstruction of Flavonoid and Isoflavonoid Biosynthesis in Yeast Using Soybean Type I and Type II Chalcone Isomerases. Plant Physiol. 137 (4), 1375–1388. doi:10.1104/pp.104.054502
Rao, M. J., Wu, S., Duan, M., and Wang, L. (2021). Antioxidant Metabolites in Primitive, Wild, and Cultivated Citrus and Their Role in Stress Tolerance. Molecules 26 (19), 5801. doi:10.3390/molecules26195801
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science 319 (5859), 64–69. doi:10.1126/science.1150646
Saitou, N., and Nei, M. (1987). The Neighbor-Joining Method: a New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 4 (4), 406–425. doi:10.1093/oxfordjournals.molbev.a040454
Scora, R. W. (1975). On the History and Origin of Citrus. Bull. Torrey Bot. Club 102 (6), 369–375. doi:10.2307/2484763
Shimada, N., Aoki, T., Sato, S., Nakamura, Y., Tabata, S., and Ayabe, S.-i. (2003). A Cluster of Genes Encodes the Two Types of Chalcone Isomerase Involved in the Biosynthesis of General Flavonoids and Legume-specific 5-Deoxy(iso)flavonoids in Lotus Japonicus. Plant Physiol. 131 (3), 941–951. doi:10.1104/pp.004820
Shirley, B. W., Hanley, S., and Goodman, H. M. (1992). Effects of Ionizing Radiation on a Plant Genome: Analysis of Two Arabidopsis Transparent Testa Mutations. Plant Cell 4 (3), 333–347. doi:10.1105/tpc.4.3.333
Smith, T. J. (1995). MOLView: A Program for Analyzing and Displaying Atomic Structures on the Macintosh Personal Computer. J. Mol. Graphics 13 (2), 122–125. doi:10.1016/0263-7855(94)00019-O
Sun, T., Wang, Y., Wang, M., Li, T., Zhou, Y., Wang, X., et al. (2015). Identification and Comprehensive Analyses of the CBL and CIPK Gene Families in Wheat (Triticum aestivum L.). BMC Plant Biol. 15, 269. doi:10.1186/s12870-015-0657-4
Szczepaniak, O., Ligaj, M., Kobus-Cisowska, J., Tichoniuk, M., Dziedziński, M., Przeor, M., et al. (2020). The Genoprotective Role of Naringin. Biomolecules 10 (5), 700. doi:10.3390/biom10050700
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING Database in 2017: Quality-Controlled Protein-Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res. 45 (D1), D362–D368. doi:10.1093/nar/gkw937
Testai, L., and Calderone, V. (2017). Nutraceutical Value of Citrus Flavanones and Their Implications in Cardiovascular Disease. Nutrients 9 (5), 502. doi:10.3390/nu9050502
Testai, L. (2015). Flavonoids and Mitochondrial Pharmacology: A New Paradigm for Cardioprotection. Life Sci. 135, 68–76. doi:10.1016/j.lfs.2015.04.017
Tocmo, R., Pena‐Fronteras, J., Calumba, K. F., Mendoza, M., and Johnson, J. J. (2020). Valorization of Pomelo ( Citrus Grandis Osbeck) Peel: A Review of Current Utilization, Phytochemistry, Bioactivities, and Mechanisms of Action. Compr. Rev. Food Sci. Food Saf. 19 (4), 1969–2012. doi:10.1111/1541-4337.12561
UniProt, C. (2019). UniProt: a Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 47 (D1), D506–D515. doi:10.1093/nar/gky1049
Wang, L., He, F., Huang, Y., He, J., Yang, S., Zeng, J., et al. (2018). Genome of Wild Mandarin and Domestication History of Mandarin. Mol. Plant 11 (8), 1024–1037. doi:10.1016/j.molp.2018.06.001
Wang, X., Xu, Y., Zhang, S., Cao, L., Huang, Y., Cheng, J., et al. (2017a). Genomic Analyses of Primitive, Wild and Cultivated Citrus Provide Insights into Asexual Reproduction. Nat. Genet. 49 (5), 765–772. doi:10.1038/ng.3839
Wang, Y., Li, J., and Xia, R. (2010). Expression of Chalcone Synthase and Chalcone Isomerase Genes and Accumulation of Corresponding Flavonoids during Fruit Maturation of Guoqing No. 4 Satsuma Mandarin (Citrus Unshiu Marcow). Scientia Horticulturae 125 (2), 110–116. doi:10.1016/j.scienta.2010.02.001
Wang, Y., Qian, J., Cao, J., Wang, D., Liu, C., Yang, R., et al. (2017b). Antioxidant Capacity, Anticancer Ability and Flavonoids Composition of 35 Citrus (Citrus Reticulata Blanco) Varieties. Molecules 22 (7), 1114. doi:10.3390/molecules22071114
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Winkel-Shirley, B. (2001). Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 126 (2), 485–493. doi:10.1104/pp.126.2.485
Wu, G. A., Terol, J., Ibanez, V., López-García, A., Pérez-Román, E., Borredá, C., et al. (2018). Genomics of the Origin and Evolution of Citrus. Nature 554 (7692), 311–316. doi:10.1038/nature25447
Wu, L.-M., Wang, C., He, L.-G., Wang, Z.-J., Tong, Z., Song, F., et al. (2020). Transcriptome Analysis Unravels Metabolic and Molecular Pathways Related to Fruit Sac Granulation in a Late-Ripening Navel Orange (Citrus Sinensis Osbeck). Plants 9 (1), 95. doi:10.3390/plants9010095
Xu, Q., Chen, L.-L., Ruan, X., Chen, D., Zhu, A., Chen, C., et al. (2013). The Draft Genome of Sweet orange (Citrus Sinensis). Nat. Genet. 45 (1), 59–66. doi:10.1038/ng.2472
Yamamoto, M., Suzuki, A., and Hase, T. (2008). Short-Term Effects of Glucosyl Hesperidin and Hesperetin on Blood Pressure and Vascular Endothelial Function in Spontaneously Hypertensive Rats. J. Nutr. Sci. Vitaminol 54 (1), 95–98. doi:10.3177/jnsv.54.95
Ye, J., McGinnis, S., and Madden, T. L. (2006). BLAST: Improvements for Better Sequence Analysis. Nucleic Acids Res. 34, W6–W9. doi:10.1093/nar/gkl164
Yin, Y.-c., Zhang, X.-d., Gao, Z.-q., Hu, T., and Liu, Y. (2019). The Research Progress of Chalcone Isomerase (CHI) in Plants. Mol. Biotechnol. 61 (1), 32–52. doi:10.1007/s12033-018-0130-3
Zhang, H.-C., Liu, J.-M., Lu, H.-Y., and Gao, S.-L. (2009). Enhanced Flavonoid Production in Hairy Root Cultures of Glycyrrhiza Uralensis Fisch by Combining the Over-expression of Chalcone Isomerase Gene with the Elicitation Treatment. Plant Cel Rep 28 (8), 1205–1213. doi:10.1007/s00299-009-0721-3
Zhao, C., Liu, X., Gong, Q., Cao, J., Shen, W., Yin, X., et al. (2021). Three AP2/ERF Family Members Modulate Flavonoid Synthesis by Regulating Type IV Chalcone Isomerase in Citrus. Plant Biotechnol. J. 19 (4), 671–688. doi:10.1111/pbi.13494
Zhao, C., Wang, F., Lian, Y., Xiao, H., and Zheng, J. (2020). Biosynthesis of Citrus Flavonoids and Their Health Effects. Crit. Rev. Food Sci. Nutr. 60 (4), 566–583. doi:10.1080/10408398.2018.1544885
Zhao, D., Tao, J., Han, C., and Ge, J. (2012). Flower Color Diversity Revealed by Differential Expression of Flavonoid Biosynthetic Genes and Flavonoid Accumulation in Herbaceous Peony (Paeonia Lactiflora Pall.). Mol. Biol. Rep. 39 (12), 11263–11275. doi:10.1007/s11033-012-2036-7
Keywords: Citrus species, chalcone-flavanone isomerase, phylogenetic analysis, gene expression pattern, flavanones
Citation: Wan Q, Bai T, Liu M, Liu Y, Xie Y, Zhang T, Huang M and Zhang J (2022) Comparative Analysis of the Chalcone-Flavanone Isomerase Genes in Six Citrus Species and Their Expression Analysis in Sweet Orange (Citrus sinensis). Front. Genet. 13:848141. doi: 10.3389/fgene.2022.848141
Received: 04 January 2022; Accepted: 15 March 2022;
Published: 12 April 2022.
Edited by:
Vinod K. K, Indian Agricultural Research Institute (ICAR), IndiaReviewed by:
Prasanth Tej Kumar Jagannadham, Central Citrus Research Institute (ICAR), IndiaChristophe F. Hano, INRA EA1207 Laboratoire de Biologie des Ligneux et des Grandes Cultures, France
Mohamed Addi, Mohamed Premier University, Morocco
Copyright © 2022 Wan, Bai, Liu, Liu, Xie, Zhang, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Wan, d2FucXVhbjEyMEAxMjYuY29t; Jinlian Zhang, anhqenpqbEAxNjMuY29t
 Quan Wan
Quan Wan Tingting Bai1
Tingting Bai1 Minmin Liu
Minmin Liu Ying Liu
Ying Liu Jinlian Zhang
Jinlian Zhang