- Crop Resources Institute of Heilongjiang Academy of Agricultural Sciences Harbin, Heilongjiang, China
Adzuki bean (Vigna angularis) is an important dietary legume crop that was first cultivated and domesticated in Asia. Currently, little is known concerning the evolution and expression patterns of the basic leucine zipper (bZIP) family transcription factors in the adzuki bean. Through the PFAM search, 72 bZIP members of adzuki bean (VabZIP) were identified from the reference genome. Most of them were located on 11 chromosomes and seven on an unknown chromosome. A comprehensive analysis, including evolutionary, motifs, gene structure, cis-elements, and collinearity was performed to identify VabZIP members. The subcellular localization results showed VabZIPs might locate on the nuclear. Quantitative real-time PCR (qRT-PCR) analysis of the relative expression of VabZIPs in different tissues at the bud stage revealed that VabZIPs had a tissue-specific expression pattern, and its expression was influenced by abiotic stress. These characteristics of VabZIPs provide insights for future research aimed at developing interventions to improve abiotic stress resistance.
Introduction
Transcription factors (TFs), which constitute approximately 8% of the protein-encoding regulators in eukaryotic genomes, are critical transcriptional regulatory factors (Pruneda-Paz et al., 2014). Therefore, functional characterization of transcription factors (TFs) is critical for understanding transcriptional regulatory networks and biological processes (Liu et al., 2014). The basic leucine zipper (bZIP) family is one of the largest and most diverse TF families (Pérez-Rodríguez et al., 2010). The bZIP domain is highly conserved and contains two structural features located on a contiguous α-helix i.e., the leucine zipper composed of several heptad repeats of Leu or other bulky hydrophobic amino acids for dimerization specificity, and the N-x7-R/K-x9 domain for specific binding (Jakoby et al., 2002; Lee et al., 2006; Nijhawan et al., 2008). Apart from the bZIP domain, several domains of the bZIP family have been found to function as transcriptional activators (Liao et al., 2008). To bind DNA, half of the basic region in the N-terminal binds double-stranded DNA, and half of the Leu zipper in the C-terminal undergoes dimerization, leading to the formation of a superimposed coiled structure (Ellenberger et al., 1992).
Members of the bZIP transcription factor family are involved in the regulation of growth and developmental processes such as seed germination, embryogenesis, flower and vascular development, hormonal control, and senescence (Jakoby et al., 2002; Schütze et al., 2008; Toh et al., 2012; Sornaraj et al., 2016). Overexpression of OsbZIP23, a member of bZIP in rice (Oryza sativa), rescued the pre-harvest budding phenotype and the decrease in expression of genes associated with ABA signaling in transgenic plants (Song et al., 2020). CAREB1, an important trans-acting factor of bZIP members, was found to regulate somatic embryogenesis in carrot (Daucus carota) (Guan et al., 2009). Eleven TabZIP genes in wheat (Triticum aestivum) were highly expressed in anthers, suggesting that they were involved in flower development (Li D. et al., 2015). In Arabidopsis, a bZIP transcription factor that control monopteros (MP) output and modulate vascular gene expression (Smit et al., 2020). The bZIP Transcription factor PERIANTHIA interacts with a variety of developmental pathways, including light and plant hormones, both of which participate in meristem formation (Maier et al., 2011). Furthermore, the bZIP members regulate response to abiotic/biotic stresses such as drought, salt, hypoxia, cold, pests, and diseases (Uno et al., 2000; Shimizu et al., 2005; Zander et al., 2012; Alves et al., 2013; E et al., 2014; Amorim et al., 2017). GmbZIP44 and GmbZIP62, the bZIP genes of soybean (Glycine max), conferred tolerance to salt and freezing stress in transgenic Arabidopsis plants (Liao et al., 2008). Overexpression of CabZIP25, a member of bZIP in pepper (Capsicum annuum), enhanced salt tolerance in transgenic Arabidopsis and promoted salt sensitivity by decreasing virus induced gene silencing (VIGS) expression in pepper (Gai et al., 2020). The study by Hsieh et al. (2010) showed that SlAREB, a member of bZIP in tomato (Solanum lycopersicum), regulated stress-responsive genes and improved water logging deficit and salt stress response. Elsewhere, it was reported that AREB1, an Arabidopsis bZIP transcription factor, conferred tolerance to water deficit (including drought and flooding stresses) in modified soybeans overexpressing AREB1 (Fuhrmann-Aoyagi et al., 2021). PPI1, a bZIP in pepper, regulated expression of genes involved in defense mechanisms (Lee et al., 2002).
The application of genome sequencing has led to identification of bZIP family members (Jakoby et al., 2002), in Arabidopsis (Jakoby et al., 2002), rice (E et al., 2014), Carthamus tinctorius (Li H. et al., 2020), Chinese jujube (Ziziphus jujuba) (Zhang Q. et al., 2020), Olive (Olea europaea) (Rong et al., 2020), common bean (Phaseolus vulgaris) (Zhang et al., 2021), and potato (Solanum tuberosum) (Herath and Verchot, 2020). However, few studies have investigated bZIP family members in adzuki bean (Vigna angularis). Adzuki bean (Vigna angularis) is an important dietary legume crop that was first cultivated in China (Han et al., 2005). Its grains have high protein content, a low-fat content, and high iron content. They contain several bioactive substances such as triterpenoids, flavonoids, and saponins. It was traditionally used as an iron supplement, to remove damp and swelling (Amarowicz et al., 2008; Yang et al., 2015). Being a sensitive species, adzuki beans are particularly vulnerable to environmental stressors such as cold, drought, salt, and heavy metals (Srivastava et al., 2018; Li W.-Y. et al., 2020). In this study, bZIP members in the adzuki bean were identified, and characterized in terms of phylogeny and evolutionary expansion in different tissues under different stress conditions such as drought (D), cold (C), salt (NaCl) and heavy metal (CdCl2). The findings will provide new insights about bZIP members which can be applied in resistance breeding.
Materials and Methods
Identification of bZIP Members in Vigna Angularis
The basic information for the reference genome (including genes, cDNAs, and proteins) in adzuki bean (Vigan1.1) was obtained from the Esembl plant’s database (https://plants.ensembl.org/Vigna angularis/Info/Index). The bZIP domain information was obtained from the PFAM database (http://pfam.xfam.org/), with PF00170 as the search key. The bZIP members in adzuki bean (Vigna angularis) were identified using the HMMER software (Finn et al., 2015) and screened using a database that included the ExPASy Proteomics Server (http://www.expasy.org) (Hoogland et al., 2008) and P3DB (http://www.p3db.org) (Yao and Xu, 2017). After deduplication, the remaining bZIP members were considered to be members of the bZIP family in adzuki bean, and were named VabZIP. VabZIPs were named according to their location in the reference genome in the Esembl database, which was determined using the TBtools software (Chen et al., 2020).
Analysis of VabZIP Members
Protein sequences of the VabZIP members were aligned using MEGA X (Kumar et al., 2018) while bootstrap values (1,000 replicates) were used for the maximum likelihood analysis. MEGA predicted the optimal model. Ten motifs from VabZIP members were identified using the MEME tool (Bailey et al., 2009), with an E-value of less than 1e−20 for motifs containing 10–50 amino acids. Gene structures for VabZIP members were analyzed using GSDS (Hu et al., 2015) and Gene-wise (Simmons et al., 2019), in which the coordinates corresponded to DNA and protein sequences. Cis-acting elements of VabZIP members were identified and their functions predicted by the plantCARE software (Lescot et al., 2002). Gene duplication events for VabZIP members were evaluated by MCScanX (Wang et al., 2013) and circus (Krzywinski et al., 2009) software. Subcellular locations for VabZIP members were predicted by the CELLO database (http://cello.life.nctu.edu.tw/) (Yu et al., 2006). Expression data for orthologous genes of VabZIP members in Arabidopsis and soybean (Glycine max) were obtained from the phytozome database (https://phytozome-next.jgi.doe.gov/).
Plant Materials, Stress Concentrations, and qRT-PCR Analysis
Plant materials for this study were “Longxiaodou 5”, which was provided by the Institute of Crop Resources, Heilongjiang Academy of Agricultural Sciences (Harbin, Heilongjiang, China). For the seedlings to bud, they were incubated at 26 °C without light (Qi et al., 2021).
During treatment, the following stressors were prioritized: drought, salt, cold, and heavy metals. Salt stress concentration was 70 mmol/L (Zhang Y. et al., 2020) while heavy metal stress concentration was 0.5 mg/L CdCl2 (Zhao et al., 2020). A temperature of 4 °C was used to induce cold stress (Wang et al., 2020) while 15% PEG was the concentrate drought stress (Ahmad et al., 2020). The stresses were separately induced on the third day, with water treatment used as the control (CK). The hypocotyl, radicle, cotyledon, and germs were collected as samples for tissue-specific analysis expressions. The radicles were collected in response to these abiotic stress treatments. The RNA Easy Fast Kit (DP452, Tiangen, Beijing) was used for sample RNA extraction, which was used for cDNA synthesis using HiScript SuperMix (R223-01, Vazyme, Nanjing). The VabZIPs primers were designed using the Primer premier5.0 software (PREMIER Biosoft, San Francisco, United States ) while Va-actin was used as the reference control gene (Li W.-Y. et al., 2020). qRT-PCR analyses for expressions of three biological replicates of each VabZIP member were performed using the Light Cycler system (Roche 480II, Roche, Switzerland) and TransStart® Top Green qPCR SuperMix (AQ131-04, TransGen Biotech, Beijing). Relative expressions were calculated as described by Livak and Schmittgen (2001).
Subcellular Localization
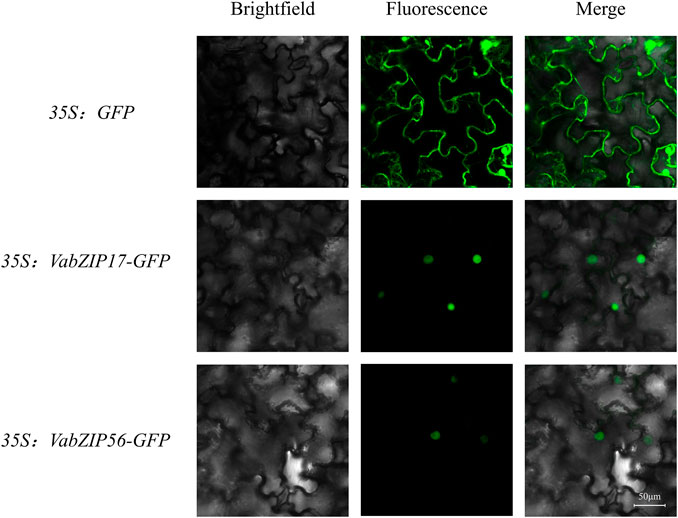
The coding sequence (CDS) of VabZIP members (VabZIP17 and VabZIP56) was PCR amplified from the cDNAs, which without stop codon. The primers used for cloning the VabZIP17 and VabZIP56 was shown in Supplementary Table S2. Then, the sequence was cloned into the vector, which had the green fluorescent protein (GFP) tag and a CaMV35S promoter. The VabZIP17-GFP and VabZIP56-GFP construct were transformed into Agrobacterium competent cells and transiently expressed in the leaves of Nicotiana benthamiana with the empty vector was used as a control. After injection for 2 days, the leaves were observed under a confocal laser microscope (TCS-SP8 Leica, Wetzlar, Germany) to find fluorescence signals (A1Si, Nikon, Japan).
Results
Identification of bZIP Members in Vigna angularis
Following a HMMER-search of the bZIP domain, 72 members of the bZIP family were identified from the reference genome in the Esembl database (Vigna angularis), which had no duplications. These members were located on all Vigna angularis chromosomes. Eight of the members were located on chromosomes 7, 8, 9, and 10 while chromosome 5 had the fewest members (2). Seven members were located on an unknown chromosome, which may be positioned on the scaffold. The bZIP members were named based on their location (VabZIP1-VabZIP72) (Figure 1). Information on the VabZIP members is presented in Supplementary Table S1. Protein lengths of VabZIPs ranged from 80 to 773, with VabZIP56 having the longest protein (773) and CDS (2,322). Isoelectric points of VabZIP members ranged from 4.76–11.56, while their molecular weights ranged from 9,438.77 Kilodalton (Kd) to 84105.11 (Kd) (Supplementary Table S3).
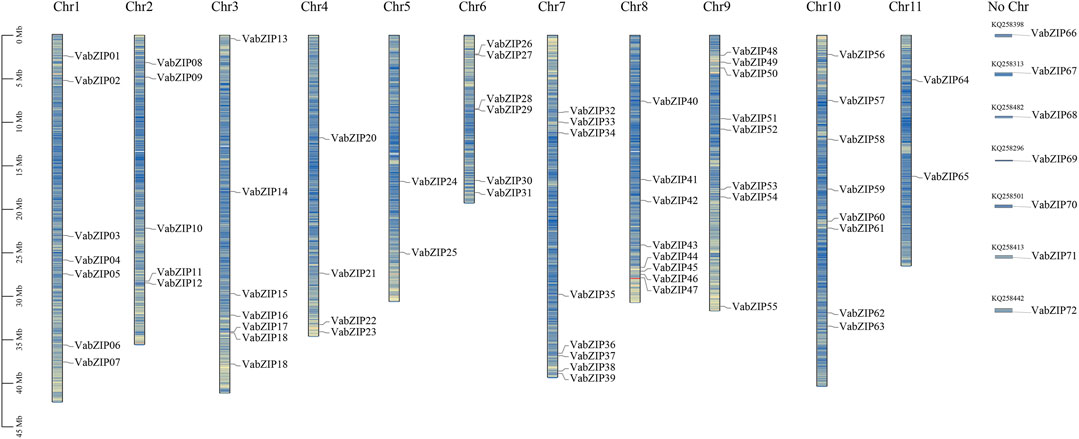
FIGURE 1. Locations of VabZIP members. The 11 pillars correspond to the 11 chromosomes, whereas No Chr depict members lacking a chromosome. The blue lines indicate gene density on chromosomes.
Evolutionary Analysis of the VabZIP Members
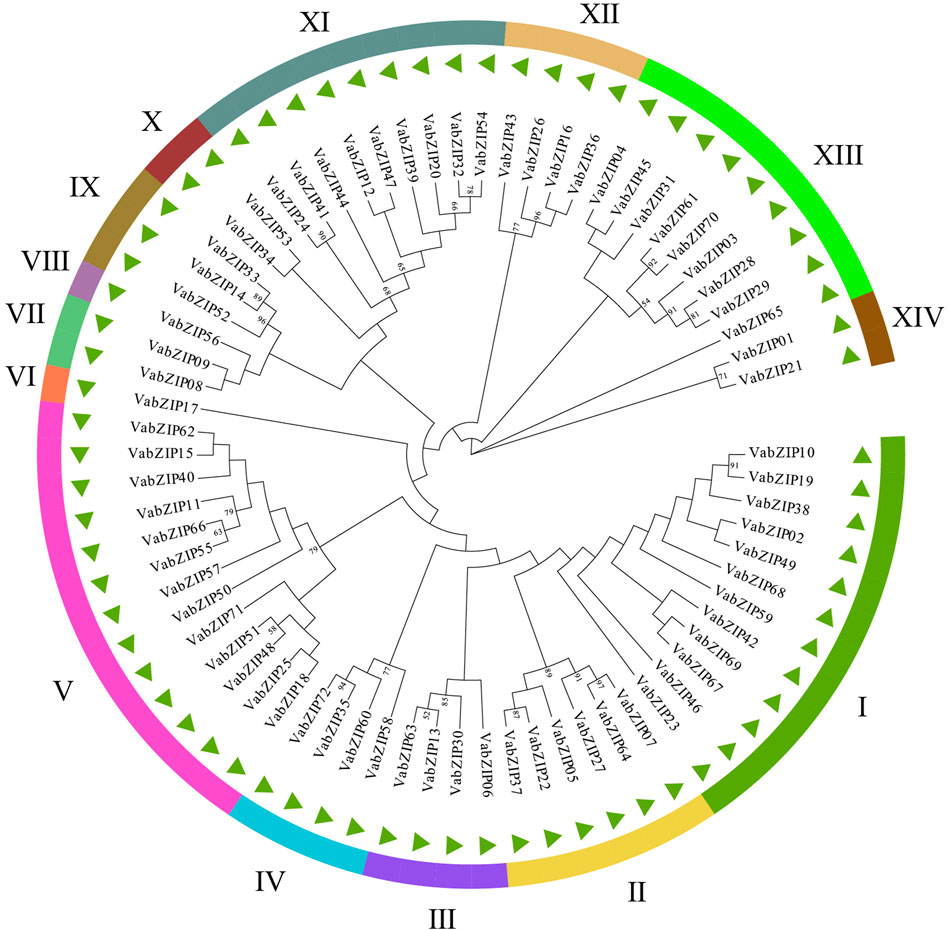
Evolutionary history of VabZIP members was determined using the Maximum Likelihoodphy (ML) analysis, with the lg + g model predicted by MEGA X software used as the model for analysis. Findings from MEGA X analysis are presented in Figure 2. These 72 members were divided into 14 subfamilies, with sub-family V having the most VabZIP members (13), while sub-families VI and VIII each had only one VabZIP member, making them the least-membered sub-families. There were 4, 2, and 9 VabZIP members in subfamily III, IV, and XII, subfamily VII, X, and XIV, and subfamily XI and XIII, respectively.
Motifs and Structure in VabZIPs
Analysis of VabZIPs using the MEME software revealed ten motifs (Figures 3A,B and Supplementary Figure S1). Apart from subfamilies VII and VIII, motif-1 was found in most VabZIPs sub-families while motif-3 and motif-5 were only found in sub-family X. Motif 9 was found only in subfamily XII. The VabZIP members in each subfamily had similar motifs. Gene structures for VabZIP members were assessed by GSDS, which revealed exon and intron structures (Figure 3C). The bZIP structure was located above all members in these VabZIPs, and sub-families I to III members had shorter introns than members of the other subfamilies.
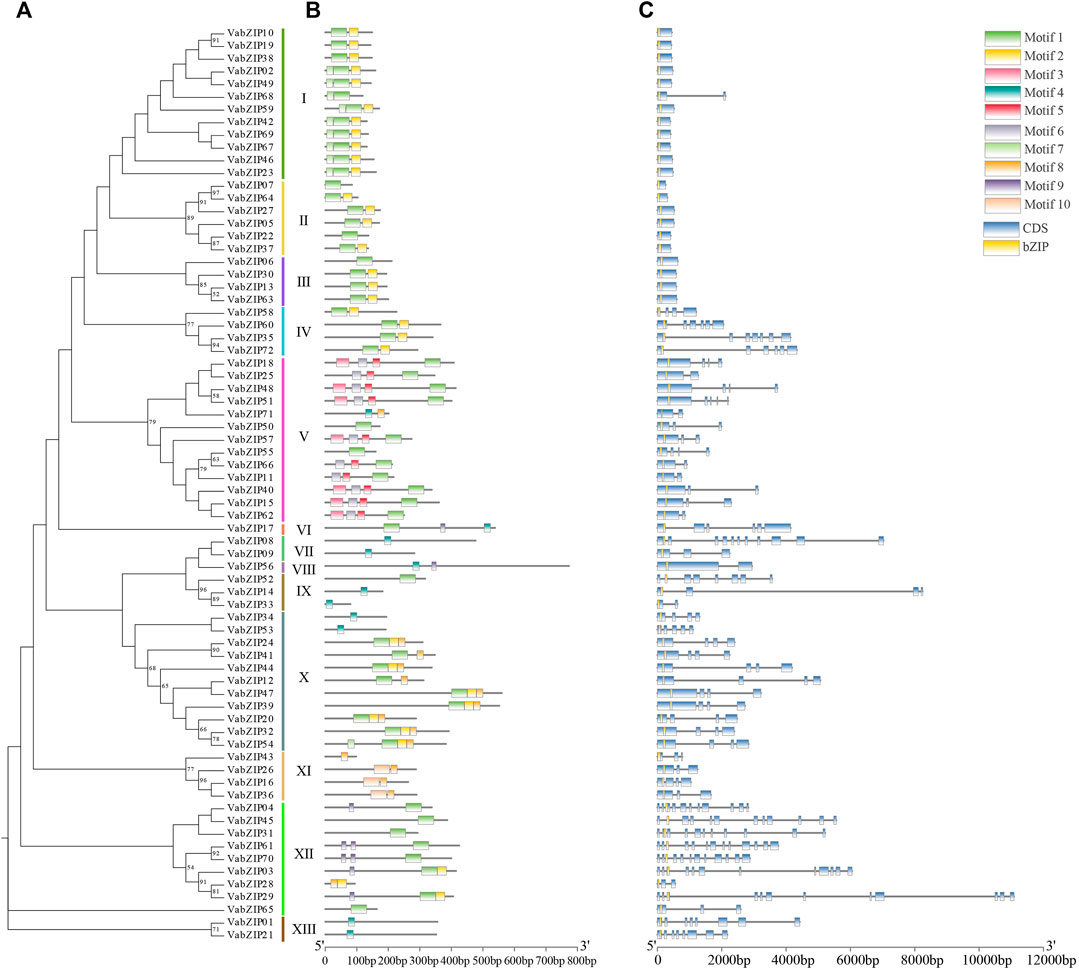
FIGURE 3. Motifs and gene structure of VabZIPs. (A) Evolutionary analysis of VabZIPs. Different colors represent different subfamilies (B) The motifs of VabZIPs (C) Gene structure of VabZIPs. Blue squares represent CDS while yellow squares represent the bZIP structure.
Evolution of bZIP Members in Two Species
The 72 VabZIPs were compared to sequences encoded by bZIP members from Arabidopsis to determine their evolutionary history, motif, and gene structure (Figure 4 and Supplementary Figure S2). MEGA X predicted the lg + g model as the best model, and 14 subfamilies were defined based on the results in these two species, consistent with the evolutionary of VabZIPs (Figure 2). Each subfamily had bZIP members of these two species. Each member of the subfamily had comparable motifs and gene structures.
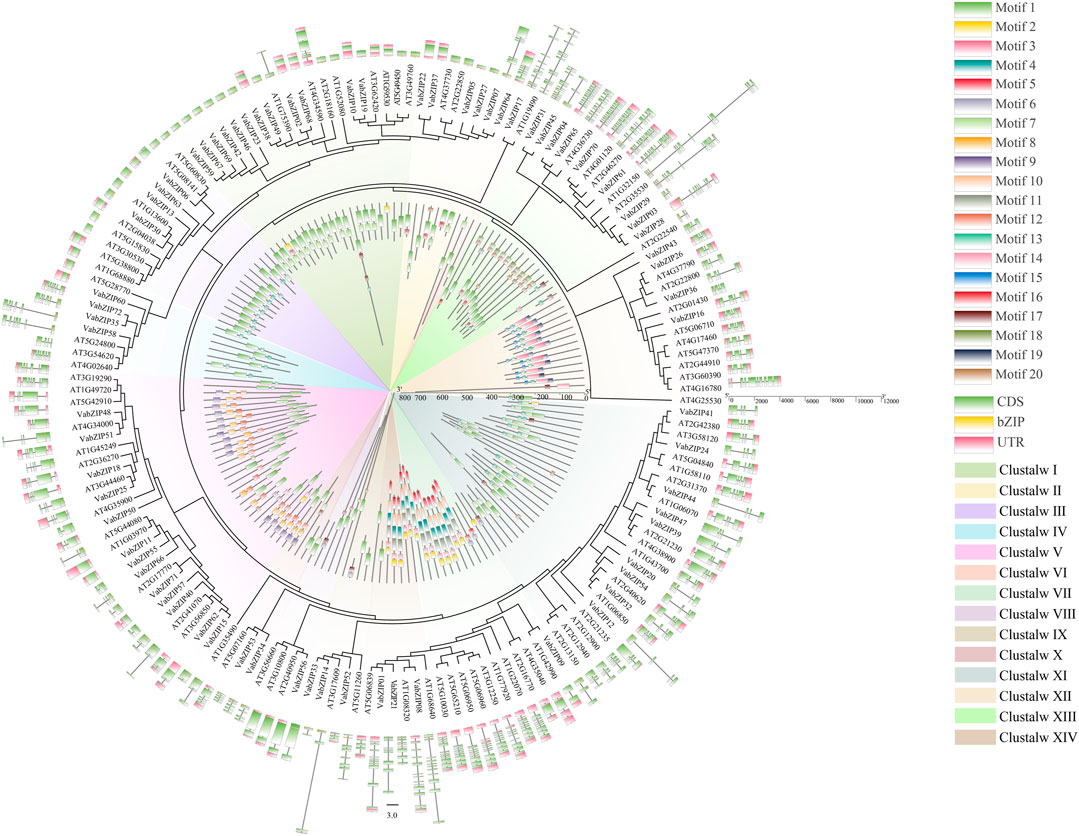
FIGURE 4. Evolution of bZIP members in Arabidopsis and VabZIPs. The inner ring indicates the motifs of bZIP members while the outer ring is the gene structure of bZIP members.
Cis-Elements of VabZIPs
Cis-elements of VabZIP members were analyzed using the plantCARE software. PlantCARE predicted the functions of the ten cis-elements (Supplementary Table S4), which were divided into three categories: hormone responsiveness (red), environmental stress (blue), and germination (yellow). Hormone responsiveness elements, including TATC-box, P-box, and GARE-motif were involved in gibberellin responsiveness, while ABRE was the cis-acting element involved in abscisic acid responsiveness. Environmental stress elements, such as LTR were involved in low-temperature responsiveness, while MBS was involved in drought-inducibility. The RY-element and NON-box elements had seed-specific regulation function. These findings indicate that VabZIP family members are involved in hormone regulation, stress resistance, and seed germination (Figure 5).
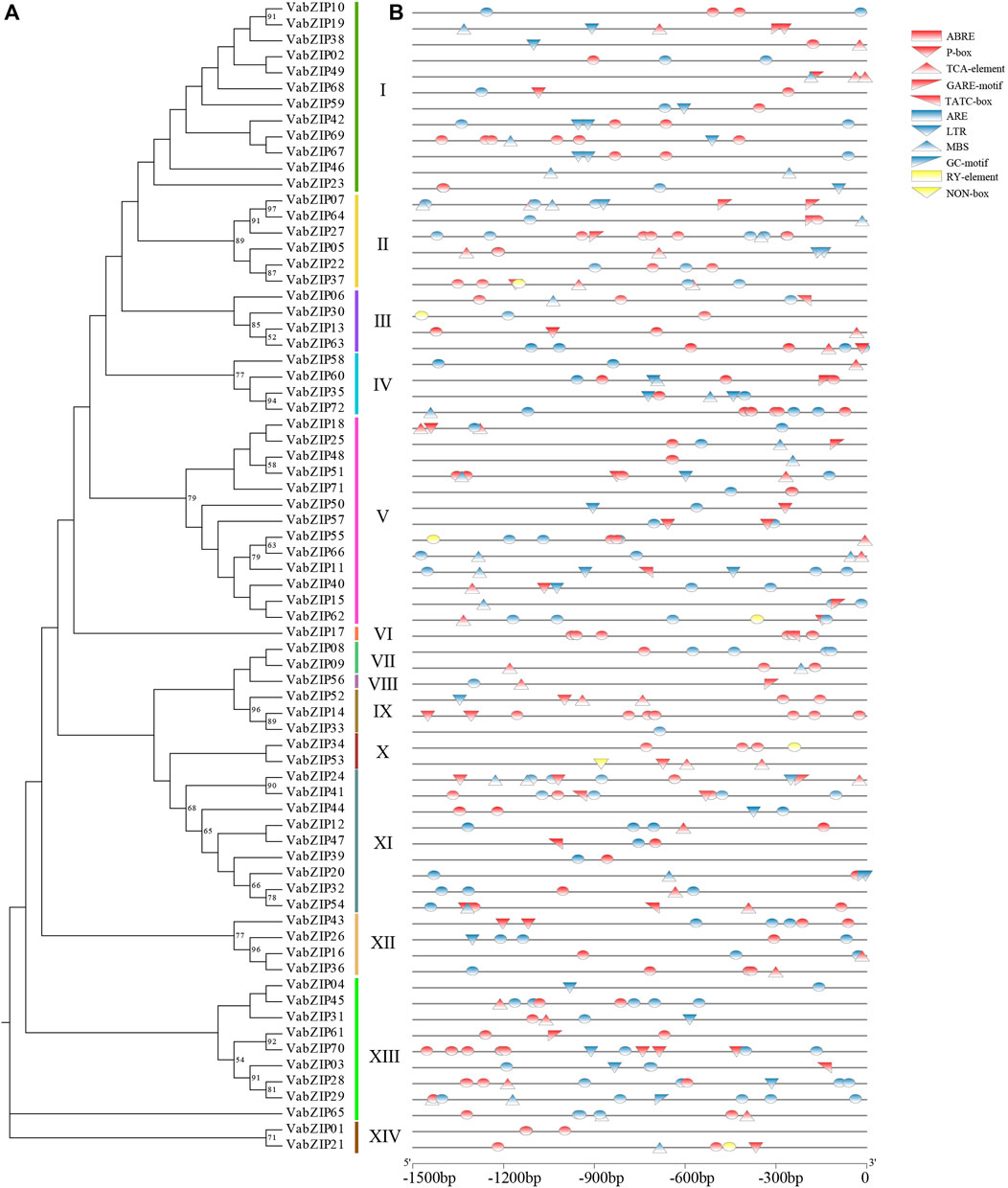
FIGURE 5. Cis-elements of VabZIPs. (A) Evolutionary analysis of VabZIPs. (B) Cis-elements of VabZIPs. The red models represent the hormone-related elements. The blue models represent the stress-related elements. The yellow models represent the elements in germination.
Collinearity Analysis of VabZIPs
There was collinearity between ten pairs of VabZIPs with VABZIP05 and VABZIP22 being the most collinear with other VabZIP members (three pairs). VaBZIP27 and VaBZIP37 had two pairs of collinearity (Figure 6A). Twenty five collinearity pairs were identified between VabZIP members and Arabidopsis, with VabZIP13, VabZIP23, VaBZIP26, VaBZIP46, and VaBZIP47 having two collinear members in Arabidopsis, implying that the 21 VabZIP members may have the same function as collinear Arabidopsis genes (Figure 6B).
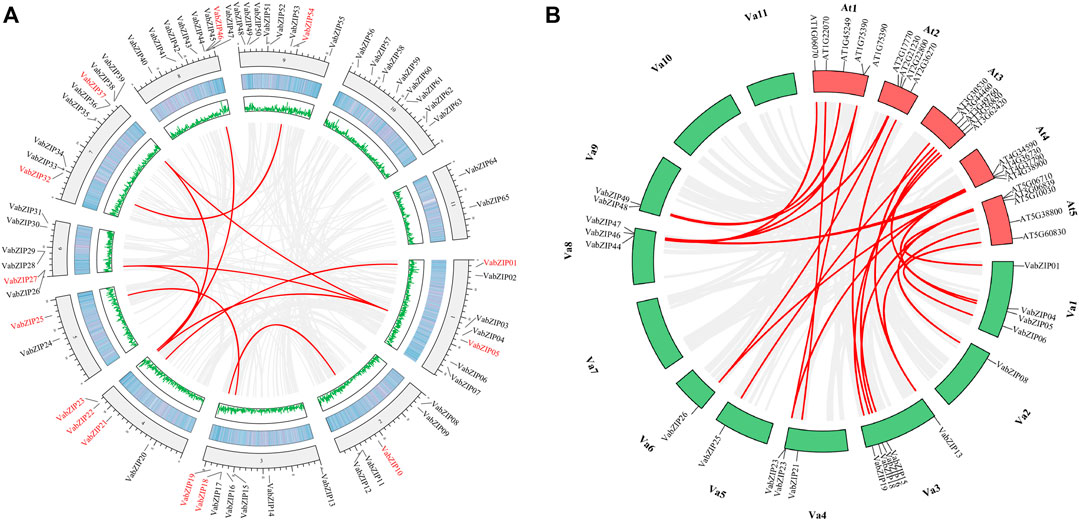
FIGURE 6. Collinearity analysis of VabZIPs. (A) Collinearity of PvHsf members. The red PvHsfs indicate collinearity while the black ones have no collinearity. The middle two rings indicate gene density. The gray background line indicates a collinear background while the green lines indicate a collinear relationship. (B) Collinearity of VabZIPs with Arabidopsis. Red boxes are the chromosomes of Arabidopsis while the green boxes are the chromosomes of the adzuki bean. The gray lines indicate the collinearity background while the red lines indicate collinearity between VabZIPs and Arabidopsis members.
Subcellular Location Analysis of VabZIPs
Subcellular locations for VabZIPs were analyzed using the CELLO database, with locations predicted by Molecular bioinformatics center. Almost all VabZIP members were predicted to be expressed in the nucleus, with only VabZIP11 predicted to be located on chloroplasts or in the cytoplasm (Supplementary Table S5).
In order to analyze the subcellular localization of VabZIP members, two members in different subfamilies were selected (VabZIP17 and VabZIP56) for subcellular location analysis. The result showed that the control (GFP) was distributed on the membrane and nuclear, while the VabZIP17-GFP and VabZIP56-GFP fusion proteins were only found on the nuclear, which indicated VabZIP17 and VabZIP56 were located on the nuclear (Figure 7).
Tissue-specific Expression Analysis at the Bud Stage
During the budding stage, the germ, cotyledon, hypocotyl, and radicle were used to investigate the expressions of bZIP members in different tissues in the adzuki bean. The twenty VabZIP members were selected randomly for qRT-PCR analysis which these twenty VabZIP members covered all of the sub-families. The VabZIP members were found to be expressed in a tissue-specific manner. VabZIP06 was abundantly expressed in the germ and cotyledon, while VabZIP11 and VabZIP26 were highly expressed in the germ. The radicle was highly enriched with VabZIP17, VabZIP30, VabZIP35, and VabZIP47, compared to the other tissues (Figure 8).
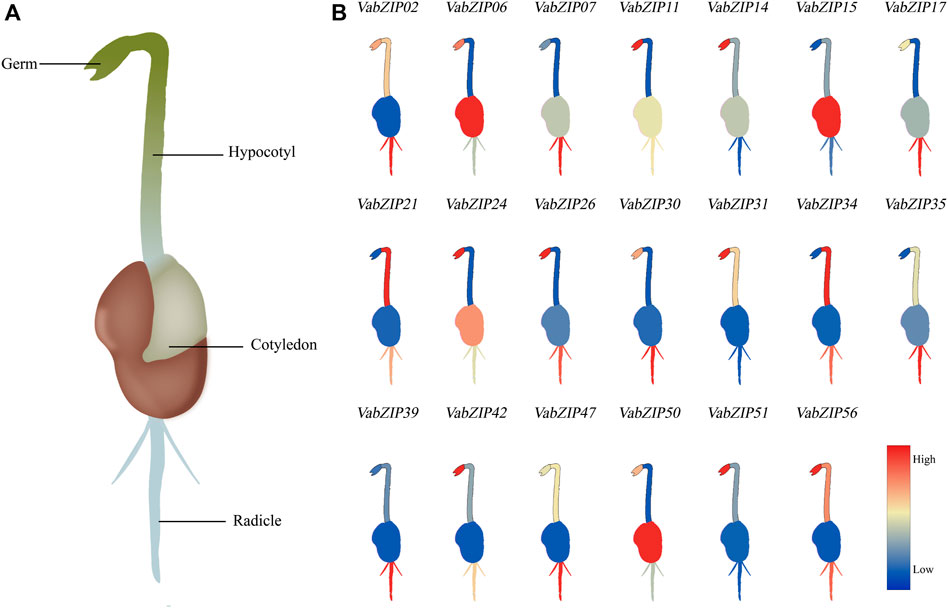
FIGURE 8. Tissue-specific expression analysis of VabZIPs at the bud stage. (A) Schematic illustration of tissues at the bud stage of adzuki bean. (B) Expressions of VabZIPs in different tissues. The change in color from red to blue indicates a high to low expression.
Expressions of VabZIPs Under Abiotic Stress at the Budding Stage
Also, these twenty VabZIP members were selected randomly for qRT-PCR analysis to determine variations in expressions in response to abiotic stress. Expressions of the nine VabZIP members varied in response to various stresses (Figure 9). Expressions of some VabZIP members (such as VabZIP06, VabZIP11, VabZIP21, VabZIP47 and VabZIP51) were up-regulated in response to drought, cold, salt, and heavy metal stress, whereas others VabZIP members (such as VabZIP24, VabZIP34, VabZIP35 and VabZIP56) were down-regulated. Differences in expressions of VabZIPs in response to various types of abiotic stress were significant, such that while VabZIP26 and VabZIP15 did not exhibit marked changes in response to heavy metal stress, they did change significantly in response to drought and cold stress, indicating that these two members may respond to other abiotic stressors other than heavy metals.
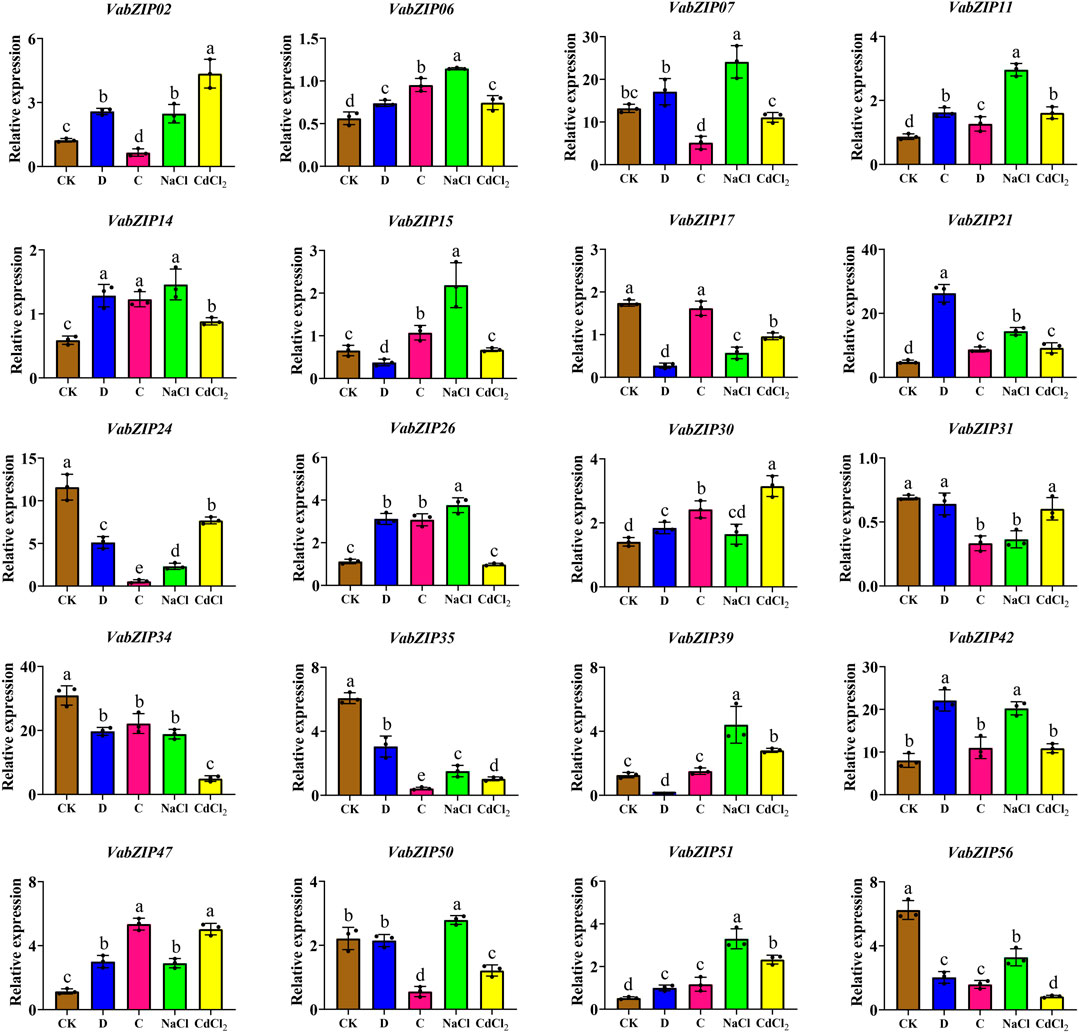
FIGURE 9. Relative expressions of VabZIPs in radicles under different abiotic stress levels at the bud stage. Brown squares denote CK treatment, whereas the blue, pink, green, and yellow squares denote drought, cold, salt, and heavy metal stress, respectively.
Discussion
The bZIP members are present in various species, and the number of members vary from one species to another. There are 78 members in Arabidopsis (Dröge-Laser et al., 2018), 80 members in potato (Solanum tuberosum) (Herath and Verchot, 2020), 160 members in soybean (Glycine max) (Zhang et al., 2018), 89 members in rice (Oryza sativa L.) (Nijhawan et al., 2008) and 69 members in tomato (Solanum lycopersicum) (Li X. et al., 2015). In this study, VabZIP members were identified from the reference genome of the adzuki bean, which contained 72 members. The number of VabZIP members was found to exceed the number of bZIP members in tomato, while the number of bZIP members was less than in Arabidopsis, potato, soybean, and rice. These findings suggest that the number of bZIP members may be related to the size of the reference genome and that after differentiation from their early ancestors, the adzuki bean may have experienced fewer genomic replication events, when compared to other species.
Evolutionary analysis revealed that VabZIP members could be divided into 14 subfamilies in the unrooted Maximum Likelihoodphy (ML) tree, and the result of evolutionary combined with bZIP members in Arabidopsis and adzuki bean was also revealed that bZIP members had 14 sub-families, which was similar to the results in wheat (Triticum aestivum) (Li D. et al., 2015) and Chinese jujube (Ziziphus jujuba Mill.) (Zhang Q. et al., 2020). With regards to motif and gene structures of VabZIP members, motif constitutions differed in different sub-families. Within the same sub-family, the motifs were similar and the motif of the coded bZIP (motif-1) was highly conserved (Zhou et al., 2017). Motif-3 and motif-5 were only found in sub-family X while motif 9 was only found in subfamily XII, which was similarly found in tartaty buckwheat (Fagopyrum tataricum) (Liu et al., 2019). VabZIP members from the same subfamily exhibited a similar gene structure, whereas VabZIPs from sub-families I, II and, III had no more than two introns, suggesting a relationship between the low number of introns and stress responses in the three sub-families (Zhao et al., 2016). The combined results of the motif and gene structure for two species of bZIP members revealed similar results.
Cis-elements in promoter regions of VabZIP members regulate the functions of VabZIPs that contain related cis-elements (Lescot et al., 2002). Similarly, bZIP members have cis-elements that are comparable across species: ABRE, TATC-box, TCA-element, and P-box are hormone-related cis-elements in VabZIP and bZIP members in common bean and potato (Wang et al., 2021; Zhang et al., 2021), implying that VabZIPs may regulate hormones involved in plant growth. Stress-related cis-elements, such as MBS and LTR, were found in VabZIPs and sesame bZIP transcription factor members (Wang et al., 2018), leading to the hypothesis that VabZIPs are involved in abiotic stress responses. Moreover, since they contain the RY element, which is similar to that found in the common bean, VabZIP members may have had an effect at the bud stage (Zhang et al., 2021).
Collinear analysis allows the transfer of functional information from a well-studied taxon to a less-studied taxon (Ghiurcuta and Moret, 2014). In this study, 25 VabZIPs pairs exhibited collinearity with an Arabidopsis member, which was found to be involved in plant growth regulation, abiotic stress responses, responses to hormones, and germination in plants. AT4G37790, AT1G45249, and AT2G36270 were the collinearity genes for VABZIP26, VaBZIP48, and VaBZIP18 respectively, which play a function in salt stress responses (Lopez-Molina et al., 2001; Nakashima et al., 2006; Liu et al., 2016). Collinearity members for Vigna angularis such as VaBZIP23, VaBZIP46, VaBZIP18, and VaBZIP47 in Arabidopsis, have a role in the bud stage of the plant: AT1G75390, the collinearity member for VabZIP23 and VabZIP46, positively regulates plant seed germination rate. Its knock-out was associated with significantly slower germination rate (Iglesias-Fernández et al., 2013). AT2G36270 had collinearity with VabZIP18, which increases proteins for preventing seed germination (Piskurewicz et al., 2008); AT4G38900, the collinearity gene for VabZIP47 was expressed in meristematic tissues and negatively modulates Arabidopsis growth (Lozano-Sotomayor et al., 2016). In Arabidopsis, VaBZIPs member collinearity genes, such as AT1G22070 (VaBZIP21) were shown to be involved in the salicylic acid (SA) signaling pathway (Zhou et al., 2000), whereas AT1G45249 (VaBZIP48) and AT2G36270 (VaBZIP18) were found to be involved in abscisic acid (ABA) responses (Fujita et al., 2005). The collinear analysis results indicate that VaBZIP members are involved in responses to hormones, coping with environmental pressures, and regulating the bud stage.
The bZIP members have previously been reported to exhibit tissue-specific expressions, including in Olive (Olea europaea) (Rong et al., 2020), radish (Raphanus sativus) (Fan et al., 2019), and poplar (Zhao et al., 2021). Expressions of VabZIPs at the bud stage revealed tissue-specificity, with the radicle having higher expressions than other tissues, indicating that the radicle could be used as a target tissue for VabZIPs’ research. Gene expression changes under abiotic stress conditions might lead to abiotic stress responses in plants, and differentially expressed genes under abiotic stress can be used as candidate genes for further research on abiotic stress responses (Qi et al., 2021). In this study, expressions of selected VabZIPs under abiotic stress indicated that VabZIPs are involved in abiotic stress responses. Expressions of VabZIP06, VabZIP11, VabZIP21, VabZIP47 and VabZIP51 were markedly up-regulates under drought, cold, salt and heavy metal stress, implying that these bZIP members are involved in abiotic stress responses. Moreover, some bZIP members are involved in abiotic stress responses in other plants, such as StbZIP25 in potato (Wang et al., 2021), TabZIP96 in wheat (Liang et al., 2022) and CabZIP25 in pepper (Gai et al., 2020). Expressions of bZIP members under abiotic stress revealed that some bZIPs can be used in plant breeding for abiotic stress resistance, such as in watermelon (Yang et al., 2019), sesame (Wang et al., 2018), and apple (Zhao et al., 2016). These results indicate that bZIPs might be useful in molecular breeding under abiotic stress and the VabZIPs that were differentially expressed under stress can be used for further research, particularly in stress-resistance breeding.
Conclusion
In this study, 72 VabZIP members were identified and divided into 14 subfamilies. The members of each sub-family had motifs and gene structures that were comparable. These VabZIP members exhibited hormonal responsiveness, environmental stress, and germination cis-elements, indicating that the VabZIPs might be involved in plant hormone and abiotic stress regulation. The VabZIPs, whose expressions were tissue specific, might be involved in abiotic stress responses. And VabZIP17 and VabZIP56 were located on the nuclear in subcellular localization ananlysis. Furthermore, expressions of VabZIPs under stress conditions such as drought, cold, salt, and heavy metal stress at the bud stage revealed that some VabZIPs (such as VabZIP06, VabZIP11, VabZIP21, VabZIP47 and VabZIP51) might regulate abiotic stresses responses in the adzuki bean. This study provides valuable information and insights into the development of VabZIPs and establishes a foundation for the use of related characteristics of VabZIPs in adzuki beans.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZY and QW conceived the study and designed and managed the experiments. XM and YG provided the plant lines. SW and YL performed the trials, collected the data and completed the statistical analysis of the data, ZY and QW wrote the paper. All authors contributed to writing the paper.
Funding
This work was supported by the Key Research and Development Program of Heilongjiang Province (GA21B009-03), The Research Funds of Heilongjiang provincial science and Technology Department (CZKYF-2-C004), the China Agriculture Research System (CARS-08-G05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.847612/full#supplementary-material
References
Ahmad, M. A., Javed, R., Adeel, M., Rizwan, M., and Yang, Y. (2020). PEG 6000-Stimulated Drought Stress Improves the Attributes of In Vitro Growth, Steviol Glycosides Production, and Antioxidant Activities in Stevia Rebaudiana Bertoni. Plants 9, 1552. doi:10.3390/plants9111552
Alves, M., Dadalto, S., Gonçalves, A., De Souza, G., Barros, V., and Fietto, L. (2013). Plant bZIP Transcription Factors Responsive to Pathogens: A Review. Int. J. Mol. Sci. 14, 7815–7828. doi:10.3390/ijms14047815
Amarowicz, R., Estrella, I., Hernández, T., and Troszyńska, A. (2008). Antioxidant Activity of Extract of Adzuki Bean and its Fractions. J. Food Lipids 15 (1), 119–136. doi:10.1111/j.1745-4522.2007.00106.x
Amorim, L., Santos, R., Neto, J., Guida-Santos, M., Crovella, S., and Benko-Iseppon, A. (2017). Transcription Factors Involved in Plant Resistance to Pathogens. Curr. Protein Pept. Sci. 18, 335–351. doi:10.2174/1389203717666160619185308
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Dröge-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP Transcription Factor Family - an Update. Curr. Opin. Plant Biol. 45, 36–49. doi:10.1016/j.pbi.2018.05.001
E, Z. G., Zhang, Y. P., Zhou, J. H., and Wang, L. (2014). Mini Review Roles of the bZIP Gene Family in Rice. Genet. Mol. Res. 13, 3025–3036. doi:10.4238/2014.April.16.11
Ellenberger, T. E., Brandl, C. J., Struhl, K., and Harrison, S. C. (1992). The GCN4 Basic Region Leucine Zipper Binds DNA as a Dimer of Uninterrupted α Helices: Crystal Structure of the Protein-DNA Complex. Cell 71, 1223–1237. doi:10.1016/s0092-8674(05)80070-4
Fan, L., Xu, L., Wang, Y., Tang, M., and Liu, L. (2019). Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus Sativus L.). Int. J. Mol. Sci. 20, 6334. doi:10.3390/ijms20246334
Finn, R. D., Clements, J., Arndt, W., Miller, B. L., Wheeler, T. J., Schreiber, F., et al. (2015). HMMER Web Server: 2015 Update. Nucleic Acids Res. 43, W30–W38. doi:10.1093/nar/gkv397
Fuhrmann-Aoyagi, M. B., de Fátima Ruas, C., Barbosa, E. G. G., Braga, P., Moraes, L. A. C., de Oliveira, A. C. B., et al. (2021). Constitutive Expression of Arabidopsis bZIP Transcription Factor AREB1 Activates Cross-Signaling Responses in Soybean under Drought and Flooding Stresses. J. Plant Physiol. 257, 153338. doi:10.1016/j.jplph.2020.153338
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 Is a Transcription Activator of Novel ABRE-Dependent ABA Signaling that Enhances Drought Stress Tolerance in Arabidopsis. The Plant cell 17, 3470–3488. doi:10.1105/tpc.105.035659
Gai, W.-X., Ma, X., Qiao, Y.-M., Shi, B.-H., Ul Haq, S., Li, Q.-H., et al. (2020). Characterization of the bZIP Transcription Factor Family in Pepper (Capsicum Annuum L.): CabZIP25 Positively Modulates the Salt Tolerance. Front. Plant Sci. 11, 139. doi:10.3389/fpls.2020.00139
Ghiurcuta, C. G., and Moret, B. M. E. (2014). Evaluating Synteny for Improved Comparative Studies. Bioinformatics 30, i9–i18. doi:10.1093/bioinformatics/btu259
Guan, Y., Ren, H., Xie, H., Ma, Z., and Chen, F. (2009). Identification and Characterization of bZIP-Type Transcription Factors Involved in Carrot (Daucus CarotaL.) Somatic Embryogenesis. Plant J. 60, 207–217. doi:10.1111/j.1365-313X.2009.03948.x
Han, O. K., Kaga, A., Isemura, T., Wang, X. W., Tomooka, N., and Vaughan, D. A. (2005). A Genetic Linkage Map for Azuki Bean [Vigna Angularis (Willd.) Ohwi & Ohashi]. TAG. Theoretical and Applied Genetics. Theor. Appl. Genet. 111, 1278–1287. doi:10.1007/s00122-005-0046-8
Herath, V., and Verchot, J. (2020). Insight into the bZIP Gene Family in Solanum Tuberosum: Genome and Transcriptome Analysis to Understand the Roles of Gene Diversification in Spatiotemporal Gene Expression and Function. Int. J. Mol. Sci. 22, 253. doi:10.3390/ijms22010253
Hoogland, C., Mostaguir, K., Appel, R. D., and Lisacek, F. (2008). The World-2DPAGE Constellation to Promote and Publish Gel-Based Proteomics Data through the ExPASy Server. J. Proteomics 71, 245–248. doi:10.1016/j.jprot.2008.02.005
Hsieh, T.-H., Li, C.-W., Su, R.-C., Cheng, C.-P., Sanjaya, S. T., Tsai, Y.-C., et al. (2010). A Tomato bZIP Transcription Factor, SlAREB, Is Involved in Water Deficit and Salt Stress Response. Planta 231, 1459–1473. doi:10.1007/s00425-010-1147-4
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 31, 1296–1297. doi:10.1093/bioinformatics/btu817
Iglesias-Fernández, R., Barrero-Sicilia, C., Carrillo-Barral, N., Oñate-Sánchez, L., and Carbonero, P. (2013). Arabidopsis T bZIP44: A Transcription Factor Affecting Seed Germination and Expression of the Mannanase-Encoding Gene AtMAN7. Plant J. 74, 767–780. doi:10.1111/tpj.12162
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP Transcription Factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi:10.1016/s1360-1385(01)02223-3
Krzywinski, M., Schein, J., Birol, İ., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 19, 1639–1645. doi:10.1101/gr.092759.109
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lee, S. C., Choi, H. W., Hwang, I. S., Choi, D. S., and Hwang, B. K. (2006). Functional Roles of the Pepper Pathogen-Induced bZIP Transcription Factor, CAbZIP1, in Enhanced Resistance to Pathogen Infection and Environmental Stresses. Planta 224, 1209–1225. doi:10.1007/s00425-006-0302-4
Lee, S. J., Lee, M. Y., Yi, S. Y., Oh, S. K., Choi, S. H., Her, N. H., et al. (2002). PPI1: A Novel Pathogen-Induced Basic Region-Leucine Zipper (bZIP) Transcription Factor from Pepper. Mol. Plant-Microbe Interactions 15, 540–548. doi:10.1094/mpmi.2002.15.6.540
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Li, D., Fu, F., Zhang, H., and Song, F. (2015a). Genome-Wide Systematic Characterization of the bZIP Transcriptional Factor Family in Tomato (Solanum lycopersicum L.). BMC Genomics 16, 771. doi:10.1186/s12864-015-1990-6
Li, H., Li, L., ShangGuan, G., Jia, C., Deng, S., Noman, M., et al. (2020a). Genome-Wide Identification and Expression Analysis of bZIP Gene Family in Carthamus Tinctorius L. Sci. Rep. 10, 15521. doi:10.1038/s41598-020-72390-z
Li, W.-Y., Wang, C., Shi, H.-H., Wang, B., Wang, J.-X., Liu, Y.-S., et al. (2020b). Genome-Wide Analysis of Ethylene-Response Factor Family in Adzuki Bean and Functional Determination of VaERF3 under Saline-Alkaline Stress. Plant Physiol. Biochem. 147, 215–222. doi:10.1016/j.plaphy.2019.12.019
Li, X., Gao, S., Tang, Y., Li, L., Zhang, F., Feng, B., et al. (2015b). Genome-Wide Identification and Evolutionary Analyses of bZIP Transcription Factors in Wheat and its Relatives and Expression Profiles of Anther Development Related TabZIP Genes. BMC Genomics 16, 976. doi:10.1186/s12864-015-2196-7
Liang, Y., Xia, J., Jiang, Y., Bao, Y., Chen, H., Wang, D., et al. (2022). Genome-Wide Identification and Analysis of bZIP Gene Family and Resistance of TaABI5 (TabZIP96) under Freezing Stress in Wheat (Triticum A). Int. J. Mol. Sci. 23, 2351. doi:10.3390/ijms23042351
Liao, Y., Zou, H.-F., Wei, W., Hao, Y.-J., Tian, A.-G., Huang, J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 Genes Function as Negative Regulator of ABA Signaling and Confer Salt and Freezing Tolerance in Transgenic Arabidopsis. Planta 228, 225–240. doi:10.1007/s00425-008-0731-3
Liu, J., Chen, N., Chen, F., Cai, B., Dal Santo, S., Tornielli, G. B., et al. (2014). Genome-Wide Analysis and Expression Profile of the bZIP Transcription Factor Gene Family in Grapevine (Vitis Vinifera). BMC Genomics 15, 281. doi:10.1186/1471-2164-15-281
Liu, M., Wen, Y., Sun, W., Ma, Z., Huang, L., Wu, Q., et al. (2019). Genome-Wide Identification, Phylogeny, Evolutionary Expansion and Expression Analyses of bZIP Transcription Factor Family in Tartary Buckwheat. BMC Genomics 20, 483. doi:10.1186/s12864-019-5882-z
Liu, T., Longhurst, A. D., Talavera-Rauh, F., Hokin, S. A., and Barton, M. K. (2016). The Arabidopsis Transcription Factor ABIG1 Relays ABA Signaled Growth Inhibition and Drought Induced Senescence. eLife 5, e13768. doi:10.7554/eLife.13768
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lopez-Molina, L., Mongrand, S., and Chua, N.-H. (2001). A Postgermination Developmental Arrest Checkpoint Is Mediated by Abscisic Acid and Requires the ABI5 Transcription Factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi:10.1073/pnas.081594298
Lozano-Sotomayor, P., Chávez Montes, R. A., Silvestre-Vañó, M., Herrera-Ubaldo, H., Greco, R., Pablo-Villa, J., et al. (2016). Altered Expression of the bZIP Transcription Factor DRINK ME Affects Growth and Reproductive Development in Arabidopsis Thaliana. Plant J. 88, 437–451. doi:10.1111/tpj.13264
Maier, A. T., Stehling-Sun, S., Offenburger, S.-L., and Lohmann, J. U. (2011). The bZIP Transcription Factor PERIANTHIA: A Multifunctional Hub for Meristem Control. Front. Plant Sci. 2, 79. doi:10.3389/fpls.2011.00079
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional Regulation of ABI3- and ABA-Responsive Genes Including RD29B and RD29A in Seeds, Germinating Embryos, and Seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi:10.1007/s11103-005-2418-5
Nijhawan, A., Jain, M., Tyagi, A. K., and Khurana, J. P. (2008). Genomic Survey and Gene Expression Analysis of the Basic Leucine Zipper Transcription Factor Family in rice. Plant Physiol. 146, 323–324. doi:10.1104/pp.107.112821
Pérez-Rodríguez, P., Riaño-Pachón, D. M., Corrêa, L. G. G., Rensing, S. A., Kersten, B., and Mueller-Roeber, B. (2010). PlnTFDB: Updated Content and New Features of the Plant Transcription Factor Database. Nucleic Acids Res. 38, D822–D827. doi:10.1093/nar/gkp805
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The Gibberellic Acid Signaling Repressor RGL2 Inhibits Arabidopsis Seed Germination by Stimulating Abscisic Acid Synthesis and ABI5 Activity. The Plant Cell 20, 2729–2745. doi:10.1105/tpc.108.061515
Pruneda-Paz, J. L., Breton, G., Nagel, D. H., Kang, S. E., Bonaldi, K., Doherty, C. J., et al. (2014). A Genome-Scale Resource for the Functional Characterization of Arabidopsis Transcription Factors. Cel Rep. 8, 622–632. doi:10.1016/j.celrep.2014.06.033
Qi, Z., Li, M., Xia, C. Y., ZhangZhang, W. J., Yin, Z. G., Zhang, Y. L., et al. (2021). Transcriptome-Based Analysis of Salt-Related Genes during the Sprout Stage of Common Bean (Phaseolus Vulgaris) under Salt Stress Conditions. Biotechnol. Biotechnol. Equip. 35, 1086–1098. doi:10.1080/13102818.2021.1954091
Rong, S., Wu, Z., Cheng, Z., Zhang, S., Liu, H., and Huang, Q. (2020). Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea Europaea L.). Genes 11, 510. doi:10.3390/genes11050510
Schütze, K., Harter, K., and Chaban, C. (2008). Post-Translational Regulation of Plant bZIP Factors. Trends Plant Sci. 13, 247–255. doi:10.1016/j.tplants.2008.03.002
Shimizu, H., Sato, K., Berberich, T., Miyazaki, A., Ozaki, R., Imai, R., et al. (2005). LIP19, a Basic Region Leucine Zipper Protein, Is a Fos-Like Molecular Switch in the Cold Signaling of rice Plants. Plant Cel Physiol. 46, 1623–1634. doi:10.1093/pcp/pci178
Simmons, M. P., Sloan, D. B., Springer, M. S., and Gatesy, J. (2019). Gene-Wise Resampling Outperforms Site-Wise Resampling in Phylogenetic Coalescence Analyses. Mol. Phylogenet. Evol. 131, 80–92. doi:10.1016/j.ympev.2018.10.001
Smit, M. E., Llavata-Peris, C. I., Roosjen, M., van Beijnum, H., Novikova, D., Levitsky, V., et al. (2020). Specification and Regulation of Vascular Tissue Identity in the Arabidopsis Embryo. Development 147 (8), dev186130. doi:10.1242/dev.186130
Song, S., Wang, G., Wu, H., Fan, X., Liang, L., Zhao, H., et al. (2020). OsMFT2 Is Involved in the Regulation of ABA Signaling‐mediated Seed Germination through Interacting with OsbZIP23/66/72 in Rice. Plant J. 103, 532–546. doi:10.1111/tpj.14748
Sornaraj, P., Luang, S., Lopato, S., and Hrmova, M. (2016). Basic Leucine Zipper (bZIP) Transcription Factors Involved in Abiotic Stresses: A Molecular Model of a Wheat bZIP Factor and Implications of its Structure in Function. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1860, 46–56. doi:10.1016/j.bbagen.2015.10.014
Srivastava, R., Kumar, S., Kobayashi, Y., Kusunoki, K., Tripathi, P., Kobayashi, Y., et al. (2018). Comparative Genome-Wide Analysis of WRKY Transcription Factors in Two Asian Legume Crops: Adzuki Bean and Mung Bean. Sci. Rep. 8, 16971. doi:10.1038/s41598-018-34920-8
Toh, S., McCourt, P., and Tsuchiya, Y. (2012). HY5is Involved in Strigolactone-Dependent Seed Germination in Arabidopsis. Plant Signaling Behav. 7, 556–558. doi:10.4161/psb.19839
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis Basic Leucine Zipper Transcription Factors Involved in an Abscisic Acid-Dependent Signal Transduction Pathway under Drought and High-Salinity Conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi:10.1073/pnas.190309197
Wang, F., Chen, X., Dong, S., Jiang, X., Wang, L., Yu, J., et al. (2020). Crosstalk of PIF4 and DELLA Modulates CBF Transcript and Hormone Homeostasis in Cold Response in Tomato. Plant Biotechnol. J. 18, 1041–1055. doi:10.1111/pbi.13272
Wang, Q., Guo, C., Li, Z., Sun, J., Wang, D., Xu, L., et al. (2021). Identification and Analysis of bZIP Family Genes in Potato and Their Potential Roles in Stress Responses. Front. Plant Sci. 12, 637343. doi:10.3389/fpls.2021.637343
Wang, Y., Li, J., and Paterson, A. H. (2013). MCScanX-Transposed: Detecting Transposed Gene Duplications Based on Multiple Colinearity Scans. Bioinformatics 29, 1458–1460. doi:10.1093/bioinformatics/btt150
Wang, Y., Zhang, Y., Zhou, R., Dossa, K., Yu, J., Li, D., et al. (2018). Identification and Characterization of the bZIP Transcription Factor Family and its Expression in Response to Abiotic Stresses in Sesame. PLoS One 13, e0200850. doi:10.1371/journal.pone.0200850
Yang, K., Tian, Z., Chen, C., Luo, L., Zhao, B., Wang, Z., et al. (2015). Genome Sequencing of Adzuki Bean (Vigna Angularis) Provides Insight into High Starch and Low Fat Accumulation and Domestication. Proc. Natl. Acad. Sci. U.S.A. 112, 13213–13218. doi:10.1073/pnas.1420949112
Yang, Y., Li, J., Li, H., Yang, Y., Guang, Y., and Zhou, Y. (2019). The bZIP Gene Family in Watermelon: Genome-Wide Identification and Expression Analysis under Cold Stress and Root-Knot Nematode Infection. PeerJ 7, e7878. doi:10.7717/peerj.7878
Yao, Q., and Xu, D. (2017). Bioinformatics Analysis of Protein Phosphorylation in Plant Systems Biology Using P3DB. Methods Mol. Biol. (Clifton, N.J.) 1558, 127–138. doi:10.1007/978-1-4939-6783-4_6
Yu, C.-S., Chen, Y.-C., Lu, C.-H., and Hwang, J.-K. (2006). Prediction of Protein Subcellular Localization. Proteins 64, 643–651. doi:10.1002/prot.21018
Zander, M., Chen, S., Imkampe, J., Thurow, C., and Gatz, C. (2012). Repression of the Arabidopsis T Jasmonic Acid/Ethylene-Induced Defense Pathway by TGA-Interacting Glutaredoxins Depends on Their C-Terminal ALWL Motif. Mol. Plant 5, 831–840. doi:10.1093/mp/ssr113
Zhang, M., Liu, Y., Shi, H., Guo, M., Chai, M., He, Q., et al. (2018). Evolutionary and Expression Analyses of Soybean Basic Leucine Zipper Transcription Factor Family. BMC Genomics 19, 159. doi:10.1186/s12864-018-4511-6
Zhang, Q., Zhang, W.-J., Yin, Z.-G., Li, W.-J., Xia, C.-Y., Sun, H.-Y., et al. (2021). Genome-Wide Identification Reveals the Potential Functions of the bZIP Gene Family in Common Bean (Phaseolus vulgaris) in Response to Salt Stress During the Sprouting Stage. J. Plant Growth Regul. doi:10.1007/s00344-021-10497-x
Zhang, Q., Zhang, W.-J., Yin, Z.-G., Li, W.-J., Zhao, H.-H., Zhang, S., et al. (2020a). Genome- and Transcriptome-Wide Identification of C3Hs in Common Bean (Phaseolus vulgaris L.) and Structural and Expression-Based Analyses of Their Functions During the Sprout Stage under Salt-Stress Conditions. Front. Genet. 11, 564607. doi:10.3389/fgene.2020.564607
Zhang, Y., Gao, W., Li, H., Wang, Y., Li, D., Xue, C., et al. (2020b). Genome-Wide Analysis of the bZIP Gene Family in Chinese Jujube (Ziziphus Jujuba Mill.). BMC genomics 21, 483. doi:10.1186/s12864-020-06890-7
Zhao, J., Guo, R., Guo, C., Hou, H., Wang, X., and Gao, H. (2016). Evolutionary and Expression Analyses of the Apple Basic Leucine Zipper Transcription Factor Family. Front. Plant Sci. 7, 376. doi:10.3389/fpls.2016.00376
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., and Jiang, T. (2021). Genome-Wide Analysis and Expression Profile of the bZIP Gene Family in poplar. BMC Plant Biol. 21, 122. doi:10.1186/s12870-021-02879-w
Zhao, Q., Wang, H., Du, Y., Rogers, H. J., Wu, Z., Jia, S., et al. (2020). MSH2 and MSH6 in Mismatch Repair System Account for Soybean (Glycine max (L.) Merr.) Tolerance to Cadmium Toxicity by Determining DNA Damage Response. J. Agric. Food Chem. 68, 1974–1985. doi:10.1021/acs.jafc.9b06599
Zhou, J.-M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1 Differentially Interacts with Members of the TGA/OBF Family of Transcription Factors that Bind an Element of the PR-1 Gene Required for Induction by Salicylic Acid. Mol. Plant-Microbe Interactions 13, 191–202. doi:10.1094/mpmi.2000.13.2.191
Keywords: adzuki bean, bZIP members, analysis, relative expression, bud stage, abiotic stress
Citation: Yin Z, Meng X, Guo Y, Wei S, Lai Y and Wang Q (2022) The bZIP Transcription Factor Family in Adzuki Bean (Vigna Angularis): Genome-Wide Identification, Evolution, and Expression Under Abiotic Stress During the Bud Stage. Front. Genet. 13:847612. doi: 10.3389/fgene.2022.847612
Received: 03 January 2022; Accepted: 05 April 2022;
Published: 25 April 2022.
Edited by:
Nasya Borisova Tomlekova, Maritsa Vegetable Crops Research Institute (MVCRI), BulgariaReviewed by:
Eleonora Cominelli, Italian National Research Council, ItalyAamir Raina, Aligarh Muslim University, India
Copyright © 2022 Yin, Meng, Guo, Wei, Lai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Wang, c2hpLnlvbmdkb3VAMTYzLmNvbQ==
 Zhengong Yin
Zhengong Yin Xianxin Meng
Xianxin Meng Qiang Wang
Qiang Wang