- 1Human Genetics Laboratory, Department of Biosciences, Jamia Millia Islamia, New Delhi, India
- 2Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, King Khalid University, Abha, Saudi Arabia
- 3Department of Laboratory Medicine, Faculty of Applied Medical Sciences, Albaha University, Albaha, Saudi Arabia
- 4Department of Surgical Oncology BRA-IRCH, All India Institute of Medical Sciences (AIIMS), New Delhi, India
Background: Forkhead box “O” one which is member of Forkhead box family of transcription factors is known to play key role in different physiological processes including cell cycle arrest, autophagy, and apoptosis. FOXO1 is defined to play tumor suppressive role in various malignancies including breast cancer and its Dysregulation is frequently reported. However, the evaluation of FOXO1 promoter methylation and its expression at mRNA and protein level in different stages of breast cancer and its association with different clinical parameters is still not studied. Therefore, for better understanding the role of FOXO1 in breast cancer, in our study we examined the FOXO1 mRNA and protein expression in Breast cancer samples of Indian breast cancer patients.
Results: Total 127 breast cancer samples along with adjacent normal tissue (n = 127) were analyzed through methylation specific PCR (MS-PCR), mRNA expression (Real-time PCR) and Immunohistochemistry (IHC). We detected 69.29% cases to be downregulated at the mRNA level, and 77.95% of cases exhibited no or low protein expression. In our data we report a significant association (p = 0.0001) between the downregulated protein expression and promoter hypermethylation of FOXO1 gene. We also found a significant correlation of FOXO1 mRNA level with Age (p = 0.008), age at first live birth (p = 0,003), tumor size (p = 0.05) and lymph node status (p = 0.01).
Conclusion: we in our study report the tumor suppressive role of FOXO1 in case of Indian breast cancer patients and our data suggest it to exhibit prognostic importance. However, further research is needed to evaluate FOXO1 significance in diagnostic and therapeutic targeting in breast cancer cases.
Introduction
According to Global Cancer statistics 2020, there were 684,996 deaths reported due to breast malignancy and it showed the highest incidence among all cancers (Globocan 2020). Breast cancer is a multifaceted disease exhibiting diverse morphological and histopathological features (Viale, 2012). To recognize the potential genes and their molecular mechanism associated with the pathogenesis of the disease still needs to be investigated (Tang et al., 2018). Lack of early diagnosis and inadequate personalized approach in treatment are chief factors in terms of poor survival of the patients (Shi et al., 2018). Hence, there is a necessity in the current time to search for more reliable molecular targets to develop a better diagnostic and therapeutic approach in the treatment of breast cancer (Chan et al., 2017; Jiang et al., 2018). Therefore, our study focuses on molecular profiling of the Forkhead box O 1 (FOXO1) gene which is a potent molecule and can exhibit promising results in the development of diagnostic and therapeutic strategies.
FOXO1 is one of the key members of the FOXO transcription factors subfamily, which is located on chromosome 13 (13q14.11) and is a chief target of insulin signalling. It is known to have a major role in the regulation of metabolic homeostasis, autophagy, apoptosis, cell cycle arrest genes, and immune regulators (Xing et al., 2018; Jiang et al., 2018; Kousteni, 2012, Gene cards). The activation of FOXO1 via binding of insulin or several growth factors to their receptors consequently activates PI3K (phosphoinositide kinase) that further triggers the activity of other kinases including Akt and SGK (serum glucocorticoid inducible kinase). However, in the absence of insulin, the FOXO1 is found to have nuclear localization and leads to cell cycle arrest. Thus, in presence of insulin or IGF-1, PI3K/Akt/SGK pathway is directly activated while FOXO1 is inhibited resulting in cell survival (Cantley, 2002; Li et al., 2015). The low levels of FOXO1 have been linked with tumor progression in several recent studies (Kaymaz et al., 2017; Procaccia et al., 2017). The decreased nuclear and cytoplasmic expressions of FOXO1 in the case of breast cancer have been reported in the previous studies (Wu et al., 2012). However, the correlation of lower levels of FOXO1 at mRNA and protein level with clinical parameters is not well known. The current study proposes to determine the correlation between methylation and expression of FOXO1 in breast cancer biopsy as compared to adjacent normal tissue.
Methodology and Materials
Collection of Biological Specimens
In our study, 127 participants were enrolled, and the cancerous tissue along with the adjacent non-cancerous tissue of the breast was obtained and stored at −20° for further experiments and analysis. The inclusion criteria for the specimens in the study included the histopathologically confirmed breast cancer patients of age group 20–79 years having at least 6 months of life expectancy.
Following clinical parameters were included for the study such as tumor size, age at diagnosis, histological grade, the status of lymph node (LN), reproductive history, and information (age at menarche and menopausal status), clinical staging or TNM stages, Estrogen receptor (ER) (+ or -), Progesterone receptor (PR) (+ or -) and Human Epidermal Growth Factor Receptor 2 (HER2) (+ or -).
The females (n = 127) included in the study were clinically confirmed with sporadic breast cancer and were genetically unrelated. Normal adjacent breast tissue was taken as control.
Inclusion Criteria
The study involved females with histopathologically confirmed primary breast cancer and having at least 6 months of life expectancy, lying between the age group 20–79 years. The participants provided consent to abide by the procedures of the study. All the females included in the study were registered in the medical record book of AIIMS, New Delhi, and their medical records were evaluated for studying various clinical and pathological parameters of the patients.
Real-Time Polymerase Chain Reaction
For the isolation of RNA, the breast cancer tissues, and the collected normal tissues were preserved in the RNAlater (Qiagen) kit, and afterward, RNA isolation was done by using TRIzo1Reagent (Invitrogen) by following the instruction provided by the manufacturer in the protocol. Thereafter, the complementary DNA (cDNA) from total RNA was synthesized using a cDNA kit (verso Thermo Fisher Scientific) and later storage was done at −20°C for further analysis. Subsequently, the above-prepared cDNA was used in the (qPCR) where amplification was carried out using Roche Light Cycler® 96 SYBR Green I Master mix. By applying the primers for FOXO1: sense 5′- CCACATTCAACAGGCAGCAG-3′ antisense 5′- GACGGAAACTGGGAGGAAGG-3′ which amplified a 152-bp. product. β actin gene was taken as an internal control and amplified in the same qPCR reaction. The primers used for qPCR reaction were sense 5′-AGATAGTGGATCAGCAAGCAG-3′ and antisense 5′-GCGAAGTTAGGTTTTGTCA-3′, which amplified a 160 bp. product. Standardized protocol of our laboratory (Real et al., 2018; Sadaf et al., 2018; Khan et al., 2020) was used to perform PCR. Measurements were taken in triplicates. The calculation was done for the relative amount of mRNA using Light Cycler 96 (Roche) equipped with Software 1.5. The calibrated normalized ratio was estimated as per the given standard formula: RQ = 2-∆∆Cq = [(Cq targeted gene–Cq βactin) calibration sample].
DNA Extraction
Phenol-chloroform Isoamyl (PCI) method was used for isolation of gDNA from Breast cancer and adjacent normal tissue (Russell and Sambrook, 2001). The quantity and quality evaluation of isolated genomic DNA was done using a Nanodrop spectrophotometer (ND1000), and agarose gel electrophoresis was further performed for validation.
Methylation Through MS-PCR
EZ DNA Methylation-Gold™ Kit was used to carry out Bisulfite conversion following the instruction given by the manufacturer. The converted product was amplified using dual sets of methylated and unmethylated FOXO1 primers. Eukaryotic promoter database was used to retrieve the FOXO1 gene promoter sequence, and MethPrimer software was used for primer designing (Figure 1). When searched by MethPrimer, the promoter region of the FOXO1 gene was found to contain two CpG islands of 600 bp. The primer pairs that were used for the detecting methylation in the promoter region of FOXO1 were as follows: sense 5′- GGAAAATCGGGTTTTATTTAGTTC-3′ and antisense 5′- GACTACTACGACTACCAAACCGC-3′, for the unmethylated detection: sense 5′- TGGAAAATTGGGTTTTATTTAGTTT-3 and antisense 5′- CAACTACTACAACTACCAAACCACC-3′. The product size for unmethylation was 172 bp, and for methylation, it was 170 bp. MS-PCR was done by the following condition: initially denaturation at 95°C for 5 min, 35 cycles of amplification at 95°C for the 30 s, annealing at 53.9°C (methylation) and 52.7°C (unmethylation) for 30 s, 72°C for 30 s and final extension at 72°C for 7 min. The pictures of the amplified product were obtained on a 2% agarose gel having EtBr, visualization was done under ultraviolet (UV) illumination with the Gel Doc (Bio-Rad Molecular Imaging System). The experiments were executed in triplicate without any disparity observed among the replicates.
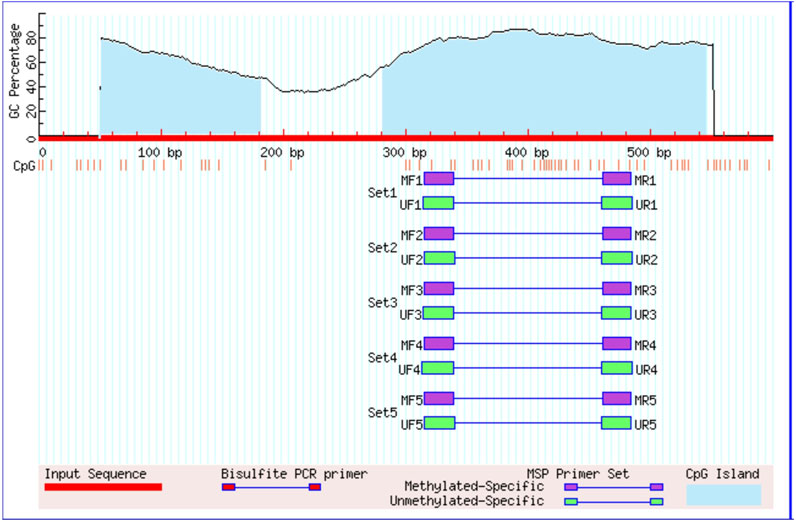
FIGURE 1. Graphical represntation of CpG islands in the foxo1 promoter region taken from Meth Primer. Criteria used: Island size>100, GC Percent >50.0, Obs/Exp>0.60/.
Immunohistochemistry
Breast cancer tissue and adjacent normal tissue stored in formalin were used for Block preparation. Poly-l-lysine slides were used, and the sections of the block were taken on slides. Subsequently, different grades of xylene were used for deparaffinization, and rehydration was done using ethanol. The quenching of internal peroxide activity was done by 0.3% hydrogen peroxide, and citrate buffer boiling resulted in Ag withdrawal. To non-specific interaction of protein was ceased using serum solution as a blocking agent, and incubation at 4°C with primary antibody (CST#2880 FOXO1, 1:100) was done overnight. Furthermore, anti-rabbit biotinylated secondary antibody and streptavidin HRP incubation was done for 20–30 min, respectively. 3,3́- 3,3′-Diaminobenzidine (DAB) method was used to visualize antibody binding sites. Also, Hematoxylin counterstaining was carried out. For positive control, normal breast tissue was considered, while for the negative control, the primary antibody was skipped following the same protocol resulting in no staining. Interpretation of the staining was carried out under the guidance expert histopathologists using light microscope (magnification ×400), and the grading was done as follows: [1] 0% tumor staining with no expression, [2] 1%–10% tumor staining with mild expression (+), [3] 10%–50% tumor staining denoting moderate expression (++) [4] >50% tumor staining indicating high expression (+++ or ++++).
Statistical Analysis
SPSS-IBM (version 22.0) was utilized to find the relevant association with the clinicopathological parameters. The current study data are represented as mean ± standard error (SE). the p-value of less than 0.005 was considered significant. To evaluate the significance of differential FOXO1 mRNA expression levels, a non-parametric test, i.e., Wilcoxon signed-rank test was used.
Results
Downregulated FOXO1 mRNA Expression in Breast Cancer Cases and its Correlation With Clinicopathological Parameters
The expression of FOXO1 at the mRNA level was detected in case of breast cancer and adjacent normal tissue. Its expression was normalized against beta-actin expression. FOXO1 mRNA expression was found to be downregulated in 69.29% cases (88/127) and out of which 73.80% cases (65/88) were categorized under histological grade I and II of breast cancer. The fold change of 88 downregulated cases was examined to be 5.16 the expression of FOXO1 in breast cancer tissue was 1.16 ± 0.02 (Mean ± SE) and in the normal tissue was 1.95 ± 0.07 (Mean ± SE) (p < 0.0001). Correlating the FOXO1 mRNA expression with different clinic pathological parameters of patients indicated significant association with Age, Age at first live birth, Tumor Size, Lymph Node Status (Table 1, Table 2, Figure 2).

TABLE 2. Correlation study of FOXO1 mRNA expression levels with clinical parameters of Breast Cancer case.
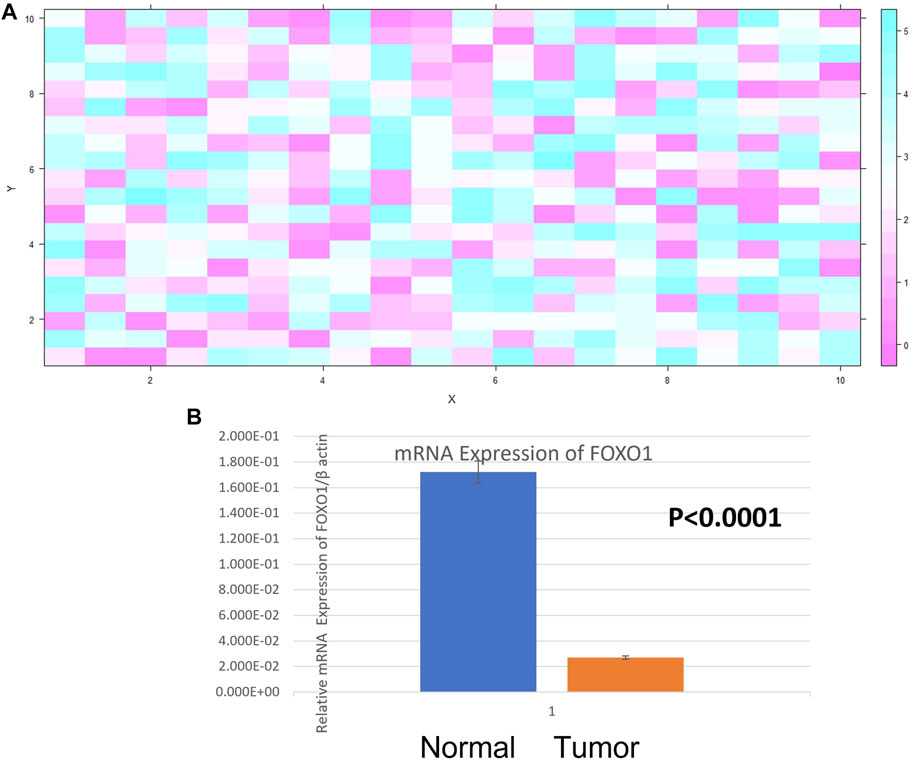
FIGURE 2. (A) Heat Map plot (analyzed by R platform version 3.6.3 64-bit) of FOXO1 mRNA relative expression (fold change) in Breast cancer cases. X-axis depicts
FOXO1 Promoter Methylation and its Correlation With Clinical Parameters of Patients
Promoter methylation study of FOXO1 promoter region was done through Methylation Specific PCR, the hypermethylated promoter region of FOXO1 was found in 61.41% (78/127) cases. The correlation of promoter methylation with clinical parameters revealed a significant association with the menopausal status of breast cancer patients. In progressive stages III and IV of breast cancer, 58/91 cases were found to be methylated (Figure 3; Table 3.)
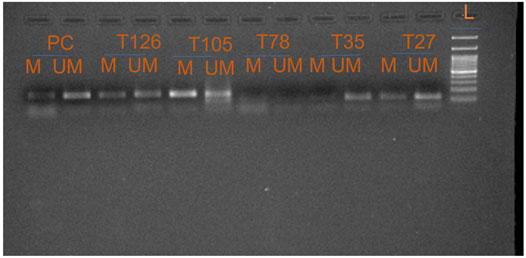
FIGURE 3. Representative gel picture of Methylation—specific PCR analysis of FOXO1 gene in Breast cancer patients: DNA methylation was assessed using two specifically designed primers to amplify either methylated DNA (M) or unmethylated DNA (UM) (L: 100 bp DNA ladder; number indicates the case number; PC: Positive Control; T: Tumour tissue).
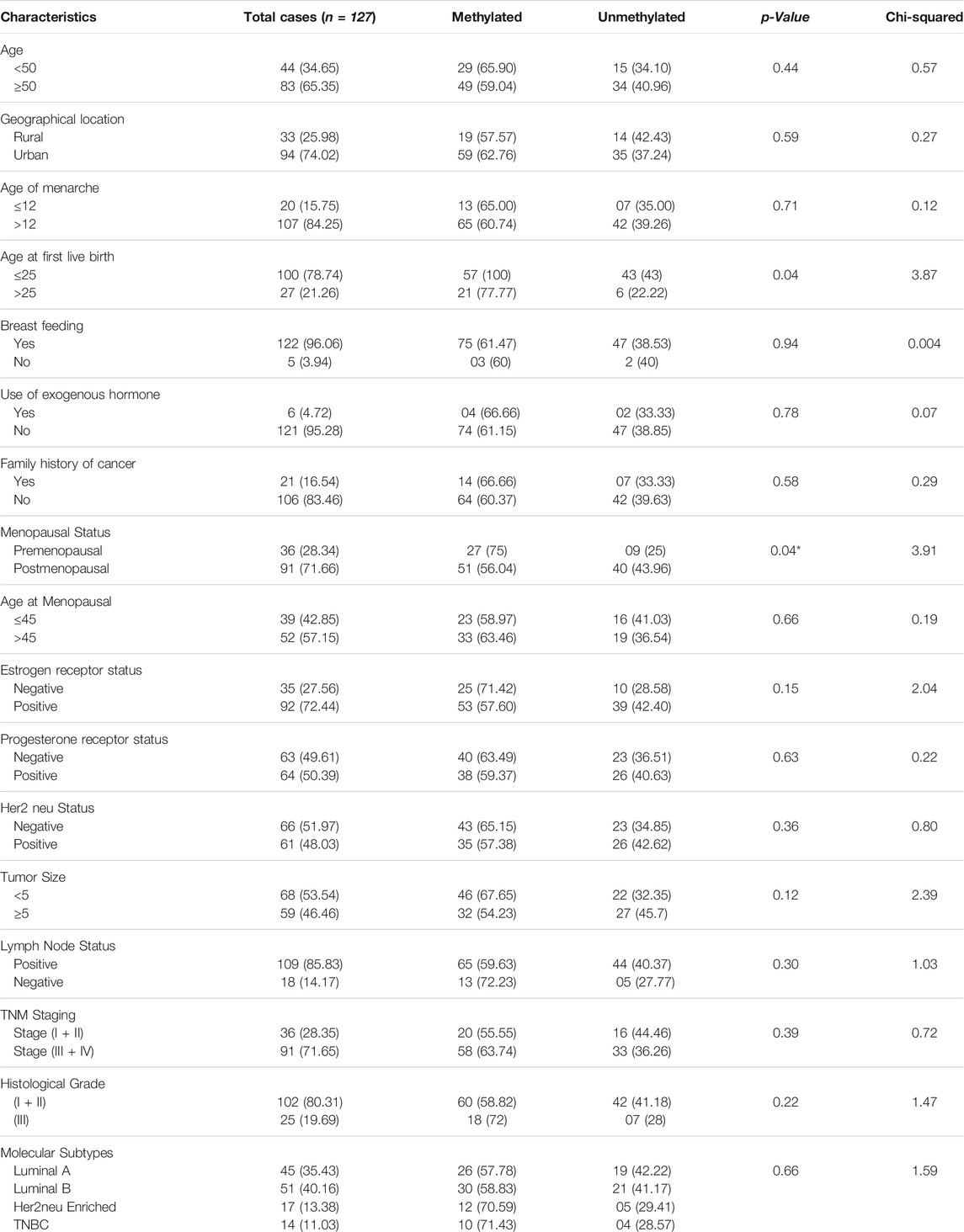
TABLE 3. Correlation study of FOXO1 Promoter Methylation status with clinical parameters of Breast Cancer Patients.
Low or No Expression of FOXO1 Protein in Breast Cancer Tissue
FOXO1 expression analysis at the protein level was done by IHC and it was found to be absent in 77.95% (99/127) cases. However, 28 Cases exhibited the moderate or high expression of FOXO1 protein. Also, the FOXO1 protein expression pattern substantiated the mRNA expression. Furthermore, the percentage of FOXO1 protein downregulation was significant with Her2 neu status, tumor size, and histological grade of breast cancer (Figure 4; Table 4.)
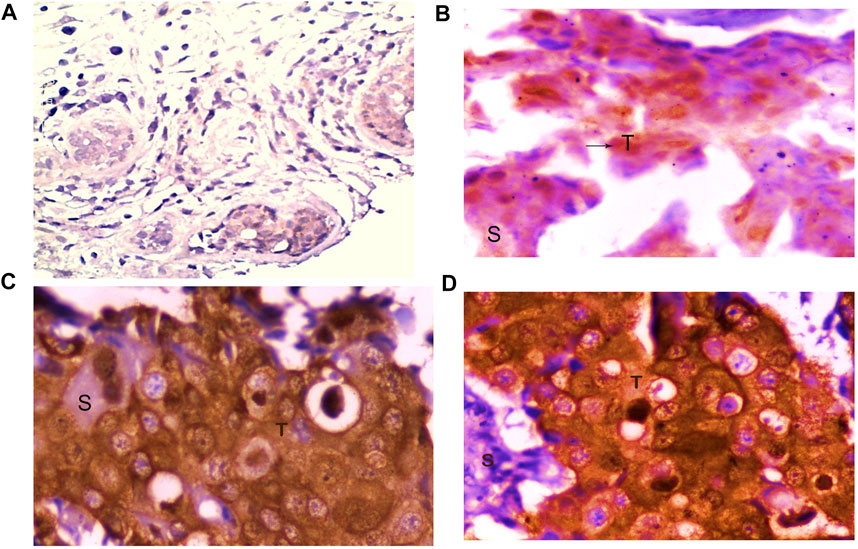
FIGURE 4. Representative picture of Immunohistochemical staining of human breast cancer tissue samples by anti-FOXO1 antibody (magnification: ×400) showing (A) no expression, (B) low (+) expression, (C) moderate (++) expression, and (D) high (+++) expression of FOXO1. S: stromal tissue, T: tumor tissue.
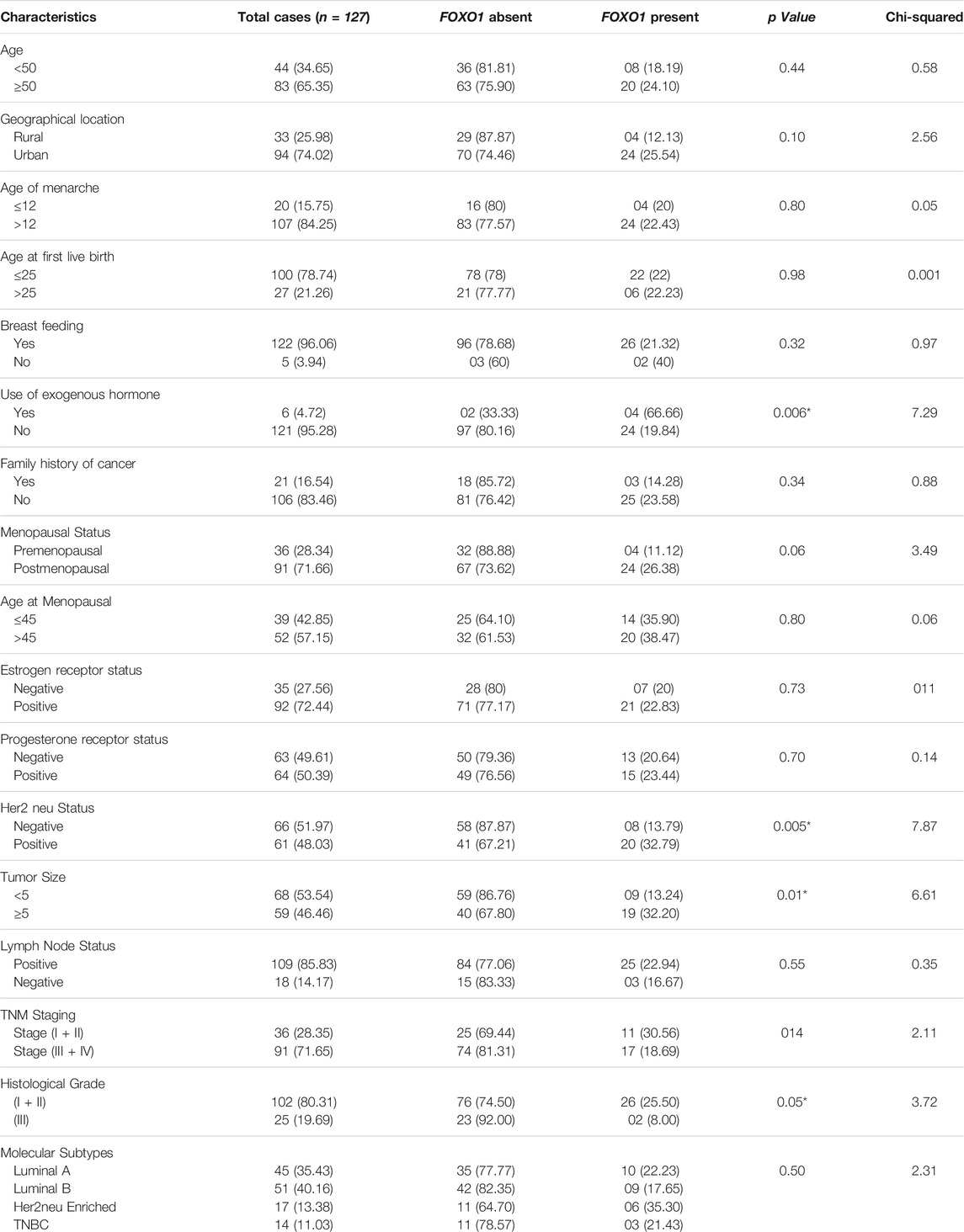
TABLE 4. Correlation of FOXO1 Protein Expression level with clinical parameters of Breast Cancer Patients.
FOXO1 Promoter Methylation and its Association With Protein Expression
The results represented a strong correlation of FOXO1 protein expression with the promoter methylation and 74 out of 78 hypermethylated cases showed low or no protein expression and 04 cases showed protein expression. In 51.02% (25/49) cases that showed no methylation had no protein expression. The cases which had downregulation of FOXO1 showed 78.40% (69/88) hypermethylation while 23.07% (9/39) cases had moderate to high-level protein expression. highly significant p-value (p= 0.0001) was found between FOXO1 methylation in the promoter region and protein expression, which represented a strong correlation Tables 5,6,7
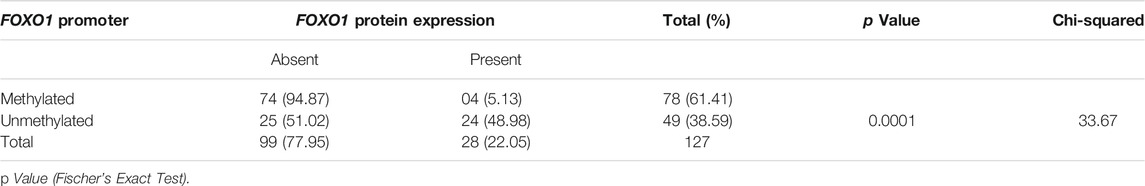
TABLE 5. Correlation study of Promoter Methylation with Protein expression in Breast Cancer Patients from North India.
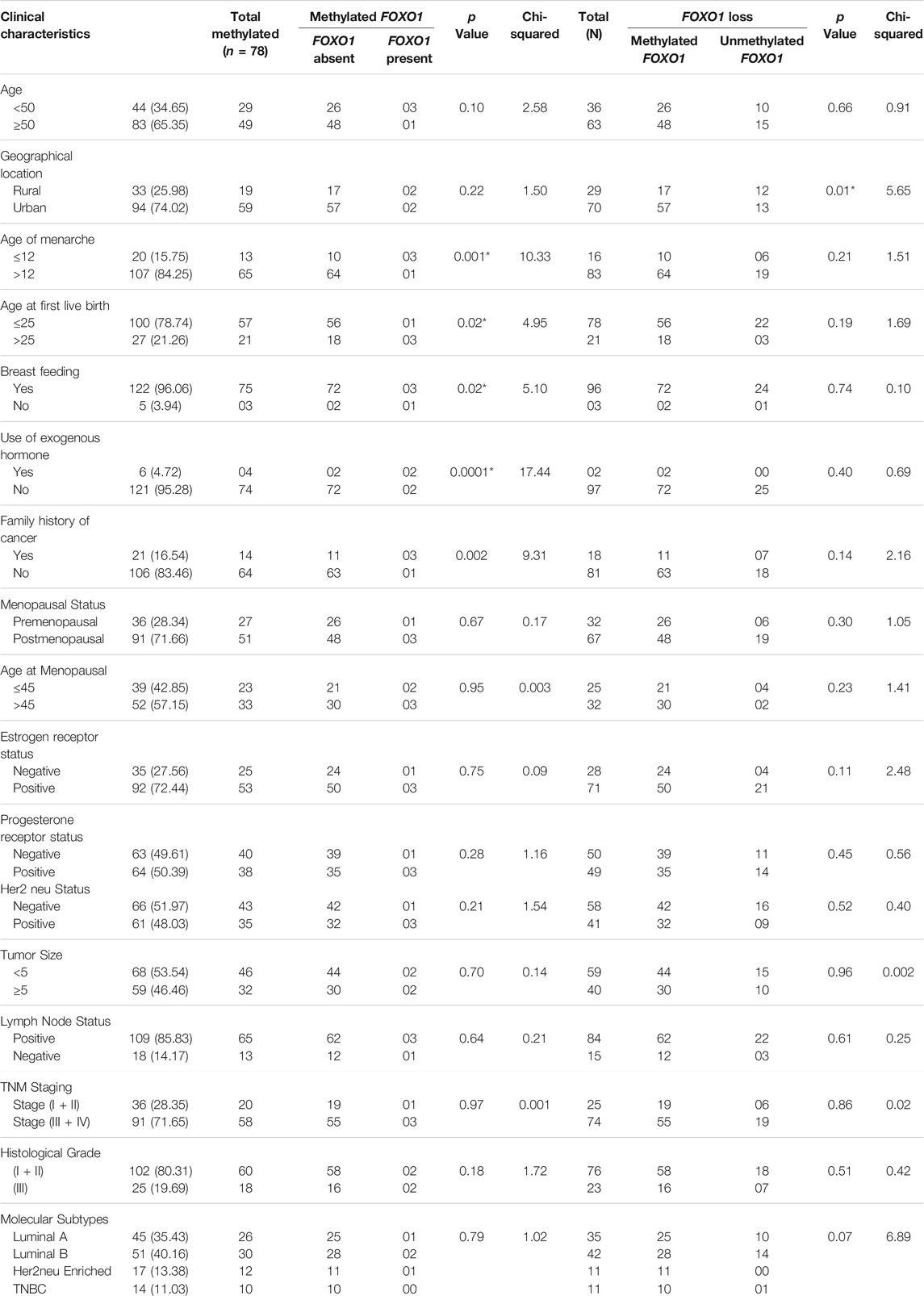
TABLE 6. Correlation study of methylation and protein expression in samples having methylated FOXO1 promoter or FOXO1 expression loss with clinical parameters of Breast cancer patients from North Indian population.
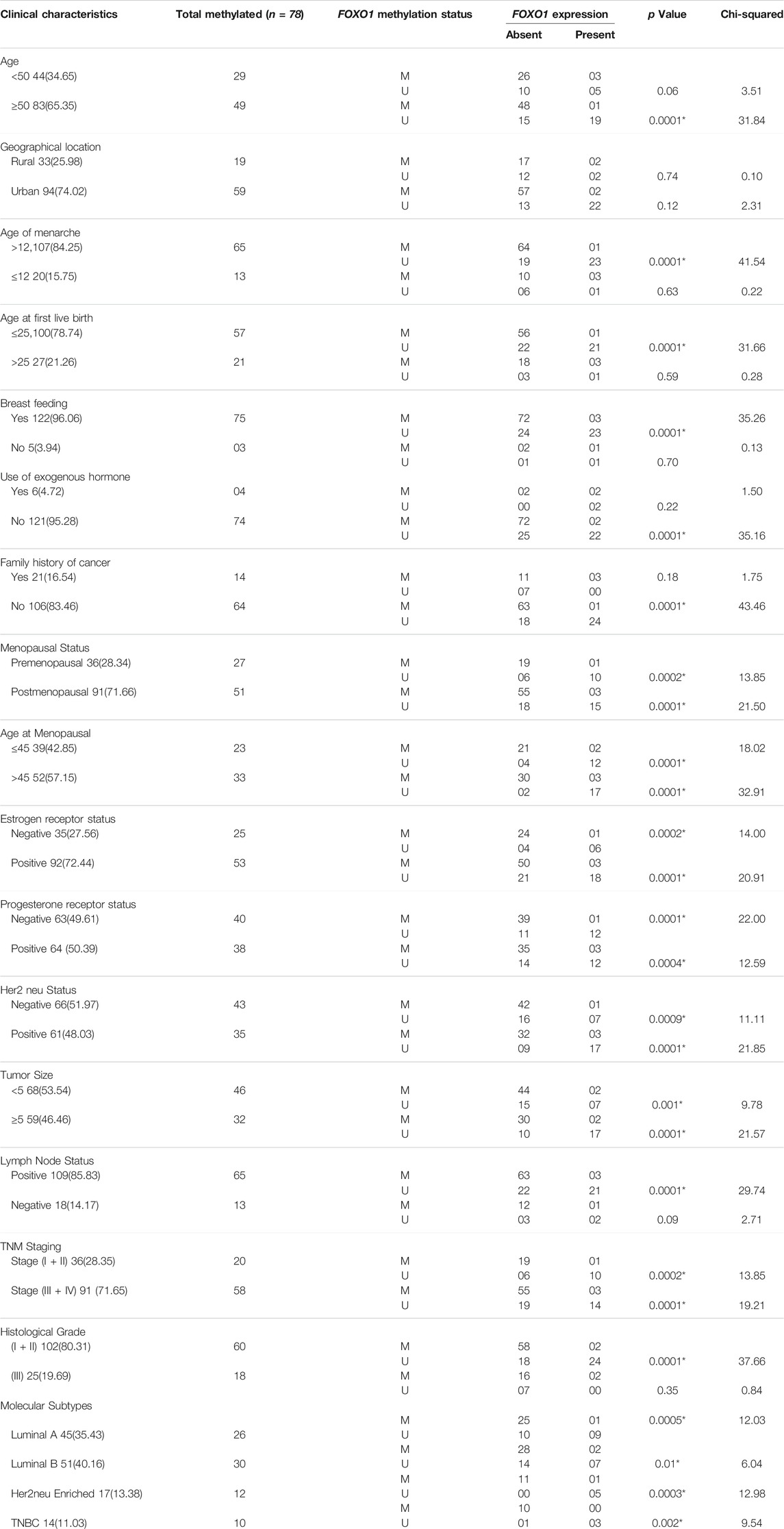
TABLE 7. Correlation analysis between FOXO1 methylation and FOXO1 protein expression in stratification by various clinical characteristics of Breast cancer patients from North India.
Discussion
FOXO subfamily of forkhead box transcription factor comprises four FOXO isoforms. FOXO1 which is a member of this subfamily is the key target of insulin that inhibits its transcriptional events through nuclear exclusion (Kousteni, 2012). The prominent role of FOXO1 is studied in the maintenance of tissue homeostasis at the time of various physiological as well as pathological conditions (Xing et al., 2018). In previous studies, the close association of lower FOXO1 levels with human cancers such as hepatocellular carcinoma, colorectal cancer, pancreatic cancer, prostate cancer, and lung cancer have been demonstrated (Wu et al., 2012; Schwartz and Cote, 2016; Lou et al., 2017; Moeinifard et al., 2017).
In the current study, we examined the expression level of FOXO1 in 127 breast cancer tissues taken along with adjacent normal tissues from the Indian female breast cancer patients. To analyze the FOXO1 mRNA expression, we performed real-time PCR. Further, we studied FOXO1 protein expression and its subcellular localization through immunohistochemistry and the epigenetic modulation in the promoter region of the FOXO1 was analyzed using MS-PCR. While investigating our data for FOXO1 mRNA expression we found 88 out of 127 cases (69.29%) exhibiting the downregulation at the FOXO1 mRNA level. The downregulation of FOXO1 mRNA in our data links positively to the previous studies which state the tumor suppressive role of the FOXO1 gene in cancer progression (Myatt et al., 2010; Prasad et al., 2014). Further, on correlating our results we found a strong correlation of FOXO1 mRNA expression with the age of the patient (p = 0.008), age at first live birth (p = 0.0003), tumor size (p = 0.05), and lymph node status (p = 0.01) of the breast cancer patients. FOXO1 mRNA downregulation was earlier reported to show a significant association with the lymph node status and age of the patients in prostate cancer cases (Yang et al., 2021), our study also reveals this strong correlation of FOXO1 mRNA expression with these clinical parameters in case of breast cancer patients.
The results of our study to detect the protein expression and localization of FOXO1 protein reveals 77.95% (99/127) cases having low or no expression of FOXO1 protein, whereas 22.05% (28/127) show moderate or high expression. The anti-proliferative role of FOXO1 has been reported in case of cervical and prostate cancer, moreover, the enforced expression of FOXO1 in endometrioid endometrial cancer cells and SiHa cells blocked the cell proliferation and decreased the tumorigenic activity (Goto et al., 2008; Zhang et al., 2015; Yang et al., 2021). Our results also suggest the tumor suppressive role of FOXO1 in breast cancer and show a strong association of low protein expression with the histological grade (p = 0.05) and tumor size (p = 0.01) of breast cancer patients. The downregulated expression of FOXO1 protein in the advanced stages (III and IV) of breast cancer suggests its repressive role in tumor progression and can be considered as the prognostic marker. Further, our investigation of FOXO1 gene promoter methylation represented the hypermethylation (61.41%) of the FOXO1 promoter region in most cases and a significant association (p = 0.0001) was found between the promoter methylation and protein expression of the FOXO1 gene. (Table 5, 6). Also, the FOXO1 promoter methylation exhibited significant association (p = 0.04) with the menopausal status of the female breast cancer patients, 65.38% (51/78) cases that showed promoter methylation were post-menopausal. Recent studies on Dysregulation of gene expression revealed the prominent role of epigenetics in gene silencing other than mutation, our study reveals FOXO1 promoter methylation to be associated with low or no expression of FOXO1 protein in breast cancer tissue in comparison to the adjacent normal tissue.
This study reports the tumor-suppressive role of FOXO1 in the case of Indian breast cancer patients and our data also suggested FOXO1 exhibited prognostic importance. Its downregulation is closely associated with the prognosis of the disease and different clinical parameters of the patients. We suggest that FOXO1 can be taken as a biomarker in the case of breast cancer and further research can be carried out to find therapeutic strategies in targeting the FOXO1 gene.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee Jamia Millia islamia and AIIMS New Delhi. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MK and SH took charge of the experimental design and execution. The experiments were executed by MK. The reagents, materials, and analysis tools were contributed by MK, Sa, SM, SD, MH, IA, NA, and SM. Manuscript preparation was carried out by MK and co-authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Author MK acknowledge ICMR for providing fellowship (Fileno 3/2/2/23/2019/NCD-III.) and the author IA, is grateful to Scientific Research Deanship at King Khalid University, Abha, Saudi Arabia for their financial support through the Small Research Group Project under grant number (RGP.01-48-42).
References
Cantley, L. C. (2002). The Phosphoinositide 3-kinase Pathway. Science 296 (5573), 1655–1657. doi:10.1126/science.296.5573.1655
Chan, C., Law, B., So, W., Chow, K., and Waye, M. (2017). Novel Strategies on Personalized Medicine for Breast Cancer Treatment: an Update. Ijms 18 (11), 2423. doi:10.3390/ijms18112423
Goto, T., Takano, M., Albergaria, A., Briese, J., Pomeranz, K. M., Cloke, B., et al. (2008). Mechanism and Functional Consequences of Loss of FOXO1 Expression in Endometrioid Endometrial Cancer Cells. Oncogene 27 (1), 9–19. doi:10.1038/sj.onc.1210626
Jiang, P., Gu, S., Pan, D., Fu, J., Sahu, A., Hu, X., et al. (2018). Signatures of T Cell Dysfunction and Exclusion Predict Cancer Immunotherapy Response. Nat. Med. 24 (10), 1550–1558. doi:10.1038/s41591-018-0136-1
Kaymaz, Y., Oduor, C. I., Yu, H., Otieno, J. A., Ong'echa, J. M., Moormann, A. M., et al. (2017). Comprehensive Transcriptome and Mutational Profiling of Endemic Burkitt Lymphoma Reveals EBV Type-specific Differences. Mol. Cancer Res. 15 (5), 563–576. doi:10.1158/1541-7786.mcr-16-0305
Khan, M. A., Tiwari, D., Dongre, A., Sadaf, S., Mustafa, S., Das, C. R., et al. (2020). Exploring the P53 Connection of Cervical Cancer Pathogenesis Involving north-east Indian Patients. PloS one 15 (9), e0238500. doi:10.1371/journal.pone.0238500
Kousteni, S. (2012). FOXO1, the Transcriptional Chief of Staff of Energy Metabolism. Bone 50 (2), 437–443. doi:10.1016/j.bone.2011.06.034
Li, X., Kover, K. L., Heruth, D. P., Watkins, D. J., Moore, W. V., Jackson, K., et al. (2015). New Insight into Metformin Action: Regulation of ChREBP and FOXO1 Activities in Endothelial Cells. Mol. Endocrinol. 29 (8), 1184–1194. doi:10.1210/me.2015-1090
Lou, W., Liu, J., Gao, Y., Zhong, G., Chen, D., Shen, J., et al. (2017). MicroRNAs in Cancer Metastasis and Angiogenesis. Oncotarget 8 (70), 115787–115802. doi:10.18632/oncotarget.23115
Moeinifard, M., Hassan, Z. M., Fallahian, F., Hamzeloo-Moghadam, M., and Taghikhani, M. (2017). Britannin Induces Apoptosis through AKT-FOXO1 Pathway in Human Pancreatic Cancer Cells. Biomed. Pharmacother. 94, 1101–1110. doi:10.1016/j.biopha.2017.08.025
Myatt, S. S., Wang, J., Monteiro, L. J., Christian, M., Ho, K.-K., Fusi, L., et al. (2010). Definition of microRNAs that Repress Expression of the Tumor Suppressor Gene FOXO1 in Endometrial Cancer. Cancer Res. 70 (1), 367–377. doi:10.1158/0008-5472.can-09-1891
Prasad, S. B., Yadav, S. S., Das, M., Govardhan, H. B., Pandey, L. K., Singh, S., et al. (2014). Down Regulation of FOXO1 Promotes Cell Proliferation in Cervical Cancer. J. Cancer 5 (8), 655–662. doi:10.7150/jca.6554
Procaccia, S., Ordan, M., Cohen, I., Bendetz-Nezer, S., and Seger, R. (2017). Direct Binding of MEK1 and MEK2 to AKT Induces FOXO1 Phosphorylation, Cellular Migration and Metastasis. Sci. Rep. 7 (1), 1–16. doi:10.1038/srep43078
Real, S. A. S., Parveen, F., Rehman, A. U., Khan, M. A., Deo, S. V. S., Shukla, N. K., et al. (2018). Aberrant Promoter Methylation of YAP Gene and its Subsequent Downregulation in Indian Breast Cancer Patients. BMC cancer 18 (1), 711–715. doi:10.1186/s12885-018-4627-8
Russell, D. W., and Sambrook, J. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 112.
Sadaf, M., Habib, M., Khan, M. A., Najm, M. Z., Mallick, M. N., Sunita, K., et al. (2018). Hypermethylated LATS2 Gene with Decreased Expression in Female Breast Cancer: A Case Control Study from North India. Gene 676, 156–163. doi:10.1016/j.gene.2018.07.033
Schwartz, A. G., and Cote, M. L. (2016). Epidemiology of Lung Cancer. Lung Cancer Personalized Med. 79, 21–41. doi:10.1007/978-3-319-24223-1_2
Shi, M., Jiang, Y., Yang, L., Yan, S., Wang, Y. G., and Lu, X. J. (2018). Decreased Levels of Serum Exosomal miR‐638 Predict Poor Prognosis in Hepatocellular Carcinoma. J. Cell. Biochem. 119 (6), 4711–4716. doi:10.1002/jcb.26650
Tang, C.-y., Zhu, L.-x., Yu, J.-d., Chen, Z., Gu, M.-c., Mu, C.-f., et al. (2018). Effect of β-elemene on the Kinetics of Intracellular Transport of D-Luciferin Potassium Salt (ABC Substrate) in Doxorubicin-Resistant Breast Cancer Cells and the Associated Molecular Mechanism. Eur. J. Pharm. Sci. 120, 20–29. doi:10.1016/j.ejps.2018.04.037
Viale, G. (2012). The Current State of Breast Cancer Classification. Ann. Oncol. 23, x207–x210. doi:10.1093/annonc/mds326
Wu, Z., Sun, H., Zeng, W., He, J., and Mao, X. (2012). Upregulation of MircoRNA-370 Induces Proliferation in Human Prostate Cancer Cells by Downregulating the Transcription Factor FOXO1. PLoS One 7 (9), e45825. doi:10.1371/journal.pone.0045825
Xing, Y.-q., Li, A., Yang, Y., Li, X.-x., Zhang, L.-n., and Guo, H.-c. (2018). The Regulation of FOXO1 and its Role in Disease Progression. Life Sci. 193, 124–131. doi:10.1016/j.lfs.2017.11.030
Yang, N., Wu, J., Zhang, T., Yang, F., Shao, J., He, C., et al. (2021). Clinical Evaluation of FOXO1 as a Tumor Suppressor in Prostate Cancer. Comput. Math. Methods Med. 2021 (1), 810. doi:10.1155/2021/8773423
Keywords: methylation, immunohistochemistry, clinical, tumor, diagnosis
Citation: Khan MA, Massey S, Ahmad I, Sadaf , Akhter N, Habib M, Mustafa S, Deo SVS and Husain SA (2022) FOXO1 Gene Downregulation and Promoter Methylation Exhibits Significant Correlation With Clinical Parameters in Indian Breast Cancer Patients. Front. Genet. 13:842943. doi: 10.3389/fgene.2022.842943
Received: 24 December 2021; Accepted: 10 February 2022;
Published: 03 March 2022.
Edited by:
Kaiser Jamil, Bhagwan Mahavir Medical Research Centre, IndiaReviewed by:
Jogendra Pawar, Purdue University, United StatesKanchan Bhatia, Arizona State University, United States
Copyright © 2022 Khan, Massey, Ahmad, Sadaf, Akhter, Habib, Mustafa, Deo and Husain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Syed Akhtar Husain, c2h1c2FpbkBqbWkuYWMuaW4=
 Mohammad Aasif Khan
Mohammad Aasif Khan Sheersh Massey1
Sheersh Massey1 Irfan Ahmad
Irfan Ahmad Sadaf
Sadaf S. V. S. Deo
S. V. S. Deo Syed Akhtar Husain
Syed Akhtar Husain