
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 11 March 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.841681
 Xiaobo Fan1,2
Xiaobo Fan1,2 Krit Pinthong3
Krit Pinthong3 Edivaldo H. C. de Oliveira4
Edivaldo H. C. de Oliveira4 Alongklod Tanomtong3
Alongklod Tanomtong3 Hongwei Chen2
Hongwei Chen2 Anja Weise2
Anja Weise2 Thomas Liehr2*
Thomas Liehr2*The chromosomal homologies of human (Homo sapiens—HSA) and Trachypithecus phayrei (TPH—Phayre’s leaf-monkey, family Cercopithecidae) have previously been studied by using classical chromosome staining/banding and fluorescence in situ hybridization (FISH) from the 1970s to 1990s. In this study, we carried out molecular cytogenetics applying human multicolor banding (MCB), locus-specific, and human heterochromatin-specific probes to establish the first detailed chromosomal map of TPH, which was not available until now. Accordingly, it was possible to precisely determine evolutionary-conserved breakpoints (ECBs) and the orientation of evolutionary-conserved segments compared to HSA. It could be shown that five chromosomes remained completely unchanged between these two species, and 16 chromosomes underwent only intrachromosomal changes. In addition, 50 ECBs that failed to be resolved in previous reports were exactly identified and characterized in this study. It could also be shown that 43.5% of TPH centromere positions were conserved and 56.5% were altered compared to HSA. Interestingly, 82% ECBs in TPH corresponded to human fragile sites. Overall, this study is an essential contribution to future studies and reviews on chromosomal evolution in Cercopithecidae.
Trachypithecus phayrei (TPH), also known as Phayre’s leaf monkey or Phayre’s lutung (Behie and Groves, 2016), belongs to old-world monkeys (OWMs), family Cercopithecidae, subfamily Colobinae—the latter including an African and an Asian clade. The genus Trachypithecus comprises 17 species with one Asian colobine—TPH (Pinthong et al., 2018). TPH is widely distributed in continental Southeast Asia including India, Bangladesh, Western Myanmar, Thailand, Laos, Vietnam, and Southern China (Muul, 2002). It is important to notice that genus TPH was initially denominated with different Latin names, such as Semnopithecus phayrei and Presbytis phayrei, before the current name came into use (Gupta and Kumar, 1994).
The pedigree and chromosomal evolution of Hominidae has been principally and roughly resolved in previous cytogenetic and molecular cytogenetic studies; however, some gaps remain, including the karyotype of TPH (Stanyon et al., 2008). The latter was first described in 1970 as 2n = 44 (Hsu and Benirschke, 1970). In 1981, G banding revealed for a male TPH the karyotype composition is as follows: 22 (M) + 18 (SM) + 2 (A), XX (SM) (Chen et al., 1981). In 1998, chromosomal homologies between human and TPH chromosomes were established by FISH applying human whole chromosome paintings. This revealed unique reciprocal translocations corresponding to chromosomes of (Homo sapiens) HSA 1 and 19, and HSA 6 and 16 as well as fusions of HSA 14 and 15 and HSA 21 and 22 (Nie et al., 1998). In 2018, the subspecies TPH crepuscula was studied by GTG-banding and NOR staining (Pinthong et al., 2018).
Accordingly, up to now, there have been few or neither really comprehensive nor high-resolution FISH-banding–based (Mrasek et al., 2001; Liehr and Claussen, 2002; Weise et al., 2008) comparative molecular cytogenetic reports on homologies between HSA and TPH chromosomes. Thus, here, the first detailed comparative chromosomal map of TPH compared to HSA is presented, established by MCB and complementary heterochromtin- and one locus-specific probe(s). Furthermore, the results obtained in TPH were compared to karyotypes of Macaques (such as Macaca fascicularis = MMU) (Fan et al., 2014) and Silvery lutung (Trachypithecus cristatus = TCR) (Xiaobo et al., 2013), which were studied by identical high-resolution molecular cytogenetic approaches. Additionally, the relationship of ECBs with human fragile sites was analyzed.
An immortalized lymphoblast cell line derived from male TPH (#KKU-THPm6) was provided by the Department of Biology Faculty of Science, Khon Kaen University, Thailand. The animal was caught for less than 30 min from wilderness, its species was determined, and blood was acquired. Afterward the animal was set free again. Ethical review and approval were waived for this study due to the use of only a cell line.
Chromosomes were prepared from B-lymphocytes of the cell line according to standard procedures. FISH was done as previously reported using 24 human chromosome-specific multicolor-banding probe sets for all chromosomes (Mrasek et al., 2001; Liehr and Claussen, 2002; Liehr et al., 2002; Weise et al., 2008). Also, single and two-color FISH techniques were performed for mapping of ECBs by one locus-specific probe for the NOR region and human heterochromatin-specific probes on a probe set described previously (Bucksch et al., 2012).
Images were captured using an Axioplan II microscope (Carl Zeiss Jena GmbH, Germany) equipped with six corresponding filter sets for multicolor-FISH evaluation (DAPI, FITC, TR, SO, Cy5, and DEAC). Image analysis was done using pseudocolor banding and fluorochrome profiles of the ISIS digital FISH-imaging system (MetaSystems Hard and Software GmbH, Altlussheim, Germany). At least, 10–20 metaphases were recorded and applied probe or probe set.
Results obtained in molecular cytogenetic studies are summarized in Figure 1 and Table 1.
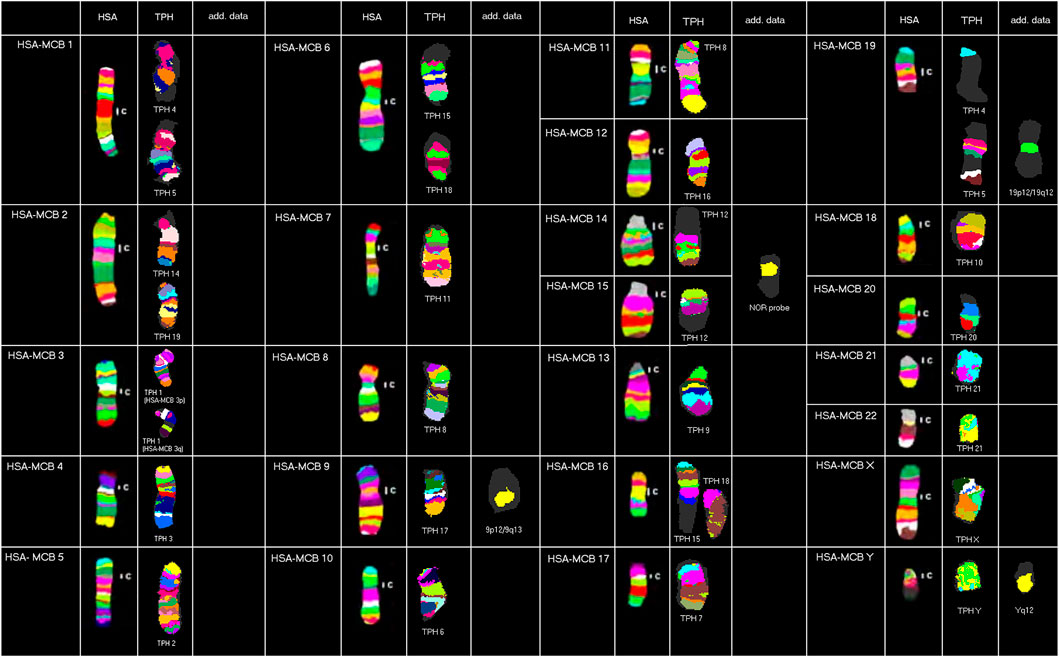
FIGURE 1. Representative results from this study using human MCB, NOR, and human heterochromatin-specific probes on TPH are depicted as pseudo-colored results for HSA and TPH (only valid for MCB results). The chromosomes are sorted here according to the HSA-chromosomes by using MCB. TPH chromosomes are numbered according to Nie et al. (1998).

TABLE 1. Homologous regions, the centromere position (C), and colocalization with human fragile sites (FS). FSs are listed acc. to Mrasek et al. (2010).
Overall, the majority of TPH chromosomes are completely homologous to one of the human chromosomes; exceptions are chromosomal exchanges that took place as follows: TPH 4 and 5 (homologous to HSA 1 and 19), TPH 12 (homologous to HSA 14 and 15), TPH 15 and 18 (homologous to HSA 6 and 16), and TPH 21 (homologous to HSA 21 and 22). The centromeric positions could be identified at the sub-band level for all 23 TPH chromosomes. In the following chromosomes, the TPH centromeric positions were the same as in HSA: TPH 2 (= HSA 5), TPH 4 (= HSA 1), TPH 5 (= HSA 19), TPH 6 (= HSA 10), TPH 7 (= HSA 17), TPH 8 (= HSA 11), TPH 11 (= HSA 7), TPH 16 (= HSA 12), TPH X (= HSA X), and TPH Y (= HSA Y). Centromere positions shifted compared to HSA as follows: TPH 1 (HSA 3q26), TPH 3 (HSA 4q21.2), TPH 9 (HSA 13q11.1), TPH 10 (HSA 18q21.32), TPH 12 (HSA 15q26.3/14q11.1), TPH 13 (HSA 8p12), TPH 14 (HSA 2q24.3), TPH 15 (HSA 16p11.2), TPH 17 (HSA 9q33.2), TPH 18 (HSA 6q24.3), TPH 19 (HSA 2p14), TPH 20 (HSA 20q13.3), and TPH 21 (HSA 21q22.3/22q11.1).
Furthermore, repetitive DNA was identified by human heterochromatin-specific probes as follows: the repetitive sequence D1Z5 located in HSA 1q11-q12 was not present in TPH 4 or TPH 5, while the region being present in human as the band 19p12/19q12 could be found in TPH 5. The human hemi-heterochromatic region 9p12/9q13 was located on the long arm of TPH 17, while D9Z3 (HSA 9q12) and D16Z3 (HSA 16q11.2) were not detectable in TPH. NOR signals can be found in the centromere region of TPH 21. Repetitive DNA in the human male in Yq12 also was observed in TPH Y. Overall, only HSA chromosomes 4 (TPH 3), 8 (TPH 13), 12 (TPH 16), X (TPH X), and Y (TPH Y) were completely unaltered during evolution between these two relatively distantly related species among OWMs.
Table 2 summarizes 50 ECBs observed in TPH in this study, which were identified according to the homologous regions in HSA. In addition, the characterized TPH breakpoints were compared with previously reported ones in TCR and in other macaque species using the MCB approach (Table 2).
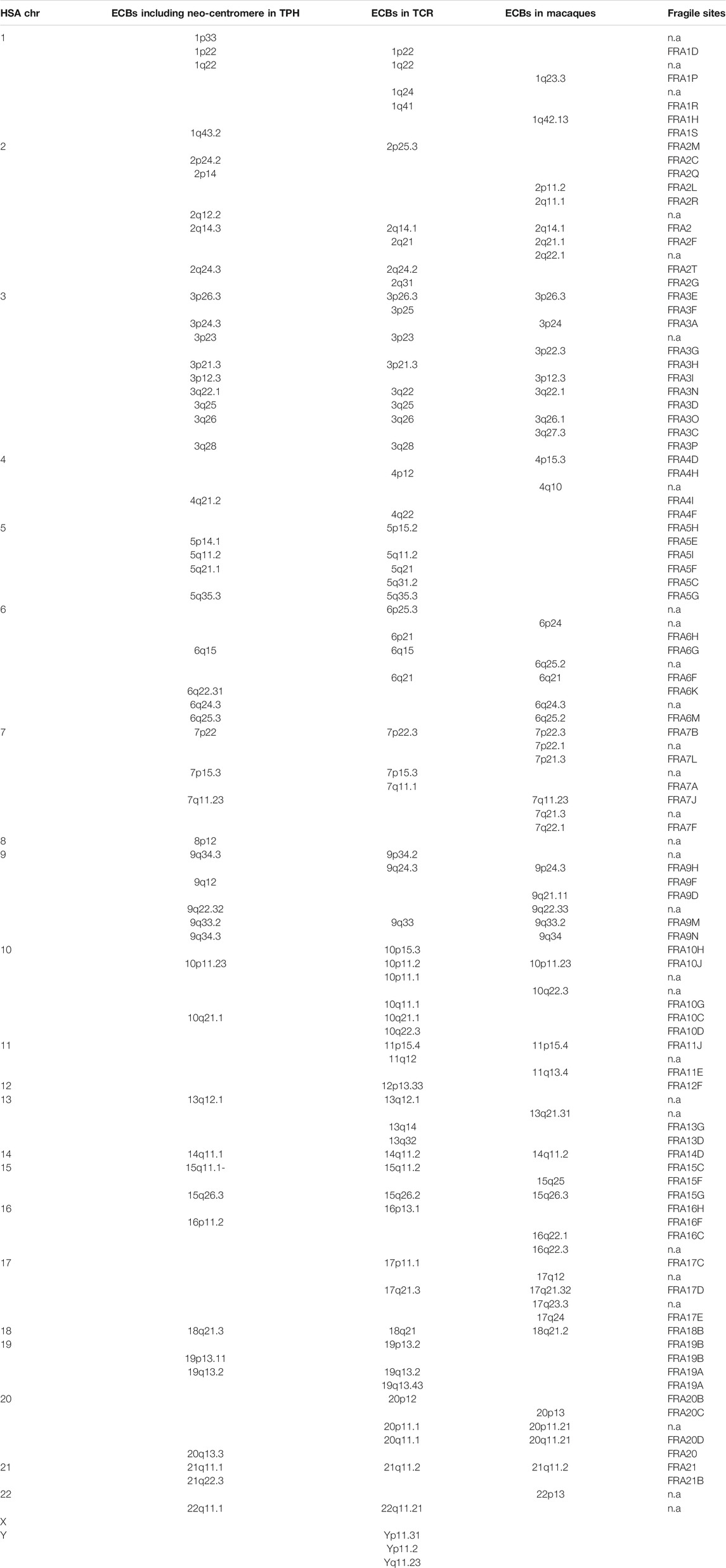
TABLE 2. Colocalization of ECBs and FSs in TPH, TCR and Macaque species. Nomenclature and data acc. to (Xiaobo et al., 2013; Fan et al., 2014; Mrasek et al., 2010).
The co-localization of ECBs among TPH, TCR, and in macaque species are listed with respect to HSA in Table 2. Out of 50 ECBs mapped in TPH, 29 (58%) and 18 (36%) coincided with ECBs in TCR and macaques, respectively (Figure 2; Table 3). Moreover, 41 (82%) reported ECBs in TPH co-localized with human fragile sites (Figure 2; Table 4).
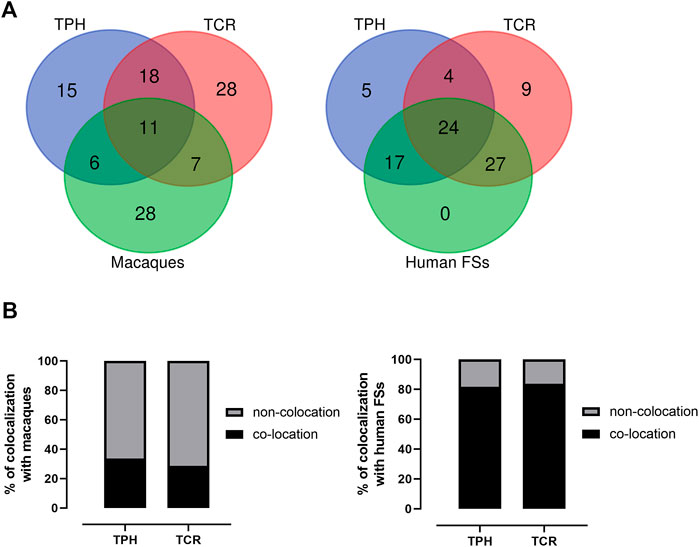
FIGURE 2. Identification of the relationship of ECBs in TPH with those in TCR, macaque species, and fragile sites. (A) Venn Diagrams depicting overlaps of TPH ECBs between TCR and macaques, and overlaps of the co-localization of ECBs in TPH with human fragile sites compared with the co-localization of ECBs in TCR with human fragile sites. (B) Left: quantification of the proportions of the co-localization of ECBs in TPH/TCR in macaques. In total, 32% of TPH ECBs and 26% of TCR ECBs were co-localizated in macaques. Right: quantification of the proportions of the co-localization of ECBs in TPH/TCR with human fragile sites. In total, 82% of TPH ECBs and 80% of TCR ECBs co-localized with human fragile sites.

TABLE 3. ECBs in TPH, TCR, and macaque species given as corresponding homologous human chromosome bands.
MCB combined with heterochromatin- and a locus-specific probe is suited best to characterize basic karyotypic features in primates, as shown in our previous studies (Mrasek et al., 2001; Fan et al., 2014; Fan et al., 2015; Xiaobo et al., 2013; Sangpakdee et al., 2018). In this study, the first comprehensive characterization of the karyotype of TPH was done; and a comparison with that in TCR and macaques was performed accordingly (Fan et al., 2014; Xiaobo et al., 2013). Our results confirmed and refined previous cytogenetic studies of TPH chromosomes, which were at a much lower resolution (Nie et al., 1998; Pinthong et al., 2018). These results extended to a detailed characterization of all TPH chromosomes aligned to HSA by MCB, that were not available before (Dutrillaux et al., 1979; Rhesus Macaque Genome Sequencing and Analysis Consortium et al., 2007). NOR was mapped to ECBs/fusion points of HSA 14 and HSA 15 (corresponding to TPH 12) confirming previous results (Pinthong et al., 2018). Compared to the basic Hominidea karyotype, five chromosomes remained unchanged in TPH, namely chromosomes 3, 13, 16, X, and Y, similar to those in TPH (Pinthong et al., 2018) and related species (Misceo et al., 2008). In addition, compared to HSA, complex chromosomal rearrangements (Table 1) first described here took place during the evolutionary process when the common ancestor of HSA and TPH diverged and may further continue.
ECBs must have undergone breaking and rejoining of double-strand breaks (Tsai and Lieber, 2010). These evolutionary conserved chromosomal changes could have been driven by several factors, such as the intrinsic instability of segmental duplications (SDs) enriched in the flanking regions of ECBs. SDs have been suggested to have a significant impact on genome plasticity during the evolution of primate chromosomes in previous studies (Kehrer-Sawatzki and Cooper, 2008). It is suggested that SDs within recombination hotspots might mediate non-allelic homologous recombination (NAHR). For example, two homologous SDs on the same chromosome, but in opposite orientation, could be the bases of an inversion. If SDs are in direct orientation, NAHR results in duplication and/or deletion as reported in human microdeletion-/microduplication syndromes and bases of copy-number variant regions (CNV’s) in human (Liehr, 2021). SDs located on different chromosomes can be the bases of NAHR-mediated chromosomal translocations (Tsai and Lieber, 2010; Gu et al., 2008).
While in previous reports, there were no detailed and characterized centromeric regions of TPH in corresponding reviews on OWMs (Ventura et al., 2004; Ventura et al., 2007; Stanyon et al., 2008), here, a first clue was possible about positions of centromeric regions in TPH (Table 1), that is, 56.5% TPH centromere positions shifted and 43.5% centromere positions were conserved compared to HSA. This is similar to the situation in TCR, that is, conserved centromeres in TPH kept their positions during evolution from common ancestors. However, these conserved centromeric regions normally do not have identical alphoid sequences as in HSA (Rocchi et al., 2012), and neo-centromeres are preferentially formed most often in gene deserts (Lomiento et al., 2008).
There are 29 identical ECBs in TPH and in TCR, and 17 ECBs are in concordance with those in macaque species. Moreover, 11 identical ECBs were identified in TPH, in TCR, and in macaque species (Tables 3 and 4). These findings are useful for the reconstruction of a common ancestral karyotype in further studies by applying, for example, locus-specific FISH-probes and/or sequencing of the TPH genome. In total, 41 (82%) of reported 50 ECBs in TPH corresponded to human fragile sites, which is in concordance to previous observations in TCR that ECB regions are highly connected to common FS locations (Francis, 2002, Mrasek et al., 2010; Fungtammasan et al., 2012). It has been suggested that FSs are low-stability regions, supporting their potential role in the formation of evolutionary chromosomal rearrangements (Mishmar et al., 1998). In this connection, others suggested the involvement of the cellular checkpoints proteins ATR and BRCA1, which are also critical for the expression of FSs (Casper et al., 2002; Arlt et al., 2006; Glover, 2006). Also, comparative analyses showed that the co-localization of ECBs in TPH/TCR with human FSs revealed no differences, indicating that Asian langurs are karyotypically closely related (Alekseyev and Pevzner, 2010).
In conclusion, the presented TPH karyotype and comparison to other langurs and macaques provided new insights into chromosomal evolution. It is another stepping stone in primate evolution research.
Raw data supporting the conclusion of this article will be made available by the authors on request, without undue reservation.
Conceptualization: AW, TL, and XF. Methodology: XF and KP. Validation: XP, KP, EdO, AW, and TL. Formal analysis: XF. Investigation: XF, AW, and HC. Resources: AT, KP, and TL. Writing—original draft preparation: XF and HC. Writing—review and editing: XF and TL. Supervision: TL. Project administration: TL. Funding acquisition: TL and XF. All authors have read and agreed to the published version of the manuscript.
This work was supported by the Natural Science Foundation of China project (Grant No. 81801512 to XF) and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 18KJD180003 to XF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alekseyev, M. A., and Pevzner, P. A. (2010). Comparative Genomics Reveals Birth and Death of Fragile Regions in Mammalian Evolution. Genome Biol. 11, R117. doi:10.1186/gb-2010-11-11-r117
Arlt, M. F., Durkin, S. G., Ragland, R. L., and Glover, T. W. (2006). Common Fragile Sites as Targets for Chromosome Rearrangements. DNA Repair 5, 1126–1135. doi:10.1016/j.dnarep.2006.05.010
Behie, A. M., and Groves, C. P. (2016). China's Primates: Preserve Wild Species. Nature 534, 179. doi:10.1038/534179c
Bucksch, M., Ziegler, M., Kosayakova, N., Mulatinho, M. V., Llerena, J. C., Morlot, S., et al. (2012). A New Multicolor Fluorescence In Situ Hybridization Probe Set Directed against Human Heterochromatin. J. Histochem. Cytochem. 60, 530–536. doi:10.1369/0022155412441708
Casper, A. M., Nghiem, P., Arlt, M. F., and Glover, T. W. (2002). ATR Regulates Fragile Site Stability. Cell 111, 779–789. doi:10.1016/s0092-8674(02)01113-3
Chen, Y., Luo, L., Shan, X., and Cao, X. (1981). Primates Chromosome in China. Beijing: Science Publishing House.
Dutrillaux, B., Biemont, M. C., Viegas-Pequignot, E., and Laurent, C. (1979). Comparison of the Karyotypes of Four Cercopithecoidae: Papio papio, P. Anubis, Macaca mulatta,and M. fascicularis. Cytogenet. Cel Genet 23, 77–83. doi:10.1159/000131305
Fan, X., Supiwong, W., Weise, A., Mrasek, K., Kosyakova, N., Tanomtong, A., et al. (2015). Comprehensive Characterization of Evolutionary Conserved Breakpoints in Four New World Monkey Karyotypes Compared to Chlorocebus Aethiops and Homo sapiens. Heliyon 1, e00042. doi:10.1016/j.heliyon.2015.e00042
Fan, X., Tanomtong, A., Chaveerach, A., Pinthong, K., Pornnarong, S., Supiwong, W., et al. (2014). High Resolution Karyotype of Thai Crab-Eating Macaque (Macaca fascicularis). Genetika 46, 877–882. doi:10.2298/gensr1403877f
Francis, C. M. (2002). A Photographic Guide to Mammals of South-East Asia Including Thailand, Malaysia, Singapore, Myanmar, Laos, Vietnam, Cambodia, Java, Sumatra, Bali and Borneo. J. Mammal. 83, 633–634. doi:10.1644/1545-1542(2002)083<0633:>2.0.co;2
Fungtammasan, A., Walsh, E., Chiaromonte, F., Eckert, K. A., and Makova, K. D. (2012). A Genome-wide Analysis of Common Fragile Sites: what Features Determine Chromosomal Instability in the Human Genome? Genome Res. 22, 993–1005. doi:10.1101/gr.134395.111
Rhesus Macaque Genome Sequencing and Analysis Consortium Gibbs, R. A., Gibbs, R. A., Rogers, J., Katze, M. G., Bumgarner, R., Weinstock, G. M., et al. (2007). Evolutionary and Biomedical Insights from the Rhesus Macaque Genome. Science 316, 222–234. doi:10.1126/science.1139247
Glover, T. W. (2006). Common Fragile Sites. Cancer Lett. 232, 4–12. doi:10.1016/j.canlet.2005.08.032
Gu, W., Zhang, F., and Lupski, J. R. (2008). Mechanisms for Human Genomic Rearrangements. Pathogenetics 1, 4–17. doi:10.1186/1755-8417-1-4
Hsu, T. C., and Benirschke, K. (1970). An Atlas of Mammalian Chromosomes. New York: Springer Science & Business Media.
Kehrer-Sawatzki, H., and Cooper, D. N. (2008). Molecular Mechanisms of Chromosomal Rearrangement during Primate Evolution. Chromosome Res. 16, 41–56. doi:10.1007/s10577-007-1207-1
Kumar Gupta, A., and Kumar, A. (1994). Feeding Ecology and Conservation of the Phayre's Leaf Monkey Presbytis Phayrei in Northeast India. Biol. Conservation 69, 301–306. doi:10.1016/0006-3207(94)90430-8
Liehr, T., and Claussen, U. (2002). Current Developments in Human Molecular Cytogenetic Techniques. Cmm 2, 283–297. doi:10.2174/1566524024605725
Liehr, T., Heller, A., Starke, H., Rubtsov, N., Trifonov, V., Mrasek, K., et al. (2002). Microdissection Based High Resolution Multicolor Banding for All 24 Human Chromosomes. Int. J. Mol. Med. 9, 335–339. doi:10.3892/ijmm.9.4.335
Lomiento, M., Jiang, Z., D’Addabbo, P., Eichler, E. E., and Rocchi, M. (2008). Evolutionary-new Centromeres Preferentially Emerge within Gene Deserts. Genome Biol. 9, R173. doi:10.1186/gb-2008-9-12-r173
Misceo, D., Capozzi, O., Roberto, R., Dell’Oglio, M. P., Rocchi, M., Stanyon, R., et al. (2008). Tracking the Complex Flow of Chromosome Rearrangements from the Hominoidea Ancestor to Extant Hylobates and Nomascus Gibbons by High-Resolution Synteny Mapping. Genome Res. 18, 1530–1537. doi:10.1101/gr.078295.108
Mishmar, D., Rahat, A., Scherer, S. W., Nyakatura, G., Hinzmann, B., Kohwi, Y., et al. (1998). Molecular Characterization of a Common Fragile Site (FRA7H) on Human Chromosome 7 by the Cloning of a Simian Virus 40 Integration Site. Proc. Natl. Acad. Sci. 95, 8141–8146. doi:10.1073/pnas.95.14.8141
Mrasek, K., Schoder, C., Teichmann, A. C., Behr, K., Franze, B., Wilhelm, K., et al. (2010). Global Screening and Extended Nomenclature for 230 Aphidicolin-Inducible Fragile Sites, Including 61 yet Unreported Ones. Int. J. Oncol. 36, 929–940. doi:10.3892/ijo_00000572
Mrasek, K., Heller, A., Rubtsov, N., Trifonov, V., Starke, H., Rocchi, M., et al. (2001). Reconstruction of the Female Gorilla gorilla Karyotype Using 25-color FISH and Multicolor Banding (MCB). Cytogenet. Genome Res. 93, 242–248. doi:10.1159/000056991
Muul, I. (2002). A Photographic Guide to Mammals of South-East Asia Including Thailand, Malaysia, Singapore, Myanmar, Laos, Vietnam, Cambodia, Java, Sumatra, Bali and Borneo. J. Mammal 83, 633. doi:10.1644/1545-1542(2002)083<0633:>2.0.CO;2
Nie, W., Liu, R., Chen, Y., Wang, J., and Yang, F. (1998). Mapping Chromosomal Homologies between Humans and Two Langurs (Semnopithecus Francoisi and S. Phayrei) by Chromosome Painting. Chromosome Res. 6, 447–453. doi:10.1023/a:1009296227460
Pinthong, K., Tanomtong, A., Khunsook, S., Patawang, I., Wonkaonoi, W., and Supanuam, P. (2018). Karyological Analysis and NOR Polymorphism of Phayre's Langur, Trachypithecus phayrei Crepuscula (Primate, Colobinae) in Thailand. Nucleus 61, 61–67. doi:10.1007/s13237-017-0220-9
Rocchi, M., Archidiacono, N., Schempp, W., Capozzi, O., and Stanyon, R. (2012). Centromere Repositioning in Mammals. Heredity 108, 59–67. doi:10.1038/hdy.2011.101
Sangpakdee, W., Tanomtong, A., Chaveerach, A., Pinthong, K., Trifonov, V., Loth, K., et al. (2018). Molecular Cytogenetic Analysis of One African and Five Asian Macaque Species Reveals Identical Karyotypes as in Mandrill. Cg 19, 207–215. doi:10.2174/1389202918666170721115047
Stanyon, R., Rocchi, M., Capozzi, O., Roberto, R., Misceo, D., Ventura, M., et al. (2008). Primate Chromosome Evolution: Ancestral Karyotypes, Marker Order and Neocentromeres. Chromosome Res. 16, 17–39. doi:10.1007/s10577-007-1209-z
Tsai, A. G., and Lieber, M. R. (2010). Mechanisms of Chromosomal Rearrangement in the Human Genome. BMC Genomics 11 (Suppl. 1), S1–S9. doi:10.1186/1471-2164-11-S1-S1
Ventura, M., Antonacci, F., Cardone, M. F., Stanyon, R., D'Addabbo, P., Cellamare, A., et al. (2007). Evolutionary Formation of New Centromeres in Macaque. Science 316, 243–246. doi:10.1126/science.1140615
Ventura, M., Weigl, S., Carbone, L., Cardone, M. F., Misceo, D., Teti, M., et al. (2004). Recurrent Sites for New Centromere Seeding. Genome Res. 14, 1696–1703. doi:10.1101/gr.2608804
Weise, A., Mrasek, K., Fickelscher, I., Claussen, U., Cheung, S. W., Cai, W. W., et al. (2008). Molecular Definition of High-Resolution Multicolor Banding Probes: First within the Human DNA Sequence Anchored FISH Banding Probe Set. J. Histochem. Cytochem. 56, 487–493. doi:10.1369/jhc.2008.950550
Keywords: chromosomal rearrangements, multicolor banding (MCB), Trachypithecus phayrei (TPH), evolutionary conserved breakpoint (ECBs), fragile sites
Citation: Fan X, Pinthong K, de Oliveira EHC, Tanomtong A, Chen H, Weise A and Liehr T (2022) First Comprehensive Characterization of Phayre’s Leaf-Monkey (Trachypithecus phayrei) Karyotype. Front. Genet. 13:841681. doi: 10.3389/fgene.2022.841681
Received: 22 December 2021; Accepted: 31 January 2022;
Published: 11 March 2022.
Edited by:
Diogo Teruo Hashimoto, São Paulo State University, BrazilReviewed by:
Darren Karl Griffin, University of Kent, United KingdomCopyright © 2022 Fan, Pinthong, de Oliveira, Tanomtong, Chen, Weise and Liehr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Liehr, VGhvbWFzLkxpZWhyQG1lZC51bmktamVuYS5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.