- 1Department of Dermatology, University of Michigan Medical School, Ann Arbor, MI, United States
- 2Department of Biostatistics, University of Michigan, Ann Arbor, MI, United States
- 3Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI, United States
Long non-coding RNAs (lncRNAs) have attracted attention for their potential roles in modulating keratinocyte differentiation and inflammatory response; however, for many identified skin-expressing lncRNAs, there is no comprehensive characterization regarding their biological roles. In addition, the reported expression profiles for lncRNAs can be ambiguous due to their low-expressing nature. The objective of this review is to utilize large scale genomic data to characterize the prominent skin-expressing lncRNAs, aiming to provide additional insights for their potential roles in the pathology of inflammatory skin of psoriasis and atopic dermatitis by integrating in vitro and in vivo data. We highlighted the different skin-expressing lncRNAs, including H19, which is significantly down-regulated in lesional skin of AD/psoriasis and upon cytokine stimulation in keratinocytes; it is also negatively correlated with CYP1A1 (r = -0.75, p = 8 × 10−73), a gene involved in drug metabolism and skin barrier homeostasis, in keratinocytes. In addition, SPRR2C, a potential regulator that modulates IL-22 stimulation, was upregulated in both atopic dermatitis and psoriasis lesional skin and was also downstream of the IL-17A and IL-17 + TNF signaling in keratinocytes. Using scRNAseq, we further revealed the cell type specificity of lncRNAs, including basal-expressing nature of H19 in the epidermis. Interestingly, instead of having cell type specific expression profile, we found few lncRNAs that are express across different cell types in skin, including MALAT1, NEAT1, and GAS5. While lncRNAs in general have lower expression, our results combining in vitro and in vivo experimental data demonstrate how some of these lncRNAs can play mediator roles in the cytokine-stimulated pathway.
Review
Long non-coding RNA (lncRNA) is a class of RNA that does not have protein-coding ability and exhibits its functions as RNA (Derrien et al., 2012; Duan et al., 2020). LncRNAs are >200 nucleotides in length and are commonly located in the chromatin and nucleus (Derrien et al., 2012; Hombach and Kretz, 2013). With the advent of next generation sequencing technologies that enable the detection of low-expressing transcripts, a substantial number of novel lncRNAs have been unraveled. According to the GENCODE consortium (Derrien et al., 2012; Harrow et al., 2012), there were approximately 9,640 lncRNAs in 2010 in human (version 7). In the latest version (v38) released in 2021, the number of lncRNAs approximately doubled with a report of 17,944 lncRNAs (Frankish et al., 2021). NONCODE, a comprehensive lncRNA resource for plants and animals, has reported >170,000 transcripts from 96,000 lncRNAs (Fang et al., 2018). Different studies have revolutionized our understanding of lncRNAs as exhibiting significant regulatory roles rather than being transcriptional waste products (Hombach and Kretz, 2013), and have demonstrated that lncRNAs exhibit stronger tissue-specific expression patterns compared to protein-coding genes (Derrien et al., 2012). LncRNAs have attracted attention due to their role in gene regulatory processes, maintaining normal tissue homeostasis, and transition to diseased states (Hombach and Kretz, 2013; Tsoi et al., 2015). They have been shown to modulate epigenetic regulation of chromatin (Rinn et al., 2007; Gupta et al., 2010; Tsai et al., 2010; Heo and Sung, 2011; Wang et al., 2011; Grote et al., 2013; Hombach and Kretz, 2013; Tsoi et al., 2015), as well as participating in promoter-specific gene regulation (Hombach and Kretz, 2013; Tsoi et al., 2015; Duan et al., 2020), X-chromosome inactivation (Tian et al., 2010; Hombach and Kretz, 2013; Tsoi et al., 2015; Duan et al., 2020), and imprinting (Lee and Bartolomei, 2013). Therefore, they are associated in various diseased states, such as neurodegenerative conditions (Wapinski and Chang, 2011), susceptibility to infection (Gomez et al., 2013), and different types cancers (Huarte et al., 2010; Prensner et al., 2011; Maruyama and Suzuki, 2012).
Skin diseases are the fourth most common cause of illness, whereby up to 33% of the population is affected (Bickers et al., 2006; Hay et al., 2014; Hay et al., 2015; Tizek et al., 2019). Many of these diseases display strong inflammatory components whose mechanisms have not been fully explained. Recent genomic studies highlight how multiple lncRNAs can be implicated in key roles for epidermal homeostasis, transition to disease state, and stress response (Hombach and Kretz, 2013; Tsoi et al., 2015) (Table 1; Supplementary Table S1). Using RNA sequencing, previous work has revealed that DANCR (anti-differentiation non-coding RNA) is significantly downregulated during terminal differentiation (Huarte et al., 2010; Kretz et al., 2012), and indicates its functional role in the transition from non-differentiated to differentiated cells (Huarte et al., 2010). DANCR may also repress differentiation of osteoblasts and keratinocyte progenitors by associating with methyltransferase EZH2, a component of the chromatin modifying protein complex PRC2, which is involved in epigenetic silencing via histone methylation (Cao et al., 2002; Hombach and Kretz, 2013; Zhu and Xu, 2013). Kretz et al. illustrated that lncRNA TINCR (terminal differentiation induced non-coding RNA) is upregulated in epidermal differentiation and forms a complex with STAU1 (staufen double-stranded RNA binding protein), which increases stabilization of differentiation (Kretz et al., 2013). Together, DANCR and TINCR are essential in maintaining epidermal homeostasis (Hombach and Kretz, 2013; Kretz et al., 2013) in cells under normal state, however their behaviors in inflammatory skin conditions are not well studied. Another study that focused on the roles of lncRNAs in cutaneous biology demonstrated that lncRNA BC020554 is downregulated while AK022798 is upregulated during keratinocyte differentiation (Tang et al., 2020). However, for the majority of the skin-expressing lncRNAs, there is limited information regarding their biological and pathological roles in cutaneous disorders.
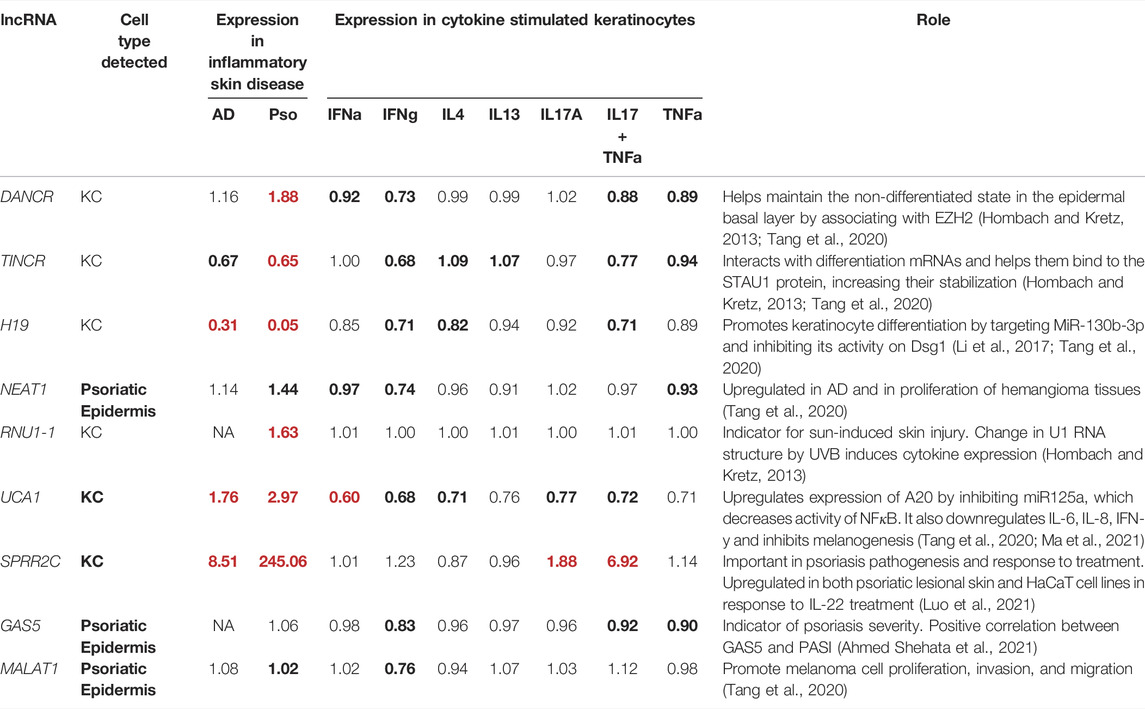
TABLE 1. The fold change for the notable lncRNAs that play roles in skin biology and/or diseases. Bold numbers denote FDR ≤0.05. Red results denote at least 1.5-fold change when comparing against control/unstimulated conditions. Expression in inflammatory skin disease represents findings in lesional skin of atopic dermatitis/psoriasis patients compared to control without skin disease.
Psoriasis is a chronic, relapsing, hyperproliferative, inflammatory skin disease whereby the epidermis is thickened secondary to abnormal differentiation of basal keratinocytes (Szegedi et al., 2010; Tang et al., 2020). LncRNA PRINS (psoriasis susceptibility related RNA gene induced by stress) has been shown to contribute to psoriasis susceptibility and is overexpressed in psoriatic uninvolved skin (Sonkoly et al., 2005; Zenz et al., 2005; Bari et al., 2011; Szegedi et al., 2012; Hombach and Kretz, 2013; Tsoi et al., 2015). It is also involved in cellular stress response and may play a role in psoriasis by decreasing sensitivity to keratinocyte apoptosis via the regulation of G1P3 (Sonkoly et al., 2005; Zenz et al., 2005; Bari et al., 2011; Szegedi et al., 2012). Ma et al. demonstrated that lncRNA UCA1 (urothelial carcinoma-associated 1) was downregulated in psoriatic lesional skin (Ma et al., 2021). Initially named for its high expression in bladder cancer cells (Roberson and Bowcock, 2010; Ma et al., 2021), UCA1 has now gained more attention regarding its role in psoriasis (Ma et al., 2021). Specifically, it upregulates the expression of cytoplasmic zinc finger protein A20 (TNFAIP3, TNFα induced protein 3) by inhibiting microRNA125a (Ma et al., 2021). Through the miRNA125a-A20 axis, UCA1 negatively regulates the activity of NF
Therefore, in this review we aim to characterize the expression profiles for the prominent skin-expressing lncRNAs, aiming to provide additional insights for their potential roles in atopic dermatitis and psoriasis pathology by integrating in vitro, in vivo, and in silico techniques. First, we identified candidate lncRNAs that were previously identified in the literature to play a role in cutaneous biology. We then evaluated the gene expression patterns in in vivo lesional atopic dermatitis and psoriasis skin to identify the differentially expressed lncRNAs (Tsoi et al., 2019a; Tsoi et al., 2019b). We evaluated the lncRNA profiles from published transcriptomes in non-lesional and lesional skin from 27 atopic dermatitis and 28 psoriatic patients, as well as from 38 healthy controls. This information was integrated with in vitro genomics data of cytokine-stimulated conditions (IFNa, IFNg, IL-4, IL-13, IL-17A, IL-17 + TNFa, TNFa) from 40 patients to infer the regulatory mechanism upon inflammatory response for keratinocytes (Tsoi et al., 2019a; Tsoi et al., 2019b). We also conducted correlation analysis for the in vivo and in vitro data to associate expression profiles of the candidate lncRNAs with the other prominent genes involved in skin inflammation. To reveal lncRNA-expressing cell type in the in vivo data, we assayed the expressions in scRNA-seq data from non-lesional and lesional skin.
Differentially Expressed lncRNAs in Cytokine Stimulated Keratinocytes and In-Vivo Lesional Skin
The summary statistics for the above lncRNAs detected in our analysis were presented in Table 1. As previously discussed, DANCR and TINCR play critical roles in maintaining epidermal homeostasis (Huarte et al., 2010; Kretz et al., 2012). DANCR is expressed in non-differentiating keratinocytes while TINCR is expressed in differentiating keratinocytes (Piipponen et al., 2020). When evaluating gene expression patterns in lesional compared to control skin, DANCR was upregulated and TINCR was downregulated in psoriasis lesional skin (Figure 1A). These findings support our current understanding of psoriasis as an inflammatory skin disease that is in part characterized by poor differentiation of keratinocytes (Ma et al., 2021). Of importance, we discovered that TINCR was also downregulated in atopic dermatitis lesional skin—a finding that has not previously been reported. Similar to TINCR, lncRNA H19 also regulates keratinocyte differentiation by targeting MiR-130b-3p and inhibiting its activity on Desmoglein 1 (Dsg1) (Li et al., 2017), which is expressed mainly in the superficial layer of the epidermis and mediates cell-cell adhesion. We found that H19 was downregulated in both atopic dermatitis (Fold Change FC =0.31; FDR = 3.26x10−6) and psoriasis (Fold Change FC = 0.05; FDR = 3.34x10−15) lesional skin, and it was also depleted in the IFN and IL17/TNF stimulated keratinocytes, further supporting our understanding of aberrant differentiation of keratinocytes in many skin diseases, such as psoriasis and atopic dermatitis (Li et al., 2017; Tang et al., 2020).
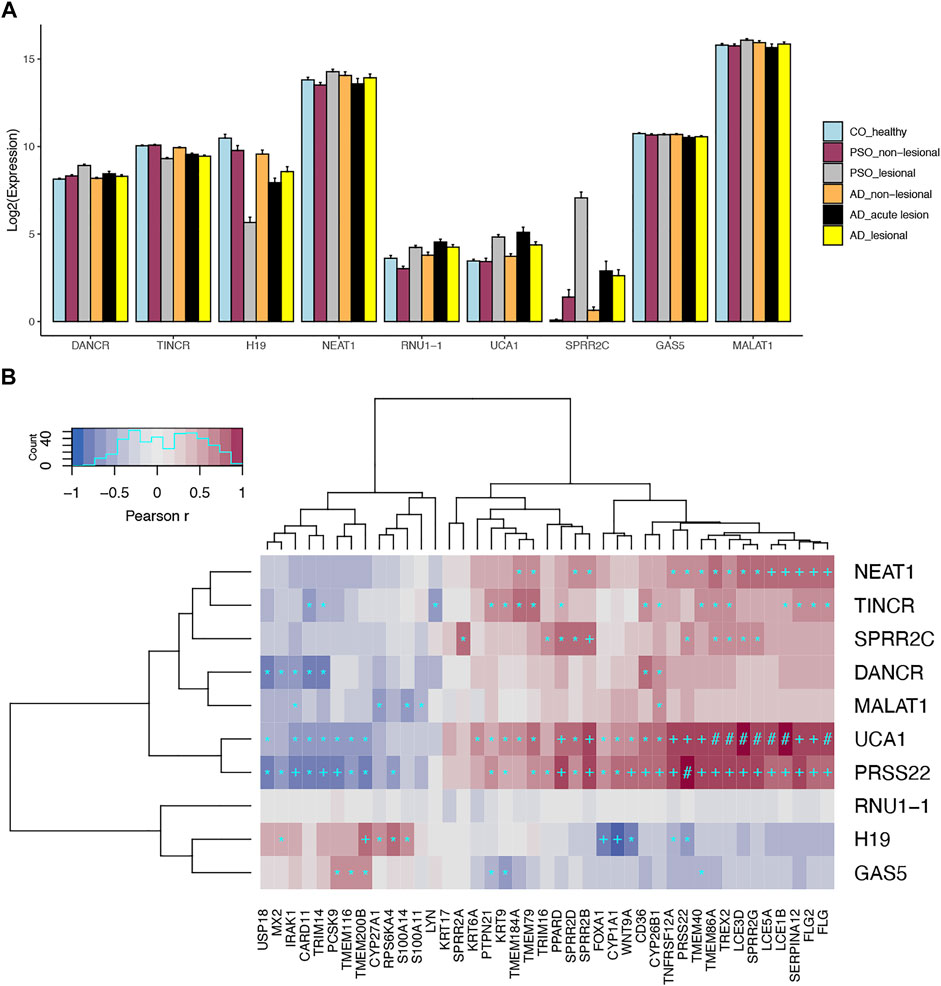
FIGURE 1. Expression profiles for skin-expressing lncRNAs. (A) Gene expression profiles for prominent skin-expressing lncRNAs in different disease conditions. (B) Top most correlated protein-coding genes for skin-expressing lncRNAs. Heatmap shows the spearman correlation in keratinocytes. *p < 1x10−20; +p < 1x10−50; #p < 1x10−100.
Prominent skin-expressing lncRNAs that were upregulated in both psoriasis and atopic dermatitis lesional skin include UCA1 and SPRR2C (small proline rich protein 2C) (Table 1). Through the miRNA125a-A20 axis, UCA1 has been shown to downregulate NF
Co-Expression Analysis Revealed Potential Biological Modules
Our previous work utilized co-expression patterns to infer potential biological roles of lncRNAs (Tsoi et al., 2015). In this study, we also found significant correlations between the aforementioned lncRNAs with other genes known to play roles in skin diseases (Figure 1B). For instances, we observed strong negative correlation (r = −0.75, p = 8 × 10−73) between H19 and CYP1A1 in keratinocytes. In addition to its important impact on drug metabolism (Mescher and Haarmann-Stemmann, 2018), CYP1A1 is involved in the AHR pathway, which controls skin barrier homeostasis (Kyoreva et al., 2021). Meanwhile, SPRR2C is positively correlated with other genes encoding for small proline-rich proteins (i.e., SPRR2D, SPRR2B, SPRR2G), which are structural components of the cornified epithelia; and it is also correlated with IL36A (r = 0.55, p = 8 × 10−33), IL36G (r = 0.58, p = 5 × 10−37) and IL36RN (r = 0.57, p = 5 × 10−36), a group of cytokines which are involved in skin inflammation (not shown in Figure 1B) (Hernández-Santana et al., 2020) and contribute to the pathogenesis of psoriasis and atopic dermatitis (Tsang et al., 2020). Notably, IL-36 signaling has been shown to induce IL-17 and IL-23 signaling (Goldstein et al., 2020), which corresponds with our finding that SPRR2C is up-regulated upon IL-17 stimulation. Other interesting correlations we discovered include UCA1 with filaggrin genes, FLG (r = 0.84, p = 4 × 10−106), FLG2 (r = 0.82, p = 3 × 10−99), as well as late cornified envelope genes, LCE1B (r = 0.89, p = 7 × 10−141), LCE3D (r = 0.87, p = 4 × 10−127), LCE3E (r = 0.85, p = 1 × 10−110), LCE5A (r = 0.85, p = 7 × 10−112). Filaggrin deficiencies are an important risk factor for atopic dermatitis, as they weaken the epidermal barrier (Weidinger et al., 2018). LCE genes, which are associated with psoriasis, have been found to have antibacterial activity (Niehues et al., 2017). Of note, UCA1 was also significantly correlated with TNFSF12A (r = 0.75, p = 2 × 10−73) in keratinocytes, which is an important regulator for the NF
lncRNA as Cell Type Signature
We looked up the expression architecture for these lncRNAs in the psoriatic and non-lesional skin scRNA-seq data. Interestingly, despite the well-known cell type specific nature of lncRNAs, we observed that some highly expressed lncRNAs, such as NEAT1, MALAT1, and GAS5 are expressed in all cell types in skin (Figure 2). Others, including UCA1, H19, and TINCR, are expressed only in keratinocytes. However, we were not able to detect SPRR2C, likely due to the lack of sensitivity in detecting low-expressing transcripts for lncRNAs. These lncRNA expression patterns allow us to better understand the cell types that these lncRNAs could exert their functionality. Notably, we observed H19 expression mostly in the basal layer of the epidermis, and the scRNA-seq data further confirmed that it is down-regulated in lesional skin. In comparison, TINCR was found to be expressed in all of the compartments in the epidermal layer. We also replicated the higher expression of UCA1 in the lesional skin of psoriasis, and its expression seems to appear throughout the epidermis while slightly higher in the keratinized layer.
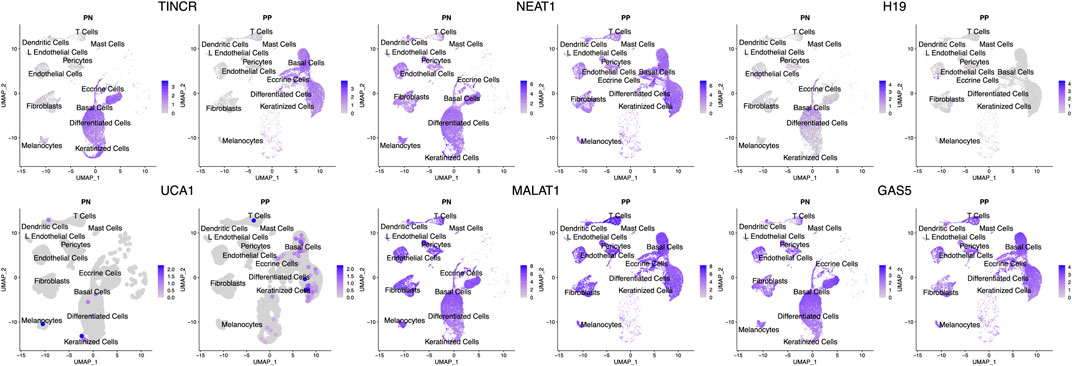
FIGURE 2. Expression of lncRNAs across different cell types. UMAP of the scRNA-seq data highlights the cell type specific/shared expression profiles for different lncRNAs. The intensity of blue corresponds to the expression level of the lncRNAs in each cell. The cell type annotations label the cell clusters nearby. PP denotes lesional psoriasis skin, while PN denotes nonlesional psoriasis skin.
Summary
With the advent of next generation sequencing technology, our understanding and characterization of lncRNAs has accelerated (Ponting et al., 2009). As a class, lncRNAs are preferentially enriched in the nucleus, do not have coding protein ability, and exhibit more tissue-specific expression patterns compared to protein-coding genes (Derrien et al., 2012). Over recent years, they have attracted attention due to their roles in epidermal homeostasis, transition to disease states, and stress response (Hombach and Kretz, 2013; Tsoi et al., 2015). In this review, we characterized the expression profiles for the prominent skin-expressing lncRNAs and provided additional insights for their potential roles in atopic dermatitis and psoriasis pathology by integrating in vitro, in vivo, and in silico techniques. We were able to better understand the behaviors for some of the reported skin-specific lncRNAs, as well as their architecture, by integrating single cell RNA-sequencing.
While TINCR is known to play a role in keratinocyte differentiation (Kretz et al., 2013), its dysregulation in atopic dermatitis and psoriasis has not yet been fully understood. Although its downregulation supports our understanding of aberrant differentiation in these diseases (Li et al., 2017), its role in modulating skin differentiation under inflammatory environment require further investigation. Our results also highlight that some of the prominent skin-expressing lncRNAs can be found in multiple different cell types, while others exhibit cell type specific expression patterns. These findings can help us to better understand the cell type specific/shared regulation programming. A higher throughput or transcript-targeted single cell transcriptomic approach can provide a better resolution in revealing expression patterns for more lncRNAs, especially for those that are low-expressing.
In conclusion, this review provided additional insights for the skin-expressing lncRNAs that contribute to atopic dermatitis and psoriasis. It further helped facilitate our understanding of the role of lncRNAs upon cytokine stimulation and how this can relate to autoimmune diseases. We discovered shared and unique expression, as well as cell type specificity, for lncRNAs in non-lesional and lesional skin of psoriatic patients. This analysis should provide valuable information for future studies that target lncRNAs for biomarker development and pharmaceutical intervention.
Author Contributions
AS and LT designed and drafted the manuscript. MP, AS, JC, RW, and LT conducted the analysis and literature review. All authors read and approved the work.
Funding
This work was supported by the National Psoriasis Foundation (LT, MP, and JG), and awards from the National Institutes of Health (K01AR072129 to LT.; 1P30AR075043 to LT, MP, and JG). MP was also supported by the Dermatology Foundation.
Conflict of Interest
JG has served as a consultant to AbbVie, Eli Lilly, Almirall, Celgene, BMS, Janssen, Prometheus, TimberPharma, Galderma, Novatis, MiRagen, AnaptysBio and has received research support from AbbVie, SunPharma, Eli Lilly, Kyowa Kirin, Almirall, Celgene, BMS, Janssen, Prometheus, and TimberPharma. LT has received research support from Janssen and Galderma.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.835740/full#supplementary-material
Supplementary Table S1 | Other previously identified lncRNAs that play roles in skin biology and/or diseases.
References
Ahmed Shehata, W., Maraee, A., Abd El Monem Ellaithy, M., Tayel, N., Abo-Ghazala, A., and Mohammed El-Hefnawy, S. (2021). Circulating Long Noncoding RNA Growth Arrest-specific Transcript 5 as a Diagnostic Marker and Indicator of Degree of Severity in Plaque Psoriasis. Int. J. Dermatol. 60, 973. doi:10.1111/ijd.15494
Bari, L., Bacsa, S., Sonkoly, E., Bata-Csörgő, Z., Kemény, L., Dobozy, A., et al. (2011). Comparison of Stress-Induced PRINS Gene Expression in normal Human Keratinocytes and HaCaT Cells. Arch. Dermatol. Res. 303 (10), 745–752. doi:10.1007/s00403-011-1162-8
Bickers, D. R., Lim, H. W., Margolis, D., Weinstock, M. A., Goodman, C., Faulkner, E., et al. (2006). The burden of Skin Diseases: 2004. J. Am. Acad. Dermatol. 55 (3), 490–500. doi:10.1016/j.jaad.2006.05.048
Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., et al. (2002). Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 298 (5595), 1039–1043. doi:10.1126/science.1076997
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 22 (9), 1775–1789. doi:10.1101/gr.132159.111
Duan, Q., Wang, G., Wang, M., Chen, C., Zhang, M., Liu, M., et al. (2020). LncRNA RP6‐65G23.1 Accelerates Proliferation and Inhibits Apoptosis via p‐ERK1/2/p‐AKT Signaling Pathway on Keratinocytes. J. Cel Biochem 121 (11), 4580–4589. doi:10.1002/jcb.29685
Fang, S., Zhang, L., Guo, J., Niu, Y., Wu, Y., Li, H., et al. (2018). NONCODEV5: a Comprehensive Annotation Database for Long Non-coding RNAs. Nucleic Acids Res. 46 (D1), D308–D314. doi:10.1093/nar/gkx1107
Frankish, A., Diekhans, M., Jungreis, I., Lagarde, J., Loveland, J. E., Mudge, J. M., et al. (2021). GENCODE 2021. Nucleic Acids Res. 49 (D1), D916–D923. doi:10.1093/nar/gkaa1087
Goldstein, J. D., Bassoy, E. Y., Caruso, A., Palomo, J., Rodriguez, E., Lemeille, S., et al. (2020). IL-36 Signaling in Keratinocytes Controls Early IL-23 Production in Psoriasis-like Dermatitis. Life Sci. Alliance 3 (6), e202000688. doi:10.26508/lsa.202000688
Gomez, J. A., Wapinski, O. L., Yang, Y. W., Bureau, J.-F., Gopinath, S., Monack, D. M., et al. (2013). The NeST Long ncRNA Controls Microbial Susceptibility and Epigenetic Activation of the Interferon-γ Locus. Cell 152 (4), 743–754. doi:10.1016/j.cell.2013.01.015
Grote, P., Wittler, L., Hendrix, D., Koch, F., Währisch, S., Beisaw, A., et al. (2013). The Tissue-specific lncRNA Fendrr Is an Essential Regulator of Heart and Body wall Development in the Mouse. Dev. Cel 24 (2), 206–214. doi:10.1016/j.devcel.2012.12.012
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long Non-coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature 464 (7291), 1071–1076. doi:10.1038/nature08975
Harrow, J., Frankish, A., Gonzalez, J. M., Tapanari, E., Diekhans, M., Kokocinski, F., et al. (2012). GENCODE: the Reference Human Genome Annotation for the ENCODE Project. Genome Res. 22 (9), 1760–1774. doi:10.1101/gr.135350.111
Hay, R. J., Augustin, M., Griffiths, C. E. M., and Sterry, W. (2015). The Global challenge for Skin Health. Br. J. Dermatol. 172 (6), 1469–1472. doi:10.1111/bjd.13854
Hay, R. J., Johns, N. E., Williams, H. C., Bolliger, I. W., Dellavalle, R. P., Margolis, D. J., et al. (2014). The Global burden of Skin Disease in 2010: an Analysis of the Prevalence and Impact of Skin Conditions. J. Invest. Dermatol. 134 (6), 1527–1534. doi:10.1038/jid.2013.446
Heo, J. B., and Sung, S. (2011). Vernalization-mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 331 (6013), 76–79. doi:10.1126/science.1197349
Hernández-Santana, Y. E., Leon, G., St Leger, D., Fallon, P. G., and Walsh, P. T. (2020). Keratinocyte Interleukin-36 Receptor Expression Orchestrates Psoriasiform Inflammation in Mice. Life Sci. Alliance 3 (4), e201900586. doi:10.26508/lsa.201900586
Hombach, S., and Kretz, M. (2013). The Non-coding Skin: Exploring the Roles of Long Non-coding RNAs in Epidermal Homeostasis and Disease. Bioessays 35 (12), 1093–1100. doi:10.1002/bies.201300068
Huarte, M., Guttman, M., Feldser, D., Garber, M., Koziol, M. J., Kenzelmann-Broz, D., et al. (2010). A Large Intergenic Noncoding RNA Induced by P53 Mediates Global Gene Repression in the P53 Response. Cell 142 (3), 409–419. doi:10.1016/j.cell.2010.06.040
Kretz, M., Siprashvili, Z., Chu, C., Webster, D. E., Zehnder, A., Qu, K., et al. (2013). Control of Somatic Tissue Differentiation by the Long Non-coding RNA TINCR. Nature 493 (7431), 231–235. doi:10.1038/nature11661
Kretz, M., Webster, D. E., Flockhart, R. J., Lee, C. S., Zehnder, A., Lopez-Pajares, V., et al. (2012). Suppression of Progenitor Differentiation Requires the Long Noncoding RNA ANCR. Genes Dev. 26 (4), 338–343. doi:10.1101/gad.182121.111
Kyoreva, M., Li, Y., Hoosenally, M., Hardman-Smart, J., Morrison, K., Tosi, I., et al. (2021). CYP1A1 Enzymatic Activity Influences Skin Inflammation via Regulation of the AHR Pathway. J. Invest. Dermatol. 141 (6), 1553–1563. doi:10.1016/j.jid.2020.11.024
Lee, J. T., and Bartolomei, M. S. (2013). X-inactivation, Imprinting, and Long Noncoding RNAs in Health and Disease. Cell 152 (6), 1308–1323. doi:10.1016/j.cell.2013.02.016
Li, C.-x., Li, H.-g., Huang, L.-t., Kong, Y.-w., Chen, F.-y., Liang, J.-y., et al. (2017). H19 lncRNA Regulates Keratinocyte Differentiation by Targeting miR-130b-3p. Cell Death Dis 8 (11), e3174. doi:10.1038/cddis.2017.516
Li, D., Kular, L., Vij, M., Herter, E. K., Li, X., Wang, A., et al. (2019). Human Skin Long Noncoding RNA WAKMAR1 Regulates Wound Healing by Enhancing Keratinocyte Migration. Proc. Natl. Acad. Sci. U.S.A. 116 (19), 9443–9452. doi:10.1073/pnas.1814097116
Li, H.-J., Li, X., Pang, H., Pan, J.-J., Xie, X.-J., and Chen, W. (2015). Long Non-coding RNA UCA1 Promotes Glutamine Metabolism by Targeting miR-16 in Human Bladder Cancer. Jpn. J. Clin. Oncol. 45 (11), 1055–1063. doi:10.1093/jjco/hyv132
Lizzul, P. F., Aphale, A., Malaviya, R., Sun, Y., Masud, S., Dombrovskiy, V., et al. (2005). Differential Expression of Phosphorylated NF-κB/RelA in Normal and Psoriatic Epidermis and Downregulation of NF-Κb in Response to Treatment with Etanercept. J. Invest. Dermatol. 124 (6), 1275–1283. doi:10.1111/j.0022-202x.2005.23735.x
Luo, M., Huang, P., Pan, Y., Zhu, Z., Zhou, R., Yang, Z., et al. (2021). Weighted Gene Coexpression Network and Experimental Analyses Identify lncRNA SPRR2C as a Regulator of the IL-22-stimulated HaCaT Cell Phenotype through the miR-330/STAT1/S100A7 axis. Cel Death Dis 12 (1), 86. doi:10.1038/s41419-020-03305-z
Ma, X. L., Wen, G. D., Yu, C., Zhao, Z., Gao, N., and Liu, Z. Y. (2021). LncRNA UCA1 Negatively Regulates NF‐kB Activity in Psoriatic Keratinocytes through the miR125a‐A20 axis. Kaohsiung J. Med. Sci. 37 (3), 172–180. doi:10.1002/kjm2.12363
Maruyama, R., and Suzuki, H. (2012). Long Noncoding RNA Involvement in Cancer. BMB Rep. 45 (11), 604–611. doi:10.5483/bmbrep.2012.45.11.227
Mescher, M., and Haarmann-Stemmann, T. (2018). Modulation of CYP1A1 Metabolism: From Adverse Health Effects to Chemoprevention and Therapeutic Options. Pharmacol. Ther. 187, 71–87. doi:10.1016/j.pharmthera.2018.02.012
Niehues, H., Tsoi, L. C., van der Krieken, D. A., Jansen, P. A. M., Oortveld, M. A. W., Rodijk-Olthuis, D., et al. (2017). Psoriasis-Associated Late Cornified Envelope (LCE) Proteins Have Antibacterial Activity. J. Invest. Dermatol. 137 (11), 2380–2388. doi:10.1016/j.jid.2017.06.003
Piipponen, M., Nissinen, L., and Kähäri, V.-M. (2020). Long Non-coding RNAs in Cutaneous Biology and Keratinocyte Carcinomas. Cell. Mol. Life Sci. 77 (22), 4601–4614. doi:10.1007/s00018-020-03554-3
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and Functions of Long Noncoding RNAs. Cell 136 (4), 629–641. doi:10.1016/j.cell.2009.02.006
Prensner, J. R., Iyer, M. K., Balbin, O. A., Dhanasekaran, S. M., Cao, Q., Brenner, J. C., et al. (2011). Transcriptome Sequencing across a Prostate Cancer Cohort Identifies PCAT-1, an Unannotated lincRNA Implicated in Disease Progression. Nat. Biotechnol. 29 (8), 742–749. doi:10.1038/nbt.1914
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 129 (7), 1311–1323. doi:10.1016/j.cell.2007.05.022
Roberson, E. D. O., and Bowcock, A. M. (2010). Psoriasis Genetics: Breaking the Barrier. Trends Genet. 26 (9), 415–423. doi:10.1016/j.tig.2010.06.006
Sonkoly, E., Bata-Csorgo, Z., Pivarcsi, A., Polyanka, H., Kenderessy-Szabo, A., Molnar, G., et al. (2005). Identification and Characterization of a Novel, Psoriasis Susceptibility-Related Noncoding RNA Gene, PRINS. J. Biol. Chem. 280 (25), 24159–24167. doi:10.1074/jbc.m501704200
Szegedi, K., Göblös, A., Bacsa, S., Antal, M., Németh, I., Bata-Csörgő, Z., et al. (2012). Expression and Functional Studies on the Noncoding RNA, PRINS. Ijms 14 (1), 205–225. doi:10.3390/ijms14010205
Szegedi, K., Sonkoly, E., Nagy, N., Németh, I. B., Bata-Csörgő, Z., Kemény, L., et al. (2010). The Anti-apoptotic Protein G1P3 Is Overexpressed in Psoriasis and Regulated by the Non-coding RNA, PRINS. Exp. Dermatol. 19 (3), 269–278. doi:10.1111/j.1600-0625.2010.01066.x
Tang, L., Liang, Y., Xie, H., Yang, X., and Zheng, G. (2020). Long Non-coding RNAs in Cutaneous Biology and Proliferative Skin Diseases: Advances and Perspectives. Cell Prolif 53 (1), e12698. doi:10.1111/cpr.12698
Tian, D., Sun, S., and Lee, J. T. (2010). The Long Noncoding RNA, Jpx, Is a Molecular Switch for X Chromosome Inactivation. Cell 143 (3), 390–403. doi:10.1016/j.cell.2010.09.049
Tizek, L., Schielein, M. C., Seifert, F., Biedermann, T., Böhner, A., and Zink, A. (2019). Skin Diseases Are More Common Than We Think: Screening Results of an Unreferred Population at the Munich Oktoberfest. J. Eur. Acad. Dermatol. Venereol. 33 (7), 1421–1428. doi:10.1111/jdv.15494
Tsai, M.-C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 329 (5992), 689–693. doi:10.1126/science.1192002
Tsang, M. S.-M., Sun, X., and Wong, C. K. (2020). The Role of New IL-1 Family Members (IL-36 and IL-38) in Atopic Dermatitis, Allergic Asthma, and Allergic Rhinitis. Curr. Allergy Asthma Rep. 20 (8), 40. doi:10.1007/s11882-020-00937-1
Tsoi, L. C., Iyer, M. K., Stuart, P. E., Swindell, W. R., Gudjonsson, J. E., Tejasvi, T., et al. (2015). Analysis of Long Non-coding RNAs Highlights Tissue-specific Expression Patterns and Epigenetic Profiles in normal and Psoriatic Skin. Genome Biol. 16, 24. doi:10.1186/s13059-014-0570-4
Tsoi, L. C., Patrick, M. T., Shuai, S., Sarkar, M. K., Chi, S., Ruffino, B., et al. (2021). Cytokine Responses in Nonlesional Psoriatic Skin as Clinical Predictor to Anti-TNF Agents. J. Allergy Clin. Immunol. 149, 640. doi:10.1016/j.jaci.2021.07.024
Tsoi, L. C., Rodriguez, E., Degenhardt, F., Baurecht, H., Wehkamp, U., Volks, N., et al. (2019). Atopic Dermatitis Is an IL-13 Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J. Invest. Dermatol. 139, 1480. doi:10.1016/j.jid.2018.12.018
Tsoi, L. C., Rodriguez, E., Stölzl, D., Wehkamp, U., Sun, J., Gerdes, S., et al. (2019). Progression of Acute-To-Chronic Atopic Dermatitis Is Associated with Quantitative rather Than Qualitative Changes in Cytokine Responses. J. Allergy Clin. Immunol. 145, 1406. doi:10.1016/j.jaci.2019.11.047
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A Long Noncoding RNA Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature 472 (7341), 120–124. doi:10.1038/nature09819
Wapinski, O., and Chang, H. Y. (2011). Long Noncoding RNAs and Human Disease. Trends Cel Biol. 21 (6), 354–361. doi:10.1016/j.tcb.2011.04.001
Weidinger, S., Beck, L. A., Bieber, T., Kabashima, K., and Irvine, A. D. (2018). Atopic Dermatitis. Nat. Rev. Dis. Primers 4 (1), 1. doi:10.1038/s41572-018-0001-z
Xia, L., Jiang, L., Chen, Y., Zhang, G., and Chen, L. (2021). ThPOK Transcriptionally Inactivates TNFRSF12A to Increase the Proliferation of T Cells with the Involvement of the NF-kB Pathway. Cytokine 148, 155658. doi:10.1016/j.cyto.2021.155658
Zenz, R., Eferl, R., Kenner, L., Florin, L., Hummerich, L., Mehic, D., et al. (2005). Psoriasis-like Skin Disease and Arthritis Caused by Inducible Epidermal Deletion of Jun Proteins. Nature 437 (7057), 369–375. doi:10.1038/nature03963
Keywords: lncRNA, atopic dermatitis, psoriasis, keratinocyte, scRNA sequencing
Citation: Shefler A, Patrick MT, Wasikowski R, Chen J, Sarkar MK, Gudjonsson JE and Tsoi LC (2022) Skin-Expressing lncRNAs in Inflammatory Responses. Front. Genet. 13:835740. doi: 10.3389/fgene.2022.835740
Received: 14 December 2021; Accepted: 08 March 2022;
Published: 26 April 2022.
Edited by:
Chi-Ming Wong, Hong Kong Polytechnic University, Hong Kong SAR, ChinaReviewed by:
Kehinde Ross, Liverpool John Moores University, United KingdomClaudia Buerger, University Hospital Frankfurt, Germany
Copyright © 2022 Shefler, Patrick, Wasikowski, Chen, Sarkar, Gudjonsson and Tsoi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lam C. Tsoi, alextsoi@umich.edu
 Alanna Shefler
Alanna Shefler