- 1School of Medicine, Tokai University, Isehara, Japan
- 2Research Institute, Tokyo Medical and Dental University, Tokyo, Japan
In viviparous mammals, genomic imprinting regulates parent-of-origin-specific monoallelic expression of paternally and maternally expressed imprinted genes (PEGs and MEGs) in a region-specific manner. It plays an essential role in mammalian development: aberrant imprinting regulation causes a variety of developmental defects, including fetal, neonatal, and postnatal lethality as well as growth abnormalities. Mechanistically, PEGs and MEGs are reciprocally regulated by DNA methylation of germ-line differentially methylated regions (gDMRs), thereby exhibiting eliciting complementary expression from parental genomes. The fact that most gDMR sequences are derived from insertion events provides strong support for the claim that genomic imprinting emerged as a host defense mechanism against the insertion in the genome. Recent studies on the molecular mechanisms concerning how the DNA methylation marks on the gDMRs are established in gametes and maintained in the pre- and postimplantation periods have further revealed the close relationship between genomic imprinting and invading DNA, such as retroviruses and LTR retrotransposons. In the presence of gDMRs, the monoallelic expression of PEGs and MEGs confers an apparent advantage by the functional compensation that takes place between the two parental genomes. Thus, it is likely that genomic imprinting is a consequence of an evolutionary trade-off for improved survival. In addition, novel genes were introduced into the mammalian genome via this same surprising and complex process as imprinted genes, such as the genes acquired from retroviruses as well as those that were duplicated by retropositioning. Importantly, these genes play essential/important roles in the current eutherian developmental system, such as that in the placenta and/or brain. Thus, genomic imprinting has played a critically important role in the evolutionary emergence of mammals, not only by providing a means to escape from the adverse effects of invading DNA with sequences corresponding to the gDMRs, but also by the acquisition of novel functions in development, growth and behavior via the mechanism of complementary monoallelic expression.
Introduction
Genomic imprinting is widely distributed in the viviparous mammals, the therians, comprising marsupials and eutherians (Reik and Walter 2001; Renfree et al., 2009; Barlow and Bartolomei 2014), yet it is an unusual biological mechanism in that it seems to runs counter to two main pillars of modern biology: it is an apparent exception to the rule of Mendelian genetics that presupposes biallelic expression from two parental alleles, and it is inconsistent with the Darwinian theory of evolution at first glance due to the apparent disadvantage (Mann and Lovell-Badge, 1984; McGrath and Solter 1984; Surani et al., 1984; Cattanach and Kirk 1985) of the monoallelic expression of certain essential/important genes in development. The evolutionary advantage it nevertheless confers as the result of a partial, functional haploidy despite such defects has long been debated. Perhaps the most widely accepted account is the conflict/kinship hypothesis (Moore and Haig, 1991; Wilkins and Haig 2003; Haig 2004), which proposes that genomic imprinting arose as a consequence of a conflict of interest between maternally and paternally derived genomes, driven by a need for prenatal resource control: PEGs promote embryonic growth while MEGs repress it. Substantial numbers of imprinted genes fit this hypothetical scenario, so it has become generally accepted and its topic covered by many reviews (Reik and Walter, 2001; Edwards et al., 2019; Hanna and Kelsey 2021). In this review, we first introduce the unique nature of genomic imprinting regulated by gDMRs, then, revisit another host defense hypothesis that proposes that genomic imprinting arose as a consequence of the DNA methylation machinery as a defense against repetitive/foreign elements (Barlow 1993). We do this from the viewpoint of the origin of gDMRs (Suzuki et al., 2011; Renfree et al., 2013; Kaneko-Ishino and Ishino 2015, 2019) and the current knowledge on the molecular mechanisms underlying the establishment of the gDMRs in germ cells (Chotalia et al., 2009; Veselovska et al., 2015; Hanna and Kelsey 2021) as well as their protection from a global DNA demethylation wave that occurs in preimplantation embryos (Li et al., 2008; Smallwood et al., 2011; Kobayashi et al., 2012; Takahashi et al., 2019). Finally, we reexamine the advantage conferred by complementary monoallelic expression (Kaneko-Ishino et al., 2003, 2006), such as functional compensation (Renfree et al., 2013; Kaneko-Ishino and Ishino 2015, 2019) and the innovation of genomic function (Kaneko-Ishino and Ishino, 2012). Recently, non-canonical genomic imprinting regulated by histone 3 lysine 27 trimethylation (H3K27me3) has been reported in the placenta in a lineage-specific manner (Okae et al., 2014; Inoue et al., 2017a, 2017b, 2018; Hanna et al., 2019; Inoue et al., 2020; Hanna and Kelsey 2021), but we shall mainly focus on the canonical genomic imprinting regulated by gDMRs that is commonly conserved in therian and eutherian mammals.
Uniqueness of Mammalian Genomic Imprinting Among Vertebrates
Pronuclear transplantation experiments demonstrated that both parthenogenetic and androgenetic embryos which exclusively possess maternally and paternally-derived genomes, respectively, cannot develop to term, and exhibit early embryonic lethality: the former exhibited severe placental defects, while the latter had severe embryonic growth retardation (Mann and Lovell-Badge, 1984; McGrath and Solter 1984; Surani et al., 1984). The complete absence of parthenogenesis is a unique feature of viviparous mammals because parthenogenesis is often observed both naturally and experimentally in vertebrates such as birds, reptiles, amphibians, and fish (Ramachandran and McDaniel 2018; Fujita et al., 2020). The differences between the paternally and maternally-derived genome have also been shown by genetic experiments using mice with partial paternal or maternal uniparental disomy (Cattanach and Kirk, 1985; Cattanach and Beechey, 1990; Beechey and Evans, 2004). The offspring exhibit lethality at various stages in pre- and postnatal development as well as growth and behavioral abnormalities, leading to the concept of chromosomal imprinted regions. Such genomic imprinting is well accounted for by the presence of paternally and maternally expressed imprinted genes (PEGs and MEGs). The early embryonic lethality of the parthenogenetic and androgenetic embryos is due to a complete lack of expression of PEGs and MEGs, respectively. The abnormal phenotypes in the partial uniparental disomy mice are due to the irregular expression of certain PEGs and MEGs in the imprinted regions in question. From these results, it is clear that several different PEGs and MEGs play essential/important roles in the current developmental system in the therian mammals, so even their monoallelic expression is advantageous compared with a complete absence of expression, leading to their widespread conservation.
The Intrinsic Nature of Reciprocal Expression by PEGs and MEGs
PEG and MEG cannot be co-expressed in the cis configuration because their expression is under the control of the gDMRs, which are also referred to as imprinting control centers (ICRs), that contain cis-regulatory elements (Figure 1). This is a critical part of the intrinsic character of the imprinted regions. In most of the canonical imprinted regions, such cis-regulatory elements have long-range effects on the expression of neighboring genes, thus regulating a number of PEGs and MEGs. In the insulator model, the insulator sequences are utilized so the cis-regulatory elements are able to inhibit downstream enhancer activity (Bell and Felsenfeld 2000; Hark et al., 2000) (Figure 1). In the antisense models, the promoters of antisense transcripts interfere with the transcription of downstream genes (Stoger et al., 1993; Wutz et al., 1997). In certain cases, two separate regulatory regions are used to control the entire imprinted region. In the bipartite model, two regulatory units are required for the activity of a bipartite imprinting center, one containing the gDMR while the other may act with the former in gametes in the cis configuration, although the precise molecular mechanism remains unknown (Nicholls and Knepper 2001; Wu et al., 2012). In other cases, both the gDMRs and secondary DMRs (sDMRs) that are established in a manner depending on the former after fertilization, play essential roles (Takada et al., 2000, 2002; Lopes et al., 2003; Kagami et al., 2008; Kagami et al., 2015; Takahashi et al., 2009; Zhou et al., 2010; Beygo et al., 2015). Regardless of which molecular mechanism model applies, DNA methylation of either one of the parental gDMRs inhibits the activity of the cis-regulatory elements, thereby undoing the off state and leading to the complementary expression of PEG and MEG (Kaneko-Ishino et al., 2003; Kaneko-Ishino et al., 2006).
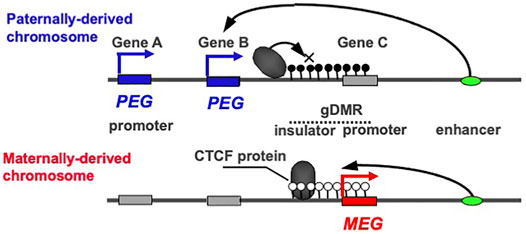
FIGURE 1. The reciprocal ON-OFF switch of PEG and MEG. In imprinted regions, PEG and MEG cannot be co-expressed in cis. In the insulator model, they are reciprocally regulated by the DNA methylation status of the gDMR including the insulator sequence. An insulator binding protein, such as CTCF, interrupts the downstream enhancer activity, leading to a repression of Genes A and B without any effect on Gene C. In contrast, DNA methylation on the insulator sequence inhibits the binding of the CTCF protein, leading to the induction of the Genes A and B concurrently with repression of the Gene C via DNA methylation on its promoter contiguous to the insulator sequence. The blue and red boxesindicate paternally and maternally active alleles, respectively, while the gray boxes indicate repressed alleles. Then, the blue and red arrows indicate PEG and MEG expression, respectively. The white and black circles indicate non-methylated and methylated CpGs, repressively. This is an updated version of Figure 2 in the previous review (Kaneko-Ishino and Ishino 2019).
DNA methylation of the gDMRs, referred to as imprinted memories, are established by DNA methyltransferase 3A (DNMT3A) and its catalytically inactive cofactor DNMT3L in either oocytes or spermatogonia (Okano et al., 1999; Bourc’his et al., 2001; Hata et al., 2002; Bourc’his and Bestor 2004; Kaneda et al., 2004), thus leading to the establishment of maternal and paternal gDMRs in the oocytes and sperm (Figure 2A). Such gDMRs are maintained in the somatic cells throughout life (Figure 2, middle), while in germ cells, it is completely erased for individuals in the next generation (Figure 2, bottom). From embryonic day 10.5 (d10.5), the erasure of genomic imprinting memory occurs in primordial germ cells (PGCs) through the passive replication-dependent DNA demethylation mechanism (Hackett et al., 2013; Yamaguchi et al., 2013a, 2013b; Kagiwada et al., 2013) and possibly together with the active mechanism catalyzed by ten-eleven translocation (TET) methylcytosine dioxygenases (Kawasaki et al., 2014). Importantly, clone embryos produced from d12.5 PGCs in the default mode of genomic imprinting (no DNA methylation of parental gDMRs) (Hajkova et al., 2002; Lee et al., 2002; Szabo et al., 2002) also exhibit early embryonic lethality (Lee et al., 2002; Yamazaki et al., 2005) similar to the parthenogenetic and androgenetic embryos. They express only MEGs in the paternally imprinted regions and only PEGs in the maternally imprinted regions, that is, the PEGs in the former and MEGs in the latter are repressed, respectively (Figure 2, middle). These results indicate that DNA methylation on the gDMRs is necessary to induce the repressed genes in the default state, indicating the strict requirement of having both paternal and maternal epigenotypes, leading to the complementary expression of PEGs and MEGs (Kaneko-Ishino et al., 2003; Kaneko-Ishino et al., 2006).
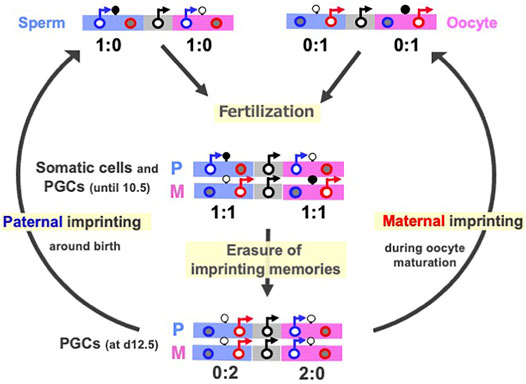
FIGURE 2. Cycle of genomic imprinting memory. Top: Sperm (left) and oocytes (right) have imprinted memories to express only PEGs and MEGs in somatic cells. Their expression patterns here represent those of androgenetic and parthenogenetic embryos, respectively. Second: The expression profiles of the imprinted genes in paternal and maternal imprinted regions in somatic cells and PGCs until at most day 10.5 (Miki et al., 2005; Yamazaki et al., 2005) This is reestablished by a combination of the sperm and oocyte patterns. Bottom: The expression profiles of imprinted genes in the default states of genomic imprinting (i.e., without any DNA methylation), such as day 12.5 PGC cloned embryos. The black, blue and red circles represent normal biallelic, paternally and maternally expressed genes, respectively. There are two types of imprinted regions, paternally (blue) and maternally imprinted (pink) reginos, in which gDMRs are methylated paternally and maternally, respectively. The erasure of imprinted memories in PGCs occurs around d10.5 and completed by d12.5, then gDMR methylation in the paternally imprinted regions (Paternal imprinting) occurs during prospermatogonia development around the time of birth, while that in the maternally imprinted regions (Maternal imprinting) occurs during oocyte maturation (arrows from the bottom to top). PEG and MEG cannot be co-expressed in cis in any stages of this cycle.
Paternal and maternal imprinting memories are then reestablished depending on the individual’s sex by DNA methylation on the paternal and maternal gDMRs in the paternal and maternal imprinted regions, respectively (Davis et al., 1999; Li et al., 2004; Lucifero et al., 2004; Hiura et al., 2006) (Figure 2, arrows from bottom to top), and as a result, the “imprinted” patterns of sperm and oocytes are completed (Figure 2, top). All of these changes in the expression pattern are consistent with the reciprocal regulation of PEGs and MEGs in a manner that depends on the DNA methylation status of the gDMRs (Figure 1). Taken together, DNA methylation at the gDMRs can be considered to serve as a major epigenetic mark in the life cycle of genomic imprinting and has recently been referred to as “canonical imprinting” in contrast to histone modification-dependent imprinting, as described later.
The Origin of gDMRs
Genomic imprinting is observed in eutherian and marsupial mammals, but the number of imprinted genes in eutherians is much larger than in marsupials (Cattanach and Beechey 1990; Killian et al., 2000; O’Neill et al., 2000; Beechey and Evans, 2004; Suzuki et al., 2005; Suzuki et al., 2007). In addition, there exist several lineage- and species-specific imprinted genes (Hayashizaki et al., 1994; Plass et al., 1996; Hagiwara et al., 1997; Smith et al., 2003; Wood et al., 2007; Das et al., 2012). In her host defense hypothesis, Denise Barlow implied that the origin of genomic imprinting lies in an existing biochemical system, such as DNA methylation, that serves to neutralize foreign invading DNA, such as retroviruses, direct repeats, and retrotransposons (Barlow et al., 1991; Neumann and Barlow. 1995). We had a similar idea and independently sought imprinted genes of exogenous origin by comprehensive screening of PEGs and MEGs (Kaneko-Ishino et al., 1995; Kuroiwa et al., 1996; Miyoshi et al., 1998; Miyoshi et al., 2000; Ono et al., 2001). Identification of paternally expressed 10 (PEG10), a gene acquired from a retrovirus (Ono et al., 2001; Ono et al., 2006), led to the finding that the PEG10-gDMR in its promoter region emerged with PEG10 itself in a common ancestor of therian mammals (Suzuki et al., 2007). Subsequent comprehensive comparative genome analysis has revealed that the DNA sequences corresponding to the gDMR in the canonical imprinted regions are derived from inserted DNA sequences (Suzuki et al., 2011; Renfree et al., 2013; Kaneko-Ishino and Ishino 2019). In most cases, such insertion events correlate well with the time when their imprinting regulation activity started as paternally and maternally imprinted regions (Figure 3, blue and red, respectively), providing strong support for the host defense hypothesis. From this perspective, the gDMRs, and most especially the cis-functional elements within them, should be recognized as the targets of the genomic imprinting mechanism rather than retroviruses, direct repeats and retrotransposons, as originally proposed (Barlow 1993; Neumann and Barlow 1995).
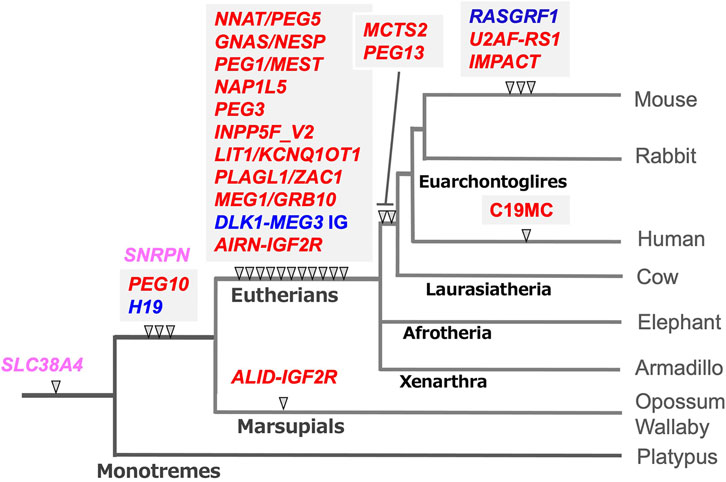
FIGURE 3. The emergence of gDMR sequences coincides with the onset of imprinted regulation in mammals. In most cases, the emergence of gDMR sequences, the DNA sequences corresponding to the gDMRs, correlate well with the establishment of imprinted regions in mammalian evolution. The arrowheads indicate when each gDMR sequence appeared in the mammalian lineage tree. Blue and red represent that imprinted regulation started as the paternally and maternally imprinted regions, respectively. Pink represents the maternally imprinted regions in which the emergence of DMR sequences preceded the onset of imprinted regulation, for example, SLC38A4-and SNRPN-DMRs (see the text for the details). It should be noted that mouse Slc38a4 has recently been recognized as an imprinted gene regulated by both canonical and non-canonical imprinting mechanisms (Okae et al., 2014; Inoue et al., 2017a; Bogutz et al., 2019). This is an updated version of Figure 6 in our previous review (Kaneko-Ishino and Ishino 2019).
For example, the PEG10-and H19-gDMR sequences emerged next to the sarcoglycan epsilon (SGCE) promotor and downstream of the Insulin-like growth factor 2 (IGF2) gene in both the marsupial and eutherian genomes, respectively (Suzuki et al., 2007; Smits et al., 2008; Kaneko-Ishino and Ishino 2010). Thus, it is certain that their insertion occurred in a common therian ancestor. They were associated with the insertion of the retrovirus-derived PEG10 and non-coding H19, respectively (Table 1). In addition, imprinted paternal expression of PEG10 (Ono et al., 2001; Suzuki et al., 2007) and reciprocal maternal/paternal expression of H19/IGF2 (DeChiara et al., 1990, 1991; Bartolomei et al., 1991; O’Neill et al., 2000; Killian et al., 2001; Weidman et al., 2004; Edwards et al., 2008) have been confirmed in both groups. Interestingly, only the PEG10 promoter is differentially methylated in marsupials. In contrast, the PEG10-gDMR extends to the SGCE promoter and controls a large imprinted region, including several MEGs, in eutherians (Ono et al., 2003; Suzuki et al., 2007).
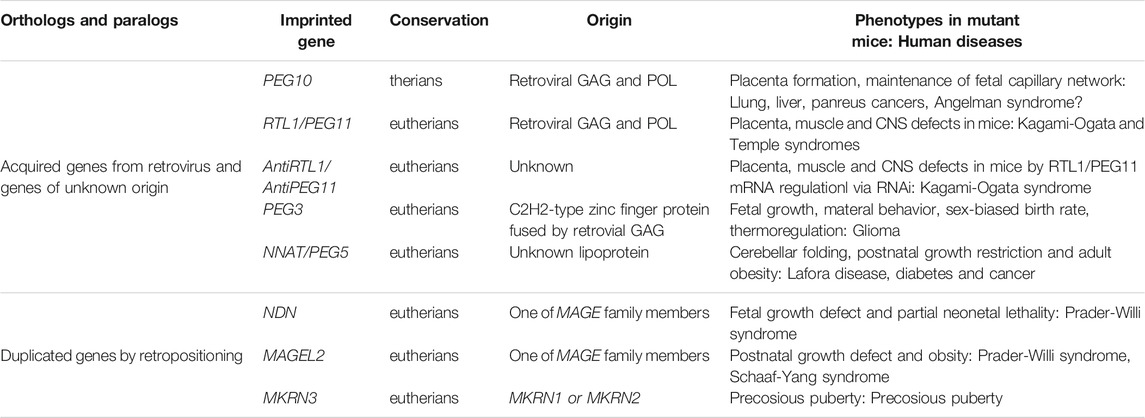
TABLE 1. Newly acquired genes in the imprinted regions. The essential/important imprinted genes that have emerged in therian- and eutherian-specific imprinted regions are summarized.
The PEG10 and H19/IGF2 imprinted regions are the first two imprinted regions in mammalian history, while most of the gDMRs emerged in the eutherian genome and constitute eutherian-specific imprinted regions. Subsequently, some lineage-specific imprinted regions appeared, indicating that genomic imprinting arose at many different time points during mammalian evolution and is still continuing to evolve (Suzuki et al., 2011; Das et al., 2012; Renfree et al., 2013). The large miRNA cluster, C19MC, in the human genome is an example of a lineage-specific imprinted region that has emerged in primates (Noguer-Dance et al., 2010; Malnou et al., 2018), implying the existence of other primate- and/or human-specific imprinted regions in the human genome.
Interestingly, the IGF2 receptor (IGF2R) is imprinted in both eutherians and marsupials (Killian et al., 2000; Szabo et al., 2000), however, it is regulated by different gDMRs. It is located on intron 2 in the former and intron 12 in the latter (Stöger et al., 1993; Suzuki et al., 2018). Both gDMRs function as promoters of antisense non-coding RNAs, a long Antisense of IGF2R non-protein coding RNA (AIRN) in the former (Stoger et al., 1993) and a short non-coding RNA, Antisense LncRNA in the IGF2R gDMR (ALID), in the latter. It is thus likely that they emerged independently and are regulated by different mechanisms. Marsupial-specific expansion of intron 12, which has occurred as the results of accumulation of a number of transposons, may have contributed to the establishment of the ALID/IGF2R-gDMR (Suzuki et al., 2018).
Two apparent exceptions are the DNA sequence corresponding to the Small nuclear ribonucleoprotein polypeptide N (SNRPN)- and the Solute carrier family 38, member 4 (SLC38A4)-gDMRs (Figure 3, pink) because their insertion did not coincide with the onset of maternal imprinted regulation. The former regulates the Prader-Willi/Angelman (PWS/AS) region as the PWS-shortest region of deletion overlap (SRO) together with the AS-SRO located 880 bp at a site 35 kb upstream of the SNRPN upstream open reading frame (SNURF)-SNRPN (Nicholls et al., 1998). The DNA sequence corresponding to the SNRPN-DMR is conserved in both marsupials and eutherians, although, in marsupials, the SNRPN, and Ubiquitin Protein Ligase E3A (UBE3A) regions exist on separate chromosomes. The PWS/AS region arose by chromosome rearrangement in eutherians, and the SNRPN-DMR located in SNURF was established in the newly reconstructed DNA sequence (Rapkins et al., 2006). Similarly, the corresponding sequence to the SLC38A4-gDMR already existed in monotremes (Suzuki et al., 2011), but it became differentially methylated only in the rodent lineage in association with the rodent-specific insertion of MT2A retrovirus (Bogutz et al., 2019; Hanna et al., 2019) (see next section). Slc38a4 has also been recognized as a member of non-canonical imprinted genes in mice, implying that an evolutionary route to the SLC38A4 imprinted region may be complicated (Okae et al., 2014; Inoue et al., 2017a).
Molecular Mechanisms Underlying the Establishment of Differential DNA Methylation Patterns in the Female and Male Gametes
So what, then, is the actual biochemical system that serves to neutralize foreign DNA that had been hypothesized by Barlow? Recent work has shown that the gDMRs are established as a consequence of differential responses to the invading DNAs in the oocytes and prospermatogonia as well as to afford subsequent protection from DNA demethylation in preimplantation development (see the next section).
Discovery of the oocyte-specific transcripts spanning the gDMRs has provided an important clue to the mechanism of the establishing of the maternal gDMRs (Chotalia et al., 2009; Veselovska et al., 2015; Smallwood et al., 2011; Hanna and Kelsey 2021). In oocytes, DNA methylation is exclusively on active gene body regions, including the maternally imprinted gDMRs (Kobayashi et al., 2012). Initially, histone lysine methylase SETD2 deposits H3K36me3 (Figure 4A), and subsequently this epigenetic mark guides DNA methylation because it promotes the binding of DNMT3A in the oocyte (Chotalia et al., 2009; Veselovska et al., 2015; Xu et al., 2019; Shirane et al., 2020) (Figure 4B). Interestingly, long terminal repeats (LTRs) of some endogenous viruses (ERVs) are used as the promoters of the oocyte-specific alternative upstream transcripts in the species-specific canonical imprinting (Brind’Amour et al., 2018; Bogutz et al., 2019). These data indicate that the oocyte-specific transcripts from the upstream promoters, such as integrated LTRs, play a critical and fundamental role in the mechanism underlying the establishment of the maternal gDMRs.
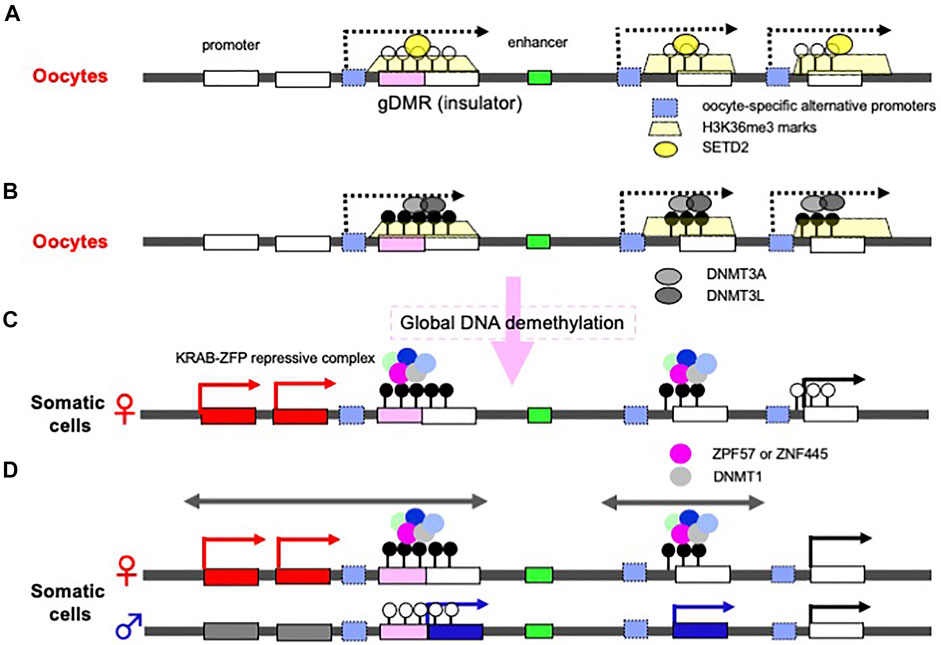
FIGURE 4. Molecular mechanisms underlying how gDMRs are established and maintained. (A) Expression of oocyte-specific transcripts from alternative promoters including LTRs. Oocyte-specific alternative upstream transcripts (dashed arrows) run through the gDMR (a pink box) within gene bodies with deposition of H3K36me3 (yellow trapezoids). In some cases, LTRs are used as the promoters for such transcripts (light blue boxes). (B) DNA methylation in gene body regions including maternal gDMRs. The H3K36me3 epigenetic mark guides DNA methylation in the oocytes, leading to the gene body DNA methylation. There are many more differential methylated CpG sequences than gDMRs in oocytes. (C) Protection of gDMR DNA methylation from a global DNA demethylation wave. Most of the differentially methylated CpG sequences disappear during preimplantation development due to a global DNA demethylation wave (right), however, both maternal and paternal gDMRs remain protected by a large complex including either ZFP57 or ZNF445, members of the KRAB-ZFPs playing an essential role in repressing invaded retroviruses (left and center). (D) The resulting canonical imprinted regions. Double-headed arrows indicate a therian-/eutherian-specific (left) and a species-specific (center) imprinted region in somatic cells. The gDMR DNA methylation is maintained during postimplantation period by symmetric CpG methylation catalyzed by DNMT1 included in the KRAB-ZFPs complex. The differential recognition mechanisms in the paternal and maternal germ cells lead to the imprinted region in somatic cells.
On the other hand, in the male germline (prospermatogonia), the lysine methyltransferase NSD1 deposits H3K36me2 in the euchromatic regions included on the paternally imprinted gDMRs, and subsequently this epigenetic mark guides DNA methylation (Shirane et al., 2020). Thus, there is a sexually dimorphic pattern of DNA methylation in mature mouse gametes via the deposition of distinct H3K36 methylation marks (Xu et al., 2019; Shirane et al., 2020). Among the three paternal gDMRs, Ras protein specific guanine nucleotide releasing factor 1 (Rasgrf1)-gDMR is known to use an LTR from the RMER4 retrotransposon as an upstream promoter to generate small RNAs that recruit the PIWI-piRNA mechanism to establish de novo DNA methylation in spermatogenesis (Watanabe et al., 2011), although the molecular mechanism remains elusive in the other two gDMRs, the H19- and DLK1-MEG3 intergenic (IG)-gDMRs. These data demonstrate that integrated retroviruses (ERVs) and LTR retrotransposons are ingeniously integrated in the DNA methylation of gDMRs that occurs in germ cells.
Molecular Mechanisms Underlying gDMR Maintenance in pre- and Postimplantation Development
Another important fact is that there are more than 1,600 differentially methylated CpG islands in oocytes and sperm, including the imprinting loci (Kobayashi et al., 2012) (Figure 4B). Most of these differences disappear during preimplantation period, while the gDMRs of the canonical imprinted regions are protected from such a global DNA demethylation wave (Figure 4C) (Kobayashi et al., 2012; Smallwood et al., 2011) and maintained in the postimplantation period (Figure 4D), indicating that the canonical genomic imprinted regions have been selected and conserved in mammals because of their evolutionary advantages. Interestingly, certain members of the Krüppel-associated box (KRAB)-containing zinc finger proteins (ZFPs), such as ZFP57 and ZNF445, play a key role in this process (Li et al., 2008; Takahashi et al., 2019). ZFP57 and ZNF445 bind a CpG-containing hexanucleotide motif present in multiple copies in most ICRs (Quenneville et al., 2011; Strogantsev et al., 2015; Anvar et al., 2016), and each forms a large complex that represses transcription by recruiting KRAB-associated protein1 [KAP1, aca tripartite motif containing 28 (TRIM28)], SET domain bifurcated histone lysine methyltransferase 1 (SETDB1), heterochromatin protein 1 (HP1) and DNMT1, and then maintain DNA methylation on the gDMRs of the newly replicated DNA by symmetric CpG methylation (Sharif et al., 2007; Quenneville et al., 2011; Liu et al., 2013; Mochizuki et al., 2021) (Figure 4C). The KRAB-ZFPs are evolutionarily diverse proteins that protect against the insertion of retroviruses (Macfarlan et al., 2012; Rowe et al., 2013), demonstrating that another host defense mechanism against retroviruses also exists and is functional in the maintenance of genomic imprinting memories (Edwards et al., 2019; Ondičova et al., 2020; Hanna and Kelsey, 2021). Thus, accumulating evidence indicates a close relationship between genomic imprinting and the insertion of retroviruses and LTR retrotransposons, as originally proposed in the host defense hypothesis. Importantly, newly invading DNAs are not completely repressed in both of the parental alleles because of the differential recognition mechanisms in the paternal and maternal germ cells, thereby leading to the complementary monoallelic expression of PEGs and MEGs (Figure 4D).
Evolutionary Advantages of Genomic Imprinting
What is the evolutionary advantage of genomic imprinting? In other words, why have the canonical genomic imprinted regions been selected and widely conserved in therian mammals? We would like to address this issue based on the complementary monoallelic expression mechanism. First, this arrangement allows for the expression of all of the genes in the imprinted regions as PEGs and MEGs by controlling for the cis-elements in the gDMRs (Figure 1). As mentioned earlier, the cis-elements involved in the newly inserted gDMR sequences (Figure 3) exert a long-range effect on nearby genes, and thus repress a substantial number of resident genes. However, DNA methylation of the cis-elements in one of the alleles allows for recovery of the expression of such resident genes, so DNA methylation is required for the expression of certain imprinted genes (Figures 1, 2). As several of the PEGs and MEGs play an essential role in the current mammalian developmental system, their monoallelic expression would be expected to be disadvantageous in a general sense. However, under a special constraint, such as described for the cis-elements, monoallelic expression affords an advantage, because even a limited monoallelic expression is preferable to the complete loss of expression (Figures 1, 2). This indicates that genomic imprinting arose as a consequence of an evolutionary trade-off for survival (Kaneko-Ishino and Ishino 2015, 2019). This may also account for why the canonical imprinted regions have been so widely conserved in the therian mammals.
As mentioned in the introduction, it is likely that the conflict over maternal resources exerts pressure on PEGs promoting and MEGs repressing embryonic growth. In most cases, at least one imprinted gene in each canonical imprinted region seems to fit the conflict/kinship hypothesis (Moore and Haig, 1991; Wilkins and Haig 2003; Haig 2004) although most imprinted genes appear to be bystanders, that is, they are simply involved in this process as bystander genes (Barlow and Bartolomei 2014). In addition, it is also likely that gene dosage regulation of the evolutionarily conserved resident imprinted genes is another important factor in selection, because it must have contributed to the formation of the current mammalian developmental system by increasing pre-and postnatal fitness (Charalambous et al., 2012; Ball et al., 2013; Wolf et al., 2014).
Other hypotheses have been proposed that suggest genomic imprinting arose for the prohibition of parthenogenetic development or protection against the development of malignant placental tissue in such parthenogenetic embryos (Solter 1988; Varmuza and Mann 1994). Although it is difficult to prove these hypotheses by experiment, these features are surely advantageous for viviparous mammals and seem to have been acquired from the beginning via genomic imprinting, because the first two imprinted regions established in a common therian ancestor were the PEG10 and H19/IGF2 regions (Figure 3), that exert potent effects on placental formation and growth, respectively. This also supports the placenta hypothesis (Hall 1990; Kaneko-Ishino et al., 2003), which proposes that genomic imprinting is deeply related to the emergence of the placenta in the course of evolution, because a considerable number of imprinted genes play important roles in the placenta. In addition, H3K27me3 based non-canonical imprinting is unique to the placenta, and contributes to placental development and growth in a species-specific manner, such as Xist, Gab1 and Sfmbt2 in mice (Itoh et al., 2000; Okae et al., 2014; Inoue et al., 2017a, 2017b, 2018; Hanna et al., 2019; Inoue et al., 2020; Hanna and Kelsey 2021).
Finally, we would like to address another benefit of the complementary monoallelic mechanism. It provides a means of expression for both newly inserted genes as well as the conserved, already resident genes, therefore, allows for the acquisition of new genes in the canonical imprinted regions, and genes like PEG10 and H19 (Figure 5). Therefore, genomic imprinting may have made a contribution to the innovation of the emergent mammalian functions. Such newcomer genes may be separated into two groups: 1) newly acquired genes from retroviruses or of unknown origin, and 2) genes duplicated by retroposition. Among these genes, several play essential/important roles in development, growth and behavior, as discussed in the following two sections.
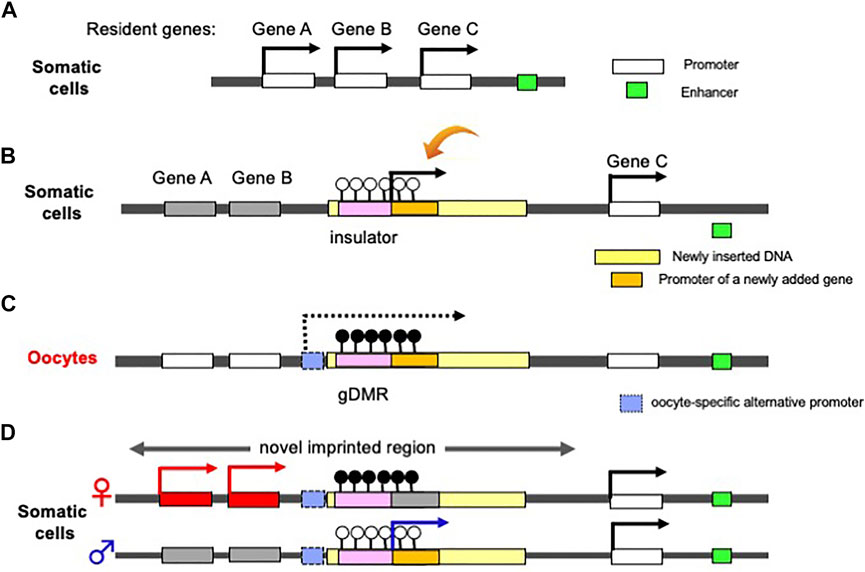
FIGURE 5. Generation of a novel canonical imprinted region. (A) Before an insertion event. A chromosomal region comprising three non-imprinted evolutionary resident Genes A, B and C. A downstream enhancer sequence (green) regulates the activation of the three genes. (b) Insertion of a large DNA fragment (yellow) containing an insulator sequence as a critical cis-element (pink) and a novel gene (orange) between Genes B and C. The insertion event leads to the repression of Genes A and B via the insulator function (see Figure 1). The novel added gene is expressed from the newly integrated DNA fragment in addition to Gene C. (C) DNA methylation on the inserted insulator in oocyte. Emergence of an upstream promotor (light blue) that expresses an oocyte-specific alternative transcript (dashed line). This transcript goes through the insulator, leading to DNA methylation on the insulator in an oocyte-specific manner. (D) Expression of a newly added gene from paternal allele of a novel imprinted region (arrow), while Genes A and B from maternal allele because the gDMR in the paternally-derived chromosome is not DNA methylated.
Novel Functions Provided by Newly Acquired Imprinted Genes in Mammals
In addition to PEG10 and H19, there are several imprinted regions that are accompanied by novel acquired genes. In the Delta like non-canonical Notch ligand 1 (DLK1)-Iodothyronine deiodinase 3 (DI O 3) region, a large insertion comprising MEG3, Retrotransposon GAG-like 1 (RTL1)/PEG11, antiRTL1/antiPEG11, and MEG8/RNA imprinted and accumulated in nucleus (RIAN) and MEG9/microRNA-containing gene (MIRG) occurred in association with the emergence of an intergenic (IG)-DMR between DLK1 and MEG3 in eutherians (Kobayashi et al., 2000; Miyoshi et al., 2000; Schmidt et al., 2000; Wylie et al., 2000; Charlier et al., 2001; Edwards et al., 2008). Thus, they are eutherian-specific genes: RTL1/PEG11 is a protein coding gene while the other maternally expressed non-coding RNAs include a cluster of siRNA and/or snoRNAs (Charlier et al., 2001; Cavaillé et al., 2002). In the PEG3 region, the PEG3-DMR localizes on the PEG3 promoter and regulates PEG3 itself and neighboring paternally expressed Ubiquitin specific peptidase 29 (USP29), as well as maternally expressed Zinc finger imprinted 1 (ZIM1) and ZIM3 (Kim et al., 2001, 2007). PEG3 is also a eutherian-specific acquired gene. PEG3 has quite unique zinc finger motif in terms of its amino acid sequence and its intervals, and no homologous C2H2-type of zinc finger protein is present in marsupials, monotremes or other vertebrates (Kuroiwa et al., 1996). The construction of the PEG3 imprinted region is polymorphic among eutherian species, that is, there are certain differences that occur in a species-specific manner (Kim et al., 2007). NEURONATIN (NNAT, aka PEG5) (Kagitani et al., 1997; Kikyo et al., 1997) is inserted within an intron of the non-imprinted Bladder cancer associated protein (BCAP) and its promoter became the NNAT/PEG5-DMR (John et al., 2001). NNAT/PEG5 is another eutherian-specific gene encoding a proteolipid of unknown origin, and there is a single imprinted gene in this locus (Evans et al., 2005) (Table 1).
Interestingly, PEG10 and RTL1/PEG11 are derived from retrovirus GAG and POL (Charlier et al., 2001; Ono et al., 2001). They exhibit a certain similarity to the sushi-ichi LTR retrotransposon, thus have come to be referred to as retrotransposon-GAG like genes. In any event, it is likely that the “gypsy” type of LTR retrotransposon to which the suchi-ichi retrotoransposon belong is derived from a retrovirus (Kim et al., 1994; Song et al., 1994). PEG3 was probably generated by fusion of the same retroviral GAG and a C2H2-type of zinc finger protein generated by gene duplication (Campillos et al., 2006). PEG10 is essential for the induction of placenta-specific cells, i.e., the trophoblast cells in the spongiotrophoblast and labyrinth layers, so its deficiency causes early embryonic lethality (Ono et al., 2006). It also plays an essential role in the maintenance of the fetal capillary network in the labyrinth layer: mutant mice with defective PEG10 protease exhibit perinatal lethality due to severe damage of the entire fetal capillary network (Shiura et al., 2021). PEG10 is also associated with progression of various cancers, such as lung, breast, liver, and pancreatic cancer (Okabe et al., 2003; Li et al., 2006). Recently, accumulation of the PEG10 protein was reported in neurons differentiated from induced pluripotent stem cells (iPSs) from Angelman syndrome (AS) patients, suggesting an important role in the brain (Pandya et al., 2021).
RTL1/PEG11 plays essential roles in the placenta, muscle and brain and is the major gene responsible for the Kagami-Ogata and Temple syndromes, two genomic imprinting diseases. It is located on human chromosome 14 (Kagami et al., 2008; Sekita et al., 2008, Kitazawa et al., 2017, 2020, 2021). Both overexpression and deficiency lead to abnormalities in the placenta as well as a variety of neuromuscular and/or psychiatric symptoms (Kitazawa et al., 2020, 2021). In the placenta, severe defects of the fetal capillaries can lead to late embryonic lethality or growth retardation (Sekita et al., 2008; Kitazawa et al., 2017). It is likely that damage to neonatal respiration-related tissues such as the intercostal, abdominal and diaphragm muscles lead to neonatal lethality (Kitazawa et al., 2020), presumably associated with abnormality of motor neurons in the descending corticonuclear tract that innervates the cranial nerves regulating the respiration-related muscles. RTL1/PEG11 is also expressed in neurons in the corpus callosum, hippocampal and anterior cerebral commissures as well as limbic system, such as in the hippocampus and amygdala, suggesting that their malfunction is related to the psychiatric symptoms that manifest in these diseases (Kitazawa et al., 2021).
PEG3 is also essential for placental functions as well as in the fetal and adult brain. Its defect causes fetal growth retardation, a problem in suckling milk in the pups, and abnormal maternal-care behaviors due to defects in the milk release process in females (Li et al., 1999) and thermoregulation (Broad et al., 2009), while biallelic expression of Peg3 leads to subtle impacts in male and female mice (Bretz and Kim 2017). Certain mouse Peg3 mutations also cause sex-biased outcomes (Kim et al., 2013; He et al., 2016). PEG3 functions as a tumor suppressor in glioma in the brain (Kohda et al., 2001; Maegawa et al., 2004; Jiang et al., 2010). The loss of NNAT/PEG5 is associated with decreased cerebellar folding in mice (Kikyo et al., 1997) and leads to postnatal growth restriction and adult obesity (Millership et al., 2018). It is reported that aggregates of the NNAT/PEG5 protein cause neuronal loss in Lafora disease, diabetes, and cancer (Joseph, 2014).
New Functions Provided by Imprinted Genes Duplicated by Retropositioning
NECDIN (NDN), MAGE Family Member L2 (MAGEL2) and Makorin Ring Finger Protein 3 (MKRN3) are located in the PWS/AS imprinted region and are retroposed genes generated from the cDNAs of their paralogs (Rapkins et al., 2006) (Table 1). The retropositioning events are sometimes associated with the generation of newly imprinted regions (Wood et al., 2007), such as the nucleosome assembly protein 1-like 5 (NAP1L5) and Inositol Polyphosphate-5-Phosphatase F (INPP5F_V2) regions in eutherians, and also the Malignant T-cell-amplified sequence 2 (MCTS2) and U2 small nuclear RNA auxiliary factor 1-related sequence 1 (U2AF1-RS1) regions in a lineage- and species-specific manner. Their own promoter regions became DMRs and interestingly, their original genes were localized in the X-chromosome (Wood et al., 2007). These genes were introduced into the eutherian genome as imprinted genes, presumably via surprising and complex processes that exploited the complementary monoallelic expression mechanism of genomic imprinting (Figure 5).
Among these genes, NDN, MAGEL2, and MKRN3 play essential roles in the brain. The former two genes are responsible for certain symptoms of PWS including neonatal lethality, prenatal growth retardation and postnatal growth abnormalities, such as, obesity (Lee et al., 2005; Bischof et al., 2007), and the latter gene is responsible for precocious puberty (Abreu et al., 2013; Li et al., 2021). MKRN3-mediated ubiquitin signaling controls expression of Gonadotropin releasing hormone 1 (GNRH1) at both transcriptional and post-transcriptional levels (Li et al., 2021). MAGEL2 mutations causes Schaaf-Yang syndrome, a rare neurodevelopmental disorder that shares multiple clinical features with the genetically related PWS (Schaaf et al., 2013).
Conclusion
Accumulating evidence has painted a picture of a close relationship between genomic imprinting and invading DNA: 1) the insertion of DNA sequences corresponding to the gDMRs (Figure 3), 2) retroviral LTRs are used as promoters of oocyte-specific transcripts for DNA methylation on the gDMRs (Figure 4B), 3) the antiviral KRAB-ZFP system protects the gDMRs from global DNA demethylation (Figure 4C), providing strong support for the host defense hypothesis. Taken together, it is likely that each genomic imprinted region arose by chance as a consequence of an evolutionary trade-off for survival (Kaneko-Ishino and Ishino 2019) using the existing DNA methylation machinery against foreign DNA. Each region has been selected and conserved in therian/eutherian mammals for the advantage(s) it confers. We have also discussed another evolutionary advantage that is exerted by the complementary monoallelic expression mechanism which is that it affords a kind of genome innovation machinery that enables the introduction of novel genes via acquisition from retroviruses and also gene duplication by retroposition (Kaneko-Ishino and Ishino 2012; Edwards et al., 2019). As discussed in this review, in addition to the “Conflict” and “Complementation” activities, there are a variety factors acting in concert that make up the biological significance of genomic imprinting which remains a fascinating and critically important theme in mammalian biology.
Author Contributions
TK-I and FI write this review.
Funding
This work was supported by funding program for Grants-in-Aid for Scientific Research (C) (21K06127) from Japan Society for the Promotion of Science (JSPS) to TK-I, Grants-in-Aid for Scientific Research (A) (19H00978) from JSPS to FI, Nanken Kyoten Program, Medical Research Institute, Tokyo Medical and Dental University (TMDU) to TK-I and FI. The funders had no role in preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank S. Suzuki for his information, genome structure reanalysis and advice on the manuscript. Pacific Edit reviewed the manuscript prior to submission.
References
Abreu, A. P., Dauber, A., Macedo, D. B., Noel, S. D., Brito, V. N., Gill, J. C., et al. (2013). Central Precocious Puberty Caused by Mutations in the Imprinted Gene MKRN3. N. Engl. J. Med. 368, 2467–2475. doi:10.1056/NEJMoa1302160
Anvar, Z., Cammisa, M., Riso, V., Baglivo, I., Kukreja, H., Sparago, A., et al. (2016). ZFP57 Recognizes Multiple And Closely Spaced Sequence Motif Variants To Maintain Repressive Epigenetic Marks In Mouse Embryonic Stem Cells. Nucl. Acids Res.. doi:10.1093/nar/gkv1059
Ball, S. T., Kelly, M. L., Robson, J. E., Turner, M. D., Harrison, J., Jones, L., et al. (2013). Gene Dosage Effects at the Imprinted Gnas Cluster. PLoS ONE 8, e65639. doi:10.1371/journal.pone.0065639
Barlow, D. P., and Bartolomei, M. S. (2014). Genomic Imprinting in Mammals. Cold Spring Harbor Perspect. Biol. 6, a018382. doi:10.1101/cshperspect.a018382
Barlow, D. P. P. (1993). Methylation and Imprinting: from Host Defense to Gene Regulation? Science 260, 309–310. doi:10.1126/science.8469984
Barlow, D. P., Stöger, R., Herrmann, B. G., Saito, K., and Schweifer, N. (1991). The Mouse Insulin-like Growth Factor Type-2 Receptor Is Imprinted and Closely Linked to the Tme Locus. Nature 349, 84–87. doi:10.1038/349084a0
Bartolomei, M. S., Zemel, S., and Tilghman, S. M. (1991). Parental Imprinting of the Mouse H19 Gene. Nature 351, 153–155. doi:10.1038/351153a0
Beechey, C. V., and Evans, E. P. (2004). MRC Harwell, Harwell, Oxfordshire. Standard Ideogram/Anomaly Breakpoints of the Mouse. Available at: https://www.har.mrc.ac.uk/about-harwell/history/imprinting-resource/.
Bell, A. C., and Felsenfeld, G. (2000). Methylation of a CTCF-dependent Boundary Controls Imprinted Expression of the Igf2 Gene. Nature 405, 482–485. doi:10.1038/35013100
Beygo, J., Elbracht, M., de Groot, K., Begemann, M., Kanber, S., Platzer, K., et al. (2015). Novel Deletions Affecting the MEG3-DMR Provide Further Evidence For A Hierarchical Regulation Of Imprinting in 14q32. Eur. J. Hum. Genet. 23, 180–188.
Bischof, J. M., Stewart, C. L., and Wevrick, R. (2007). Inactivation of the Mouse Magel2 Gene Results in Growth Abnormalities Similar to Prader-Willi Syndrome. Hum. Mol. Genet. 16, 2713–2719. doi:10.1093/hmg/ddm225
Bogutz, A. B., Brind’Amour, J., Kobayashi, H., Jensen, K. N., Nakabayashi, K., Imai, H., et al. (2019). Evolution of Imprinting via Lineage-specific Insertion of Retroviral Promoters. Nat. Commun. 10, 5674. doi:10.1038/s41467-019-13662-9
Bourc'his, D., Xu, G. L., Lin, C. S., Bollman, B., and Bestor, T. H. (2001). Dnmt3L and the Establishment of Maternal Genomic Imprints. Science 294, 2536–2539. doi:10.1126/science.1065848
Bourc’his, D., and Bestor, T. H. (2004). Meiotic Catastrophe and Retrotransposon Reactivation in Male Germ Cells Lacking Dnmt3L. Nature 431, 96–99.
Bretz, C. L., and Kim, J. (2017). Transcription-driven DNA Methylation Setting on the Mouse Peg3 Locus. Epigenetics 12, 945–952. doi:10.1080/15592294.2017.1377869
Brind’Amour, J., Kobayashi, H., Albert, J. R., Shirane, K., Sakashita, A., Kamio, A., et al. (2018). LTR Retrotransposons Transcribed In Oocytes Drive Species-Specific And Heritable Changes in DNA Methylation. Nat. Commun. 9, 3331. doi:10.1038/s41467-018-05841-x
Broad, K. D., Curley, J. P., and Keverne, E. B. (2009). Increased Apoptosis during Neonatal Brain Development Underlies the Adult Behavioral Deficits Seen in Mice Lacking a Functionalpaternally Expressed Gene 3(Peg3). Devel Neurobio 69, 314–325. doi:10.1002/dneu.20702
Campillos, M., Doerks, T., Shah, P., and Bork, P. (2006). Computational Characterization of Multiple Gag-like Human Proteins. Trends Genet. 22, 585–589. doi:10.1016/j.tig.2006.09.006
Cattanach, B. M., and Beechey, C. V. (1990). Autosomal and X-Chromosome Imprinting. DevelopmentSuppl 108, 63–72. doi:10.1242/dev.108.supplement.63
Cattanach, B. M., and Kirk, M. (1985). Differential Activity of Maternally and Paternally Derived Chromosome Regions in Mice. Nature 315, 496–498. doi:10.1038/315496a0
Cavaillé, J., Seitz, H., Paulsen, M., Ferguson-Smith, A. C., and Bachellerie, J.-P. (2002). Identification of Tandemly-Repeated C/D snoRNA Genes at the Imprinted Human 14q32 Domain Reminiscent of Those at the Prader-Willi/Angelman Syndrome Region. Hum. Mol. Genet. 11, 1527–1538. doi:10.1093/hmg/11.13.1527
Charalambous, M., Ferron, S. R., da Rocha, S. T., Murray, A. J., Rowland, T., Ito, M., et al. (2012). Imprinted Gene Dosage Is Critical for the Transition to Independent Life. Cel Metab. 15, 209–221. doi:10.1016/j.cmet.2012.01.006
Charlier, C., Segers, K., Wagenaar, D., Karim, L., Berghmans, S., Jaillon, O., et al. (2001). Human-Ovine Comparative Sequencing of a 250-kb Imprinted Domain Encompassing the Callipyge (Clpg) Locus and Identification of Six Imprinted Transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11, 850–862. doi:10.1101/gr.172701
Chotalia, M., Smallwood, S. A., Ruf, N., Dawson, C., Lucifero, D., Frontera, M., et al. (2009). Transcription Is Required For Establishment Of Germline Methylation Marks At Imprinted Genes. Genes Dev. 23, 105–117.
Das, R., Anderson, N., Koran, M. I., Weidman, J. R., Mikkelsen, T. S., Kamal, M., et al. (2012). Convergent and Divergent Evolution of Genomic Imprinting in the Marsupial Monodelphis Domestica. BMC Genomics 13, 394. doi:10.1186/1471-2164-13-394
Davis, T. L., Trasler, J. M., Moss, S. B., Yang, G. J., and Bartolomei, M. S. (1999). Acquisition of theH19Methylation Imprint Occurs Differentially on the Parental Alleles during Spermatogenesis. Genomics 58, 18–28. doi:10.1006/geno.1999.5813
DeChiara, T. M., Efstratiadis, A., and Robertsen, E. J. (1990). A Growth-Deficiency Phenotype in Heterozygous Mice Carrying an Insulin-like Growth Factor II Gene Disrupted by Targeting. Nature 345, 78–80. doi:10.1038/345078a0
DeChiara, T. M., Robertson, E. J., and Efstratiadis, A. (1991). Parental Imprinting of the Mouse Insulin-like Growth Factor II Gene. Cell 64, 849–859. doi:10.1016/0092-8674(91)90513-x
Edwards, C. A., Mungall, A. J., Matthews, L., Ryder, E., Gray, D. J., Pask, A. J., et al. (2008). The Evolution of the DLK1-DIO3 Imprinted Domain in Mammals. Plos Biol. 6:e135.
Edwards, C. A., Takahashi, N., Corish, J. A., and Ferguson-Smith, A. C. (2019). The Origins of Genomic Imprinting in Mammals. Reprod. Fertil. Dev. 31, 1203–1218. doi:10.1071/rd18176
Evans, H. K., Weidman, J. R., Cowley, D. O., and Jirtle, R. L. (2005). Comparative Phylogenetic Analysis of Blcap/nnat Reveals Eutherian-specific Imprinted Gene. Mol. Biol. Evol. 22, 1740–1748. doi:10.1093/molbev/msi165
Fujita, M. K., Singhal, S., BrunesMaldonado, T. O. J. A., and Maldonado, J. A. (2020). Evolutionary Dynamics and Consequences of Parthenogenesis in Vertebrates. Annu. Rev. Ecol. Evol. Syst. 51, 191–214. doi:10.1146/annurev-ecolsys-011720-114900
Hackett, J. A., Sengupta, R., Zylicz, J. J., Murakami, K., Lee, C., Down, T. A., et al. (2013). Germline DNA Demethylation Dynamics and Imprint Erasure through 5-hydroxymethylcytosine. Science 339, 448–452. doi:10.1126/science.1229277
Hagiwara, Y., Hirai, M., Nishiyama, K., Kanazawa, I., Ueda, T., Sakaki, Y., et al. (1997). Screening for Imprinted Genes by Allelic Message Display: Identification of a Paternally Expressed Gene Impact on Mouse Chromosome 18. Proc. Natl. Acad. Sci. 94, 9249–9254. doi:10.1073/pnas.94.17.9249
Haig, D. (2004). Genomic Imprinting and Kinship: How Good Is the Evidence? Annu. Rev. Genet. 38, 553–585. doi:10.1146/annurev.genet.37.110801.142741
Hajkova, P., Erhardt, S., Lane, N., Haaf, T., El-Maarri, O., Reik, W., et al. (2002). Epigenetic Reprogramming in Mouse Primordial Germ Cells. Mech. Development 117, 15–23. doi:10.1016/s0925-4773(02)00181-8
Hall, J. G. (1990). Genomic Imprinting: Review and Relevance to Human Diseases. Am. J. Hum. Genet. 46, 857–873.
Hanna, C. W., and Kelsey, G. (2021). Features and Mechanisms of Canonical and Noncanonical Genomic Imprinting. Genes Dev. 35, 821–834. doi:10.1101/gad.348422.121
Hanna, C. W., Pérez-Palacios, R., Gahurova, L., Schubert, M., Krueger, F., Biggins, L., et al. (2019). Endogenous Retroviral Insertions Drive Non-canonical Imprinting in Extra-embryonic Tissues. Genome Biol. 20, 225. doi:10.1186/s13059-019-1833-x
Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M., and Tilghman, S. M. (2000). CTCF Mediates Methylation-Sensitive Enhancer-Blocking Activity at the H19/Igf2 Locus. Nature 405, 486–489. doi:10.1038/35013106
Hata, K., Okano, M., Lei, H., and Li, E. (2002). Dnmt3L Cooperates with the Dnmt3 Family of De Novo DNA Methyltransferases to Establish Maternal Imprints in Mice. Development 129, 1983–1993. doi:10.1242/dev.129.8.1983
Hayashizaki, Y., Shibata, H., Hirotsune, S., Sugino, H., Okazaki, Y., Sasaki, N., et al. (1994). Identification of an Imprinted U2af Binding Protein Related Sequence on Mouse Chromosome 11 Using the RLGS Method. Nat. Genet. 6, 33–40. doi:10.1038/ng0194-33
He, H., Perera, B. P. U., Ye, A., and Kim, J. (2016). Parental and Sexual Conflicts over the Peg3 Imprinted Domain. Sci. Rep. 6, 38136. doi:10.1038/srep38136
Hiura, H., Obata, Y., Komiyama, J., Shirai, M., and Kono, T. (2006). Oocyte Growth-dependent Progression of Maternal Imprinting in Mice. Genes Cells 11, 353–361. doi:10.1111/j.1365-2443.2006.00943.x
Inoue, A., Chen, Z., Yin, Q., and Zhang, Y. (2018). Maternal Eed Knockout Causes Loss of H3K27me3 Imprinting and Random X Inactivation in the Extraembryonic Cells. Genes Dev. 32, 1525–1536. doi:10.1101/gad.318675.118
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017a). Maternal H3K27me3 Controls DNA Methylation-independent Imprinting. Nature 547, 419–424. doi:10.1038/nature23262
Inoue, A., Jiang, L., Lu, F., and Zhang, Y. (2017b). Genomic Imprinting of Xist by Maternal H3K27me3. Genes Dev. 31, 1927–1932. doi:10.1101/gad.304113.117
Inoue, K., Ogonuki, N., Kamimura, S., Inoue, H., Matoba, S., Hirose, M., et al. (2020). Loss of H3K27me3 Imprinting in the Sfmbt2 miRNA Cluster Causes Enlargement of Cloned Mouse Placentas. Nat. Commun. 11, 2150. doi:10.1038/s41467-020-16044-8
Itoh, M., Yoshida, Y., Nishida, K., Narimatsu, M., Hibi, M., and Hirano, T. (2000). Role of Gab1 in Heart, Placenta, and Skin Development and Growth Factor- and Cytokine-Induced Extracellular Signal-Regulated Kinase Mitogen-Activated Protein Kinase Activation. Mol. Cel Biol 20, 3695–3704. doi:10.1128/mcb.20.10.3695-3704.2000
Jiang, X., Yu, Y., Yang, H. W., Agar, N. Y. R., Frado, L., and Johnson, M. D. (2010). The Imprinted Gene PEG3 Inhibits Wnt Signaling and Regulates Glioma Growth. J. Biol. Chem. 285, 8472–8480. doi:10.1074/jbc.m109.069450
John, R. M., Aparicio, S. A. J. R., Ainscough, J. F.-X., Arney, K. L., Khosla, S., Hawker, K., et al. (2001). Imprinted Expression of Neuronatin from Modified BAC Transgenes Reveals Regulation by Distinct and Distant Enhancers. Developmental Biol. 236, 387–399. doi:10.1006/dbio.2001.0327
Joseph, R. M. (2014). Neuronatin Gene: Imprinted and Misfolded. Genomics 103, 183–188. doi:10.1016/j.ygeno.2013.12.001
Kagami, M., Sekita, Y., Nishimura, G., Irie, M., Kato, F., Okada, M., et al. (2008). Deletions and Epimutations Affecting the Human 14q32.2 Imprinted Region in Individuals with Paternal and Maternal Upd(14)-like Phenotypes. Nat. Genet. 40, 237–242. doi:10.1038/ng.2007.56
Kagami, M., Kurosawa, K., Miyazaki, O., Ishino, F., Matsuoka, K., and Ogata, T. (2015). Comprehensive Clinical Studies In 34 Patients With Molecularly Defined UPD(14)Pat And Related Conditions (Kagami-Ogata Syndrome). Eur. J. Hum. Genet. 23, 1488–1498.
Kagitani, F., Kuroiwa, Y., Wakana, S., Shiroishi, T., Kobayashi, S., Kohda, T., et al. (1997). Peg5/Neuronatin Is an Imprinted Gene Located on Sub-distal Chromosome 2 in the Mouse. Nucl. Acids Res. 25, 3428–3432. doi:10.1093/nar/25.17.3428
Kagiwada, S., Kurimoto, K., Hirota, T., Yamaji, M., and Saitou, M. (2013). Replication-coupled Passive DNA Demethylation for the Erasure of Genome Imprints in Mice. EMBO J. 32, 340–353. doi:10.1038/emboj.2012.331
Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E., et al. (2004). Essential Role for De Novo DNA Methyltransferase Dnmt3a in Paternal and Maternal Imprinting. Nature 429, 900–903. doi:10.1038/nature02633
Kaneko-Ishino, T., and Ishino, F. (2019). Evolution of Viviparity in Mammals: what Genomic Imprinting Tells Us about Mammalian Placental Evolution. Reprod. Fertil. Dev. 31, 1219–1227. doi:10.1071/rd18127
Kaneko-Ishino, T., and Ishino, F. (2015). Mammalian-specific Genomic Functions: Newly Acquired Traits Generated by Genomic Imprinting and LTR Retrotransposon-Derived Genes in Mammals. Proc. Jpn. Acad. Ser. B: Phys. Biol. Sci. 91, 511–538. doi:10.2183/pjab.91.511
Kaneko-Ishino, T., and Ishino, F. (2010). Retrotransposon Silencing by DNA Methylation Contributed to the Evolution of Placentation and Genomic Imprinting in Mammals. Dev. Growth Differ. 52, 533–543. doi:10.1111/j.1440-169x.2010.01194.x
Kaneko-Ishino, T., and Ishino, F. (2012). The Role of Genes Domesticated from LTR Retrotransposons and Retroviruses in Mammals. Front. Microbio. 3, 262. doi:10.3389/fmicb.2012.00262
Kaneko-Ishino, T., Kohda, T., and Ishino, F. (2003). The Regulation and Biological Significance of Genomic Imprinting in Mammals. J. Biochem. 133, 699–711. doi:10.1093/jb/mvg090
Kaneko-Ishino, T., Kohda, T., Ono, R., and Ishino, F. (2006). Complementation Hypothesis: the Necessity of a Monoallelic Gene Expression Mechanism in Mammalian Development. Cytogenet. Genome Res. 113, 24–30. doi:10.1159/000090811
Kaneko-Ishino, T., Kuroiwa, Y., Miyoshi, N., Kohda, T., Suzuki, R., Yokoyama, M., et al. (1995). Peg1/Mest Imprinted Gene on Chromosome 6 Identified by cDNA Subtraction Hybridization. Nat. Genet. 11, 52–59. doi:10.1038/ng0995-52
Kawasaki, Y., Lee, J., Matsuzawa, A., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2014). Active DNA Demethylation Is Required for Complete Imprint Erasure in Primordial Germ Cells. Sci. Rep. 4, 3658. doi:10.1038/srep03658
Kikyo, N., Williamson, C. M., John, R. M., Barton, S. C., Beechey, C. V., Ball, S. T., et al. (1997). Genetic and Functional Analysis of Neuronatin in Mice with Maternal or Paternal Duplication of Distal Chr 2. Developmental Biol. 190, 66–77. doi:10.1006/dbio.1997.8681
Killian, J. K., Byrd, J. C., Jirtle, J. V., Munday, B. L., Stoskopf, M. K., MacDonald, R. G., et al. (2000). M6P/IGF2R Imprinting Evolution in Mammals. Mol. Cel 5, 707–716. doi:10.1016/s1097-2765(00)80249-x
Killian, J. K., Nolan, C. M., Stewart, N., Munday, B. L., Andersen, N. A., Nicol, S., et al. (2001). MonotremeIGF2 Expression and Ancestral Origin of Genomic Imprinting. J. Exp. Zool. 291, 205–212. doi:10.1002/jez.1070
Kim, A., Terzian, C., Santamaria, P., Pélisson, A., Purd'homme, N., and Bucheton, A. (1994). Retroviruses in Invertebrates: The Gypsy Retrotransposon Is Apparently an Infectious Retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. 91, 1285–1289. doi:10.1073/pnas.91.4.1285
Kim, J., Bergmann, A., Choo, J. H., and Stubbs, L. (2007). Genomic Organization and Imprinting of the Peg3 Domain in Bovine. Genomics 90, 85–92. doi:10.1016/j.ygeno.2007.03.012
Kim, J., Bergmann, A., Wehri, E., Lu, X., and Stubbs, L. (2001). Imprinting and Evolution of Two Kruppel-type Zinc-finger Genes, ZIM3 and ZNF264, Located in the PEG3/USP29 Imprinted Domain. Genomics 77, 91–98. doi:10.1006/geno.2001.6621
Kim, J., Frey, W. D., He, H., Kim, H., Ekram, M. B., Bakshi, A., et al. (2013). Peg3 Mutational Effects on Reproduction and Placenta-specific Gene Families. PLoS ONE 8, e83359. doi:10.1371/journal.pone.0083359
Kitazawa, M., Hayashi, S., Imamura, M., Takeda, S., Oishi, Y., Kaneko-Ishino, T., et al. (2020). Deficiency and Overexpression of Rtl1 in the Mouse Cause Distinct Muscle Abnormalities Related to Temple and Kagami-Ogata Syndromes. Development 147, dev185918. doi:10.1242/dev.185918
Kitazawa, M., Sutani, A., Kaneko‐Ishino, T., and Ishino, F. (2021). The Role of Eutherian‐specific RTL1 in the Nervous System and its Implications for the Kagami‐Ogata and Temple Syndromes. Genes Cells 26, 165–179. doi:10.1111/gtc.12830
Kitazawa, M., Tamura, M., Kaneko-Ishino, T., and Ishino, F. (2017). Severe Damage to the Placental Fetal Capillary Network Causes Mid- to Late Fetal Lethality and Reduction in Placental Size inPeg11/Rtl1KO Mice. Genes Cells 22, 174–188. doi:10.1111/gtc.12465
Kobayashi, H., Sakurai, T., Imai, M., Takahashi, N., Fukuda, A., Yayoi, O., et al. (2012). Contribution of Intragenic DNA Methylation in Mouse Gametic DNA Methylomes to Establish Oocyte-specific Heritable marks. Plos Genet. 8, e1002440. doi:10.1371/journal.pgen.1002440
Kobayashi, S., Wagatsuma, H., Ono, R., Ichikawa, H., Yamazaki, M., Tashiro, H., et al. (2000). MousePeg9/Dlk1and humanPEG9/DLK1are Paternally Expressed Imprinted Genes Closely Located to the Maternally Expressed Imprinted Genes: mouseMeg3/Gtl2and humanMEG3. Genes Cells 5, 1029–1037. doi:10.1046/j.1365-2443.2000.00390.x
Kohda, T., Asai, A., Kuroiwa, Y., Kobayashi, S., Aisaka, K., Nagashima, G., et al. (2001). Tumour Suppressor Activity of Human Imprinted Gene PEG3 in a Glioma Cell Line. Genes Cells 6, 237–247. doi:10.1046/j.1365-2443.2001.00412.x
Kuroiwa, Y., Kaneko-Ishino, T., Kagitani, F., Kohda, T., Li, L.-L., Tada, M., et al. (1996). Peg3 Imprinted Gene on Proximal Chromosome 7 Encodes for a Zinc finger Protein. Nat. Genet. 12, 186–190. doi:10.1038/ng0296-186
Lee, J., Inoue, K., Ono, R., Ogonuki, N., Kohda, T., Kaneko-Ishino, T., et al. (2002). Erasing Genomic Imprinting Memory in Mouse Clone Embryos Produced from Day 11.5 Primordial Germ Cells. Development 129, 1807–1817. doi:10.1242/dev.129.8.1807
Lee, S., Walker, C. L., Karten, B., Kuny, S. L., Tennese, A. A., O'Neill, M. A., et al. (2005). Essential Role for the Prader-Willi Syndrome Protein Necdin in Axonal Outgrowth. Hum. Mol. Genet. 14, 627–637. doi:10.1093/hmg/ddi059
Li, C., Han, T., Li, Q., Zhang, M., Guo, R., Yang, Y., et al. (2021). Hu. MKRN3-Mediated Ubiquitination of Poly(A)-binding Proteins Modulates the Stability and Translation of GNRH1 mRNA in Mammalian Puberty. Nucl. Acids Res. 49, 2396–2813. doi:10.1093/nar/gkab155
Li, C.-M., Margolin, A. A., Salas, M., Memeo, L., Mansukhani, M., Hibshoosh, H., et al. (2006). PEG10 Is a C-MYC Target Gene in Cancer Cells. Cancer Res. 66, 665–672. doi:10.1158/0008-5472.can-05-1553
Li, J.-Y., Lees-Murdock, D. J., Xu, G.-L., and Walsh, C. P. (2004). Timing of Establishment of Paternal Methylation Imprints in the Mouse. Genomics 84, 952–960. doi:10.1016/j.ygeno.2004.08.012
Li, L.-L., Keverne, E. B., Aparicio, S. A., Ishino, F., Barton, S. C., and Surani, M. A. (1999). Regulation of Maternal Behavior and Offspring Growth by Paternally Expressed Peg3. Science 284, 330–334. doi:10.1126/science.284.5412.330
Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., et al. (2008). A Maternal-Zygotic Effect Gene, Zfp57, Maintains Both Maternal and Paternal Imprints. Developmental Cel 15, 547–557. doi:10.1016/j.devcel.2008.08.014
Liu, X., Gao, Q., Li, P., Zhao, Q., Zhang, J., Li, J., et al. (2013). UHRF1 Targets DNMT1 for DNA Methylation Through Cooperative Binding Of Hemi-Methylated DNA And Methylated H3K9. Nat. Commun. 4, 1563. doi:10.1038/ncomms2562
Lopes, S., Lewis, A., Hajkova, P., Dean, W., Oswald, J., Forne, T., et al. (2003). Epigenetic Modifications in an Imprinting Cluster Are Controlled by a Hierarchy of DMRs Suggesting Long-Range Chromatin Interactions. Hum. Mol. Genet. 12, 295–305. doi:10.1093/hmg/ddg022
Lucifero, D., Mann, M. R. W., Bartolomei, M. S., and Trasler, J. M. (2004). Gene-specific Timing and Epigenetic Memory in Oocyte Imprinting. Hum. Mol. Genet. 13, 839–849. doi:10.1093/hmg/ddh104
Macfarlan, T. S., Gifford, W. D., Driscoll, S., Lettieri, K., Rowe, H. M., Bonanomi, D., et al. (2012). Embryonic Stem Cell Potency Fluctuates with Endogenous Retrovirus Activity. Nature 487, 57–63. doi:10.1038/nature11244
Maegawa, S., Itaba, N., Otsuka, S., Kamitani, H., Watanabe, T., Tahimic, C. G., et al. (2004). Coordinate Downregulation of a Novel Imprinted Transcript ITUP1 with PEG3 in Glioma Cell Lines. DNA Res. 11, 37–49. doi:10.1093/dnares/11.1.37
Malnou, E. C., Umlauf, D., Mouysset, M., and Cavaillé, J. (2018). Imprinted microRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 9, 706. doi:10.3389/fgene.2018.00706
Mann, J. R., and Lovell-Badge, R. H. (1984). Inviability of Parthenogenones Is Determined by Pronuclei, Not Egg Cytoplasm. Nature 310, 66–67. doi:10.1038/310066a0
McGrath, J., and Solter, D. (1984). Completion of Mouse Embryogenesis Requires Both the Maternal and Paternal Genomes. Cell 37, 179–183. doi:10.1016/0092-8674(84)90313-1
Miki, H., Inoue, K., Kohda, T., Honda, A., Ogonuki, N., Yuzuriha, M., et al. (2005). Birth of Mice Produced by Germ Cell Nuclear Transfer. Genesis 41, 81–86. doi:10.1002/gene.20100
Millership, S. J., Tunster, S. J., Van de Pette, M., Choudhury, A. I., Irvine, E. E., Christian, M., et al. (2018). Neuronatin Deletion Causes Postnatal Growth Restriction and Adult Obesity in 129S2/Sv Mice. Mol. Metab. 18, 97–106. doi:10.1016/j.molmet.2018.09.001
Miyoshi, N., Kuroiwa, Y., Kohda, T., Shitara, H., Yonekawa, H., Kawabe, T., et al. (1998). Identification of the Meg1/Grb10 Imprinted Gene on Mouse Proximal Chromosome 11, a Candidate for the Silver-Russell Syndrome Gene. Proc. Natl. Acad. Sci. 95, 1102–1107. doi:10.1073/pnas.95.3.1102
Miyoshi, N., Wagatsuma, H., Wakana, S., Shiroishi, T., Nomura, M., Aisaka, K., et al. (2000). Identification of an Imprinted Gene, Meg3/Gtl2 and its Human Homologue MEG3 , First Mapped on Mouse Distal Chromosome 12 and Human Chromosome 14q. Genes to Cells 5, 211–220. doi:10.1046/j.1365-2443.2000.00320.x
Mochizuki, K., ShiraneUranishi, K. K., JanssenSuzuki, S. M. A., KosekiLorincz, H. M. C., Bogutz, A. B., Janssen, S. M., et al. (2021). “Repression of Germline Genes by PRC1.6 and SETDB1 in the Early Embryo Precedes DNA Methylation-Mediated Silencing,”. Nat. Commun. 12, 7020. doi:10.1038/s41467-021-27345-x
Moore, T., and Haig, D. (1991). Genomic Imprinting in Mammalian Development: a Parental Tug-Of-War. Trends Genet. 7, 45–49. doi:10.1016/0168-9525(91)90040-w
Neumann, B., Kubicka, P., and Barlow, D. P. (1995). Characteristics of Imprinted Genes. Nat. Genet. 9, 12–13. doi:10.1038/ng0195-12
Nicholls, R. D., and Knepper, J. L. (2001). Genome Organization, Function, and Imprinting in Prader-Willi and Angelman Syndromes. Annu. Rev. Genom. Hum. Genet. 2, 153–175. doi:10.1146/annurev.genom.2.1.153
Nicholls, R. D., Saitoh, S., and Horsthemke, B. (1998). Imprinting in Prader-Willi and Angelman Syndromes. Trends Genet. 14, 194–200. doi:10.1016/s0168-9525(98)01432-2
Noguer-Dance, M., Abu-Amero, S., Al-Khtib, M., Lefèvre, A., Coullin, P., Moore, G. E., et al. (2010). The Primate-specific microRNA Gene Cluster (C19MC) Is Imprinted in the Placenta. Hum. Mol. Genet. 19, 3566–3582. doi:10.1093/hmg/ddq272
O'Neill, M. J., Ingram, R. S., Vrana, P. B., and Tilghman, S. M. (2000). Allelic Expression of IGF2 in Marsupials and Birds. Dev. Genes Evol. 210, 18–20. doi:10.1007/pl00008182
Okabe, H., Satoh, S., Furukawa, Y., Kato, T., Hasegawa, S., Nakajima, Y., et al. (2003). Involvement of PEG10 in Human Hepatocellular Carcinogenesis through Interaction with SIAH1. Cancer Res. 63, 3043–3048.
Okae, H., Chiba, H., Hiura, H., Hamada, H., Sato, A., Utsunomiya, T., et al. (2014). Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development. Plos Genet. 10, e1004868. doi:10.1371/journal.pgen.1004868
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 99, 247–257. doi:10.1016/s0092-8674(00)81656-6
Ondičová, M., Oakey, R. J., and Walsh, C. P. (2020). Is Imprinting the Result of “Friendly Fire” by the Host Defense System? Plos Genet. 16, e1008599. doi:10.1371/journal.pgen.1008599
Ono, R., Kobayashi, S., Wagatsuma, H., Aisaka, K., Kohda, T., Kaneko-Ishino, T., et al. (2001). A Retrotransposon-Derived Gene, PEG10, Is a Novel Imprinted Gene Located on Human Chromosome 7q21. Genomics 73, 232–237. doi:10.1006/geno.2001.6494
Ono, R., Nakamura, K., Inoue, K., Naruse, M., Usami, T., Wakisaka-Saito, N., et al. (2006). Deletion of Peg10, an Imprinted Gene Acquired from a Retrotransposon, Causes Early Embryonic Lethality. Nat. Genet. 38, 101–106. doi:10.1038/ng1699
Ono, R., Shiura, H., Aburatani, H., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2003). Identification of a Large Novel Imprinted Gene Cluster on Mouse Proximal Chromosome 6. Genome Res. 13, 1696–1705. doi:10.1101/gr.906803
Pandya, N. J., CostaLopatta, V. P., ZampetaPunt, F. I. A. M., GrossenDistler, P. T., MartíBanfai, Y. B., RasmussenHoener, S. M., et al. (2021). “Secreted Retrovirus-like GAG-Domain-Containing Protein PEG10 Is Regulated by UBE3A and Is Involved in Angelman Syndrome Pathophysiology,”. Cel Rep. Med. 2, 100360. doi:10.1016/j.xcrm.2021.100360
Plass, C., Shibata, H., Kalcheva, I., Mullins, L., Kotelevtseva, N., Mullins, J., et al. (1996). Identification of Grf1 on Mouse Chromosome 9 as an Imprinted Gene by RLGS-M. Nat. Genet. 14, 106–109. doi:10.1038/ng0996-106
Quenneville, S., Verde, G., Corsinotti, A., Kapopoulou, A., Jakobsson, J., Offner, S., et al. (2011). In Embryonic Stem Cells, ZNP57/KAP1 Recognize A Methylated Hexanucleotide To Affect Chromatin And DNA Methylation Of Imprinting Control Regions. Mol. Cell 44, 361–372. doi:10.1016/j.molcel.2011.08.032
Ramachandran, R., and McDaniel, C. D. (2018). Parthenogenesis in Birds: a Review. Reproduction 155, R245–R257. doi:10.1530/rep-17-0728
Rapkins, R. W., Hore, T., Smithwick, M., Ager, E., Pask, A. J., Renfree, M. B., et al. (2006). Recent Assembly of an Imprinted Domain from Non-imprinted Components. Plos Genet. 2, e182. doi:10.1371/journal.pgen.0020182
Reik, W., and Walter, J. (2001). Genomic Imprinting: Parental Influence on the Genome. Nat. Rev. Genet. 2, 21–32. doi:10.1038/35047554
Renfree, M. B., Hore, T. A., Shaw, G., Marshall Graves, J. A., and Pask, A. J. (2009). Evolution of Genomic Imprinting: Insights from Marsupials and Monotremes. Annu. Rev. Genom. Hum. Genet. 10, 241–262. doi:10.1146/annurev-genom-082908-150026
Renfree, M. B., Suzuki, S., and Kaneko-Ishino, T. (2013). The Origin and Evolution of Genomic Imprinting and Viviparity in Mammals. Phil. Trans. R. Soc. B 368, 20120151. doi:10.1098/rstb.2012.0151
Rowe, H. M., Friedli, M., Offner, S., Verp, S., Mesnard, D., Marquis, J., et al. (2013). De Novo DNA Methylation of Endogenous Retroviruses Is Shaped by KRAB-ZFPs/KAP1 and ESET. Development 140, 519–529. doi:10.1242/dev.087585
Schaaf, C. P., Gonzalez-Garay, M. L., Xia, F., Potocki, L., Gripp, K. W., Zhang, B., et al. (2013). Truncating Mutations of MAGEL2 Cause Prader-Willi Phenotypes and Autismcause Prader-Willi Phenotypes and Autism. Nat. Genet. 45, 1405–1408. doi:10.1038/ng.2776
Schmidt, J. V., Matteson, P. G., Jones, B. K., Guan, X.-J., and Tilghman, S. M. (2000). The Dlk1 and Gtl2 Genes Are Linked and Reciprocally Imprinted. Genes Dev. 14, 1997–2002. doi:10.1101/gad.14.16.1997
Sekita, Y., Wagatsuma, H., Nakamura, K., Ono, R., Kagami, M., Wakisaka, N., et al. (2008). Role of Retrotransposon-Derived Imprinted Gene, Rtl1, in the Feto-Maternal Interface of Mouse Placenta. Nat. Genet. 40, 243–248. doi:10.1038/ng.2007.51
Sharif, J., Muto, M., Takebayashi, S., Suetake, I., Iwamatsu, A., Endo, T. A., et al. (2007). The SRA Protein Np95 Mediates Epigenetic Inheritance By Recruiting Dnmt1 to Methylated DNA. Nature 450, 908–912.
Shirane, K., Miura, F., Ito, T., and Lorincz, M. C. (2020). NSD1-deposited H3k36me2 Directs De Novo Methylation in the Mouse Male Germline and Counteracts Polycomb-Associated Silencing. Nat. Genet. 52, 1088–1098. doi:10.1038/s41588-020-0689-z
Shiura, H., Ono, R., Tachibana, S., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2021). PEG10 Viral Aspartic Protease Domain Is Essential for the Maintenance of Fetal Capillary Structure in the Mouse Placenta. Development 148, dev199564. doi:10.1242/dev.199564
Smallwood, S. A., Tomizawa, S.-i., Krueger, F., Ruf, N., Carli, N., Segonds-Pichon, A., et al. (2011). Dynamic CpG Island Methylation Landscape in Oocytes and Preimplantation Embryos. Nat. Genet. 43, 811–814. doi:10.1038/ng.864
Smith, R. J., Dean, W., Konfortova, G., and Kelsey, G. (2003). Identification of Novel Imprinted Genes in a Genome-wide Screen for Maternal Methylation. Genome Res. 13, 558–569. doi:10.1101/gr.781503
Smits, G., Mungall, A. J., Mungall, A. J., Griffiths-Jones, S., Smith, P., Beury, D., et al. (2008). Conservation of the H19 Noncoding RNA and H19-IGF2 Imprinting Mechanism in Therians. Nat. Genet. 40, 971–976. doi:10.1038/ng.168
Strogantsev, R., Krueger, F., Yamazawa, K., Shi, H., Gould, P., Goldman-Roberts, M., et al. (2015). Allele-Specific Binding Of ZFP57 in the Epigenetic Regulation Of Imprinted And Non-Imprinted Monoallelic Expression. Genome Biol. 16, 112. doi:10.1186/s13059-015-0672-7
Solter, D. (1988). Differential Imprinting and Expression of Maternal and Paternal Genomes. Annu. Rev. Genet. 22, 127–146. doi:10.1146/annurev.ge.22.120188.001015
Song, S. U., Gerasimova, T., Kurkulos, M., Boeke, J. D., and Corces, V. G. (1994). An Env-like Protein Encoded by a Drosophila Retroelement: Evidence that Gypsy Is an Infectious Retrovirus. Genes Dev. 8, 2046–2057. doi:10.1101/gad.8.17.2046
Stöger, R., Kubicka, P., Liu, C. G., Kafri, T., Razin, A., Cedar, H., et al. (1993). Maternal-specific Methylation of the Imprinted Mouse Igf2r Locus Identifies the Expressed Locus as Carrying the Imprinting Signal. Cell 73, 61–71. doi:10.1016/0092-8674(93)90160-r
Surani, M. A. H., Barton, S. C., and Norris, M. L. (1984). Development of Reconstituted Mouse Eggs Suggests Imprinting of the Genome during Gametogenesis. Nature 308, 548–550. doi:10.1038/308548a0
Suzuki, S., Ono, R., Narita, T., Pask, A. J., Shaw, G., Wang, C., et al. (2007). Retrotransposon Silencing by DNA Methylation Can Drive Mammalian Genomic Imprinting. Plos Genet. 3, e55. doi:10.1371/journal.pgen.0030055
Suzuki, S., Renfree, M. B., Pask, A. J., Shaw, G., Kobayashi, S., Kohda, T., et al. (2005). Genomic Imprinting of IGF2, p57KIP2 and PEG1/MEST in a Marsupial, the Tammar Wallaby. Mech. Development 122, 213–222. doi:10.1016/j.mod.2004.10.003
Suzuki, S., Shaw, G., Kaneko-Ishino, T., Ishino, F., and Renfree, M. B. (2011). The Evolution of Mammalian Genomic Imprinting Was Accompanied by the Acquisition of Novel CpG Islands. Genome Biol. Evol. 3, 1276–1283. doi:10.1093/gbe/evr104
Suzuki, S., Shaw, G., and Renfree, M. B. (2018). Identification of a Novel Antisense Noncoding RNA, ALID, Transcribed from the Putative Imprinting Control Region of Marsupial IGF2R. Epigenetics & Chromatin 11, 55. doi:10.1186/s13072-018-0227-8
Szabó, P. E., Hübner, K., Schöler, H., and Mann, J. R. (2002). Allele-specific Expression of Imprinted Genes in Mouse Migratory Primordial Germ Cells. Mech. Development 115, 157–160. doi:10.1016/s0925-4773(02)00087-4
Szabó, P. E., Tang, S.-H. E., Rentsendorj, A., Pfeifer, G. P., and Mann, J. R. (2000). Maternal-specific Footprints at Putative CTCF Sites in the H19 Imprinting Control Region Give Evidence for Insulator Function. Curr. Biol. 10, 607–610. doi:10.1016/s0960-9822(00)00489-9
Takada, S., Paulsen, M., Tevendale, M., Tsai, C. E., Kelsey, G., Cattanach, B. M., et al. (2002). Epigenetic Analysis of the Dlk1-Gtl2 Imprinted Domain on Mouse Chromosome 12: Implications for Imprinting Control from Comparison with Igf2-H19. Hum. Mol. Genet. 11, 77–86. doi:10.1093/hmg/11.1.77
Takada, S., Tevendale, M., Baker, J., Georgiades, P., Campbell, E., Freeman, T., et al. (2000). Delta-like and Gtl2 Are Reciprocally Expressed, Differentially Methylated Linked Imprinted Genes on Mouse Chromosome 12. Curr. Biol. 10, 1135–1138. doi:10.1016/s0960-9822(00)00704-1
Takahashi, N., Coluccio, A., Thorball, C. W., Planet, E., Shi, H., Offner, S., et al. (2019). ZNF445 Is a Primary Regulator of Genomic Imprinting. Genes Dev. 33, 49–54. doi:10.1101/gad.320069.118
Takahashi, N., Okamoto, A., Kobayashi, R., Shirai, M., Obata, Y., Ogawa, H., et al. (2009). Deletion of Gtl2 , Imprinted Non-coding RNA, with its Differentially Methylated Region Induces Lethal Parent-origin-dependent Defects in Mice. Hum. Mol. Genet. 18, 1879–1888. doi:10.1093/hmg/ddp108
Veselovska, L., Smallwood, S. A., Saadeh, H., Stewart, K. R., Krueger, F., Maupetit-Me9;houas, S., et al. (2015). Deep Sequencing And De Novo Assembly Of The Mouse Oocyte Transcriptome Define Thecontribution Of Transcription To The DNA Methylation Landscape. Genome Biol. 16, 209. doi:10.1186/s13059-015-0769-z
Varmuza, S., and Mann, M. (1994). Genomic Imprinting - Defusing the Ovarian Time Bomb. Trends Genet. 10, 118–123. doi:10.1016/0168-9525(94)90212-7
Watanabe, T., Tomizawa, S.-i., Mitsuya, K., Totoki, Y., Yamamoto, Y., Kuramochi-Miyagawa, S., et al. (2011). Role for piRNAs and Noncoding RNA in De Novo DNA Methylation of the Imprinted Mouse Rasgrf1 Locus. Science 332, 848–852. doi:10.1126/science.1203919
Weidman, J. R., Murphy, S. K., Nolan, C. M., Dietrich, F. S., and Jirtle, R. L. (2004). Phylogenetic Footprint Analysis of IGF2 in Extant Mammals. Genome Res. 14, 1726–1732. doi:10.1101/gr.2774804
Wilkins, J. F., and Haig, D. (2003). What Good Is Genomic Imprinting: The Function of Parent-specific Gene Expression. Nat. Rev. Genet. 4, 359–368. doi:10.1038/nrg1062
Wolf, J. B., Oakey, R. J., and Feil, R. (2014). Imprinted Gene Expression in Hybrids: Perturbed Mechanisms and Evolutionary Implications. Heredity 113, 167–175. doi:10.1038/hdy.2014.11
Wood, A. J., Roberts, R. G., Monk, D., Moore, G. E., Schulz, R., and Oakey, R. J. (2007). A Screen for Retrotransposed Imprinted Genes Reveals an Association between X Chromosome Homology and Maternal Germ-Line Methylation. Plos Genet. 3, e20. doi:10.1371/journal.pgen.0030020
Wu, M.-Y., Jiang, M., Zhai, X., Wu, A. L., and au, R.-C. (2012). An Unexpected Function of the Prader-Willi Syndrome Imprinting center in Maternal Imprinting in Mice. PLoS ONE 7, e34348. doi:10.1371/journal.pone.0034348
Wutz, A., Smrzka, O. W., Schweifer, N., Schellander, K., Wagner, E. F., and Barlow, D. P. (1997). Imprinted Expression of the Igf2r Gene Depends on an Intronic CpG Island. Nature 389, 745–749. doi:10.1038/39631
Wylie, A. A., Murphy, S. K., Orton, T. C., and Jirtle, R. L. (2000). Novel Imprinted DLK1/GTL2 Domain on Human Chromosome 14 Contains Motifs that Mimic Those Implicated in IGF2/H19 Regulation. Genome Res. 10, 1711–1718. doi:10.1101/gr.161600
Xu, Q., Xiang, Y., Wang, Q., Wang, L., Brind’Amour, J., Bogutz, A. B., et al. (2019). SETD2 Regulates the Maternal Epigenome, Genomic Imprinting and Embryonic Development. Nat. Genet. 51, 844–856. doi:10.1038/s41588-019-0398-7
Yamaguchi, S., Hong, K., Liu, R., Inoue, A., Shen, L., Zhang, K., et al. (2013a). Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during Germ Cell Reprogramming. Cell Res 23, 329–339. doi:10.1038/cr.2013.22
Yamaguchi, S., Shen, L., Liu, Y., Sendler, D., and Zhang, Y. (2013b). Role of Tet1 in Erasure of Genomic Imprinting. Nature 504, 460–464. doi:10.1038/nature12805
Yamazaki, Y., Low, E. W., Marikawa, Y., Iwahashi, K., Bartolomei, M. S., McCarrey, J. R., et al. (2005). Adult Mice Cloned from Migrating Primordial Germ Cells. Proc. Natl. Acad. Sci. 102, 11361–11366. doi:10.1073/pnas.0504943102
Keywords: complementation, genome innovation, evolutionary trade-off, insertion of exogenous DNA, host defense hypothesis
Citation: Kaneko-Ishino T and Ishino F (2022) The Evolutionary Advantage in Mammals of the Complementary Monoallelic Expression Mechanism of Genomic Imprinting and Its Emergence From a Defense Against the Insertion Into the Host Genome. Front. Genet. 13:832983. doi: 10.3389/fgene.2022.832983
Received: 10 December 2021; Accepted: 11 February 2022;
Published: 03 March 2022.
Edited by:
Julie Demars, Institut national de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Hisato Kobayashi, Nara Medical University, JapanPhilippe Arnaud, GRED CNRS-Université Clermont Auvergne-INSERM, France
Rebecca J. Oakey, King’s College London, United Kingdom
Copyright © 2022 Kaneko-Ishino and Ishino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoko Kaneko-Ishino, dGthbmVrb2lAaXMuaWNjLnUtdG9rYWkuYWMuanA=; Fumitoshi Ishino, ZmlzaGluby5lcGduQG1yaS50bWQuYWMuanA=
 Tomoko Kaneko-Ishino
Tomoko Kaneko-Ishino Fumitoshi Ishino
Fumitoshi Ishino